ABSTRACT
The gp120/gp41 HIV-1 envelope glycoprotein (Env) is highly glycosylated, with up to 50% of its mass consisting of N-linked glycans. This dense carbohydrate coat has emerged as a promising vaccine target, with its glycans acting as epitopes for a number of potent and broadly neutralizing antibodies (bnAbs). Characterizing the glycan structures present on native HIV-1 Env is thus a critical goal for the design of Env immunogens. In this study, we used a complementary, multistep approach involving ion mobility mass spectrometry and high-performance liquid chromatography to comprehensively characterize the glycan structures present on HIV-1 gp120 produced in peripheral blood mononuclear cells (PBMCs). The capacity of different expression systems, including pseudoviral particles and recombinant cell surface trimers, to reproduce native-like glycosylation was then assessed. A population of oligomannose glycans on gp120 was reproduced across all expression systems, supporting this as an intrinsic property of Env that can be targeted for vaccine design. In contrast, Env produced in HEK 293T cells failed to accurately reproduce the highly processed complex-type glycan structures observed on PBMC-derived gp120, and in particular the precise linkage of sialic acid residues that cap these glycans. Finally, we show that unlike for gp120, the glycans decorating gp41 are mostly complex-type sugars, consistent with the glycan specificity of bnAbs that target this region. These findings provide insights into the glycosylation of native and recombinant HIV-1 Env and can be used to inform strategies for immunogen design and preparation.
IMPORTANCE Development of an HIV vaccine is desperately needed to control new infections, and elicitation of HIV bnAbs will likely be an important component of an effective vaccine. Increasingly, HIV bnAbs are being identified that bind to the N-linked glycans coating the HIV envelope glycoproteins gp120 and gp41, highlighting them as important targets for vaccine design. It is therefore important to characterize the glycan structures present on native, virion-associated gp120 and gp41 for development of vaccines that accurately mimic native-Env glycosylation. In this study, we used a number of analytical techniques to precisely study the structures of both the oligomannose and complex-type glycans present on native Env to provide a reference for determining the ability of potential HIV immunogens to accurately replicate the glycosylation pattern on these native structures.
INTRODUCTION
The HIV-1 envelope glycoprotein (Env) consists of a noncovalently linked trimer of gp120/gp41 heterodimers. Interaction of this trimer with CD4 and its subsequent rearrangement is essential for infection of target cells and therefore is the sole target for broadly neutralizing antibodies (bnAbs). The surface protein, gp120, is extensively modified with host-derived glycans, having a median of 25 potential N-linked glycosylation sites (PNGSs) (1) and thus making it one of the most densely glycosylated proteins known. The transmembrane protein, gp41, is less densely glycosylated, having a mean of 4 PNGSs. The N-linked glycans on Env play important roles in correct protein folding, disease transmission through interaction with host receptors, and shielding conserved regions of the protein from the immune system. It is important to characterize the glycosylation of virion-associated gp120, as a large number of HIV-1 bnAbs are dependent on Env glycans for neutralization (2–6).
It has been shown that glycosylation of gp120 is divergent from that typically found on glycoproteins produced by the host cell (7). Two main subpopulations of glycans have been identified on gp120, underprocessed oligomannose glycans and processed complex-type glycans (7–14). The dense clustering of glycans limits the accessibility to the endoplasmic reticulum (ER) and Golgi α-mannosidase enzymes, resulting in a patch of underprocessed oligomannose glycans which cannot be further processed to complex glycans in the Golgi (7). This patch of oligomannose glycans arising from steric restriction is referred to here as the intrinsic mannose patch (IMP). We have previously shown that the IMP is conserved across recombinant gp120, recombinant trimers (including SOSIP trimers [14, 15]), pseudovirions, and human peripheral blood mononuclear cell (PBMC)-derived virions (7, 9). The remaining processed complex-type glycans on Env are determined by the host cell glycosylation machinery and are therefore thought to be dependent upon the cell type used for production of the recombinant protein or the virus.
Despite the glycans on Env often being referred to as the “glycan shield,” a number of HIV-1 bnAbs have been isolated that bind to these glycans (3–5, 16–21). These bnAbs target three main regions on Env, the N332 glycan (e.g., 2G12, PGT121, PGT128, and PGT135) (3, 4, 16–18), the N160 glycan (e.g., PG9, PGT145, and CH04) (4, 5, 19, 20), and the glycans at the gp120-gp41 interface (e.g., PGT151) (21). Glycan microarray analysis has shown these bnAbs to have differing specificities for N-linked glycan structures; e.g., PGT128 is specific for Man8GlcNAc2 and Man9GlcNAc2 glycans (22), whereas PGT151 preferentially binds α2,3-linked sialylated tri- and tetra-antennary complex-type glycans (21). PGT151 has been shown to distinguish cleaved from uncleaved Env trimers and therefore provides a useful reagent to characterize the glycosylation of native envelope spike from pseudovirions (21). A high-resolution crystal structure of PGT151 in complex with its epitope has not yet been solved, but mapping studies have suggested that the epitope includes highly processed glycans on gp41 at positions N611 and N637 (21, 23).
In this study, we used ion mobility mass spectrometry (MS), negative-ion collision-induced dissociation (CID), and high-performance liquid chromatography (HPLC) (24–26) to compare, in detail, the glycan structures present on JR-CSF native Env from two viral systems commonly used for determining HIV-1 bnAb neutralizing activity (27) with a recombinant JR-CSF gp120 protein typically used in vaccine trials. We show that virion-associated gp120 produced in human PBMCs and gp120 isolated from pseudoviral particles using cleavage-specific bnAb PGT151 bear very similar glycosylation patterns. Using these systems, we showed that native Env glycosylation includes a dominant population of oligomannose-type glycans with a population of complex-type glycans that are predominantly highly processed bi-, tri-, and tetra-antennary sialylated structures. Interestingly, the PBMC-derived material has predominantly α2,6-linked sialic acids, which have a strong immunomodulatory role (28), whereas the HEK 293T produced pseudoviral material is entirely α2,3 linked. Recombinant gp120 produced in HEK 293T cells bears the same oligomannose isomers as native Env but a different distribution of complex-type sugars. Finally, we showed using HPLC analysis that unlike for gp120, the glycosylation of virion-associated gp41 is highly heterogeneous, with only a small population of oligomannose glycans, suggesting that gp41 is not under the same steric constraints as gp120 despite its close proximity to the membrane. These results provide new insights into the relative contributions of the producer cell and the intrinsic properties of Env in determining native Env glycosylation, and they provide an important reference for analyzing potential HIV-1 Env immunogens.
MATERIALS AND METHODS
PBMC virus preparation.
Human PBMCs were obtained from healthy donors and isolated and stimulated as previously described (29). HIV-1 JR-CSF virus was grown in and titers were determined on CD8-depleted PBMCs (30). Virus production was monitored by p24 enzyme-linked immunosorbent assay (ELISA) (Aalto Bioreagents, Dublin, Ireland). Samples were obtained from The Normal Blood Donor service at The Scripps Research Institute; sample collection and subsequent propagation of HIV-1 was approved by the Institutional Review Board at The Scripps Research Institute (protocol number HSC-06-4604).
Pseudovirus production in 293T cells.
Pseudovirus was generated in HEK 293T cells as previously described (31). HEK 293T cells were cotransfected with plasmids carrying the HIV backbone (pSG3ΔEnv) and the envelope clone (pSVIII-JR-CSF) at a ratio of 2:1 (total DNA, 60 μg) using polyethylenimine (PEI; Polysciences Europe, GmbH) according to the manufacturer's instructions. Virus supernatant was harvested 72 h later and filtered before concentration by ultracentrifugation. Virus titers were determined on TZM-bl cells.
Env isolation. (i) bnAb cocktail.
Concentrated virus was lysed in the presence of protease inhibitor cocktail (Roche) with 1% NP-40. The lysate was cleared by centrifugation and incubated with a cocktail of monoclonal antibodies (5 μg each of F425-b4e8, b6, b12, 4E10, and 2F5) for 4 h at 4°C. Protein A beads were added and incubated at 4°C overnight. The beads were washed and the protein was eluted by heating in loading buffer (containing dithiothreitol) for 10 min at 100°C and resolved by SDS-PAGE. The envelope bands were confirmed by Western blotting (primary antibodies 2G12, b6, 2F5, 4E10, and F425-b4e8) and cut to use directly in glycan analysis. Where discrete bands for gp160, gp120, and gp41 were detected, each was analyzed separately.
(ii) PGT151 virus Env isolation.
Concentrated virus was incubated with PGT151 (at 10 μg/ml) for 4 h at 4°C. Virus was then lysed in the presence of protease inhibitor cocktail with 1% NP-40. The lysate was cleared by centrifugation and incubated with protein A beads at 4°C overnight. The beads were washed and processed as described above.
(iii) PGT151 isolation of cell surface trimers.
HEK 293T cells used for preparing pseudovirus were washed and incubated with PGT151 (at 10 μg/ml) for 1 h at 4°C. The cells were washed with phosphate-buffered saline (PBS) and lysed with 0.5% Triton-X in the presence of protease inhibitor cocktail. The lysate was cleared by centrifugation and incubated with protein A beads overnight at 4°C. The beads were washed and processed as described above.
gp120 expression and purification.
gp120 was expressed by transient transfection in HEK 293T cells. Protein was purified by Galanthus nivalis lectin (GNL) affinity chromatography followed by size exclusion chromatography (SEC).
In-gel PNGase F release of N-glycans.
N-glycans were released from Env species following SDS-PAGE using peptide-N-glycosidase F (PNGase F; New England BioLabs). Coomassie-stained gel bands were excised and washed alternately with acetonitrile and water before being dried under vacuum. Gel pieces were rehydrated in 20 mM sodium bicarbonate buffer, pH 7.0, and incubated with PNGase F (1 μl) for 16 h at 37°C. Released glycans were extracted from the gel matrix by 3 washing steps with water.
Ion mobility ESI-MS/MS analysis.
All glycan samples were cleaned with a Nafion 117 membrane as described earlier by Börnsen et al. (32) before examination by mass spectrometry (MS). They were then dissolved in a solution of methanol-water (1:1, vol/vol) containing ammonium phosphate (0.5 M, to maximize formation of [M+H2PO4]− ions, the ions usually encountered from biological samples) and spun at 10,000 rpm (9,503 × g) for 1 min to sediment any particulates. Traveling-wave ion mobility spectrometry (TWIMS) experiments were carried out in negative-ion mode with a Waters Synapt G2 traveling-wave ion mobility mass spectrometer (Waters MS-Technologies, Manchester, United Kingdom) (33) fitted with an electrospray (ESI) ion source. Samples were infused through Waters thin-wall nanospray capillaries. The ESI capillary voltage was 1.2 kV, the cone voltage was 120 V, and the ion source temperature was 80°C. The traveling (T)-wave mobility cell contained nitrogen and was operated at a pressure of 0.55 mbar. The T-wave velocity was 450 m/s, and the peak height was 40 V. Collision-induced dissociation (CID) was performed after mobility separation in the transfer cell with argon as the collision gas. The instrument was externally mass-calibrated with sialylated N-glycans released from bovine fetuin. Data acquisition and processing were carried out using the Waters DriftScope (version 2.1) software and MassLynx (version 4.0). The scheme devised by Domon and Costello (34) was used to name the fragment ions, with the exception that the subscripts R and R-1 are used for the reducing-terminal and penultimate GlcNAc residues of the di-N-acetylchitobiose core when the subscript numbers change as the result of differing chain lengths. Structural determination from the negative-ion spectra was as described in earlier publications (35–38).
Fluorescent labeling of N-glycans.
N-Glycans released by PNGase F were fluorescently labeled using 2-aminobenzoic acid (2-AA). The labeling mixture comprised 2-AA (30 mg/ml) and sodium cyanoborohydride (45 mg/ml) dissolved in a solution of sodium acetate trihydrate (4%, wt/vol) and boric acid (2%, wt/vol), in methanol. The labeling mixture (80 μl) was added to each sample (in 30 μl of water) and incubated at 80°C for 1 h. Labeled oligosaccharides were purified using Spe-ed Amide-2 columns (Applied Separations, Allentown, PA) preequilibrated with acetonitrile. Before loading, 97% acetonitrile (vol/vol) (1 ml) was added to each sample. Loaded samples were then washed with 95% acetonitrile (vol/vol) (2 ml) and eluted with water (1.5 ml). Glycans were dried under vacuum prior to HPLC analysis or glycosidase treatment.
HPLC.
Fluorescently labeled N-glycans were analyzed using a LudgerSep N2 amide column (4.6 by 140 mm; Ludger Ltd., Abingdon, United Kingdom) and an Alliance high-performance liquid chromatography (HPLC) system (Waters, Elstree, United Kingdom) run using the following gradient: time (t) of 0 min, 35% solvent A and 65% solvent B; t of 26 min, 46% solvent A and 54% solvent B; t of 26.5 min, 80% solvent A and 20% solvent B; t of 28.5 min, 80% solvent A and 20% solvent B; t of 30 min, 35% solvent A and 65% solvent B; and t of 46 min, 35% solvent A and 65% solvent B, at a flow rate of 1 ml/min, where solvent A was 50 mM ammonium formate, pH 4.4, and solvent B was acetonitrile. Fluorescence was measured using an excitation wavelength of 330 nm and a detection wavelength of 420 nm.
Fractionation of charged and neutral N-glycans.
Total pools of N-glycans were separated into neutral and charged (sialylated) fractions by anion-exchange chromatography as previously described (39). Briefly, 2-AA-labeled glycans in water were passed over QAE-Sephadex A-25 resin and washed with water. Neutral glycans were first eluted with 2 ml of 0.5 M acetic acid, followed by elution of charged N-glycans using 2 ml of 0.5 M ammonium acetate. N-Glycans were dried by lyophilization and resuspended in water.
Glycosidase treatment of free N-glycans.
Glycosidase digestions of free N-glycans were performed to help with structural assignments of the HPLC-resolved glycans. 2-AA-labeled glycans were resuspended in water and digested with the relevant glycosidase in a total volume of 20 μl for 16 h at 37°C. Digested glycans were purified using a polyvinylidene difluoride (PVDF) protein-binding membrane plate prior to HPLC analysis. For sialic acid linkage analysis, neuraminidase digestions were performed on total glycan pools, or charged fractions only, using an α2,3-specific neuraminidase from Salmonella enterica serovar Typhimurium and a nonspecific neuraminidase from Clostridium perfringens. Additionally, a β1,4-galactosidase from Bacteroides fragilis and endoglycosidase H (Endo H) from Streptomyces picatus (New England BioLabs) were used to help with glycan assignments and quantitation.
RESULTS
Oligomannose isomers are the same on PBMC-derived Env and recombinant gp120.
CD4+ T cells are one of the main sources of HIV-1 virions in infected patients, and therefore, viruses produced in human PBMCs are a good physiological representation to study the glycosylation of native HIV-1 Env. We wanted to compare this directly to the glycosylation of recombinant monomeric gp120, as this has been a common immunogen in vaccine trials to date (40). Our previous matrix-assisted laser desorption ionization (MALDI) analysis of glycans on PBMC-derived gp120 had not fully assigned the glycan structures present, as MALDI provides only information on the overall composition of a glycan and not structural information regarding linkage and stereochemistry of individual monosaccharides (7, 9). Therefore, we used ion mobility mass spectrometry (MS) and negative-ion collision-induced dissociation (CID) to compare the isomers present on physiologically relevant PBMC-derived virion-associated gp120 with those on recombinant gp120 prepared in HEK 293T cells. Ion mobility MS facilitates analysis of low-abundance, complex mixture samples by using ion mobility capability to separate sugars based on shape, size, and charge (Fig. 1B). Specific populations of ions can then be analyzed individually, as shown in Fig. 1D, giving greater resolution compared to analyzing the full mixture. Furthermore, negative-ion CID fragmentation discriminates between isomers and allows characterization of some glycosidic linkages.
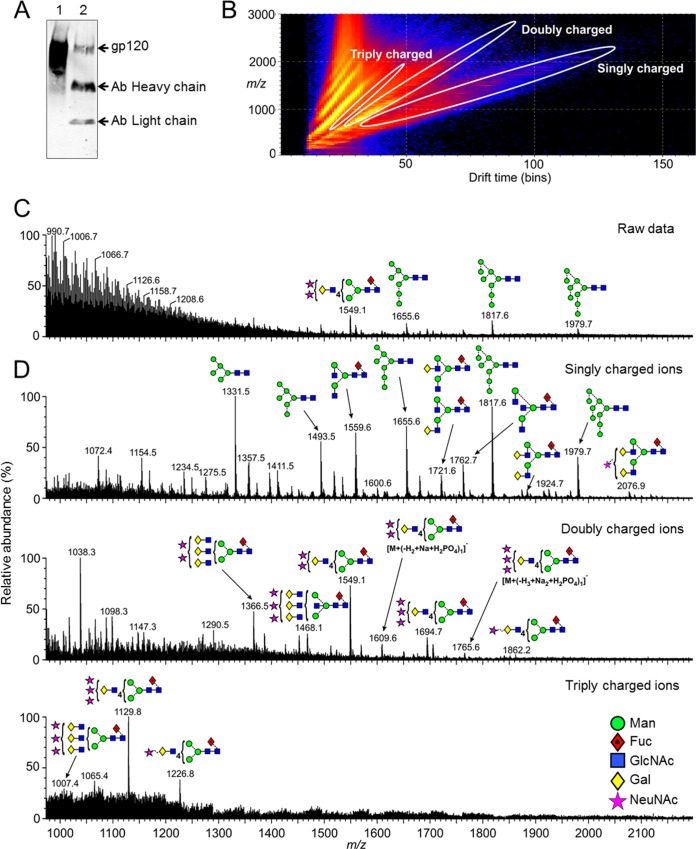
FIG 1.
Ion mobility mass spectrometric analysis of glycans released from PBMC-derived gp120. (A) Western blot analysis of JR-CSF Env isolated from PBMC produced (lane 2) virus compared to recombinant gp120JR-CSF (lane 1). Env was isolated with a cocktail of bnAbs (b12, b6, F425-b4e8, 4E10, and 2F5). (B) DriftScope plot (drift time:m/z) of the ions (negative mode) from the N-glycans obtained from the virion-associated gp120 produced in PBMCs. Ions in different charge states are enclosed in the white ovals. (C) Negative-ion ESI spectrum of the ions from the N-glycans obtained from the virion-associated gp120 without ion extraction. (D) Spectrum of the singly, doubly, and triply charged ions extracted from the highlighted regions of the DriftScope plot (B). Structures are drawn with the symbols and linkage as proposed by Harvey et al. (70) using the colors adopted by the Consortium for Functional Glycomics as shown in panel D.
Replication-competent full-length JR-CSF was prepared in CD8-depleted PBMCs and the Env isolated by immunoprecipitation (IP) with a cocktail of bnAbs (b12, b6, F425-b4e8, 2F5, and 4E10). The sample was further purified by SDS-PAGE, revealing one band corresponding to gp120 that was confirmed by Western blotting (Fig. 1A; see also Fig. S1A in the supplemental material). An in-gel PNGase F digest was used to release the N-linked glycans; Fig. 1B shows the negative-ion plot of ion mobility drift time against m/z of N-glycans from the PBMC-derived gp120 (see Fig. S2 for recombinant gp120). The total negative-ion ESI spectrum is shown in Fig. 1C, and the remaining graphs show the spectra of the singly, doubly, and triply charged ions computationally extracted from the DriftScope plot (Fig. 1D), as outlined by the ovals in Fig. 1B.
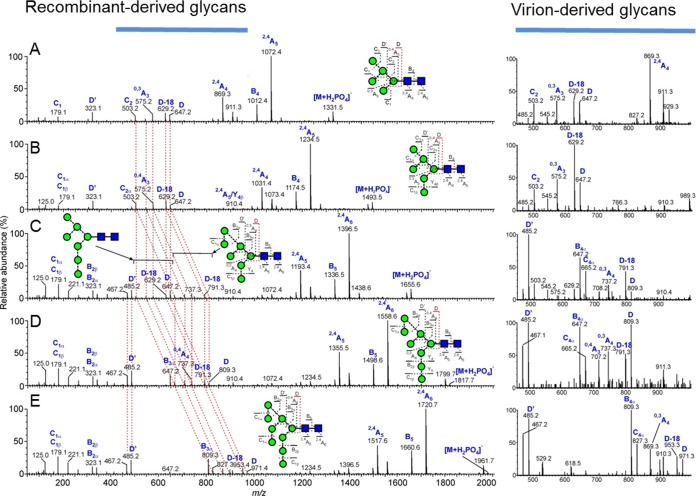
The oligomannose-type glycans on gp120 are the target of a number of HIV-1 bnAbs (4, 16–18, 22). The masses of the singly charged ions were typical of those from the oligomannose glycans Man5GlcNAc2 to Man9GlcNAc2, and as previously observed, these glycans were identified in both the recombinant and virion-derived PBMC samples. Analysis of the fragmentation patterns in the negative-ion CID spectra (Fig. 2) shows the same oligomannose isomers present in both sources. The spectrum of Man8GlcNAc2 shows a single set of ions for the D1, D3 isomer, consistent with the known preference for α-mannosidase I to remove the D2 mannose from Man9GlcNAc2 during biosynthesis of these glycans (Fig. 2D). However, the Man7GlcNAc2 spectrum (Fig. 2C) showed two sets of ions. The less abundant set exhibited m/z values showing substitution of the seventh mannose on D1 arm whereas the second, major, set showed m/z values showing substitution on the D3 arm. Man5GlcNAc2, Man6GlcNAc2, and Man9GlcNAc2 appeared to be single isomers. In summary, the oligomannose isomers present on recombinant gp120 and PBMC-derived Env are highly similar, and therefore, recombinant gp120 produced in HEK 293T cells is sufficient to reproduce the intrinsic mannose patch found on virion-associated gp120.
FIG 2.
Oligomannose isomers are the same on PBMC-derived Env and recombinant gp120. Shown are negative-ion CID spectra of oligomannose glycans from the recombinant gp120JR-CSF. The panels correspond to Man5GlcNAc2 (A), Man6GlcNAc2 (B), a mixture of Man7GlcNAc2 isomers (C), Man8GlcNAc2 (D), and Man9GlcNAc2 (E). The red dashed lines connect the fragment ions derived from the antenna linked to the 6-position of the core branching mannose residue (termed the 6-antenna), showing their shifts with increasing mannose residues in the 6-antenna of the larger glycans. The images on the right show spectra of the diagnostic region (m/z 450 to 1,000) from the virion-associated gp120 derived from PBMCs, confirming similarity in the glycans within the two samples.
PBMC-derived Env has large highly sialylated structures.
We next analyzed the complex and hybrid-type glycans present on the PBMC-derived Env and recombinant gp120. The use of negative-ion MS allows the analysis of complex-type glycans displaying sialic acid residues, whereas our previous MALDI analysis used the positive mode. The masses of the singly charged ions were typical of those from nonsialylated complex-type (mainly biantennary) glycans, whereas those in the spectra of doubly and triply charged ions corresponded to sialylated versions of bi-, tri-, and tetra-antennary glycans together (Fig. 1). Analysis of the fragmentation patterns showed that the complex-type glycans were mostly α1,6-core fucosylated on the reducing-terminal GlcNAc residue (Fig. 3). The complex-type glycans present on recombinant gp120 were mostly similar to those on the PBMC-derived virion sample, although differences in abundance could not be determined using this method (see Fig. S2 in the supplemental material).
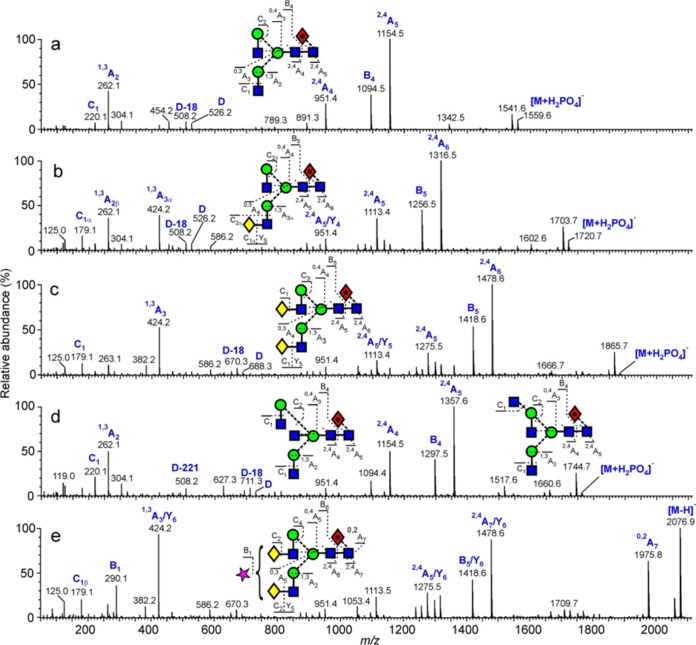
FIG 3.
Example of negative-ion CID spectra of biantennary complex glycans from PBMC-derived Env sample. (a) Man3GlcNAc4Fuc1. (b) Gal1Man3GlcNAc4Fuc1. The single set of D and D-18 ions (m/z 526 and 508, respectively) shows galactose substitution only on the 3-antenna. (c) Gal2Man3GlcNAc4Fuc1. D and D-18 ions are at m/z 688 and 670, respectively, reflecting the presence of the galactose residue in the 6-antenna. (d). Mixture of a bisected biantennary glycan and a triantennary glycan. The bisected glycan produced the prominent D-221 ion at m/z 508, whereas the triantennary glycan produced the D, D-18, and D-36 triplet at m/z 629, 611, and 693, respectively, together with the cross-ring cleavage ion at m/z 627. These compounds did not separate by ion mobility. (e) Monosialylated biantennary glycan Neu5Ac1Gal2Man3GlcNAc4Fuc1. The spectrum is similar to that of the biantennary glycan shown in panel c with the fragments having lost the sialic acid residue. This residue produced the B1 fragment at m/z 290.
PBMC-derived Env has predominantly α-2,6-linked sialic acids.
The interaction of sialic acid residues with host receptors can play an important role in immune modulation (28, 41), and the linkage of sialic acid can impact its function (28). For example, interaction of α2,6-linked sialic acid residues with CD22 on the surface of B cells plays an important role in regulating multiple B cell functions, including cellular activation, and B cell survival and homeostasis (28). The glycosylation of HIV Env has also been shown to be important for complement-mediated enhancement of HIV infection in cell lines (42, 43), and disialylation with neuraminidase or using glycosidase inhibitors can lead to activation of the alternate complement pathway (44, 45). However, the exact sialic acid linkages of complex-type glycans on virion-associated gp120 prepared in PBMCs has not previously been reported (8, 10). Therefore, we next characterized the linkage of the sialylated N-glycans on PBMC-derived Env. The sialylated structures were of too low abundance to use CID for linkage analysis, so we used a combination of high-performance liquid chromatography (HPLC) and neuraminidase enzymatic digestion to assign sialic acid linkage. The PNGase F-released N-linked glycans were fluorescently labeled using 2-AA and analyzed by HPLC. Peaks were assigned by exoglycosidase digestion (see Fig. S3 in the supplemental material). This analysis of the released glycans confirmed a high proportion of oligomannose as observed in previous studies (7, 9) (Fig. 4A). Due to the limited sample and the poor-quality trace baseline, a percentage of oligomannose-type glycans could not be accurately determined; however, a particularly dominant peak corresponding to Man9GlcNAc2 was evident.
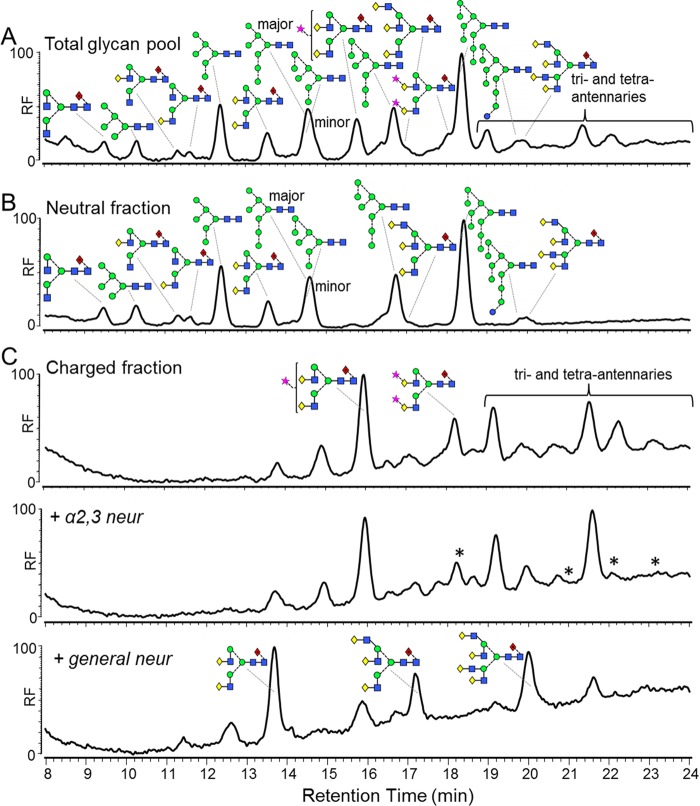
FIG 4.
Complex-type glycans on virion-associated gp120 produced in PBMCs have predominantly α2,6-linked sialic acids. (A) HPLC trace of bulk glycans released from PBMC-derived gp120 using PNGase F. The bulk glycans were fractionated into neutral glycans (B) and charged glycans (C) for subsequent neuraminidase digestion. The charged fraction was digested first with α2,3-specific neuraminidase and then with a general neuraminidase that cleaves α2,3-, α2,6-, and α2,8-linked sialic acids. Analysis by HPLC showed that the sialic acids were predominantly α2,6 linked. Asterisks indicate peaks which have been digested by the α2,3-specific neuraminidase and thus represent glycans containing at least one α2,3-linked sialic acid. Glycan structures were assigned using exoglycosidase digestions (see Fig. S2 in the supplemental material). Although we used a general neuraminidase that cleaves α2,3-, α2,6-, and α2,8-linked sialic acids, terminal α2,8-linked sialic acid linkages are found mainly in the brain (46) and were therefore not considered in this analysis.
To assess the linkage of the sialic acid residues present, the charged fraction (containing sialylated glycans) was separated from the neutral species using anion-exchange chromatography (Fig. 4B and C). The charged fraction was treated with an α2,3-specific neuraminidase, and HPLC analysis was used to look for changes in glycan structure; however, very little change was observed (Fig. 4C). Treatment with a general neuraminidase, which cleaves α2,3-, α2,6-, and α2,8-linked sialic acid residues, resulted in the majority of the peaks collapsing to 3 major peaks corresponding to bi-, tri-, and tetra-antennary nonsialylated glycans. As α2,8-linked sialic acids are mainly found in the brain (46), these data indicate that the sialic acids present on PBMC-derived Env are predominantly α2,6 linked.
Cleavage-specific PGT151-purified pseudovirion Env has a glycan profile similar to that of the PBMC-derived gp120.
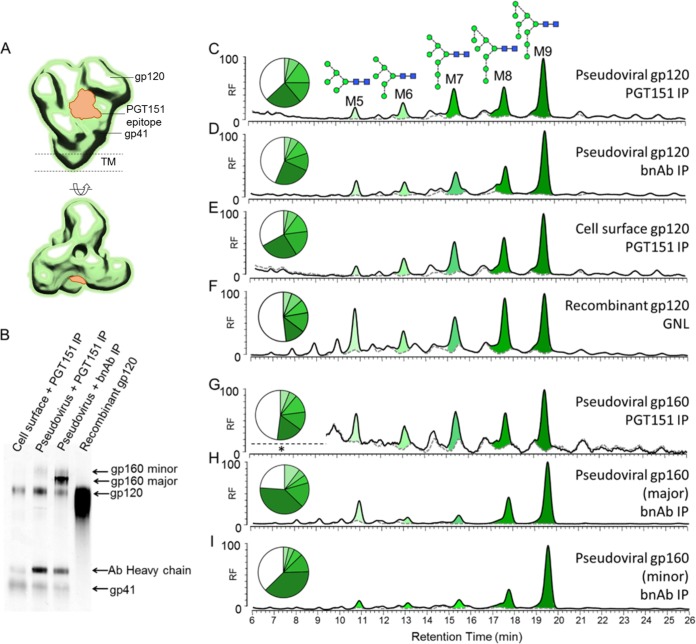
We next looked at the glycosylation of Env isolated from pseudovirions produced in HEK 293T cells, as this virus system is commonly used to determine neutralization activity of HIV bnAbs (27). Our previous studies had highlighted differences in the glycosylation of PBMC-derived gp120 and pseudovirion-derived gp120 (7, 9). However, a number of studies have also shown that pseudovirions display not only native trimers but also various nonfunctional forms, including uncleaved gp160 and gp41 stumps (47–51). As mentioned above, the recently isolated bnAb PGT151 is able to distinguish cleaved from uncleaved Env trimers (Fig. 5A) (21). Therefore, to further our understanding of native Env glycosylation, we used PGT151 to isolate only fully cleaved native trimers from the pseudovirion surface and analyzed their glycosylation compared to Env isolated using a cocktail of bnAbs (b12, b6, F425-b4e8, 2F5, and 4E10). JR-CSF virus was incubated with bnAb PGT151 before lysis with NP-40, and the Env-Ab complex was then isolated with protein A beads. SDS-PAGE analysis of the two IP conditions showed clear differences (Fig. 5B; see also Fig. S1B and C in the supplemental material). PGT151 alone isolated gp120 and gp41 bands that were confirmed with Western blot analysis. Additionally, a very faint gp160 band was observed in the Western blot (Fig. 5B, lane 2), which may have come from trimers consisting of two cleaved protomers and one uncleaved protomer. In contrast, the bnAb cocktail isolated gp120, gp41, and two dominant species of nonnative gp160 (Fig. 5B, lane 3). The glycans present on these samples were removed with PNGase F and analyzed using HPLC, as this method provides the best overall quantitative analysis of different glycan types.
FIG 5.
HPLC analysis of pseudovirion-associated gp120JR-CSF produced in HEK 293T cells. (A) Structural representation of the PGT151 epitope (23). (B) Western blot analysis of Env proteins isolated using the methods described below. (C to I) HPLC traces for gp120 isolated by IP with cleavage-specific PGT151 (C), gp120 isolated by IP with a cocktail of bnAbs (b12, b6, F425-b4e8, 2F5, and 4E10) (D), cell surface-associated gp120 isolated by IP with cleavage-specific PGT151 (E), recombinant gp120JR-CSF expressed in HEK 293T cells and GNL purified (F), gp160 isolated by IP with cleavage-specific PGT151 (G), and major (H) and minor (I) populations gp160 isolated by IP with a cocktail of bnAbs (b12, b6, F425-b4e8, 2F5, and 4E10). The black line outlining the peaks represents all N-linked glycans released after PNGase F digestion, and the gray dashed line shows the sample after Endo H digestion. Peak areas corresponding to oligomannose-type glycans are colored in shades of green, with their abundances (% to total glycans) depicted in the pie charts. Remaining hybrid- and complex-type glycans are presented in white.
Glycosylation patterns of the gp120 bands were very similar, consisting of predominantly oligomannose-type glycans (approximately 60%) and a series of highly processed bi-, tri-, and tetra-antennary glycans (Fig. 5C and D) similar to those seen on the PBMC-derived sample (Fig. 4A). This differed from the complex-type glycans found on recombinant gp120 that had a larger population of smaller, less processed complex-type glycans compared to the highly processed complex-type sugars (Fig. 5F). The uncleaved gp160 band, observed as a very minor population in the PGT151 IP, had a profile similar to that of pseudovirion gp120 (Fig. 5G). However, the nonnative uncleaved gp160 bands observed from the bnAb cocktail IP had a greater abundance of oligomannose-type glycans (>80%) (Fig. 5H and I). This pattern is consistent with the observations by Crooks et al. of a predominantly Endo H-sensitive gp160 species in pseudoviral particles (47). This form of gp160 may bypass the normal processing pathway through the Golgi and be trafficked directly from the ER to the cell membrane (52, 53). The PGT151-reactive cell surface-associated Env was also isolated and analyzed (Fig. 5B, lane 1; see also Fig. S1D in the supplemental material). The glycan profile of this material was almost identical to that from the pseudoviral particles, also containing highly processed sialylated complex-type sugars (Fig. 5E). Overall, the glycosylations of native and nonnative pseudovirion Env differ, and PGT151 is a useful tool for isolating Env with a native-like glycosylation pattern from pseudovirions and cell surface trimers.
We next performed neuraminidase digests to analyze and compare the sialic acid residues present on the complex-type sugars of pseudovirus with that of PBMC-derived Env. HEK 293T cells are able to add both α2,3- and α2,6-linked sialic acids; however, it has been reported that the majority of HEK 293T expressed proteins have mostly α2,6-linked sialic acids (54–57). When the glycans released from gp120 were treated with the α2,3-specific neuraminidase, all sialylated glycans were shown to be sensitive and no further trimming was observed upon treatment with a general, non-linkage-specific neuraminidase (see Fig. S4 in the supplemental material). Therefore, in contrast to the PBMC-derived Env that has predominantly α2,6-linked sialic acid residues, pseudoviral Env has α2,3-linked sialic acids. This difference highlights the role of the producer cell in complex-type glycosylation.
Glycans decorating gp41 are mostly highly processed complex-type sugars, with a small proportion of Endo H-sensitive structures.
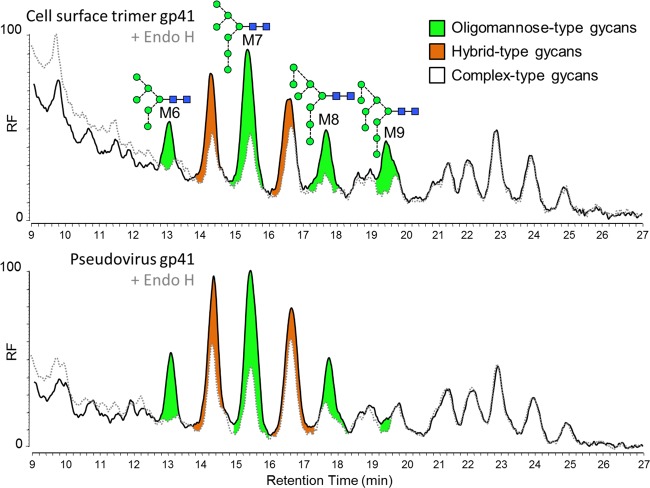
There has been very little reported on the specific glycan structures present on gp41, with most information being provided by SDS-PAGE gel shift differences after enzymatic digestion (58, 59). Pal et al. showed that gp41 from viruses prepared in MOLT-3 cells was Endo H resistant, but when prepared in the presence of an ER mannosidase inhibitor, 1-deoxymannojirimycin, gp41 became sensitive to Endo H digestion (59). More recent work by us and by Go and colleagues has shown that the gp41 glycans on recombinant gp140 trimers (13, 60, 61), including the SOSIP trimer (14), are predominantly complex type and that these structures are generally more dependent on producer cells. In this study, we were able to record the chromatograms of labeled glycans from PGT151-isolated pseudovirion-derived material and cell surface-associated Env to provide a more detailed description of glycan structures present on native virion-derived gp41. Unfortunately, we were unable to obtain equivalent spectra from PBMC-derived gp41 due to a lack of sample abundance. The glycans released from the gp41 band on PGT151-isolated pseudovirion-derived material and cell surface-associated Env were found to be predominantly complex type, although with a minor population of both oligomannose and hybrid-type glycans (Fig. 6). HPLC analysis of the released glycans before and after Endo H digestion showed very minor differences between the chromatograms of cell surface and virion-derived gp41. The glycans include a significant population of highly processed sialylated bi-, tri-, and tetra-antennary complex-type glycans. The glycan profiles of pseudovirion gp41 and cell surface-associated gp41 were very similar, suggesting that the glycan processing of gp41 is not under the same steric restriction as that on gp120. In fact, in the recent crystal structures of the BG505 SOSIP.664 trimers, the N611 and N637 glycan sites were revealed to be adjacent in space but highly exposed (62, 63).
FIG 6.
Glycosylation of virion-associated gp41 is highly heterogeneous and consists of highly processed complex-type glycans. HPLC traces for gp41 isolated using cleavage-specific PGT151 from the cell surface and pseudovirus. The black line represents all N-linked glycans released after PNGase F digestion, and the gray dashed line shows the sample after Endo H digestion. Peak areas corresponding to oligomannose-type glycans are colored in green, Endo H-sensitive hybrid-type glycans are colored in orange, and complex-type glycans are colored in white.
DISCUSSION
As many of the most broad and potent HIV-1 neutralizing antibodies bind to or are dependent on Env glycosylation, including both gp120 and gp41, it is critical to characterize the glycan structures on native virion-associated Env to design immunogens that accurately mimic the native trimer. In this study, we characterized in detail the glycosylation of native Env from PBMC-derived virus and determined the ability of different Env expression systems to replicate certain aspects of native Env glycosylation. Although Env is entirely processed by the glycosylation machinery of the host cell, we show that there are various degrees of protein-directed and cell-directed processing that determine the overall glycosylation profile of native Env. The data presented here represent a reference to compare the glycosylation of potential HIV-1 immunogens and thus can be used to inform strategies for Env immunogen preparation.
The protein-directed element of Env glycosylation has previously been described (7, 9) and arises from the steric restriction of glycan processing leading to the presence of oligomannose glycans, in particular Man8GlcNAc2 and Man9GlcNAc2, forming the so-called IMP (14). These glycans are a key component of the epitope of the N332-dependent bnAbs (4, 18, 22). Our CID analysis showed for the first time that identical isomers of the oligomannose series exist on both recombinant gp120 prepared in HEK 293T cells and virion-associated gp120 prepared in PBMCs. Thus, HEK 293T cells are sufficient to recreate the IMP, confirming that this feature of Env glycosylation is consistent regardless of producer cell. The ion mobility mass spectrometry of PBMC-derived Env showed that the remaining glycans on gp120 were highly processed di-, tri-, and tetra-antennary sialylated complex-type sugars, and HPLC analysis confirmed their abundance. Using PGT151, we were able to isolate a component of pseudovirion Env that had a range of complex-type sugars similar to that isolated from PBMC-derived Env, except for a different linkage of terminal sialic acids. These highly processed complex-type glycans might be indicative of a prolonged exposure in the Golgi or retromer-mediated sorting of virion-associated Env from the cell surface back to the Golgi, as has recently been demonstrated (64). In contrast, recombinant gp120, which is not membrane bound, displayed an additional range of smaller complex-type glycans, suggesting faster transit through the Golgi apparatus.
In contrast to that of gp120, the heterogeneous glycosylation of gp41 indicates a lesser degree of protein-directed processing, presumably due to the lower density of glycans in this region. The gp41 glycan structures are predominantly Endo H resistant, and the complex-type glycans present are mostly sialylated tri- and tetra-antennary glycans. PGT151 neutralization is dependent on N611 and N637 and sensitive to changes in glycan structure, and neutralization plateaus as low as 50% have been observed for some viruses (21). Furthermore, PGT151 has been shown to bind tri- and tetra-antennary complex-type glycans on glycan microarrays (21). These observations, in addition to the heterogeneous nature of gp41, suggest that the incomplete neutralization of some strains by PGT151 may arise from virus particles displaying glycans at positions N611 and N637 that PGT151 cannot bind (although incomplete site occupancy could also play a role). However, it should be noted that PGT151 neutralizes JR-CSF to >95% and will therefore have isolated the majority of native Env from JR-CSF pseudovirions. As the glycan processing on gp41 is not under the same steric restriction as that on gp120, this heterogeneity may highlight a limitation of targeting the glycans on gp41 for vaccine design.
The cell-specific influences on Env glycosylation for recombinant gp120, gp140, and SOSIP trimers have been analyzed previously (14, 65, 66). However, these differences have not been explored in detail for virion-associated gp120, in particular the sialic acid linkages of complex-type sugars. The cell-specific influences on virion-associated gp120 are clearly evident in the differences observed between sialic acid linkages for virion-associated gp120 derived in PBMCs, compared to pseudovirion material derived in HEK 293T cells. This may have implications for the choice of cell for vaccine production. Studies of both human and mouse CD4+ T cells have shown that activation leads to a dramatic remodeling of N-linked glycans (67). In contrast to freshly isolated cells that have predominantly complex-type glycans with terminal α2,6-linked sialic acids, activated cells exhibit a dramatic decrease in sialylated glycans (67, 68). This has been correlated with a decrease in ST6GAL1 mRNA. However, interestingly, we observed in this study that activated HIV-1-infected CD4+ T cells produce virion-associated Env that contains highly sialylated complex glycans, where the sialic acid is predominantly, but not exclusively, α2,6-linked. This may suggest that HIV-1 infection can lead to a remodeling of cellular N-linked glycans, somehow reversing any downregulation of ST6GAL1. Indeed, it has been reported that the glycosylation of CD43 and CD45 is altered upon HIV-1 infection (69). In contrast to the case with PBMC-derived gp120, the sialylated complex-type sugars on pseudovirion gp120 are α2,3 linked. Several studies have reported that some glycan-reactive bnAbs bind specific sialic acid linkages. For example, both PGT121 and PG9 have been shown to preferentially bind α2,6-linked biantennary glycans (17, 57), whereas PGT151 preferentially binds α2,3-sialylated tetra-antennary glycans (21). However, all bnAbs neutralize both HEK 293T-derived pseudovirions and PBMC-derived virus, with very similar potencies (17, 21, 57; unpublished data), yet the implications for the difference in sialic acid preference have not been addressed. Our data may indicate that in the context of a glycan array, where bnAbs are relying on only a portion of their full glycan-protein epitope for binding, the sialic acid linkage is critical for binding affinity. However, when the complete and optimal epitope is present both sialic acid linkages can be tolerated, allowing neutralization of both PBMC- and HEK 293T-derived viruses. Although HEK 293T cells are sufficient to recreate the mannose patch, they are unable to mimic the α2,6-linked sialic acid extensions displayed by the PBMC-derived Env. However, given that the HIV-1 bnAbs identified thus far tolerate both α2,3 and α2,6 linkages, this suggests that HEK 293T cells may still represent a suitable choice for vaccine production.
Our data support observations that pseudovirions display various forms of nonfunctional Env in addition to native Env trimers (47) and further reveal differences in the glycosylation of the native and nonnative forms of Env. We isolated three forms of uncleaved gp160 using the different IP methods. The minor gp160 population isolated through IP with PGT151 has a glycan profile similar to that of gp120 from cleaved, functional trimers, suggesting that this form of virion-associated uncleaved Env is restricted from the glycan processing machinery in the same way as fully cleaved Env. Given that this species was isolated using PGT151, this form of gp160 likely derives from a minor population of trimers comprising cleaved and uncleaved protomers whose glycans are processed similarly, but where furin cleavage is not 100% efficient. The dominant gp160 forms on pseudovirions, isolated using the bnAb cocktail, have greatly increased levels of oligomannose glycans compared to those of virion-associated gp120. This additional level of oligomannose is unlikely to arise from steric constraints of Man5GlcNAc2 glycosyltransferase extension, as these glycans are not similarly protected on the minor gp160 form. As we have recently shown that uncleaved recombinant gp140 trimers have a much higher degree of processing than their cleaved equivalents (14), the elevated level of oligomannose on these membrane-bound gp160 forms may suggest that they bypass the Golgi apparatus and are trafficked straight to the cell surface (52). Indeed, alternate pathways of secretion of SIV Env have been observed (53). Therefore, the glycosylation of pseudovirion nonnative Env could also be impacted by how Env traffics from the ER to the cell membrane. These forms of uncleaved Env were not observed in the virus produced in PBMCs.
In summary, we have shown that the glycosylation of native Env trimers isolated from PBMC-derived virions consists of predominantly protein-directed oligomannose glycans with a population of highly processed sialylated bi-, tri-, and tetra-antennary glycans that are capped with mostly α2,6-linked sialic acids. We have shown that HEK 293T cell-produced pseudoviral Env isolated using PGT151 is largely capable of reproducing this glycan profile; however, the sialic acid linkage of complex-type sugars differs due to a dependence upon the host cell. As these differences do not appear to impact bnAb recognition, PGT151 is therefore a useful tool for isolating Env with a native-like glycosylation pattern from pseudovirions. This will allow easier preparation of virion-associated Env for future glycan analysis and could be applied to purification of Env for vaccine development. Finally, we show that the glycosylation of virion-associated gp41 is not under the same steric constraints as that of gp120 and therefore is highly heterogeneous, with a low abundance of oligomannose and hybrid-type glycans. Overall, the analyses presented demonstrate the protein-directed and cell-directed effects upon native Env glycosylation and provide an important reference for analyzing potential HIV-1 Env immunogens.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dennis Burton for providing the human PBMCs and Khoa Le for technical assistance in preparing the PBMC virus sample.
K.J.D. is supported by an MRC Career Development Fellowship (MR/K024426/1), and M.C. is supported by the Center for HIV/AIDS Vaccine Immunology and Immunogen Discovery Grant (UM1AI100663) and the International AIDS Vaccine Initiative through the Neutralizing Antibody Consortium and Bill and Melinda Gates Center for Vaccine Discovery (glycan characterization and outer domain glycoform design).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01190-15.
REFERENCES
- 1.Korber B, Gaschen B, Yusim K, Thakallapally R, Kesmir C, Detours V. 2001. Evolutionary and immunological implications of contemporary HIV-1 variation. Br Med Bull 58:19–42. doi: 10.1093/bmb/58.1.19. [DOI] [PubMed] [Google Scholar]
- 2.Doores KJ, Burton DR. 2010. Variable loop glycan dependency of the broad and potent HIV-1-neutralizing antibodies PG9 and PG16. J Virol 84:10510–10521. doi: 10.1128/JVI.00552-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scanlan CN, Pantophlet R, Wormald MR, Ollmann Saphire E, Stanfield R, Wilson IA, Katinger H, Dwek RA, Rudd PM, Burton DR. 2002. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of alpha1→2 mannose residues on the outer face of gp120. J Virol 76:7306–7321. doi: 10.1128/JVI.76.14.7306-7321.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong CH, Phogat S, Wrin T, Simek MD, Koff WC, Wilson IA, Burton DR, Poignard P. 2011. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW, Kaminsky S, Zamb T, Moyle M, Koff WC, Poignard P, Burton DR. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crispin M, Doores KJ. 2015. Targeting host-derived glycans on enveloped viruses for antibody-based vaccine design. Curr Opin Virol 11C:63–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doores KJ, Bonomelli C, Harvey DJ, Vasiljevic S, Dwek RA, Burton DR, Crispin M, Scanlan CN. 2010. Envelope glycans of immunodeficiency virions are almost entirely oligomannose antigens. Proc Natl Acad Sci U S A 107:13800–13805. doi: 10.1073/pnas.1006498107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geyer H, Holschbach C, Hunsmann G, Schneider J. 1988. Carbohydrates of human immunodeficiency virus. Structures of oligosaccharides linked to the envelope glycoprotein 120. J Biol Chem 263:11760–11767. [PubMed] [Google Scholar]
- 9.Bonomelli C, Doores KJ, Dunlop DC, Thaney V, Dwek RA, Burton DR, Crispin M, Scanlan CN. 2011. The glycan shield of HIV is predominantly oligomannose independently of production system or viral clade. PLoS One 6:e23521. doi: 10.1371/journal.pone.0023521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mizuochi T, Matthews TJ, Kato M, Hamako J, Titani K, Solomon J, Feizi T. 1990. Diversity of oligosaccharide structures on the envelope glycoprotein gp 120 of human immunodeficiency virus 1 from the lymphoblastoid cell line H9. Presence of complex-type oligosaccharides with bisecting N-acetylglucosamine residues. J Biol Chem 265:8519–8524. [PubMed] [Google Scholar]
- 11.Mizuochi T, Spellman MW, Larkin M, Solomon J, Basa LJ, Feizi T. 1988. Structural characterization by chromatographic profiling of the oligosaccharides of human immunodeficiency virus (HIV) recombinant envelope glycoprotein gp120 produced in Chinese hamster ovary cells. Biomed Chromatogr 2:260–270. [DOI] [PubMed] [Google Scholar]
- 12.Mizuochi T, Spellman MW, Larkin M, Solomon J, Basa LJ, Feizi T. 1988. Carbohydrate structures of the human-immunodeficiency-virus (HIV) recombinant envelope glycoprotein gp120 produced in Chinese-hamster ovary cells. Biochem J 254:599–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Go EP, Irungu J, Zhang Y, Dalpathado DS, Liao HX, Sutherland LL, Alam SM, Haynes BF, Desaire H. 2008. Glycosylation site-specific analysis of HIV envelope proteins (JR-FL and CON-S) reveals major differences in glycosylation site occupancy, glycoform profiles, and antigenic epitopes' accessibility. J Proteome Res 7:1660–1674. doi: 10.1021/pr7006957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pritchard LK, Vasiljevic S, Ozorowski G, Seabright GE, Cupo A, Ringe RP, Kim HJ, Sanders RW, Doores KJ, Burton DR, Wilson IA, Ward AB, Moore JP, Crispin M. 2015. Structural constraints determine the glycoyslation of HIV-1 envelope trimers. Cell Rep 11:1604–1613. doi: 10.1016/j.celrep.2015.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Pena AT, Korzun J, Golabek M, de Los Reyes K, Ketas TJ, van Gils MJ, King CR, Wilson IA, Ward AB, Klasse PJ, Moore JP. 2013. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog 9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doores KJ, Kong L, Krumm SA, Le KM, Sok D, Laserson U, Garces F, Poignard P, Wilson IA, Burton DR. 2015. Two classes of broadly neutralizing antibodies within a single lineage directed to the high-mannose patch of HIV envelope. J Virol 89:1105–1118. doi: 10.1128/JVI.02905-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Julien JP, Sok D, Khayat R, Lee JH, Doores KJ, Walker LM, Ramos A, Diwanji DC, Pejchal R, Cupo A, Katpally U, Depetris RS, Stanfield RL, McBride R, Marozsan AJ, Paulson JC, Sanders RW, Moore JP, Burton DR, Poignard P, Ward AB, Wilson IA. 2013. Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans. PLoS Pathog 9:e1003342. doi: 10.1371/journal.ppat.1003342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kong L, Lee JH, Doores KJ, Murin CD, Julien JP, McBride R, Liu Y, Marozsan A, Cupo A, Klasse PJ, Hoffenberg S, Caulfield M, King CR, Hua Y, Le KM, Khayat R, Deller MC, Clayton T, Tien H, Feizi T, Sanders RW, Paulson JC, Moore JP, Stanfield RL, Burton DR, Ward AB, Wilson IA. 2013. Supersite of immune vulnerability on the glycosylated face of HIV-1 envelope glycoprotein gp120. Nat Struct Mol Biol 20:796–803. doi: 10.1038/nsmb.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonsignori M, Hwang KK, Chen X, Tsao CY, Morris L, Gray E, Marshall DJ, Crump JA, Kapiga SH, Sam NE, Sinangil F, Pancera M, Yongping Y, Zhang B, Zhu J, Kwong PD, O'Dell S, Mascola JR, Wu L, Nabel GJ, Phogat S, Seaman MS, Whitesides JF, Moody MA, Kelsoe G, Yang X, Sodroski J, Shaw GM, Montefiori DC, Kepler TB, Tomaras GD, Alam SM, Liao HX, Haynes BF. 2011. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J Virol 85:9998–10009. doi: 10.1128/JVI.05045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McLellan JS, Pancera M, Carrico C, Gorman J, Julien JP, Khayat R, Louder R, Pejchal R, Sastry M, Dai K, O'Dell S, Patel N, Shahzad-ul Hussan S, Yang Y, Zhang B, Zhou T, Zhu J, Boyington JC, Chuang GY, Diwanji D, Georgiev I, Kwon YD, Lee D, Louder MK, Moquin S, Schmidt SD, Yang ZY, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Burton DR, Koff WC, Walker LM, Phogat S, Wyatt R, Orwenyo J, Wang LX, Arthos J, Bewley CA, Mascola JR, Nabel GJ, Schief WR, Ward AB, Wilson IA, Kwong PD. 2011. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Falkowska E, Le KM, Ramos A, Doores KJ, Lee JH, Blattner C, Ramirez A, Derking R, van Gils MJ, Liang CH, McBride R, von Bredow B, Shivatare SS, Wu CY, Chan-Hui PY, Liu Y, Feizi T, Zwick MB, Koff WC, Seaman MS, Swiderek K, Moore JP, Evans D, Paulson JC, Wong CH, Ward AB, Wilson IA, Sanders RW, Poignard P, Burton DR. 2014. Broadly neutralizing HIV antibodies define a glycan-dependent epitope on the prefusion conformation of gp41 on cleaved envelope trimers. Immunity 40:657–668. doi: 10.1016/j.immuni.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pejchal R, Doores KJ, Walker LM, Khayat R, Huang PS, Wang SK, Stanfield RL, Julien JP, Ramos A, Crispin M, Depetris R, Katpally U, Marozsan A, Cupo A, Maloveste S, Liu Y, McBride R, Ito Y, Sanders RW, Ogohara C, Paulson JC, Feizi T, Scanlan CN, Wong CH, Moore JP, Olson WC, Ward AB, Poignard P, Schief WR, Burton DR, Wilson IA. 2011. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science 334:1097–1103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blattner C, Lee JH, Sliepen K, Derking R, Falkowska E, de la Pena AT, Cupo A, Julien JP, van Gils M, Lee PS, Peng W, Paulson JC, Poignard P, Burton DR, Moore JP, Sanders RW, Wilson IA, Ward AB. 2014. Structural delineation of a quaternary, cleavage-dependent epitope at the gp41-gp120 interface on intact HIV-1 Env trimers. Immunity 40:669–680. doi: 10.1016/j.immuni.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harvey DJ, Sobott F, Crispin M, Wrobel A, Bonomelli C, Vasiljevic S, Scanlan CN, Scarff CA, Thalassinos K, Scrivens JH. 2011. Ion mobility mass spectrometry for extracting spectra of N-glycans directly from incubation mixtures following glycan release: application to glycans from engineered glycoforms of intact, folded HIV gp120. J Am Soc Mass Spectrom 22:568–581. doi: 10.1007/s13361-010-0053-0. [DOI] [PubMed] [Google Scholar]
- 25.Mariño K, Bones J, Kattla JJ, Rudd PM. 2010. A systematic approach to protein glycosylation analysis: a path through the maze. Nat Chem Biol 6:713–723. doi: 10.1038/nchembio.437. [DOI] [PubMed] [Google Scholar]
- 26.Domann PJ, Pardos-Pardos AC, Fernandes DL, Spencer DI, Radcliffe CM, Royle L, Dwek RA, Rudd PM. 2007. Separation-based glycoprofiling approaches using fluorescent labels. Proteomics 7(Suppl 1):S70–S76. [DOI] [PubMed] [Google Scholar]
- 27.Montefiori DC. 2005. Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. Curr Protoc Immunol Chapter 12:Unit 12.11. [DOI] [PubMed] [Google Scholar]
- 28.Crocker PR, Paulson JC, Varki A. 2007. Siglecs and their roles in the immune system. Nat Rev Immunol 7:255–266. doi: 10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- 29.Mann AM, Rusert P, Berlinger L, Kuster H, Gunthard HF, Trkola A. 2009. HIV sensitivity to neutralization is determined by target and virus producer cell properties. AIDS 23:1659–1667. doi: 10.1097/QAD.0b013e32832e9408. [DOI] [PubMed] [Google Scholar]
- 30.Rusert P, Fischer M, Joos B, Leemann C, Kuster H, Flepp M, Bonhoeffer S, Gunthard HF, Trkola A. 2004. Quantification of infectious HIV-1 plasma viral load using a boosted in vitro infection protocol. Virology 326:113–129. doi: 10.1016/j.virol.2004.05.022. [DOI] [PubMed] [Google Scholar]
- 31.Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, Bilska M, Kothe DL, Salazar-Gonzalez JF, Wei X, Decker JM, Hahn BH, Montefiori DC. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol 79:10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Börnsen KO, Mohr MD, Widmer HM. 1995. Ion exchange and purification of carbohydrates on a Nafion membrane as a new sample pretreatment for matrix-assisted laser desorption-ionization mass spectrometry. Rapid Commun Mass Spectrom 9:1031–1034. doi: 10.1002/rcm.1290091112. [DOI] [Google Scholar]
- 33.Giles K, Pringle SD, Worthington KR, Little D, Wildgoose JL, Bateman RH. 2004. Applications of a travelling wave-based radio-frequency-only stacked ring ion guide. Rapid Commun Mass Spectrom 18:2401–2414. doi: 10.1002/rcm.1641. [DOI] [PubMed] [Google Scholar]
- 34.Domon B, Costello CE. 1988. A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconj J 5:397–409. doi: 10.1007/BF01049915. [DOI] [Google Scholar]
- 35.Harvey DJ. 2005. Fragmentation of negative ions from carbohydrates: part 2. Fragmentation of high-mannose N-linked glycans. J Am Soc Mass Spectrom 16:631–646. [DOI] [PubMed] [Google Scholar]
- 36.Harvey DJ. 2005. Fragmentation of negative ions from carbohydrates: part 1. Use of nitrate and other anionic adducts for the production of negative ion electrospray spectra from N-linked carbohydrates. J Am Soc Mass Spectrom 16:622–630. [DOI] [PubMed] [Google Scholar]
- 37.Harvey DJ. 2005. Fragmentation of negative ions from carbohydrates: part 3. Fragmentation of hybrid and complex N-linked glycans. J Am Soc Mass Spectrom 16:647–659. [DOI] [PubMed] [Google Scholar]
- 38.Harvey DJ, Royle L, Radcliffe CM, Rudd PM, Dwek RA. 2008. Structural and quantitative analysis of N-linked glycans by matrix-assisted laser desorption ionization and negative ion nanospray mass spectrometry. Anal Biochem 376:44–60. doi: 10.1016/j.ab.2008.01.025. [DOI] [PubMed] [Google Scholar]
- 39.Neville DC, Dwek RA, Butters TD. 2009. Development of a single column method for the separation of lipid- and protein-derived oligosaccharides. J Proteome Res 8:681–687. doi: 10.1021/pr800704t. [DOI] [PubMed] [Google Scholar]
- 40.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, Benenson M, Gurunathan S, Tartaglia J, McNeil JG, Francis DP, Stablein D, Birx DL, Chunsuttiwat S, Khamboonruang C, Thongcharoen P, Robb ML, Michael NL, Kunasol P, Kim JH. 2009. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med 361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 41.Macauley MS, Crocker PR, Paulson JC. 2014. Siglec-mediated regulation of immune cell function in disease. Nat Rev Immunol 14:653–666. doi: 10.1038/nri3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Montefiori DC, Robinson WE Jr, Mitchell WM. 1989. Antibody-independent, complement-mediated enhancement of HIV-1 infection by mannosidase I and II inhibitors. Antiviral Res 11:137–146. doi: 10.1016/0166-3542(89)90025-9. [DOI] [PubMed] [Google Scholar]
- 43.Montefiori DC, Murphey-Corb M, Desrosiers RC, Daniel MD. 1990. Complement-mediated, infection-enhancing antibodies in plasma from vaccinated macaques before and after inoculation with live simian immunodeficiency virus. J Virol 64:5223–5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fearon DT. 1978. Regulation by membrane sialic acid of beta1H-dependent decay-dissociation of amplification C3 convertase of the alternative complement pathway. Proc Natl Acad Sci U S A 75:1971–1975. doi: 10.1073/pnas.75.4.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hirsch RL, Wolinsky JS, Winkelstein JA. 1986. Activation of the alternative complement pathway by mumps infected cells: relationship to viral neuraminidase activity. Arch Virol 87:181–190. doi: 10.1007/BF01315298. [DOI] [PubMed] [Google Scholar]
- 46.Sato C, Fukuoka H, Ohta K, Matsuda T, Koshino R, Kobayashi K, Troy FA II, Kitajima K. 2000. Frequent occurrence of pre-existing alpha 2→8-linked disialic and oligosialic acids with chain lengths up to 7 Sia residues in mammalian brain glycoproteins. Prevalence revealed by highly sensitive chemical methods and anti-di-, oligo-, and poly-Sia antibodies specific for defined chain lengths. J Biol Chem 275:15422–15431. [DOI] [PubMed] [Google Scholar]
- 47.Crooks ET, Tong T, Osawa K, Binley JM. 2011. Enzyme digests eliminate nonfunctional Env from HIV-1 particle surfaces, leaving native Env trimers intact and viral infectivity unaffected. J Virol 85:5825–5839. doi: 10.1128/JVI.00154-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moore PL, Crooks ET, Porter L, Zhu P, Cayanan CS, Grise H, Corcoran P, Zwick MB, Franti M, Morris L, Roux KH, Burton DR, Binley JM. 2006. Nature of nonfunctional envelope proteins on the surface of human immunodeficiency virus type 1. J Virol 80:2515–2528. doi: 10.1128/JVI.80.5.2515-2528.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leaman DP, Kinkead H, Zwick MB. 2010. In-solution virus capture assay helps deconstruct heterogeneous antibody recognition of human immunodeficiency virus type 1. J Virol 84:3382–3395. doi: 10.1128/JVI.02363-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Herrera C, Klasse PJ, Michael E, Kake S, Barnes K, Kibler CW, Campbell-Gardener L, Si Z, Sodroski J, Moore JP, Beddows S. 2005. The impact of envelope glycoprotein cleavage on the antigenicity, infectivity, and neutralization sensitivity of Env-pseudotyped human immunodeficiency virus type 1 particles. Virology 338:154–172. doi: 10.1016/j.virol.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 51.Poignard P, Moulard M, Golez E, Vivona V, Franti M, Venturini S, Wang M, Parren PW, Burton DR. 2003. Heterogeneity of envelope molecules expressed on primary human immunodeficiency virus type 1 particles as probed by the binding of neutralizing and nonneutralizing antibodies. J Virol 77:353–365. doi: 10.1128/JVI.77.1.353-365.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nickel W, Seedorf M. 2008. Unconventional mechanisms of protein transport to the cell surface of eukaryotic cells. Annu Rev Cell Dev Biol 24:287–308. doi: 10.1146/annurev.cellbio.24.110707.175320. [DOI] [PubMed] [Google Scholar]
- 53.Spies CP, Compans RW. 1993. Alternate pathways of secretion of simian immunodeficiency virus envelope glycoproteins. J Virol 67:6535–6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Otto VI, Schurpf T, Folkers G, Cummings RD. 2004. Sialylated complex-type N-glycans enhance the signaling activity of soluble intercellular adhesion molecule-1 in mouse astrocytes. J Biol Chem 279:35201–35209. doi: 10.1074/jbc.M404947200. [DOI] [PubMed] [Google Scholar]
- 55.Sánchez-Felipe L, Villar E, Munoz-Barroso I. 2012. alpha2-3- and alpha2-6-N-linked sialic acids allow efficient interaction of Newcastle disease virus with target cells. Glycoconj J 29:539–549. doi: 10.1007/s10719-012-9431-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guo Y, Rumschlag-Booms E, Wang J, Xiao H, Yu J, Guo L, Gao GF, Cao Y, Caffrey M, Rong L. 2009. Analysis of hemagglutinin-mediated entry tropism of H5N1 avian influenza. Virol J 6:39. doi: 10.1186/1743-422X-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pancera M, Shahzad-Ul-Hussan S, Doria-Rose NA, McLellan JS, Bailer RT, Dai K, Loesgen S, Louder MK, Staupe RP, Yang Y, Zhang B, Parks R, Eudailey J, Lloyd KE, Blinn J, Alam SM, Haynes BF, Amin MN, Wang LX, Burton DR, Koff WC, Nabel GJ, Mascola JR, Bewley CA, Kwong PD. 2013. Structural basis for diverse N-glycan recognition by HIV-1-neutralizing V1-V2-directed antibody PG16. Nat Struct Mol Biol 20:804–813. doi: 10.1038/nsmb.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dewar RL, Vasudevachari MB, Natarajan V, Salzman NP. 1989. Biosynthesis and processing of human immunodeficiency virus type 1 envelope glycoproteins: effects of monensin on glycosylation and transport. J Virol 63:2452–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pal R, Hoke GM, Sarngadharan MG. 1989. Role of oligosaccharides in the processing and maturation of envelope glycoproteins of human immunodeficiency virus type 1. Proc Natl Acad Sci U S A 86:3384–3388. doi: 10.1073/pnas.86.9.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Go EP, Hewawasam G, Liao HX, Chen H, Ping LH, Anderson JA, Hua DC, Haynes BF, Desaire H. 2011. Characterization of glycosylation profiles of HIV-1 transmitted/founder envelopes by mass spectrometry. J Virol 85:8270–8284. doi: 10.1128/JVI.05053-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Go EP, Chang Q, Liao HX, Sutherland LL, Alam SM, Haynes BF, Desaire H. 2009. Glycosylation site-specific analysis of clade C HIV-1 envelope proteins. J Proteome Res 8:4231–4242. doi: 10.1021/pr9002728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, Huang J, Acharya P, Chuang GY, Ofek G, Stewart-Jones GB, Stuckey J, Bailer RT, Joyce MG, Louder MK, Tumba N, Yang Y, Zhang B, Cohen MS, Haynes BF, Mascola JR, Morris L, Munro JB, Blanchard SC, Mothes W, Connors M, Kwong PD. 2014. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature 514:455–461. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ward AB, Wilson IA. 2015. Insights into the trimeric HIV-1 envelope glycoprotein structure. Trends Biochem Sci 40:101–107. doi: 10.1016/j.tibs.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Groppelli E, Len AC, Granger LA, Jolly C. 2014. Retromer regulates HIV-1 envelope glycoprotein trafficking and incorporation into virions. PLoS Pathog 10:e1004518. doi: 10.1371/journal.ppat.1004518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Go EP, Liao HX, Alam SM, Hua D, Haynes BF, Desaire H. 2013. Characterization of host-cell line specific glycosylation profiles of early transmitted/founder HIV-1 gp120 envelope proteins. J Proteome Res 12:1223–1234. doi: 10.1021/pr300870t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Raska M, Takahashi K, Czernekova L, Zachova K, Hall S, Moldoveanu Z, Elliott MC, Wilson L, Brown R, Jancova D, Barnes S, Vrbkova J, Tomana M, Smith PD, Mestecky J, Renfrow MB, Novak J. 2010. Glycosylation patterns of HIV-1 gp120 depend on the type of expressing cells and affect antibody recognition. J Biol Chem 285:20860–20869. doi: 10.1074/jbc.M109.085472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Comelli EM, Sutton-Smith M, Yan Q, Amado M, Panico M, Gilmartin T, Whisenant T, Lanigan CM, Head SR, Goldberg D, Morris HR, Dell A, Paulson JC. 2006. Activation of murine CD4+ and CD8+ T lymphocytes leads to dramatic remodeling of N-linked glycans. J Immunol 177:2431–2440. doi: 10.4049/jimmunol.177.4.2431. [DOI] [PubMed] [Google Scholar]
- 68.Toscano MA, Bianco GA, Ilarregui JM, Croci DO, Correale J, Hernandez JD, Zwirner NW, Poirier F, Riley EM, Baum LG, Rabinovich GA. 2007. Differential glycosylation of TH1, TH2 and TH-17 effector cells selectively regulates susceptibility to cell death. Nat Immunol 8:825–834. doi: 10.1038/ni1482. [DOI] [PubMed] [Google Scholar]
- 69.Lantéri M, Giordanengo V, Hiraoka N, Fuzibet JG, Auberger P, Fukuda M, Baum LG, Lefebvre JC. 2003. Altered T cell surface glycosylation in HIV-1 infection results in increased susceptibility to galectin-1-induced cell death. Glycobiology 13:909–918. doi: 10.1093/glycob/cwg110. [DOI] [PubMed] [Google Scholar]
- 70.Harvey DJ, Merry AH, Royle L, Campbell MP, Dwek RA, Rudd PM. 2009. Proposal for a standard system for drawing structural diagrams of N- and O-linked carbohydrates and related compounds. Proteomics 9:3796–3801. doi: 10.1002/pmic.200900096. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.