ABSTRACT
Salmonella enteric serovar Typhimurium, a major cause of food-borne illness, is capable of using a variety of carbon and nitrogen sources. Fructoselysine and glucoselysine are Maillard reaction products formed by the reaction of glucose or fructose, respectively, with the ε-amine group of lysine. We report here that S. Typhimurium utilizes fructoselysine and glucoselysine as carbon and nitrogen sources via a mannose family phosphotransferase (PTS) encoded by gfrABCD (glucoselysine/fructoselysine PTS components EIIA, EIIB, EIIC, and EIID; locus numbers STM14_5449 to STM14_5454 in S. Typhimurium 14028s). Genes coding for two predicted deglycases within the gfr operon, gfrE and gfrF, were required for growth with glucoselysine and fructoselysine, respectively. GfrF demonstrated fructoselysine-6-phosphate deglycase activity in a coupled enzyme assay. The biochemical and genetic analyses were consistent with a pathway in which fructoselysine and glucoselysine are phosphorylated at the C-6 position of the sugar by the GfrABCD PTS as they are transported across the membrane. The resulting fructoselysine-6-phosphate and glucoselysine-6-phosphate subsequently are cleaved by GfrF and GfrE to form lysine and glucose-6-phosphate or fructose-6-phosphate. Interestingly, although S. Typhimurium can use lysine derived from fructoselysine or glucoselysine as a sole nitrogen source, it cannot use exogenous lysine as a nitrogen source to support growth. Expression of gfrABCDEF was dependent on the alternative sigma factor RpoN (σ54) and an RpoN-dependent LevR-like activator, which we designated GfrR.
IMPORTANCE Salmonella physiology has been studied intensively, but there is much we do not know regarding the repertoire of nutrients these bacteria are able to use for growth. This study shows that a previously uncharacterized PTS and associated enzymes function together to transport and catabolize fructoselysine and glucoselysine. Knowledge of the range of nutrients that Salmonella utilizes is important, as it could lead to the development of new strategies for reducing the load of Salmonella in food animals, thereby mitigating its entry into the human food supply.
INTRODUCTION
Salmonella enterica serovar Typhimurium is one of the leading causes of food-borne illness. Salmonella causes 11% of all infections caused by bacterial food-borne pathogens in the United States and is responsible for an estimated 1 million cases of salmonellosis in the United States each year, which result in approximately 19,000 hospitalizations and 400 deaths (1). S. Typhimurium causes gastroenteritis in humans but is capable of colonizing a wide range of animals with few or no disease symptoms. S. Typhimurium utilizes a broad array of carbon and nitrogen sources (2), which may contribute to the wide host range of this bacterium. Chadhuri and coworkers found that genes encoding enzymes in a variety of catabolic processes appear to be important for S. Typhimurium colonization of the chicken, pig, and calf (3). Among those catabolic genes, some encoded phosphoenolpyruvate-dependent sugar phosphotransferase systems (PTS) of unknown function.
PTS, which are major routes for carbohydrate transport in bacteria, couple the phosphorylation of sugar substrates with their translocation across the cell membrane. The general mechanism of PTS-mediated transport is as follows. A phosphoryl group from phosphoenolpyruvate is transferred to enzyme I (EI), which in turn transfers it to a phosphoryl carrier, the histidine protein (HPr). From HPr, the phosphoryl group is transferred to a sugar-specific permease, a membrane-bound complex known as enzyme 2 (EII), which phosphorylates the sugar as it is transported across the cell membrane. EII complexes are associated with the cell membrane and are specific for a single substrate or set of structurally related substrates. EII complexes consist of at least three distinct domains (IIA, IIB, and IIC), which are fused into a single polypeptide or exist as multiple interacting polypeptides. Mannose family PTS have an additional domain, EIID (4, 5). The best-described mannose family PTS is ManXYZ, which transports mannose and other hexoses (6). Enzyme I and HPr are cytoplasmic proteins used in conjunction with most PTS EII complexes in Salmonella, although some complexes (e.g., FruA and FrwBCD fructose-specific PTS permeases) have dedicated enzyme I and/or HPr homologs (7).
One of the PTS identified by Chadhuri and coworkers as important in colonization of the chicken, pig, and calf was homologous to a PTS encoded in the Enterococcus faecium gfrABCDEF operon. Wiame and coworkers showed that gfrE and gfrF encode deglycases involved in fructoselysine and glucoselysine catabolism, respectively (8).
Fructoselysine is formed by the reaction of glucose with the ε-amine of lysine followed by a spontaneous isomerization of the sugar, referred to as an Amadori rearrangement (9). Glucoselysine is formed similarly from the reaction of fructose with the ε-amine of lysine followed by the spontaneous isomerization of the sugar, referred to as Heyns rearrangements. Multiple products can be formed via Heyns rearrangements, as 2-amino-aldose or 2-amino-3-hexulose derivatives of the sugar are formed, each of which can be in either a glucosamine or mannosamine configuration (10). Fructosamines resulting from the reaction of glucose with free amino acids or proteins is common in nature. Fructoselysine occurs in dehydrated fruits, grains, and vegetables, including raisins, prunes, dates, figs, apricots, onion, cereal, and carrots (11). Other fructosamines are found in rotting fruits and vegetables, accounting for as much as 7% of the fresh mass (12).
We show here that the predicted gfrABCDEF operon, encoded by STM14_5449 to STM14_5454, is responsible for utilization of both fructoselysine and glucoselysine in S. Typhimurium 14028s. Fructoselysine and glucoselysine can serve as both the sole carbon and nitrogen source for S. Typhimurium. Fructoselysine is a good nitrogen source for S. Typhimurium, with the growth rate of S. Typhimurium cultures grown in a minimal medium containing fructoselysine as the sole nitrogen source comparable to that of cultures grown in medium containing ammonium as the nitrogen source. Interestingly, although lysine derived from fructoselysine and glucoselysine can be used as a nitrogen source for S. Typhimurium, exogenous lysine does not support growth of S. Typhimurium.
The S. Typhimurium gfrABCDEF operon was previously predicted to be under the control of the alternative sigma factor RpoN (σ54) (13, 14). Transcription from RpoN-dependent promoters requires an activator to stimulate isomerization of a closed complex between σ54-RNA polymerase holoenzyme and the promoter to an open complex that is competent to initiate transcription (15). A gene encoding an RpoN-dependent LevR-like activator is located immediately upstream of the gfrABCDEF operon (Fig. 1), which we found is required for utilization of fructoselysine and glucoselysine and designate gfrR.
FIG 1.
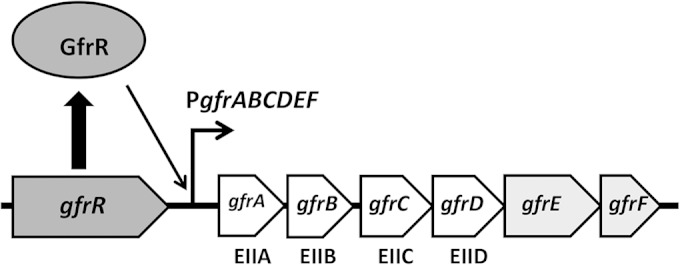
S. Typhimurium gfr locus. The EIIA, EIIB, EIIC, and EIID components of the fructoselysine/glucoselysine PTS permease are encoded by gfrA, gfrB, gfrC, and gfrD, respectively. The other genes in the operon, gfrE and gfrF, encode glucoselysine-6-phosphate deglycase and fructoselysine-6-phosphate deglycase, respectively. GfrR is an RpoN-dependent activator required for transcriptional activation of gfrABCDEF. The intergenic region between gfrR and gfrA is 101 bp and contains a predicted RpoN-dependent promoter (PgfrABCDEF; 5′-TGGCAGGCCGCTTGCT-3′) located 37 bp upstream of gfrA.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and reagents.
Strains were maintained in Luria-Bertani (LB) broth or agar supplemented with 100 μg/ml of ampicillin or 50 μg/ml kanamycin as needed. Growth of S. Typhimurium 14028s on different carbon and nitrogen sources was carried out using the basal morpholinepropanesulfonic acid (MOPS) minimal medium (16) with the modifications described by Maloy et al. (17). Various carbon and nitrogen sources were added to the minimal medium at the concentrations indicated. Strains used in this study are listed in Table S1 in the supplemental material.
Construction of plasmids.
Plasmids used for this study are listed in Table S2 in the supplemental material. Both gfrE and gfrF were amplified by PCR from S. Typhimurium 14028s genomic DNA using Phusion high-fidelity DNA polymerase (New England BioLabs), and appropriate primer sets are listed in Table S3. Adenine residues were added to the 3′ ends of the amplicons using Taq DNA polymerase (Thermo Fisher Scientific). The amplicons were cloned into pCR2.1-TOPO (Invitrogen), and the sequences of the cloned fragments were confirmed using DNA sequencing (Genewiz). NdeI and HindIII sites were introduced by the primer sets and were used to clone gfrE and gfrF into the expression vector pET21a (Novagen). The resulting plasmids, designated pET21a+gfrE and pET21a+gfrF, expressed the proteins with a C-terminal hexahistidine tag.
For all transformations involving S. Typhimurium 14028s, DNA was introduced by electroporation at 2.4 kV, 25 μF, and 400 Ω. Following electroporation, cells were allowed to recover in SOC broth (0.5% yeast extract, 2% tryptone, 10 mM NaCl, 2.5 mM KCl, 10 mM MgCl2, 10 mM MgSO4, and 20 mM glucose) for 1 h at 37°C. Chemically competent cells were prepared using the calcium chloride method and were used for all transformations involving Escherichia coli. Chemically competent cells were allowed to recover in LB broth for 1 h at 37°C following the heat shock step.
Expression and purification of proteins.
Plasmids pET21a+gfrE and pET21a+gfrF were transformed into E. coli BL21 (DE3λ) containing the plasmid pLysE (18). E. coli BL21 (DE3λ) expresses T7 RNA polymerase, which is needed for transcription of gfrE and gfrF from pET21a+gfrE and pET21a+gfrF. Overnight cultures were subcultured into 1 liter of LB broth with appropriate antibiotics and grown at 30°C until an optical density at 650 nm (OD650) of 0.4 to 0.6. Cells then were moved to ice for 30 min, after which isopropyl β-d-1-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM to the medium to induce the expression of the desired proteins. Cells then were allowed to grow at 18°C for 48 h. Cells were harvested by centrifugation at 8,000 × g for 5 min and then resuspended in 30 ml 50 mM 3-(N-morpholino)propanesulfonic acid (MOPS) buffer (pH 7) containing 100 mM potassium thiocyanate (buffer A). Cells were lysed by sonication using a Branson Sonifier 450 with 100% output and four 1-min intervals, maintaining the cell extract on ice to prevent heating. The cell lysate was clarified by centrifugation at 2,500 × g for 90 min. The resulting supernatant was applied to a nickel-nitrilotriacetic acid–agarose column (10 ml; Sigma-Aldrich) and washed with buffer A until the absorbance at 280 nm reached the baseline. The His-tagged proteins were eluted from the column with 100 ml of buffer A containing 0.15 M imidazole. Peak protein fractions were pooled, concentrated using a centrifugal concentrator (YM-30 membrane), and dialyzed against 50 mM MOPS buffer (pH 7).
Determination of growth rates and Ks.
S. Typhimurium was grown aerobically at 37°C in MOPS minimal medium containing various carbon and nitrogen sources. Cell growth was measured using a Klett colorimeter (model 900-3) with a green (520- to 580-nm) glass filter. Generation (g) time was calculated using the equation g = ln (2)/b, where b is the best-fit constant from a plot of log values of Klett units versus incubation time. The substrate concentration at a half-maximal growth rate (Ks) for fructoselysine was estimated by growing cells in minimal media containing 20 mM l-arabinose as the primary carbon source and fructoselysine at concentrations ranging from 0 to 4 mM as the sole nitrogen source. For experiments in which a second carbon source was included in the growth medium, arabinose was used, since it is a non-PTS sugar and would not compete with fructoselysine (or glucoselysine) for the phosphoryl group from phosphorylated HPr. Cells were grown in 96-well plates (0.2 ml medium per well) at 37°C in a BioTek ELx808 absorbance microplate reader (BioTek, Winooski, VT). Each culture condition was performed with three biological replicates. Absorbance was measured every 30 min for 24 h, and absorbance versus incubation time was plotted using Prism software. Generation times for each condition were calculated as described above, after plotting log values of absorbance versus incubation times. The Ks value for fructoselysine was estimated from a double-reciprocal plot (Lineweaver-Burk plot) of g values versus fructoselysine concentrations using the SigmaPlot 12.5 software package.
Sugar-amine synthesis.
Sugar-amines were synthesized as described in references 19 and 20. For the synthesis of fructoselysine, d-glucose (3.3 gm, 18.3 mmol) and Nα-(tert-butoxycarbonyl)-l-lysine (Boc-Lys-OH; Sigma-Aldrich) (1 g, 4 mmol) were refluxed in 50 ml of methanol for 5 h under argon gas. The solvent was evaporated in vacuo, and the resulting solids were resuspended in 16.7 ml of 1 M HCl to remove the tert-butoxycarbonyl protecting group. The HCl was removed under vacuum, and the product was resuspended in 20 ml distilled water. The sample was incubated with 5 g of AG 50W X-8 (Bio-Rad) cation exchange resin for 1 h to separate unreacted sugar from the desired product. After the separation, the resin was washed with water and then incubated in 30 ml NH4OH to remove the product from the resin. The sample was decolorized with charcoal, filtered, and then freeze-dried. The yield for the synthesis ranged from 32 to 53%. The synthesis of glucoselysine was essentially the same as that described for fructoselysine, except fructose (3.3 gm, 18.3 mmol) was used in place of glucose. The Heyns rearrangement resulted in several products following the reaction of fructose with Boc-Lys-OH, and we did not determine what proportion of the final product was glucoselysine.
Ribuloselysine was formed by reacting d-ribose (0.6 g, 4 mmol) with Boc-Lys-OH (0.22 g, 0.9 mmol) for 5 h at 50°C in 50 ml methanol as described previously (20). The Boc protecting group was removed using HCl, and ribuloselysine was separated from the unreacted d-ribose using the cation exchange resin as described for the synthesis of fructoselysine. Erythruloselysine was synthesized by reacting d-erythrose (0.12 g, 1 mmol) with Doc-Lys-OH (0.22 g, 0.9 mmol) in 50 ml methanol for 1 h at 40°C as described previously (20), the Boc protecting group was removed, and erythruloselysine was separated using the cation exchange resin as described above. The yields for ribuloselysine and erythruloselysine both were ∼56%. Other sugar amines were synthesized and purified by following the same general procedure as that used for fructoselysine synthesis. Tagatoselysine was synthesized by reacting d-galactose (3.3 g, 18.3 mmol) with Boc-Lys-OH (1 g, 4 mmol), and the final yield was ∼25%. Fructose–d-lysine was formed by reacting glucose (3.3 g, 18.3 mmol) with Boc-d-Lys-OH (1 g, 4 mmol), and the final yield was ∼27%. Fructose-ornithine was formed by reacting d-glucose (3.3 g, 18.3 mmol) with Nα-tert-butoxycarbonyl-l-ornithine (0.94 g, 4 mmol; Sigma-Aldrich), and the final yield was ∼44%. All products were verified using 1H nuclear magnetic resonance (NMR) and mass spectrometry (see the supplemental material).
Construction of mutant strains.
The S. Typhimurium mutants were constructed using the λ Red recombineering method (21). Plasmid pKD4, which contains a kanamycin resistance (kan) cassette flanked by two Flp recognition sights (FRT sites), was used to amplify linear DNA which included sequences at the ends that were homologous to the genes targeted for deletion. Primer sets used to generate the amplicons for targeted mutagenesis are listed in Table S3 in the supplemental material. The amplicons were introduced into S. Typhimurium 14028s bearing the plasmid pKD46, which carries the λ phage recombinase genes under the control of the araBAD promoter, by electroporation. DNA isolated from the resulting kanamycin-resistant colonies was checked by PCR, using primers that flanked the target gene deletion (primers are listed in Table S3), to confirm that the target gene had been deleted. Mutant alleles in which the target genes had been replaced with the kanamycin resistance (kan) cassette were moved into a clean genetic background by transduction using P22 HT int. The kan cassette then was removed by introducing plasmid pCP20 into the mutant strains by electroporation. Plasmid pCP20 expresses the Flp recombinase, which recognized the FRT sites, and excised the kan cassette. Loss of the kan cassette was confirmed by susceptibility to kanamycin and by PCR using the same flanking primers (listed in Table S3).
Fructoselysine-6-phosphate deglycase assay.
Deglycase assays were carried out as previously described (8). Fructoselysine-6-phosphate was generated by incubating 5 mM fructoselysine, 10 mM ATP, 10 mM MgCl2, 50 mM HEPES, pH 7, and 5 U/ml of purified E. coli fructoselysine kinase (YhfQ; kindly provided by E. Van Schaftingen and E. Wiame) for 30 min at 30°C. Fructoselysine kinase was purified as described previously (22). Deglycase activity was measured spectrophotometrically at 340 nm in a coupled assay through the formation of glucose-6-phosphate. The mixture contained 50 mM HEPES, pH 7.1, 50 mM KCl, 5 mM MgCl2, 0.25 mM NADP, 0.1 mM fructoselysine-6-phosphate, 5 μg glucose-6-phosphate dehydrogenase (Sigma-Aldrich), and various concentrations of purified GfrF to obtain linear slopes for changes in absorbance.
Testing fructoselysine analogs for growth inhibition.
The fructoselysine analogs fructose–d-lysine and fructose–l-ornithine were examined for their ability to inhibit growth of S. Typhimurium 14028s in MOPS minimal medium containing 20 mM l-arabinose and either 10 mM ammonium chloride or 5 mM fructoselysine. S. Typhimurium cultures grown aerobically overnight in either medium were used to inoculate fresh medium containing various concentrations of the fructoselysine analogs (0 to 20 mM) in 96-well plates (0.2 ml growth medium per well) at 37°C in a BioTek ELx808 absorbance microplate reader. Each culture condition was performed with three biological replicates. Absorbance was measured every 30 min for 24 h, and absorbance versus incubation time was plotted using Prism software.
qRT-PCR assays.
Transcript levels of gfrAB in wild-type S. Typhimurium 14028s cultured in minimal medium that contained 20 mM l-arabinose and either 5 mM fructoselysine or 10 mM ammonium were assessed by quantitative reverse transcription-PCR (qRT-PCR). Ten milliliters of cell culture grown to mid-log phase was used for RNA extraction. RNA was isolated using a boil method (23). RNA then was treated with RQ1 DNase (Promega). cDNA was synthesized using an RT Superscript III synthesis kit (Invitrogen), after which qRT-PCR was carried out using an iCycler iQ system (Bio-Rad). Primer specificity was confirmed by PCR using a genomic DNA template. Three biological replicates and three technical replicates were assessed for expression of gfrAB. All expression of gfrAB was normalized to levels of rpoD, an internal control for S. Typhimurium gene expression. Reaction volumes were 20 μl and included 10 μl of iQ SYBR green Supermix (Bio-Rad), 5 μl off 100-fold-diluted cDNA, and 200 nM each primer specific for either gfrAB or rpoD. Gene expression levels were determined using the 2−ΔΔCT equation (24). The fold change of gfr expression of cells grown with fructoselysine compared to that with ammonium for all three biological replicates were averaged, and standard deviations were calculated. Statistical significance was determined using the two-sample t test.
Mass spectrometry analysis.
To examine intracellular metabolites in S. Typhimurium grown with fructoselysine, S. Typhimurium cultures were grown overnight in MOPS minimal media containing either 20 mM l-arabinose and 13 mM ammonium or 20 mM fructoselysine at 37°C. One milliliter of overnight culture was subjected to 10 cycles of freezing and thawing using a dry ice-ethanol bath. After the last thaw, cell debris was removed by centrifugation at 6,000 × g for 1 min, and the resulting supernatant was removed for analysis by electrospray mass spectrometry using a Burker Esquire 3000 Plus mass spectrometer.
RESULTS
A mannose family PTS permease in S. Typhimurium is required for utilization of fructoselysine and glucoselysine.
Wiame and coworkers identified deglycases in E. faecium that use fructoselysine-6-phosphate and glucoselysine-6-phosphate as substrates (8). The genes encoding these enzymes appeared to be part of an operon with genes encoding a mannose family PTS. They proposed that the genes in the operon were involved in the transport and catabolism of fructoselysine and glucoselysine. Additionally, they noted that S. Typhimurium possesses homologs to the PTS and deglycase genes and postulated that the S. Typhimurium homologs were similarly involved in fructoselysine and glucoselysine transport and catabolism (8). We wished to test this hypothesis by seeing if S. Typhimurium could use fructoselysine and glucoselysine as carbon or nitrogen sources and, if so, whether the PTS and deglycase genes were required for the utilization of these compounds.
At 37°C under aerobic conditions, wild-type S. Typhimurium grew in a minimal medium with fructoselysine as the sole carbon source with an estimated generation time of 88 ± 5 min (Table 1). The minimal medium contained 13 mM ammonium chloride as a nitrogen source, and S. Typhimurium grew somewhat better when ammonium chloride was withheld from the medium (80 ± 3 min generation time). Including l-arabinose in the minimal medium with fructoselysine further improved the growth rate of S. Typhimurium (55 ± 2 min generation time). By comparison, the doubling time of S. Typhimurium in minimal medium containing l-arabinose as a carbon source and ammonium chloride as a nitrogen source was 44 ± 2 min (Table 1). Using minimal medium that contained l-arabinose and fructoselysine, we estimated the Ks (concentration that gives half maximal growth rate) for fructoselysine to be 0.44 mM. Taken together, these data indicate that fructoselysine is a good nitrogen source for S. Typhimurium and is a physiologically relevant substrate.
TABLE 1.
Growth rates of S. Typhimurium on various carbon and nitrogen sources
| Nitrogen source | Generation time (min) for carbon sourcea: |
||
|---|---|---|---|
| l-Arabinose | Fructoselysine | Glucoselysine | |
| NH4Cl | 44 ± 2 | 88 ± 5 | 131 ± 13 |
| Fructoselysine | 55 ± 2 | 80 ± 3 | ND |
| Glucoselysine | 124 ± 12 | ND | 140 ± 8 |
ND, not determined.
Glucoselysine was used similarly by S. Typhimurium as a carbon and nitrogen source, although it did not support the same rapid growth observed with fructoselysine (Table 1). In minimal medium in which glucoselysine was the sole carbon source (glucoselysine plus ammonium), sole carbon and nitrogen source (glucoselysine only), or sole nitrogen source (l-arabinose plus glucoselysine), generation times for S. Typhimurium cultures were 131 ± 13 min, 140 ± 8 min, and 124 ± 12 min, respectively. The reason for the lower growth rate of S. Typhimurium with glucoselysine compared to that with fructoselysine could be due to a couple of reasons. One reason is that fructoselysine may be transported or catabolized by S. Typhimurium more efficiently than glucoselysine. Alternatively, the synthesis of glucoselysine likely resulted in a mixture of Heyns products, and glucoselysine may not have been the major end product or some of the other Heyns products may have inhibited the growth of S. Typhimurium.
Genes encoding the EIIAB components (gfrAB homologs) or the deglycases (gfrE and grfF homologs) were deleted to assess their requirement for growth on fructoselysine and glucoselysine. The gfrABCDEF operon has a predicted RpoN-dependent promoter and a gene encoding a putative RpoN-dependent activator immediately upstream, and we also deleted this gene (gfrR). The ΔgfrAB, ΔgfrR, ΔgfrE, and ΔgfrF mutants were examined for their ability to grow in minimal media with fructoselysine or glucoselysine as the sole carbon or nitrogen source. The ΔgfrAB and ΔgfrR mutants failed to grow in minimal media with either fructoselysine or glucoselysine as the sole carbon or nitrogen source, indicating that the gfr locus is responsible for both fructoselysine and glucoselysine utilization. The ΔgfrE mutant failed to grow in minimal medium in which glucoselysine was the sole carbon or nitrogen source but grew normally in minimal medium that contained fructoselysine as the sole carbon and nitrogen source (generation time was 83 ± 4 min). Conversely, the ΔgfrF mutant failed to grow in minimal medium that contained fructoselysine as the sole carbon or nitrogen source but grew normally in minimal medium containing glucoselysine as the sole carbon and nitrogen source (generation time of 113 ± 13 min). These results were consistent with the prediction that GrfE and GrfF are deglycases for glucoselysine-6-phosphate and fructoselysine-6-phosphate, respectively (8).
Consistent with the prediction that transcription of the gfrABCDEF operon is dependent on RpoN, a ΔrpoN mutant failed to grow in minimal media containing fructoselysine or glucoselysine as the sole carbon source. For these phenotypic assays the medium was supplemented with 5 mM l-glutamine, as the deletion of rpoN results in glutamine auxotrophy (25). l-Glutamine is a poor carbon and nitrogen source and does not support growth of S. Typhimurium at the concentration used for the experiment. A ΔptsH mutant (ptsH encodes HPr) likewise failed to grow in minimal medium containing either fructoselysine or glucoselysine. S. Typhimurium 14028s possesses a paralog of HPr (STM14_4558), two additional HPr homologs fused to other PTS components (FruF and PtsA), and an HPr-like nitrogen-regulatory protein (NPr). The inability of the ΔptsH mutant to utilize fructoselysine or glucoselysine indicates that these other HPr homologs are either unable to substitute for HPr in the transport of fructoselysine and glucoselysine or are not expressed at sufficient levels under the assay conditions.
Regulated expression of the gfr locus.
Similar to Bacillus subtilis LevR, GfrR is predicted to possess an N-terminal DNA-binding domain, a central transcriptional activation domain, and a C-terminal regulatory domain that contains two PTS regulation domains (PRD) separated by EIIA(Man)- and EIIB(Gat)-like domains (26). Conserved histidine residues within the PRDs are phosphorylated by PTS components which, depending on the PRD, either stimulate or inhibit the activity of the protein (27). LevR regulates expression of the levDEFGsacC operon, which is required for fructose transport and catabolism (27). In the absence of exogenous fructose, the conserved histidine in the second LevR PRD (i.e., one at the C terminus) is phosphorylated by EI, HPr, and the EIIBC component of the fructose-specific PTS (LevEF), which inhibits the activity of LevR (27, 28). When fructose is present in the medium and transported across the membrane by the fructose-specific PTS, LevEF phosphorylates fructose instead of the LevR PRD (27, 28). To determine if fructoselysine induces expression of the gfrABCDEF operon, gfrAB transcript levels in S. Typhimurium grown in minimal medium containing 20 mM l-arabinose and either 5 mM fructoselysine or 10 mM ammonium chloride were measured by qRT-PCR. For these assays, gfrAB transcripts were normalized to rpoD transcript levels. Levels of gfrAB transcripts were 13-fold higher (13 ± 4.6; P < 0.01) in cultures grown with fructoselysine as a nitrogen source compared to cultures in which ammonium was the nitrogen source, indicating that fructoselysine induces expression of the gfrABCDEF operon.
Operons regulated by LevR-like activators often are subject to carbon catabolite repression. LevR is activated by the EI- and HPr-dependent phosphorylation of a conserved histidine (His-506) within the first PRD (28). The presence of glucose or other efficiently metabolized carbon sources in the growth medium prevents phosphorylation of His-506 and LevR remains inactive. GfrR has a tyrosine residue (Tyr-487) in the first PRD where the conserved histidine normally would be found. Joyet and coworkers recently reported that the PRD-containing activator MtlR from Lactobacillus casei similarly has a tyrosine in place of the conserved histidine which allows the protein to be active without being phosphorylated by EI and HPr; in contrast, MtlR from B. subtilis contains the conserved histidine residue within the PRD and needs to be activated by EI- and HPr-mediated phosphorylation (29). Taken together, these observations suggested that GfrR is not subject to carbon catabolite repression as occurs with other LevR-like activators. To test this hypothesis, we determined if S. Typhimurium could utilize fructoselysine in the presence of glucose. S. Typhimurium grew robustly in minimal medium containing glucose and fructoselysine as the sole nitrogen source (generation time, 38 ± 4 min), indicating that expression of the gfr locus is not significantly repressed by glucose.
GfrF has a fructoselysine-6-phosphate deglycase activity.
Wiame and coworkers showed that E. faecium GfrE and GfrF have glucoselysine-6-phosphate deglycase and fructoselysine-6-phosphate deglycase activity, respectively, and proposed that the S. Typhimurium counterparts have the same activities (8). Phenotypes of the S. Typhimurium gfrE and gfrF mutants were consistent with the activities suggested by Wiame and coworkers (8). To verify the proposed activities of S. Typhimurium GfrE and GfrF, we expressed His-tagged versions of the proteins in E. coli and attempted to purify the proteins by immobilized nickel-affinity chromatography. Unfortunately, we were unable to purify GfrE, as it was insoluble under all of the conditions we tested for inducing its expression, including induction at temperatures as low as 4°C. Problems with solubility of E. faecium GfrE were reported by Wiame and coworkers as well, but these researchers were able to purify the protein by overproducing it at low growth temperatures (8).
We were able to purify GfrF, however, and assay the purified protein for fructoselysine-6-phosphate deglycase activity using a coupled enzyme assay with glucose-6-dehydrogenase. In this coupled assay, the formation of glucose-6-phosphate from fructoselysine-6-phosphate was monitored by including glucose-6-phosphate dehydrogenase in the reaction mixture and monitoring the reduction of NADP at 340 nm. The specific activity of the purified S. Typhimurium GfrF was 0.78 μmol/min/mg, which was comparable to the specific activity reported for E. faecium GfrF (0.4 μmol/min/mg) (8).
Examining the substrate specificity of the fructoselysine/glucoselysine PTS permease and associated deglycases in vivo.
To examine the substrate specificities of the fructoselysine/glucoselysine PTS permease and GfrF, we synthesized analogs of fructoselysine and other sugar amines. In addition, we determined if the fructoselysine analogs inhibited growth of S. Typhimurium when fructoselysine was present in the medium as a nitrogen source. Fructoselysine analogs were synthesized using either d-lysine or l-ornithine, which has a side chain that is one carbon shorter than that of lysine. S. Typhimurium was able to use fructose–d-lysine as a carbon source but not as a nitrogen source. These data indicated that fructose–d-lysine is transported and phosphorylated by GfrABCD and the resulting fructose–d-lysine-6-phosphate is recognized by GfrF, but that d-lysine is not utilized as a nitrogen source. S. Typhimurium was unable to use fructose-ornithine as a carbon or nitrogen source, suggesting that it is not recognized as a substrate by the fructoselysine/glucoselysine PTS and/or GfrF.
We tested the ability of the fructoselysine analogs to inhibit growth of S. Typhimurium in minimal medium that contained l-arabinose as the primary carbon source and either ammonium chloride (10 mM) or fructoselysine (5 mM) as the nitrogen source. Cells grown with fructoselysine as the nitrogen source were not inhibited by fructose–d-lysine or fructose-ornithine at concentrations ranging from 2 to 20 mM the analog.
Ribuloselysine, erythuloselysine, and tagatoselysine were synthesized and tested for their ability to support growth of S. Typhimurium. These compounds were chosen since they are derived from the reaction of three relatively common sugars (ribose, erythrose, and galactose) with the ε-amine of lysine. None of these compounds supported the growth of S. Typhimurium when included in the medium as either the sole carbon or nitrogen source, indicating that the fructoselysine/glucoselysine PTS and/or GfrF (or GfrE) does not recognize these compounds as substrates.
S. Typhimurium uses lysine derived from fructoselysine as a nitrogen source but not exogenous lysine.
Gutnick and Ames reported that S. Typhimurium LT2 is unable to use l-lysine as a nitrogen source (2). We confirmed that S. Typhimurium 14028s is unable to grow in minimal medium with l-lysine as the sole nitrogen source. We postulated that lysine accumulates to high levels inside the cell when S. Typhimurium is grown on fructoselysine, which allows the bacterium to use lysine as a nitrogen source, and such high intracellular levels of lysine cannot be achieved by the lysine transporters. We reasoned that one or more transaminases in the cell may be able to use lysine as a substrate when intracellular levels of lysine are high, which would allow the bacterium to use lysine derived from fructoselysine as a nitrogen source. Deamination of l-lysine results in (S)-2-amino-6-oxohexanoate, which undergoes a spontaneous intramolecular dehydration to form Δ1-piperideine-6-carboxylate. To test our hypothesis, we used mass spectrometry to examine S. Typhimurium grown with fructoselysine for the accumulation of extracellular or intracellular Δ1-piperideine-6-carboxylate.
S. Typhimurium cultures were grown to stationary phase in a minimal medium that contained 20 mM fructoselysine as the sole carbon and nitrogen source, at which point the cells were removed from the medium by centrifugation (i.e., extracellular fraction) and the resulting supernatant liquid was analyzed. Alternatively, part of the culture was subjected to multiple freeze-thaw cycles to lyse the cells, which then were removed by centrifugation (i.e., combined intracellular and extracellular fractions), and the resulting supernatant liquid was analyzed. We did not observe an obvious peak for the expected mass of Δ1-piperideine-6-carboxylate [m/z 128.1, (M + 1)+] in the mass spectra of either the extracellular-only fraction or the combined extracellular and intracellular fractions. From the mass spectra, however, we noted that the fructoselysine had been depleted from the spent medium and observed a prominent peak with the mass expected for lysine [m/z 147.2, (M + 1)+] in the mass spectrum of the sample of the combined extracellular and intracellular fractions (see Fig. S1 in the supplemental material). We also observed a prominent peak with the mass expected for histidine [m/z 156.1, (M + 1)+] in the mass spectrum of the sample of the combined extracellular and intracellular fractions (see Fig. S1). Neither peak was evident in the mass spectrum of extracts of S. Typhimurium grown in minimal medium with arabinose as the carbon source and ammonium as the nitrogen source (data not shown).
In the histidine biosynthesis pathway, histidinol-phosphate aminotransferase (HisC) catalyzes the conversion of imidazole acetol-phosphate to histidinol-phosphate using l-glutamate as an amino donor. We hypothesized that HisC uses lysine as an amino donor when intracellular concentrations of lysine are high, and this reaction allows S. Typhimurium to use fructoselysine as a nitrogen source. HisC is known to be promiscuous, reacting with phenylalanine and tyrosine as an amino donor, as well as glutamate (30). To test the hypothesis that HisC is required for S. Typhimurium to use fructoselysine as a nitrogen source, we deleted hisC in S. Typhimurium 14028s and examined the growth of the resulting mutant in medium that contained fructoselysine (20 mM) as the primary carbon and nitrogen source. Since the ΔhisC mutant is a histidine auxotroph, the medium was supplemented with 140 μM l-histidine, an amount sufficient to allow some growth of the mutant but insufficient to support a significant growth yield. Compared to the wild-type strain, the ΔhisC mutant grew significantly slower (116 ± 13 min generation time for the mutant versus 80 ± 3 min for the wild type), suggesting that the ΔhisC mutant was able to utilize fructoselysine as a nitrogen source but not as efficiently as the wild-type strain. Growth rates of the wild-type strain and ΔhisC mutant were essentially the same in minimal medium containing l-arabinose plus ammonium and supplemented with 140 μM histidine (generation times for the wild type and ΔhisC mutant were 54 ± 4 min and 47 ± 3 min, respectively). Taken together, these data indicate that loss of HisC interferes with S. Typhimurium's ability to utilize fructoselysine but does not abolish growth on fructoselysine.
The results with the ΔhisC mutant indicated that S. Typhimurium uses multiple pathways for catabolism of lysine derived from fructoselysine, so we examined other pathways through which the bacterium might utilize lysine. S. Typhimurium GfrD was reported to modulate the activity of CadC (31), a positive regulator of the cadBA genes which encode a lysine/cadaverine antiporter and lysine decarboxylase, respectively, and are involved in acid stress response in S. Typhimurium. We postulated that growth of S. Typhimurium on fructoselysine upregulates the expression of cadA and allows the bacterium to use lysine derived from fructoselysine as a nitrogen source. S. Typhimurium possesses a second lysine decarboxylase, LdcC, so we constructed a mutant strain that lacked both cadA and ldcC. Growth of the cadA-ldcC double mutant with fructoselysine as the sole carbon and nitrogen source was impaired but not abrogated (generation time of 99 ± 14 min for the mutant versus 80 ± 3 min for the wild type), suggesting that decarboxylation of lysine is another route by which S. Typhimurium can utilize lysine derived from fructoselysine.
DISCUSSION
We verified that the S. Typhimurium gfr locus encodes a mannose family PTS and associated deglycases required for the transport and catabolism of fructoselysine and glucoselysine (Fig. 2). The gfr locus allows S. Typhimurium to use fructoselysine and glucoselysine as both carbon and nitrogen sources, although these compounds support more rapid growth of S. Typhimurium as nitrogen sources than as carbon sources. In the initial steps of the Maillard reaction, Amadori products and Heyns products are formed by the spontaneous reaction of the carbonyl groups of reducing sugars with the free amino groups of proteins or amino acids. Microorganisms likely encounter such products frequently in their environments, since a variety of microorganisms are able to utilize these compounds. Enteric bacteria may encounter fructoselysine from glycated proteins in its hosts' diets as the passage of fructosamines through the intestinal mucosa is very limited (12). Other Amadori products encountered by enteric bacteria could result from the glycation of peptides or amino acids during digestion. In soil environments, the Maillard reaction takes place during the formation of humic substances (12). Significant levels of Amadori products also are present in rotting fruits and vegetables (11), as well as in dried fruits, nuts, and grains (12).
FIG 2.
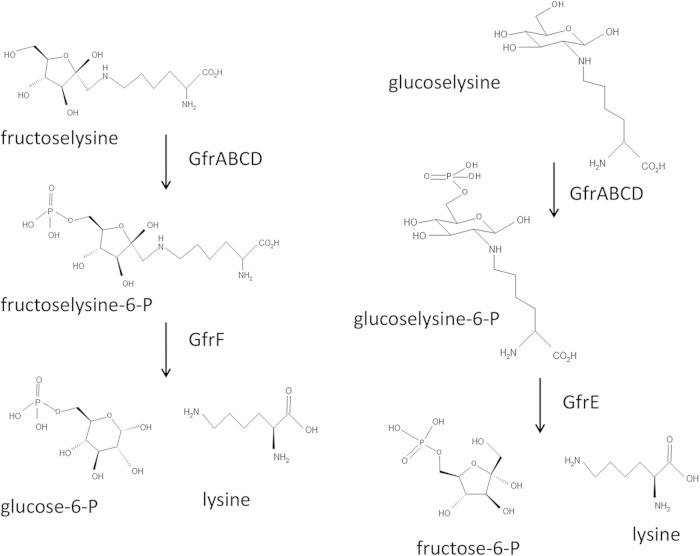
Proposed pathway for utilization of fructoselysine and glucoselysine. GfrABCD is a PTS permease that transports both fructoselysine and glucoselysine, phosphorylating the sugar moieties of these compounds at the C-6 position as they are transported across the membrane. GfrF and GfrE are deglycases that cleave fructoselysine 6-phosphate and glucoselysine 6-phosphate, respectively, to generate the corresponding hexose-6-phosphates and lysine.
A couple of pathways for the catabolism of fructosamines have been described to date. Fructosyl amino acid oxidases (also known as Amadoriases) catalyze the oxidative degradation of fructosamines to generate the corresponding amino acid, glucosone, and hydrogen peroxide (32). Amadoriases have been found in fungi such as Aspergillus and Penicillium, as well as in bacteria such as Arthrobacter, Pseudomonas, and Corynebacterium (32). A second type of pathway is found in E. coli and B. subtilis and involves phosphorylation of the fructosamine by a fructosamine 6-kinase (FrlD) and a subsequent cleavage by a deglycase (FrlB) to generate the corresponding amino acid and glucose-6-phosphate (22). The E. coli and B. subtilis FrlB/FrlD systems share about 30% amino acid identity but have different substrate specificities. The E. coli FrlB/FrlD system is specific for ε-glycated lysine, whereas the B. subtilis enzymes catalyze reactions with a number of different α-glycated amino acids (33). The E. coli frl locus includes a gene that encodes a fructoselysine 3-epimerase (frlC) which catalyzes the reversible interconversion of fructoselysine with its C-3 epimer, psicoselysine, and allows the bacterium to utilize this Amadori product (34).
Ali and coworkers recently showed that S. Typhimurium possesses an FrlB/FrlD-like system that is required for utilization of α-glycated asparagine (i.e., fructose-asparagine) (35). The S. Typhimurium operon that contains the genes encoding the FrlB/FrlD-like system (fraB and fraD) also contains genes predicted to encode a transporter (fraA) and an asparaginase (fraE). FraE is thought to be localized to the periplasm, where it converts fructose-asparagine to fructose-aspartate, which subsequently is transported across the cell membrane by FraA. Once inside the cell, fructose-aspartate is thought to be converted to aspartate and glucose-6-phosphate by the sequential action of the FraB kinase and FraD deglycase (35). The fra locus was found to be essential for Salmonella fitness in an inflamed mouse intestine model, suggesting that fructose-asparagine represents a significant nutrient for Salmonella in the inflamed intestine (35).
The enzymes encoded by the S. Typhimurium and E. faecium gfr loci represent a variation on the FrlB/FrlD-like system. In E. coli, fructoselysine and psicoselysine are transported across the cell membrane by the permease FrlA, while in B. subtilis the α-glycated amino acid substrates are transported across the membrane by an ABC transport system (FrlONM) (33). These Amadori products subsequently are phosphorylated by the FrlB kinase once inside the cell. In S. Typhimurium and E. faecium, fructoselysine and glucoselysine are transported by a mannose-type PTS which phosphorylates the sugar moieties of these compounds at the C-6 position as they are translocated across the cell membrane. As with the FrlD enzymes, the GfrE and GfrF deglycases in S. Typhimurium and E. faecium then cleave the phosphorylated products to generate lysine and either glucose-6-phosphate or fructose-6-phosphate.
Although GfrR is a LevR-like activator, it lacks the conserved histidine that is required for activation of LevR by EI- and HPr-mediated phosphorylation (26). Similar to L. casei MtlR (29), GfrR has a tyrosine residue at this position which appears to render the protein insensitive to carbon catabolite repression and allows S. Typhimurium to simultaneously metabolize glucose and fructoselysine. Consistent with the apparent lack of carbon catabolite repression in fructoselysine utilization, there does not appear to be a Crp-binding site (TGTGA-N6-TCACA) upstream of gfrA. In addition to GfrR, S. Typhimurium has two other LevR-like activators. One of these activators, DgaR, regulates expression of the dgaABCDEF operon, which encodes a PTS and enzymes for the transport and catabolism d-glucosaminate (36). DgaR has the conserved histidine required for activation (His-497), and glucose inhibits d-glucosaminate utilization in S. Typhimurium (36). The other LevR-like activator (SL1344_0559) presumably stimulates transcription of a PTS operon of unknown function, and like GfrR, this activator has a tyrosine residue (Tyr-476) in place of the activating histidine; therefore, its activity likely is insensitive to carbon catabolite repression.
Paradoxically, S. Typhimurium grows well with fructoselysine as a nitrogen source but not with lysine. We postulate that S. Typhimurium must accumulate lysine to high intracellular levels to use it as a nitrogen source, and such concentrations cannot be achieved through the lysine permeases. S. Typhimurium has at least two transport systems for lysine, the lysine-specific permease LysP and the lysine/arginine/ornithine periplasmic transport protein (ArgT), which functions in concert with the HisQMP ABC transporter (37). Consistent with the hypothesis that high intracellular levels of lysine are required to support growth of S. Typhimurium as a nitrogen source, lysine was readily detected in cell extracts of S. Typhimurium grown in minimal medium containing fructoselysine but not in extracts of cells grown in medium lacking fructoselysine (see Fig. S1 in the supplemental material). We postulate that S. Typhimurium uses multiple pathways to catabolize endogenously generated lysine, as we observed that both the hisC mutant and cadA-ldcC double mutant were impaired, but not completely blocked, in their ability to utilize fructoselysine as a nitrogen source.
Supplementary Material
ACKNOWLEDGMENTS
We thank Emile Van Schaftingen and Elsa Wiame for providing E. coli fructoselysine kinase as well as other materials. We thank Josef Deutscher for helpful discussions. We thank Ashley Bono for providing S. Typhimurium strain ACB01, Diana Downs for the use of equipment and providing S. Typhimurium strains, and Anna Karls for helpful discussions.
This research was supported by grant MCB-1051175 from the National Science Foundation to T.R.H. and by Agriculture and Food Research Initiative competitive grant 2012-67011-19934 from the USDA National Institute of Food and Agriculture to K.A.M.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00339-15.
REFERENCES
- 1.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States–major pathogens. Emerg Infect Dis 17:7–15. doi: 10.3201/eid1701.091101p1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gutnick D, Calvo JM, Klopotowski T, Ames BN. 1969. Compounds which serve as the sole source of carbon or nitrogen for Salmonella typhimurium LT-2. J Bacteriol 100:215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chaudhuri RR, Morgan E, Peters SE, Pleasance SJ, Hudson DL, Davies HM, Wang J, van Diemen PM, Buckley AM, Bowen AJ, Pullinger GD, Turner DJ, Langridge GC, Turner AK, Parkhill J, Charles IG, Maskell DJ, Stevens MP. 2013. Comprehensive assignment of roles for Salmonella typhimurium genes in intestinal colonization of food-producing animals. PLoS Genet 9:e1003456. doi: 10.1371/journal.pgen.1003456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Erni B, Zanolari B, Graff P, Kocher HP. 1989. Mannose permease of Escherichia coli. Domain structure and function of the phosphorylating subunit. J Biol Chem 264:18733–18741. [PubMed] [Google Scholar]
- 5.Erni B, Zanolari B, Kocher HP. 1987. The mannose permease of Escherichia coli consists of three different proteins. Amino acid sequence and function in sugar transport, sugar phosphorylation, and penetration of phage lambda DNA. J Biol Chem 262:5238–5247. [PubMed] [Google Scholar]
- 6.Plumbridge J. 1998. Control of the expression of the manXYZ operon in Escherichia coli: Mlc is a negative regulator of the mannose PTS. Mol Microbiol 27:369–380. doi: 10.1046/j.1365-2958.1998.00685.x. [DOI] [PubMed] [Google Scholar]
- 7.Geerse RH, Izzo F, Postma PW. 1989. The PEP:fructose phosphotransferase system in Salmonella typhimurium: FPr combines enzyme IIIFru and pseudo-HPr activities. Mol Gen Genet 216:517–525. doi: 10.1007/BF00334399. [DOI] [PubMed] [Google Scholar]
- 8.Wiame E, Lamosa P, Santos H, Van Schaftingen E. 2005. Identification of glucoselysine-6-phosphate deglycase, an enzyme involved in the metabolism of the fructation product glucoselysine. Biochem J 392:263–269. doi: 10.1042/BJ20051183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baynes JW, Watkins NG, Fisher CI, Hull CJ, Patrick JS, Ahmed MU, Dunn JA, Thorpe SR. 1989. The Amadori product on protein: structure and reactions. Prog Clin Biol Res 304:43–67. [PubMed] [Google Scholar]
- 10.Suarez G, Rajaram R, Oronsky AL, Gawinowicz MA. 1989. Nonenzymatic glycation of bovine serum albumin by fructose (fructation). Comparison with the Maillard reaction initiated by glucose. J Biol Chem 264:3674–3679. [PubMed] [Google Scholar]
- 11.Mossine VV, Mawhinney TP. 2010. 1-Amino-1-deoxy-d-fructose (“fructosamine”) and its derivatives. Adv Carbohydr Chem Biochem 64:291–402. [DOI] [PubMed] [Google Scholar]
- 12.Deppe VM, Bongaerts J, O'Connell T, Maurer KH, Meinhardt F. 2011. Enzymatic deglycation of Amadori products in bacteria: mechanisms, occurrence and physiological functions. Appl Microbiol Biotechnol 90:399–406. doi: 10.1007/s00253-010-3083-4. [DOI] [PubMed] [Google Scholar]
- 13.Studholme DJ. 2002. Enhancer-dependent transcription in Salmonella enterica Typhimurium: new members of the sigmaN regulon inferred from protein sequence homology and predicted promoter sites. J Mol Microbiol Biotechnol 4:367–374. [PubMed] [Google Scholar]
- 14.Samuels DJ, Frye JG, Porwollik S, McClelland M, Mrazek J, Hoover TR, Karls AC. 2013. Use of a promiscuous, constitutively-active bacterial enhancer-binding protein to define the sigma54 (RpoN) regulon of Salmonella Typhimurium LT2. BMC Genomics 14:602. doi: 10.1186/1471-2164-14-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu H, Hoover TR. 2001. Transcriptional regulation at a distance in bacteria. Curr Opin Microbiol 4:138–144. doi: 10.1016/S1369-5274(00)00179-X. [DOI] [PubMed] [Google Scholar]
- 16.Neidhardt FC, Bloch PL, Smith DF. 1974. Culture medium for Enterobacteria. J Bacteriol 119:736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maloy SR, Stewart VJ, Taylor RK. 1996. Genetic analysis of pathogenic bacteria: a laboratory manual. Cold Spring Harbor Laboratory Press, Plainview, NY. [Google Scholar]
- 18.Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol 185:60–89. [DOI] [PubMed] [Google Scholar]
- 19.Delpierre G, Collard F, Fortpied J, Van Schaftingen E. 2002. Fructosamine 3-kinase is involved in an intracellular deglycation pathway in human erythrocytes. Biochem J 365:801–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fortpied J, Gemayel R, Stroobant V, van Schaftingen E. 2005. Plant ribulosamine/erythrulosamine 3-kinase, a putative protein-repair enzyme. Biochem J 388:795–802. doi: 10.1042/BJ20041976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wiame E, Delpierre G, Collard F, Van Schaftingen E. 2002. Identification of a pathway for the utilization of the Amadori product fructoselysine in Escherichia coli. J Biol Chem 277:42523–42529. doi: 10.1074/jbc.M200863200. [DOI] [PubMed] [Google Scholar]
- 23.Stead MB, Marshburn S, Mohanty BK, Mitra J, Pena Castillo L, Ray D, van Bakel H, Hughes TR, Kushner SR. 2011. Analysis of Escherichia coli RNase E and RNase III activity in vivo using tiling microarrays. Nucleic Acids Res 39:3188–3203. doi: 10.1093/nar/gkq1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat Prot 3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 25.Garcia E, Bancroft S, Rhee SG, Kustu S. 1977. The product of a newly identified gene, glnF, is required for synthesis of glutamine synthetase in Salmonella. Proc Natl Acad Sci U S A 74:1662–1666. doi: 10.1073/pnas.74.4.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deutscher J, Francke C, Postma PW. 2006. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol Mol Biol Rev 70:939–1031. doi: 10.1128/MMBR.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stülke J, Martin-Verstraete I, Charrier V, Klier A, Deutscher J, Rapoport G. 1995. The HPr protein of the phosphotransferase system links induction and catabolite repression of the Bacillus subtilis levanase operon. J Bacteriol 177:6928–6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin-Verstraete I, Charrier V, Stülke J, Galinier A, Erni B, Rapoport G, Deutscher J. 1998. Antagonistic effects of dual PTS-catalysed phosphorylation on the Bacillus subtilis transcriptional activator LevR. Mol Microbiol 28:293–303. doi: 10.1046/j.1365-2958.1998.00781.x. [DOI] [PubMed] [Google Scholar]
- 29.Joyet P, Derkaoui M, Bouraoui H, Deutscher J. 2015. PTS-mediated regulation of the transcription activator MtlR from different species: surprising differences despite strong sequence conservation. J Mol Microbiol Biotechnol 25:94–105. [DOI] [PubMed] [Google Scholar]
- 30.Mavrides C, Comerton M. 1978. Aminotransferases for aromatic amino acids and aspartate in Bacillus subtilis. Biochim Biophys Acta 524:60–67. doi: 10.1016/0005-2744(78)90103-1. [DOI] [PubMed] [Google Scholar]
- 31.Lee YH, Kim S, Kim JH, Bang IS, Lee IS, Bang SH, Park YK. 2013. A phosphotransferase system permease is a novel component of CadC signaling in Salmonella enterica. FEMS Microbiol Lett 338:54–61. doi: 10.1111/1574-6968.12025. [DOI] [PubMed] [Google Scholar]
- 32.Wu X, Monnier VM. 2003. Enzymatic deglycation of proteins. Arch Biochem Biophys 419:16–24. doi: 10.1016/j.abb.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 33.Wiame E, Duquenne A, Delpierre G, Van Schaftingen E. 2004. Identification of enzymes acting on alpha-glycated amino acids in Bacillus subtilis. FEBS Lett 577:469–472. doi: 10.1016/j.febslet.2004.10.049. [DOI] [PubMed] [Google Scholar]
- 34.Wiame E, Van Schaftingen E. 2004. Fructoselysine 3-epimerase, an enzyme involved in the metabolism of the unusual Amadori compound psicoselysine in Escherichia coli. Biochem J 378:1047–1052. doi: 10.1042/BJ20031527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ali MM, Newsom DL, Gonzalez JF, Sabag-Daigle A, Stahl C, Steidley B, Dubena J, Dyszel JL, Smith JN, Dieye Y, Arsenescu R, Boyaka PN, Krakowka S, Romeo T, Behrman EJ, White P, Ahmer BM. 2014. Fructose-asparagine is a primary nutrient during growth of Salmonella in the inflamed intestine. PLoS Pathol 10:e1004209. doi: 10.1371/journal.ppat.1004209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller KA, Phillips RS, Mrazek J, Hoover TR. 2013. Salmonella utilizes D-glucosaminate via a mannose family phosphotransferase system permease and associated enzymes. J Bacteriol 195:4057–4066. doi: 10.1128/JB.00290-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wissenbach U, Six S, Bongaerts J, Ternes D, Steinwachs S, Unden G. 1995. A third periplasmic transport system for L-arginine in Escherichia coli: molecular characterization of the artPIQMJ genes, arginine binding and transport. Mol Microbiol 17:675–686. doi: 10.1111/j.1365-2958.1995.mmi_17040675.x. [DOI] [PubMed] [Google Scholar]
- 38.Martin-Verstraete I, Stülke J, Klier A, Rapoport G. 1995. Two different mechanisms mediate catabolite repression of the Bacillus subtilis levanase operon. J Bacteriol 177:6919–6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.