ABSTRACT
RNA interference (RNAi) is a process of eukaryotic posttranscriptional gene silencing that functions in antiviral immunity in plants, nematodes, and insects. However, recent studies provided strong supports that RNAi also plays a role in antiviral mechanism in mammalian cells. To combat RNAi-mediated antiviral responses, many viruses encode viral suppressors of RNA silencing (VSR) to facilitate their replication. VSRs have been widely studied for plant and insect viruses, but only a few have been defined for mammalian viruses currently. We identified a novel VSR from coronaviruses, a group of medically important mammalian viruses including Severe acute respiratory syndrome coronavirus (SARS-CoV), and showed that the nucleocapsid protein (N protein) of coronaviruses suppresses RNAi triggered by either short hairpin RNAs or small interfering RNAs in mammalian cells. Mouse hepatitis virus (MHV) is closely related to SARS-CoV in the family Coronaviridae and was used as a coronavirus replication model. The replication of MHV increased when the N proteins were expressed in trans, while knockdown of Dicer1 or Ago2 transcripts facilitated the MHV replication in mammalian cells. These results support the hypothesis that RNAi is a part of the antiviral immunity responses in mammalian cells.
IMPORTANCE RNAi has been well known to play important antiviral roles from plants to invertebrates. However, recent studies provided strong supports that RNAi is also involved in antiviral response in mammalian cells. An important indication for RNAi-mediated antiviral activity in mammals is the fact that a number of mammalian viruses encode potent suppressors of RNA silencing. Our results demonstrate that coronavirus N protein could function as a VSR through its double-stranded RNA binding activity. Mutational analysis of N protein allowed us to find out the critical residues for the VSR activity. Using the MHV-A59 as the coronavirus replication model, we showed that ectopic expression of SARS-CoV N protein could promote MHV replication in RNAi-active cells but not in RNAi-depleted cells. These results indicate that coronaviruses encode a VSR that functions in the replication cycle and provide further evidence to support that RNAi-mediated antiviral response exists in mammalian cells.
INTRODUCTION
RNA interference (RNAi) is originally regarded as a mechanism of eukaryotic posttranscriptional gene regulation mediated by small interfering RNA (siRNA)-induced sequence-specific RNA degradation (1). It is also well known to exert as an important antiviral defense mechanism in a wide range of organisms, from plants to invertebrates (2). During the virus infection, the virus-derived long double-stranded RNA (dsRNA) is cleaved by RNAIII-like endonuclease (named Dicer) into approximately 21- to 23-nucleotide (nt) siRNA, which is incorporated into the RNA-induced silencing complex (RISC) and activates the antiviral RNAi for viral RNA degradation. In mammalian cells, although the activation of RNAi by synthetic siRNA or short hairpin RNA (shRNA) is widely used as a tool for gene knockdown and antiviral treatment, the RNAi-mediated antiviral mechanism has been debated for a long time (3), because the interferon (IFN) response of the innate immune system is well known as the dominant antiviral mechanism (4). However, more and more evidence has provided strong support for the existence of a natural RNAi-mediated antiviral response in mammals (5). Moreover, recent studies showed that in undifferentiated cells and immature mice, the RNAi-mediated antiviral response is essential (6–8).
To overcome the RNAi-mediated antiviral defense, viruses have evolved to encode a viral suppressor of RNA silencing (VSR) (9, 10). For example, in plant viruses, rice hoja blancavirus NS3, tombusvirus P19, and tomato aspermy virus 2b bind to long dsRNA or siRNA to block RNAi (11–13). Turnip crinkle virus P38 and cauliflower mosaic virus P6 disrupt the components of RNAi machinery (14, 15). In insect viruses, flock house virus (FHV) B2 blocks RNAi by dsRNA binding (16, 17), and Wuhan nodavirus (WhNV) B2 was identified as a VSR by targeting both dsRNAs and Dicer-2 (18, 19). Although the majority of VSRs have been identified in plant and invertebrate viruses, several mammalian viruses were shown to encode VSRs. For instance, Ebola virus VP35, influenza A virus NS1, vaccinia virus E3L, and Nodamura virus (NoV) B2 act as VSRs by binding dsRNA (20–23). Hepatitis C virus core and HIV-1 Tat block RNAi by inhibiting the activity of Dicer (24, 25). Interestingly, all VSRs identified from mammalian viruses possess IFN or protein kinase R antagonistic properties and are essential for replication and pathogenesis, suggesting that RNAi and other innate antiviral responses are interrelated (26–28).
Coronaviruses (CoVs) are the largest positive single-stranded RNA (ssRNA) viruses carrying an RNA genome of 26.2 to 31.7 kb that infect a wide range of mammalian and avian species (29). It is reported that coronaviruses generate significant amount of dsRNAs as replicative and transcriptive intermediates (30, 31). Therefore, it might be the target of Dicer and thus induce RNAi-mediated antiviral responses. An indirect evidence was shown that severe acute respiratory syndrome coronavirus (SARS-CoV) accessory protein 7a was identified as a VSR (32). However, 7a protein is not essential for viral replication and transcription at least in cell culture and tested animal models and is unique to SARS-CoV (33, 34). Consequently, it would be interesting to determine whether there is another VSR commonly encoded among coronaviruses family. In the present study, we screened the viral proteins of SARS-CoV as a representative by a reversal-of-silencing assay and identified the nucleocapsid (N) protein as a novel VSR, which is conserved and encoded by all of the coronaviruses.
N protein is a basic protein (with typical pIs of ∼10) and has nonspecific binding activity toward nucleic acids, including ssRNA, ssDNA, and dsDNA (35, 36). It encapsulates viral genomic RNA (gRNA) to protect the genome and enters the host cell together with the viral RNA to facilitate its replication (37–40). Furthermore, we have reported that the N protein antagonizes IFN-β by targeting the initial pattern recognition receptor/RNA recognition step and that the C-terminal domain (CTD) is critical for this antagonism (41). Other studies also revealed that the SARS-CoV N protein contains two distinct RNA-binding domains (the N-terminal domain [NTD] and the CTD) linked by a poorly structured linkage region (Linker) containing a serine/arginine-rich (SR-rich) domain (SRD) (42–45). The CTD spanning residues 248 to 365 shows stronger nucleic acid-binding activity than the NTD (36, 45, 46), and the basic region between residues 248 to 280 of CTD forms a positively charged groove that represents a likely binding region for RNA (46). Here, we demonstrated that the N protein of CoVs could efficiently inhibit Dicer-mediated dsRNA cleavage and post-Dicer activities by sequestering dsRNAs and siRNAs. Furthermore, we show that N protein deficient in RNAi inhibition activity was unable to promote the replication of mouse hepatitis virus (MHV) compared to the wild-type N protein and that knockdown of Dicer1 or Ago2 enhanced MHV replication. Our studies identified a novel coronaviral VSR and provide new evidence on the existence of RNAi-mediated antiviral response in mammalian cells.
MATERIALS AND METHODS
Plasmids and RNAs.
For RNAi suppression assays in mammalian cells, the plasmid eGFP-C1 (Clontech) was used to express enhanced green fluorescent protein (eGFP). The eGFP-specific shRNA (shGFP) and control shRNA targeting luciferase (shLuc) with the indicated sequences in Table S1 in the supplemental material were cloned to vector pSuperRetro driven by H1 polymerase III promoter. The plasmid pCMV-tag2b-N expressing SARS-CoV N protein and the deletion mutants with Flag tag were constructed in our previous work (41). The open reading frame 6 (ORF6) expression plasmid with a hemagglutinin (HA) tag was kindly provided by Stanley Perlman. Point mutations were introduced into the N coding region by PCR mediated mutagenesis, with appropriate primers containing the desired nucleotide changes (see Table S2 in the supplemental material) and subsequently selected by DpnI digestion. The coding sequences of Middle East respiratory syndrome coronavirus (MERS-CoV) N protein were chemically synthesized and cloned into the pCMV-tag2b vector. Plasmids expressing N protein of coronaviruses MHV, porcine epidemic diarrhea virus (PEDV), and transmissible gastroenteritis virus (TGEV) were gifts from Shaobo Xiao. The NoV B2-expressing plasmid with Myc tag was provided by Christopher S. Sullivan. For RNAi suppression assays in Drosophila S2 cells, the eGFP reporter gene and the FHV B2 were constructed into the insect expression vector pAc5.1/V5-HisB. SARS-CoV N protein and ORF6 were inserted into the EcoRI/NotI sites of pAc5.1/V5-HisB. Nonstructural protein 14 (nsp14) cloned in pAc5.1/V5-HisB was inserted into the NotI/XhoI sites. The primers are shown in Table S2 in the supplemental material. Full-length cDNA of FHV RNA1 and RNA1-ΔB2 (T2739C and C2910A) were described previously (17). In addition, the siRNAs targeting eGFP (siGFP) were prepared by chemical synthesis (Invitrogen), whereas siRNAs targeting mouse Dicer1 and Ago2 were obtained from Qiagen. The oligonucleotides are shown in Table S3 in the supplemental material. The 244-bp dsRNA for eGFP silencing in Drosophila S2 cells was generated by in vitro transcription using MEGAscript kits (Ambion).
Cell culture and transfection.
Human embryonic kidney 293T cells (HEK293T), mouse Neuro-2a cells (gifts from Yan Zhou) and L2 cells (gifts from Rong Ye) were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum, 100 U of penicillin/ml, and 100 μg of streptomycin/ml. Drosophila S2 cells were cultured in semi suspension at 27°C in Schneider's insect medium (Gibco, Carlsbad, CA) supplemented with 10% fetal bovine serum (Gibco) (18). HEK293T cells were seeded on 12-well dishes and grown overnight to reach 50% confluence, followed by transfection with standard calcium phosphate precipitation method. Transfection of Drosophila S2 cells was conducted by using FuGene HD reagent (Roche, Basel, Switzerland) when the cells were grown to reach 80% confluence, according to the manufacturer's protocol. Neuro-2a and L2 cells were seeded on 12-well dishes and grown overnight to reach 106, followed by Lipofectamine 2000 (Invitrogen) transfection. In dose-dependent experiments, empty control plasmid was added to ensure that each transfection received the same amount of total DNA.
Western blotting.
Cells were harvested in cell lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% NP-40, 0.25% deoxycholate, and a protease inhibitor cocktail [Roche]), and the extracts were then subjected to SDS-PAGE and Western blotting, according to our standard procedures (47). The blots were exposed to luminescent image analyzer LAS4000 (Fuji Film). The antibodies used here were as follows: anti-β-actin (Proteintech Group), horseradish peroxidase-conjugated anti-eGFP (Santa Cruz Biotechnology; 1:2,000), anti-Flag and anti-HA (Sigma; 1:5,000), and anti-Myc (Roche; 1:2,000).
Northern blotting.
Total RNA was extracted from cells using TRIzol reagent (Invitrogen), according to the manufacturer's protocol. For eGFP mRNA detection, 5 μg of RNA was subjected to electrophoresis in 1.2% denaturing agarose gels containing 2.2 M formaldehyde. The separated RNAs were transferred onto a Hybond N+ nylon membrane (GE Healthcare, Waukesha, WI) and then cross-linked by exposure to UV light. For siRNA detection, 10 μg of low-molecular-weight RNAs extracted from cells using RNAiso (TaKaRa) were separated on a 12% polyacrylamide gel with 7 M urea and transferred to Hybond N+ nylon membranes by electroblotting using a semidry blotting apparatus. The hybridization with digoxigenin (DIG)-labeled probes and DIG chemiluminescent detection were conducted with DIG Northern Starter kit (Roche Diagnostics, Indianapolis, IN) according to the manufacturer's instruction. The blots were exposed to luminescent image analyzer LAS4000 (Fuji Film). The probe for detection of eGFP mRNA was complementary to the eGFP ORF region of nucleotides 1 to 500 (for experiments in mammalian cells) or 501 to 720 (for experiments in insect cells). The probe for detection of FHV RNA1 and subgenomic RNA3 specifically targets the B2 coding region from nt 2738 to nt 3058. For eGFP shRNA and siRNA detection, the sense sequence of siGFP was used to probe the antisense moiety of shRNA and siRNA of eGFP. All probes were labeled with DIG-UTP by in vitro transcription using DIG Northern starter kit. The templates were made from PCR amplification or annealing with the oligonucleotides listed in Table S1 in the supplemental material. rRNAs or low-molecular-weight RNAs were visualized by staining with ethidium bromide.
Expression and purification of recombinant proteins.
The coding sequences of SARS-CoV N protein and WhNV B2 were PCR amplified and inserted into the BamHI/NotI sites of pGEX-6P-1. Escherichia coli BL21 (Invitrogen) transformed with the expression plasmids was grown to the log phase and induced with 0.6 mM IPTG (isopropyl-β-d-thiogalactopyranoside), followed by incubation at 16°C for 12 h. After harvesting by centrifugation, the bacterial pellet was lysed with lysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol [DTT], 0.1 mg of lysozyme/ml, 0.05% NP-40), and the recombinants proteins were purified with glutathione resin (GenScript) according to the manufacturer's instructions and stored at −80°C.
Gel shift assay and RNase III-mediated cleavage assays.
We generated 244-bp DIG-labeled dsRNA, 500-bp dsRNA, and 500-nt ssRNA by in vitro transcription using DIG RNA labeling mix (Roche). Gel shift assays for RNA binding were performed using 15 μM glutathione S-transferase (GST), 15 μM GST-WhNV B2, or increasing concentrations of GST-N up to 15 μM and 0.2 pmol of DIG-labeled RNAs in a 20-μl reaction system containing 50 mM Tris-HCl (pH 7.4), 75 mM NaCl, 1 mM EDTA, 1 mM DTT, and 20 U of RNA inhibitor (Fermentas). After incubation for 30 min at 25°C, the reaction mixtures were separated on 1.2% Tris-borate-EDTA (TBE)–agarose gel and subjected to Northern blotting for DIG signal detection. In a gel shift assay for siRNA binding, 0.2 pmol of 5′Hex-labeled siRNA was incorporated into the reaction. The reaction mixtures were separated using 4% native polyacrylamide gel electrophoresis, followed by fluorescent detection with a Typhoon 9200 (Amersham Biosciences).
RNase III cleavage inhibition assays were conducted using DIG-labeled 500-bp dsRNA and RNase III (Invitrogen) as described previously (19). Each assay was performed in a 20-μl reaction system containing 0.2 pmol of DIG-labeled 500-bp dsRNA, 2 μl of 10× RNase III reaction buffer (Invitrogen), and 15 μM concentrations of either GST, GST-WhNV B2, or GST-N. After 30 min of preincubation at 25°C, 1 U of RNase III was added, and the reaction mixtures were incubated at 37°C for 30 min. Reaction products were resolved by 1.2% TBE–agarose gel electrophoresis and then subjected to Northern blotting for DIG signal detection.
Sequence alignment and analysis of coronavirus N protein.
The CLUSTAL X program V2.0 was used to align the sequences of coronavirus N protein N. The resulting file was transferred to GENDOC to prepare for the graphic figures. Sequences for SARS-CoV N proteins were collected from the following genome sequences: SARS-CoV, strain Tor2, NC_004718; MERS-CoV-Jeddah-human-1, (KF958702.1); Bat-CoV strain HKU9-1 BF_005I, NC_009021; MHV-A59, NC_001846; IBV strain Beaudette, NC_001451; TGEV strain PUR46-MAD, NC_002306; PEDV strain CV777, NC_003436; and H-CoV strain 229E, NC_002645.
Virus infection and real-time PCR.
At 24 or 48 h posttransfection, Neuro-2a cells or L2 cells were infected with MHV strain A59 at a multiplicity of infection (MOI) of 0.1. At 16 h postinfection, the supernatants were collected, and the virus titer was determined by plaque assay on L2 cells (48). For mRNA detection, total RNA was isolated from cells with TRIzol reagent. The RNA was reverse transcribed to first-strand cDNA using Moloney murine leukemia virus reverse transcriptase (Promega). The SYBR green master mix (Roche) was used for real-time PCR. Mouse GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA was used as an internal control. The primers used are shown in Table S3 in the supplemental material.
RESULTS
Identification of coronaviral N protein as a VSR in mammalian cells.
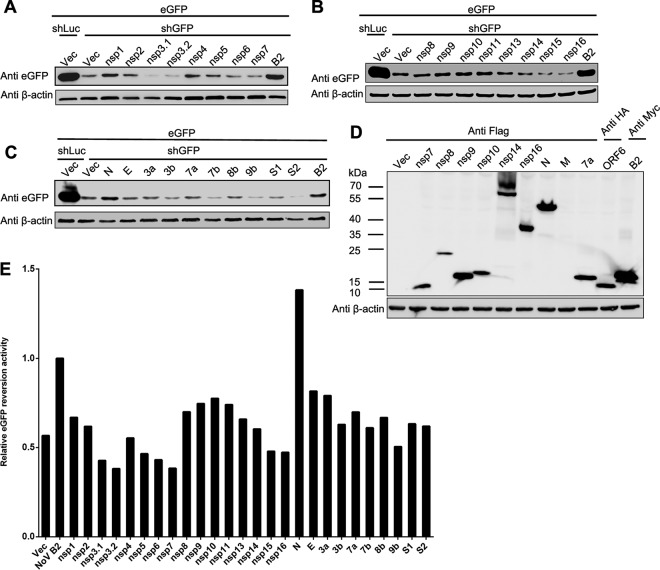
To evaluate whether coronaviruses encode a VSR, we screened the proteins encoded by SARS-CoV using the reversal-of-silencing assay in HEK293T cells (Fig. 1A, B, and C). NoV B2, a well-known VSR in mammalian cells (23), was used as a positive control. The expression level of eGFP reporter gene was tested by Western blotting and fluorescence microscopy, and the relative eGFP reversion activities of each protein toward NoV B2 were calculated. As shown in Fig. 1E, SARS-CoV N protein could efficiently revert the expression of RNAi-silenced eGFP, as well as NoV B2. In contrast, the accessory protein 7a, which was reported as a VSR of SARS-CoV (32), showed extremely limited VSR activity in the screening assays. We detected the protein expression level of some constructs, including nsp7-16, N, 7a, ORF6, M, and B2, and the results showed that most of these proteins were readily detectable but at variable levels (Fig. 1D). Therefore, the VSR activity and its strength observed in the initial screening were not conclusive and had to be verified by further systematic experiments. Because SARS-CoV N showed relatively higher VSR activity in the reversal-of-silencing assay, we continued to characterize SARS-CoV N as a potential VSR.
FIG 1.
Screening of potential VSR of SARS-CoV by reversal-of-silencing assays. 293T cells were cotransfected with plasmids encoding eGFP reporter (125 ng), eGFP-specific shRNA (shGFP) (1 μg), or luciferase-specific shRNA (shLuc) (1 μg) and viral protein of SARS-CoV (500 ng), respectively. (A to C) The expression level of eGFP was determined by Western blotting at 72 h posttransfection. Empty vector (Vec) and NoV B2 (B2) were used as a mock control and a positive control. The shLuc was used as an irrelevant silencing control. β-Actin was used as a loading control. (D) The expression of SARS-CoV-encoded proteins as indicated was detected by Western blotting. (E) The relative eGFP reversion activity of different viral proteins in panels A to C was normalized by that of typical VSR NoV B2 control and is shown in a bar diagram.
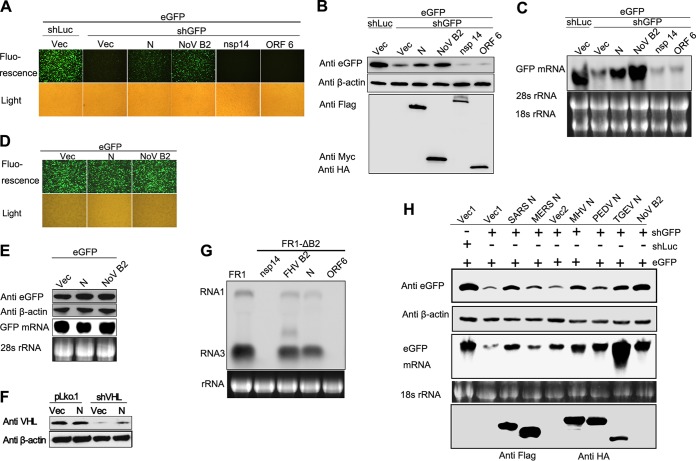
To confirm the RNAi suppressor activity of SARS-CoV N protein, we assayed the capability of N protein in suppressing shRNA-induced eGFP silencing at protein and mRNA level. The eGFP-specific shRNA (shGFP) caused a strong decrease of eGFP expression (Fig. 2A and B) and transcription (Fig. 2C) compared to the irrelevant shRNA that targets luciferase (shLuc). The nsp14 with nonspecific RNA binding activity (49) and ORF6 protein with IFN antagonistic activity (50) were used as negative controls. Notably, transfection of the N expression plasmid remarkably resurrected the expression of RNAi-silenced eGFP at both protein and mRNA levels, as well as B2, but ORF6 and nsp14 could not (Fig. 2A, B, and C), indicating that N suppressed the shRNA-induced RNA silencing of eGFP. Importantly, N protein did not affect the expression efficiency of eGFP in the absence of shRNA (Fig. 2D and E), confirming that N protein inhibited the effects of shGFP-mediated RNAi rather than promoted eGFP transcription or translation. To exclude that the observed effects specifically exists in the eGFP reporter system, the RNAi suppression activity of N protein was determined in an endogenous RNAi system (Fig. 2F). We demonstrated that the presence of N protein restored the endogenous VHL expression silenced by VHL-specific shRNA (shVHL), whereas the VHL expression level was not affected in mock vector (pLko.1)-transfected cells (Fig. 2F). Since the RNAi pathway is conserved from plants to animals, some VSRs of mammalian viruses are also functional in insect cells such as influenza A virus NS1 and vaccinia virus E3L (22). We further confirmed the VSR activity of SARS-CoV N protein using FHV RNA1 replication system (Fig. 2G). Transfection of pRNA1 led to the self-replication of FHV RNA1 and the transcription of RNA3 in S2 cells. FHV B2 was well known to inhibit RNA silencing induced by virus RNA replication. B2-deficient mutant (pRNA1-ΔB2) failed to accumulate FHV RNA1 and RNA3. This defect could be partially rescued by cotransfection with plasmids either expressing SARS-CoV N protein or FHV B2 (Fig. 2G). To explore whether the RNAi inhibition activity of N protein is universal among coronaviruses, the N proteins from alpha coronavirus (PEDV and TGEV) and beta coronavirus (SARS-CoV, MERS-CoV, and MHV) were tested. As shown in Fig. 2H, the indicated coronavirus N proteins inhibited shRNA-induced RNAi to various degrees in the reversal-of-silencing system, wherein MERS-CoV N showed notably low VSR activity.
FIG 2.
N protein of coronaviruses represses shRNA-induced RNAi in mammalian cells. (A to C) HEK293T cells were cotransfected with plasmids encoding eGFP reporter (125 ng), shGFP, or shLuc (1 µg) and plasmids encoding viral proteins as indicated (300 ng). The expression of eGFP reporter was analyzed 72 h after cotransfection. (A) The intensity of eGFP was observed under fluorescence microscopy. (B) Cell lysates were harvested and analyzed by Western blotting. (C) Cellular total mRNAs were harvested and analyzed by Northern blotting. (D and E) HEK293T cells were cotransfected with plasmids encoding eGFP and SARS-CoV N protein or NoV B2, respectively. eGFP fluorescence, protein, and mRNA levels were determined by fluorescence microscopy (D) and Western and Northern blotting (E), respectively. (F) HEK293T cells were cotransfected with plasmids of mock vector pLko.1 (1 μg) or shRNA targeting endogenous gene VHL (shVHL) in the presence or absence of SARS-CoV N protein (300 ng). At 72 h posttransfection, cells were harvested and subjected to Western blotting to determine the endogenous VHL expression. (G) S2 Cells were transfected with 0.03 μg of pFR1 or 0.6 μg of pFRNA1-ΔB2 and with SARS-CoV N or FHV B2 as indicated above. At 48 h posttransfection, FHV RNA transcription was induced by incubation with CuSO4 at 0.5 mM. At 24 h after induction, the cellular total mRNA was harvested for Northern blot analysis by a probe recognizing FHV RNA1 and RNA3. The band between RNA1 and RNA3 represents the mRNA transcribed from B2 expression plasmid. (H) HEK293T cells were cotransfected with plasmids encoding eGFP, shLuc, or shGFP and N proteins of different coronaviruses (Flag-tagged N proteins of SARS-CoV and MERS-CoV and HA-tagged N proteins of MHV, PEDV, and TGEV), respectively. The eGFP expression level and mRNA level were determined by Western blotting and Northern blotting. Empty vector (Vec, Vec1, or Vec2), nsp14, and ORF6 were used as negative controls, while NoV B2 was used as a positive control. The shLuc was irrelevant control shRNA. β-Actin and rRNAs were used as loading controls for Western and Northern blotting, respectively.
To investigate whether the VSR activity is dependent on the protein expression levels, the increasing amounts of SARS-CoV N were tested using the reversal-of-silencing assay (Fig. 3A and B). The reversal effect of eGFP silencing increased progressively at both protein and mRNA levels, along with the gradual increase of the transfected N-expressing plasmid (Fig. 3A and B). We also detected eGFP expression at different time points, and the results showed that the reversal of eGFP silencing could be observed as early as 24 h posttransfection and was more effective at 48 and 72 h (Fig. 3C), indicating that the VSR activity was dependent on the expression level of SARS-CoV N protein.
FIG 3.
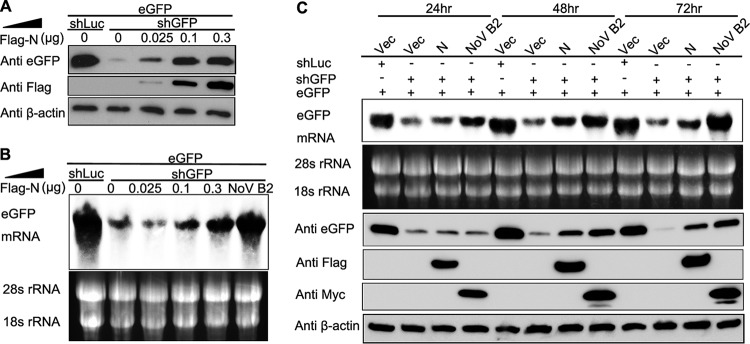
N protein represses shRNA-induced RNAi in a dose-dependent and time-dependent manner in mammalian cells. Increasing amounts of the plasmid expressing SARS-CoV N protein were transfected into HEK293T cells, as indicated in the upper panel in the reversal-of-silencing assay. At 72 h after transfection, Western blotting (A) and Northern blotting (B) were performed to determine the eGFP protein and mRNA levels, respectively. (C) Northern blotting (upper panel) and Western blotting (lower panel) were performed 24, 48, and 72 h posttransfection, respectively.
N protein inhibits Dicer-mediated siRNA generation and siRNA-induced RNAi.
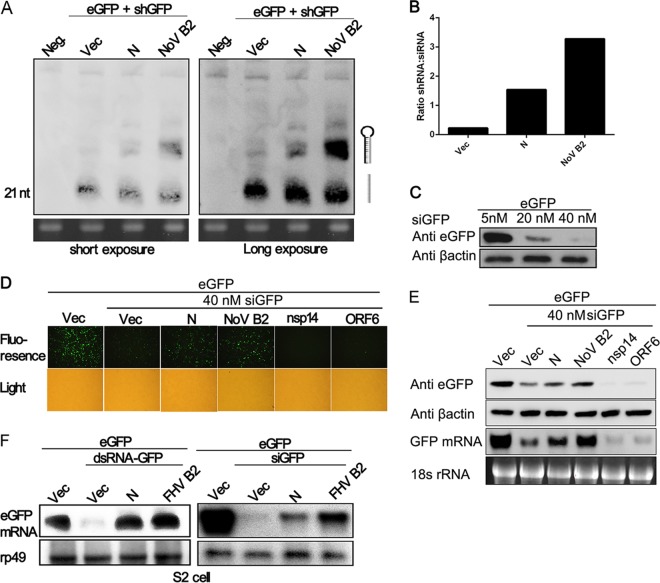
The shRNA-induced RNAi pathway requires Dicer-mediated dsRNA cleavage into siRNA. To find out whether N protein blocks the aforementioned step, small RNAs harvested from HEK293T cells were subjected to Northern blotting with DIG-labeled probes which recognized both shRNA and siRNA (Fig. 4A and B). In the presence of SARS-CoV N protein, an increase in the ratio of shRNA to 21-nt Dicer-processed siRNA was detected. However, the increase was more obvious in the presence of NoV B2 (Fig. 4A and B). This phenomenon is consistent with the eGFP expression and its mRNA level observed in the reversal-of-silencing assays (Fig. 2). To further demonstrate whether N protein could block RNAi induced by siRNA (the post-Dicer product), 40 nM eGFP-specific siRNA (siGFP) was used to induce a remarkable reduction of eGFP expression (Fig. 4C). As shown in Fig. 4D and E, the decrease in eGFP expression and its mRNA level in HEK293T cells was partially inhibited in the presence of SARS-CoV N protein comparing with NoV B2. We investigated the ability of N protein to suppress RNA silencing in insect cells. Consistent with the phenomenon observed in cells, the presence of SARS-CoV N protein efficiently abolished the eGFP-specific RNAi induced by dsRNA (dsRNA-GFP) while remarkably inhibiting the siGFP-induced RNAi in Drosophila S2 cells (Fig. 4F). Thus, the N protein could inhibit the RNA silencing in mammalian and insect cells at both Dicer-processing and post-Dicer stages with different efficiencies.
FIG 4.
N protein inhibits the production of siRNA and RNAi in both mammalian and insect cells. (A) HEK293T cells were cotransfected with plasmid as indicated above, 72 h after transfection, and small RNAs were harvested from the cells and probed with DIG-labeled oligonucleotides that correspond to the target sites of siRNA produced from shGFP. The locations of bands corresponding to shRNA and siRNA are indicated with diagrams on the right side. Short exposures and long exposures are shown on the left and right, respectively. An ethidium bromide-stained gel of low-molecular-weight RNA is shown as a loading control. Neg., mock control transfected with shLuc. (B) The ratios of shRNA to siRNA in panel A were quantified based on the corresponding exposure signals and are shown as a bar diagram. (C) eGFP expression plasmid (200 ng) was cotransfected with multiple concentrations (5 to 40 nM) of synthetic eGFP-specific siRNA (siGFP) to confirm the effective siGFP concentration in HEK293T cells. At 72 h after transfection, cell lysates were harvested to determine the reduction in eGFP expression by Western blotting. (D and E) SARS-CoV N protein inhibits siRNA-induced RNAi. HEK293T cells were transfected with plasmids as indicated above. At 72 h after transfection, eGFP fluorescence (D), protein (E), and mRNA (F) were detected as described in Fig. 2. (F) SARS-CoV N protein inhibits RNAi in Drosophila S2 cells. eGFP-specific dsRNA (dsRNA-GFP) and siGFP were used to induce RNAi in S2 cells. The mRNA of eGFP was detected by Northern blotting. Empty vector (Vec), nsp14, and ORF6 were used as negative controls. NoV B2 and FHV B2 were used as positive controls. rp49 was used as a loading control.
N protein directly binds to RNAs and prevents dsRNA from RNase III-mediated cleavage in vitro.
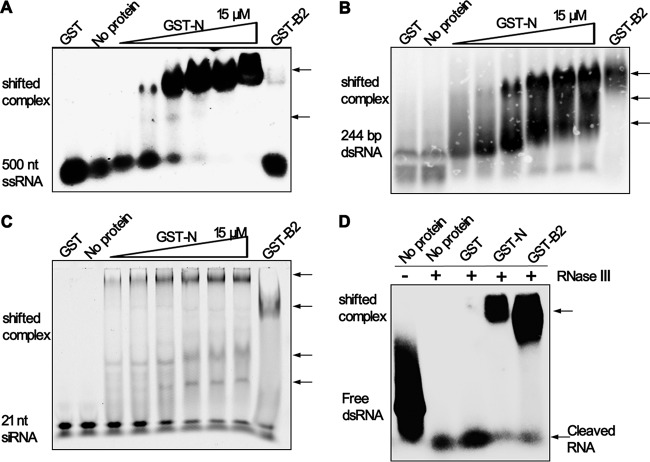
A number of VSRs could sequester RNA duplexes from Dicer cleavage and the incorporation of siRNA into RISC by dsRNA binding activity, such as NoV B2, EBoV VP35, and influenza virus NS1 (20, 22, 23). To investigate the mechanism of N protein as a VSR, we used a gel shift assay after the incubation of recombinant GST-tagged SARS-CoV N protein (GST-N) and WhNV B2 (GST-B2) with DIG-labeled 500-nt ssRNA (Fig. 5A), DIG-labeled 244-bp dsRNA (Fig. 5B), and synthetic 5′-hexachlorofluorescein phosphoramidite (HEX)-labeled 21-nt siRNA (Fig. 5C), which mimic viral ssRNA, cellular pre-Dicer dsRNA, and post-Dicer siRNA, respectively. As shown in Fig. 5A to C, the shifting amount of all labeled RNAs increased as more SARS N protein was used in the reaction up to 15 μM, while the WhNV B2 could not bind ssRNA. Interestingly, the mobility of labeled ssRNA (Fig. 5A) and dsRNA (Fig. 5B) decreased as more SARS-COV N protein was used, except that of labeled siRNA (Fig. 5C), suggesting that one molecule of long RNAs (ssRNA and dsRNA) was bound by the multiple N proteins. Subsequently, we examined the possibility of SARS-COV N protein to protect dsRNA from RNase III-mediated cleavage. RNase III is a Dicer homolog and was widely used as the Dicer substitute in vitro as previously reported (51). As shown in Fig. 5D, in a gel shift assay, 500-bp dsRNA formed a high-molecular-weight complex with GST-N, as well as GST-B2, and therefore was protected from RNase III digestion. However, without the protection of GST-N or GST-B2, dsRNA was digested into 21-to 23-nt siRNA. These data illustrated that N protein could directly bind dsRNA to prevent the digestion of Dicer and partially sequester siRNA at post-Dicer stages.
FIG 5.
N protein binds to RNAs and inhibits the Dicer-like RNase III cleavage reaction in vitro. Increasing amounts of purified GST-tagged SARS-CoV N protein (GST-N) from 0 to 15 μM were incubated with 0.2 pmol of 500-nt DIG-labeled ssRNA (A) or 244-bp DIG-labeled dsRNA (B) at 25°C for 30 min. Complexes were separated on 1.2% TBE–agarose gel and subjected to Northern blotting. The free ssRNA and dsRNA are indicated on the left side. (C) GST-N up to 15 μM was incubated with 0.2 pmol of 5′-HEX-labeled 21-nt siRNA as described in panels A and B. Complexes were applied to 4% native polyacrylamide gel, and the fluorescent signal was visualized by using a Typhoon 9200. The free siRNA is indicated on the left side. (D) DIG-labeled 500-bp dsRNA was incubated with purified proteins, as indicated above, at 25°C for 30 min before the processing of Dicer-like RNase III at 37°C for 30 min. The reaction products were separated on 1.2% TBE–agarose gel and subjected to Northern blotting. The free dsRNA is indicated on the left side, and the cleaved dsRNA is indicated on the right side. The shifted protein-RNA complexes are indicated by black arrows on the right side. The protein GST and GST-tagged WhNV B2 (GST-B2) were used as a mock control and a positive control.
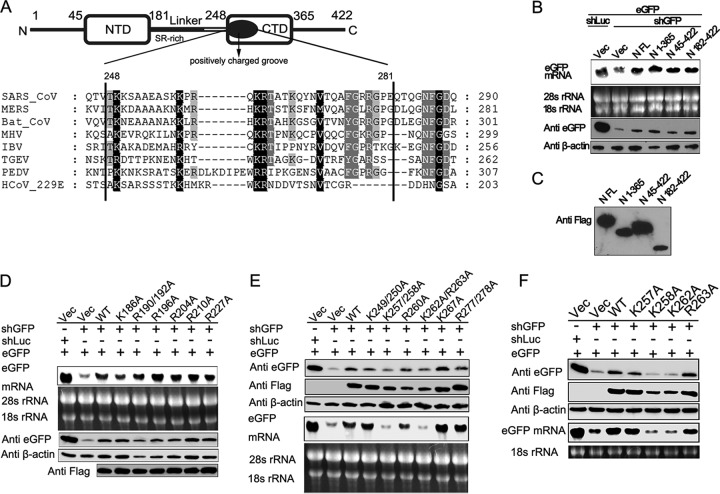
Positively charged residues Lys 258 and Lys 262 of SARS-CoV N protein are critical for the RNAi repression activity.
Mutagenesis and alanine scanning analyses were performed to identify the critical domain or amino acid (aa) required for the VSR activity of SARS-CoV N protein. The domain architecture of SARS-CoV N protein is shown in Fig. 6A. Based on domain truncation analysis, we found that the N-terminal (1 to 181 aa) and the C-terminal (365 to 422 aa) of SARS-CoV N protein are not required for RNAi inhibition activity (Fig. 6B), suggesting that the functional VSR region might be located at the linkage region (Linker) and CTD of SARS-CoV N protein (182 to 365 aa). The expression of truncated proteins was evaluated (Fig. 6C). Previous studies revealed that this region possesses an SR-rich domain (182 to 227 aa) and a positively charged groove (248 to 280 aa) (Fig. 6A), which might use positively charged residues arginine and lysine to mediate electrostatic interaction with the phosphate backbone of RNAs (36, 46, 52, 53). Therefore, multiple sequence alignments of the SR-rich domain (data not shown) and the positively charged groove of the CTD of the coronavirus N protein (Fig. 6A) were performed, and the single or double mutations of conserved arginine and lysine were analyzed by the reversal-of-silencing assay in HEK293T cells, as shown in Fig. 6D and E. Further mutational analysis revealed that mutations of conserved Lys 258 and Lys262 in CTD of N protein (K258A and K262A) completely abolished its ability to repress RNAi induced by shRNA in HEK293T cells (Fig. 6F). Taken together, these data show that the positively charged residues Lys 258 and Lys 262 are critical for the RNAi repression activity of SARS-CoV N protein, and these results support the importance of the RNA-binding activity of protein N in RNAi suppression.
FIG 6.
Conserved residues LysK258 and LysK262 of SARS-CoV N protein are critical for RNAi activity. (A) Schematic diagram of the domain architecture of the SARS-CoV N protein and multiple-sequence alignment of CTD spanning residues 248 to 281 of coronavirus N proteins. The conserved residues are indicated by solid black boxes. NTD, N-terminal domain; CTD, C-terminal domain; SR-rich, rich in serine and arginine; Linker, linkage region. (B and C) Mapping of critical residues of SARS-CoV N protein for VSR activity. The RNAi repression activity of Flag-tagged truncations of SARS-CoV N protein was analyzed by a reversal-of-silencing assay as described for Fig. 2B. The expression of truncated SARS-CoV N proteins was detected by Western blotting. (D to F) The RNAi repression activity of Flag-tagged mutants of SARS-CoV N protein was analyzed by a reversal-of-silencing assay, as described for Fig. 2. (F) The K257A, K258A, K262A, and R263A mutants were further analyzed based on the results from panel E.
SARS-CoV N protein and NoV B2 promote the replication of coronavirus MHV.
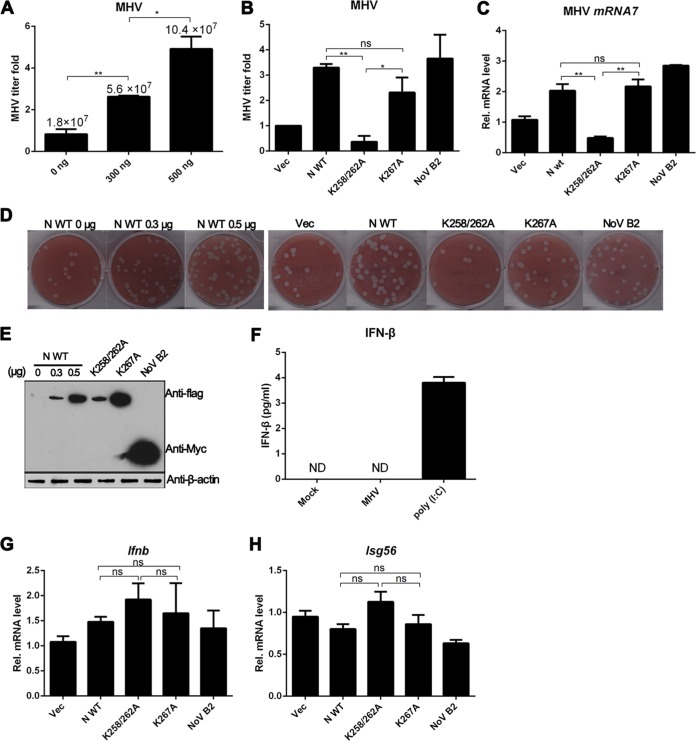
To demonstrate the physiologic function of coronavirus N protein as a VSR in vivo, we used coronavirus MHV strain A59, which belongs to the same virus group as SARS-CoV, as a model to investigate the influence of N protein and its mutants on virus replication. We and others have reported that the multiple functional N protein is essential for coronavirus, and the expression of N protein either in cis or in trans could facilitate the viral replication (40, 54, 55). Therefore, SARS-CoV N protein, N mutants and NoV B2 provided in trans were tested in the in vivo system. As shown in Fig. 7A to D, the MHV titer was upregulated 3- to 5-fold by SARS-CoV N protein in a dosage-dependent manner in Neuro-2a cells (Fig. 7A), whereas the VSR inactive mutant K258/262A failed to promote the replication of MHV (Fig. 7B). In contrast, the VSR activity irrelevant mutant K267A (Fig. 6E) exhibited promotion similar to that of the wild-type N protein (Fig. 7B). Impressively, the typical VSR NoV B2 could also facilitate the replication of MHV (Fig. 7B), further suggesting that the increase of MHV replication may be due to the general suppression of RNAi in host cells. The MHV titers were checked by the plaque assays in L2 cells (Fig. 7D). These results were also confirmed by analyzing the synthesis of viral gene 7 mRNA (mRNA 7) using reverse transcription-PCR (RT-PCR) in Neuro-2a cells (Fig. 7C), and the expression of corresponding proteins was evaluated (Fig. 7E). As shown in Fig. 7D and E, the effect of SARS-CoV N protein on the enhancement of MHV growth was also dose dependent, a finding consistent with the previous observation (Fig. 3A and B). In the above-described assays, the steady-state protein level of K258/262A mutant was similar to that of wild-type N protein with the transfection dosage of 300 ng of DNA (Fig. 7E); however, the mutant K258/262A could not promote MHV growth, but 300 ng of wild-type N protein could still significantly enhance virus replication (Fig. 7A, D, and E). Therefore, the deficiency of K258/262A mutant in VSR activity was not due to reduced expression level.
FIG 7.
SARS-CoV N protein promotes MHV replication when provided in trans. (A and B) Mouse Neuro-2a cells were transfected with plasmids as indicated and infected with MHV strain A59 at an MOI of 0.1 at 24 h posttransfection. At 16 h after infection, culture supernatants were collected and subjected to plaque assay on L2 cells to determine the MHV titers. The relative titer fold of MHV was quantified, as shown in bar diagrams. The actual virus titers are in indicated in panel A to show MHV replication efficiency. (C) Total RNAs were extracted from the transfected cells and subjected to RT-PCR using primers targeting the subgenomic RNA7 of MHV. The data were normalized to the abundance of endogenous mouse GAPDH mRNA. (D) Virus plaque formation was analyzed on L2 cells at a dilution of 10−6. (E) Protein expression levels in transfected cell lysates were detected by Western blotting with the indicated antibodies. β-Actin was used as a loading control. (F) Neuro-2a cells were infected with MHV strain A59 at an MOI of 0.1 or transfected with 2 μg of poly(I·C). At 16 h postinfection or transfection, IFN-β production was determined by enzyme-linked immunosorbent assay (ELISA). (G and H) Total RNAs were extracted from the transfected cells as described in panel B and subjected to RT-PCR to detect Ifn-β and Isg56 mRNA. Error bars indicate the means and standard deviations for triplicate experiments. *, P < 0.05; **, P < 0.01; ns, not significant (unpaired Student t test).
Coronavirus N proteins have been reported to have IFN-β response inhibition activity in IFN induction systems (41, 50, 56), suggesting that N might also promote MHV replication through antagonizing IFN responses in host cells. However, previous studies on coronavirus infections showed that MHV induces a minimal type I IFN response in several cell types, and the expression of IFN-β is undetectable (57–60). When Neuro-2a cells were treated with dsRNA analog poly(I·C), low level of IFN-β was produced, but MHV infection did not induce expression of IFN-β (Fig. 7F). We also analyzed the transcription of the Ifn-β mRNA and its downstream gene Isg56 mRNA that represent the expression level of type I IFN. As shown in Fig. 7G and H, the mRNA levels of Ifn-β and Isg56 are neither reduced compared to Nov B2 and vector control nor significantly changed with N protein and its mutants, which ruled out the possibility that the N protein increased MHV production by antagonizing the IFN-β signaling pathway. These results demonstrated that SARS-CoV N protein could promote the virus replication independently of IFN signaling pathway and that the critical residues Lys 258 and Lys 262 of SARS-CoV N protein for VSR activity are involved in facilitating MHV replication by inhibiting the RNAi-mediated antiviral mechanism. Moreover, the irrelevant viral protein NoV B2 could also promote the replication of MHV, indicating that the MHV replication might be sensitive to the RNAi-mediated antiviral response.
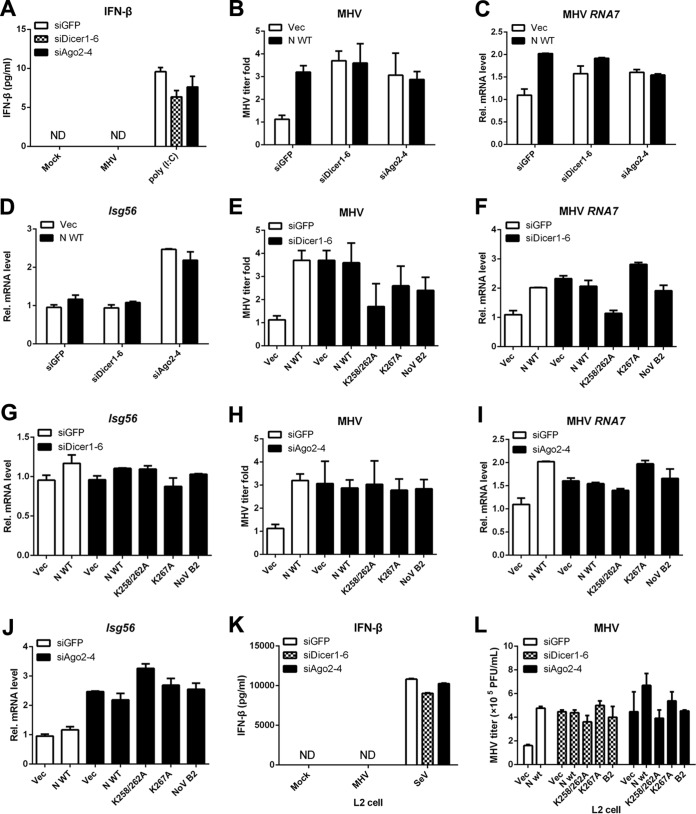
Knockdown of either Dicer1 or Ago2 facilitated the replication of MHV.
To further investigate the function of RNAi-mediated antiviral response when the IFN-mediated antiviral response is highly repressed by coronaviruses, the Dicer1 and Ago2 transcripts in mouse Neuro-2a and L2 cells were knocked down by synthetic siRNAs (siDicer1 and siAgo2). As shown in Fig. 8, the mRNA levels of Dicer1 and Ago2 were markedly reduced in cells transfected with siDicer1-6 and siAgo2-4, respectively (Fig. 8A and C), whereas the Ifn-β mRNA was not significantly affected (Fig. 8B and D). The RNAi-deficient Neuro-2a cells were still competent in IFN production (Fig. 9A), indicating that RNAi depletion did not nonspecifically abolish the IFN pathway. In RNAi-deficient Neuro-2a cells, either in the presence or in the absence of SARS-CoV N protein, the replications of MHV were increased to the same level as in RNAi pathway healthy cells provided with SARS N protein (Fig. 9B and C). Accordingly, there was no reduction in the mRNA level of Isg56 observed in the presence of SARS-CoV N (Fig. 9D), indicating that the increase of MHV replication was affected by the attenuated RNAi pathway but not by suppression of type I IFN response. In fact, the mRNA level of Isg56 increased to some extent, whereas the replication of MHV was still promoted in Ago2-knockdown cells (Fig. 9B to D), strongly suggesting that RNAi knockdown was involved in these cells. Moreover, the differences of MHV replication promotion between wild-type SARS-CoV N protein, NoV B2, and mutant K285/262A of SARS-COV N protein observed in Neuro-2a cells (Fig. 7) were eliminated in RNAi knockdown cells by siDicer1 or siAgo2, respectively (Fig. 9E, F, H, and I), whereas the expression of these proteins did not result in different IFN responses (Fig. 9G and J). The VSR activity was also analyzed in L2 cells that are highly IFN sensitive. Both healthy and RNAi pathway-deficient L2 cells could produce high levels of IFN-β (up to 10,000 pg/ml) when infected by Sendai virus (SeV), but the production of IFN-β was still undetectable when infected by MHV (Fig. 9K). In RNAi-competent L2 cells (treated with siGFP), N protein could significantly increase MHV growth (Fig. 9L). Interestingly, when the RNA pathway was depleted by siDicer1-6 or siAgo2-4, the increment of MHV growth was kept in the absence of N or in the presence of VSR-deficient mutant K258/262A (Fig. 9L). Together, the observations in L2 cells indicate that either suppression of RNAi by SARS-CoV N or depletion of RNAi pathway by siRNAs could lead to increment of MHV growth. These results were consistent with that of Neuro-2a cells. Taken together, these results demonstrate that SARS-CoV N protein is a novel VSR and the RNAi pathway was involved in anticoronavirus response in mammalian cells.
FIG 8.
Screen for siRNAs targeting mouse Dicer1 and Ago2. Mouse Neuro-2a cells were transfected with 40 nM siDicer1 (A and B) or siAgo2 (C and D). At 48 h posttransfection, total cellular mRNAs were extracted and subjected to RT-PCR to determine the mRNA levels of Dicer1 (A), Ago2 (C), and Ifn-β (B and D). siGFP was used as a negative control. The data were normalized to the abundance of internal mouse GAPDH mRNA. Error bars indicate the means and standard deviations for triplicate experiments.
FIG 9.
Knockdown of Dicer1 or Ago2 facilitated MHV replication. (A) Mouse Neuro-2a cells were transfected with siRNAs as indicated above and, at 48 h posttransfection, the cells were infected with MHV at an MOI of 0.1 or transfected with 2 μg of poly(I·C). At 16 h postinfection or posttransfection, IFN-β production was determined by ELISA. (B to J) Mouse Neuro-2a cells were cotransfected with 40 nM siRNA and 500-ng protein expression plasmids as indicated and infected with MHV strain A59 at an MOI of 0.1 at 48 h posttransfection. At 16 h postinfection, the culture supernatants were collected and subjected to plaque assays on L2 cells to determine the MHV titers. (B, E, and H) The relative titer folds of MHV were quantified and are shown in the bar diagrams. Total RNAs were extracted from the transfected cells and subjected to RT-PCR to determine the indicated mRNA levels. (C, F, and I) MHV RNA7 level. (D, G, and J) Isg56 mRNA level. (K) L2 cells were transfected with siRNAs as indicated above and, at 48 h posttransfection, the cells were infected with MHV or SeV. At 16 h postinfection, IFN-β production was determined by ELISA. (L) L2 was transfected and infected similarly as described for Neuro-2a cells. The MHV titers were determined by plaque assay. Empty vector (Vec) and siGFP were used as the negative control. The data were normalized to the abundance of endogenous mouse GAPDH mRNA. Error bars indicate the means and standard deviations for triplicate experiments.
DISCUSSION
Mammalian RNA viruses have evolved many mechanisms to protect their genomic RNAs and dsRNAs generated as replicative and transcriptive intermediates (30, 31, 59, 61) from host cell recognition. For example, they associate the replication-transcription complex with the double-membrane vesicles to form a protective microenvironment which prevents viral RNA from being detected by host cell sensors (62, 63), they encode several RNA processing enzymes to undergo RNA modifications such as RNA capping to mimic the host cell RNAs (49, 64), and they adopt RNA binding proteins to protect viral RNAs (65, 66). However, virus-derived small RNAs were still detectable in 41 human cell lines infected with six different RNA viruses, respectively (5). Moreover, several reports showed that virus-specific siRNAs accumulated in infected mammalian cells, and recently the specific siRNAs of encephalomyocarditis virus and Nodamura virus (NoV) were deep sequenced (6, 7). Therefore, in mammalian cells that possess a functional RNAi pathway, the sequence-specific RNA degradation would still threaten the abundant viral mRNA in cytoplasm if without VSR during virus infection, especially for coronavirus, the largest RNA virus as known. We present here evidence that coronaviruses also possess VSR, which was identified as N protein that shares similar structural architecture and gRNA/subgenomic RNA (sgRNA) binding activity to enhance the viral transcription, replication, and assembly among coronaviruses (37, 38, 40, 65, 66). It is abundantly produced in infected cells to support its multiple functions, as well as VSR. In addition to the ssRNA binding activity, we showed that N protein also possesses dsRNA and siRNA binding activities. Moreover, SARS-CoV N protein could bind more efficiently with long ssRNA and dsRNA than siRNAs (Fig. 5), which may be resulted from cooperative binding of multiple N protein monomers. Coronaviruses possess the largest single-stranded genomic RNA and abundant sgRNAs and dsRNA intermediates (29–31, 61). Therefore, we propose a hypothesis that the coronavirus N protein might protect viral RNA from RNAi-mediated gene silencing at three stages (Fig. 10): (i) binding viral ssRNAs to prevent the positive- and negative-sense genomic or subgenomic RNAs from formation of unnecessary intramolecular and intermolecular dsRNA; (ii) shielding virus-derived dsRNA from Dicer cleavage through dsRNA binding activity; and (iii) binding to virus-derived siRNA to interfere with RISC assembly.
FIG 10.
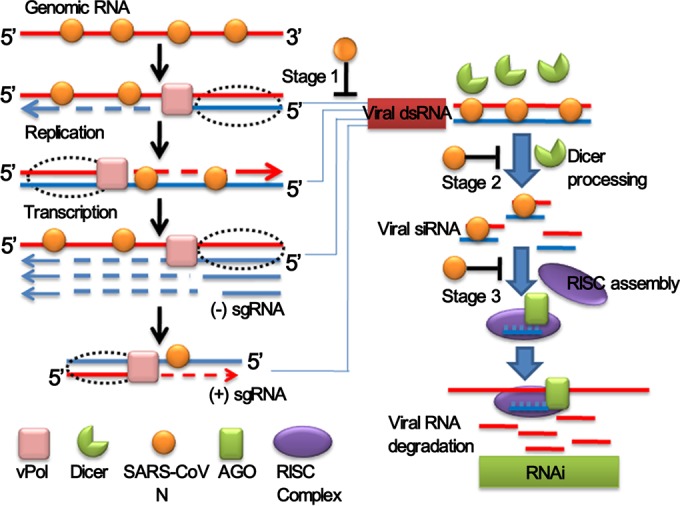
Model for the suppression of RNAi in mammalian cells by coronavirus N protein. After the entry and un coating of coronaviral virions, the single-stranded genomic RNA (gRNA) is protected by N proteins and serves as a template for the synthesis of negative-strand gRNA and a set of subgenomic RNA (sgRNA). The (−)gRNA and (−)sgRNA are replicated to generate full-length gRNA and a set of (+)sgRNA. The virus-derived dsRNA could be generated during viral transcription and replication. sgRNA and gRNA sequences may also form an intramolecular hairpin structure. These viral dsRNAs may be recognized by Dicer, consequently triggering antiviral RNAi. Coronavirus N proteins may repress the RNAi at three different stages as indicated.
In the present study, we showed that the VSR activity was highly dependent on the protein expression levels (Fig. 3A and B; Fig. 7D and E). In the initial screening assays for putative VSRs (Fig. 1), we did not evaluate the protein expression levels of different constructs, and therefore effects observed in the reversal-of-RNA silencing could not be directly compared and may result in either false-positive or false-negative outcomes. For example, SARS-CoV 7a, previously reported as a VSR (32), did not show apparent VSR activity in our assay system. Notably, the MERS-CoV N protein expression level is higher than that of SARS-CoV N protein, while the VSR activity of MERS-CoV N protein is less than that of SARS-CoV N protein (Fig. 2H). These results suggest that the N proteins of coronaviruses may have different intrinsic activities in suppressing RNAi. We speculate that the low VSR activity of MERS-CoV N might be compensated for by a second VSR and one potential candidate could be the dsRNA-binding IFN antagonist protein 4a (67).
Previous studies revealed that the three distinct and highly conserved domains of N proteins (NTD, SRD, and CTD) could bind with viral RNAs in different coronaviruses (42, 66, 68, 69). The NTD has been found to associate with a stem-loop structure located at the 3′ end of RNA genome, and the CTD was involved in the specific binding of coronaviral packaging signal within nonstructural protein nsp15 region (43, 70, 71). The critical residues Lys 258 and Lys 262 of SARS-CoV N protein for VSR identified here are located in the positively charged groove of CTD (Fig. 6). Moreover, Lys 258 was previously reported as a significant determinant of SARS-CoV N protein CTD binding affinity toward oligonucleotides (36). Here, we chose SARS-CoV N protein to promote MHV because SARS-CoV is closely related to MHV on one side, while the CTD of SARS-CoV N protein cannot enhance the specific selective packaging of MHV gRNA to facilitate the MHV replication (71). Therefore, the fact that the VSR inactive mutant K258/262A of SARS-CoV N protein led to the attenuation of MHV replication might imply that the sufficient VSR activity is important for coronavirus replication.
Although the IFN-mediated antiviral response is important for mammalian cells, a lot of studies have indicated that MHV is a poor inducer of type I IFN response (59). Moreover, MHV is resistant to the pretreatment of IFN-α/β and does not induce the generation of IFN-β in various cell types, suggesting that it can suppress IFN signaling pathway or its downstream gene effects at multiple levels (56, 58). In the present study, we showed that MHV infection did not induce the production of IFN-β in both mouse Neuro-2a and L2 cells (Fig. 7F and 9A). In primary cells, IFN is induced by MHV infection in plasmacytoid dendritic cells and macrophages (57, 72, 73), but not in neurons, astrocytes, and hepatocytes (57). Therefore, we used coronavirus MHV strain A59 as a replication model, which might to some extent avoid the impact of IFN response, to investigate the influence of N protein and its mutants on virus replication. We demonstrated that SARS-CoV N protein could promote MHV replication in both Neuro-2a cells and L2 cells, the former being not very responsive to IFN induction, whereas the latter were highly sensitive (Fig. 7 and 9). We also tested the VSR activity of N protein in IFN-deficient insect cells (Fig. 2G). The results indicated that the VSR activity of SARS-CoV N protein is independent of either the cell type or the IFN pathway. Moreover, the depletion of critical components Dicer1 or Ago2 of the RNAi pathway in Neuro-2a and L2 cells results in a significant upregulation of MHV replication, which was not dependent on the changes in IFN response (Fig. 9), further strengthen the role of SARS-CoV N protein in IFN-independent RNAi suppression.
In a study by Schelle et al. (74), the observed role of CoV N protein in the transcomplementation was most likely related to the virus recovery step before viral replication, whereas in our experimental system, we are dealing with a wild-type virus that does not require transcomplementation for virus recovery, and thus the role of SARS-CoV N protein that we observed is its effect on virus replication. The modest increment of MHV replication by SARS-CoV N protein suggests that the natural N protein of MHV may already exert rather effective VSR function and that exogenous SARS-CoV N protein may further increase the anti-RNAi effect. Interestingly, NoV B2, which has no correlation and no structural or sequence similarities with SARS-CoV N, exhibited the same ability in stimulating MHV replication. These results indicated that the MHV replication is sensitive to the RNAi-mediated antiviral response. However, there has been no direct evidence for existence of CoV-derived siRNAs in infected cells. Generation of a VSR-defective MHV mutant may help to detect MHV-derived siRNAs in mammalian cells. Currently, viral siRNAs have been discovered in cell culture for six RNA viruses and in vivo for encephalomyocarditis virus and NoV (5–7). Our studies provide new evidence for the functional RNAi-mediated antiviral response in addition to the IFN-mediated innate immunity in mammalian cells. These two important antiviral mechanisms might work in turn in different situations and locations to defend against virus infection, which would constantly provide a positive selection toward the VSR and viral IFN antagonist during the tachytelic evolution of viruses.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Christopher S. Sullivan from the University of Texas, Stanley Perlman from the University of Iowa, Shaobo Xiao from Huazhong Agricultural University, Rong Ye from Fudan University, and Yan Zhou from Wuhan University for generously providing experimental materials. We thank Ruidong Hao, Ruangang Pan, Zeng Cong, Xiaolu Lu, Yang Qiu, and Yujie Liu from Wuhan University for helpful discussions.
This study was supported by the China 973 Basic Research Program (2013CB911101) and China NSFC grants (81130083, 81271817, 31170152, and 31221061).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01331-15.
REFERENCES
- 1.Carthew RW, Sontheimer EJ. 2009. Origins and mechanisms of miRNAs and siRNAs. Cell 136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ding SW. 2010. RNA-based antiviral immunity. Nat Rev Immunol 10:632–644. doi: 10.1038/nri2824. [DOI] [PubMed] [Google Scholar]
- 3.Umbach JL, Cullen BR. 2009. The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev 23:1151–1164. doi: 10.1101/gad.1793309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takeuchi O, Akira S. 2009. Innate immunity to virus infection. Immunol Rev 227:75–86. doi: 10.1111/j.1600-065X.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parameswaran P, Sklan E, Wilkins C, Burgon T, Samuel MA, Lu R, Ansel KM, Heissmeyer V, Einav S, Jackson W, Doukas T, Paranjape S, Polacek C, FBdos Santos Jalili R, Babrzadeh F, Gharizadeh B, Grimm D, Kay M, Koike S, Sarnow P, Ronaghi M, Ding SW, Harris E, Chow M, Diamond MS, Kirkegaard K, Glenn JS, Fire AZ. 2010. Six RNA viruses and forty-one hosts: viral small RNAs and modulation of small RNA repertoires in vertebrate and invertebrate systems. PLoS Pathog 6:e1000764. doi: 10.1371/journal.ppat.1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y, Lu J, Han Y, Fan X, Ding SW. 2013. RNA interference functions as an antiviral immunity mechanism in mammals. Science 342:231–234. doi: 10.1126/science.1241911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maillard PV, Ciaudo C, Marchais A, Li Y, Jay F, Ding SW, Voinnet O. 2013. Antiviral RNA interference in mammalian cells. Science 342:235–238. doi: 10.1126/science.1241930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sagan SM, Sarnow P. 2013. Molecular biology: RNAi, antiviral after all. Science 342:207–208. doi: 10.1126/science.1245475. [DOI] [PubMed] [Google Scholar]
- 9.Voinnet O. 2005. Induction and suppression of RNA silencing: insights from viral infections. Nat Rev Genet 6:206–220. doi: 10.1038/nrg1555. [DOI] [PubMed] [Google Scholar]
- 10.Bivalkar-Mehla S, Vakharia J, Mehla R, Abreha M, Kanwar JR, Tikoo A, Chauhan A. 2011. Viral RNA silencing suppressors (RSS): novel strategy of viruses to ablate the host RNA interference (RNAi) defense system. Virus Res 155:1–9. doi: 10.1016/j.virusres.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hemmes H, Lakatos L, Goldbach R, Burgyan J, Prins M. 2007. The NS3 protein of Rice hoja blanca tenuivirus suppresses RNA silencing in plant and insect hosts by efficiently binding both siRNAs and miRNAs. RNA 13:1079–1089. doi: 10.1261/rna.444007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gonzalez I, Martinez L, Rakitina DV, Lewsey MG, Atencio FA, Llave C, Kalinina NO, Carr JP, Palukaitis P, Canto T. 2010. Cucumber mosaic virus 2b protein subcellular targets and interactions: their significance to RNA silencing suppressor activity. Mol Plant-Microbe Interact 23:294–303. doi: 10.1094/MPMI-23-3-0294. [DOI] [PubMed] [Google Scholar]
- 13.Lakatos L, Szittya G, Silhavy D, Burgyan J. 2004. Molecular mechanism of RNA silencing suppression mediated by p19 protein of tombusviruses. EMBO J 23:876–884. doi: 10.1038/sj.emboj.7600096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haas G, Azevedo J, Moissiard G, Geldreich A, Himber C, Bureau M, Fukuhara T, Keller M, Voinnet O. 2008. Nuclear import of CaMV P6 is required for infection and suppression of the RNA silencing factor DRB4. EMBO J 27:2102–2112. doi: 10.1038/emboj.2008.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomas CL, Leh V, Lederer C, Maule AJ. 2003. Turnip crinkle virus coat protein mediates suppression of RNA silencing in Nicotiana benthamiana. Virology 306:33–41. doi: 10.1016/S0042-6822(02)00018-1. [DOI] [PubMed] [Google Scholar]
- 16.Chao JA, Lee JH, Chapados BR, Debler EW, Schneemann A, Williamson JR. 2005. Dual modes of RNA-silencing suppression by Flock House virus protein B2. Nat Struct Mol Biol 12:952–957. doi: 10.1038/nsmb1005. [DOI] [PubMed] [Google Scholar]
- 17.Li H, Li WX, Ding SW. 2002. Induction and suppression of RNA silencing by an animal virus. Science 296:1319–1321. doi: 10.1126/science.1070948. [DOI] [PubMed] [Google Scholar]
- 18.Qi N, Zhang L, Qiu Y, Wang Z, Si J, Liu Y, Xiang X, Xie J, Qin CF, Zhou X, Hu Y. 2012. Targeting of dicer-2 and RNA by a viral RNA silencing suppressor in Drosophila cells. J Virol 86:5763–5773. doi: 10.1128/JVI.07229-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qi N, Cai D, Qiu Y, Xie J, Wang Z, Si J, Zhang J, Zhou X, Hu Y. 2011. RNA binding by a novel helical fold of b2 protein from Wuhan nodavirus mediates the suppression of RNA interference and promotes b2 dimerization. J Virol 85:9543–9554. doi: 10.1128/JVI.00785-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haasnoot J, de Vries W, Geutjes EJ, Prins M, de Haan P, Berkhout B. 2007. The Ebola virus VP35 protein is a suppressor of RNA silencing. PLoS Pathog 3:e86. doi: 10.1371/journal.ppat.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bucher E, Hemmes H, de Haan P, Goldbach R, Prins M. 2004. The influenza A virus NS1 protein binds small interfering RNAs and suppresses RNA silencing in plants. J Gen Virol 85:983–991. doi: 10.1099/vir.0.19734-0. [DOI] [PubMed] [Google Scholar]
- 22.Li WX, Li H, Lu R, Li F, Dus M, Atkinson P, Brydon EW, Johnson KL, Garcia-Sastre A, Ball LA, Palese P, Ding SW. 2004. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc Natl Acad Sci U S A 101:1350–1355. doi: 10.1073/pnas.0308308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sullivan CS, Ganem D. 2005. A virus-encoded inhibitor that blocks RNA interference in mammalian cells. J Virol 79:7371–7379. doi: 10.1128/JVI.79.12.7371-7379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bennasser Y, Le SY, Benkirane M, Jeang KT. 2005. Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immunity 22:607–619. doi: 10.1016/j.immuni.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 25.Chen W, Zhang Z, Chen J, Zhang J, Zhang J, Wu Y, Huang Y, Cai X, Huang A. 2008. HCV core protein interacts with Dicer to antagonize RNA silencing. Virus Res 133:250–258. doi: 10.1016/j.virusres.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 26.Basler CF, Wang X, Muhlberger E, Volchkov V, Paragas J, Klenk HD, Garcia-Sastre A, Palese P. 2000. The Ebola virus VP35 protein functions as a type I IFN antagonist. Proc Natl Acad Sci U S A 97:12289–12294. doi: 10.1073/pnas.220398297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brand SR, Kobayashi R, Mathews MB. 1997. The Tat protein of human immunodeficiency virus type 1 is a substrate and inhibitor of the interferon-induced, virally activated protein kinase, PKR. J Biol Chem 272:8388–8395. doi: 10.1074/jbc.272.13.8388. [DOI] [PubMed] [Google Scholar]
- 28.Geiss GK, Salvatore M, Tumpey TM, Carter VS, Wang X, Basler CF, Taubenberger JK, Bumgarner RE, Palese P, Katze MG, Garcia-Sastre A. 2002. Cellular transcriptional profiling in influenza A virus-infected lung epithelial cells: the role of the nonstructural NS1 protein in the evasion of the host innate defense and its potential contribution to pandemic influenza. Proc Natl Acad Sci U S A 99:10736–10741. doi: 10.1073/pnas.112338099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perlman S, Netland J. 2009. Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol 7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knoops K, Kikkert M, Worm SH, Zevenhoven-Dobbe JC, van der Meer Y, Koster AJ, Mommaas AM, Snijder EJ. 2008. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol 6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR. 2006. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J Virol 80:5059–5064. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karjee S, Minhas A, Sood V, Ponia SS, Banerjea AC, Chow VT, Mukherjee SK, Lal SK. 2010. The 7a accessory protein of severe acute respiratory syndrome coronavirus acts as an RNA silencing suppressor. J Virol 84:10395–10401. doi: 10.1128/JVI.00748-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yount B, Roberts RS, Sims AC, Deming D, Frieman MB, Sparks J, Denison MR, Davis N, Baric RS. 2005. Severe acute respiratory syndrome coronavirus group-specific open reading frames encode nonessential functions for replication in cell cultures and mice. J Virol 79:14909–14922. doi: 10.1128/JVI.79.23.14909-14922.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tan YJ, Lim SG, Hong W. 2006. Understanding the accessory viral proteins unique to the severe acute respiratory syndrome (SARS) coronavirus. Antivir Res 72:78–88. doi: 10.1016/j.antiviral.2006.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang TK, Wu MP, Chen ST, Hou MH, Hong MH, Pan FM, Yu HM, Chen JH, Yao CW, Wang AH. 2005. Biochemical and immunological studies of nucleocapsid proteins of severe acute respiratory syndrome and 229E human coronaviruses. Proteomics 5:925–937. doi: 10.1002/pmic.200401204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takeda M, Chang CK, Ikeya T, Guntert P, Chang YH, Hsu YL, Huang TH, Kainosho M. 2008. Solution structure of the c-terminal dimerization domain of SARS coronavirus nucleocapsid protein solved by the SAIL-NMR method. J Mol Biol 380:608–622. doi: 10.1016/j.jmb.2007.11.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baric RS, Nelson GW, Fleming JO, Deans RJ, Keck JG, Casteel N, Stohlman SA. 1988. Interactions between coronavirus nucleocapsid protein and viral RNAs: implications for viral transcription. J Virol 62:4280–4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grossoehme NE, Li L, Keane SC, Liu P, Dann CE III, Leibowitz JL, Giedroc DP. 2009. Coronavirus N protein N-terminal domain (NTD) specifically binds the transcriptional regulatory sequence (TRS) and melts TRS-cTRS RNA duplexes. J Mol Biol 394:544–557. doi: 10.1016/j.jmb.2009.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tylor S, Andonov A, Cutts T, Cao JX, Grudesky E, Van Domselaar G, Li XG, He RT. 2009. The SR-rich motif in SARS-CoV nucleocapsid protein is important for virus replication. Can J Microbiol 55:254–260. doi: 10.1139/W08-139. [DOI] [PubMed] [Google Scholar]
- 40.Almazan F, Galan C, Enjuanes L. 2004. The nucleoprotein is required for efficient coronavirus genome replication. J Virol 78:12683–12688. doi: 10.1128/JVI.78.22.12683-12688.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu X, Pan J, Tao J, Guo D. 2011. SARS-CoV nucleocapsid protein antagonizes IFN-beta response by targeting initial step of IFN-beta induction pathway, and its C-terminal region is critical for the antagonism. Virus Genes 42:37–45. doi: 10.1007/s11262-010-0544-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang CK, Sue SC, Yu TH, Hsieh CM, Tsai CK, Chiang YC, Lee SJ, Hsiao HH, Wu WJ, Chang WL, Lin CH, Huang TH. 2006. Modular organization of SARS coronavirus nucleocapsid protein. J Biomed Sci 13:59–72. doi: 10.1007/s11373-005-9035-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang Q, Yu L, Petros AM, Gunasekera A, Liu Z, Xu N, Hajduk P, Mack J, Fesik SW, Olejniczak ET. 2004. Structure of the N-terminal RNA-binding domain of the SARS CoV nucleocapsid protein. Biochemistry 43:6059–6063. doi: 10.1021/bi036155b. [DOI] [PubMed] [Google Scholar]
- 44.Luo H, Ye F, Chen K, Shen X, Jiang H. 2005. SR-rich motif plays a pivotal role in recombinant SARS coronavirus nucleocapsid protein multimerization. Biochemistry 44:15351–15358. doi: 10.1021/bi051122c. [DOI] [PubMed] [Google Scholar]
- 45.Luo H, Chen J, Chen K, Shen X, Jiang H. 2006. Carboxyl terminus of severe acute respiratory syndrome coronavirus nucleocapsid protein: self-association analysis and nucleic acid binding characterization. Biochemistry 45:11827–11835. doi: 10.1021/bi0609319. [DOI] [PubMed] [Google Scholar]
- 46.Chen CY, Chang CK, Chang YW, Sue SC, Bai HI, Riang L, Hsiao CD, Huang TH. 2007. Structure of the SARS coronavirus nucleocapsid protein RNA-binding dimerization domain suggests a mechanism for helical packaging of viral RNA. J Mol Biol 368:1075–1086. doi: 10.1016/j.jmb.2007.02.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu D, Wu A, Cui L, Hao R, Wang Y, He J, Guo D. 2014. Hepatitis B virus polymerase suppresses NF-κB signaling by inhibiting the activity of IKKs via interaction with Hsp90β. PLoS One 9:e91658. doi: 10.1371/journal.pone.0091658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chang GH, Luo BJ, Lu P, Lin L, Wu XY, Li J, Hu Y, Zhu QY. 2011. Construction and genetic analysis of murine hepatitis virus strain A59 Nsp16 temperature-sensitive mutant and the revertant virus. Virol Sin 26:19–29. doi: 10.1007/s12250-011-3145-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen Y, Su C, Ke M, Jin X, Xu L, Zhang Z, Wu A, Sun Y, Yang Z, Tien P, Ahola T, Liang Y, Liu X, Guo D. 2011. Biochemical and structural insights into the mechanisms of SARS coronavirus RNA ribose 2′-O-methylation by nsp16/nsp10 protein complex. PLoS Pathog 7:e1002294. doi: 10.1371/journal.ppat.1002294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kopecky-Bromberg SA, Martinez-Sobrido L, Frieman M, Baric RA, Palese P. 2007. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J Virol 81:548–557. doi: 10.1128/JVI.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ji X. 2008. The mechanism of RNase III action: how dicer dices. Curr Top Microbiol Immunol 320:99–116. [DOI] [PubMed] [Google Scholar]
- 52.Lingel A, Simon B, Izaurralde E, Sattler M. 2005. The structure of the flock house virus B2 protein, a viral suppressor of RNA interference, shows a novel mode of double-stranded RNA recognition. EMBO Rep 6:1149–1155. doi: 10.1038/sj.embor.7400583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chang CK, Hsu YL, Chang YH, Chao FA, Wu MC, Huang YS, Hu CK, Huang TH. 2009. Multiple Nucleic acid binding sites and intrinsic disorder of severe acute respiratory syndrome coronavirus nucleocapsid protein: implications for ribonucleocapsid protein packaging. J Virol 83:2255–2264. doi: 10.1128/JVI.02001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McBride R, van Zyl M, Fielding BC. 2014. The coronavirus nucleocapsid is a multifunctional protein. Viruses 6:2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pan J, Peng X, Gao Y, Li Z, Lu X, Chen Y, Ishaq M, Liu D, Dediego ML, Enjuanes L, Guo D. 2008. Genome-wide analysis of protein-protein interactions and involvement of viral proteins in SARS-CoV replication. PLoS One 3:e3299. doi: 10.1371/journal.pone.0003299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ye Y, Hauns K, Langland JO, Jacobs BL, Hogue BG. 2007. Mouse hepatitis coronavirus A59 nucleocapsid protein is a type I interferon antagonist. J Virol 81:2554–2563. doi: 10.1128/JVI.01634-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roth-Cross JK, Bender SJ, Weiss SR. 2008. Murine coronavirus mouse hepatitis virus is recognized by MDA5 and induces type I interferon in brain macrophages/microglia. J Virol 82:9829–9838. doi: 10.1128/JVI.01199-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roth-Cross JK, Martinez-Sobrido L, Scott EP, Garcia-Sastre A, Weiss SR. 2007. Inhibition of the alpha/beta interferon response by mouse hepatitis virus at multiple levels. J Virol 81:7189–7199. doi: 10.1128/JVI.00013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rose KM, Weiss SR. 2009. Murine coronavirus cell type dependent interaction with the type I interferon response. Viruses 1:689–712. doi: 10.3390/v1030689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou H, Perlman S. 2007. Mouse hepatitis virus does not induce beta interferon synthesis and does not inhibit its induction by double-stranded RNA. J Virol 81:568–574. doi: 10.1128/JVI.01512-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Versteeg GA, Bredenbeek PJ, van den Worm SH, Spaan WJ. 2007. Group 2 coronaviruses prevent immediate-early interferon induction by protection of viral RNA from host cell recognition. Virology 361:18–26. doi: 10.1016/j.virol.2007.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gosert R, Kanjanahaluethai A, Egger D, Bienz K, Baker SC. 2002. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J Virol 76:3697–3708. doi: 10.1128/JVI.76.8.3697-3708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Snijder EJ, van der Meer Y, Zevenhoven-Dobbe J, Onderwater JJ, van der Meulen J, Koerten HK, Mommaas AM. 2006. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J Virol 80:5927–5940. doi: 10.1128/JVI.02501-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen Y, Cai H, Pan J, Xiang N, Tien P, Ahola T, Guo D. 2009. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc Natl Acad Sci U S A 106:3484–3489. doi: 10.1073/pnas.0808790106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.de Haan CA, Rottier PJ. 2005. Molecular interactions in the assembly of coronaviruses. Adv Virus Res 64:165–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Masters PS. 2006. The molecular biology of coronaviruses. Adv Virus Res 66:193–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Niemeyer D, Zillinger T, Muth D, Zielecki F, Horvath G, Suliman T, Barchet W, Weber F, Drosten C, Muller MA. 2013. Middle East respiratory syndrome coronavirus accessory protein 4a is a type I interferon antagonist. J Virol 87:12489–12495. doi: 10.1128/JVI.01845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hsieh PK, Chang SC, Huang CC, Lee TT, Hsiao CW, Kou YH, Chen IY, Chang CK, Huang TH, Chang MF. 2005. Assembly of severe acute respiratory syndrome coronavirus RNA packaging signal into virus-like particles is nucleocapsid dependent. J Virol 79:13848–13855. doi: 10.1128/JVI.79.22.13848-13855.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Masters PS. 1992. Localization of an RNA-binding domain in the nucleocapsid protein of the coronavirus mouse hepatitis virus. Arch Virol 125:141–160. doi: 10.1007/BF01309634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hurst KR, Koetzner CA, Masters PS. 2013. Characterization of a critical interaction between the coronavirus nucleocapsid protein and nonstructural protein 3 of the viral replicase-transcriptase complex. J Virol 87:9159–9172. doi: 10.1128/JVI.01275-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kuo L, Koetzner CA, Hurst KR, Masters PS. 2014. Recognition of the murine coronavirus genomic RNA packaging signal depends on the second RNA-binding domain of the nucleocapsid protein. J Virol 88:4451–4465. doi: 10.1128/JVI.03866-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cervantes-Barragan L, Zust R, Weber F, Spiegel M, Lang KS, Akira S, Thiel V, Ludewig B. 2007. Control of coronavirus infection through plasmacytoid dendritic-cell-derived type I interferon. Blood 109:1131–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cervantes-Barragan L, Kalinke U, Zust R, Konig M, Reizis B, Lopez-Macias C, Thiel V, Ludewig B. 2009. Type I IFN-mediated protection of macrophages and dendritic cells secures control of murine coronavirus infection. J Immunol 182:1099–1106. doi: 10.4049/jimmunol.182.2.1099. [DOI] [PubMed] [Google Scholar]
- 74.Schelle B, Karl N, Ludewig B, Siddell SG, Thiel V. 2005. Selective replication of coronavirus genomes that express nucleocapsid protein. J Virol 79:6620–6630. doi: 10.1128/JVI.79.11.6620-6630.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.