ABSTRACT
Siderophores, small iron-binding molecules secreted by many microbial species, capture environmental iron for transport back into the cell. Vibrio cholerae synthesizes and uses the catechol siderophore vibriobactin and also uses siderophores secreted by other species, including enterobactin produced by Escherichia coli. E. coli secretes both canonical cyclic enterobactin and linear enterobactin derivatives likely derived from its cleavage by the enterobactin esterase Fes. We show here that V. cholerae does not use cyclic enterobactin but instead uses its linear derivatives. V. cholerae lacked both a receptor for efficient transport of cyclic enterobactin and enterobactin esterase to promote removal of iron from the ferrisiderophore complex. To further characterize the transport of catechol siderophores, we show that the linear enterobactin derivatives were transported into V. cholerae by either of the catechol siderophore receptors IrgA and VctA, which also transported the synthetic siderophore MECAM [1,3,5-N,N′,N″-tris-(2,3-dihydroxybenzoyl)-triaminomethylbenzene]. Vibriobactin is transported via the additional catechol siderophore receptor ViuA, while the Vibrio fluvialis siderophore fluvibactin was transported by all three catechol receptors. ViuB, a putative V. cholerae siderophore-interacting protein (SIP), functionally substituted for the E. coli ferric reductase YqjH, which promotes the release of iron from the siderophore in the bacterial cytoplasm. In V. cholerae, ViuB was required for the use of vibriobactin but was not required for the use of MECAM, fluvibactin, ferrichrome, or the linear derivatives of enterobactin. This suggests the presence of another protein in V. cholerae capable of promoting the release of iron from these siderophores.
IMPORTANCE Vibrio cholerae is a major human pathogen and also serves as a model for the Vibrionaceae, which include other serious human and fish pathogens. The ability of these species to persist and acquire essential nutrients, including iron, in the environment is epidemiologically important but not well understood. In this work, we characterize the ability of V. cholerae to acquire iron by using siderophores produced by other organisms. We resolve confusion in the literature regarding its ability to use the Escherichia coli siderophore enterobactin and identify the receptor and TonB system used for the transport of several siderophores. The use of some siderophores did not require the ferric reductase ViuB, suggesting that an uncharacterized ferric reductase is present in V. cholerae.
INTRODUCTION
Vibrio cholerae is a Gram-negative pathogen that causes the severe diarrheal disease cholera. The ongoing cholera pandemic causes more than 1 million cases each year with mortality in the tens of thousands, including those from the current epidemic in Haiti (1, 2). Transmission of cholera is fecal-oral and is frequently associated with untreated sewage entering an aquatic environment. V. cholerae may persist for extended periods of time in these environments, where it must compete with other microbes for nutrients, including iron, for which it has an absolute requirement.
Although iron is abundantly present in the earth's crust, its bioavailability is limited by its poor solubility at physiological pH in the presence of oxygen. Pathogenic bacteria face additional challenges caused by iron being sequestered inside cells or bound to carriers with high affinity for iron, including transferrin, lactoferrin, heme, hemopexin, and haptoglobin (3–5). Bacteria have evolved a number of strategies to efficiently acquire this essential element (6). A common mechanism for iron acquisition is the synthesis and export of small molecules, termed siderophores, which bind ferric iron with extremely high affinity. The ferrisiderophore complex is then transported back into the cell, where the iron is released for use in cellular processes. In Gram-negative bacteria, siderophore transport requires an outer membrane receptor, which is relatively specific for its ligand. When it binds its ferrisiderophore ligand, the receptor physically interacts with the TonB protein. TonB, together with ExbB and ExbD, transduces energy from the proton motive force of the inner membrane to the receptor to allow transport of the ligand across the outer membrane (7, 8). In most cases, the transported siderophore ligand is then bound by a ligand-specific periplasmic binding protein that delivers the ferrisiderophore to an ATP binding cassette (ABC) transport protein in the inner membrane for subsequent delivery to the cytoplasm. The iron is then removed from the siderophore in a process that involves degradation of the siderophore; reduction of the bound iron to the ferrous form, which has lower affinity for the siderophore; or both. There is a metabolic cost associated with the synthesis of siderophores, and many microorganisms transport and use siderophores secreted by the other species in their environments (referred to as xenosiderophores).
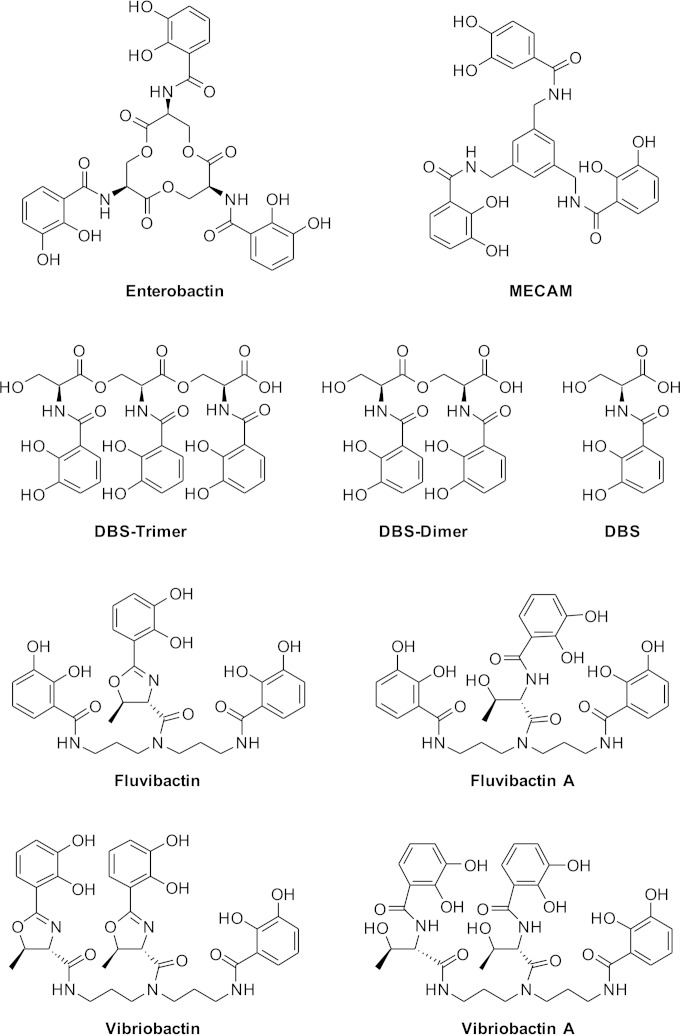
Siderophores fall into several chemical classes, including catechols, hydroxamates, carboxylates, and mixed siderophores containing more than one of the above structures (9). The archetype catecholate siderophore enterobactin is produced by Escherichia coli, Salmonella enterica, and related bacteria. It consists of three dihydroxybenzoate moieties, each linked through an amide bond to serine, with the serines joined through ester linkages to form a cycle (Fig. 1). E. coli transports ferrienterobactin across the outer membrane using the receptor FepA and through the periplasm and across the inner membrane by the periplasmic binding protein-dependent ABC transporter FepBDGC. It then enters the cytoplasm, where two enzymes participate in the release of the iron. Fes reduces the affinity of the siderophore for the iron by cleaving the ester bonds linking the serines in the siderophore, yielding dihydroxybenzoylserine (DBS) trimers, dimers, and monomers (10). The affinity of the siderophore for its iron ligand is lowered further by enzymatic reduction of the siderophore-bound ferric iron to its ferrous form by YqjH, a flavin adenine dinucleotide (FAD)-containing, cytosolic protein with ferric iron reductase activity (11, 12). In addition to cyclic enterobactin, the culture supernatant from wild-type E. coli also contains a mixture of linear trimers, dimers, and monomers of dihydroxybenzoylserine (Fig. 1). E. coli has two receptors, CirA and Fiu, for the transport of these ligands across the outer membrane (13). As with enterobactin, its linear derivatives are transported across the inner membrane by the FepBDGC system, and the iron is removed by the joint action of Fes and YqjH.
FIG 1.
Siderophore structures. Structures of siderophores and related compounds used in this report.
V. cholerae synthesizes and exports the linear catechol siderophore vibriobactin (14, 15), which is synthesized by the action of VibABCDEFH (16). Vibriobactin consists of three dihydroxybenzoate moieties joined to a norspermidine backbone, with one directly linked to the norspermidine and two connected through a threonine that is cyclized to form an oxazoline (Fig. 1) (14). The ferrivibriobactin is imported across the V. cholerae outer membrane by the receptor ViuA (17), and energy can be supplied to this receptor by either of the two TonB systems TonB1 and TonB2 (18). Vibriobactin is bound in the periplasm and transported across the inner membrane by either of the two ABC transport systems ViuPDGC (19) and VctPDGC (20). Unlike enterobactin, vibriobactin does not have cleavable ester bonds, and no homologue of Fes or other potential siderophore-degrading enzyme has been identified in V. cholerae, suggesting that the hydrolysis of vibriobactin is not required for removal of the iron. Utilization of iron from vibriobactin by V. cholerae requires ViuB, and expression of viuB from a high-copy-number vector suppressed an E. coli fes mutation (21). ViuB has 23% sequence identity and 16% similarity with E. coli YqjH, and based on the recent characterization of that enzyme (12), it is believed that ViuB promotes release of iron from vibriobactin by reducing the bound ferric iron to ferrous iron.
V. cholerae can also transport and use the iron from siderophores that it does not itself produce, including the hydroxamate ferrichrome (22), and the catechols agrobactin (14, 23) and fluvibactin (24, 25). There has been conflicting evidence on the use of the xenosiderophore enterobactin, since purified enterobactin is only weakly bound and used by V. cholerae (14, 26); however, V. cholerae readily uses the siderophore secreted by enterobactin-producing strains of E. coli (20). In this work, we show that V. cholerae most efficiently uses the linear DBS dimer and DBS trimer derivatives of enterobactin and that efficient use of cyclic enterobactin requires an enterobactin-specific outer membrane receptor, such as FepA, and the enterobactin esterase Fes. To further characterize the range of siderophores that can be used by V. cholerae, we also show that it will use vibriobactin and fluvibactin derivatives in which the oxazoline rings have been cleaved, and this cleavage affected neither the receptor specificity for the ligand nor the requirement for viuB. We also show that V. cholerae will use the synthetic, uncleavable enterobactin analog MECAM [1,3,5-N,N′,N″-tris-(2,3-dihydroxybenzoyl)-triaminomethylbenzene] and that, surprisingly, the use of iron from this ligand does not require ViuB. Further, we show that viuB and its E. coli homologue yqjH can functionally substitute for each other.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and iron utilization assays.
The strains and plasmids used in this study are listed in Table 1. The iron chelator ethylenediamine di(ortho-hydroxyphenylacetic acid) (EDDA) was deferrated by the method of Rogers (27). The antibiotic concentrations used were 250 μg ampicillin per ml, 50 μg kanamycin per ml, and 50 μg/ml (E. coli) or 5 μg/ml (V. cholerae) tetracycline. The bioassay for siderophore utilization was performed as previously described (28). Briefly, 10 μl of overnight culture was mixed with 20 ml molten LB agar containing 100 μg EDDA per ml (V. cholerae) or 150 μg EDDA per ml (E. coli). Then, 5 μl of siderophore solution or overnight culture of the indicated bacterial strain was pipetted onto the surface of the solidified agar. The plates were incubated at 37°C for 24 h, and the diameter of the zone of growth stimulated by the siderophore was measured. The siderophores used were at the following concentrations: 10 μM vibriobactin, 10 μM vibriobactin A, 10 μM enterobactin, 100 μM MECAM, 100 μM DBS monomer, 100 μM DBS dimer, 100 μM DBS trimer, 200 μM fluvibactin, 100 μM fluvibactin A, and 200 μM ferrichrome. When used, the concentration of FeSO4 was 10 mM. The concentration of the ligands used was determined empirically and reflects both efficiency of bacterial siderophore transport and ability of the siderophore to compete with the EDDA chelator. Enterobactin was used at the concentration that stimulated a well-defined zone of growth on an E. coli enterobactin biosynthesis mutant. All other siderophores were used at the concentration that stimulated growth of a vibriobactin synthesis mutant of V. cholerae. The siderophore biosynthetic mutants were used as the test strains to reduce background growth in the assay.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| V. cholerae | ||
| O395 | Classical biotype | 47 |
| ALV101 | O395 ΔvibB | 48 |
| JRB9 | O395 ΔviuB | 21 |
| CA40130N | Classical biotype; Vib− Nalr | 29 |
| ARM316 | CA40130N irgA::Camr | 20 |
| AGO310 | CA40130N vct::Kanr | 20 |
| ARM616 | CA40130N irgA::Kanr vctA::Camr | 20 |
| EWV136 | CA40130N irgA::Kanr vctA::Camr ΔviuA | This study |
| AMV527 | CA40130N tonB1::Kanr | 29 |
| ARM320 | CA40130N exbB2::Camr | 49 |
| ARM330 | CA40130N tonB1::Kanr exbB2::Camr | A. R. Mey, unpublished data |
| Lou15 | El Tor biotype | 50 |
| E. coli | ||
| DH5α | Cloning strain; Ent+ | 51 |
| BW25113 | K-12 Ent+ | 52 |
| W3110N | Wild-type K-12 W3110; Nalr | 53 |
| ARM110 | W3110N entF::Camr | 53 |
| EWE1018 | ARM110 yqjH::Kanr | This study |
| EWE1020 | ARM110 fes::Kanr | This study |
| EWE1024 | ARM110 Δfes | This study |
| EWE1026 | ARM110 Δfes yqjH::Kanr | This study |
| EWE1046 | ARM110 fepA::Kanr | This study |
| JW3041 | BW25113 yqjH::Kanr | 54 |
| JW0576 | BW25113 fes::Kanr | 54 |
| JW0587 | BW25113 entB::Kanr | 54 |
| JW5086 | BW25113 fepA::Kanr | 54 |
| Plasmids | ||
| pWKS30 | Low-copy-number cloning vector | 55 |
| pCP20 | Flp recombinase vector | 56 |
| pFepA | pACYC184 carrying E. coli fepA | 49 |
| pFes1 | pWKS30 carrying E. coli fes | This study |
| pYqjH2 | pWKS30 carrying E. coli yqjH | This study |
| pViuB4 | pWKS30 carrying V. cholerae viuB | This study |
| pCat119 | pWKS30 carrying V. cholerae vctA | 20 |
| pCat121 | pWKS30 carrying V. cholerae irgAB | 20 |
| pViuA1 | pWKS30 carrying V. cholerae viuA | This study |
| pACYC184 | Medium-copy-number cloning vector | 57 |
| pRK1013 | Helper plasmid for triparental mating | 58 |
| pCVD442N | Suicide vector pCVD442 with NotI adaptor | 48 |
| pSViuA | pCVD442N with V. cholerae sequences for deleting viuA | E. Peng, unpublished data |
Plasmid construction.
The plasmids pFes1, pYqjH2, pViuA1, and pViuB4 consist of PCR products amplified with Pfx polymerase (Life Technologies) and inserted into the SmaI site of pWKS30. For pFes1, the primers were Fes.for (5′-TCCCCAGATTGACCAACAAGGC) and Fes.rev (5′-GGCGACCAAAGGTAAATGCTG) and the template was E. coli strain W3110. For pYqjH2, the primers were yqjH.for (5′-ACACCCTTCGTGATGATGGCTC) and yqjH.rev (5′-AATCGCTTGGTCGCTGGTTC) and the template was E. coli strain DH5α. For pViuA1, the primers were viuA.3006 (5′-GTGGTAAGCGAGTCATCAAGTAAGG) and viuA.5964.rev (5′GAGGCGTTGTTTTGTTTCAGTCC) and the template was V. cholerae strain Lou15. For pViuB4, the primers were viu.2009 (5′-TGCCAATGCTCAGTTAGCCTATG) and viu.3546.rev (5′-CCCACAGATTCATCCCTTTACTCG) and the template was V. cholerae strain Lou15.
Strain construction.
E. coli strains EWE1018, EWE1020, and EWE1046 were constructed by bacteriophage P1 transduction of the yqjH::Kanr mutation from strain JW3041, the fes::Kanr mutation from strain JW0576, or the fepA::Kanr mutation from strain JW5086, respectively, into strain ARM110. The Flp recombinase encoded on plasmid pCP20 was used to delete the kanamycin cassette from the fes gene in EWE1020, and the yqjH::Kanr mutation was then introduced by P1 transduction from strain JW3041 to yield the fes yqjH double mutant strain EWE1026.
V. cholerae strain EWV136 (CA40130 aro irgA::Kanr vctA::Camr ΔviuA) was constructed by introducing the plasmid pSViuA into ARM616 by triparental mating. Allelic exchange was performed as previously described (29).
Siderophore synthesis.
Enterobactin, MECAM, DBS monomer, fluvibactin, fluvibactin A, vibriobactin, and vibriobactin A were synthesized as previously published (30–33). The synthesis of DBS dimers and DBS trimers is reported in the supplemental material.
RESULTS
Use of enterobactin derivatives by V. cholerae.
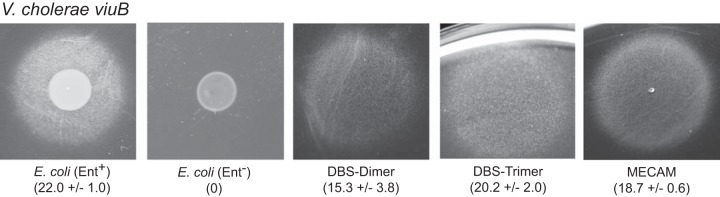
It was previously reported that V. cholerae does not efficiently use ferrienterobactin as a source of iron (14, 26). However, its growth is strongly stimulated in iron-restricted medium by compounds secreted by wild-type E. coli but not by E. coli strains defective in enterobactin biosynthesis (18, 20). This apparently contradictory result is shown again here in a bioassay (Fig. 2A). However, wild-type E. coli secretes not just cyclic enterobactin but also enterobactin derivatives that are monomers, dimers, and trimers of DBS (Fig. 1) (34), which may also function as siderophores (13). When we tested these compounds in the siderophore utilization assay, we found that both the DBS dimer and DBS trimer supplied iron to V. cholerae, but the DBS monomer did not (Fig. 2A), suggesting that the DBS dimer and DBS trimer are the compounds secreted by enterobactin-producing strains of E. coli that are used by V. cholerae.
FIG 2.
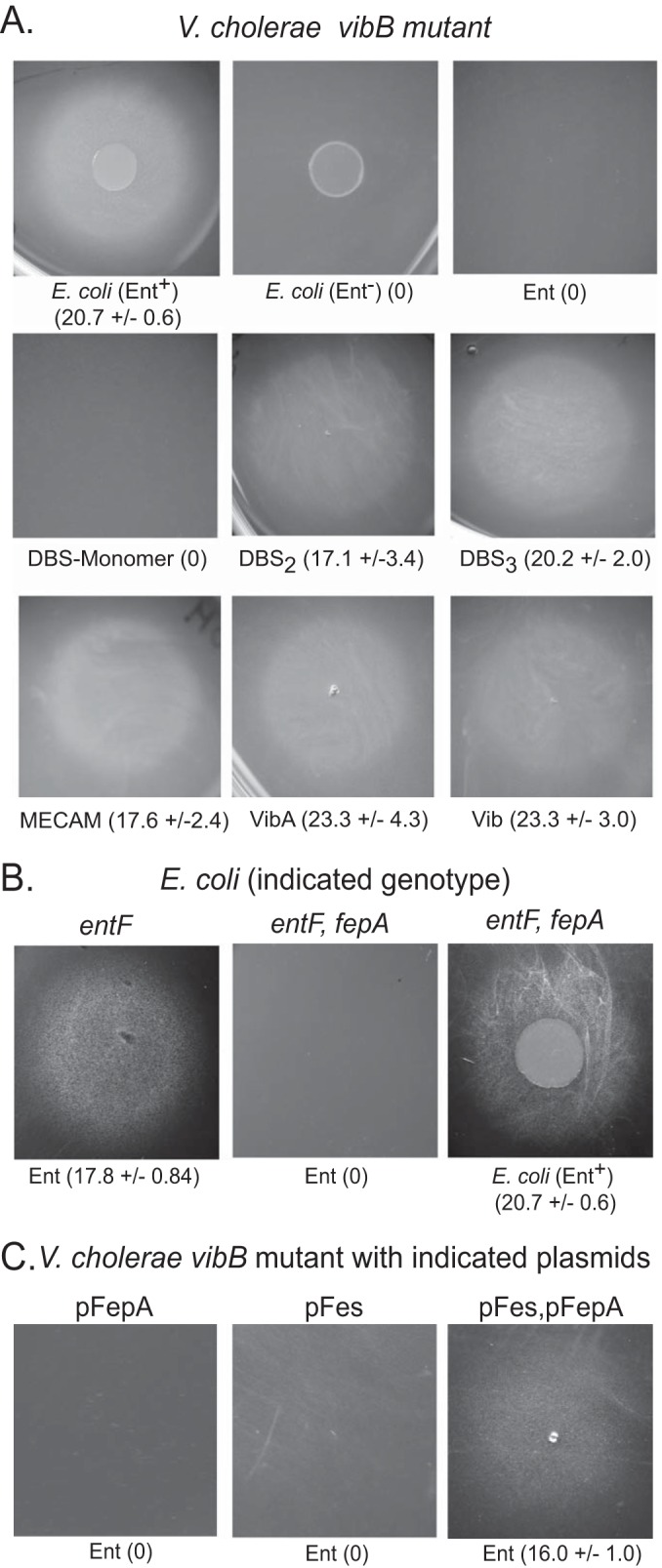
Siderophore utilization bioassays. The indicated strains were seeded into LB agar containing the iron chelator EDDA. The siderophore or E. coli strain spotted on the agar is noted below each picture. The siderophore then diffuses onto the agar, and to give a positive signal in the assay, it must bind iron efficiently in competition with the chelator and be transported and used by the bacteria embedded in the agar. The growth around each siderophore was photographed after 24 h of incubation at 37°C. The numbers in parentheses are the average diameter in millimeters of the zone of growth ± standard deviation measured in at least three independent assays. The agar was seeded with the following strains: V. cholerae vibB mutant ALV101 (A), E. coli strain ARM110 (entF) or EWE1046 (entF fepA) (B), and V. cholerae strain ALV101 (vibB) carrying the plasmid pFepA, pFes1, or pFepA and pFes1 (C). The vibB (V. cholerae) and entF (E. coli) mutants were used to reduce the background growth in the assay.
This phenotype is similar to an E. coli strain carrying a mutation in the gene for the cyclic enterobactin receptor fepA, which fails to use cyclic enterobactin but transports the linear enterobactin derivatives through the receptors CirA and Fiu (13). It is anticipated that growth of an E. coli fepA mutant would be stimulated by an enterobactin-producing strain of E. coli but not by purified enterobactin, and that result was observed in our assays (Fig. 2B). In the positive control, purified enterobactin stimulated growth of the FepA+ E. coli strain (Fig. 2B), indicating that the enterobactin used in these assays is functional.
This similarity between the enterobactin utilization phenotypes of V. cholerae and the E. coli fepA mutant suggests that the failure of V. cholerae to use enterobactin may be due to lack of an appropriate receptor. To test this, a plasmid containing fepA was introduced into V. cholerae. In most assays with V. cholerae carrying this plasmid, no growth was observed around enterobactin (Fig. 2C), although in a few of the assays there was a low level of growth that was too faint to be reproducibly quantified. The lack of robust growth of V. cholerae carrying fepA in the presence of enterobactin may indicate that FepA has limited transport function in V. cholerae or that something in addition to functional FepA is needed for efficient enterobactin utilization by V. cholerae.
Another difference between V. cholerae and E. coli is that V. cholerae lacks a gene for the enterobactin esterase Fes. Supplying E. coli fes on a plasmid to V. cholerae did not stimulate use of enterobactin (Fig. 2C); however, supplying both fepA and fes on compatible plasmids promoted efficient utilization of enterobactin (Fig. 2C). We conclude that use of enterobactin in V. cholerae is limited both by lack of an outer membrane receptor for cyclic enterobactin and by lack of the ability to cleave the cyclic enterobactin intracellularly to efficiently remove the iron. It does not appear that failure to transport ferrienterobactin across the inner membrane is a significant factor, since the V. cholerae inner membrane ABC transporter VctPDGC was able to transport purified enterobactin across the inner membrane in an E. coli strain defective in FepB, a component of the E. coli inner membrane enterobactin transport system (see Table S1 in the supplemental material).
Use of structurally related catechol siderophores by V. cholerae.
To further characterize the specificity of siderophore utilization by V. cholerae, its ability to use structurally distinct catechols was tested. MECAM is a cyclic catechol similar to enterobactin, except that the three dihydroxybenzoate (DHB) moieties are joined through a benzene ring rather than through three serines (Fig. 1). MECAM is efficiently used as a siderophore by E. coli (35), and we found that MECAM is also used efficiently by V. cholerae (Fig. 2A). This synthetic catechol is frequently used in structural and enzymatic studies of siderophore transport and utilization, and to our knowledge it was not previously known whether V. cholerae can use this for iron uptake.
Vibriobactin includes two oxazoline rings formed from threonine (Fig. 1) (14), but the role of these oxazoline rings in vibriobactin function is unknown. To explore this, a modified form of vibriobactin, termed vibriobactin A, in which the oxazoline rings have been opened (Fig. 1), was tested. Like native vibriobactin, vibriobactin A stimulated iron acquisition by V. cholerae (Fig. 2A), indicating that intact oxazoline rings are not required for use of this siderophore.
V. cholerae uses the siderophore fluvibactin, which is normally produced by Vibrio fluvialis (24, 25). Fluvibactin is similar to vibriobactin in that three DHB moieties are carried on a norspermidine backbone, but in fluvibactin, only the central DHB is linked through an oxazoline ring, while the DHB moieties on the ends are directly attached to the norspermidine (Fig. 1) (25). In our bioassays, fluvibactin gave an unusual pattern when tested for stimulation of growth of V. cholerae. There was a dense zone of growth, surrounded by a larger, more diffuse zone (Table 2). Fluvibactin A, a fluvibactin-related compound in which the single oxazoline ring has been opened (Fig. 1), stimulated a single dense zone of growth, and no diffuse zone of growth was observed (Table 2).
TABLE 2.
Ligand transport by V. cholerae outer membrane receptorsa
| Ligand | Zone of stimulation (mm) ± SD on CA40130N (vib) | Zone of stimulation (mm) ± SD on EWV136b carrying plasmid gene: |
|||
|---|---|---|---|---|---|
| None | viuA | vctA | irgA | ||
| Vibriobactin | 20.0 ± 3.1 | 0 | 20.3 ± 1.5 | 0 | 0 |
| Vibriobactin A | 21.0 ± 2.9 | 0 | 23.3 ± 3.2 | 0 | 0 |
| DH5α | 19.6 ± 2.8 | 0 | 0 | 22.0 ± 3.0 | 21.3 ± 2.1 |
| DBS trimer | 23.5 ± 2.1 | 0 | 17.3 ± 2.5c | 22.3 ± 1.2 | 19.0 ± 1.0 |
| DBS dimer | 22.0 ± 2.3 | 0 | 0 | 21.0 ± 5.2 | 15.7 ± 0.6 |
| MECAM | 25.3 ± 0.58 | 0 | 0 | 24.0 ± 2.0 | 19.3 ± 1.1 |
| Fluvibactin (diffuse)d | 18.7 ± 1.2 (49.0 ± 2.0) | 0 (0) | 0 (39.7 ± 8.4) | 19.3 ± 1.5 (0) | 14.3 ± 3.1 (0) |
| Fluvibactin A | 24.4 ± 2.2 | 0 | 28.3 ± 1.5 | 27.0 ± 2.0 | 16.0 ± 1.7 |
| Ferrichrome | 19.0 ± 2.4 | 22.0 ± 1.9 | 22.6 ± 2.4 | 21.9 ± 1.6 | 21.4 ± 1.6 |
Cultures of the indicated strains were seeded into LB agar containing 100 μM EDDA per ml, and the indicated siderophores or bacterial strain was spotted on the surface. The zone of growth was measured after 24 h of growth. The numbers are the average ± standard deviation for at least three independent assays. CA40130N is the parental strain of EWV136. The vibriobactin biosynthesis defect was included to reduce background growth in the assay.
EWV136 is CA40130N vib viuA irgA vctA. The plasmids used are pViuA1 (viuA), pCat119 (vctA), and pCat121 (irgA).
This was a diffuse zone of growth.
The numbers not in parentheses are the diameter of the compact zone of growth, while the numbers in parentheses are the diameter of the more diffuse zone.
Defining the outer membrane receptor used by the siderophore ligands.
In previous work, it was found that ViuA is specifically required for the transport of vibriobactin (17), while the siderophore secreted by E. coli could be transported by either VctA or IrgA but not by ViuA (20). To identify the outer membrane receptors required for transport of the siderophores described here, a V. cholerae vib irgA vctA viuA quadruple mutant was constructed and transformed individually with low-copy-number plasmids carrying one each of the catechol receptor genes (Table 2). As expected, vibriobactin was used only when viuA was supplied on a plasmid. Vibriobactin A also specifically required viuA, indicating that opening the oxazoline rings did not change the receptor specificity. As observed with the siderophore secreted by E. coli, the DBS dimer and DBS trimer were transported by either VctA or IrgA. There was also a weak but reproducible zone of growth of the strain carrying viuA around the DBS trimer, which may reflect a low level of transport by this receptor. This was surprising, given that no zone was seen when the enterobactin-producing E. coli was the source of the siderophore, but this may reflect a quantitative difference between the amount of pure DBS trimer spotted on the plate and the amount secreted by E. coli. MECAM was also transported by either VctA or IrgA (Table 2).
Fluvibactin was transported by all three receptors, but interestingly, ViuA was required for the larger, diffuse zone of growth, while either VctA or IrgA was needed for the more dense, compact zone (Table 2). Since the concentration of fluvibactin should decrease over distance from the spot as it diffuses though the agar, we speculate that ViuA may have higher affinity for fluvibactin than VctA and IrgA but that the rate of transport of this ligand may be lower. Fluvibactin A was also transported by all three receptors, but the sizes and the densities of the zones of growth were similar for all three receptors (Table 2). To our knowledge, this is the first example of siderophores that can be efficiently transported by all three of the V. cholerae catechol siderophore receptors. The positive control, ferrichrome, which is transported independently of the catechol-specific systems (22), was used by all strains (Table 2).
TonB requirements for transport of the ligands.
V. cholerae has two TonB systems with partially redundant functions. The TonB2 system is required for transport of the siderophore secreted by E. coli, while either TonB system can function in the transport of vibriobactin and ferrichrome (18, 36). Each of the ligands studied here was tested for transport by V. cholerae tonB1 and tonB2 single mutants and the tonB1 tonB2 double mutant (Table 3). Both vibriobactin and vibriobactin A were transported by strains with either an active TonB1 or an active TonB2 system, as previously observed for ligands transported by ViuA (36). In contrast, the E. coli-produced siderophore, the DBS dimer, and MECAM specifically required TonB2 for transport, again consistent with transport by either VctA or IrgA (18). The DBS trimer was transported most efficiently when TonB2 was present, but a weak zone of growth was sometimes observed in the presence of TonB1, consistent with a low level of transport by ViuA. The TonB requirements for fluvibactin utilization are consistent with the receptor data. The larger diffuse zone of growth was observed in both the tonB1 and the tonB2 single mutants, as expected given its association with the ViuA receptor. In contrast, the small, compact zone of growth required either VctA or IrgA and here specifically required TonB2. Fluvibactin A transport could be facilitated by either TonB1 or TonB2 (Table 3).
TABLE 3.
TonB requirement for transport of ligands
| Ligand | Zone of stimulation (mm) ± SD on straina: |
|||
|---|---|---|---|---|
| CA40130N (Vib−) | AMV527 (Vib− TonB1−) | ARM320 (Vib− TonB2−) | ARM330 (Vib− TonB1− TonB2−) | |
| Vibriobactin | 20.0 ± 3.1 | 22.0 ± 5.1 | 21.3 ± 3.1 | 0 |
| Vibriobactin A | 21.0 ± 2.9 | 22.5 ± 2.4 | 23.5 ± 1.9 | 0 |
| DH5α | 19.6 ± 2.8 | 21.7 ± 2.3 | 0 | 0 |
| DBS trimer | 23.5 ± 2.1 | 22.0 ± 1.0 | 0b | 0 |
| DBS dimer | 22.0 ± 2.3 | 23.0 ± 1.7 | 0 | 0 |
| MECAM | 25.3 ± 0.58 | 15.3 ± 0.6 | 0 | 0 |
| Fluvibactin (diffuse)c | 18.7 ± 1.2 (49.0 ± 2.0) | 19.7 ± 1.2 (50 ± 2.6) | 0 (45.3 ± 7.2) | 0 |
| Fluvibactin A | 24.4 ± 2.2 | 26.3 ± 1.5 | 20.3 ± 3.2 | 0 |
| Ferrichrome | 19.0 ± 2.4 | 19.8 ± 3.3 | 19.4 ± 1.7 | 0 |
| FeSO4 | 16.0 ± 4.9 | 14.9 ± 3.6 | 15.9 ± 4.7 | 12.4 ± 3.9 |
Cultures of the indicated strains were seeded into LB agar containing 100 μM EDDA per ml, and the indicated siderophores or bacterial strain was spotted on the surface. The zone of growth was measured after 24 h of growth. The numbers are the average ± standard deviation from at least three independent assays. CA40130N is the parental strain of the tonB mutants.
A faint zone of growth was seen in some assays.
The numbers not in parentheses are the diameter of the compact zone of growth, while the numbers in parentheses are the diameter of the more diffuse zone.
In control experiments, either TonB system could function in ferrichrome transport, as previously observed (18). None of the siderophores was transported in a strain lacking both the TonB1 and TonB2 systems, indicating that there was no detectable TonB-independent transport of these ligands. Ferric sulfate, which can be transported by TonB-independent systems, stimulated the growth of the strain defective in both TonB systems, showing that failure of the siderophores to stimulate growth of this strain was due to their inability to supply iron to the cell in the absence of a functional TonB.
ViuB and YqjH can functionally substitute for each other.
ViuB was initially identified as an apparent cytosolic protein required for vibriobactin utilization by V. cholerae. The observation that viuB suppressed an E. coli fes mutation when expressed from a high-copy-number vector led to the model that it is involved in the removal of iron from the ferrisiderophore following its transport into the cytoplasm (21). More recently, a screen for iron-regulated genes in E. coli identified yqjH, which encodes a protein with 23% amino acid identity with ViuB (37). Subsequent structural and functional studies of YqjH have shown that it is an FAD-containing enzyme that uses NADPH to reduce ferric iron in triscatecholates and ferric dicitrate (11, 12, 38). This class of proteins has been termed siderophore-interacting proteins (SIPs), and though it is generally assumed that ViuB also functions as a SIP, to our knowledge this was not previously supported by experimental data. To test whether yqjH can functionally substitute for viuB, viuB and yqjH were separately cloned onto the low-copy-number vector pWKS30 and transformed into the V. cholerae viuB mutant, which is also defective in the synthesis of vibriobactin. Plasmids carrying either yqjH or viuB allowed the use of vibriobactin and vibriobactin A as iron sources (Fig. 3A), indicating that yqjH can substitute for viuB in V. cholerae. To determine whether viuB can functionally substitute for yqjH in E. coli, an E. coli entF yqjH fes mutant was constructed. This mutant carrying the empty plasmid vector failed to use purified enterobactin, MECAM, or the siderophore secreted by wild-type E. coli in a cross-feeding assay, consistent with published data (12). As expected, providing fes allowed use of enterobactin and the siderophore secreted by E. coli but not the uncleavable synthetic siderophore MECAM (Fig. 3B). When either yqjH or viuB was supplied on a plasmid, MECAM, in addition to enterobactin and the E. coli siderophore, was used (Fig. 3B). This shows that ViuB can functionally substitute for YqjH.
FIG 3.
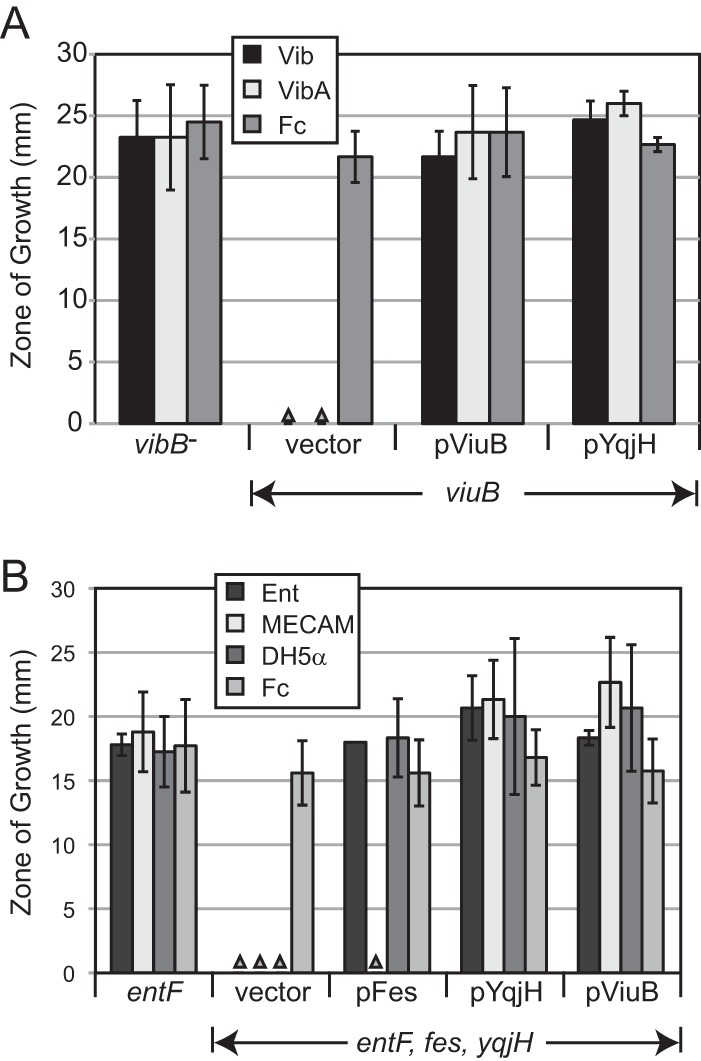
ViuB and YqjH can functionally substitute for each other. V. cholerae (A) or E. coli (B) of the indicated genotypes and carrying the plasmid noted below each set of bars was seeded in EDDA agar, and vibriobactin (Vib), vibriobactin A (VibA), ferrichrome (Fc), enterobactin (Ent), MECAM, or E. coli strain DH5α was spotted on the surface. The zone of growth was measured after 24 h of incubation at 37°C. Each bar represents the average from at least three independent experiments ± standard derivation. The triangles indicate no growth. The strains used in the assay were ALV101 (vibB), JRB9 (viuB), ARM110 (entF), and EWE1026 (entF fes yqjH).
Requirement for ViuB.
To further characterize the role of ViuB in V. cholerae, we tested siderophore utilization by the viuB mutant JRB9 (21). As previously observed for vibriobactin, utilization of vibriobactin A also required viuB (Fig. 3A). The viuB mutant efficiently used the compounds secreted by wild-type E. coli, and consistent with this, the use of the DBS dimer and DBS trimer did not require ViuB (Fig. 4). ViuB was not required for utilization of MECAM (Fig. 4), which is distinct from the observation for E. coli, where use of MECAM requires YqjH (12). ViuB was also not required for the use of fluvibactin, fluvibactin A, or ferrichrome (the zones of stimulation for the viuB mutant JRB9 were 21.0 ± 3.5, 26.3 ± 3.1, and 22.2 ± 2.9 mm, respectively).
FIG 4.
Siderophore utilization by a V. cholerae viuB mutant. The V. cholerae viuB mutant strain JRB9 was seeded into EDDA-containing LB agar, and the E. coli strain or the siderophore indicated below each picture was spotted on the surface. The numbers in parentheses are the average diameter in millimeters of the zone of growth ± standard deviation from at least three independent experiments. All zones were measured and photographed after 24 h of growth at 37°C.
DISCUSSION
It was previously observed that V. cholerae efficiently used a siderophore secreted by enterobactin-producing strains of E. coli, but it was not known which of the enterobactin-related compounds secreted by E. coli can be used by V. cholerae. We show here that it is primarily the linear DBS dimer and DBS trimer that are used. Efficient use of enterobactin by V. cholerae occurred only when both E. coli fepA and fes were supplied on plasmids (Fig. 2C), indicating that both transport and utilization of the cytoplasmic ferrienterobactin are deficient in V. cholerae. The requirement for Fes is of interest, because plasmid-carried viuB was sufficient to allow utilization of enterobactin in an E. coli strain deficient in Fes and YqjH (Fig. 3), indicating that ViuB can reduce ferric iron bound to enterobactin. It is not clear why ViuB allowed use of iron from enterobactin in the absence of Fes in E. coli but not in V. cholerae, but possibilities include that in E. coli, the level of ViuB is higher due to expression from the plasmid (albeit having a low copy number), more efficient transport of enterobactin by FepA with its native TonB partner, or some unidentified difference in the genetic background of the two species.
Consistent with published data (14), we observed weak usage of enterobactin when high concentrations of cyclic enterobactin were provided, and this level was similar to that observed when elevated concentrations of enterobactin were spotted on an E. coli fepA mutant (data not shown). In these experiments, it is not possible to determine whether this is due to a low level of usage of the cyclic enterobactin, or whether the enterobactin is breaking down to its linear derivatives during the 24-h growth of the bacteria.
The genetic requirements for use of the siderophore ligands investigated in this study are summarized in Table 4. V. cholerae transports primarily linear siderophores, which is consistent with its lack of a gene encoding the esterase Fes. Oddly, the sole cyclic siderophore used by V. cholerae in our assays was MECAM. At this time, there are no structural studies of IrgA or VctA, so the basis for their ability to transport MECAM but not enterobactin is unknown. MECAM lacks ester bonds that can be hydrolyzed by Fes, and its use in E. coli requires reduction of the iron by the YqjH reductase (12). By analogy, we had anticipated that use of MECAM by V. cholerae would require ViuB, but this was not the case (Fig. 4). Similarly, YqjH was required for use of the DBS trimer by an E. coli fes mutant (12), but ViuB was not needed for use of the DBS trimer in V. cholerae (Fig. 4). We also show that fluvibactin, fluvibactin A, ferrichrome, and the E. coli-produced siderophores can all be used by a V. cholerae viuB mutant (Fig. 4) (in the test described above). The molecular basis of this ViuB-independent siderophore utilization is not known, but we speculate that V. cholerae may have an unidentified reductase that has sufficient reducing potential to reduce ferric iron bound to the ViuB-independent siderophores but not to vibriobactin or vibriobactin A. We are not aware of a candidate for that reductase. V. cholerae does not have a homologue of FhuF, which removes iron from hydroxamate siderophores in E. coli (39), so it is also unknown how iron is reduced from ferrichrome in V. cholerae.
TABLE 4.
Summary of V. cholerae proteins participating in siderophore utilization
| Ligand | Transport by catechol siderophore receptor: |
Utilization via: |
ViuB requirement | |||
|---|---|---|---|---|---|---|
| ViuA | VctA | IrgA | TonB1 | TonB2 | ||
| E. colia | No | Yes | Yes | No | Yes | No |
| DBS dimer | No | Yes | Yes | No | Yes | No |
| DBS trimer | Nob | Yes | Yes | No | Yes | No |
| MECAM | No | Yes | Yes | No | Yes | No |
| Vibriobactin | Yes | No | No | Yes | Yes | Yes |
| Vibriobactin A | Yes | No | No | Yes | Yes | Yes |
| Fluvibactin | Yesc | Yesd | Yesd | Yes | Yesd | No |
| Fluvibactin A | Yes | Yes | Yes | Yes | Yes | No |
The ligand is the siderophore secreted when 5 μl of an overnight culture of Ent+ E. coli was spotted on the surface of the medium.
A faint zone of growth was seen in some assays.
Diffuse zone only.
Compact zone only.
Outer membrane receptors for siderophores are generally considered to be quite specific for their ligands, while the inner membrane ABC transporters recognize a variety of closely related siderophores. In this work, we show that the catechol receptors of V. cholerae each possess considerable functional plasticity. This is especially true for IrgA and VctA, both of which transported at least five distinct ligands, including the cyclic siderophore MECAM, and linear siderophores in which the dihydroxybenzoyl moieties are joined either through serines or through norspermidine (Fig. 1; Table 4). ViuA was more specific, in that it transported four different siderophores; however, their structures were more similar, all consisting of three dihydroxybenzoyl moieties joined on a linear norspermidine backbone (Fig. 1; Table 4). V. cholerae uses one additional catechol siderophore, agrobactin, in which the dihydroxybenzoyl moieties are connected through a spermidine backbone (14, 23). It is likely to be transported by one or more of these catechol siderophore receptors, but this has not been determined. Recent work in Campylobacter jejuni shows that the ability of a single outer membrane receptor to transport multiple, related ligands is not limited to V. cholerae receptors (40), and this may represent an underappreciated aspect of siderophore biology.
The biological relevance of the ability of V. cholerae to use the linear forms of enterobactin is unknown. It is unlikely that V. cholerae would encounter significant numbers of E. coli bacteria during infection of the human host, since V. cholerae primarily colonizes the small intestine, while E. coli is mostly located in the colon. However, it is likely that V. cholerae would encounter E. coli in sewers and in waters contaminated with sewage. The ability to use the enterobactin breakdown products may contribute to its ability to persist in these aquatic environments and cause periodic outbreaks in regions where it is endemic.
Information about the specificities of the siderophore receptors has implications beyond V. cholerae. Several members of the Vibrionaceae have been shown to transport enterobactin, including Vibrio parahaemolyticus (41), Aeromonas hydrophila (42), and Vibrio anguillarum (43), and homologues of IrgA and/or VctA are found in the sequenced genomes of a number of members of the Vibrionaceae (Table 5 shows selected homologues). V. fluvialis, Vibrio vulnificus, Vibrio alginolyticus, and V. parahaemolyticus have been reported to use vibriobactin (24), and the vulnibactin receptors of V. vulnificus (44) and V. fluvialis have significant sequence similarity with ViuA (Table 5). The structure of vulnibactin is the same as that of vibriobactin, except that the two dihydroxybenzoate residues attached to the norspermidine through the oxazoline moieties in vibriobactin are replaced with salicylate in vulnibactin (45). We speculate that these vulnibactin receptors may also function in the transport of vibriobactin. The V. alginolyticus genome contains a receptor gene with moderate similarity with ViuA (Table 5), while we were not able to identify a candidate receptor for vibriobactin in the V. parahaemolyticus genome. It is not known whether the receptors of these other Vibrionaceae strains show functional plasticity similar to that of the V. cholerae receptors. However, vibrios are primarily aquatic species that live in complex associations with each other and with varied zooplankton and phytoplankton species (46). In these environments, there may be strong selection to adapt existing siderophore acquisition systems for the use of the varied available siderophores.
TABLE 5.
Homologies of V. cholerae catechol transporters to selected Vibrionaceae proteins
| Protein (ORF)a and species | Identity (%) | Similarity (%) | GI no. |
|---|---|---|---|
| IrgA (VC0475) | |||
| V. alginolyticus | 68 | 80 | 491523372 |
| V. parahaemolyticus | 67 | 80 | 646358207 |
| V. anguillarum (FetA) | 67 | 79 | 516401775 |
| A. hydrophila | 45 | 64 | 500026927 |
| V. fluvialis | 43 | 59 | 521097610 |
| VctA (VCA0232) | |||
| V. fluvialis | 70 | 82 | 520908277 |
| V. parahaemolyticus | 64 | 78 | 639559333 |
| V. alginolyticus | 64 | 78 | 491524981 |
| ViuA (VC2211) | |||
| V. vulnificus (VuuA) | 76 | 87 | 516401775 |
| V. fluvialis (VuuA) | 46 | 61 | 521097987 |
| V. alginolyticus | 30 | 48 | 305379311 |
Open reading frame (ORF) number is according to the V. cholerae strain N16961 (27).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants AI091957 (to S.M.P.) and AI011744 (to K.N.R.) from the National Institute of Allergy and Infectious Diseases.
We thank Alexandra Mey for helpful discussions, strains, and critical review of the manuscript and Benjamin Koestler for comments on the manuscript. We are also grateful to Jide Xu for assistance with siderophore synthesis, Eric Peng for strains, and Carolyn Fisher for technical assistance. We thank NBRP (NIG, Japan) for E. coli strains obtained from the Keio collection.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00417-15.
REFERENCES
- 1.Ali M, Lopez AL, You YA, Kim YE, Sah B, Maskery B, Clemens J. 2012. The global burden of cholera. Bull World Health Organ 90:209–218A. doi: 10.2471/BLT.11.093427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moore S, Thomson N, Mutreja A, Piarroux R. 2014. Widespread epidemic cholera caused by a restricted subset of Vibrio cholerae clones. Clin Microbiol Infect 20:373–379. doi: 10.1111/1469-0691.12610. [DOI] [PubMed] [Google Scholar]
- 3.Chu BC, Garcia-Herrero A, Johanson TH, Krewulak KD, Lau CK, Peacock RS, Slavinskaya Z, Vogel HJ. 2010. Siderophore uptake in bacteria and the battle for iron with the host; a bird's eye view. Biometals 23:601–611. doi: 10.1007/s10534-010-9361-x. [DOI] [PubMed] [Google Scholar]
- 4.Parrow NL, Fleming RE, Minnick MF. 2013. Sequestration and scavenging of iron in infection. Infect Immun 81:3503–3514. doi: 10.1128/IAI.00602-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cassat JE, Skaar EP. 2013. Iron in infection and immunity. Cell Host Microbe 13:509–519. doi: 10.1016/j.chom.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andrews SC, Robinson AK, Rodriguez-Quinones F. 2003. Bacterial iron homeostasis. FEMS Microbiol Rev 27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 7.Krewulak KD, Vogel HJ. 2011. TonB or not TonB: is that the question? Biochem Cell Biol 89:87–97. doi: 10.1139/O10-141. [DOI] [PubMed] [Google Scholar]
- 8.Postle K, Larsen RA. 2007. TonB-dependent energy transduction between outer and cytoplasmic membranes. Biometals 20:453–465. doi: 10.1007/s10534-006-9071-6. [DOI] [PubMed] [Google Scholar]
- 9.Miethke M. 2013. Molecular strategies of microbial iron assimilation: from high-affinity complexes to cofactor assembly systems. Metallomics 5:15–28. doi: 10.1039/C2MT20193C. [DOI] [PubMed] [Google Scholar]
- 10.Brickman TJ, McIntosh MA. 1992. Overexpression and purification of ferric enterobactin esterase from Escherichia coli. Demonstration of enzymatic hydrolysis of enterobactin and its iron complex. J Biol Chem 267:12350–12355. [PubMed] [Google Scholar]
- 11.Wang S, Wu Y, Outten FW. 2011. Fur and the novel regulator YqjI control transcription of the ferric reductase gene yqjH in Escherichia coli. J Bacteriol 193:563–574. doi: 10.1128/JB.01062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miethke M, Hou J, Marahiel MA. 2011. The siderophore-interacting protein YqjH acts as a ferric reductase in different iron assimilation pathways of Escherichia coli. Biochemistry 50:10951–10964. doi: 10.1021/bi201517h. [DOI] [PubMed] [Google Scholar]
- 13.Hantke K. 1990. Dihydroxybenzoylserine—a siderophore for E coli. FEMS Microbiol Lett 67:5–8. [DOI] [PubMed] [Google Scholar]
- 14.Griffiths GL, Sigel SP, Payne SM, Neilands JB. 1984. Vibriobactin, a siderophore from Vibrio cholerae. J Biol Chem 259:383–385. [PubMed] [Google Scholar]
- 15.Wyckoff EE, Mey AR, Payne SM. 2007. Iron acquisition in Vibrio cholerae. Biometals 20:405–416. doi: 10.1007/s10534-006-9073-4. [DOI] [PubMed] [Google Scholar]
- 16.Keating TA, Marshall CG, Walsh CT. 2000. Reconstitution and characterization of the Vibrio cholerae vibriobactin synthetase from VibB, VibE, VibF, and VibH. Biochemistry 39:15522–15530. doi: 10.1021/bi0016523. [DOI] [PubMed] [Google Scholar]
- 17.Butterton JR, Stoebner JA, Payne SM, Calderwood SB. 1992. Cloning, sequencing, and transcriptional regulation of viuA, the gene encoding the ferric vibriobactin receptor of Vibrio cholerae. J Bacteriol 174:3729–3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seliger SS, Mey AR, Valle AM, Payne SM. 2001. The two TonB systems of Vibrio cholerae: redundant and specific functions. Mol Microbiol 39:801–812. doi: 10.1046/j.1365-2958.2001.02273.x. [DOI] [PubMed] [Google Scholar]
- 19.Wyckoff EE, Valle AM, Smith SL, Payne SM. 1999. A multifunctional ATP-binding cassette transporter system from Vibrio cholerae transports vibriobactin and enterobactin. J Bacteriol 181:7588–7596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mey AR, Wyckoff EE, Oglesby AG, Rab E, Taylor RK, Payne SM. 2002. Identification of the Vibrio cholerae enterobactin receptors VctA and IrgA: IrgA is not required for virulence. Infect Immun 70:3419–3426. doi: 10.1128/IAI.70.7.3419-3426.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Butterton JR, Calderwood SB. 1994. Identification, cloning, and sequencing of a gene required for ferric vibriobactin utilization by Vibrio cholerae. J Bacteriol 176:5631–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rogers MB, Sexton JA, DeCastro GJ, Calderwood SB. 2000. Identification of an operon required for ferrichrome iron utilization in Vibrio cholerae. J Bacteriol 182:2350–2353. doi: 10.1128/JB.182.8.2350-2353.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ong SA, Peterson T, Neilands JB. 1979. Agrobactin, a siderophore from Agrobacterium tumefaciens. J Biol Chem 254:1860–1865. [PubMed] [Google Scholar]
- 24.Andrus CR, Walter M, Crosa JH, Payne S. 1983. Synthesis of siderophores by pathogenic Vibrio species. Curr Microbiol 9:209–214. doi: 10.1007/BF01567583. [DOI] [Google Scholar]
- 25.Yamamoto S, Okujo N, Fujita Y, Saito M, Yoshida T, Shinoda S. 1993. Structures of two polyamine-containing catecholate siderophores from Vibrio fluvialis. J Biochem 113:538–544. [DOI] [PubMed] [Google Scholar]
- 26.Rutz JM, Abdullah T, Singh SP, Kalve VI, Klebba PE. 1991. Evolution of the ferric enterobactin receptor in gram-negative bacteria. J Bacteriol 173:5964–5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rogers HJ. 1973. Iron-binding catechols and virulence in Escherichia coli. Infect Immun 7:445–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wyckoff EE, Stoebner JA, Reed KE, Payne SM. 1997. Cloning of a Vibrio cholerae vibriobactin gene cluster: identification of genes required for early steps in siderophore biosynthesis. J Bacteriol 179:7055–7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mey AR, Payne SM. 2001. Haem utilization in Vibrio cholerae involves multiple TonB-dependent haem receptors. Mol Microbiol 42:835–849. [DOI] [PubMed] [Google Scholar]
- 30.Harris WR, Weitl FL, Raymond KN. 1979. Synthesis and evaluation of an enterobactin model compound. 1,3,5-Tris-(2,3-dihydroxybenzoylaminomethyl)benzene and its iron(III) complex. J Chem Soc Chem Commun 1979:177–178. [Google Scholar]
- 31.Marinez ER, Salmassian EK, Lau TT, Gutierrez CG. 1996. Enterobactin and enantioenterobactin. J Org Chem 61:3548–3550. doi: 10.1021/jo9520194. [DOI] [Google Scholar]
- 32.Rastetter WH, Erickson TJ, Venuti MC. 1981. Synthesis of iron chelators—enterobactin, enantioenterobactin, and a chiral analog. J Org Chem 46:3579–3590. doi: 10.1021/jo00331a001. [DOI] [Google Scholar]
- 33.Allred BE, Correnti C, Clifton MC, Strong RK, Raymond KN. 2013. Siderocalin outwits the coordination chemistry of vibriobactin, a siderophore of Vibrio cholerae. ACS Chem Biol 8:1882–1887. doi: 10.1021/cb4002552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winkelmann G, Cansier A, Beck W, Jung G. 1994. HPLC separation of enterobactin and linear 2,3-dihydroxybenzoylserine derivatives: a study on mutants of Escherichia coli defective in regulation (fur), esterase (fes) and transport (fepA). Biometals 7:149–154. [DOI] [PubMed] [Google Scholar]
- 35.Heidinger S, Braun V, Pecoraro VL, Raymond KN. 1983. Iron supply to Escherichia coli by synthetic analogs of enterochelin. J Bacteriol 153:109–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Occhino DA, Wyckoff EE, Henderson DP, Wrona TJ, Payne SM. 1998. Vibrio cholerae iron transport: haem transport genes are linked to one of two sets of tonB, exbB, exbD genes. Mol Microbiol 29:1493–1507. [DOI] [PubMed] [Google Scholar]
- 37.McHugh JP, Rodriguez-Quinones F, Abdul-Tehrani H, Svistunenko DA, Poole RK, Cooper CE, Andrews SC. 2003. Global iron-dependent gene regulation in Escherichia coli. A new mechanism for iron homeostasis. J Biol Chem 278:29478–29486. [DOI] [PubMed] [Google Scholar]
- 38.Bamford VA, Armour M, Mitchell SA, Cartron M, Andrews SC, Watson KA. 2008. Preliminary X-ray diffraction analysis of YqjH from Escherichia coli: a putative cytoplasmic ferri-siderophore reductase. Acta Crystallogr Sect F Struct Biol Cryst Commun 64:792–796. doi: 10.1107/S174430910802352X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matzanke BF, Anemuller S, Schunemann V, Trautwein AX, Hantke K. 2004. FhuF, part of a siderophore-reductase system. Biochemistry 43:1386–1392. doi: 10.1021/bi0357661. [DOI] [PubMed] [Google Scholar]
- 40.Naikare H, Butcher J, Flint A, Xu J, Raymond KN, Stintzi A. 2013. Campylobacter jejuni ferric-enterobactin receptor CfrA is TonB3 dependent and mediates iron acquisition from structurally different catechol siderophores. Metallomics 5:988–996. doi: 10.1039/c3mt20254b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanabe T, Funahashi T, Shiuchi K, Okajima N, Nakao H, Miyamoto K, Tsujibo H, Yamamoto S. 2012. Characterization of Vibrio parahaemolyticus genes encoding the systems for utilization of enterobactin as a xenosiderophore. Microbiology 158:2039–2049. doi: 10.1099/mic.0.059568-0. [DOI] [PubMed] [Google Scholar]
- 42.Funahashi T, Tanabe T, Miyamoto K, Tsujibo H, Maki J, Yamamoto S. 2013. Characterization of a gene encoding the outer membrane receptor for ferric enterobactin in Aeromonas hydrophila ATCC 7966(T). Biosci Biotechnol Biochem 77:353–360. [DOI] [PubMed] [Google Scholar]
- 43.Naka H, Crosa JH. 2012. Identification and characterization of a novel outer membrane protein receptor FetA for ferric enterobactin transport in Vibrio anguillarum 775 (pJM1). Biometals 25:125–133. doi: 10.1007/s10534-011-9488-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Webster AC, Litwin CM. 2000. Cloning and characterization of vuuA, a gene encoding the Vibrio vulnificus ferric vulnibactin receptor. Infect Immun 68:526–534. doi: 10.1128/IAI.68.2.526-534.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Okujo N, Saito M, Yamamoto S, Yoshida T, Miyoshi S, Shinoda S. 1994. Structure of vulnibactin, a new polyamine-containing siderophore from Vibrio vulnificus. Biometals 7:109–116. [DOI] [PubMed] [Google Scholar]
- 46.Takemura AF, Chien DM, Polz MF. 2014. Associations and dynamics of Vibrionaceae in the environment, from the genus to the population level. Front Microbiol 5:38. doi: 10.3389/fmicb.2014.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mekalanos JJ, Swartz DJ, Pearson GD, Harford N, Groyne F, de Wilde M. 1983. Cholera toxin genes: nucleotide sequence, deletion analysis and vaccine development. Nature 306:551–557. doi: 10.1038/306551a0. [DOI] [PubMed] [Google Scholar]
- 48.Wyckoff EE, Mey AR, Leimbach A, Fisher CF, Payne SM. 2006. Characterization of ferric and ferrous iron transport systems in Vibrio cholerae. J Bacteriol 188:6515–6523. doi: 10.1128/JB.00626-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mey AR, Payne SM. 2003. Analysis of residues determining specificity of Vibrio cholerae TonB1 for its receptors. J Bacteriol 185:1195–1207. doi: 10.1128/JB.185.4.1195-1207.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sigel SP, Payne SM. 1982. Effect of iron limitation on growth, siderophore production and expression of outer membrane proteins of Vibrio cholerae. J Bacteriol 150:148–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166:557–580. doi: 10.1016/S0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 52.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mey AR, Wyckoff EE, Hoover LA, Fisher CR, Payne SM. 2008. Vibrio cholerae VciB promotes iron uptake via ferrous iron transporters. J Bacteriol 190:5953–5962. doi: 10.1128/JB.00569-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang RF, Kushner SR. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195–199. doi: 10.1016/0378-1119(91)90366-J. [DOI] [PubMed] [Google Scholar]
- 56.Cherepanov PP, Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14. doi: 10.1016/0378-1119(95)00193-A. [DOI] [PubMed] [Google Scholar]
- 57.Chang ACY, Cohen SN. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol 134:1141–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Figurski DH, Helinski DR. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A 76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.