ABSTRACT
The infectious process of human papillomaviruses (HPVs) has been studied considerably, and many cellular components required for viral entry and trafficking continue to be revealed. In this study, we investigated the role of the nonreceptor tyrosine kinase Pyk2 during HPV16 pseudovirion infection of human keratinocytes. We found that Pyk2 is necessary for infection and appears to be involved in the intracellular trafficking of the virus. Small interfering RNA-mediated reduction of Pyk2 resulted in a significant decrease in infection but did not prevent viral entry at the plasma membrane. Pyk2 depletion resulted in altered endolysosomal trafficking of HPV16 and accelerated unfolding of the viral capsid. Furthermore, we observed retention of the HPV16 pseudogenome in the trans-Golgi network (TGN) in Pyk2-depleted cells, suggesting that the kinase could be required for the viral DNA to exit the TGN. While Pyk2 has previously been shown to function during the entry of enveloped viruses at the plasma membrane, the kinase has not yet been implicated in the intracellular trafficking of a nonenveloped virus such as HPV. Additionally, these data enrich the current literature on Pyk2's function in human keratinocytes.
IMPORTANCE In this study, we investigated the role of the nonreceptor tyrosine kinase Pyk2 during human papillomavirus (HPV) infection of human skin cells. Infections with high-risk types of HPV such as HPV16 are the leading cause of cervical cancer and a major cause of genital and oropharyngeal cancer. As a nonenveloped virus, HPV enters cells by interacting with cellular receptors and established cellular trafficking routes to ensure that the viral DNA reaches the nucleus for productive infection. This study identified Pyk2 as a cellular component required for the intracellular trafficking of HPV16 during infection. Understanding the infectious pathways of HPVs is critical for developing additional preventive therapies. Furthermore, this study advances our knowledge of intracellular trafficking processes in keratinocytes.
INTRODUCTION
Papillomaviridae is a family of small, nonenveloped DNA viruses with tropism for epithelial tissue. Low-risk human papillomavirus (HPV) types cause benign warts, and high-risk types cause anogenital or oropharyngeal area cancers (1–3). Studying the tactics used by viruses during infection has a 2-fold importance: (i) the potential to identify drug targets and (ii) understanding processes of endocytosis and intracellular trafficking.
The HPV capsid is composed of the major capsid protein L1 and the minor capsid protein L2. Infection begins when the virus capsid binds to heparan sulfate proteoglycans (HSPGs) at the cell surface (in vitro) or on the basement membrane (in vivo) (4–8). The HSPG syndecan-1 has been shown to be unnecessary in a mouse model; however, it was suggested that other HSPGs may compensate (9). Conformational changes to the viral capsid mediated by cyclophilin B (a peptidyl-prolyl cis-trans isomerase) and furin or PC5/6 (proprotein convertase) occur during initial binding (10–12). It is thought that the virus is translocated to a secondary receptor(s) for internalization, candidates for which are integrins, growth factor receptors (GFRs), annexin A2, and tetraspanins (4, 13–16). After entry, virions travel through early endosomes and late endosomes/lysosomes, where acidification and proteases facilitate further unfolding of the viral capsid (17, 18). After acidification, the encapsidated genome and L2 have been shown to separate from L1, a process facilitated by retromer components, with the majority of L1 found in the lysosome (19, 20). It has been demonstrated that trafficking through the trans-Golgi network (TGN), through the Golgi complex, and possibly through the endoplasmic reticulum (ER) is necessary for HPV infection (21–26).
Pathways shown to be activated upon HPV binding include phosphoinositide 3-kinase (PI3K)/Akt/mTOR through epidermal GFR (EGFR), PI3K, and focal adhesion kinase (FAK) through integrins (4, 27, 28). The α6β4 integrin complex has been shown to be necessary for HPV16 infection of human keratinocytes and can activate FAK (4, 13, 29, 30). Previously, we showed that the binding of HPV16 pseudovirions (PsVs) to HaCaT cells (human basal keratinocytes) induced the phosphorylation of FAK at Y397, activating the kinase (4). In that study, we demonstrated that treatment with the FAK inhibitor TAE226 significantly reduced HPV16 infection of HaCaT cells. Although TAE226 was initially designated a FAK-specific inhibitor, studies revealed that the compound equally inhibits Pyk2, a close family member of FAK (31).
Here we describe Pyk2 functioning in HPV16 (the most commonly found genotype in human cancers) endolysosomal trafficking and exit from the TGN in HaCaT cells (32). Pyk2 has been shown to be necessary for the entry of various enveloped viruses, such as Kaposi's sarcoma-associated herpesvirus and HIV-1, at the plasma membrane (33, 34). To our knowledge, Pyk2 has not been implicated in the infectious pathway of nonenveloped viruses. Pyk2 has been reported to play a role in cell migration, proliferation, and survival and to be upregulated in various human tumors (31, 35–37). Studies of Pyk2's function in keratinocytes are limited, but the kinase has been shown to play roles during differentiation and inflammation (38, 39). In terms of intracellular trafficking, Pyk2 has most recently been shown to be necessary for sustained EGFR signaling in breast cancer cells, where Pyk2 depletion resulted in early EGFR degradation via lysosomal trafficking (40). Additionally, studies have suggested that Pyk2 is involved in the regulation of some vesicular transport from the Golgi complex (41, 42). Taken together, our results suggest novel roles for a nonreceptor tyrosine kinase in the HPV16 infectious pathway and expand the literature of Pyk2's function in keratinocytes.
MATERIALS AND METHODS
Cell culture and PsV production.
HaCaT cells, purchased from AddexBio (San Diego, CA), were described by Boukamp et al. (43). Cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Thermo Scientific, Somerset, NJ) supplemented with 10% BenchMark fetal bovine serum (Gemini Bio-Products, West Sacramento, CA). HPV16 PsVs were produced in 293TT cells as described by Buck et al. (44). The p8fwb plasmid was used as a reporter genome and is referred to as the pseudogenome. The cultures were incubated with 10 μM 5-ethynyl-2′-deoxyuridine (EdU) at 6 h posttransfection.
Determination of number of viral particles.
We determined percent infectivity by measuring green fluorescent protein (GFP) expression after 48 h in HaCaT cells via flow cytometry (Accuri C6; BD, Franklin Lakes, NJ). We infected all of our experimental cells to achieve 15% GFP expression (0.15 infectious unit per cell). A multiplicity of infection (MOI) of 0.15 corresponds to approximately 800 pseudogenome-containing or viral genome equivalents per cell (measured via quantitative PCR of p8fwb with the StepOne real-time PCR system [Thermo Scientific]).
Flow cytometry and infection assays.
HaCaT cells were seeded into the wells of 24-well plates at 50,000 per well and in triplicate for each condition. After 24 h, the cells were placed on ice for 30 min in serum-free DMEM. HPV16 PsVs were allowed to bind for 1 h on ice. Excess virus was washed off, and cultures were placed at 37°C for 48 h. Cells were harvested with trypsin and washed three times with phosphate-buffered saline (PBS) with centrifugation (3,000 rpm for 5 min) before GFP flow cytometry analysis. Ten thousand events were collected for each sample.
siRNA transfections and TAE226 treatment.
Accell small interfering RNA (siRNA) SMARTpools (each a set of four siRNAs) for FAK and Pyk2 were purchased from GE Healthcare Dharmacon Inc. (Lafayette, CO). Nontargeting control siRNA was purchased from IDT (Coralville, IA). RNA interference (RNAi) Max Lipofectamine was purchased from Life Technologies (Norwalk, CT). HaCaT cells were seeded in six-well plates at 200,000 cells per well. Transfections were prepared in serum-free DMEM before being added to wells. Transfections proceeded for 48 h at 37°C, after which the cells were harvested with trypsin, counted, and replated for experiments to be performed the following day (72 h posttransfection). Optimization of the siRNA knockdowns was performed with 50 or 100 pmol of siRNA for 24, 48, and 72 h. The most efficient knockdown of FAK and Pyk2 was obtained at 72 h posttransfection with 100 pmol of siRNA per well. Cells were treated with TAE226 at 2 μM for 1 h at 37°C before infection. TAE226 (NVP-TAE226) was purchased from Selleck Chemicals (Houston, TX).
Western blotting and internalization assay.
The antibodies used in Western blot assays at a 1:1,000 dilution were Pyk2 antibody (number 3292; Cell Signaling Technology, Danvers, MA), FAK antibody (ab98961; Abcam, Cambridge, United Kingdom), actin antibody (A3853; Sigma-Aldrich, St. Louis, MO), and anti-HPV antibody (BPV-1/1H8 + CAMVIR, ab2417; Abcam). IRDye secondary antibodies (anti-rabbit and anti-mouse 680 and 800) from Li-Cor (Lincoln, NE) were used at 1:30,000 for 680 nm and at 1:20,000 for 800 nm. Cell lysates were harvested with Nonidet P-40 (United States Biological, Salem, MA) lysis buffer containing Halt Protease and Phosphatase Inhibitor Cocktail (Life Technologies). Samples were subjected to SDS-PAGE and transferred to nitrocellulose membranes (Thermo Scientific). The membranes were blocked for 30 min in 10% milk in TNET wash buffer. Primary antibody incubations were carried out at 4°C overnight or at room temperature for 4 h. Secondary antibody incubations were carried out at room temperature for 30 min. Proteins were visualized and band intensities were measured with the Li-Cor Odyssey Imaging System. To measure virus internalization, PsVs were allowed to bind to cells on ice for 1 h. Excess virus was washed off, and the cells were incubated at 37°C for 40 min to allow the virus to be internalized. After 40 min, one set of samples was harvested with NP-40 lysis buffer. The second set of samples was trypsinized for 7 min; the cells were washed three times with PBS and centrifuged before lysis buffer was added. Actin protein levels were used for normalization in Western blot assays.
Immunofluorescence analysis.
PsVs were allowed to bind to HaCaT cells on glass coverslips on ice for 1 h. Excess virus was washed off, and the cells were incubated at 37°C. Cells were fixed with 4% paraformaldehyde (PFA) and permeabilized and blocked for 30 min, 1 h, or 1.5 h with a buffer containing 0.02% Triton X-100 (Sigma-Aldrich) and 0.02% Fish Skin Gelatin (Electron Microscopy Sciences, Hatfield, PA). The Click-iT reaction was performed with the Click-iT Alexa Fluor 647 Imaging kit (Life Technologies) for 30 min before or after primary antibody incubation. The samples were incubated with primary antibodies for 1 h and with secondary antibodies at 1:2,000 for 30 min. PBS washes were performed between incubation steps. Coverslips were mounted on slides with ProLong Gold Antifade Mountant with 4′,6-diamidino-2-phenylindole (DAPI; Life Technologies).
The following antibodies were used for immunofluorescence at a 1:100 dilution: anti-EEA1 antibody (C-15, sc-6414; Santa Cruz Biotechnologies, Dallas, TX), anti-HPV16 L1 (H16.V5) antibody (a kind gift from Neil Christensen, Penn State University, Hershey, PA), Alexa Fluor 488 phalloidin (A12379; Life Technologies), anti-LAMP1 antibody (D2D11, number 9091; Cell Signaling Technologies), anti-TGN46 antibody (AHP500GT; AbD Serotec, Kidlington, United Kingdom), and Pyk2 antibody (number 3292; Cell Signaling Technology). Anti-HPV16 L1 (33L1-7) antibody, a kind gift from Martin Sapp (Louisiana State University, Shreveport, LA), was used at 1:50. Secondary antibodies (donkey anti-rabbit 568, donkey anti-sheep 568, donkey anti-goat 568, and donkey anti-mouse 555 and 647) were purchased from Life Technologies.
Colocalization and color pixel analysis.
Confocal image analysis was performed in ImageJ (45). The JACoP plugin (46) was used to calculate the fraction of virus signal overlapping the organelle signal (Manders coefficient M1). Thresholds were set manually and kept constant across all of the scans measured in each experiment. The Color Pixel Counter plugin (47) was used to measure the number of red pixels in each scan, which was normalized to the number of nuclei. Analysis was performed on the center slice of at least three separate Z-stacks scanned from each coverslip. The average M1 coefficient or average red pixel count per cell is presented with standard deviation. Representative data are shown from one of three separate experiments performed, except for Fig. 6, where data from three independent experiments (a total of nine scans) are shown.
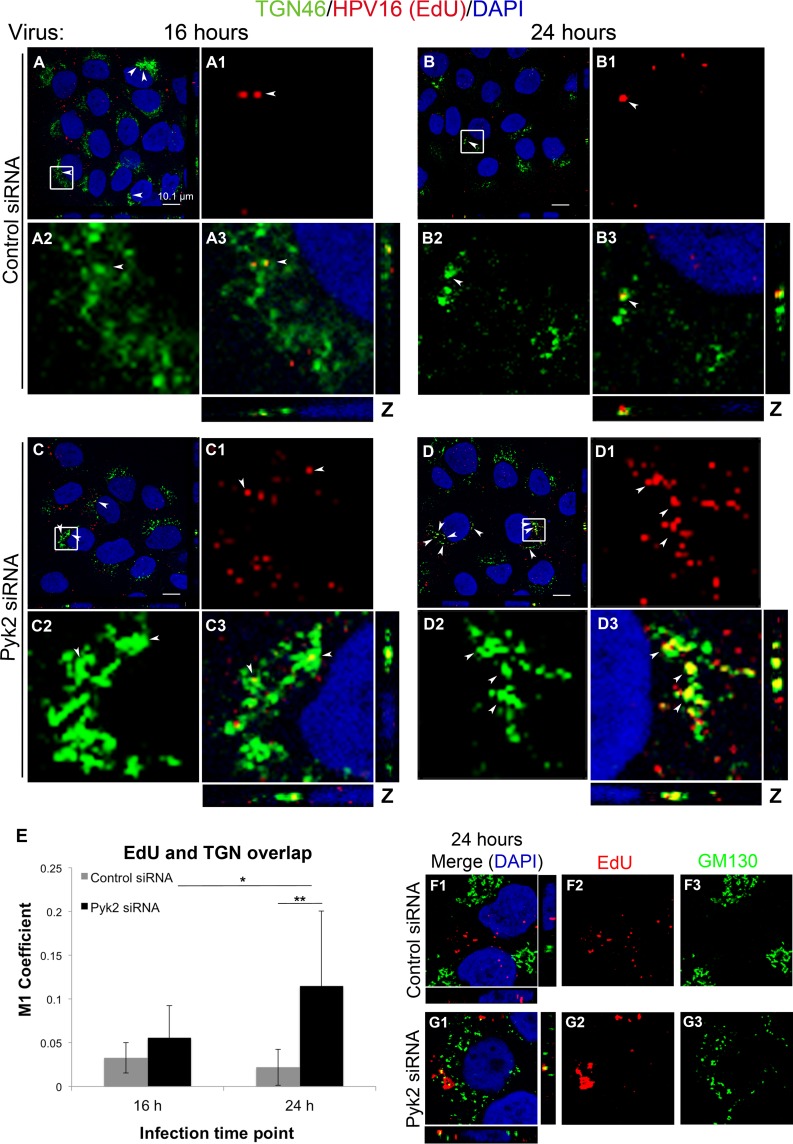
FIG 6.
siRNA-mediated reduction of Pyk2 results in the accumulation of the HPV16 pseudogenome in the TGN in HaCaT cells. (A, B) Cells transfected with control nontargeting siRNA. (C, D) Cells transfected with Pyk2 siRNA. Cells were fixed at 16 and 24 h postinfection with HPV16 PsVs. The Click-iT reaction was performed to label the EdU-labeled pseudogenome (red) and anti-TGN46 antibody (green), and nuclei were stained with DAPI (blue). Colocalization of EdU and TGN46 appears yellow. (E) The JACoP plugin for ImageJ was used to measure the M1 coefficient (fraction of red overlapping green) with three confocal scans for each condition in three independent experiments (nine scans total). *, P < 0.05; **, P < 0.005 (paired one-tailed t test). (F and G) siRNA-transfected cells were fixed 24 h postinfection with HPV16 PsVs. The Click-iT reaction was performed to label the EdU-labeled pseudogenome (red) and anti-GM130 antibody (green), and nuclei were stained with DAPI (blue). Colocalization of EdU and GM130 appears yellow.
RESULTS
siRNA-mediated reduction of Pyk2 decreases HPV16 infection in HaCaT cells.
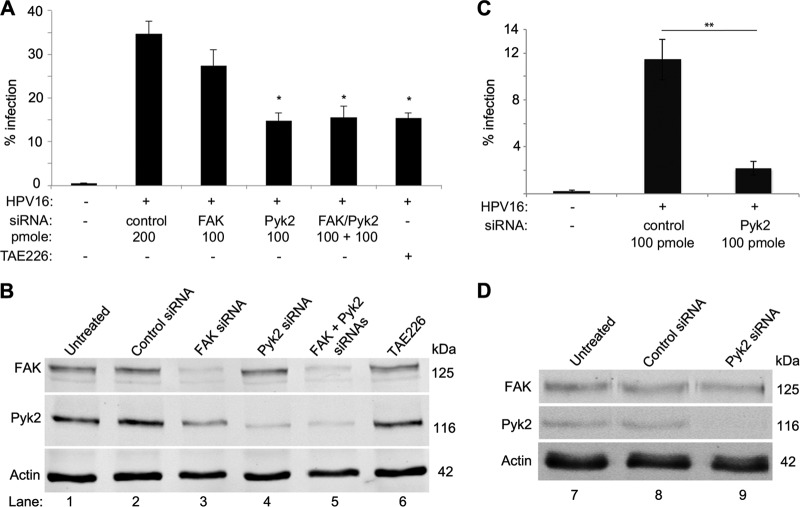
We had previously shown that TAE226 interfered with HPV16 PsV infection in HaCaT cells (4). Here we focused on two targets of TAE226, FAK and Pyk2. Infection levels in HaCaT cells transfected with FAK, Pyk2, or both siRNAs were compared with those in TAE226-treated cells (Fig. 1). Infection levels in cells transfected with 200 pmol of nontargeting (control) siRNA were compared with those in cells transfected with 100 pmol of FAK or PyK2 siRNA or 100 pmol each of the FAK and Pyk2 siRNAs (i.e., 200 pmol total) and in cultures treated with 2 μM TAE226 (Fig. 1A). We observed a 20% decrease in infection in FAK siRNA-transfected cells and a 60% decrease in Pyk2 and Pyk2-FAK siRNA-transfected cells and TAE226-treated cells. Compared to those in untreated cells or control siRNA-transfected cells, the FAK and Pyk2 protein levels were not affected by TAE226 (Fig. 1B, lanes 1, 2, and 6). FAK siRNA reduced the total FAK level by 83% but surprisingly reduced the total Pyk2 level by 40% (Fig. 1B, lane 3). Pyk2 siRNA reduced Pyk2 protein levels by 81% and caused a 17% decrease in FAK protein levels (Fig. 1B, lane 4). Dual transfection reduced FAK by 71% and Pyk2 by 83% (Fig. 1B, lane 5). These experiments showed that 81% depletion of Pyk2 resulted in a significant 60% decrease in viral infection (P < 0.005). We optimized the experimental design with equimolar siRNA levels (100 pmol total) and a PsV MOI of 0.15. Results showed a >80% loss of infection in Pyk2 siRNA-transfected cells (Fig. 1C, P < 0.0005). There was a 90% loss of Pyk2 protein expression under these conditions (Fig. 1D, lane 9; described in Materials and Methods). These data suggested that the loss of Pyk2 had a more significant effect on PsV infection than did the loss of FAK. Additionally, the FAK siRNA pool affected Pyk2 protein levels (Fig. 1B, lane 4).
FIG 1.
siRNA-mediated reduction of Pyk2 results in decreased HPV16 infection in HaCaT cells. (A) Infection levels in HaCaT cells transfected with nontargeting (control), FAK, or Pyk2-FAK siRNA or treated with 2 μM TAE226. Percent infection was measured via flow cytometry. (B) Western blot analysis of FAK, Pyk2, and actin protein levels in cells used in the experiment shown in panel A. (C) Infection levels in cells transfected with 100 pmol of control or Pyk2-specific siRNA. (D) Western blot analysis of FAK, Pyk2, and actin protein levels in cells used in the experiment shown in panel C. *, P < 0.005; **, P < 0.0005 (paired one-tailed t test).
Pyk2 depletion does not interfere with HPV16 PsV binding and internalization.
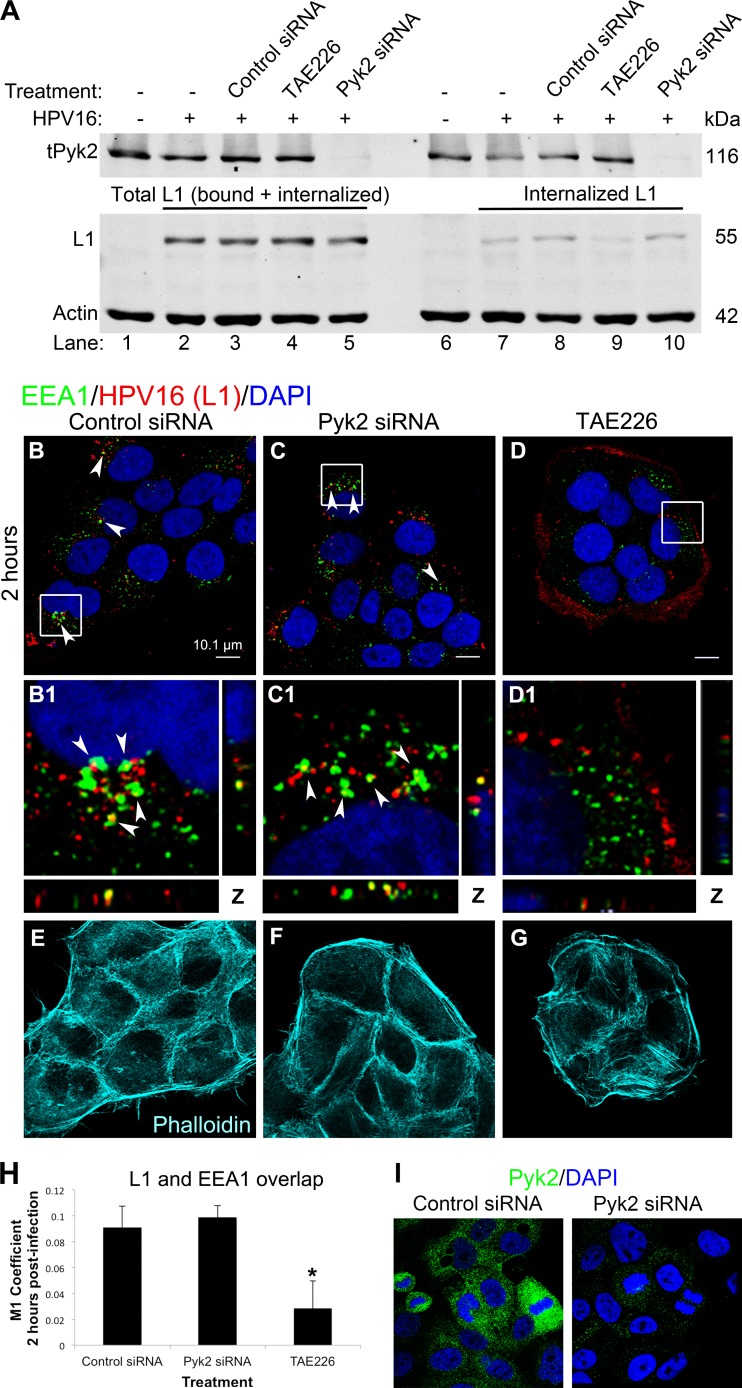
Having shown that Pyk2 depletion reduced HPV16 PsV infection, we assessed if Pyk2 depletion prevented binding, internalization, and initial trafficking of PsVs into an endosome (Fig. 2). Equal levels of L1 protein in our cultures after 40 min of infection suggested no differences in PsV binding (Fig. 2A, lanes 1 to 5). To test for internalization, cell cultures were treated with trypsin 40 min after infection to remove virions that were bound but not internalized (Fig. 2A, lanes 6 to 10). We observed comparable L1 levels in untreated, control, and Pyk2 siRNA-transfected cells, indicating internalized PsVs (Fig. 2B, lanes 7, 8, and 10, respectively). In TAE226-treated cells, there was a >50% decrease in internalized L1 levels, indicating reduced viral internalization (Fig. 2A, lane 10). Total levels of Pyk2 are shown.
FIG 2.
HPV16 internalization and early trafficking events are not disrupted in Pyk2-depleted HaCaT cells. (A) Western blot analysis of Pyk2, L1, and actin levels in HaCaT cells transfected with control or Pyk2 siRNA (lanes 3, 5, 8, and 10) or treated with 2 μM TAE226 and infected for 40 min with HPV16 PsVs. Cells were harvested directly for lysates (lanes 1 to 5) or treated with trypsin prior to harvesting (lanes 6 to 10). (B to G) Immunofluorescence analysis of EEA1 (green), L1 (red), and phalloidin (cyan) in cells transfected with control or Pyk2 siRNA or treated with 2 μM TAE226. Nuclei are stained with DAPI (blue). Colocalization of EEAI and L1 appears yellow. (H) The JACoP plugin for ImageJ was used to measure the M1 coefficient (fraction of red overlapping green) with three confocal scans for each condition. *, P < 0.05 (paired one-tailed t test). (I) Confirmation of siRNA-mediated Pyk2 knockdown via immunofluorescence analysis. HaCaT cells were transfected for 72 h with control or Pyk2 siRNA. Immunofluorescence analysis was performed with anti-Pyk2 antibody (green), and nuclei were stained with DAPI (blue).
We analyzed siRNA-transfected or TAE226-treated cells by confocal immunofluorescence microscopy. Viral particles were detected with the conformation-dependent antibody H16.V5 against the L1 capsid protein (red) (48), endosomes were identified with the early endosome marker EEA1 (green), and nuclei were stained with DAPI (blue). At 2 h after infection, our data showed similar levels of L1 signal colocalizing with EEA1 in control and Pyk2-depleted cells (Fig. 2B, C, and H). In TAE226-treated cells, colocalization of viral particles with EEA1 was minimal (Fig. 2D and H). Previously, we had observed a loss of filopodia with TAE226 treatment (43). Here we stained our samples with phalloidin and observed almost complete loss of filopodia in TAE226-treated cells (Fig. 2G). Filopodia were observed in control and Pyk2 siRNA-transfected cells, although there were fewer filopodia in Pyk2-depleted cells (Fig. 2E and F). Pyk2 knockdown was confirmed via microscopy (Fig. 2I).
Taken together, these data showed that Pyk2 depletion did not hinder viral binding, internalization, or trafficking into an endosome. These data suggested that although Pyk2 depletion interfered with filopodium formation, the loss of filopodia did not correlate with viral internalization levels (Fig. 2A, lane 5, C, and H). Furthermore, the reductions of infection after TAE226 treatment or Pyk2 depletion appeared to work through different mechanisms because TAE226 prevented viral internalization, whereas Pyk2 depletion did not.
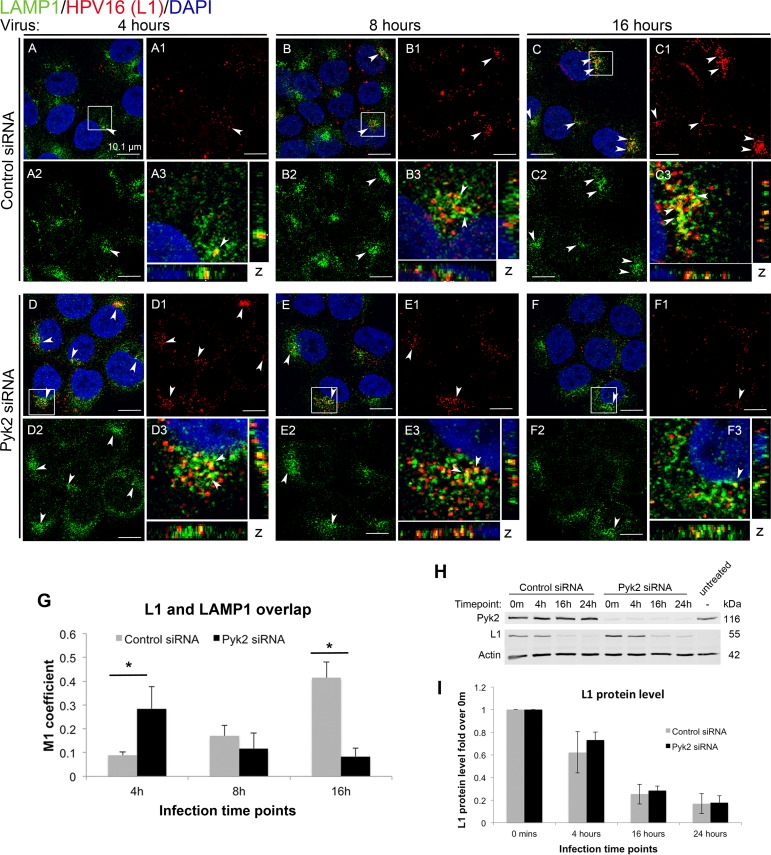
Depletion of Pyk2 resulted in earlier lysosomal detection of HPV16 PsVs.
Models of HPV infection suggest that after the endosome, viral particles are diverted to the lysosome or to the TGN (49, 50). An antibody to LAMP1 was used as a lysosome marker (green), H16.V5 was used to detect PsVs (red), and nuclei were stained with DAPI (blue) (Fig. 3). In control cells, virus colocalization with LAMP1 was minimal at 4 h and increased at 8 h, with maximum overlap observed at 16 h (Fig. 3A to C and G). In contrast, colocalization of the virus and LAMP1 signals in Pyk2-depleted cells was maximal at 4 h and then decreased at 8 and 16 h (Fig. 3D to F and G).
FIG 3.
Depletion of Pyk2 results in earlier lysosomal targeting of the L1 protein in HaCaT cells. (A to C) Cells transfected with control nontargeting siRNA. (D to F) Cells transfected with Pyk2 siRNA. Cells were fixed at 4, 8, and 16 h postinfection with HPV16 PsVs. Immunofluorescence analysis was performed with anti-L1 H16.V5 (red) and anti-LAMP1 (green) antibodies. Nuclei were stained with DAPI (blue). Colocalization of L1 and LAMP1 appears yellow. (G) The JACoP plugin for ImageJ was used to measure the M1 coefficient (fraction of red overlapping green) with three confocal scans for each condition. *, P < 0.05 (paired one-tailed t test). (H) Western blot analysis of Pyk2, L1, and actin protein levels in control siRNA-transfected or Pyk2 siRNA-transfected cells at time points postinfection with HPV16 PsVs. (I) Measurement of the integrated density of L1 protein bands in Western blot assays from three independent experiments. All L1 levels were normalized to actin levels and compared to the levels at 0 min.
These data suggested that HPV16 PsVs trafficked to the lysosome earlier in Pyk2-depleted cells. Compared to that of the control, the H16.V5 signal decreased by 16 h in the Pyk2-depleted cells (Fig. 3, compare panels F and C). We wondered if the early trafficking to the lysosome caused degradation of the L1 protein. Western blotting results indicated that the rates of L1 degradation in control and Pyk2-depleted cells were similar, suggesting that the decrease in the H16.V5 signal was not due to early degradation of L1 (Fig. 3H and I).
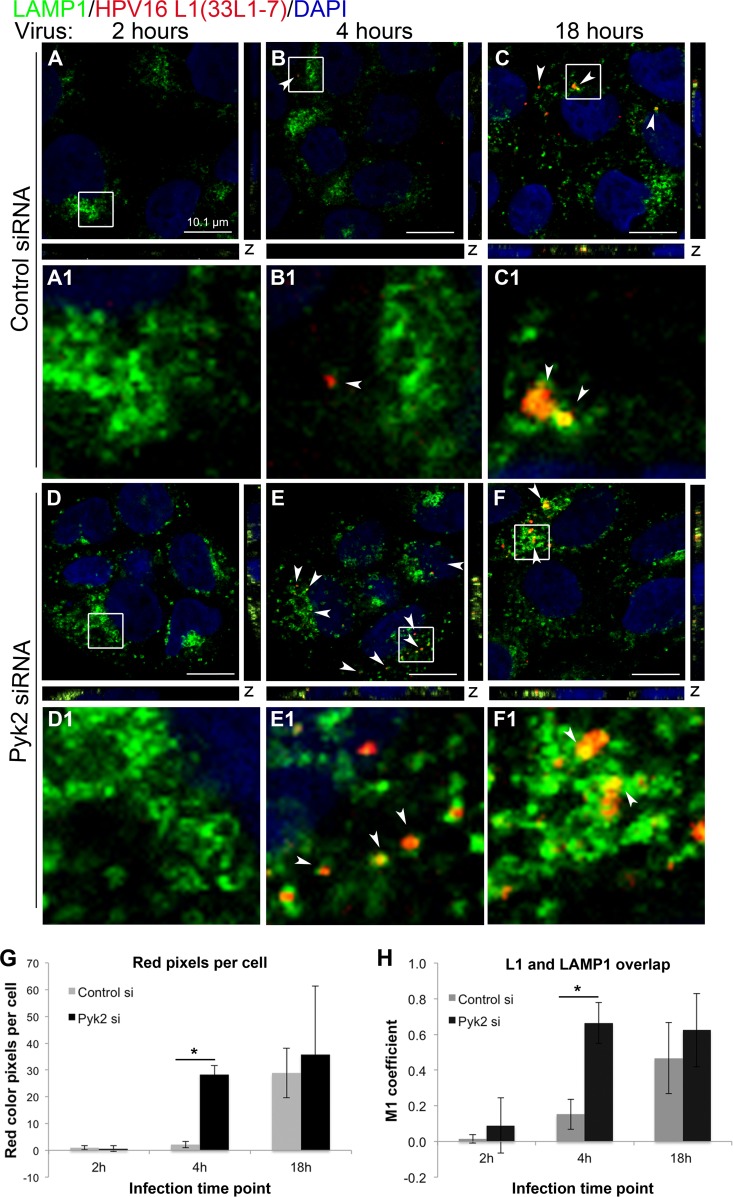
Early unfolding of the HPV16 capsid occurs in Pyk2 siRNA-transfected HaCaT cells.
We next determined if a conformational change in the viral capsid occurred earlier in Pyk2-depleted HaCaT cells by performing an immunofluorescence assay with the antibody 33L1-7, against a hidden epitope of L1 (Fig. 4) (51). At 2 h postinfection, there was no detectable L1-7 signal in the control or Pyk2 siRNA-transfected cells (Fig. 4A, D, and G). We observed a minimal L1-7 signal in the control cells at 4 h (Fig. 4B and B1) but a strong and consistent appearance in the Pyk2-depleted cells (Fig. 4E, E1, and G). The L1-7 signal was visible at 18 h in the control cells (Fig. 4C and C1), similar to the timing of L1-7 signal appearance reported by Bienkowska-Haba et al. (52). The mean L1-7 signal level at 18 h in the control cells was comparable to that in Pyk2-depleted cells (Fig. G). At least three scans, each with 10 to 20 nuclei per field of view, were used to measure L1-7 signal levels.
FIG 4.
Depletion of Pyk2 results in early unfolding of the HPV16 capsid in HaCaT cells. (A to C) Cells transfected with control nontargeting siRNA. (D to F) Cells transfected with Pyk2 siRNA. Cells were fixed at 2, 4, and 18 h postinfection with HPV16 PsVs. Immunofluorescence analysis was performed with anti-L1 33L1-7 (red) and anti-LAMP1 (green) antibodies. Nuclei were stained with DAPI (blue). Colocalization of L1 and LAMP1 appears yellow. (G) The color pixel counter plugin for ImageJ was used to measure red pixels, and the results were normalized to the number of cells in each scan. (H) The JACoP plugin for ImageJ was used to measure the M1 coefficient (fraction of red overlapping green) with three confocal scans for each condition. *, P < 0.05 (paired one-tailed t test).
We stained with a LAMP1 antibody to detect if the L1-7 signal localized to lysosomes (Fig. 4). At 4 h in the control cells, there was minimal colocalization of L1-7 and LAMP1, resulting in an M1 coefficient (fraction of L1 overlapping with LAMP1) of <0.2 (Fig. 4H). In contrast, at 4 h in the Pyk2-depleted cells, there was distinct colocalization of L1-7 and LAMP1, resulting in an M1 coefficient of approximately 0.8 (Fig. 4E, E1, and H). At 18 h, there was an increase in L1-7 colocalization with LAMP1 in the control siRNA-transfected cells and no observable difference from the Pyk2-depleted cells (Fig. 4C, C1, F, F1, and H). These results suggested that unfolding of the viral capsid in the lysosome occurred earlier in Pyk2-depleted cells.
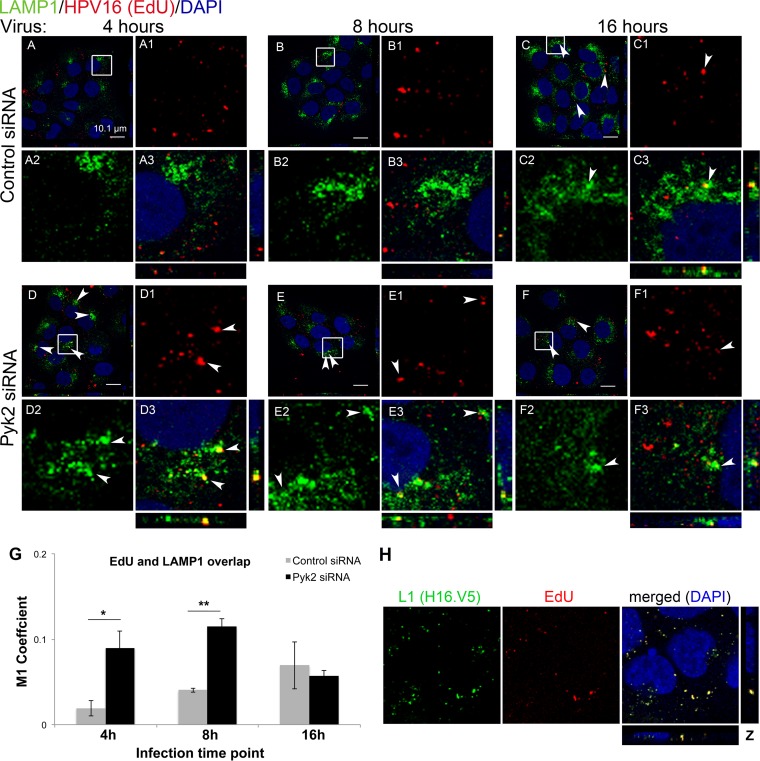
The HPV16 pseudogenome localizes to the lysosome in Pyk2-depleted HaCaT cells at early time points.
Next we wanted to determine the trafficking of the encapsidated pseudogenome in these cells. We infected siRNA-transfected HaCaT cells with HPV16 PsVs containing an EdU-labeled pseudogenome. We used the Click-iT reaction to visualize the EdU-labeled DNA (red), a LAMP1 antibody for the lysosome (green), and DAPI for the nuclei (blue) (Fig. 5). At 4 h postinfection, there was no visible colocalization of the pseudogenome and LAMP1 in control siRNA-transfected cells (Fig. 5A and A3). Significant colocalization was observed in Pyk2-depleted cells (Fig. 5D, D3, and G). The colocalization increased slightly at 8 h under both conditions; however, there was a significantly higher level in the Pyk2-depleted cells (Fig. 5G). At 16 h postinfection, the control cells showed a slight increase in the localization of the pseudogenome to the lysosome, but we did not see an accumulation in the lysosome of the Pyk2-depleted cells (Fig. 5C, F, and G). The fraction of the EdU-labeled pseudogenome in Pyk2-depleted cells colocalizing with LAMP1 was minimal at approximately 10% (indicated by M1 coefficients; Fig. 5G). The latter data suggested that a portion of the viral DNA could be degraded in the lysosome in the Pyk2-depleted cells, although this may not account for the ≥80% loss of viral infection (Fig. 1C). To verify that the EdU signal specifically labeled the encapsidated genomes, we stained with the H16.V5 antibody before the EdU/Click-iT reaction (a reaction that denatures the capsid, as Day et al. have noted) (22). We observed all of the EdU signal overlapping the H16.V5 signal; a fraction of the H16.V5 signal stained empty viral capsids or capsids containing the pseudogenome that did not incorporate EdU (Fig. 5H).
FIG 5.
Depletion of Pyk2 results in early and increased localization of the HPV16 pseudogenome to the lysosome in HaCaT cells. (A to C) Cells transfected with nontargeting control siRNA. (D to F) Cells transfected with Pyk2 siRNA. Cells were fixed at 4, 8, and 16 h postinfection with HPV16 PsVs. The Click-iT reaction was used to label the EdU-labeled genome (red) and anti-LAMP1 antibody (green), and nuclei were stained with DAPI (blue). Colocalization of L1 and LAMP1 appears yellow. (G) The JACoP plugin for ImageJ was used to measure the M1 coefficient (fraction of red overlapping green) with three confocal scans for each condition. *, P < 0.05; **, P < 0.005 (paired one-tailed t test). (H) EdU signal corresponds to viral particles. HaCaT cells were infected for 4 h before fixation. The Click-iT reaction was performed to label the EdU-labeled pseudogenome (red) after immunofluorescence analysis with anti-L1 H16.V5 antibody (green). Nuclei were stained with DAPI (blue).
The HPV16 pseudogenome accumulates in the TGN in Pyk2 siRNA-transfected HaCaT cells.
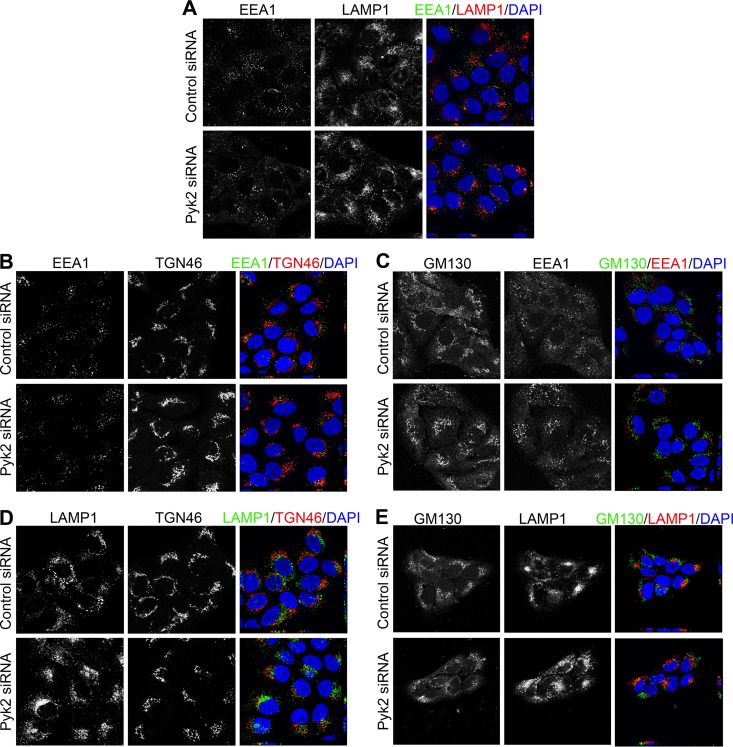
We considered if the viral pseudogenome was able to reach the TGN in Pyk2-depleted cells. We visualized the EdU-labeled pseudogenome with Click-iT (red), the TGN with the anti-TGN46 antibody (green), and nuclei with DAPI (blue). At 16 h postinfection, similar levels of colocalization of the pseudogenome and TGN46 were observed in the control and Pyk2-depleted cells (Fig. 6A, A1, C, C1, and E). At 24 h, we observed a decrease in pseudogenome localization to the TGN in control cells (Fig. 6B, B1, and E). In contrast, at 24 h in Pyk2-depleted cells, we observed a marked increase in pseudogenome and TGN colocalization (Fig. 6D, D1, and E). These results suggested that the viral pseudogenome did reach the TGN in Pyk2-depleted cells. However, as pseudogenome levels decreased in the TGN in the control cells at 24 h, the levels increased in Pyk2-depleted cells, suggesting that the pseudogenome accumulated in the TGN when Pyk2 levels were decreased. We confirmed these results with the Golgi complex marker GM130 and observed less colocalization of the EdU-labeled pseudogenome with GM130 at 24 h in control cells than in Pyk2-depleted cells (Fig. 6F and G). To verify that Pyk2 depletion in HaCaT cells did not alter the localization of the organelle markers used in our immunofluorescence experiments, we costained each of the markers in siRNA-transfected cells (Fig. 7). We did not observe any alterations in the staining of the organelle markers in the Pyk2-depleted cells.
FIG 7.
siRNA-mediated reduction of Pyk2 and localization of organelle markers for early endosomes, lysosomes, or the Golgi compartment in HaCaT cells. (A to E) Cells transfected with control nontargeting siRNA or Pyk2-specific siRNA for 72 h before 4% PFA fixation. Nuclei in all panels were stained with DAPI (blue). Immunofluorescence analysis was performed with anti-EEA1 (green) and anti-LAMP1 (red) antibodies (A), anti-EEA1 (green) and anti-TGN46 (red) antibodies (B), anti-GM130 (green) and anti-EEA1 (red) antibodies (C), anti-GM130 (green) and anti-LAMP1 (red) antibodies (E), or anti-LAMP1 (green) and anti-TGN46 (red) antibodies (D).
DISCUSSION
In this study, using siRNAs, we found that FAK family member Pyk2 appears to be involved in HPV16 PsV trafficking in HaCaT cells. The alterations to intracellular trafficking under Pyk2-depleted conditions affect unfolding of the viral capsid, localization of the pseudogenome to the lysosome, and exit from the TGN. To our knowledge, this is the first report implicating Pyk2 in the later trafficking and sorting processes of HPV16 infection.
We had previously observed that treatment with the FAK inhibitor TAE226 prevented filopodium formation and reduced virus entry (4). We hypothesized that Pyk2 could mediate these rearrangements that have been shown to be necessary for virus internalization (53, 54). Our findings here showed that although there was disruption of filopodia in Pyk2-depleted cells, there was no significant reduction of virus binding or localization to early endosomes (Fig. 2).
During the preparation of this report, depletion of Pyk2 was reported to result in premature targeting of EGFR to the lysosome for degradation, thus halting EGFR downstream signaling in breast cancer cells (40). We observed a similar acceleration of the endolysosomal pathway during HPV16 infection in Pyk2-depleted HaCaT cells, resulting in early capsid unfolding. However, we did not observe premature degradation of the L1 protein in Pyk2-depleted cells. We consistently observed earlier localization of the pseudogenome to the lysosome in Pyk2-depleted cells, and at higher levels than in control cells, which may contribute to the decreased infection seen. However, our data showed that the viral pseudogenome is able to reach the TGN in Pyk2-depleted cells, suggesting that accelerated lysosomal localization is not the primary reason for the decreased infection seen.
Studies of HPV16 infection have reported the localization of viral particles to the TGN (20, 22, 23, 25, 26). In HeLa cells, Popa et al., Lipovsky et al., and Zhang et al. detected L1 and L2 localizing at the TGN and Golgi complex at 16 h (20, 25, 26). In HaCaT cells, DiGiuseppe et al. detected the EdU-labeled pseudogenome localizing to the TGN at 18 h and a decrease in colocalization at 36 h, suggesting that by 36 h the pseudogenome had trafficked out of the TGN (23). Here we similarly observed colocalization of the pseudogenome (Fig. 6) and L1 protein (data not shown) with the TGN in control siRNA-transfected HaCaT cells at 16 h and a decrease in this localization at 24 h (Fig. 6). The reduction in staining at 24 h in control cells differs from a study by Day et al. where colocalization of L1 or the EdU-labeled pseudogenome and the TGN was observed at 24 h postinfection in untreated HeLa cells (22). The difference may be due to the cell type used, the PsV amounts used, or the experimental design used. In contrast to the results observed in our control cells, pseudogenome localization to the TGN increased at 24 h in Pyk2-depleted cells. The increase in Pyk2-depleted cells suggests an accumulation of the pseudogenome in the TGN.
Our data and published previously literature support a model in which Pyk2 mediates the movement of the pseudogenome out of the TGN. Two identified substrates of Pyk2, Papα and ASAP1, were demonstrated to have Arf-GAP (GTPase-activating protein) activity in HeLa, 293, and mouse embryo fibroblast (MEF) cells (41, 42). Interestingly, Arfs are known to recruit coat protein I (COPI) to the Golgi membrane for vesicle formation (55). Previously, we used brefeldin A to prevent COPI vesicle formation and suggested that HPV16 might use COPI-mediated movement to reach the ER (24). We are currently investigating whether Pyk2 may regulate Arf1 recruitment of COPI for vesicle formation. Alternatively, Pyk2 may be involved in cell cycle regulation or cell cycle progression, two events that have been shown to be necessary for HPV16 DNA to access the nucleus during infection or in general vesicle trafficking (41, 56–58).
In summary, we have shown that depletion of Pyk2 significantly reduces HPV16 PsV infection in human keratinocytes. Taken together, our results suggest a multifunctional role for Pyk2 in the HPV16 infectious pathway. First, the presence of endogenous Pyk2 protein in HaCaT cells may delay unfolding of the viral capsid, potentially preventing viral DNA from being degraded in the lysosome. Second, Pyk2 function may be necessary for viral DNA to exit the TGN for productive infection to occur. Further studies are required to determine how Pyk2 functions in these two processes. To our knowledge, these are novel functions for a kinase to perform during viral infection. Additional studies of Pyk2's role in the endolysosomal pathway and regulation of vesicular transport will provide valuable knowledge about intracellular trafficking in human keratinocytes.
ACKNOWLEDGMENTS
We thank C. B. Buck, N. D. Christensen, M. A. Ozbun, and M. Sapp for essential reagents.
This work was supported by RSG-12-021-01-MPC from the American Cancer Society and R21CA153096 from the National Cancer Institute, National Institutes of Health.
REFERENCES
- 1.Chaturvedi AK, Engels EA, Pfeiffer RM, Hernandez BY, Xiao W, Kim E, Jiang B, Goodman MT, Sibug-Saber M, Cozen W, Liu L, Lynch CF, Wentzensen N, Jordan RC, Altekruse S, Anderson WF, Rosenberg PS, Gillison ML. 2011. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol 29:4294–4301. doi: 10.1200/JCO.2011.36.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doorbar J, Quint W, Banks L, Bravo IG, Stoler M, Broker TR, Stanley MA. 2012. The biology and life-cycle of human papillomaviruses. Vaccine 30(Suppl 5):F55–F70. doi: 10.1016/j.vaccine.2012.06.083. [DOI] [PubMed] [Google Scholar]
- 3.zur Hausen H. 1991. Human papillomaviruses in the pathogenesis of anogenital cancer. Virology 184:9–13. doi: 10.1016/0042-6822(91)90816-T. [DOI] [PubMed] [Google Scholar]
- 4.Abban CY, Meneses PI. 2010. Usage of heparan sulfate, integrins, and FAK in HPV16 infection. Virology 403:1–16. doi: 10.1016/j.virol.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giroglou T, Florin L, Schäfer F, Streeck RE, Sapp M. 2001. Human papillomavirus infection requires cell surface heparan sulfate. J Virol 75:1565–1570. doi: 10.1128/JVI.75.3.1565-1570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson KM, Kines RC, Roberts J N, Lowy DR, Schiller J T, Day PM. 2009. Role of heparan sulfate in attachment to and infection of the murine female genital tract by human papillomavirus. J Virol 83:2067–2074. doi: 10.1128/JVI.02190-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kines RC, Thompson CD, Lowy DR, Schiller J T, Day PM. 2009. The initial steps leading to papillomavirus infection occur on the basement membrane prior to cell surface binding. Proc Natl Acad Sci U S A 106:20458–20463. doi: 10.1073/pnas.0908502106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumar A, Jacob T, Abban CY, Meneses PI. 2014. Intermediate heparan sulfate binding during HPV-16 infection in HaCaTs. Am J Ther 21:331–342. doi: 10.1097/MJT.0000000000000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang H-S, Lambert PF. 2012. Use of an in vivo animal model for assessing the role of integrin α(6)β(4) and syndecan-1 in early steps in papillomavirus infection. Virology 433:395–400. doi: 10.1016/j.virol.2012.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bienkowska-Haba M, Patel HD, Sapp M. 2009. Target cell cyclophilins facilitate human papillomavirus type 16 infection. PLoS Pathog 5:e1000524. doi: 10.1371/journal.ppat.1000524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richards RM, Lowy DR, Schiller J T, Day PM. 2006. Cleavage of the papillomavirus minor capsid protein, L2, at a furin consensus site is necessary for infection. Proc Natl Acad Sci U S A 103:1522–1527. doi: 10.1073/pnas.0508815103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seidah NG, Prat A. 2012. The biology and therapeutic targeting of the proprotein convertases. Nat Rev Drug Discov 11:367–383. doi: 10.1038/nrd3699. [DOI] [PubMed] [Google Scholar]
- 13.Aksoy P, Abban CY, Kiyashka E, Qiang W, Meneses PI. 2014. HPV16 infection of HaCaTs is dependent on β4 integrin, and α6 integrin processing. Virology 449:45–52. doi: 10.1016/j.virol.2013.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spoden G, Freitag K, Husmann M, Boller K, Sapp M, Lambert C, Florin L. 2008. Clathrin- and caveolin-independent entry of human papillomavirus type 16—involvement of tetraspanin-enriched microdomains (TEMs). PLoS One 3:e3313. doi: 10.1371/journal.pone.0003313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Surviladze Z, Dziduszko A, Ozbun MA. 2012. Essential roles for soluble virion-associated heparan sulfonated proteoglycans and growth factors in human papillomavirus infections. PLoS Pathog 8:e1002519. doi: 10.1371/journal.ppat.1002519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woodham AW, Da Silva DM, Skeate J G, Raff AB, Ambroso MR, Brand HE, Isas J M, Langen R, Kast WM. 2012. The S100A10 subunit of the annexin A2 heterotetramer facilitates L2-mediated human papillomavirus infection. PLoS One 7:e43519. doi: 10.1371/journal.pone.0043519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dabydeen SA, Meneses PI. 2009. The role of NH4Cl and cysteine proteases in human papillomavirus type 16 infection. Virol J 6:109. doi: 10.1186/1743-422X-6-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith JL, Campos SK, Wandinger-Ness A, Ozbun MA. 2008. Caveolin-1-dependent infectious entry of human papillomavirus type 31 in human keratinocytes proceeds to the endosomal pathway for pH-dependent uncoating. J Virol 82:9505–9512. doi: 10.1128/JVI.01014-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kämper N, Day PM, Nowak T, Selinka H-C, Florin L, Bolscher J, Hilbig L, Schiller J T, Sapp M. 2006. A membrane-destabilizing peptide in capsid protein L2 is required for egress of papillomavirus genomes from endosomes. J Virol 80:759–768. doi: 10.1128/JVI.80.2.759-768.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Popa A, Zhang W, Harrison MS, Goodner K, Kazakov T, Goodwin EC, Lipovsky A, Burd CG, DiMaio D. 2015. Direct binding of retromer to human papillomavirus type 16 minor capsid protein L2 mediates endosome exit during viral infection. PLoS Pathog 11:e1004699. doi: 10.1371/journal.ppat.1004699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bossis I, Roden RBS, Gambhira R, Yang R, Tagaya M, Howley PM, Meneses PI. 2005. Interaction of tSNARE syntaxin 18 with the papillomavirus minor capsid protein mediates infection. J Virol 79:6723–6731. doi: 10.1128/JVI.79.11.6723-6731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Day PM, Thompson CD, Schowalter RM, Lowy DR, Schiller JT. 2013. Identification of a role for the trans-Golgi network in human papillomavirus 16 pseudovirus infection. J Virol 87:3862–3870. doi: 10.1128/JVI.03222-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DiGiuseppe S, Bienkowska-Haba M, Hilbig L, Sapp M. 2014. The nuclear retention signal of HPV16 L2 protein is essential for incoming viral genome to transverse the trans-Golgi network. Virology 458-459:93–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laniosz V, Dabydeen SA, Havens MA, Meneses PI. 2009. Human papillomavirus type 16 infection of human keratinocytes requires clathrin and caveolin-1 and is brefeldin A sensitive. J Virol 83:8221–8232. doi: 10.1128/JVI.00576-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lipovsky A, Popa A, Pimienta G, Wyler M, Bhan A, Kuruvilla L, Guie M-A, Poffenberger AC, Nelson CDS, Atwood WJ, DiMaio D. 2013. Genome-wide siRNA screen identifies the retromer as a cellular entry factor for human papillomavirus. Proc Natl Acad Sci U S A 110:7452–7457. doi: 10.1073/pnas.1302164110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang W, Kazakov T, Popa A, DiMaio D. 2014. Vesicular trafficking of incoming human papillomavirus 16 to the Golgi apparatus and endoplasmic reticulum requires γ-secretase activity. mBio 5(5):e01777–01714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fothergill T, McMillan NAJ. 2006. Papillomavirus virus-like particles activate the PI3-kinase pathway via alpha-6 beta-4 integrin upon binding. Virology 352:319–328. doi: 10.1016/j.virol.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 28.Surviladze Z, Sterk RT, DeHaro SA, Ozbun MA. 2013. Cellular entry of human papillomavirus type 16 involves activation of the phosphatidylinositol 3-kinase/Akt/mTOR pathway and inhibition of autophagy. J Virol 87:2508–2517. doi: 10.1128/JVI.02319-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.David FS, Zage PE, Marcantonio EE. 1999. Integrins interact with focal adhesions through multiple distinct pathways. J Cell Physiol 181:74–82. [DOI] [PubMed] [Google Scholar]
- 30.Evander M, Frazer IH, Payne E, Qi YM, Hengst K, McMillan NA. 1997. Identification of the alpha6 integrin as a candidate receptor for papillomaviruses. J Virol 71:2449–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu T-J, LaFortune T, Honda T, Ohmori O, Hatakeyama S, Meyer T, Jackson D, J de Groot Yung WKA. 2007. Inhibition of both focal adhesion kinase and insulin-like growth factor-I receptor kinase suppresses glioma proliferation in vitro and in vivo. Mol Cancer Ther 6:1357–1367. doi: 10.1158/1535-7163.MCT-06-0476. [DOI] [PubMed] [Google Scholar]
- 32.Doorbar J. 2005. The papillomavirus life cycle. J Clin Virol 32(Suppl 1):S7–S15. [DOI] [PubMed] [Google Scholar]
- 33.Harmon B, Ratner L. 2008. Induction of the Galpha(q) signaling cascade by the human immunodeficiency virus envelope is required for virus entry. J Virol 82:9191–9205. doi: 10.1128/JVI.00424-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krishnan HH, Sharma-Walia N, Streblow DN, Naranatt PP, Chandran B. 2006. Focal adhesion kinase is critical for entry of Kaposi's sarcoma-associated herpesvirus into target cells. J Virol 80:1167–1180. doi: 10.1128/JVI.80.3.1167-1180.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lipinski CA, Tran NL, Menashi E, Rohl C, Kloss J, Bay RC, Berens ME, Loftus JC. 2005. The tyrosine kinase pyk2 promotes migration and invasion of glioma cells. Neoplasia 7:435–445. doi: 10.1593/neo.04712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang S, Qiu X, Gu Y, Wang E. 2008. Up-regulation of proline-rich tyrosine kinase 2 in non-small cell lung cancer. Lung Cancer 62:295–301. doi: 10.1016/j.lungcan.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y, Moschetta M, Huynh D, Tai Y-T, Zhang Y, Zhang W, Mishima Y, Ring J E, Tam WF, Xu Q, Maiso P, Reagan M, Sahin I, Sacco A, Manier S, Aljawai Y, Glavey S, Munshi NC, Anderson KC, Pachter J, Roccaro AM, Ghobrial IM. 2014. Pyk2 promotes tumor progression in multiple myeloma. Blood 124:2675–2686. doi: 10.1182/blood-2014-03-563981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Halfter UM, Derbyshire ZE, Vaillancourt RR. 2005. Interferon-gamma-dependent tyrosine phosphorylation of MEKK4 via Pyk2 is regulated by annexin II and SHP2 in keratinocytes. Biochem J 388:17–28. doi: 10.1042/BJ20041236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schindler EM, Baumgartner M, Gribben EM, Li L, Efimova T. 2007. The role of proline-rich protein tyrosine kinase 2 in differentiation-dependent signaling in human epidermal keratinocytes. J Investig Dermatol 127:1094–1106. doi: 10.1038/sj.jid.5700662. [DOI] [PubMed] [Google Scholar]
- 40.Verma N, Keinan O, Selitrennik M, Karn T, Filipits M, Lev S. 2015. PYK2 sustains endosomal-derived receptor signalling and enhances epithelial-to-mesenchymal transition. Nat Commun 6:6064. doi: 10.1038/ncomms7064. [DOI] [PubMed] [Google Scholar]
- 41.Andreev J, Simon J P, Sabatini DD, Kam J, Plowman G, Randazzo PA, Schlessinger J. 1999. Identification of a new Pyk2 target protein with Arf-GAP activity. Mol Cell Biol 19:2338–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kruljac-Letunic A, Moelleken J, Kallin A, Wieland F, Blaukat A. 2003. The tyrosine kinase Pyk2 regulates Arf1 activity by phosphorylation and inhibition of the Arf-GTPase-activating protein ASAP1. J Biol Chem 278:29560–29570. doi: 10.1074/jbc.M302278200. [DOI] [PubMed] [Google Scholar]
- 43.Boukamp P, Petrussevska RT, Breitkreutz D, Hornung J, Markham A, Fusenig NE. 1988. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J Cell Biol 106:761–771. doi: 10.1083/jcb.106.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buck CB, Pastrana DV, Lowy DR, Schiller JT. 2005. Generation of HPV pseudovirions using transfection and their use in neutralization assays. Methods Mol Med 119:445–462. [DOI] [PubMed] [Google Scholar]
- 45.Rasband WS. 2014. ImageJ. National Institutes of Health, Bethesda, MD. [Google Scholar]
- 46.Bolte S, C F. 2006. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc 224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 47.Pichette B. 2010. Color pixel counter. http://imagejdocu.tudor.lu/doku.php?id=plugin:color:color_pixel_counter:start. [Google Scholar]
- 48.Christensen ND, Cladel NM, Reed CA, Budgeon LR, Embers ME, Skulsky DM, McClements WL, Ludmerer SW, Jansen KU. 2001. Hybrid papillomavirus L1 molecules assemble into virus-like particles that reconstitute conformational epitopes and induce neutralizing antibodies to distinct HPV types. Virology 291:324–334. doi: 10.1006/viro.2001.1220. [DOI] [PubMed] [Google Scholar]
- 49.Day PM, Schelhaas M. 2014. Concepts of papillomavirus entry into host cells. Curr Opin Virol 4:24–31. doi: 10.1016/j.coviro.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schiller JT, Day PM, Kines RC. 2010. Current understanding of the mechanism of HPV infection. Gynecol Oncol 118:S12–S17. doi: 10.1016/j.ygyno.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sapp M, Kraus U, Volpers C, Snijders PJ, Walboomers J M, Streeck RE. 1994. Analysis of type-restricted and cross-reactive epitopes on virus-like particles of human papillomavirus type 33 and in infected tissues using monoclonal antibodies to the major capsid protein. J Gen Virol 75(Pt 12):3375–3383. doi: 10.1099/0022-1317-75-12-3375. [DOI] [PubMed] [Google Scholar]
- 52.Bienkowska-Haba M, Williams C, Kim SM, Garcea RL, Sapp M. 2012. Cyclophilins facilitate dissociation of the human papillomavirus type 16 capsid protein L1 from the L2/DNA complex following virus entry. J Virol 86:9875–9887. doi: 10.1128/JVI.00980-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schelhaas M, Ewers H, Rajamäki M-L, Day PM, Schiller JT, Helenius A. 2008. Human papillomavirus type 16 entry: retrograde cell surface transport along actin-rich protrusions. PLoS Pathog 4:e1000148. doi: 10.1371/journal.ppat.1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith JL, Lidke DS, Ozbun MA. 2008. Virus activated filopodia promote human papillomavirus type 31 uptake from the extracellular matrix. Virology 381:16–21. doi: 10.1016/j.virol.2008.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shiba K, Inaba K. 2014. Distinct roles of soluble and transmembrane adenylyl cyclases in the regulation of flagellar motility in Ciona sperm. Int J Mol Sci 15:13192–13208. doi: 10.3390/ijms150813192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aydin I, Weber S, Snijder B, Samperio Ventayol P, Kühbacher A, Becker M, Day PM, Schiller JT, Kann M, Pelkmans L, Helenius A, Schelhaas M. 2014. Large scale RNAi reveals the requirement of nuclear envelope breakdown for nuclear import of human papillomaviruses. PLoS Pathog 10:e1004162. doi: 10.1371/journal.ppat.1004162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pyeon D, Pearce SM, Lank SM, Ahlquist P, Lambert PF. 2009. Establishment of human papillomavirus infection requires cell cycle progression. PLoS Pathog 5:e1000318. doi: 10.1371/journal.ppat.1000318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao J, Zheng C, Guan J. 2000. Pyk2 and FAK differentially regulate progression of the cell cycle. J Cell Sci 113(Pt 17):3063–3072. [DOI] [PubMed] [Google Scholar]