ABSTRACT
The infectivity of hepadnavirus virions produced during either acute or chronic stages of infection was compared by testing the ability of the virions of woodchuck hepatitis virus (WHV) to induce productive acute infection in naive adult woodchucks. Serum WHV collected during acute infection was compared to virions harvested from WHV-infected woodchucks during either (i) early chronic infection, when WHV-induced hepatocellular carcinoma (HCC) was not yet developed, or (ii) late chronic infection, when established HCC was terminal. All tested types of WHV inoculum were related, because they were collected from woodchucks that originally were infected with standardized WHV7 inoculum. Despite the individual differences between animals, the kinetics of accumulation of serum relaxed circular DNA of WHV demonstrated that the virions produced during early or late chronic infection are fully capable of inducing productive acute infection with long-lasting high viremia. These findings were further supported by the analysis of such intrahepatic markers of WHV infection as replicative intermediate DNA, covalently closed circular DNA, pregenomic RNA, and the percentage of WHV core antigen-positive hepatocytes measured at several time points over the course of 17.5 weeks after the inoculation. In addition, the observed relationship between the production of antibodies against WHV surface antigens and parameters of WHV infection appears to be complex. Taken together, the generated data suggest that in vivo hepadnavirus virions produced during different phases of chronic infection did not demonstrate any considerable deficiencies in infectivity compared to that of virions generated during the acute phase of infection.
IMPORTANCE The generated data suggest that infectivity of virions produced during the early or late stages of chronic hepadnavirus infection is not compromised. Our novel results provided several lines of further evidence supporting the idea that during the state of chronic infection in vivo, the limitations of hepadnavirus cell-to-cell spread/superinfection (observed recently in the woodchuck model) are not due to the diminished infectivity of the virions circulating in the blood and likely are (i) related to the properties of hepatocytes (i.e., their capacity to support hepadnavirus infection/replication) and (ii) influenced by the immune system. The obtained results further extend the understanding of the mechanisms regulating the persistence of hepadnavirus infection. Follow-up studies that will further investigate hepadnavirus cell-to-cell spread as a potential regulator of the chronic state of the infection are warranted.
INTRODUCTION
Hepatitis B virus (HBV) is a very significant human pathogen that causes transient and chronic liver infections. Approximately 400 million individuals around the world are chronic carriers of HBV. Chronic HBV infection is the number one risk factor for development of hepatocellular carcinoma (HCC) (1–3). Woodchuck hepatitis virus (WHV) is another member of the Hepadnaviridae family, for which HBV is the prototype hepadnavirus. WHV is closely related to HBV and is frequently employed as a surrogate virus for studying the mechanisms of HBV replication and infection and for testing anti-HBV drugs. It is more carcinogenic than HBV. Most woodchucks chronically infected with WHV usually develop WHV-induced HCC between 1 and 3 years after infection (4–7). Better understanding of the determinants of the maintenance of the chronic state of hepadnavirus infection would benefit the future design of advanced anti-HBV interventions that aim to suppress chronic infection and, as a result, decrease the risk of HCC development. A long-standing argument in the HBV research field suggests that during chronic infection, virus spread is at least very inefficient (if it occurs at all); that superinfection with a hepadnavirus is an unlikely event; and that the state of chronicity is being maintained by the division of already infected hepatocytes (in the absence of virus cell-to-cell spread) (8–14). It became apparent that several important issues need to be resolved in order to better understand the complex mechanism of maintenance of chronic hepadnavirus infection. Recently, we showed that woodchuck livers chronically infected with strain WHV7 can be superinfected in vivo with a different WHV strain, WHVNY. Our results suggested (i) that hepadnavirus cell-to-cell spread and superinfection continue during chronic hepadnavirus infection and (ii) that spread and superinfection, while being limited, still may represent the determinants of maintenance of chronic infection (15). In a separate study, we compared strains WHV7 and WHVNY and concluded that the two strains were quite similar in terms of their replication parameters and the ability to induce productive acute infection in naive adult woodchucks (16). This result was consistent with the interpretation that the observed limited efficiency of WHVNY superinfection in vivo (15) was not mediated by the properties of WHVNY virions but rather reflected the properties of hepatocytes residing in livers chronically infected with WHV7. In another study, we compared the infectivity of different types of HDV virions that differed only by the nature of the envelope proteins which coated the virions. To assemble these different HDV types, we used the envelope proteins, sequences of which were found in HBV isolates collected during either acute or chronic HBV infections. The obtained results did not support the hypothesis that during chronic HBV infection, the accumulated envelope proteins of HBV are responsible for the assembly of the virions, most of which would have diminished infectivity (17). In the current study, we performed a side-by-side comparison of WHV virions harvested from WHV-infected woodchucks during either acute or chronic WHV infection. We included in the comparison WHV virions collected during early chronic infection, when WHV-induced HCC was not yet developed, and virions obtained during late chronic infection, at the time when established HCC was terminal. We examined the ability of these collected WHV virions to induce a productive acute infection of the livers of naive (WHV-negative) woodchucks. We used three woodchucks per type of WHV inoculum tested and monitored animals for 17.5 weeks after inoculation. We measured and compared the accumulation of serum relaxed circular DNA (rcDNA) of WHV in collected serum samples. In addition, we quantified intrahepatic replicative intermediate DNA (RI-DNA), covalently closedcircular DNA (cccDNA), and pregenomic RNA (pgRNA) of WHV in harvested liver tissue samples. In addition, we analyzed the percentages of hepatocytes positive for WHV core antigen (WHcAg), as well as the accumulation of surface antigens (WHsAg) and antibodies against WHsAg in serum samples. Taken together, our data suggest that hepadnavirus virions generated during early and late chronic infection did not display any considerable deficiencies of infectivity compared to that of the virions produced during acute hepadnavirus infection.
MATERIALS AND METHODS
WHV inocula and infections.
Twelve adult WHV-seronegative (naive) woodchucks were used. All woodchucks were maintained and infected at Northeastern Wildlife facilities. All experimental manipulations of animals have been performed under protocols approved by the Institutional Animal Care and Use Committee. Woodchucks F7386, M7297, and M7391 received as inoculum serum-derived WHV collected from woodchuck F6693 (16) at the peak of viremia during acute infection (week +7 after inoculation). Animals M7393, F7238, and M7379 were infected with serum WHV harvested from woodchuck M1001 at the stage of early chronic infection (12 months postinoculation), when HCC was not yet developed. Woodchucks M7296, F7237, and F7384 were injected with serum WHV collected from animal M1001 during the late stage of chronic infection (17 months postinoculation), at the time when terminal HCC was established. The other three woodchucks (M7392, F7394, and M7249) were inoculated with serum WHV obtained from woodchuck F1017 during the late stage of chronic infection (16 months postinoculation), when the animal developed terminal HCC. Each animal was inoculated with 1.66 × 108 genome equivalents (GE) of WHV. The above-mentioned four different types of WHV inoculum were collected from woodchucks that originally were infected with standardized cWHV7P2 inoculum. The inoculum cWHV7P2 was derived from original standardized inoculum cWHV7P1 (5) and has the same biological and virological characteristics as cWHV7P1. The infected woodchucks subsequently were monitored for 17.5 weeks after the inoculation (which is later referred to as week +18). The serum samples were collected weekly during the time course, including week +16. The biopsy specimens of liver tissues were harvested at 2 weeks prior to inoculation (week −2) and then during weeks +5, +9, and +13 postinoculation. The blood and liver tissues also were harvested during necropsy (week +18).
Assay for rcDNA of WHV in serum samples.
For extraction of serum rcDNA, we used a modification of the previously described procedure (18). A volume of 450 μl of lysis buffer (0.01 M Tris-HCl, pH 8.0, 0.01 M EDTA, 0.1 M NaCl, 0.2% SDS) containing 1 mg/ml of Pronase (Roche) was added to each aliquot of collected woodchuck serum (50 μl). The resulting mixture was incubated at 37°C for 2 h. After the incubation, DNA was extracted first using an equal volume of phenol and then with a mixture of chloroform-isoamyl alcohol (24:1). The adding of 2 μl of dextran (molecular weight of 35,000 to 50,000; 10 μg/μl solution in DNase-/RNase-free water), 0.1 volume of 3.0 M sodium acetate, pH 5.5, and 2 volumes of 100% ethanol was used to precipitate DNA. The real-time quantitative PCR (qPCR) was employed to measure the copy numbers of rcDNA of WHV. The calibration of the assay was performed using 10-fold dilution series of plasmid pUC-CMVWHV, which was linearized with NheI. A range from 20 × 107 to 2.0 × 107 GE of WHV was used to obtain the calibration curve (16, 19). The qPCR employed primer 23 (2504-AGAAGACGCACTCCCTCTCCT-2524), primer 24 (2579-TGGCAGATGGAGATTGAGAGC-2559), and TaqMan probe 28 (2531-/6-FAM/AGAAGATCTCAATCACCGCGTCGCAG/3BHQ_1/-2556). The 5′ end of the probe was labeled with 6-carboxyfluorescein (6-FAM). The 3′ end of the probe contained Black Hole Quencher 1 (BHQ_1). The positions of the primers and probes throughout the paper are shown based on the sequence of strain WHV7 (15). qPCR was performed using the TaqMan gene expression mastermix on a 7500 Real-Time PCR system (Applied Biosystems) as previously described (19).
Assays for WHsAg and for antibodies that recognize WHsAg (anti-WHsAg antibodies).
WHsAg and anti-WHsAg antibodies in sera were measured using enzyme-linked immunosorbent assays (ELISA) as previously described (20, 21). The detection limit of the assay for WHsAg was 50 ng/ml, while for analysis of anti-WHsAg antibodies it was 100 U/ml (U stands for standard units).
Immunohistochemical analysis.
The harvested liver tissues were prepared and stained as previously described using the antibodies against the core antigen of WHV (WHcAg) (16, 19). The WHcAg-positive and WHcAg-negative cells were identified manually, and then TMARKER software was employed for counting the fraction of WHcAg-positive cells (22).
Assays for RI-DNA and cccDNA of WHV.
Liver tissue samples first were homogenized in glass grinders using 0.6 to 1.5 ml of TE buffer (10:10) (10 mM Tris-HCl pH 7.6, 10 mM EDTA) (15). To isolate total DNA, 390 μl of TE buffer (10:10) and 600 μl of a buffer containing 10 mM Tris-HCl, pH 7.6, 10 mM EDTA, 0.2% SDS, and 4 mg/ml Pronase were added to 210 μl of the resulting homogenate. The obtained mixture was incubated at 37°C for 2 h. After the incubation, total DNA was extracted two times using an equal volume of phenol-chloroform, and then DNA was precipitated by adding 2 volumes of 100% ethanol and 0.1 volumes of 3.0 M sodium acetate, pH 5.5. DNA next was sedimented by centrifugation at 13,000 rpm at 4°C for 30 min. The DNA pellets were washed using 100% ethanol and then resuspended in 25 to 50 μl of DNase-/RNase-free water (23). Measurements of WHV RI-DNA were conducted by qPCR as described above for rcDNA.
For isolation of cccDNA, 930 μl of TE buffer (10:10) and 60 μl of 10% SDS were mixed with 210 μl of the above-mentioned tissue homogenate. The obtained mixtures were vortexed, and then 300 μl of 2.5 M KCl was added to each mixture. The resulting mixtures were vortexed again and then were incubated at room temperature for at least 30 min. After the incubation, the mixtures were centrifuged at 13,000 rpm for 30 min at 4°C. The supernatants were extracted first using phenol, and a second extraction was done using a phenol-chloroform mixture. Each aqueous phase obtained after the second extraction then was precipitated by adding two volumes of 100% ethanol and 2 μl of dextran solution (10 μg/μl) in DNase-/RNase-free water. The samples were incubated at room temperature overnight and then were centrifuged to precipitate DNA. Resulting DNA pellets were washed using 70% ethanol two times, and then one additional wash was performed using 100% ethanol. Each obtained DNA pellet was resuspended using 50 μl of DNase-/RNase-free water. After the resuspension, 40 μl of each DNA sample was incubated at 37°C overnight in a 500-μl reaction mixture, which contained 1× NEB (New England BioLabs) buffer 4, 1 mM ATP, 40 μg/ml of RNase A, and 50 U of plasmid-safe ATP-dependent DNase (Epicentre). Upon completion of the incubation, cccDNA was further isolated by extraction with phenol and a second extraction with a phenol-chloroform mixture followed by ethanol precipitation (15). The measurements of cccDNA amounts were conducted using qPCR. The assay employed primer 290 (1701-GGTCCGTGTTGCTTGGTCT-1719), primer 291 (1977-GGACATGGAACACAGGCAAAAACA-1954), and TaqMan probe 292 (1846-/6-FAM/AATGGGAGGAGGGCAGCATTGATCCT/3BHQ_1/-1871). A 10-fold dilution series of plasmid pUC-CMVWHV linearized with NheI (range, 20 × 107 to 2.0 × 107 GE of WHV) was used to generate a calibration curve for the quantification. The concentration of DNA in total DNA preparations was quantified with the DNA-binding dye Hoechst 33258 and then used for normalization of copy numbers of RI-DNA and cccDNA (15). The DNA copy numbers are expressed as WHV GE per microgram of total DNA (GE/µg) (in the corresponding total DNA preparation).
Assay for pgRNA of WHV.
Total RNA was isolated from harvested liver tissues using TRI Reagent (Molecular Research Center) and then treated using Turbo DNase (Life Technologies) with the primary goal of eliminating WHV DNA replication intermediates. The WHV-specific cDNA synthesis was done using the primer 850 (2602-TGTACCCATTGAAGATCAGCAGTT-2579) and employing a high-capacity cDNA reverse transcription kit (Life Technologies). The levels of pgRNA were measured using qPCR, which employed the primers 23 and 24 and TaqMan probe 28 as described above for rcDNA. The quantification of copy numbers was done using a 10-fold dilution series of in vitro-transcribed and gel-purified RNA standard (WHV7 RNA was transcribed from the plasmid pJSWHV7-2C6, which was linearized with XhoI) within a range of 20 × 106 to 2.0 × 106 GE of WHV RNA (15). The pgRNA numbers were expressed per microgram of total RNA.
Assay for the portion of WHV virions that are bound to anti-WHsAg antibodies.
The quantification of the percentage of serum WHV virions bound to anti-WHsAg antibodies in the inocula used was conducted as previously described (16). Briefly, each serum aliquot was incubated with 100 μl of Pansorbin (Calbiochem) in Williams' E medium (the total volume of the mixture was 1 ml) using a rocking platform. The incubation was conducted at 4°C for 3 h. The WHV virions bound to anti-WHsAg antibodies that became attached to Pansorbin during the incubation were sedimented by centrifugation for 1 min at 13,000 rpm. The precipitates were washed four times with ice-cold phosphate-buffered saline (PBS) containing 0.5% Nonidet P-40 (Fisher) and then subjected to isolation of DNA. The amounts of WHV rcDNA were measured in the isolated DNA samples using qPCR as described above (16, 19).
RESULTS
Groups of infected woodchucks.
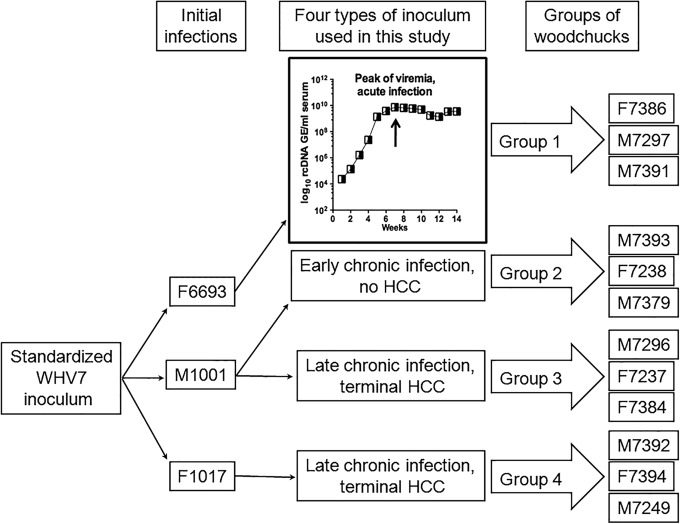
Four groups of WHV-seronegative (i.e., naive) adult woodchucks (three woodchucks per group) were experimentally inoculated with four different types of WHV inoculum. The information on the inocula and groups of infected animals is summarized in Fig. 1. Group 1, which consisted of woodchucks F7386, M7297, and M7391, was infected with serum WHV that had been collected from woodchuck F6693 during acute infection, when the viremia reached the highest levels (week +7 after inoculation) (Fig. 1). The other three groups were infected with serum WHV collected at the chronic stage of WHV infection. Group 2 (M7393, F7238, and M7379) was infected with WHV harvested from woodchuck M1001 during early chronic infection, at a time when WHV-induced HCC had not yet been developed (Fig. 1). Group 3 (M7296, F7237, and F7384) also was infected with serum WHV collected from M1001. However, this third type of WHV inoculum was harvested during the late stage of chronic infection, when terminal HCC was developed (Fig. 1). Woodchucks of group 4 (M7392, F7394, and M7249) were inoculated with serum WHV obtained from woodchuck F1017 late in chronic infection, when the established HCC was terminal. All four different types of WHV inoculum were related, because they were obtained from woodchucks that had been inoculated in the first place with the standardized WHV7 inoculum, cWHV7P2 (Fig. 1) (24). All four groups of infected woodchucks were monitored for 17.5 weeks postinoculation. The liver tissue samples for analysis were collected at week −2 prior to inoculation, weeks +5, +9, and +13 postinoculation, and at necropsy (week +18). Blood was drawn on a weekly basis, including the samples collected prior to infection.
FIG 1.
Groups of naive adult woodchucks infected with four different types of WHV inoculum obtained during either acute or chronic infection. The figure depicts the outline of the current study. Four different types of WHV inocula were used. All four types were related, because they had been harvested from woodchucks that originally were infected with standardized inoculum cWHV7P2, which was derived from original standardized inoculum cWHV7P1 (5). Twelve naive adult woodchucks were used for infections. Each animal was inoculated with 1.66 × 108 genome equivalents (GE) of WHV. Group 1 consisted of woodchucks F7386, M7297, and M7391. These animals were inoculated with serum WHV harvested from woodchuck F6693 during acute infection at the high-viremia phase (at week +7 postinoculation). The time course of acute infection in F6693 depicting the kinetics of serum rcDNA values over 14 weeks postinoculation is shown in the inset. The arrow indicates the time of harvesting the first type of WHV inoculum used in the current study. The other three types of WHV inoculum were obtained from woodchucks during the chronic stage of WHV infection. Group 2 included animals M7393, F7238, and M7379, which were inoculated with serum WHV collected from woodchuck M1001 during early chronic infection (12 months postinoculation), when HCC was not yet developed. The woodchucks of group 3 (M7296, F7237, and F7384) were inoculated with serum WHV obtained from M1001 during late chronic infection (17 months postinoculation), when terminal WHV-induced HCC was already developed. Group 4 included woodchucks M7392, F7394, and M7249. These woodchucks were infected with serum WHV, obtained from woodchuck F1017 late in chronic infection, when developed HCC was terminal (16 months postinoculation).
WHV rcDNA, WHsAg, and antibodies against surface antigens (anti-WHsAg antibodies) in serum samples.
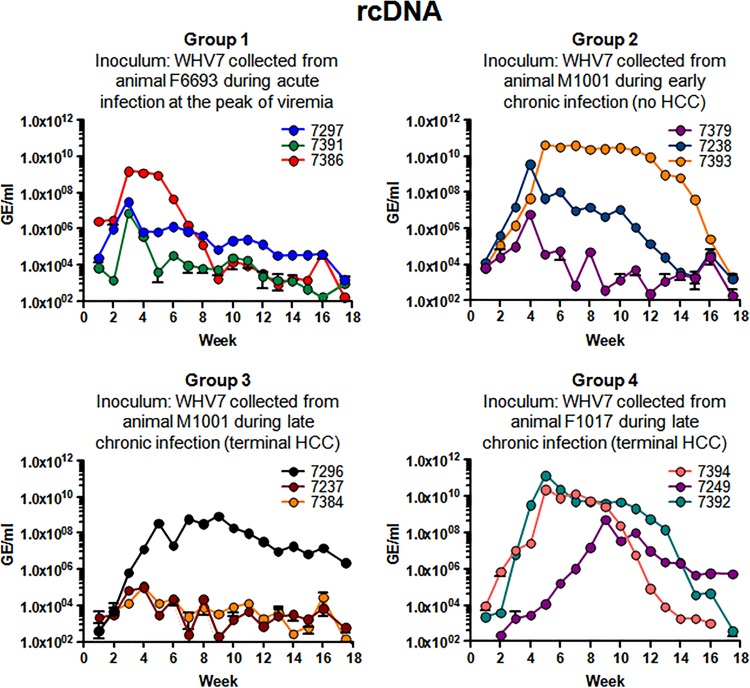
The kinetics of the accumulation of WHV rcDNA in sera following inoculation of the animals in groups 1 to 4 is presented in Fig. 2. The amounts of rcDNA were quantified using a real-time PCR assay (qPCR) as described previously (16, 19). No rcDNA was detected in serum samples collected prior to inoculation (not shown). WHV rcDNA was detected in serum samples obtained from animals infected with the virus collected during acute WHV infection (group 1) beginning at week +1. Although all three animals became WHV positive, none of them demonstrated a relatively long period of very high viremia (Fig. 2). Only for this group of animals were the highest titers observed as early as week +3. The highest concentrations of rcDNA were measured for F7386. The rcDNA titer at week +3 was 1.54 × 109 GE/ml. It remained at about 1.0 × 109 GE/ml for the next 2 weeks, then went down to 1.78 × 103 GE/ml by week +9, and eventually reached the level of 1.72 × 102 GE/ml at week +18. The other two woodchucks had a peak of rcDNA accumulation at week +3 as well. The highest rcDNA concentrations observed were 3.15 × 107 GE/ml for M7297 and 7.61 × 106 GE/ml for M7391. After week +3, the titers of WHV were declining for both M7297 and M7391 and reached very low levels (below 1.60 × 103 GE/ml) at week +18. Woodchuck F7386 had the widest peak of viremia in this group, which was observed between weeks +3 and +6 postinoculation (Fig. 2).
FIG 2.
Serum titers of WHV (rcDNA profiles). All four groups of infected animals are described in detail in the legend to Fig. 1 and in Materials and Methods. The data are arranged by groups. For each group, the description of WHV inoculum used is provided. The rcDNA profile for each animal is displayed using a unique color. The color codings representing individual woodchucks are indicated in each panel. The isolation of rcDNA and titer measurements using qPCR are described in Materials and Methods. The time, in weeks postinoculation, is shown on the x axis of each graph. For each panel, the y axis represents the titer values (rcDNA concentrations) using a logarithmic scale, starting at 1.0 × 102 WHV GE/ml. The standard deviations are indicated.
In group 2, the most productive acute infection was developed by woodchuck M7393. Viremia peaked at week +5 (4.25 × 1010 GE/ml). The titers were relatively high (between 4.25 × 1010 GE/ml and 4.03 × 107 GE/ml) until week +15. Between weeks +5 and +11, the concentrations of rcDNA were above 2.00 × 1010 GE/ml. The other two woodchucks displayed peaks of viremia by week +4. For F7238, a profile of moderate viremia was observed. After week +4, the titers of F7238 were declining from 3.60 × 109 GE/ml and finally were measured at the level of 1.57 × 103 GE/ml at week +18. Between weeks +5 and +18, rcDNA concentrations for M7393 and F7238 were higher than those of M7379. For most of the time, M7379 demonstrated rcDNA numbers below 9.84 × 104 GE/ml (this level was observed at week +3). The only exception was week +4, at which a titer of 6.26 × 106 GE/ml was measured. For all three animals, the titers declined below 2.10 × 103 GE/ml by week +18 (Fig. 2).
In group 3, the most efficient acute infection profile was observed for woodchuck M7296. Viremia became relatively high by week +5 (3.54 × 108 GE/ml), reached the highest level (8.83 × 108 GE/ml) at week +9, and then gradually declined to 2.31 × 106 GE/ml at week +18, which was considered a quite moderate level of rcDNA. The other two animals did not develop a productive infection, and their rcDNA titers were below 1.20 × 105 GE/ml for the entire observation period after inoculation. By week +18, their titers were below 6.10 × 102 GE/ml (Fig. 2).
All three animals in group 4 developed productive acute infection profiles with relatively high rcDNA titers. M7392 and F7394 displayed similar viremia patterns between weeks +1 and +9. For both animals, viremia peaked at week +5. The titers observed at this time point were 1.41 × 1011 GE/ml (M7392) and 2.37 × 1010 GE/ml (F7394). For animal F7394, the titers began to decline progressively starting at week +10, while for M7392, the decline was somewhat delayed. For F7394, rcDNA could not be detected by week +18, while for M7392, the rcDNA level at this time point was 3.92 × 102 GE/ml. In the case of M7249, the viremia peaked relatively late at week +9 (5.22 × 108 GE/ml) and then gradually decreased to a moderate level of 5.48 × 105 GE/ml at week +18 (Fig. 2).
Interestingly, as can be judged by the kinetics of WHV rcDNA in serum samples harvested over the course of 17.5 weeks postinoculation, the virions collected during chronic WHV infection (groups 2, 3, and 4) overall performed better than virions produced during acute infection (group 1) in terms of induction of productive acute infection in naive adult woodchucks, which resulted in relatively high and long-lasting viremia (Fig. 2). Clearly, there were individual variabilities in viremia between infected woodchucks. The observation that some animals infected with the same inoculum did not develop infection with relatively high viremia is not unexpected. Similar observations were reported previously for woodchucks infected with standardized WHV7 inoculum (24).
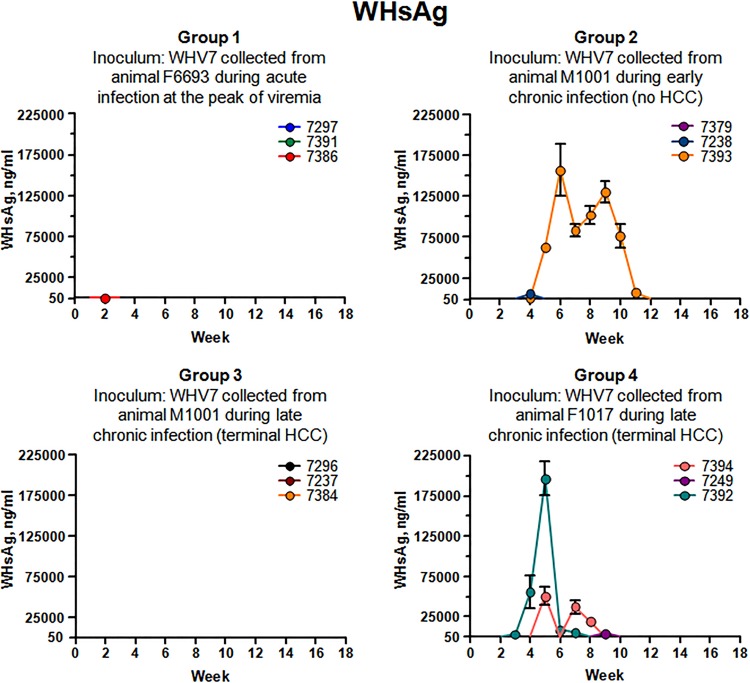
The accumulation of serum WHsAg was analyzed next. The results of the measurements are summarized in Fig. 3. Interestingly, regardless of the levels of rcDNA in sera (Fig. 2), the levels of WHsAg stayed below detection limits of ELISA (i.e., 50 ng/ml) for animals of groups 1 and 3 for the entire observation period. The only exception was woodchuck F7386, for which only at a single time point, at week +2 postinoculation, a concentration of 345 ng/ml of WHsAg was measured. For group 2, considerable WHsAg accumulation was observed only for M7393. The amounts of WHsAg were within the range from 312 ng/ml to 160,000 ng/ml, with the highest concentration of 156,758 ng/ml at week +6. The levels of WHsAg were above 62,000 ng/ml between weeks +5 and +10. For M7379, WHsAg was undetectable, while for F7238, 5,743 ng/ml of WHsAg was detected only at week +4 (Fig. 3). In group 4, a sharp peak of WHsAg accumulation was observed only for M7392 between weeks +3 and +6, with a maximum concentration of 196,333 ng/ml at week +5. For M7249, WHsAg (4,184 ng/ml) was measured only once at week +9. For F7394, WHsAg was detectable at weeks +5, +7, and +8, with a maximum of 50,674 ng/ml at week +5 (Fig. 3). Overall, the levels of serum rcDNA and concentrations of serum WHsAg apparently did not correlate well. However, in some cases, relatively high concentrations of WHsAg were detectable for infection profiles with high viremia. This was the case for M7393 and M7392. The same tendency, however, was not observed for M7249, M7296, and F7394 (Fig. 2 and 3). It became apparent that in different woodchucks (even for those infected with the same type of WHV inoculum, such as group 4 animals, for example), the ratio of virions/subviral particles varied significantly and appeared not to be determined by the characteristics of the inoculum.
FIG 3.
Accumulation of WHsAg in serum samples. The groups of infected woodchucks are described in the legend to Fig. 1 and in Materials and Methods. The data are arranged by groups. Each panel corresponds to one group. For each panel, the group number is indicated and a description of the inoculum used is provided. The color codings for individual woodchucks are shown in each panel. The time after inoculation is indicated in weeks on each x axis. The concentration of WHsAg is shown on each y axis in nanograms of WHsAg per milliliter of serum. The assay for quantification of WHsAg is described in Materials and Methods. The cutoff value of the ELISA for detection of WHsAg was 50 ng/ml. The standard deviations are indicated.
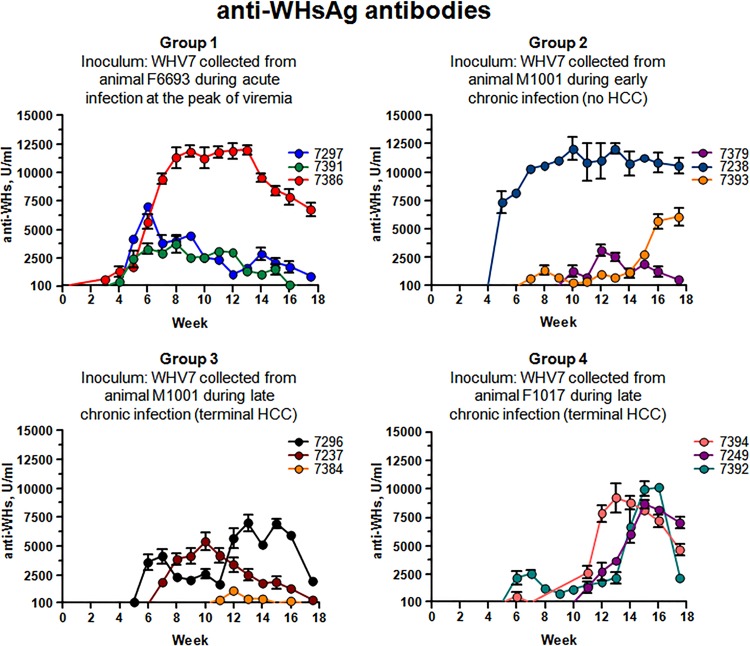
The analysis was continued further by measuring the levels of anti-WHsAg antibodies in sera of infected animals. The production of anti-WHsAg antibodies was detected for each of the infected woodchucks. The results of quantification of the levels of anti-WHsAg antibodies are presented in Fig. 4. The timing and the extent of the observed B-cell-mediated adaptive immune responses were diverse. For example, significant levels of anti-WHsAg antibodies were produced in response to infection in woodchuck F7386, which developed the most productive acute infection within group 1 (Fig. 2 and 4). The antibody response observed in woodchuck F7386 was the most pronounced among the animals in group 1. The amounts of anti-WHsAg antibodies in sera were above 10,000 U/ml between weeks +8 and +13. This observation may explain that only relatively small amounts of WHsAg were detected at just one time point (Fig. 3 and 4). For this woodchuck, a significant increase in the accumulation of serum antibodies may at least in part be accountable for the corresponding decline in viral titer values after week +7 postinoculation (Fig. 2 and 4). For the other two animals in group 1, anti-WHsAg antibodies were readily detectable starting at week +5, while their levels did not measure up to those of F7386. For these woodchucks (M7297 and M7391), the concentrations of the antibodies declined along with the decline in WHV titers (Fig. 2 and 4). For animal F7238 of group 2, the relationship between WHsAg and anti-WHsAg antibodies was somewhat similar to that of F7386 (Fig. 3 and 4). This woodchuck demonstrated the most efficient antibody accumulation within group 2, with levels above 10,000 U/ml between weeks +7 and +18. However, unlike F7386, animal F7238 maintained considerable moderate rcDNA titers for the large part of the monitoring period, regardless of the very high levels of anti-WHsAg antibodies. Only at later times, a decline in WHV titers correlated with significant accumulation of the antibodies (Fig. 2 and 4). In another situation, while considerable amounts of WHsAg were detected for M7393 (group 2) between weeks +5 and +10, there was no significant antibody response detected within the same time period (Fig. 3 and 4). The antibody concentration was rising at later times and reached levels above 5,000 U/ml by week +16, which could facilitate the considerable decline in rcDNA titers observed at the same time (Fig. 2 and 4). In group 2, the weakest antibody response was observed for M7379, which developed the least productive WHV infection (Fig. 2 and 4). The highest concentrations of anti-WHsAg antibodies among the woodchucks of group 3 were observed for M7296 starting at week +12. As noted above, this animal developed the most productive acute WHV infection within the group. However, the antibodies' levels did not exceed 8,000 U/ml and did not exhibit a profound effect on the kinetics of serum rcDNA (Fig. 2 and 4). The production of anti-WHsAg antibodies, however, may have participated in the control of WHV infection in woodchuck F7237 (Fig. 2 and 4). For group 4, the situation was rather more uniform. For all three animals, the antibody response was on the rise after week +12. This was in agreement with the observation that WHsAg was detected only at early times (with much higher WHsAg levels for F7394 and M7392 than for M7249) (Fig. 3 and 4). For M7249 and M7392, the highest antibody production was during weeks +15 and +16. For F7394, the peak of antibody accumulation was approximately between weeks +12 and +16. At these times, the levels of antibodies were above 7,000 U/ml (Fig. 4). In this group, the decrease of serum WHV titers to a certain extent coincided with mounting antibodies levels in sera. However, despite significant concentrations of anti-WHsAg antibodies, WHV titers of M7249 displayed a plateau-like pattern with moderate rcDNA values between weeks +15 and +18 (Fig. 2 and 4). Overall, the relationship between the production of anti-WHsAg antibodies and levels of serum WHsAg and rcDNA titers appears to be rather complex, suggesting that in agreement with previously published findings (25), the timing and magnitude of the adaptive immune response are critical in regulation of the outcomes of hepadnavirus infection.
FIG 4.
Accumulation of anti-WHsAg antibodies in serum samples. The woodchucks and infections are described in Materials and Methods and in the legend to Fig. 1. The results are organized by groups. In each panel, the group number is shown and the inoculum used is described. For each woodchuck, the data are displayed using a unique color. The color coding for each woodchuck is indicated. On the x axis, the time after inoculation is shown in weeks. The y axis represents the scale for the amounts of anti-WHsAg antibodies in units per milliliter of serum. The measurements of anti-WHsAg are described in Materials and Methods. The cutoff value of the ELISA for detection of anti-WHsAg antibodies was 100 U/ml. The standard deviations are indicated.
Expansion and contraction of the virus replication space in infected livers.
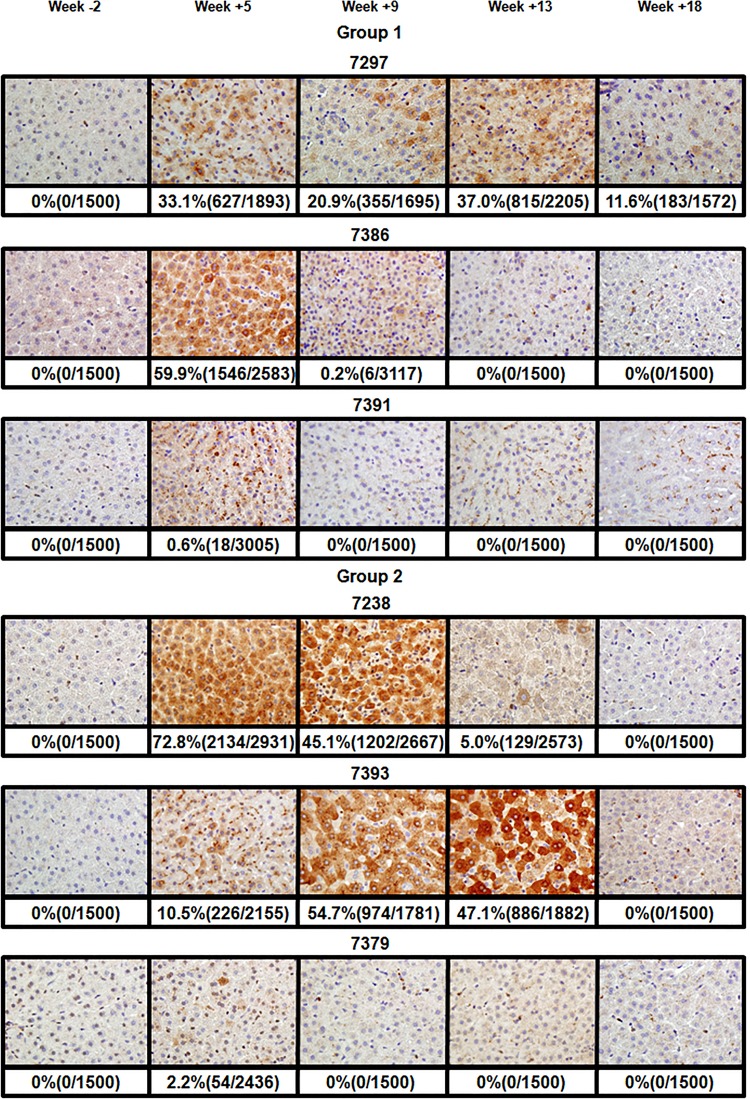
The spread of the virus through the liver, as well as expansion and contraction of the virus replication space, was analyzed by quantifying the fraction of hepatocytes positive for WHcAg using immunohistochemistry (19). The results of the staining of the tissues for WHcAg are summarized in Fig. 5 (groups 1 and 2) and Fig. 6 (groups 3 and 4). As expected, for all animals, no core-positive hepatocytes were found in tissues biopsied prior to inoculation (Fig. 5 and 6). In group 1, for animal F7386, which demonstrated the pattern of most productive acute infection in this group, the highest fraction of core-positive hepatocytes, 59.9%, was found at week +5 and corresponded to the peak of viremia. The decline of virus serum titers correlated with the rapid drop in the number of core-positive cells to 0.2% at week +9 and to 0% at week +13 (Fig. 2 and 5). For animal M7391, with the lowest rcDNA titers observed in group 1, a small fraction of WHcAg-positive cells was detected only at week +5 (0.6%). No core-positive cells were detected at other time points. In the case of M7297, the number of core-positive hepatocytes remained between 20 and 37% between weeks +5 and +13 despite developing just a moderate viremia. The somewhat diminished percentage of positive cells at week +9 (20.9%) could be explained by a smaller fraction of positive cells in the particular area of the liver chosen for the biopsy (Fig. 2 and 5). The absence of uniformity of hepadnavirus infection in the livers, which is reflected by clustering of the hepatocytes positive for core antigen, have been noted previously (8, 23, 26). By week +18, the number of positive hepatocytes decreased to about 12% (Fig. 5). In this animal, the relatively moderate rcDNA titers observed between weeks +5 and +13 in the absence of apparent profound loss of core-positive hepatocytes may be explained at least in part by noncytopathic effects of cytokines that suppressed WHV replication (27, 28).
FIG 5.
Analysis of WHcAg-positive hepatocytes in infected livers of woodchucks of groups 1 and 2. The immunostaining for the core antigen of WHV in infected liver tissue samples was done as described previously (19). The core-positive cells displayed characteristic brown staining resulting from the peroxidase reaction. Tissues were collected during biopsies at 2 weeks prior to inoculation, at weeks +5, +9, +13 after inoculation with WHV, and at necropsy at week +18 postinfection. The results are organized by the groups of infected woodchucks. Each row of images represents data for a single woodchuck. The identification number for each woodchuck is shown above the corresponding row of images. One representative image is shown for each time point. The percentages of core-positive hepatocytes along with the total number of positive and counted cells are presented below the corresponding images. The immunostaining and counting of WHcAg-positive cells are described in Materials and Methods.
FIG 6.
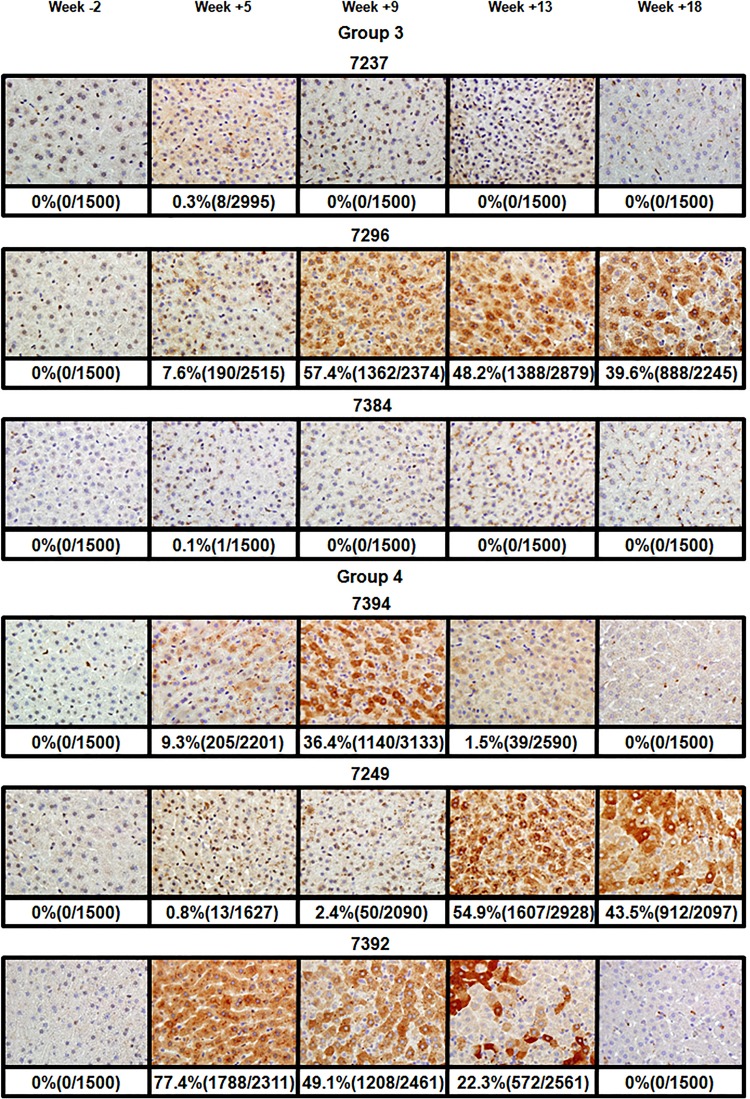
Analysis for WHcAg-positive hepatocytes in infected livers of woodchucks of groups 3 and 4. The immunostaining for WHcAg in infected liver tissues was performed as previously described (19). The WHcAg-positive cells demonstrated characteristic brown staining resulting from the peroxidase reaction. Tissues were harvested during biopsies at 2 weeks prior to infection (week −2), at weeks +5, +9, and +13 after inoculation with WHV, and at necropsy at week +18 postinfection. The data are organized by the groups of infected animals. Each row of images displays results for a single woodchuck. The identification number for each woodchuck is indicated above the corresponding row of images. One representative image is shown per time point. The percentages of WHcAg-positive hepatocytes along with the total number of counted and core-positive cells are shown below the corresponding images. The immunostaining and counting of core-positive cells are described in Materials and Methods.
In group 2, for M7379, which did not develop a productive acute infection with long-lasting viremia, a very small fraction of 2.2% of WHcAg-positive cells was detected only once at week +5. For F7238 (moderately high viremia), core-positive hepatocytes were detected at weeks +5, +9, and +13. The fraction of stained cells declined gradually from 72.8% (week +5) to 45.1% (week +9) and then to 5% at week +13. No positive hepatocytes were found at week +18. These observations were in agreement with the rcDNA accumulation (Fig. 2 and 5). For M7393, which displayed a long-lasting high-titer viremia, the results were a bit surprising. At week +5, only 10.5% of cells stained positive for WHcAg, while the viremia already reached its highest level. The number of positive cells remained significantly high at week +9 (54.7%) and at week +13 (47.1%). However, at week +18, no WHcAg-positive cells were observed. This pattern of core-positive cell percentages was mostly in agreement with the virus titer kinetics in serum samples (Fig. 2 and 5).
In group 3, in agreement with the very low virus titers observed, only a small portion of core-positive cells was found only once at week +5 for F7384 (0.1%) and F7237 (0.3%). For M7296, the percentage of WHcAg-positive cells increased from 7.6% at week +5 to 57.4% at week +9, and later it remained relatively high at levels of 48.2% (week +13) and 39.6% (week +18). Therefore, the highest percentage of core-positive hepatocytes correlated with the highest rcDNA titer at week +9. At later time points the percentage of WHcAg-positive hepatocytes also was in agreement with moderately high virus titers (Fig. 2 and 6).
In group 4, for M7392 (the animal with the longest-lasting high viremia in this group), the number of core-positive cells was already at 77.4% at week +5. It declined to 49.1% (week +9), then to 22.3% (week +13), and finally to 0% at week +18. These numbers were in agreement with the serum rcDNA profile (Fig. 2 and 6). The second most productive viremia in this group was developed by animal F7394 (Fig. 2). Similar to M7393 (group 2), only 9.3% of cells were core positive at week +5, while the highest virus titer was already reached at this point. At week +9, 36.4% of cells were positive. At week +13, this number went down to 1.5% and then to 0% at week +18. Therefore, for woodchuck F7394, most of the time the percentage of WHcAg-positive hepatocytes also correlated with the virus titer values in serum samples (Fig. 2 and 6). Similarly, for the third animal in this group, M7249, the fraction of core antigen-positive cells mostly correlated with the titer measurements. Although the highest rcDNA titer was observed at week +9, the number of WHcAg-positive cells was just 2.4% at that time. This observation was similar to that of M7393 and F7394. Later on, in agreement with the delayed viremia, relatively high numbers of WHcAg-positive cells were found at week +13 (54.9%) and week +18 (43.5%). These numbers were reflective of relatively high virus titers at these late time points (Fig. 2, 5, and 6).
Overall, in groups 1 to 4, most of the time (with the above-described exceptions) the fraction of core-positive cells mostly correlated with the profile of serum rcDNA (Fig. 2, 5, and 6).
Markers of intrahepatic replication of WHV.
The analysis first considered the measurements of intracellular RI-DNA of WHV. The isolation of DNA and quantification of RI-DNA was conducted as described previously (16, 19). The results are presented in Table 1. No RI-DNA of WHV was detected in tissue samples collected prior to infection (not shown). In group 1, the highest values for RI-DNA were measured for week +5, which corresponded to the peak of viremia for all three woodchucks. In agreement with the rcDNA titer kinetics (Fig. 2), the lowest RI-DNA numbers were quantified for M7391. The levels of RI-DNA reached 1.18 × 106 GE/μg of total DNA at week +5 and then declined and stayed within a range between 1.17 × 104 and 3.33 × 104 GE/μg of total DNA during the remainder of the monitoring period. For the other two animals, accumulation of RI-DNA was higher and mostly correlated with the serum rcDNA levels and percentage of core-positive hepatocytes (Fig. 2 and 5 and Table 1). For F7386, RI-DNA was at 6.06 × 108 GE/μg of total DNA at week +5 and then gradually declined to a level of 3.29 × 104 GE/μg of total DNA by week +18. In the case of M7297, RI-DNA levels stayed between 1.71 × 107 and 4.36 × 107 GE/μg of total DNA, with little variations between weeks +5 and +13, and then decreased to 3.97 × 105 GE/μg of total DNA at week +18 (Table 1).
TABLE 1.
Quantity of WHV RI-DNA at different time points postinoculationa
| Group-woodchuckb | RI-DNA (GE/μg) level by week postinoculation |
|||
|---|---|---|---|---|
| +5 | +9 | +13 | +18 | |
| 1-F7386 | (6.06 ± 2.25) × 108 | (2.61 ± 1.08) × 106 | (1.05 ± 0.54) × 105 | (3.29 ± 1.59) × 104 |
| 1-M7297 | (4.36 ± 1.20) × 107 | (1.75 ± 0.64) × 107 | (1.71 ± 0.65) × 107 | (3.97 ± 2.49) × 105 |
| 1-M7391 | (1.18 ± 0.52) × 106 | (1.70 ± 0.83) × 104 | (1.17 ± 0.25) × 104 | (3.33 ± 2.66) × 104 |
| 2-M7393 | (1.07 ± 0.53) × 108 | (6.82 ± 3.48) × 108 | (6.36 ± 1.34) × 108 | (6.47 ± 2.76) × 105 |
| 2-F7238 | (1.77 ± 0.67) × 108 | (9.75 ± 3.76) × 107 | (4.63 ± 2.47) × 106 | (2.51 ± 0.82) × 104 |
| 2-M7379 | (3.41 ± 1.91) × 106 | (3.19 ± 1.33) × 104 | (1.10 ± 0.48) × 104 | (2.33 ± 1.69) × 104 |
| 3-M7296 | (2.07 ± 0.94) × 107 | (1.91 ± 0.63) × 108 | (1.58 ± 0.26) × 108 | (1.07 ± 0.19) × 108 |
| 3-F7237 | (6.51 ± 3.30) × 104 | (3.95 ± 0.76) × 103 | (4.60 ± 2.45) × 102 | (1.74 ± 2.65) × 103 |
| 3-F7384 | (3.08 ± 0.11) × 104 | (3.06 ± 2.55) × 102 | (2.03 ± 0.96) × 103 | (3.75 ± 2.92) × 103 |
| 4-M7392 | (4.51 ± 2.57) × 108 | (1.70 ± 0.36) × 108 | (2.69 ± 0.88) × 108 | (3.24 ± 1.16) × 105 |
| 4-F7394 | (2.42 ± 1.83) × 107 | (6.29 ± 2.60) × 108 | (9.15 ± 2.47) × 105 | (4.51 ± 1.90) × 104 |
| 4-M7249 | (1.04 ± 0.66) × 104 | (5.84 ± 2.43) × 106 | (1.29 ± 0.54) × 108 | (7.23 ± 4.06) × 107 |
The DNA isolation procedure and qPCR assay used for measurements of RI-DNA in collected liver tissue samples are described in Materials and Methods. The RI-DNA values are expressed in WHV genome equivalents (GE) per microgram of total DNA (± standard deviations). The results of measurements of DNA concentrations in total DNA preparations were used for calculations.
Groups of animals, woodchucks used for inoculation, and collection of tissue samples for analysis are detailed in Materials and Methods.
In group 2, RI-DNA numbers were in agreement with rcDNA titer profiles (Fig. 2 and Table 1). As expected, the highest RI-DNA levels were measured for M7393. Between weeks +5 and +13, the numbers ranged between 1.07 × 108 GE/μg of total DNA and 6.82 × 108 GE/μg of total DNA. Only at week +18 did the RI-DNA level decline to 6.47 × 105 GE/μg of total DNA. For F7238, RI-DNA values reached 1.77 × 108 GE/μg of total DNA by week +5 and then gradually decreased to a level of 2.51 × 104 GE/μg of total DNA at week +18. For M7379, as anticipated, the numbers were the lowest in the group, and only at week +5 did they reach 3.41 × 106 GE/μg of total DNA. At all other time points RI-DNA levels were below 3.20 × 104 GE/μg of total DNA (Table 1).
Regardless of the fact that for most of the time the rcDNA titer values for M7296 (group 3) were lower than those of M7393 (group 2) (Fig. 2), the intracellular accumulation of RI-DNA in the liver of M7296 was relatively high and somewhat comparable to that of M7393. The RI-DNA level was 2.07 × 107 GE/μg of total DNA at week +5 and then remained between 1.07 × 108 and 1.91 × 108 GE/μg of total DNA between weeks +9 and +18. These findings were in agreement with the observation that for M7296, the rcDNA titer at week +18 was higher than that of M7393 (Fig. 2 and Table 1). As anticipated, the rest of the animals in group 3 displayed low RI-DNA levels between 3.06 × 102 and 6.51 × 104 GE/μg of total DNA, with the highest values measured at week +5. The observed variations in RI-DNA values, like decreased levels for F7384 at week +9 (compared to that of week +13) and for F7237 at week +13 (compared to that of week +18), could be due to the areas of the liver used for biopsy specimens and also to the facts that hepadnavirus infection of the liver is not uniform (8, 23, 26) and that very small numbers of core-positive hepatocytes were observed for both of these woodchucks during the entire monitoring period (Table 1 and Fig. 6).
In group 4, for M7392, RI-DNA levels stayed between 1.70 × 108 GE/μg and 4.51 × 108 GE/μg of total DNA from week +5 until week +13 (regardless of the significant drop in rcDNA levels [Fig. 2]) and only then decreased to 3.24 × 105 GE/μg of total DNA by week +18. This RI-DNA profile was similar to that of M7393 (group 2). In the case of F7394, RI-DNA reached a level above 1.00 × 108 GE/μg of total DNA (i.e., 6.29 × 108 GE/μg of total DNA) only by week +9. This observation correlated with the fraction of core-positive hepatocytes but did not correlate with rcDNA profile, because the highest rcDNA values were achieved already by week +5. Thereafter, a rather significant decrease of RI-DNA numbers to 9.15 × 105 GE/μg of total DNA (week +13) and to 4.51 × 104 GE/μg of total DNA (week +18) was observed, which was in agreement with the significant decrease of (i) serum rcDNA levels and (ii) percentages of core-positive cells later in infection (Fig. 2 and 6 and Table 1). For M7249, in correlation with the portion of WHcAg-positive cells, RI-DNA levels reached 1.29 × 108 GE/μg of total DNA by week +13 and only slightly decreased to 7.23 × 107 GE/μg of total DNA by week +18. The latter finding also was somewhat in agreement with the observation that at late times (weeks +13 through +18), the titers of serum rcDNA ranged between 4.58 × 105 GE/ml and 2.25 × 106 GE/ml (Fig. 2 and 6 and Table 1).
The next replication marker analyzed was cccDNA of WHV (Table 2). In group 1, in correlation with the viremia pattern (Fig. 2), for animal F7386, the highest cccDNA value was measured at week +5 (1.64 × 105 GE/μg of total DNA). At the other time points, a dramatic decline was observed, resulting in cccDNA levels being below 5.60 × 102 GE/μg of total DNA during weeks +9 to +18. For M7297, at weeks +5 and +9, cccDNA was measured in a narrow range between 1.55 × 104 and 1.65 × 104 GE/μg of total DNA. For the rest of the monitoring period, cccDNA levels were below 7.70 × 103 GE/μg of total DNA. As expected, the lowest cccDNA numbers, below 3.35 × 103 GE/μg of total DNA, were observed for M7391. The cccDNA profiles generally followed the kinetics of rcDNA titers (Fig. 2 and Table 2).
TABLE 2.
Quantity of WHV cccDNA at different time points postinoculationa
| Group-woodchuckb | cccDNA (GE/μg) level by week postinoculation |
|||
|---|---|---|---|---|
| +5 | +9 | +13 | +18 | |
| 1-F7386 | (1.64 ± 0.80) × 105 | (5.50 ± 3.56) × 102 | (5.59 ± 3.49) × 102 | (3.27 ± 0.36) × 102 |
| 1-M7297 | (1.65 ± 0.79) × 104 | (1.55 ± 1.06) × 104 | (7.69 ± 3.11) × 103 | (4.99 ± 2.51) × 103 |
| 1-M7391 | (2.26 ± 1.03) × 103 | (1.96 ± 1.36) × 103 | (1.76 ± 0.84) × 103 | (3.30 ± 1.69) × 103 |
| 2-M7393 | (4.84 ± 0.21) × 105 | (4.36 ± 2.35) × 105 | (7.10 ± 3.29) × 105 | (2.97 ± 0.97) × 102 |
| 2-F7238 | (3.06 ± 2.35) × 105 | (4.43 ± 3.99) × 104 | (1.40 ± 0.85) × 104 | (2.59 ± 0.63) × 102 |
| 2-M7379 | (8.19 ± 2.42) × 104 | (2.66 ± 0.17) × 103 | (3.52 ± 0.08) × 103 | (5.22 ± 1.46) × 103 |
| 3-M7296 | (1.32 ± 0.95) × 105 | (1.76 ± 1.48) × 105 | (9.79 ± 5.55) × 104 | (1.54 ± 0.22) × 105 |
| 3-F7237 | (5.35 ± 1.13) × 101 | (1.37 ± NA) × 101 | (3.23 ± 1.98) × 101 | (4.08 ± 1.56) × 101 |
| 3-F7384 | (2.63 ± 1.23) × 101 | (6.10 ± 1.62) × 101 | (1.63 ± 0.11) × 101 | (5.55 ± 2.95) × 101 |
| 4-M7392 | (6.68 ± 5.53) × 105 | (2.39 ± 1.73) × 105 | (3.24 ± 0.82) × 105 | (3.43 ± 0.51) × 102 |
| 4-F7394 | (1.75 ± 1.31) × 105 | (1.97 ± 0.01) × 106 | (2.54 ± 1.11) × 103 | (5.46 ± 1.02) × 102 |
| 4-M7249 | (1.80 ± NA) × 101 | (2.49 ± 1.08) × 104 | (1.13 ± 0.07) × 105 | (3.75 ± 2.09) × 104 |
The DNA isolation procedure and qPCR assay used for measurements of cccDNA in liver tissue samples are described in Materials and Methods. The cccDNA levels are shown in WHV genome equivalents (GE) per microgram of total DNA (± standard deviations). The results of measurements of DNA concentrations in the corresponding total DNA preparations were used for calculations. NA stands for not applicable, and it was used on only two occasions. For each of these occasions, the cccDNA level was quantified during only one of the independent repeats, so that standard deviation values could not be established.
Groups of animals, woodchucks used for inoculation, and harvesting of liver tissue samples are detailed in Materials and Methods.
The tendency of correlation with rcDNA profiles was observed for groups 2, 3, and 4 as well. As anticipated, in group 2, the highest cccDNA values were observed for M7393. They were above 4.35 × 105 GE/μg of total DNA during weeks +5 to +13 and then went down to 2.97 × 102 GE/μg of total DNA. For F7238, the level of 3.06 × 105 GE/μg of total DNA was measured at the peak of viremia (week +5). At later times, cccDNA accumulation decreased to below 4.45 × 104 GE/μg of total DNA for weeks +9 and +13 and to 2.59 × 102 GE/μg of total DNA at week +18. For M7379, cccDNA levels were below 5.25 × 103 GE/μg of total DNA, with the exception of week +5, for which a level of 8.19 × 104 GE/μg of total DNA was observed (Fig. 2 and Table 2).
In group 3, as expected, considerable cccDNA accumulation between 9.79 × 104 GE/μg of total DNA and 1.76 × 105 GE/μg of total DNA was observed for the entire monitoring period only for M7296. The other two infected woodchucks displayed cccDNA levels below 6.15 × 101 GE/μg of total DNA (Table 2).
In group 4, similar to RI-DNA numbers (Table 1), the pattern of cccDNA accumulation for M7392 was similar to that of M7393 (group 2). The levels of cccDNA were within a narrow range between 2.39 × 105 GE/μg of total DNA and 6.68 × 105 GE/μg of total DNA for weeks +5 through +13, with a subsequent decline to 3.43 × 102 GE/μg of total DNA at week +18. For F7394, considerable cccDNA accumulation was found at week +5 (1.75 × 105 GE/μg of total DNA) and week +9 (1.97 × 106 GE/μg of total DNA). The cccDNA levels then declined to 5.46 × 102 GE/μg of total DNA at week +18. As expected, for M7249, the accumulation of cccDNA displayed a delay earlier and then reached a relatively high level of 1.13 × 105 GE/μg of total DNA at week +13. Later, it somewhat declined to 3.75 × 104 GE/μg of total DNA at week +18. In this case, at week +9, the accumulation of cccDNA apparently still was behind the rise of rcDNA levels (Fig. 2 and Table 2). Overall, a good level of correlation was observed between the profiles of rcDNA, RI-DNA, and cccDNA for groups 1 to 4 (Fig. 2 and Tables 1 and 2).
The levels of pgRNA of WHV were evaluated next (Table 3). No pgRNA of WHV was detected in the tissues collected prior to infection (not shown). The pgRNA numbers generally followed tendencies similar to those of WHV DNA replication markers (Tables 1 to 3). The accumulation profiles of pgRNA also mostly correlated with the kinetics of rcDNA in sera (Fig. 2 and Table 3). Thus, in group 1, the accumulation of pgRNA in all woodchucks peaked at week +5, in agreement with the peaks of viremia. For F7386, the numbers gradually decreased from 5.14 × 108 GE/μg of total RNA (week +5) to 4.41 × 104 GE/μg of total RNA at week +18. For M7297, pgRNA levels declined from 2.63 × 107 GE/μg of total RNA at week +5 to 2.83 × 105 GE/μg of total RNA at week +13. At week +18, the level of pgRNA did not change significantly (i.e., it was about 3.7 times higher than that of week +13). For woodchuck M7391, after the peak at week +5 (2.45 × 106 GE/μg of total RNA), the accumulation of pgRNA relatively quickly declined considerably to the range between 1.87 × 102 and 4.35 × 102 GE/μg of total RNA (Fig. 2 and Table 3).
TABLE 3.
Quantity of WHV pgRNA at different time points postinoculationa
| Group-woodchuckb | pgRNA (GE/μg) level by week postinoculation |
|||
|---|---|---|---|---|
| +5 | +9 | +13 | +18 | |
| 1-F7386 | (5.14 ± 0.71) × 108 | (2.95 ± 1.27) × 105 | (7.89 ± 2.61) × 104 | (4.41 ± 1.09) × 104 |
| 1-M7297 | (2.63 ± 0.71) × 107 | (1.96 ± 1.90) × 107 | (2.83 ± 2.00) × 105 | (1.05 ± 0.81) × 106 |
| 1-M7391 | (2.45 ± 0.01) × 106 | (3.25 ± 1.07) × 102 | (4.35 ± 2.57) × 102 | (1.87 ± 0.42) × 102 |
| 2-M7393 | (3.71 ± 0.91) × 107 | (2.68 ± 1.50) × 108 | (6.23 ± 0.61) × 108 | (1.87 ± 0.16) × 105 |
| 2-F7238 | (1.49 ± 0.41) × 108 | (2.50 ± 1.46) × 108 | (1.54 ± 0.13) × 107 | (7.13 ± 4.24) × 104 |
| 2-M7379 | (8.91 ± 4.38) × 106 | (2.14 ± 1.14) × 104 | (2.71 ± 2.10) × 102 | (3.23 ± 2.64) × 102 |
| 3-M7296 | (6.06 ± 2.12) × 106 | (3.20 ± 0.76) × 108 | (2.08 ± 1.41) × 108 | (2.75 ± 0.82) × 108 |
| 3-F7237 | (7.50 ± 0.67) × 104 | (1.76 ± 0.95) × 101 | (1.97 ± 0.12) × 101 | (2.32 ± 1.03) × 101 |
| 3-F7384 | (7.37 ± 1.49) × 104 | (1.99 ± 0.33) × 102 | (5.08 ± 0.23) × 101 | (1.79 ± 2.15) × 103 |
| 4-M7392 | (1.22 ± 0.95) × 108 | (1.64 ± 0.28) × 108 | (2.78 ± 0.48) × 108 | (5.06 ± 0.27) × 104 |
| 4-F7394 | (2.33 ± 0.84) × 107 | (1.21 ± 0.42) × 109 | (1.03 ± 0.36) × 106 | (2.40 ± 0.75) × 104 |
| 4-M7249 | (1.11 ± 0.02) × 103 | (5.96 ± 2.14) × 106 | (3.57 ± 0.39) × 108 | (2.20 ± 0.57) × 107 |
The isolation of RNA and qPCR assay used for measurements of pgRNA in harvested liver tissue samples are described in Materials and Methods. The measured pgRNA values are expressed in WHV genome equivalents (GE) per microgram of total RNA (± standard deviations).
Groups of animals, woodchucks used for inoculation, and collection of liver tissue samples for subsequent analysis are detailed in Materials and Methods.
As expected, group 2 performed better than group 1. For M7393, pgRNA numbers were above 2.65 × 108 GE/μg of total RNA at weeks +9 and +13 and then decreased considerably to 1.87 × 105 GE/μg of total RNA at week +18. For F7238, regardless of the lower viremia compared to that of M7393 (Fig. 2), pgRNA levels remained relatively high between 1.54 × 107 GE/μg of total RNA and 2.50 × 108 GE/μg of total RNA between weeks +5 and +13 and then decreased significantly to 7.13 × 104 GE/μg of total RNA at week +18. The pgRNA levels for M7379 were the lowest, with the highest number of 8.91 × 106 GE/μg of total RNA observed at week +5 and with the lowest values within the range from 2.71 × 102 to 3.23 × 102 GE/μg of total RNA between weeks +13 and +18 (Table 3).
As anticipated, in group 3, the highest pgRNA values were measured for M7296. At week +5, pgRNA only reached 6.06 × 106 GE/μg of total RNA. pgRNA levels then considerably increased and stayed within the range between 2.08 × 108 and 3.20 × 108 GE/μg of total RNA for the rest of the monitoring period. This profile of pgRNA accumulation was in agreement with the observed percentages of the core-positive hepatocytes at different time points postinoculation and with the rcDNA titers (Fig. 2 and 6 and Table 3). For F7237 and F7384, the pgRNA levels were much lower at all times and did not exceed 7.55 × 104 GE/μg of total RNA, with the highest values observed at week +5 (Table 3).
In group 4, for animal M7392, levels of pgRNA were considerably high (within the range from 1.22 × 108 to 2.78 × 108 GE/μg of total RNA) between weeks +5 and +13 and then declined by week +18 to 5.06 × 104 GE/μg of total RNA. For woodchuck F7394, pgRNA reached a peak of 1.21 × 109 GE/μg of total RNA at week +9, which was the highest level of pgRNA observed in this study. Later on, the amounts of pgRNA decreased to 2.40 × 104 GE/μg of total RNA by week +18, which was similar to that of M7392. Similar to the RI-DNA and cccDNA profiles, in the case of M7249, the level of pgRNA was low (1.11 × 103 GE/μg of total RNA) at week +5, reached a peak at week +13 (3.57 × 108 GE/μg of total RNA), and remained relatively high (2.20 × 107 GE/μg of total RNA) at week +18 (Tables 1 to 3).
Overall, for the cases when a relatively productive acute infection was developed, the accumulation of RI-DNA, cccDNA, and pgRNA in the livers of animals infected with WHV inocula collected during chronic infection was either similar to or often was higher than that of woodchucks inoculated with the virus obtained during acute WHV infection (i.e., compare woodchucks M7393, F7238, M7296, F7394, M7249, and M7392 to F7386 and M7297) (Tables 1 to 3).
DISCUSSION
This study compared the profiles of acute WHV infection induced in livers of naive adult woodchucks that were inoculated with serum WHV harvested either during the acute or chronic stages of WHV infection. Four different types of WHV inoculum were tested. Group 1 was infected with the virus collected at the peak of viremia during the acute phase of infection of naive adult woodchuck. The other three types of inoculum were harvested from adult woodchucks during chronic WHV infection. Two types of WHV inoculum were from the same animal but were collected either during early chronic infection prior to development of WHV-induced HCC or late in chronic infection when developed HCC was terminal. The fourth type of WHV inoculum was collected from a different woodchuck during late chronic infection at the time of development of terminal HCC. It is important to emphasize that all four types of inoculum were related, because they all were collected from woodchucks infected with standardized WHV7 inoculum (cWHV7P2), which was derived from original standardized inoculum cWHV7P1 (Fig. 1) (5, 24). All animals were inoculated with the same amount of WHV GE per animal. Under these conditions, the profiles of induced WHV infections should be reflective of the overall infectivity of WHV virions in inocula. Therefore, the infectivity of virions produced at the acute phase of infection could be adequately compared to the infectivity of virions accumulated either during the early or late stages of chronic infection.
The infected woodchucks were monitored for 17.5 weeks postinoculation. The serum samples were obtained weekly, whereas liver tissue samples were collected at five different time points (at weeks −2, +5, +9, +13, and +18). The infection profiles were compared by measuring serum concentrations of rcDNA; intrahepatic accumulation of RI-DNA, cccDNA, and pgRNA; as well as the numbers of WHcAg-positive hepatocytes (Fig. 2, 5, and 6 and Tables 1 to 3). The quantification of the parameters described above along with the measurements of the levels of WHsAg and anti-WHsAg antibodies (Fig. 3 and 4) allowed us to obtain a fairly complete impression of induced infection profiles. Generally (with the above-described exceptions), the measured intracellular replication markers of WHV were in agreement with kinetics of serum rcDNA and profiles of expansion/contraction of the infection that were reflected by the detectable fractions of hepatocytes positive for the core antigen of WHV. Detailed analysis of the data presented in Results allowed us to conclude that WHV virions harvested during the early and late stages of chronic infection did not display any significant deficiency in terms of the overall infectivity of virus particles. In fact, based on the above-mentioned quantifications, it became apparent that virions collected during chronic infection were able to induce productive acute WHV infection profiles in naive woodchuck livers with parameters that were either similar to or exceeding those of the infection profiles induced by the virions harvested during the acute phase of WHV infection (Fig. 2, 5, and 6 and Tables 1 to 3).
Obviously, there were variabilities in the infection profiles between individual woodchucks. The fact that a few woodchucks did not develop productive acute infection was not surprising. Previously, similar observations were reported for naive adult woodchucks inoculated with the standardized WHV7 inocula, cWHV7P1 or cWHV7P2 (16, 24). Variability in infection profiles was rather expected given the outbred nature of woodchucks used and the anticipated differences in the immune responses to WHV infection. However, six out of nine woodchucks infected with WHV virions collected during chronic infection displayed profiles of acute infection with relatively high and long-lasting viremia. Each WHV inoculum generated during chronic infection resulted in at least 1 out of 3 infected animals with a clear profile of productive infection. In groups 2 (WHV inoculum was generated during early chronic infection) and 4 (WHV inoculum was harvested during late chronic infection), altogether 5 out of 6 woodchucks demonstrated a productive acute infection with relatively high WHV titers. In group 1, which was infected with WHV collected during acute infection at the peak of viremia (Fig. 1), considerable WHV replication and percentage of core-positive cells usually were observed relatively early within the first half of the monitoring period, although there were two exceptions. First, for woodchuck M7391, no significant fraction of WHV core-positive hepatocytes was detected during the entire observation period. Second, for animal M7297, a significant number of core-positive cells still was seen at week +13. Following the early peak, the infection parameters generally decreased faster than that of animals from the other groups that were able to develop productive acute infection. The reasons for the observed relatively rapid decrease in replication/infection markers in animals inoculated with WHV virions produced during the acute phase of infection are not immediately apparent and could be addressed in follow-up studies. Furthermore, none of the woodchucks of group 1 displayed long-lasting and considerably high viremia (Fig. 2, 5, and 6 and Tables 1 to 3).
In addition, we noted that in group 1, the viremia peaked at week +3 for all three animals, which was an earlier time than for the rest of the infected woodchucks that displayed peaks of viremia at weeks +4, +5, and +9 (Fig. 2). However, if the early peak of viremia could be considered an advantage for the virions collected during the acute infection, in the long run it did not result in any further advantages in terms of development of long-lasting high-viremia profiles and prolonged and significantly high accumulation of other replication markers/high percentages of core-positive hepatocytes (Fig. 2, 5, and 6 and Tables 1 to 3).
Furthermore, it became clear that the levels of acute WHV infection induced by the virions collected during chronic infection in a number of animals described in the current study are comparable to the profiles of productive acute infection previously described for naive adult woodchucks infected with different WHV strains (4, 5, 15, 16, 24). In addition, it is important to emphasize that virions harvested during the early and late stages of chronic WHV infection did not display any profound differences in infectivity (Fig. 2, 5, and 6 and Tables 1 to 3). Therefore, several lines of evidence presented in the current study strongly suggest that virions generated during chronic infection are not defective in terms of overall infectivity and, therefore, should be able to facilitate efficient hepadnavirus transmission if the ability of the hepatocytes to support productive infection (and to facilitate cell-to-cell spread and superinfection) is not altered.
The data generated in the current study are not in agreement with the findings previously published by others that (i) assayed the infectivity of serum HBV virions collected from infected chimpanzee by using the infection of the chimeric mice with transplanted human hepatocytes as a model system and (ii) reported that infectivity of HBV virions harvested at day 244 postinoculation (i.e., longer than 6 months after inoculation) was severely compromised compared to that of virions harvested earlier at day 57 postinoculation (29). The reason for these results was unclear, and no further investigation was conducted to understand the basis of profoundly suppressed infectivity at a relatively late time postinoculation. However, it has been speculated that diminished HBV infectivity could be due to formation of immune complexes between virions and serum antibodies against HBV surface antigens, but no evidence was provided to substantiate this hypothesis (29, 30). To further evaluate our findings and test the hypothesis described above, we conducted additional experiments to determine what fraction of WHV in the inocula used in the current study was in the context of immune complexes with anti-WHsAg antibodies. We found that virions collected during the acute phase of infection contained 5.76% of such immune complexes (used for infection of group 1 animals). The virions harvested during chronic WHV infection actually contained larger amounts of the immune complexes. Thus, virions used for infection of group 2 contained 27.36% of WHV in complexes with anti-WHsAg antibodies. The virions used for infection of group 3 had 45.51% antibody-bound WHV, while WHV used for infection of group 4 contained 22.94% of the virus in the context of the immune complexes. Therefore, our data regarding the infectivity of WHV virions harvested during different stages of WHV infection do not demonstrate an apparent correlation with the relative content of the immune complexes found in the inocula. In fact, based on the content of antibody-bound virions, the virus harvested during the acute infection should be the most infectious, which was not the case (Fig. 2, 5, and 6 and Tables 1 to 3). The only correlation to be suggested could be the expectation of a lesser overall infectivity of the inoculum used for group 3 infection, which had the highest content of the immune complexes. As described above, in group 3, only 1 out of 3 animals developed a productive acute WHV infection, which was the lowest percentage among the three groups of woodchucks that were infected with WHV collected during chronic infection. Our data also are not consistent with another theory suggesting the accumulation of mutations in the virus pool late in infection or a higher rate of replication of hepadnavirus early in the infection (29, 30). All three types of inocula that were produced during chronic hepadnavirus infection and were used in the current study for infection of groups 2, 3, and 4 clearly demonstrated better overall infectivity than the virions collected during the acute phase of infection (Fig. 2, 5, and 6 and Tables 1 to 3). If any of the inoculum that was harvested during chronic infection and tested in the current study contained defective virions, which could have the potential to alter overall infectivity of the inoculum because these virions bore mutated genomes or were not infectious for other reasons, we should have observed (in all animals infected with a particular inoculum) either failure to induce productive acute infection or at least substantially delayed expansion of the virus infection through the infected liver. In the latter case, we would expect to observe delayed infection of a substantial fraction of hepatocytes in the livers and low values of other replication/infection markers for a period of several weeks after the inoculation with a subsequent somewhat “catching-up” stage and development of a relatively efficient infection profile at later times after inoculation. The scenarios described above were not observed in the current study. However, we did not investigate the variants/mutants of WHV that could be present in the inocula used and did not conduct titration experiments, which would examine the overall infectivity of inocula at different multiplicities of infection (MOI). These could be considered limitations of our approach. Although it is reasonable to speculate that if a particular inoculum tested in the current study had a substantial presence of defective virus particles, which could considerably diminish the overall infectivity of the virus pool, we would have obtained results consistent with this assumption in our experimental settings.
Our results are consistent with our earlier report, which (i) included the examination of how the envelope proteins, sequences of which were found in HBV variants obtained during chronic HBV infection, facilitated the infectivity of human hepatitis delta virus (HDV) particles, and (ii) concluded that no sufficient evidence was found to suggest that HBV envelope proteins produced during chronic infection are responsible for the formation of virions, most of which would have diminished infectivity and would not support efficient virus transmission (17). In addition, because in the current study the observed relationship between the timing/levels of the production of anti-WHsAg antibodies and the regulation of acute WHV infection appears to be somewhat complex, future studies examining how the percentage of hepadnavirus virions bound to anti-envelope antibodies influences the overall infectivity of the virions circulating in the blood (which may affect the kinetics of virus expansion through the liver) are warranted.
The results obtained in the present study further advance the understanding of the mechanism of chronic hepadnavirus infection and especially the determinants of the maintenance of the chronic state of the infection. It has been suggested previously (i) that during the chronic stage of hepadnavirus infection, the cell-to-cell spread of hepadnavirus and superinfection are unlikely events, and (ii) that chronic infection is maintained exclusively by the division of hepadnavirus-infected hepatocytes in the absence of virus spread (8–14). However, our recent findings obtained during the study in which woodchucks chronically infected with strain WHV7 were superinfected with another strain, WHVNY, suggested that limited hepadnavirus spread and superinfection do continue during the chronic stage of hepadnavirus infection (15). The presence in infected livers of several categories of hepatocytes that may not actively participate in hepadnavirus replication/infection (i.e., the hepatocytes that are either uninfected, cleared the infection, or experienced profound loss of cccDNA [28, 31, 32]) suggests that the infection/reinfection of these hepatocytes might be needed for the maintenance of the chronic state of hepadnavirus infection (15). Our new data produced in the present study suggest that hepadnavirus virions generated during the early and late stages of chronic infection do not have diminished infectivity compared to that of the virions produced during the acute stage of infection (Fig. 2, 5, and 6 and Tables 1 to 3), and therefore, virions generated during chronic hepadnavirus infection should be capable of infecting susceptible hepatocytes (in chronically infected livers). Therefore, our novel results further extend the recent findings and provide new lines of evidence that support the previously suggested notion that maintenance of chronic hepadnavirus infection could be mediated by a complex mechanism which includes both division of hepadnavirus-infected hepatocytes and hepadnavirus cell-to-cell spread and superinfection (15).
ACKNOWLEDGMENTS
S.O.G. and S.M. were supported by NIH grant R01CA166213. S.O.G. was also supported by NIH grants R21AI097647 and R21AI099696.
We are grateful to the Histopathology and Tissue Shared Resource (HTSR) of Georgetown University for assistance with the staining of woodchuck liver tissues.
REFERENCES
- 1.Dienstag JL. 2008. Hepatitis B virus infection. N Engl J Med 359:1486–1500. doi: 10.1056/NEJMra0801644. [DOI] [PubMed] [Google Scholar]
- 2.Seeger C, Mason WS. 2000. Hepatitis B virus biology. Microbiol Mol Biol Rev 64:51–68. doi: 10.1128/MMBR.64.1.51-68.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lupberger J, Hildt E. 2007. Hepatitis B virus-induced oncogenesis. World J Gastroenterol 13:74–81. doi: 10.3748/wjg.v13.i1.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Menne S, Cote PJ. 2007. The woodchuck as an animal model for pathogenesis and therapy of chronic hepatitis B virus infection. World J Gastroenterol 13:104–124. doi: 10.3748/wjg.v13.i1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cote PJ, Korba BE, Miller RH, Jacob JR, Baldwin BH, Hornbuckle WE, Purcell RH, Tennant BC, Gerin JL. 2000. Effects of age and viral determinants on chronicity as an outcome of experimental woodchuck hepatitis virus infection. Hepatology 31:190–200. doi: 10.1002/hep.510310128. [DOI] [PubMed] [Google Scholar]
- 6.Schaefer S. 2005. Hepatitis B virus: significance of genotypes. J Viral Hepat 12:111–124. doi: 10.1111/j.1365-2893.2005.00584.x. [DOI] [PubMed] [Google Scholar]
- 7.Schaefer S. 2007. Hepatitis B virus taxonomy and hepatitis B virus genotypes. World J Gastroenterol 13:14–21. doi: 10.3748/wjg.v13.i1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mason WS, Litwin S, Xu C, Jilbert AR. 2007. Hepatocytes turnover in transient and chronic hepadnavirus infection. J Viral Hepat 14(Suppl 1):S22–S8. [DOI] [PubMed] [Google Scholar]
- 9.Mason WS, Litwin S, Jilbert AR. 2008. Immune selection during chronic hepadnavirus infection. Hepatol Intl 2:3–16. doi: 10.1007/s12072-008-9051-8,10.1007/s12072-007-9024-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mason WS, Liu C, Aldrich CE, Litwin S, Yeh MM. 2010. Clonal expansion of normal-appearing human hepatocytes during chronic hepatitis B virus infection. J Virol 84:8308–8315. doi: 10.1128/JVI.00833-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mason WS, Low HC, Xu C, Aldrich CE, Scougall CA, Grosse A, Clouston A, Chavez D, Litwin S, Peri S, Jilbert AR, Lanford RE. 2009. Detection of clonally expanded hepatocytes in chimpanzees with chronic hepatitis B virus infection. J Virol 83:8396–8408. doi: 10.1128/JVI.00700-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Litwin S, Toll E, Jilbert AR, Mason WS. 2005. The competing roles of virus replication and hepatocyte death rates in the emergence of drug-resistant mutants: theoretical consideration. J Clin Virol 34(Suppl 1):S96–S107. doi: 10.1016/S1386-6532(05)80018-6. [DOI] [PubMed] [Google Scholar]
- 13.Walters KA, Joyce MA, Addison WR, Fischer KP, Tyrrell DL. 2004. Superinfection exclusion in duck hepatitis virus infection is mediated by large surface antigen. J Virol 78:7925–7937. doi: 10.1128/JVI.78.15.7925-7937.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mason WS, Jilbert AR, Summers J. 2005. Clonal expansion of hepatocytes during chronic woodchuck hepatitis virus infection. Proc Natl Acad Sci U S A 102:1139–1144. doi: 10.1073/pnas.0409332102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodrigues L, Freitas N, Kallakury BV, Menne S, Gudima SO. 2015. Superinfection with woodchuck hepatitis virus strain WHVNY of the livers chronically infected with strain WHV7. J Virol 89:384–405. doi: 10.1128/JVI.02361-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freitas N, Lukash T, Dudek M, Litwin S, Menne S, Gudima SO. Capacity of a natural strain of woodchuck hepatitis virus, WHVNY, to induce acute infection in naive adult woodchucks. Virus Res 205:12–21. doi: 10.1016/j.viruses.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freitas N, Abe K, Cunha C, Menne S, Gudima SO. 2014. Support of the infectivity of hepatitis delta virus particles by the envelope proteins of different genotypes of hepatitis B virus. J Virol 88:6255–6267. doi: 10.1128/JVI.00346-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou T, Saputelli J, Aldrich CE, Deslauriers M, Mason WS. 1999. Emergence of drug-resistant populations of woodchuck hepatitis virus in woodchucks treated with the antiviral nucleoside lamivudine. Antimicrob Agents Chemother 43:1947–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freitas N, Salisse J, Cunha C, Toshkov I, Menne S, Gudima SO. 2012. Hepatitis delta virus infects the cells of hepadnavirus-induced hepatocellular carcinoma in woodchucks. Hepatology 56:76–85. doi: 10.1002/hep.25663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cote PJ, Roneker C, Cass K, Schödel F, Peterson D, Tennant B, De Noronha F, Gerin J. 1993. New enzyme immunoassays for the serologic detection of woodchuck hepatitis virus infection. Viral Immunol 6:161–169. doi: 10.1089/vim.1993.6.161. [DOI] [PubMed] [Google Scholar]
- 21.Liu KH, Ascenzi MA, Bellezza CA, Bezuidenhout AJ, Cote PJ, Gonzalez-Aseguinolaza G, Hannaman D, Luxembourg A, Evans CF, Tennant BC, Menne S. 2011. Electroporation enhances immunogenicity of a DNA vaccine expressing woodchuck hepatitis surface antigen in woodchucks. J Virol 85:4853–4862. doi: 10.1128/JVI.02437-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schüffler PJ, Fuchs TJ, Ong CS, Wild PJ, Rupp NJ, Buhmann JM. 2013. TMARKER: a free software toolkit for histopathological cell counting and staining estimation. J Pathol Inform 4(Suppl):S2. doi: 10.4103/2153-3539.109804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mason W, Xu C, Low HC, Saputelli J, Aldrich CE, Scougall C, Grosse A, Colonno R, Litwin S, Jilbert AR. 2009. The amount of hepatocytes turnover that occurred during resolution of transient hepadnavirus infections was lower when virus replication was inhibited with entecavir. J Virol 83:1778–1789. doi: 10.1128/JVI.01587-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glebe D, Lorenz H, Gerlich WH, Butler SD, Tochkov IA, Tennant BC, Cote P, Menne S. 2009. Correlation of virus and host response markers with circulating immune complexes during acute and chronic woodchuck hepatitis virus infection. J Virol 83:1579–1591. doi: 10.1128/JVI.01934-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Asabe S, Wieland SF, Chattopadhyay PK, Roederer M, Engle RE, Purcell RH, Chisari FV. 2009. The size of the viral inoculum contributes to the outcome of hepatitis B virus infection. J Virol 83:9652–9662. doi: 10.1128/JVI.00867-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu C, Yamamoto T, Zhou T, Aldrich CE, Frank K, Cullen JM, Jilbert AR, Mason WS. 2007. The liver of woodchucks chronically infected with the woodchuck hepatitis virus contains foci of virus core antigen-negative hepatocytes with both altered and normal morphology. Virology 359:283–294. doi: 10.1016/j.virol.2006.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wieland S, Thimme R, Purcell RH, Chisari FV. 2004. Genomic analysis of the host response to hepatitis B virus infection. Proc Natl Acad Sci U S A 101:6669–6674. doi: 10.1073/pnas.0401771101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang HC, Kao JH. 2014. Persistence of hepatitis B virus covalently closed circular DNA in hepatocytes: molecular mechanisms and clinical significance. Emerg Microbes Infect 3:e64. doi: 10.1038/emi.2014.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tabuchi A, Tanaka J, Katayama K, Mizui M, Matsukara H, Shimada T, Miyakawa Y, Yoshizawa H. 2008. Titration of hepatitis B virus infectivity in the sera of preacute and late acute phases of HBV infection: transmission experiments to chimeric mice with human liver repopulated hepatocytes. J Med Virol 80:2064–2068. doi: 10.1002/jmv.21320. [DOI] [PubMed] [Google Scholar]
- 30.Prince AM, Lee DH, Brotman B. 2001. Infectivity of blood from PCR-positive, HBsAg-negative, anti-HBs-positive cases of resolved hepatitis B infection. Transfusion 41:329–332. doi: 10.1046/j.1537-2995.2001.41030329.x. [DOI] [PubMed] [Google Scholar]
- 31.Petersen J, Lutgehetmann M, Volz T, Dandri M. 2007. What is the role of cccDNA in chronic HBV infection? Impact on HBV therapy. Hepatology Rev 4:9–13. [Google Scholar]
- 32.Levrero M, Pollicino T, Petersen J, Belloni L, Raimondo G, Dandri M. 2009. Control of cccDNA function in hepatitis B virus infection. J Hepatol 51:581–592. doi: 10.1016/j.jhep.2009.05.022. [DOI] [PubMed] [Google Scholar]