ABSTRACT
Resistance to methicillin and other β-lactam antibiotics in staphylococci is due to mecA, which is carried on a genomic island, staphylococcal cassette chromosome mec (SCCmec). The chromosomal excision and integration of SCCmec are mediated by the site-specific recombinase CcrAB or CcrC, encoded within this element. A plasmid-borne system was constructed to assess the activities of CcrA and CcrB in the excision and integration of SCCmec in Escherichia coli and Staphylococcus aureus. The excision frequency in E. coli mediated by CcrAB from methicillin-resistant S. aureus (MRSA) strain N315 was only 9.2%, while the integration frequency was 31.4%. In S. aureus the excision and integration frequencies were 11.0% and 18.7%, respectively. Truncated mutants identified the N-terminal domain of either CcrB or CcrA to be necessary for both integration and excision, while the C-terminal domain was important for recombination efficiency. Site-directed mutagenesis of the N-terminal domain identified S11 and R79 of CcrA and S16, R89, T149, and R151 of CcrB to be residues essential for catalytic activities, and the critical location of these residues was consistent with a model of the tertiary structure of the N terminus of CcrA and CcrB. Furthermore, CcrAB and CcrC, cloned from a panel of 6 methicillin-resistant S. aureus strains and 2 methicillin-resistant Staphylococcus epidermidis strains carrying SCCmec types II, IV, and V, also catalyzed integration at rates 1.3 to 10 times higher than the rates at which they catalyzed excision, similar to the results from N315. The tendency of SCCmec integration to be favored over excision may explain the low spontaneous excision frequency seen among MRSA strains.
IMPORTANCE Spontaneous excision of the genomic island (SCCmec) that encodes resistance to beta-lactam antibiotics (methicillin resistance) in staphylococci would convert a methicillin-resistant strain to a methicillin-susceptible strain, improving therapy of difficult-to-treat infections. This study characterizes a model system by which the relative frequencies of excision and integration can be compared. Using a plasmid-based model for excision and integration mediated by the recombinases CcrA and CcrB, integration occurred at a higher frequency than excision, consistent with the low baseline excision frequency seen in most strains. This model system can now be used to study conditions and drugs that may raise the SCCmec excision frequency and generate strains that are beta-lactam susceptible.
INTRODUCTION
Staphylococcus aureus is a leading cause of infection in hospitals and communities throughout the world. The treatment for such infections has become increasingly difficult as a result of the development of resistance to multiple antibiotics. The multiresistance phenotype in S. aureus is built around the acquisition of resistance to beta-lactam antibiotics (resulting in methicillin-resistant S. aureus [MRSA]), mediated by the gene mecA, which encodes a beta-lactam target (PBP 2a) resistant to beta-lactam inactivation. mecA is carried on a mobile genomic island, staphylococcal cassette chromosome mec (SCCmec) (1). At least 11 types of SCCmec (types I to XI), ranging from 22 kb to >60 kb, have been described by typing based on the mec and ccr complexes (2; http://www.sccmec.org/Pages/SCC_TypesEN.html). SCCmec integrates into and excises from the chromosome at specific DNA sites (att), with integration and excision being catalyzed by a group of serine recombinases, CcrAB and CcrC, carried by the element (3–6). Two pairs of att sites are required for SCCmec integration (attB and attS) and excision (attR and attL) (6). All of the att sites share a conserved 8-base core region, required for CcrB DNA binding, with variable flanking sequences that determine the insertion frequency (6).
Serine recombinases catalyze site-specific recombination and play an essential role in the movement of genetic elements, such as bacteriophages, transposons, and large genomic islands (7, 8). For some serine recombinases, there are two different genes for excision and integration, such as phage integrase and phage directionality factor Xis for excision (9–14). Other recombinases, such as TndX from the Clostridium difficile tetracycline resistance element (15, 16), mediate both excision and integration. CcrAB and CcrC are examples of large serine recombinases that belong to the latter group. While CcrC is a single protein that carries out both activities, the two proteins CcrA and CcrB must interact in a precise ratio to mediate either integration or excision (4–6). Our previous data showed that CcrB is the major DNA-binding protein, while CcrA is required to direct the complex to the attB integration site (5, 6). However, recent data from Misiura et al. (17) demonstrated weak DNA-binding activity for CcrA as well. Most of our previous work has focused on the roles of CcrA and CcrB in integration by putting a gene(s) encoding one or both of these proteins on a temperature-sensitive plasmid, measuring the frequency of integration into chromosomal att sites, and scoring for antibiotic resistance after growth at the nonpermissive temperature for plasmid replication. However, we have not previously assessed the relative frequency of integration versus excision. Stojanov et al. (18) found a very low frequency of spontaneous SCCmec excision (10−5 to 10−6) in S. aureus, a frequency that we have also observed (L. Wang, unpublished observations). This suggests either that excision rarely occurred or that excision occurred frequently but was followed by immediate reintegration. The following study was performed in an attempt to measure the relative rates of SCCmec excision and integration mediated by CcrAB and CcrC, using a two-plasmid system in both Escherichia coli and S. aureus, and the contribution of the various domains of each of the two proteins CcrA and CcrB to the process.
MATERIALS AND METHODS
Strains and media.
All of the strains used in this work are listed in Table S1 in the supplemental material. Staphylococcal strains were cultured in tryptic soy broth (TSB; Thermo Scientific, Waltham, MA). E. coli TOP10 (Invitrogen, Carlsbad, CA) was used for gene cloning. The antibiotics and concentrations used were as follows: 100 μg/ml ampicillin (Ap) for selection of E. coli strains following transformation and 10 μg/ml chloramphenicol (Cml) and 5 μg/ml tetracycline (Tet) for selection of S. aureus strains following electroporation (19).
Plasmid model for excision and integration.
Each excision/integration model plasmid contained a tetracycline resistance gene (tetM) flanked by a pair of att sites in direct orientation and by a copy of a ccr gene driven by its own promoter (ccrA, ccrB, ccrAB, or ccrC) (Fig. 1B and D). The ccrAB genes for all studies were from strain N315 (see Table S1 in the supplemental material), unless otherwise specified. The ccrC gene was from strain WBG8318 (see Table S1 in the supplemental material). All of the att site-containing fragments were 600 bp, with the att core sequence being in the middle. Recombination between attR and attL was defined as excision, and that between attB and attS was defined as integration. Recombination mediated by Ccr proteins was identified by the deletion of the tetM gene. As recombination occurred, a 3.1-kb fragment containing tetM was deleted, reducing the size of the plasmid. Recombination was identified and quantified by first cleaving the plasmid with BamHI, releasing the ccr gene(s), and leaving the backbone containing either the unexcised tetM-containing fragment (9 kb) or the excised fragment (6 kb). This size shift was easily detected by agarose gel electrophoresis. The relative recombination frequency (RF) was determined by the following equation: RF (in percent) = molecular number of excised bands/(molecular number of excised bands + molecular number of unexcised bands).
FIG 1.
Schematic diagram of the excision and integration of chromosome and model plasmids. (A and C) Schematic diagrams of excision and integration of SCCmec. Black and red double lines, the chromosome and SCCmec, respectively; arrows, the orfX gene; arrowheads, att sites. (B and D) Schematic diagrams of excision and integration of model plasmids. Both of the plasmids were derived from a chloramphenicol-resistant E. coli-S. aureus shuttle vector, pCN38. Green, a tetracycline resistance gene (tetM), flanked with a pair of 0.6-kb att site-containing fragments in a direct orientation; purple arrows, a copy of ccrAB genes from MRSA or MRSE strains; blue arrows, a chloramphenicol resistance gene (cat194); yellow arrows, an ampicillin resistance gene (amp).
The molecular number of each DNA fragment was calculated to be the relative band density divided by the molecular weight. The relative band density on the gel image was determined by ImageJ software (http://imagej.nih.gov/ij/), and the molecular weight was calculated as follows (20): approximate molecular weight of double-stranded DNA (dsDNA; in grams per mole) = numbers of nucleotides × 660 g/mol.
The loss of tetracycline resistance was not a useful marker for recombination because in a multicopy plasmid system recombination could occur in some fraction of the plasmid population but not the entire population, resulting in lower numbers for excision and integration. However, the results were in the same direction and the differences mirrored those seen with the E. coli plasmid system. Thus, since the plasmid size system did not work well in S. aureus, we used tetracycline susceptibility to confirm in S. aureus some of the observations made in E. coli.
The plasmids described above were constructed as follows. A chloramphenicol-resistant E. coli-S. aureus shuttle vector, pCN38 (21), was the backbone for all constructs. The ∼0.6-kb fragments containing attR and attL sites were amplified from the N315 genome with primers attR-F-BamHI and attR-R-KpnI and primers attL-F-KpnI and attL-R-EcoRI, respectively. The 0.6-kb attB and attS fragments were amplified from N315/pWA46 (5) with primers attB-F-BamHI and attB-R-KpnI and primers attS-F-KpnI and attS-R-EcoRI, respectively. A 2.5-kb tetM gene was amplified from pCN36 (21) using the primers tetM-F-KpnI and tetM-R-KpnI. pWA340/pWA341 were constructed by the use of three PCR products cloned into pCN38 one by one: an attR- or attB-containing fragment at the BamHI/KpnI site, an attL- or attS-containing fragment at the KpnI/EcoRI site, and then a tetM fragment at the KpnI site. The same strategy was used to construct pWA362/pWA363, in which the att sites are from the MW2 genome, and pWA376/pWA377, in which the att sites are from the WBG8318 (WIS) genome. ccrA, ccrB, ccrAB, or ccrC was amplified from the genomic DNA of S. aureus strains N315, MW2, C98-370, C99-529, J28, J35, FPR3757, and WBG8318 and Staphylococcus epidermidis strains SE7 and SE50. PCR products were cloned into the BamHI site of pWA340/pWA341, pWA362/pWA363, or pWA376/pWA377, yielding a set of excision/integration model plasmids. All of the primers used are shown in Table S2 in the supplemental material; all of the plasmids and their characteristics are shown in Table S1 in the supplemental material.
Construction of truncated CcrA and CcrB on model plasmids.
To delete the N terminus of CcrA (from amino acids [aa] 119 to 177), two amplified fragments were ligated together after digestion by KpnI: one 0.6-kb fragment amplified with primers ccrAB-BamHI-2 and ccrA-SR-3 included the promoter and first 118 aa of ccrA; the other 1.7-kb fragment of intact ccrB was amplified with ccrB-SR-1 and ccrAB-BamHI-1. The ligated product was amplified with primers ccrAB-BamHI-2 and ccrAB-BamHI-1 and then inserted into pWA340/pWA341 after it was digested by BamHI, yielding pWA533/pWA534. To delete the C terminus of CcrA (from aa 299 to 450), two amplified fragments were ligated together after digestion by KpnI: one 1-kb fragment amplified with primers ccrAB-BamHI-2 and ccrA-TR-5 and another 1.7-kb fragment of intact ccrB amplified with primers ccrB-SR-1 and ccrAB-BamHI-1. The ligated product was amplified with primers ccrAB-BamHI-1 and ccrAB-BamHI-2 and then inserted into pWA340/pWA341 after digestion by BamHI, yielding pWA535/pWA536. To delete the N terminus of CcrB (from aa 1 to 245), two amplified fragments were ligated together after digestion by KpnI: one 1.7-kb fragment including the promoter and intact ccrA amplified with primers ccrAB-BamHI-2 and ccrA-TR-6, and another 1-kb fragment including the last 313 aa of CcrB amplified with primers ccrB-SR-9 and ccrAB-BamHI-1. The ligated product was amplified with primers ccrAB-BamHI-1 and ccrAB-BamHI-2 and then inserted into pWA340/pWA341 after it was digested by BamHI, yielding pWA526/pWA527. To get three C-terminal deletion mutants of CcrB (from aa 399 to 543, aa 301 to 543, and aa 241 to 543), three fragments were amplified with three pairs of primers, ccrAB-BamHI-2 and ccrB-SR-3, ccrAB-BamHI-2 and ccrB-SR-4, and ccrAB-BamHI-2 and ccrB-SR-8, respectively. These three fragments were digested by BamHI and inserted into pWA340/pWA341, generating pWA537/pWA538, pWA539/pWA540, and pWA541/pWA542, respectively.
Site-directed mutation of CcrA and CcrB proteins on model plasmids.
The primers for site-directed mutagenesis were designed by the online software primerX (http://bioinformatics.org/primerx/cgi-bin/DNA_1.cgi) and are shown in Table S2 in the supplemental material. Site-directed mutation was performed according to the manufacturer's protocol (Stratagene, La Jolla, CA). Briefly, 25 ng of template plasmid was used to prepare the PCR mixture. The amplification was performed as follows: 95°C for 2 min; 95°C for 30 s, 60°C for 30 s, and 68°C for 6.5 min repeated for 18 cycles; and 68°C for 5 min. Then, 2 μl of the DpnI restriction enzyme provided with the kit was added to each amplification reaction mixture. The reaction mixtures were incubated at 37°C for 5 min to digest the parental supercoiled dsDNA. The DpnI-treated DNA was transferred into the XL10-Gold ultracompetent cells provided with the kit. The transformants were confirmed by DNA sequencing.
Statistical analysis.
The results of independent repeat experiments were averaged, and standard deviations were calculated by the use of Microsoft Excel software. The statistical significance of differences between samples was determined by Student's t test, and a P value of <0.05 was considered statistically significant. DNA level differences were evaluated densitometrically using ImageJ software. Each experiment was performed at least three times, unless otherwise described in the text.
Structural modeling.
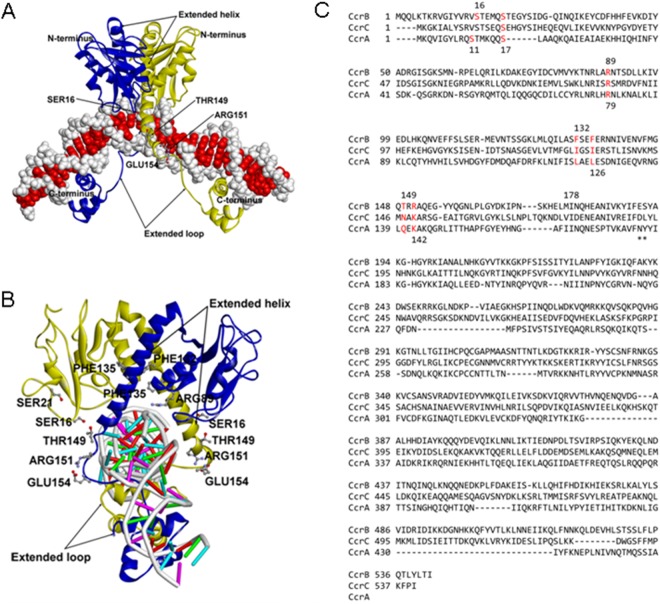
The N-terminal domain of the CcrB (amino acids 1 to 205) sequence was submitted to the HHpred interactive server, using default settings, in order to find a suitable template from the Protein Data Bank (PDB) database (22). A list of templates and query-template alignments by hidden Markov matrix (HMM) profiling was generated, with the best being the γδ resolvase (PDB accession number 1GDT) from Escherichia coli (27% sequence identity; E value, <3.5 × 10−34) (23). The E value is a parameter that describes the number of hits expected to be seen by chance, which decreases exponentially as the score of the match increases. In other words, the E value describes the random background noise, with lower values indicating a better match. The resulting alignment of the γδ resolvase and the CcrB N-terminal domain sequences was provided to the Modeller program for the actual model construction, which is done on the basis of the satisfaction of spatial restraints derived from the template (24). The subsequent CcrB N-terminal domain model was validated using Ramachandran plots and was found to be within acceptable limits (data not shown). The coordinates of the DNA fragment cocrystallized with the γδ resolvase were then merged with the coordinates of the CcrB N-terminal domain model, and the sequence of the DNA fragment was mutated in silico to match the CcrB cognitive DNA sequence using the Discovery Studio Visualizer program (version 3.5; Accelrys, San Diego, CA). After addition of hydrogen atoms, the complete CcrB-DNA model was subjected to energy minimization using the Tripos force field with Gasteiger-Hückel charges and a distance-dependent dielectric to a gradient of 0.01 kcal mol−1 Å−1 in the Sybyl program (version 8.1; Tripos LP, St. Louis, MO).
RESULTS
Recombination frequency in E. coli and S. aureus determined using plasmid models.
The plasmid model was first tested in E. coli. Figure 2A demonstrates the N315 CcrAB and N315 att site-mediated loss of the 3.1-kb tetM-containing fragment from pWA479 (excision) and pWA480 (integration) compared to that in the control plasmids, pWA340 and pWA341, the plasmids with att sites and tetM but without ccrAB. The time course of recombination was monitored through 20 h of growth for pWA479 and pWA480. Plasmid DNA was extracted at 8, 12, 16, and 20 h, digested by BamHI, and separated on a 0.7% agarose gel. As shown in Fig. 2B, the frequency of excision and integration doubled over 20 h, but since the relative frequencies remained approximately the same over this time period, the frequency at 16 h was chosen for comparison for all subsequent studies. In the experiment whose results are illustrated in Fig. 2B, the excision frequency at 16 h was 9.2% ± 1.6%, while the integration frequency was 3-fold greater at 31.4% ± 2.7%. Furthermore, DNA sequencing of the smaller plasmids generated following excision showed that 87.5% (7 out of 8) of excision events occurred between attL and attR, with only 12.5% (1 out of 8) taking place between attL and attR2. This is of interest because we have previously shown that excision from attR and integration into attB require the activity of both CcrA and CcrB, while CcrB alone can mediate excision from an accessory att site within SCCmec, attR2 (6). The model plasmid system shows that excision from attR is favored over excision from attR2.
FIG 2.
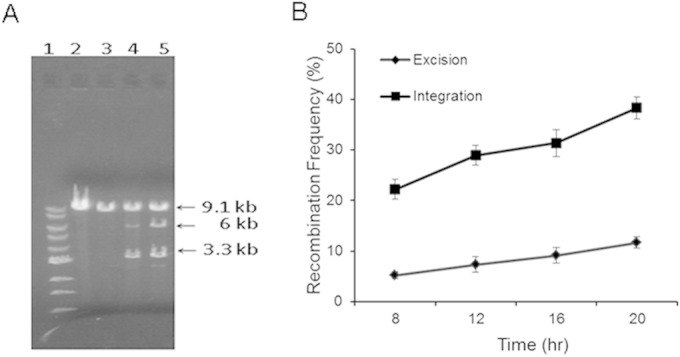
(A) Detection of recombination after plasmid digestion. Lane 1, 1-kb DNA ladder; lane 2, pWA340; lane 3, pWA341; lane 4, pWA479; lane 5, pWA480. All of the plasmids were extracted from E. coli TOP10. The plasmids in lanes 2 and 3 are the model plasmids for excision (pWA340) and integration (pWA341). The plasmids in lanes 4 and 5 are the model plasmids following the addition of ccrAB and cleavage with BamHI, releasing the 3.3-kb ccrAB genes and leaving the plasmid backbone. Recombination frequency was determined by the difference in intensity between the 9.1-kb intact plasmid band and the 6-kb fragment produced when recombination released the tetracycline resistance gene. A complete description of the plasmids is available in Table S1 in the supplemental material. (B) Recombination frequency monitored throughout 8 to 20 h of E. coli growth and analyzed for excision (pWA479) and integration (pWA480). The value at each time point represents the mean from four separate experiments, with bars showing the standard deviation.
To validate the plasmid system in its natural host, S. aureus, the model plasmids pWA479 and pWA480 were electroporated into the strain, AW8, from which attB was deleted (6) and in which no chromosomal integration could occur. Tetracycline-resistant colonies were picked following electroporation, incubated overnight in TSB without tetracycline, and then grown in 25 ml of fresh TSB to an optical density of 0.6. Following plasmid extraction and BamHI digestion, plasmid DNA was examined on agarose gels as described above for E. coli. In the S. aureus assay, the excision frequency was 11.0% ± 2.8%, while the integration frequency was 18.7% ± 2.6%. In addition, the loss of tetracycline resistance could also be assessed in staphylococci. Strains containing each plasmid were inoculated into fresh TSB without tetracycline on each of three consecutive days to maximize recombination on all plasmid copies. Colonies growing on agar without tetracycline overnight were picked and placed on tetracycline plates, and the number susceptible to tetracycline was determined. Among over 600 colonies, 4.7% ± 1.0% of the colonies were tetracycline susceptible for the excision model, but 8.5% ± 1.2% were susceptible for the integration model. As described in Materials and Methods, these values are probably underestimates because in some bacterial cells recombination would not have been complete for all plasmid copies, leaving the colony tetracycline resistant. On the basis of both gel examination of the plasmids and the susceptibility of colonies to tetracycline, the integration frequency was approximately twice as high as the excision frequency in S. aureus. These results are somewhat lower than the 3-fold increase of the integration frequency over the excision frequency in E. coli but are in the same direction, validating the model in both E. coli and the native host. Since the manipulation to determine the frequency of recombination was easier in E. coli, most of the following studies were done only in E. coli.
Activities of CcrA/CcrB mutants on excision and integration.
Although both CcrA and CcrB belong to the large serine recombinase family, they have different contributions to the process of SCCmec excision and integration. Incorporation of either CcrA or CcrB but not both on model plasmids confirmed our previous observations (5) that CcrA alone could not mediate either excision or integration and that CcrB alone mediated excision only and only between attL and attR2 at a low frequency (1.2%; Fig. 3), a result also confirmed by determining the DNA sequence of the deletion junction of plasmids as described above.
FIG 3.

Changes of recombination abilities mediated by CcrAB truncated mutants. CcrA, CcrB, and CcrA CcrB, intact ccrA, ccrB, and ccrAB operons, respectively; CcrAc CcrB, a mutant with deletion of 58 aa of CcrA (from aa 119 to 177) in the ccr operon; CcrAn(1–298) CcrB, a mutant with deletion of the C-terminal domain of CcrA (from aa 299 to 450) in the ccr operon; CcrA CcrBc, a mutant with deletion of the N-terminal domain of CcrB (from aa 1 to 245) in the ccr operon; CcrA CcrBn(1–398), a mutant with deletion of the C-terminal domain of CcrB (from aa 399 to 543) in the ccr operon; CcrA CcrBn(1–300), a mutant with deletion of the C-terminal domain of CcrB (from aa 301 to 543) in the ccr operon; CcrA CcrBn(1–240), a mutant with deletion of the C-terminal domain of CcrB (from aa 241 to 543) in the ccr operon. At least three separate experiments were performed for each mutant. *, P < 0.05 versus the excision frequency of wild-type CcrA CcrB; #, P < 0.05 versus the integration frequency of wild-type CcrA CcrB.
CcrA and CcrB have two domains: a conserved N-terminal catalytic domain (referred to as CcrAn and CcrBn, respectively) and a variant large C-terminal domain (referred to as CcrAc and CcrBc, respectively). In order to assess the roles of these two domains in excision and insertion, deletion mutants of CcrA or CcrB were constructed. The recombination frequency was determined as described above. As shown in Fig. 3, a deletion of only 58 aa of CcrA in the N-terminal domain (aa 119 to 177; yielding the CcrAc CcrB mutant) led to the failure of integration. In this mutant, excision took place only between attL and attR2, a result found only in the complete absence of CcrA. The loss of the C terminus of CcrA [aa 299 to 450; referred to as the CcrAn(1–298) (CcrAn from aa 1 to 298) CcrB mutant] decreased both excision and integration by 90%. Loss of either the N terminus of CcrB (aa 1 to 245; referred to as the CcrA CcrBc mutant) or 303 aa of the C terminus [aa 241 to 543; referred to as the CcrA CcrBn(1–240) (CcrBn from aa 1 to 240) mutant] abolished recombination completely, while deletion of only 243 aa or 145 aa of the C terminus [referred to as the CcrA CcrBn(1–300) (CcrBn from aa 1 to 300) mutant and the CcrA CcrB(1–398) (CcrB from aa 1 to 398) mutant, respectively] was less disruptive, resulting in a 22 to 43% decrease in excision and a 70% decrease in integration. Thus, the N-terminal domains of both Ccr proteins are required for recombination, while the C-terminal domain determines catalytic efficiency.
In order to assess the role of specific amino acids in CcrA and CcrB activity, the ∼220-aa N-terminal CcrB-DNA complex model (Fig. 4A) was constructed using the published structure of similar serine recombinases (PDB accession number 1GDT) (23). The protein structure of the first 220 aa of CcrB is made up of an N-terminal catalytic domain (NTD) and C-terminal helix-turn-helix DNA-binding domain (CTD) that are linked by an extended helix loop arm. In the complex with DNA, the NTD forms the dimer interface, with the CTD being located in the major groove of the DNA, while the extended helix loop arm makes contact with the minor groove of the DNA. It is assumed that CcrA and CcrC form structures similar to those formed by CcrB, as suggested by models of the N-terminal portion of these proteins. Critical residues were identified from the structure model and compared among the aligned sequences of CcrA, CcrB, and CcrC, as shown in Fig. 4B and C. Point mutations were introduced into the model plasmids, substituting glycine or alanine for the existing amino acids, and activities were investigated as described above. The effects of the mutations on excision and integration are shown in Fig. 5. S16A, R89G, T149A, and R151G abolished all recombination activity, as predicted because the first three residues are located at the dimer interface, while R151 is likely to be in the DNA-binding loop. Ser16, Arg89, and Thr149 appear to make intersubunit interactions with the opposite subunit (Ser16 to Glu142 and/or Thr149; Arg89 to Asn139), including the flexible long extended helix loop arm (formed by amino acids 123 to 152). Arg151 is located at the extended helix loop arm that connects the NTD and CTD, and although it is not involved in nucleotide binding, it makes a salt bridge/hydrogen bond interaction with Glu154 that should stabilize the loop in the correct orientation for DNA binding. Thus, these mutations could affect not only the dimer stability but also the proper orientation of the extended helix loop arm for DNA binding. Mutations in similar locations in CcrA had less dramatic effects: both S11G (similar to S16 in CcrB) and R79G (similar to R89 in CcrB) abolished integration but only reduced excision compared with that for the control containing both functional proteins. However, the excision occurred only between attL and attR2 and not between attL and attR, an excision event seen only when CcrB acts alone without any CcrA activity. Thus, it is likely that these two mutations abolished the contribution of CcrA that guided the recombination complex to the usual excision site. The S17A and K142A mutations in CcrA, in locations similar to S21 and R151 in CcrB, respectively, reduced but did not eliminate either excision or integration. It is interesting that the L125A mutation in CcrA markedly increased the level of excision compared to that for the positive control, with much of the increase in excision occurring between attL and attR2.
FIG 4.
Homology model of the first 205 N-terminal amino acids of CcrB in complex with DNA. (A) Two CcrB N terminus units (in yellow and blue ribbon representations) forming a complex with the DNA fragment (backbone in white and side chains in red, space-filling representation). (B) A 90° rotation of the complex shown in panel A. DNA is in stick representation (backbone in white; side chains of A, C, G, and T in red, magenta, green, and cyan, respectively). (C) Sequence alignment of CcrA, CcrB, and CcrC. Red residues, residues in the N-terminal domain identified from the structural model to be critical for excision and/or integration.
FIG 5.
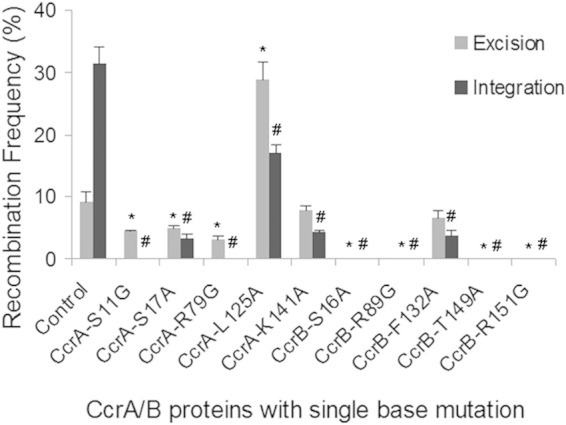
Alteration of catalytic function by introducing single base mutations into CcrAB. For each set, one mutation was introduced into the ccr operon. At least three separate experiments were performed for each mutant. *, P < 0.05 versus the excision frequency of wild-type CcrA and CcrB; #, P < 0.05 versus the integration frequency of wild-type CcrA and CcrB.
Some of the mutants were examined in S. aureus using the change in plasmid size used in E. coli, but because of the low recombination frequency and lower yield of plasmid DNA from S. aureus, differences were not seen. We therefore evaluated tetracycline susceptibility as a measure of recombination. After 10 days of passage, no tetracycline-susceptible colonies of mutants CcrB S16A and CcrB R89G were seen, confirming the results seen with E. coli. The CcrB F132 mutation produced 1.8% ± 0.3% tetracycline-susceptible colonies for excision and 1.0% ± 0.2% for integration, mirroring, and in the same direction as, the difference seen for E. coli. Similarly, the CcrA S11G and R79G mutations produced no tetracycline-susceptible colonies for the integration model but 0.7% ± 0.2% and 0.6% ± 0.1% tetracycline-susceptible colonies, respectively, for the excision model. Thus, while the results for S. aureus were not as robust as those for E. coli, they precisely duplicated the changes in both excision and integration observed in the heterologous host.
Activities of CcrA and CcrB from MRSA and MRSE strains.
An earlier study from the Archer lab showed that the ccrAB genes found in SCCmec types II and IV varied by up to 5% at the amino acid level, yet all of the tested recombinases mediated SCCmec excision in MRSA strains (25). To further identify the activities of CcrAB from a panel of strains, the ccrAB genes and the ccrC gene of seven MRSA strains and two methicillin-resistant S. epidermidis (MRSE) strains were examined using the model plasmid system to determine their abilities to mediate excision and integration, and the results were compared with those presented above for N315 (Fig. 6A). CcrC and most CcrAB proteins catalyzed integration at a higher frequency than they did excision, with the frequency of integration exceeding the frequency of excision by from 1.3-fold (FPR3757) to 10-fold (J28). The exception was CcrAB from MW2, in which excision and integration frequencies were the same. The protein sequences of CcrAB in each of the S. aureus and S. epidermidis strains for which the results are shown in Fig. 6A were compared (see Fig. S1 in the supplemental material). For CcrA, none of the amino acid differences were seen only in MW2. However, there were nine amino acid changes in the CcrB sequence of MW2 not seen in the CcrB sequences of the other strains. All but one (E73D) of the changes were in the carboxy terminus (see Fig. S1 in the supplemental material). All of the changes were relatively conservative, except for the change at aa 385, at which glycine was changed to cysteine in MW2. We repaired the mutation in MW2 from a cysteine to a glycine and exchanged the glycine for the mutant cysteine in N315, yielding the result seen in Fig. 6B. The MW2 C385G change reduced excision by 47%, causing integration to exceed excision by 2.32-fold, a difference similar to that seen in N315. In contrast, the G385C exchange in N315 both decreased integration and increased excision so that the recombination frequency seen in mutant N315 resembled the recombination frequency seen in MW2. These results suggest both that this is a critical mutation that affects the direction of recombination and that the carboxy terminus of CcrB plays an important role in determining the frequency of integration versus that of excision.
FIG 6.
(A) Excision and integration catalyzed by CcrAB or CcrC (WBG8318) from strains carrying SCCmec types II, IV, and V in E. coli TOP10 cells. The ccrAB or ccrC genes were from a series of MRSA or MRSE strains which are described in Table S1 in the supplemental material. Comparison of the amino acid sequences of the CcrAB proteins from the S. aureus strains described here can be found in Fig. S1 in the supplemental material. (B) Excision and integration catalyzed by CcrAB from N315 and MW2 with a change of aa 385 in CcrB. At least three repeats were done for each set of CcrA and CcrB proteins. *, P < 0.01 for integration versus excision.
DISCUSSION
In the current study, we have demonstrated that the recombinational activity of CcrA and CcrB favors integration over excision, demonstrated by plasmid models in both S. aureus and E. coli and in a panel of clinical S. aureus and S. epidermidis isolates. This is consistent with our observation that spontaneous excision in vitro is an extremely rare event but does not support either one of the two potential explanations: rare excision associated with a stably integrated element, as postulated by others (18, 26), or frequent excision followed by immediate reintegration, events associated with more instability of the integrated element. The observation that in one isolate, S. aureus MW2, plasmid-modeled excision and integration were equal yet spontaneous excision in vitro was at the same low rate seen for other isolates (Wang, unpublished) suggests that excision with reinsertion may be occurring in some isolates. Our previous data have shown that excision can be favored over integration when the concentrations of CcrB and CcrA are increased by their inclusion on a high-copy-number plasmid or driven by an inducible promoter (5). Thus, the concentrations and ratios of the two recombinases are critical for determining stable integration versus excision, and anything that increases ccrAB expression may favor excision. A previous study found that the ccrAB promoter was active in only a small percentage of cells in a population and that the activity, determined by unknown genes elsewhere in the chromosome, was strain dependent (26). An earlier report suggesting that various antibiotics increased ccrAB expression (27) and our observation that some isolates colonizing patient nares contained excised variants occurring at rates higher than those seen in vitro may point to in vivo conditions that could favor excision over integration (28). While the current study suggests that under stable in vitro conditions integration is favored and, thus, that passage of SCCmec to progeny is the rule, excision is likely to occur under certain conditions in order for horizontal transmission of SCCmec to occur.
CcrA and CcrB are large serine recombinases that consist of an N-terminal domain and a C-terminal domain. The model that we generated (Fig. 4) is based on the similarity of the N terminus (amino acid 1 to ∼220) to smaller serine recombinases (23) and contains both a catalytic domain and the DNA-binding domain. The function of the C terminus (amino acids ∼220 to 543 for CcrB and ∼210 to 450 for CcrA) is unknown. However, as shown in Fig. 3, while an N-terminal deletion of either CcrA or CcrB abolishes both excision and integration, a C-terminal deletion does not eliminate but markedly reduces both activities. Thus, the C terminus seems to have a role in the efficiency of recombination. Rutherford et al. (29) recently reported the X-ray structure of the C-terminal domain of listeria phage integrase bound to its attP half site, which, in conjunction with recombination and DNA-binding experimental data, led the authors to propose a model for the recombination pathway that explains how large serine recombinases regulate site selectivity and directionality. Interestingly, using the HHpred structural homology program (22), we found the CTD of CcrB to be structurally homologous to the CTD of listeria phage integrase. Moreover, using the C-terminal amino acid sequence of CcrB for a search, we also identified a recombinase, the zinc beta ribbon domain from the Pfam database (30), which is known to bind DNA. These observations lead us to speculate that the CTD of CcrB might also associate with the DNA half site, orienting it in the correct position for integration/excision operations conducted through the NTD.
An understanding of the dynamics of SCCmec excision and integration could have therapeutic implications. We have previously shown (31) that when isogenic MRSA strains with and without SCCmec compete in vitro, the strain without SCCmec is more fit, becoming the predominant species in the culture within 4 days. Thus, if SCCmec excision could be induced to exceed integration in vivo, the excised variant may be more fit and become predominant, converting an MRSA strain into a susceptible strain, broadening therapeutic options.
Supplementary Material
ACKNOWLEDGMENTS
Nucleic acid sequence determination was performed at the Virginia Commonwealth University Nucleic Acid Research Facility.
This study was supported by grant 5R01AI035705-20 from the National Institute of Allergy and Infectious Diseases.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00078-15.
REFERENCES
- 1.Crossley K, Jefferson K, Archer GL, Fowler VG Jr (ed). 2009. Staphylococci in human disease. Wiley-Blackwell, West Sussex, United Kingdom. [Google Scholar]
- 2.Ito T, Hiramatsu K, Oliveira DC, de Lencastre H, Zhang K, Westh H, O'Brien F, Giffard PM, Coleman D, Tenover FC, Boyle-Vavra S, Skov RL, Enright MC, Kreiswirth B, Ko KS, Grundmann H, Laurent F, Sollid JE, Kearns AM, Goering R, John JF, Daum R, Soderquist B, International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements (IWG-SCC) . 2009. Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob Agents Chemother 53:4961–4967. doi: 10.1128/AAC.00579-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ito T, Ma XX, Takeuchi F, Okuma K, Yuzawa H, Hiramatsu K. 2004. Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC. Antimicrob Agents Chemother 48:2637–2651. doi: 10.1128/AAC.48.7.2637-2651.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Katayama Y, Ito T, Hiramatsu K. 2000. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother 44:1549–1555. doi: 10.1128/AAC.44.6.1549-1555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang L, Archer GL. 2010. Roles of CcrA and CcrB in excision and integration of staphylococcal cassette chromosome mec, a Staphylococcus aureus genomic island. J Bacteriol 192:3204–3212. doi: 10.1128/JB.01520-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang L, Safo M, Archer GL. 2012. Characterization of DNA sequences required for the CcrAB-mediated integration of staphylococcal cassette chromosome mec, a Staphylococcus aureus genomic island. J Bacteriol 194:486–498. doi: 10.1128/JB.05047-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuhstoss S, Rao RN. 1991. Analysis of the integration function of the streptomycete bacteriophage ϕC31. J Mol Biol 222:897–908. doi: 10.1016/0022-2836(91)90584-S. [DOI] [PubMed] [Google Scholar]
- 8.Smith MCM, Brown WRA, McEwan AR, Rowley PA. 2010. Site-specific recombination by phiC31 integrase and other large serine recombinases. Biochem Soc Trans 38:388–394. doi: 10.1042/BST0380388. [DOI] [PubMed] [Google Scholar]
- 9.Bibb LA, Hancox MI, Hatfull GF. 2005. Integration and excision by the large serine recombinase ϕφRv1 integrase. Mol Microbiol 55:1896–1910. doi: 10.1111/j.1365-2958.2005.04517.x. [DOI] [PubMed] [Google Scholar]
- 10.Christiansen B, Brøndsted L, Vogensen FK, Hammer K. 1996. A resolvase-like protein is required for the site-specific integration of the temperate lactococcal bacteriophage TP901-1. J Bacteriol 178:5164–5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghosh P, Bibb LA, Hatfull GF. 2008. Two-step site selection for serine-integrase-mediated excision: DNA-directed integrase conformation and central dinucleotide proofreading. Proc Natl Acad Sci U S A 105:3238–3243. doi: 10.1073/pnas.0711649105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghosh P, Wasil LR, Hatfull GF. 2006. Control of phage Bxb1 excision by a novel recombination directionality factor. PLoS Biol 4:e186. doi: 10.1371/journal.pbio.0040186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim AI, Ghosh P, Aaron MA, Bibb LA, Jain S, Hatfull GF. 2003. Mycobacteriophage Bxb1 integrates into the Mycobacterium smegmatis groEL1 gene. Mol Microbiol 50:463–473. doi: 10.1046/j.1365-2958.2003.03723.x. [DOI] [PubMed] [Google Scholar]
- 14.Lyras D, Adams V, Lucet I, Rood JI. 2004. The large resolvase TnpX is the only transposon-encoded protein required for transposition of the Tn4451/3 family of integrative mobilizable elements. Mol Microbiol 51:1787–1800. doi: 10.1111/j.1365-2958.2003.03950.x. [DOI] [PubMed] [Google Scholar]
- 15.Wang H, Mullany P. 2000. The large resolvase TndX is required and sufficient for integration and excision of derivatives of the novel conjugative transposon Tn5397. J Bacteriol 182:6577–6583. doi: 10.1128/JB.182.23.6577-6583.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang H, Smith MCM, Mullany P. 2006. The conjugative transposon Tn5397 has a strong preference for integration into its Clostridium difficile target site. J Bacteriol 188:4871–4878. doi: 10.1128/JB.00210-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Misiura A, Pigli YZ, Boyle-Vavra S, Daum RS, Boocock MR, Rice PA. 2013. Roles of two large serine recombinases in mobilizing the methicillin-resistance cassette SCCmec. Mol Microbiol 88:1218–1229. doi: 10.1111/mmi.12253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stojanov M, Moreillon P, Sakwinska O. 2012. Dynamics of excision of the staphylococcal cassette chromosome mec (SCCmec) in methicillin-resistant Staphylococcus aureus (MRSA), abstr C1-1746, p 169 Abstr 52nd Intersci Conf Antimicrob Agents Chemother, San Francisco, CA. American Society for Microbiology, Washington, DC. [Google Scholar]
- 19.Schenk S, Laddaga RA. 1992. Improved method for electroporation of Staphylococcus aureus. FEBS Microbiol Lett 94:1333–1338. [DOI] [PubMed] [Google Scholar]
- 20.Stephenson FH. 2010. Recombinant DNA, p 313–368. In Calculations for molecular biology and biotechnology. Academic Press, Amsterdam, Netherlands. [Google Scholar]
- 21.Charpentier E, Anton AI, Barry P, Alfonso B, Fang Y, Novick RP. 2004. Novel cassette-based shuttle vector system for gram-positive bacteria. Appl Environ Microbiol 70:6076–6085. doi: 10.1128/AEM.70.10.6076-6085.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soding J, Biegert A, Lupas AN. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33(Web Server issue):W244–W248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang W, Steitz TA. 1995. Crystal structure of the site-specific recombinase gamma delta resolvase complexed with a 34 bp cleavage site. Cell 82:193–207. doi: 10.1016/0092-8674(95)90307-0. [DOI] [PubMed] [Google Scholar]
- 24.Fiser A, Sali A. 2003. Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol 374:461–491. [DOI] [PubMed] [Google Scholar]
- 25.Noto MJ, Archer GL. 2006. A subset of Staphylococcus aureus strains harboring staphylococcal cassette chromosome mec (SCCmec) type IV is deficient in CcrAB-mediated SCCmec excision. Antimicrob Agents Chemother 50:2782–2788. doi: 10.1128/AAC.00032-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stojanov M, Sakwinska O, Moreillon P. 2013. Expression of SCCmec cassette chromosome recombinases in methicillin-resistant Staphylococcus aureus and Staphylococcus epidermidis. J Antimicrob Chemother 68:749–757. doi: 10.1093/jac/dks494. [DOI] [PubMed] [Google Scholar]
- 27.Higgins PG, Rosato AE, Seifert H, Archer GL, Wisplinghoff H. 2009. Differential expression of ccrA in methicillin-resistant Staphylococcus aureus strains carrying staphylococcal cassette chromosome mec type II and IVa elements. Antimicrob Agents Chemother 53:4556–4558. doi: 10.1128/AAC.00395-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boundy S, Zhao Q, Fairbanks C, Folgosa L, Climo MW, Archer GL. 2012. Spontaneous SCCmec excision in Staphylococcus aureus nasal carriers. J Clin Microbiol 50:469–471. doi: 10.1128/JCM.01063-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rutherford K, Yuan P, Perry K, Sharp R, Van Duyne GD. 2013. Attachment site recognition and regulation of directionality by the serine integrases. Nucleic Acids Res 41:8341–8356. doi: 10.1093/nar/gkt580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer ELL, Tate J, Punta M. 2014. The Pfam protein families database. Nucleic Acids Res 42(Database issue):D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noto MB, Fox PM, Archer GL. 2008. Spontaneous deletion of the methicillin resistance determinant, mecA, partially compensates for the fitness cost associated with high level vancomycin resistance in Staphylococcus aureus. Antimicrob Agents Chemother 52:1221–1229. doi: 10.1128/AAC.01164-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.