ABSTRACT
Accumulating evidence indicates a role for Fc receptor (FcR)-mediated effector functions of antibodies, including antibody-dependent cell-mediated cytotoxicity (ADCC), in prevention of human immunodeficiency virus type 1 (HIV-1) acquisition and in postinfection control of viremia. Consequently, an understanding of the molecular basis for Env epitopes that constitute effective ADCC targets is of fundamental interest for humoral anti-HIV-1 immunity and for HIV-1 vaccine design. A substantial portion of FcR effector function of potentially protective anti-HIV-1 antibodies is directed toward nonneutralizing, transitional, CD4-inducible (CD4i) epitopes associated with the gp41-reactive region of gp120 (cluster A epitopes). Our previous studies defined the A32-like epitope within the cluster A region and mapped it to the highly conserved and mobile layers 1 and 2 of the gp120 inner domain within the C1-C2 regions of gp120. Here, we elucidate additional cluster A epitope structures, including an A32-like epitope, recognized by human monoclonal antibody (MAb) N60-i3, and a hybrid A32-C11-like epitope, recognized by rhesus macaque MAb JR4. These studies define for the first time a hybrid A32-C11-like epitope and map it to elements of both the A32-like subregion and the seven-layered β-sheet of the gp41-interactive region of gp120. These studies provide additional evidence that effective antibody-dependent effector function in the cluster A region depends on precise epitope targeting—a combination of epitope footprint and mode of antibody attachment. All together these findings help further an understanding of how cluster A epitopes are targeted by humoral responses.
IMPORTANCE HIV/AIDS has claimed the lives of over 30 million people. Although antiretroviral drugs can control viral replication, no vaccine has yet been developed to prevent the spread of the disease. Studies of natural HIV-1 infection, simian immunodeficiency virus (SIV)- or simian-human immunodeficiency virus (SHIV)-infected nonhuman primates (NHPs), and HIV-1-infected humanized mouse models, passive transfer studies in infants born to HIV-infected mothers, and the RV144 clinical trial have linked FcR-mediated effector functions of anti-HIV-1 antibodies with postinfection control of viremia and/or blocking viral acquisition. With this report we provide additional definition of the molecular determinants for Env antigen engagement which lead to effective antibody-dependent effector function directed to the nonneutralizing CD4-dependent epitopes in the gp41-reactive region of gp120. These findings have important implications for the development of an effective HIV-1 vaccine.
INTRODUCTION
Antibodies (Abs) must bind conserved domains on viral envelope (Env) glycoproteins during key points in retroviral replication in order to broadly protect against human immunodeficiency virus type 1 (HIV-1) infection. Their contribution to protection may result from a variety of antiviral mechanisms, including direct neutralization of virus and Fc receptor-dependent effector functions such as antibody-dependent cell-mediated cytotoxicity (ADCC) or antibody-mediated phagocytosis (1–4). Antibodies that directly neutralize HIV can provide protection, as evidenced in several nonhuman primate studies with passively transferred monoclonal antibodies (MAbs) (5–8), although their role in preventing natural HIV transmission remains equivocal (reviewed in reference 9). On the other hand, a growing body of evidence indicates that direct neutralizing activity is not an absolute requirement for humoral protection against HIV infection. The RV144 vaccine trial in humans (10–13), vaccine trials in nonhuman primates (14–17), early passive immunization studies against simian immunodeficiency virus (SIV) using polyclonal sera (18, 19), and a breast milk transmission study of mother-infant pairs (20, 21) have linked Fc receptor-mediated effector functions with control or prevention of infection, often in the absence of neutralization. Finally, the Fc effector functions' contribution to the blocking of viral entry, the suppression of viremia, and the therapeutic activity of several different anti-Env broadly neutralizing Abs (bnAbs) was confirmed recently in both a mouse model of HIV-1 entry and a model of MAb-mediated therapy using HIV-1-infected humanized mice (22). Overall, these findings suggest that a vaccine capable of generating both neutralizing and nonneutralizing humoral responses will provide the broadest measure of protection at the population level.
While the neutralizing epitopes have been examined in much detail (23–34), relatively little is known about epitopes that are targets for antibodies acting through Fc receptor-dependent effector functions, their degree of overlap with neutralizing epitopes, the immunological rules underlying their selection during anti-Env antibody responses, or their precise locus of action (e.g., transmission blocking or postinfection viral control). While neutralization and Fc receptor-dependent processes of antibodies can be coincident for a given specificity, as has been reported for antibodies targeting the gp120 variable loops, the coreceptor binding site, or the V2 loop region (35–38), they can also be dissociated. Epitopes on both gp120 and gp41 are known that are targeted by antibodies lacking neutralizing activity but capable of potent Fc-mediated effector function (reviewed in references 37 38). In this group, nonneutralizing, CD4-inducible (CD4i) epitopes in the C1-C2 region of gp120 (A32-like epitopes) have recently received much attention as potent ADCC targets (39–42). RV144 analyses implicated this gp120 region as a target of ADCC responses that correlated with reduced infection. In addition, a number of MAbs specific for A32-blockable epitopes were recovered from vaccinated subjects (43) which mediated cross-clade ADCC activity and synergized with V2-specific MAbs to mediate ADCC against the tier 2 isolate AE.CM235 (44). Lastly, the protective vaccine efficacy due to ADCC responses of C1-region-specific MAbs was greatly attenuated by the presence of IgA MAbs incapable of NK cell-mediated effector function but competing for the same Env binding sites (45).
Previously, we designated C1-C2 epitopes mapping to the gp41-reactive face of gp120 cluster A, the canonical examples being MAbs A32 and C11 (41). These epitopes are exposed after envelope trimers engage target cell CD4 and persist on freshly infected cell surfaces for extended periods of time postinfection (46–48). They are also exposed on the surfaces of persistently infected cells. In general, cluster A epitopes are naturally immunogenic as HIV-1-infected individuals frequently elicit C1-C2-specific antibodies (39, 40, 42, 49). We along with others have shown that these epitopes become major targets for ADCC responses during HIV-1 infection (39, 41, 42, 50), and ADCC responses to this region are also subject to immune escape early in infection (51). Recently, it was also shown that exposure of cluster A epitopes is modulated by downregulation of CD4 on the surface of the infected target cell by host factors Nef and Vpu (42, 52). This points toward the possibility of Nef and Vpu evolving as viral defenses against the exposure of these epitope targets during virion release and as an ADCC evasion mechanism preventing antibody-mediated clearance of virus-infected cells (42, 52).
We previously reported that cluster A is comprised of at least three epitope subregions, as defined by enzyme-linked immunosorbent assay (ELISA) competition with MAbs A32 and C11 for binding to CD4 triggered gp120 (41). One subgroup competes with only A32 (A32-like epitopes), the second competes with only C11 (C11-like epitopes), and the third competes with both A32 and C11 (hybrid A32-C11-like epitopes). Recently, we defined the A32-like epitope subregion at the atomic level by describing structures of Fab fragments of two A32-like antibodies in complexes with the CD4-triggered gp120 cores (53). These studies mapped the A32-like epitope to the mobile layers 1 and 2 of the gp120 inner domain within the C1-C2 regions. They also pointed toward a role of precise epitope targeting and mode of antibody binding in the Fc-mediated effector functions of antibodies against HIV-1. Here, we elucidate two more epitope structures within the cluster A region and provide a more comprehensive understanding of how these epitopes are recognized by a human MAb and a rhesus macaque MAb, both capable of potent ADCC function.
MATERIALS AND METHODS
Protein purification.
JR4 and N60-i3 monoclonal antibodies (MAbs) were purified by HiTrap protein A column (GE Healthcare) chromatography from 293T supernatants prepared by transfecting plasmids carrying the heavy- and light-chain genes of the respective Abs. The Fabs of both the MAbs were prepared from the purified IgGs (10 mg/ml) by proteolytic digestion with immobilized papain (Pierce, Rockford, IL) and purified using a protein A column to remove Fc (GE Healthcare, Piscataway, NJ), followed by gel filtration chromatography on a Superdex 200 16/60 column (GE Healthcare, Piscataway, NJ). The elution peak of each of the Fabs corresponded to a molecular mass of approximately 50 kDa and was collected and concentrated for use in the crystallization trials.
For crystallographic studies, the gp120 extended core (coree) protein of clade A/E strain 93TH057 (gp12093TH057 coree; gp120 lacking the N and C termini and variable loops 1, 2, and 3 [V1V2V3]) and the CD4-mimetic miniprotein M48 (F23M47) or M48U1 (54, 55) were used to prepare the ternary complexes of JR4 and N60-i3, respectively. gp12093TH057 coree was prepared and purified as previously described (53). Deglycosylated gp12093TH057 coree was first mixed with the CD4-mimetic peptide M48 or M48U1 at a molar ratio of 1:1.5 and purified through gel filtration chromatography using a Superdex 200 16/60 column (GE Healthcare, Piscataway, NJ). After concentration, the gp12093TH057 coree-M48 or gp12093TH057 coree-M48U1 complex was mixed with a 20% molar excess of JR4 Fab or N60-i3 Fab, respectively, and passed again through the gel filtration column equilibrated with 25 mM Tris-HCl buffer, pH 7.2, with 0.35 M NaCl for the JR4 Fab-gp12093TH057 coree-M48 complex and with 0.15 M NaCl for the N60-i3 Fab-gp12093TH057 coree-M48U1 complex. The purified complexes were concentrated to ∼10 mg/ml for crystallization experiments.
Crystallization.
Initial crystal screens were done in robotic vapor diffusion sitting-drop trials using commercially available sparse-matrix crystallization screens and then reproduced and optimized using the hanging-drop vapor diffusion method (drops of 0.5 μl of protein and 0.5 μl of precipitant solution equilibrated against 700 μl of reservoir solution). JR4 Fab crystals were obtained from a solution containing 0.2 M ammonium sulfate, 1.0 M sodium cacodylate trihydrate, pH 6.5, and 30% (wt/vol) polyethylene glycol (PEG) 5000. Prior to being frozen, the crystals were transferred into a crystallization solution containing 15% (vol/vol) glycerol. Crystals of JR4 Fab-gp12093TH057 coree-M48 were grown from 16.6% PEG 400, 13.3% PEG 3350, 0.1 M MgCl2, and 0.1 M Tris (pH 8.5) and soaked in mother liquor supplemented with 20% 2-methyl-2,4-pentanediol (MPD) prior to being frozen for data collection. Crystals of N60-i3 Fab-gp12093TH057-M48U1 were grown in 10 to 16% PEG 8000 or PEG 10000, 0.065 M NaCl, and 0.1 M Tris-HCl (pH 8.5) at 22°C and cryoprotected in 18% MPD, 16% PEG 8000 or 10000, 0.1 M Tris-HCl (pH 8.5), and 0.065 M sodium chloride.
Data collection and structure solution.
Diffraction data were collected at the Stanford Synchrotron Radiation Light Source (SSRL) at the beam lines BL9-2 (JR4 Fab), BL12-2 (JR4 Fab-gp12093TH057 coree-M48), and BL7-1 (N60-i3 Fab-gp12093TH057 coree-M48U1), equipped with MAR325, Pilatus 6M PAD, and ADSC Quantum 315 area detectors, respectively. All data were processed and reduced with HKL2000 (56). Structures were solved by molecular replacement with Phaser (57) from the CCP4 suite (58) based on the coordinates of gp120 (PDB accession number 3TGT) and the N5-i5 Fab (PDB 4H8W) and the coordinates of the CD4-mimetic peptide M48 (PDB 4K0A) and M48U1 (PDB 4JZW). Refinement was carried out with Refmac (59) and/or Phenix (60). Refinement was coupled with manual refitting and rebuilding with COOT (61). Data collection and refinement statistics are shown in Table 1.
TABLE 1.
Crystallographic data collection and refinement statistics
| Parameter | Value for:a |
||
|---|---|---|---|
| Fab N60-i3-gp12093TH057 coree-M48U1 | Fab JR4 | Fab JR4-gp12093TH057 coree-M48 | |
| Data collection | |||
| Wavelength (Å) | 1.127 | 1.045 | 1.127 |
| Space group | P2(1)2(1)2 | P1 | P2(1) |
| Cell parameters | |||
| a, b, c (Å) | 98.3, 102.6, 108.0 | 79.4, 79.6, 82.0 | 110.3 77.8 127.6 |
| α, β, γ (°) | 90, 90, 90 | 78.8, 82.9, 65.2 | 90, 114.3, 90 |
| No. of molecules/AUb | 4 | 8 | 8 |
| Resolution (Å) | 50–3.20 (3.26–3.20) | 50.0–1.91 (1.95–1.91) | 50–3.17 (3.23–3.17) |
| No. of reflections | |||
| Total | 66,561 | 220,077 | 145,898 |
| Unique | 17,516 | 129,457 | 39,432 |
| Rmerg (%)c | 12.6 (89.9) | 12.5 (60.1) | 25.2 (87.1) |
| I/σ | 10.7 (1.3) | 7.35 (1.5) | 6.9 (1.4) |
| Completeness (%) | 93.7 (96.0) | 93.6 (95.5) | 99.8 (100) |
| Redundancy | 3.8 (3.8) | 1.7 (1.6) | 3.7 (3.7) |
| Refinement statistics | |||
| Resolution (Å) | 36.01–3.2 | 40.2–1.91 | 45.0–3.21 |
| R (%)d | 22.0 | 19.2 | 27.3 |
| Rfree (%)e | 27.7 | 24.6 | 33.2 |
| No. of atoms | |||
| Protein | 6,010 | 12,736 | 12,060 |
| Water | 1 | 1456 | |
| Ligand | 158 | 25 | 278 |
| Overall B value (Å2) | |||
| Protein | 117.1 | 21.1 | 94.6 |
| Water | 44.1 | 29.6 | |
| No. of ligands/No. of ions | 112.0 | 40.9 | 96.7 |
| Root mean square deviation | |||
| Bond lengths (Å) | 0.007 | 0.018 | 0.007 |
| Bond angle (°) | 1.41 | 1.77 | 1.48 |
| Ramachandran plotf | |||
| Favored (%) | 72.9 | 90.4 | 90.2 |
| Allowed (%) | 24.0 | 9.1 | 9.3 |
| Outliers (%) | 3.1 | 0.5 | 0.5 |
Values in parentheses are for highest-resolution shell.
AU, asymmetric unit.
Rmerge = Σ |I − <I>|/ΣI, where I is the observed intensity and <I> is the average intensity obtained from multiple observations of symmetry-related reflections after rejections.
R = Σ ||Fo| − |Fc||/Σ |Fo|, where Fo and Fc are the observed and calculated structure factors, respectively.
Rfree, calculated as defined by Brünger (73).
Calculated with MolProbity (62).
Structure validation and analysis.
The quality of the final refined models was monitored using the program MolProbity (62). Structural alignments were performed using the Dali server and the program lsqkab from the CCP4 suite (58). PISA (63) and PIC (64) web servers were used to determine contact surfaces and residues. All illustrations were prepared with the PyMol molecular graphics suite (http://pymol.org) (DeLano Scientific, San Carlos, CA, USA).
FRET-FCS competition assay.
Alexa 488-Alexa 568 donor-acceptor pairs were used for competition assays using fluorescence resonance energy transfer (FRET)-fluorescence correlation spectroscopy (FCS). For FRET measurements, the Fabs (C11, A32, N60i3, and JR4) were labeled with either donor (Alexa 488) or acceptor (Alexa 568) probes (Invitrogen MAb labeling kit). Briefly, the Alexa Fluor 488 or 568 reactive dye has a succinimidyl ester moiety that reacts efficiently with primary amines of Fab to form stable dye-protein conjugates. The dye-labeled Fabs were purified using 10-kDa cutoff spin columns. Purified Alexa 488- or 568-labeled Fab was quantified by a UV-visible light (UV-Vis) spectrometer (NanoDrop 2000). Dye-to-protein ratios were determined by measuring absorbance at 280 nm (protein) versus 488 or 577 nm (dye). The dye-to-protein ratios were between 1 and 2. We specifically aimed to keep this low level of dye labeling as we were using a single-molecule fluorescence method to minimally perturb the functionality of the protein. FRET measurements were performed in a confocal microscope (MicroTime 200; PicoQuant). PicoQuant Symphotime software was used to generate the FRET histograms and for further analyses. FRET measurements were performed after an immune complex with full-length single-chain gp120BaL-soluble CD4 (sCD4) (FLSC; gp120BaL is gp120 of the HIV-1 BaL isolate) was formed with donor-labeled Fab and acceptor-labeled Fab. In all of our measurements each Fab concentration was 1 μg/ml, and the FLSC concentration was 1.5 μg/ml. The immune complexes were made by incubating Fabs with the FLSC at 20°C for 1 h. Fluorescence responses from the donor and the acceptor molecules were separated by a 50/50 beam splitter and detected by two avalanche photodiode detectors (APD) using the method of time-correlated single-photon counting and the time-tagged time-resolved (TTTR) mode of the PicoHarp 300 board. High-quality bandpass (Chroma) filters were used for recording donor and acceptor fluorescence in two separate detection channels. The collected single-photon data were binned by a 1-ms bin in each channel (donor or acceptor), which resulted in intensity-time traces and count-rate histograms. Threshold values in each channel were used to identify the single-molecule bursts from the corresponding background signal level. Fluorescence bursts were recorded simultaneously in donor and acceptor channels, and FRET efficiencies were calculated using E = IA/(IA + γID) where ID and IA are the sums of donor counts and acceptor counts for each burst, respectively, taking into account the possible difference in the detection efficiency (γ) values in two separate channels (65). The donor-to-acceptor distance (r) in terms of efficiency of energy transfer (E) and Förster distance (R0) is given by r = R0(1/E − 1)1/6. We have used an R0 value of 62 Å for the Alexa 488 (donor) and Alexa 568 (acceptor) pair for estimating the donor-to-acceptor distances. In addition to FRET measurements, we have also performed FCS measurements to assess in vitro binding of single or multiple Fab fragments to the FLSC. We determined translational diffusion coefficients of Alexa 488- or 568-labeled Fabs and the corresponding immune complexes from FCS measurements. The FCS measurements and analyses were performed as previously reported (47).
SPR competition analysis.
The binding footprints of MAb N60-i3 and JR4 in relation to MAb C11 and A32 were assessed by surface plasmon resonance (SPR) competition on a Biacore T-100 (GE Healthcare) at 25°C with buffer HBS-EP (0.01 M HEPES, pH 7.4, 0.15 M NaCl, 3 mM EDTA, and 0.05% surfactant P-20). Protein A was first immobilized onto the second of two flow cells on a CM5 chip to ∼3,000 response units (RU), and the first flow cell was blocked with a standard amine coupling protocol (GE Healthcare). IgGs to be evaluated were then captured onto the second flow cell by flowing a 5 to 10 nM solution of MAb at a 10-μl/min flow rate for 30 s. The antibody concentration was varied to give an RU in the range of 150 to 400. The single-chain gp120BaL-sCD4 complex (FLSC) (66) was then passed over the same flow cell at a flow rate of 10 μl/min for 30 s. An FLSC concentration was chosen to give an RU in the range of 150 to 400, comparable to the RU for the antibody. Various concentrations of MAb Fab were then passed over both flow cells at a flow rate of 30 μl/min for 200 s and allowed to dissociate by passing the buffer over both cells at the same flow rate for 800 s. The cells were regenerated between concentrations with a 30-s injection of 0.1 M glycine, pH 3.0, at a flow rate of 100 μl/min. The antibody and FLSC were then reloaded onto the second flow cell for the next concentration. Blank sensorgrams were obtained by injection of HBS-EP buffer in place of Fab. Sensorgrams of the concentration series (flow cell two minus flow cell one) were corrected with a corresponding blank.
ADCC assays.
ADCC assays were carried out using the rapid fluorescence ADCC method (67) modified to reduce prozone effects. All ADCC studies used CEM-NKr-CCR5 target cells sensitized with recombinant gp120 from the HIV-1 BaL (HIV-1BaL) isolate or spinoculated with AT-2-inactivated BaL HIV-1 virus (kindly supplied by Jeffrey Lifson, National Cancer Institute) at 3,000 rpm for 2 h at 12°C. gp120-sensitized or virus-spinoculated cells were then washed twice and added to a 96-well V-bottom plate (5,000 cells/well). The gp120-sensitized or virion-bound target cells were incubated with MAb dilutions for 15 min and washed with culture medium before the addition of peripheral blood mononuclear effector cells from healthy donors at a final ratio of 50:1. The effector and target cells were pelleted by centrifugation and incubated for 2 to 3 h at 37°C, followed by fixation and cytolysis determined by flow cytometry as described in Gomez-Roman et al. (67). The absolute cytotoxicity values were normalized using the MAb C11 as described previously (41).
Protein structure accession numbers.
Structures of Fab N60-i3-gp12093TH057-M48U1, Fab JR4, and Fab JR4-gp12093TH057-M48 were deposited in Protein Data Bank with accession codes 4RFO, 4RFE, and 4RFN, respectively.
RESULTS
MAb origin and epitope cluster A assignment.
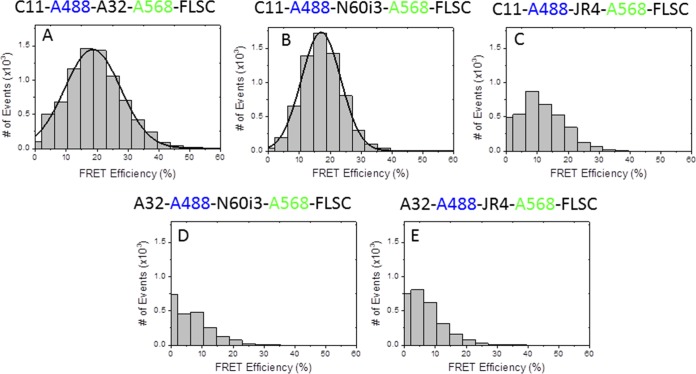
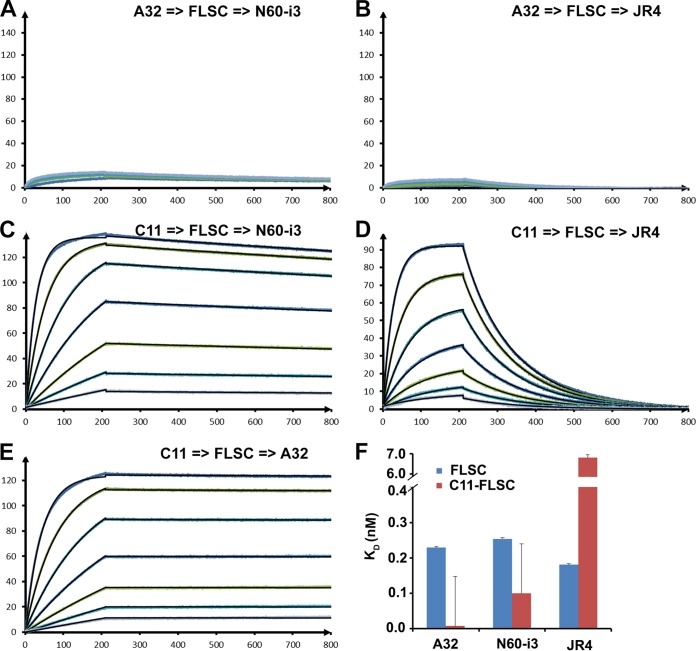
MAb N60-i3 was isolated from B cells of an HIV-1-infected individual and characterized for initial reactivity using recombinant proteins based on the HIV-1BaL isolate as described previously for other cluster A MAbs (41). MAb JR4 was derived from the peripheral blood B cells of a rhesus macaque infected with a simian-human immunodeficiency virus (SHIV) KB9 mutant with deletions of glycosylation sites in gp41. The detailed description of MAbs N60-i3 and JR4, JR4 isolation, germ line gene usage, and degree of somatic hypermutation will be published elsewhere. MAbs N60-i3 and JR4, similar to other CD4-inducible (CD4i) MAbs of cluster A (41), show preferential binding to gp120-CD4 complexes compared with monomeric gp120 and no binding to Env trimers expressed on the cell surface in the presence or absence of soluble CD4 (sCD4; domains d1 to d4 of CD4) (see Fig. S1 in the supplemental material) (47, 48, 53). The initial epitope assignments were accessed based on competition of N60-i3 and JR4 binding to the single-chain gp120BaL-sCD4 complex (FLSC) (66) by MAbs A32 and C11, two antibodies specific for distinct (nonoverlapping) epitopes in the cluster A region (41). Recently, we defined epitopes in the A32-like region by describing epitope structures of two A32-like MAbs, N5-i5 and 2.2c, and mapped them to the C1-C2 regions of gp120 (53). In contrast, the binding site for MAb C11 is still unresolved, but it has been mapped to the seven-stranded β-sandwich of gp120 and a residue in the extended C terminus of gp120 by mutagenesis studies (68). To precisely access competition due to the overlaps of epitope footprints and eliminate a possibility of avidity effects or steric clashes outside the antigen-antibody binding interface, we have developed a new fluorescence resonance energy transfer-fluorescence correlation spectroscopy (FRET-FCS)-based competition assay in which antigen-binding fragments (Fabs) of tested antibodies compete in solution for binding to the Env antigen (Fig. 1). Additionally, we also tested the capacities of MAbs A32 and C11 to block Fab N60-i3 and JR4 binding to the FLSC in an SPR competition assay (69) (Fig. 2). Shown in Fig. 1 are the FRET histograms of Fab pairs labeled with Alexa 488 (A488) or Alexa 568 (A568) of MAbs N60-i3, JR4, A32, and C11 bound to the FLSC. When binding of C11-A488 Fab and A32-A568 Fab was tested, the data could be fitted well with a Gaussian profile showing ∼20% FRET efficiency (Fig. 1A), clearly confirming the coexistence of A32 and C11 Fabs bound to a single FLSC protein molecule with a stoichiometry of C11/A32/FLSC of 1:1:1. A similar FRET profile was observed for C11-A488 Fab and N60-i3-A568 Fab binding to the FLSC with ∼18% FRET efficiency (Fig. 1B), confirming that the C11 Fab and N60-i3 Fab bind to the nonoverlapping FLSC epitopes. Additionally, mean FRET efficiencies translated to an average distance of 78 Å between C11 and A32 Fabs and a distance of 79.8 Å between C11 and N60-i3 Fabs. It is important to note that the Fabs are not fluorescently labeled at a specific position; hence, the distance derived from our FRET measurements represents an average value between the donor- and acceptor-labeled epitope probes. However, the binding of epitope probes to the FLSC is further confirmed by the diffusion coefficients derived from the FCS measurements. In contrast, when binding of C11-A488 Fab and JR4-A568 Fab was analyzed (Fig. 1C), the efficiency of FRET was below 10%, and a Gaussian distribution could not be obtained. The autocorrelation measurements in the C11 channel also showed the presence (25%) of unbound C11. This indicated that JR4 Fab, in contrast to N60-i3 Fab, is capable of partially blocking C11 Fab binding to the FLSC antigen. On the other hand, in the same assay both N60-i3 Fab and JR4 Fab blocked the binding of A32 Fab to the FLSC, as indicated by the lack of FRET signal for the mixtures of A32-A488 N60-i3-A568-FLSC and A32-A488 JR4-A568 Fab-FLSC (Fig. 1D and E). The FRET-FCS competition data are in good agreement with the results of the SPR competition assay. With A32 IgG bound to the FLSC, neither the N60-i3 nor the JR4 Fab could bind, indicative of complete competition for the same binding site on the FLSC. With C11 IgG bound to the FLSC, N60-i3 Fab could also bind with an apparent KD (equilibrium dissociation constant) similar to that of N60-i3 binding to the FLSC alone, suggesting no overlap in the binding sites. A32 Fab showed a similar result with C11 IgG bound to the FLSC. However, with C11 IgG bound to the FLSC, JR4's apparent KD was approximately 38-fold lower than that of JR4 binding to the FLSC alone, suggesting a partial overlap in their binding sites (Fig. 2). Altogether, these data suggest that MAb N60-i3 recognizes an A32-like epitope, whereas MAb JR4 may recognize an A32-C11 hybrid epitope.
FIG 1.
FRET histograms of donor (A488)-labeled Fabs, acceptor (A568)-labeled Fabs, and full-length single-chain gp120BaL-CD4 complex (FLSC) in solution as determined by the FRET-FCS approach (see Materials and Methods for details). Data in panels A and B indicate ∼20% FRET efficiency for A488-labeled C11 Fab and A568-labeled A32 Fab or N60-i3 Fab bound to FLSC, respectively. Panels D and E do not show detectable FRET signals for A488-labeled A32 Fab and A568-labeled N60-i3 Fab or JR4 Fab in the presence of FLSC in solution. From the FCS measurements, the diffusion coefficient of dye-labeled Fab is ∼83 μm2/s. This diffusion has been significantly decreased upon binding two Fabs to FLSC. The diffusion coefficient for this immune complex is ∼36 μm2/s.
FIG 2.
MAb A32/C11 competition of N60-i3/JR4 Fab binding as measured by a surface plasmon resonance competition assay. MAb A32 (A and B) and MAb C11 (B, C, and D) were immobilized on a protein A chip, and FLSC antigen was loaded to form a competitor antibody-antigen complex. Various concentrations of Fab MAb were tested. N60-i3 (A and C), JR4 (B and D), and A32 (E) were then passed over the chip. (F) Sensorgrams of the concentration series are shown. Binding affinities for MAbs A32, N60-i3, and JR4 to FLSC in the absence (blue bars) and presence (red bars) of C11 IgG were calculated with the BIAevaluation software.
MAbs N60-i3 and JR4 show potent ADCC activity.
We tested the ADCC potency of MAb N60-i3 and MAb JR4 using CEM-Nkr-CCR5 target cells sensitized with gp120 (Fig. 3A) or with AT-2-inactivated BaL virions (Fig. 3B) of the HIV-1BaL isolate, as described in Materials and Methods. MAbs N60-i3 and JR4 are potent mediators of ADCC in both assay formats using the potency criteria described previously (41).
FIG 3.
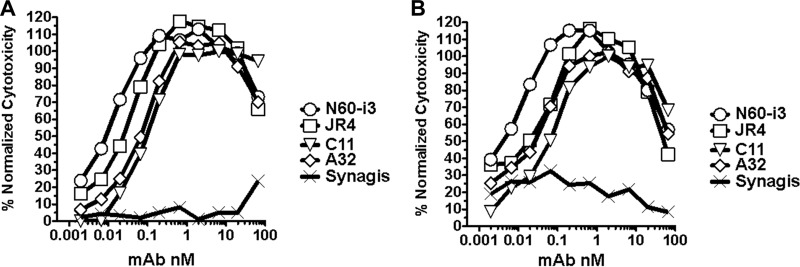
Representative ADCC curves for MAbs N60-i3 and JR4. ADCC assays were performed as described in Materials and Methods using CEM-NKr-CCR5 target cells sensitized with gp120 of HIV-1BaL isolate (A) or spinoculated with AT-2-inactivated BaL viruses (B). MAb palivizumab (Synagis; MedImmune, Inc.) was included as a control.
Structures of MAb N60-i3- and JR4-Env antigen complexes.
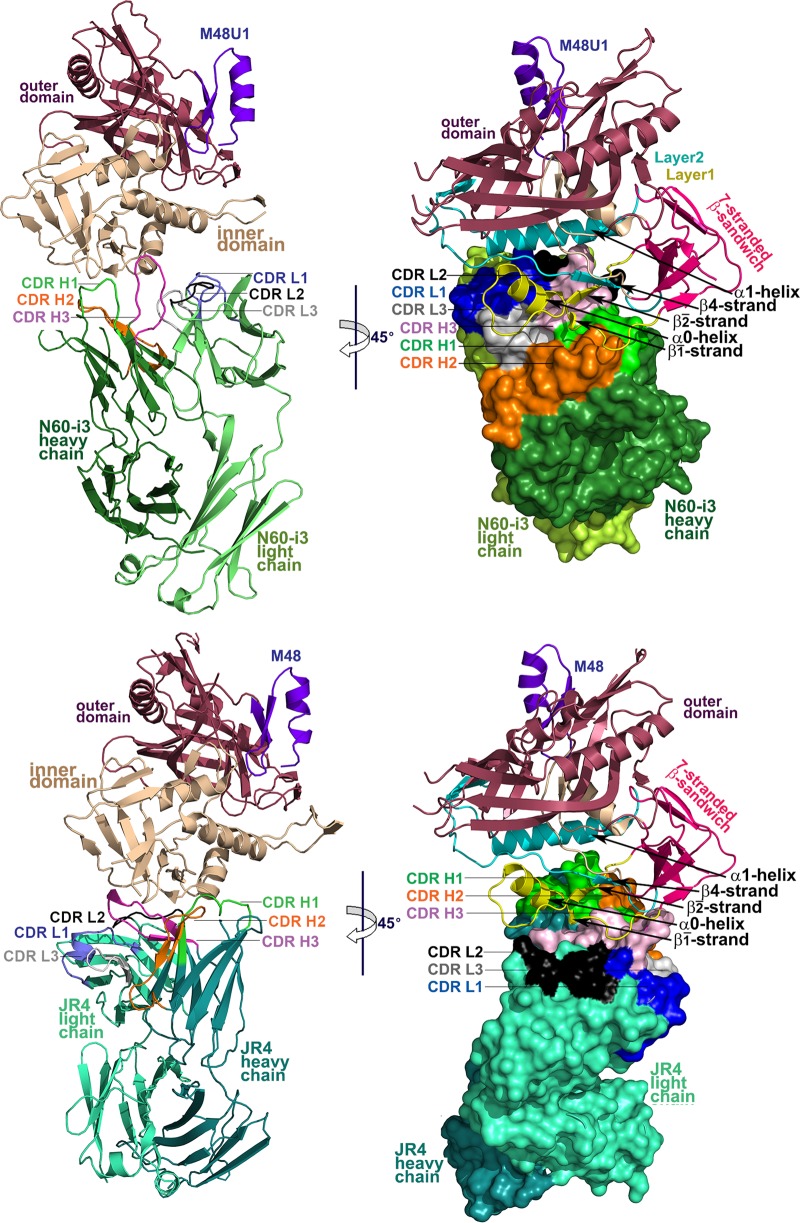
In an effort to elucidate the epitopes of MAbs N60-i3 and JR4 and differences in epitope footprints, if any, that could explain differences in their A32/C11 cross-competition, we determined the crystal structures of the complexes formed between their antigen-binding fragments (Fabs) and CD4-triggered gp120 antigen. Both complexes were formed using the gp120 extended core protein (residues 44 to 492 with V1V2V3 loops deleted) (27) of the clade A/E 93TH057 isolate (gp12093TH057 coree) and CD4 peptide-mimetic M48U1 (54) (N60-i3 complex) or M48 (JR4 complex). His375 of gp12093TH057 coree in the N60-i3 Fab-gp12093TH057 coree-M48U1 complex was mutated to Ser to accommodate ligands such as M48U1 that penetrate the gp120 Phe43 cavity, as described previously (70). M48U1 is a derivative of M48 and is identical, with the exception that the phenylalanine at position 23 has been replaced with a phenyl cyclohexylmethoxy moiety, increasing its affinity for gp120 by roughly 10-fold or more, based on 50% effective concentrations (EC50s), depending on the HIV-1 strain used (54). The N60-i3 Fab-gp12093TH057 coree-M48U1 assembly crystallized in space group P2(1)2(1)2 with one complex in the asymmetric unit (Table 1). The JR4 Fab-gp12093TH057 coree-M48 assembly crystallized in space group P2(1) with two almost identical copies of complex present in the asymmetric unit (see Fig. S2 in the supplemental material). Structures were solved by molecular replacement at resolutions of 3.2 Å for the N60-i3 Fab complex and 3.21 Å for the JR4 Fab complex and refined to a final R/Rfree of 22.0/27.7% and 27.3/33.2%, respectively. The data collection and refinement statistics for the structures are summarized in Table 1, and the overall structures of complexes are shown in Fig. 4.
FIG 4.
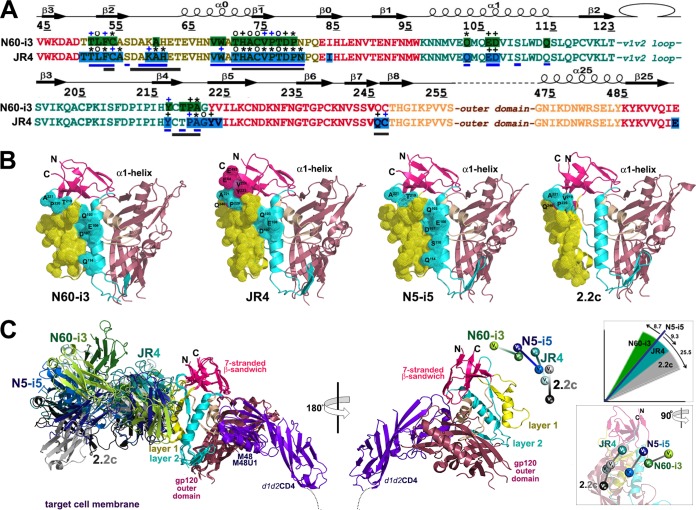
Crystal structures of N60-i3 Fab-gp12093TH057 coree-M48U1 and JR4 Fab-gp12093TH057 coree-M48 complex. Complexes are shown in ribbon representation (right panels), or the molecular surface is displayed over Fab molecules (left panels), with the light/heavy chain of N60-i3 Fab and JR4 Fab shown in light green/dark green and light cyan/dark cyan, respectively. The complementarity-determining regions (CDRs) are shown in slate (CDR L1), black (CDR L2), gray (CDR L3), green (CDR H1), orange (CDR H2), and pink (CDR H3). The gp120 outer domain is shown in raspberry, and the inner domain is shown in wheat (left panels) or colored in a layered color scheme (right panels), with the seven-stranded β-sandwich in magenta, layer 1 in yellow, layer 2 in cyan, and layer 3 in light wheat. The mimetic peptides M48U1 and M48 are shown in violet.
The N60-i3 Fab-gp12093TH057 coree-M48U1 and JR4 Fab-gp12093TH057 coree-M48 complex structures revealed that both MAb N60-i3 and MAb JR4 bind at largely overlapping sites in the C1-C2 gp120 region shown previously to be recognized by A32-like MAbs N5-i5 and 2.2c (53). In both cases, a conformational epitope is formed by bridging mobile layers 1 and 2 of the gp120 inner domain involving residues of the α0- and α1-helices, the β2̄-, β1̄-, and β4-strands, and the β2̄-α0-, β1̄-β0-, and β4-β5-connecting coils (Fig. 4). The interactive surface that becomes buried due to N60-i3 Fab-gp12093TH057 coree interaction encompasses 1,464 Å2 (770 Å2 contributed by Fab and 694 Å2 from gp120) (see Table S1 in the supplemental material) and is roughly half (63%) the buried surface area (BSA) of the JR4 Fab-gp12093TH057 coree interface (BSA of 2,340 Å2 with 1,145 Å2 buried by Fab and 1,195 Å2 buried by gp120) (see Table S1). Despite the differences in the surface areas buried at the N60-i3 and JR4 complex interfaces, these antibodies show the same affinity for gp120 in a CD4-bound conformation, as confirmed by SPR analysis (see Fig. S1 in the supplemental material) and isothermal titration calorimetry (ITC) (see Fig. S3). The paratope terrains of N60-i3 and JR4 Fab are flat and electropositive, with the only protruding areas contributed by complementarity-determining regions (CDRs) of heavy chains 1 and 3 (CDR H1 and CDR H3), respectively, making the contacts with the α1-helix of layer 2. In both complexes, the heavy chain contributes most of the Fab binding surface (approximately 85% and 83% of BSA of N60-i3 Fab and JR4 Fab, respectively), with all three CDRs (CDRs H1 to H3) involved in the interaction (see Table S1 in the supplemental material). CDR H3s of 12 and 14 residues for N60-i3 and JR4, respectively, provide most of their heavy chain BSA (approximately 57% and 44% as calculated for N60-i3 and JR4, respectively) (see Tables S1 and S2 and Fig. S4 in the supplemental material). In both cases, the contribution of the light chain to the antibody-antigen interface is minimal, with only two CDRs (CDR light chain 1 [L1] and CDR L3 for N60-i3 and CDR L1 and CDR L2 for JR4) engaged in binding (Fig. 5; see also Fig. S4). A total of 56 (7 H bonds) and 54 (15 H bonds) contacts as defined by a 5-Å cutoff are formed at the heavy chain-gp12093TH057 coree interfaces of N60-i3 and JR4, respectively (Fig. 5, right panel). By comparison, the light chain contributes 4 (0 H bonds) and 5 (1 H bond) contacts to the complex interface of N60-i3 and JR4, respectively (Fig. 5, right panel).
FIG 5.
Binding of MAbs N60-i3 and JR4 to gp120 antigen. On the left, the N60-i3 Fab-gp12093TH057 coree and the JR4 Fab-gp12093TH057 coree interfaces are shown with the molecular surfaces displayed at the gp120 molecules and with CDRs of Fabs shown as ribbons. Only CDRs contributing to the binding are shown. The 45° rotations show close-up views of the binding interfaces of Fabs and the α1-helix of layer 2. H bonds are shown as dashes in blue. Colors are as defined in the legend of Fig. 4. On the right, networks of interactions formed between Fabs and gp120 antigen as defined by a 5-Å distance criterion cutoff are shown as solid lines. H bonds are shown as dashes in blue.
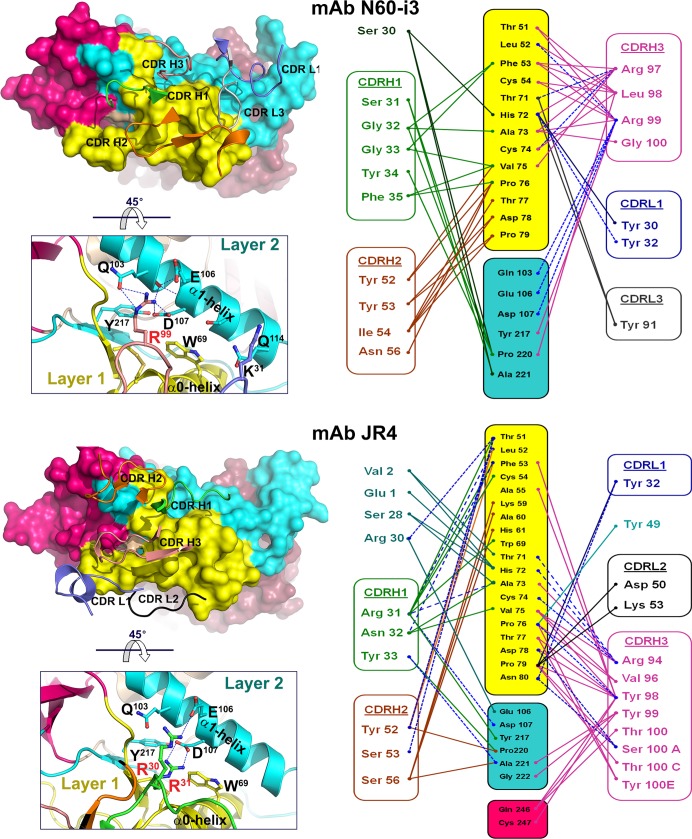
MAb N60-i3 and JR4 epitope footprints.
The epitope footprint of MAb N60-i3 maps exclusively to layers 1 and 2 of the C1-C2 region of the gp120 inner domain. MAb JR4 largely overlaps MAb N60-i3 in targeting layers 1 and 2 but also involves contacts within the seven-stranded β-sandwich and the C terminus of gp120 (Fig. 6).
FIG 6.
Structural basis for interaction of cluster A MAbs with gp120 antigen. (A) Mapping of the N60-i3 and JR4 contact residues on the primary sequence of the gp120 inner domain of the 93TH057 isolate. The topology diagram depicting a distribution of secondary structure elements is shown above the gp120 sequences. Buried residues are highlighted in green and blue. Main chain (o), side chain (+), and both side and main chain (*) interactions are shown immediately above the residues as defined by a 5-Å distance criterion cutoff and colored based on contact type: hydrophobic, blue; hydrophilic, green; or both, black. Residues forming the N5-i5 and 2.2c epitopes as described in reference 53 are indicated by blue and gray lines below the gp120 sequence, respectively. (B) Epitope footprints of MAb N60-i3, JR4, N5-i5, and 2.2c. The Cα atoms of the gp120 residues involved in Fab binding are shown as spheres and displayed over the gp120 ribbon diagram. The selected residues of layer 2 and of the seven-stranded β-sandwich contribution to Fab binding and all the residues in the α1-helix involved in N60-i3, JR4, and N5-i5 binding are labeled. (C) Comparison of binding of MAb N60-i3, JR4, N5-i5, and 2.2c to CD4-triggered gp120 antigen. The N60-i3 Fab-gp12093TH057 coree-M48, JR4 Fab-gp12093TH057 coree-M48U1, and 2.2c Fab-gp120YU2 coree-M48U1 complexes were superimposed based on the gp120 outer domain onto the N5-i5 Fab-gp12093TH057 coree-d1d2CD4 complex and oriented relative to the target cell membrane. In the 180° view only the gp12093TH057 coree and d1d2CD4 from N5-i5 Fab-gp12093TH057 coree-d1d2CD4 complex (53) are shown, with variable heavy and light (VH and VL) domains of Fabs displayed as balls. Insets show rotation angles calculated using gp120's center of mass as an origin and the average α-carbon position for the heavy chain framework region 2 (residues 36 to 49) as a reference point for each antibody (top) and a 90°rotation of the 180°view (bottom).
Layer 1 (C1 gp120 region) contacts.
Layer 1 of the C1 gp120 region makes up the majority of gp120 contact surface engaged in MAb N60-i3 and JR4 binding (79% and 73% of BSA for N60-i3 and JR4, respectively) (see Table S1 in the supplemental material). Layer 1 contacts for both MAbs are similar and include residues 51 to 54, 60, 68 to 69, and 71 to 79 buried at the N60-i3 Fab-gp12093TH057 coree interface and residues 50 to 55, 59 to 61, 68 to 69, 71 to 80, and 82 buried at the JR4 Fab-gp12093TH057 coree interface (Fig. 6A). There are two anchoring points that provide most of the hydrophobic surface utilized by both antibodies to attach to layer 1. These include a Thr51LeuPheCys motif of the β2̄-strand of gp120 and a Thr71HisAlaCysValPro motif at the C termini of the α0-helix and β1̄-strand. JR4 utilizes CDRs H1 to H3 to make hydrophobic contacts with the Thr51LeuPheCys motif, whereas N60-i3 contacts in this region include contributions of CDR H1 and H3 only (Fig. 5; see also Table S2, and Fig. S4 in the supplemental material). In contrast, five CDRs of N60-i3 Fab (CDRs H1 to H3 and CDRs L1 and L3) contribute to its attachment to the Thr71HisAlaCysValPro motif, whereas JR4 utilizes almost exclusively CDR H3 (with a few contacts from residues of the framework region H1 [FWRH1] and CDR H1) to contact this region (Fig. 5, right panel; see also Table S2 and Fig. S4). The Leu53PheCys and Thr71HisAlaCysValPro motifs are coupled by a disulfide bond, Cys54 to Cys74, connecting these two anchor points. The Cys54-Cys74 disulfide bond plays a functional role in stabilizing the native conformation of gp120 and is highly conserved among HIV-1 isolates across clades. With the exception of His72 (97.8% of conservation across clades), most residues of these two motifs are invariant in greater than 99% of HIV-1 sequences, with some like Pro76 and Pro79 invariant in greater than 99.9% of sequences, as determined by the HIV Sequence Database Compendia (http://www.hiv.lanl.gov). JR4's reach into layer 1 is slightly longer than that of N60-i3 and continues to the edge of layer 1 residues, Asn80, Pro81, and Gln82 (Fig. 5, right panel, and 6A).
Layer 2 (C1 and C2 gp120 region) contacts.
MAb N60-i3 and JR4 largely overlap in binding to layer 2 (Fig. 5, right panel, and 6A). These contacts contribute 21% and 16% of BSA for the N60-i3 and JR4 complexes, respectively (see Table S1 in the supplemental material). Layer 2 residues buried at the N60-i3 Fab-gp12093TH057 coree interface include residues 103, 106 to 107, 114, 217, and 219 to 221, whereas the JR4 Fab-gp12093TH057 coree complex buries residues 103, 106 to 107, 217, and 220 to 222. Residues Gln103, Glu106, and Asp107 of the α1-helix serve as the major anchor points for MAbs N60-i3 and JR4 in layer 2 of the C1 gp120 region. N60-i3 coordinates these three residues exclusively through Arg99 of CDR H3, forming a salt bridge with Asp107 and multiple H bonds with Gln103, and Glu106 JR4 uses Arg31 (CDR H1) to coordinate Asp107 through a salt bridge and Arg30 (FWRH1) to establish an H bond with Glu106 (Fig. 5 and 6A; see also Fig. S4 in the supplemental material). The N60-i3 Fab-gp12093TH057 coree interaction also buries Gln114 of the α1-helix, but the contribution of this residue to binding is minimal (see Table S2). Interestingly, CDR H3 of N60-i3 and CDR H1 of JR4, by providing contacts to both the α0-helix of layer 1 and the α1-helix of layer 2, span these two layers and form a single binding surface. This mode of cross-layer attachment closely resembles binding of the potent ADCC mediator MAb N5-i5, which uses its CDR H2 to contact both the α0- and α1-helices (53). While electrostatic interactions play a major role in MAb N60-i3 and JR4 attachment to the α1-helix, the rest of the binding contacts with layer 2 are predominantly hydrophobic. These contacts center on the Tyr217 and around the Thr219ProAla motif of the β4-strand and β4-β5-connecting coil of gp120 (Fig. 5; see also Tables S2 in the supplemental material). The main layer 2 contact residues used by N60-i3 and JR4 are highly conserved; Gln103, Asp107, and Pro220 are invariant in greater than 99.9% of HIV-1 sequences, and Tyr217, Thr219, and Ala221 are present in 99.8% of sequences. Thus, MAbs N60-i3 and JR4 are similar to the ADCC potent cluster A MAb N5-i5 (53) and target highly conserved elements of the HIV-1 envelope within both layers of the C1-C2 gp120 region.
Seven-stranded β-sandwich (C2 region).
Analysis of the JR4 epitope footprint (Fig. 6A) indicates that JR4 also reaches residues in the seven-stranded β-sandwich (residues 84, 223 to 224, and 246 to 247) and residue 492 of the C terminus of gp120. These contacts contribute to approximately 11% to the BSA of the JR4 complex and are not present in the N60-i3 Fab-gp12093TH057 coree interface (see Tables S1 and S2 in the supplemental material). CDR H3 of JR4 is anchored deeper in this area than in N60-i3 and makes multiple contacts with Gln246 and Cys247 of the seven-stranded β-sandwich. In addition Tyr223, Val224, and Glu492 are buried in the JR4 Fab-gp12093TH057 coree interface (Fig. 5). When both N60-i3 and JR4 epitopes are mapped on the gp120 antigen and displayed over the gp120 coree ribbon diagram (Fig. 6B), the shift of the JR4 epitope toward the seven-stranded β-sandwich and the N and C termini of gp120 is evident. In FCS-FRET and SPR cross-competition assays, MAb JR4, but not N60-i3, cross-competes with MAb C11 binding to the gp120 antigen (Fig. 1 and 2). Since the N60-i3 and JR4 epitopes in layers 1 and 2 largely overlap, we can speculate that the JR4 contacts to the seven-stranded β-sandwich account for its hybrid phenotype and its ability to cross-compete with MAb C11 binding to gp120. In this regard, the JR4 epitope represents a hybrid A32-C11 epitope within the cluster A region.
Structural basis for ADCC potency to cluster A region.
We recently reported the atomic-level definition of the A32-like region by providing epitope footprints of two human A32-like antibodies, N5-i5 and 2.2c, both specific for largely overlapping epitope surfaces in the C1-C2 region but varying in their abilities to mediate ADCC, with N5-i5 75-fold more potent than 2.2c (53). These studies pointed toward a dominant role of precise epitope targeting and mode of antibody attachment in ADCC responses when largely overlapping epitopes in the A32-like region are involved. MAb N5-i5, which engages both the α0- and α1-helices of the inner domain layers 1 and 2, respectively, was shown to cross-link antigen better on target cells and to be more effective at ADCC. On the other hand, the impaired ability of 2.2c to mediate effective Fc effector function resulted from suboptimal positioning of its CH2 domain for Fc receptor interaction within the immune complex and from poor accessibility of its epitope for antibody avidity interactions, as judged by cell surface binding and saturation studies, with the positioning having a greater impact. As shown in Fig. 3, MAbs N60-i3 and JR4 represent very potent ADCC mediators in the cluster A region capable of Fc-dependent effector function against target cells sensitized with gp120 of the HIV-1BaL isolate, with an effectiveness comparable to that of MAbs A32 and C11 (41). To better understand the structural basis for the ADCC potency in the cluster A region, we compared the epitope footprints (Fig. 6A and B) and modes of attachment (Fig. 6C) of MAbs N60-i3 and JR4 to the previously described potent and weaker ADCC mediators, MAbs N5-i5 and 2.2c. As expected, the comparison revealed close similarities between epitope footprints of MAbs N60-i3 and JR4 to the MAb N5-i5 footprint as their epitopes largely overlap in layers 1 and 2 (Fig. 6A). Most importantly, MAbs N60-i3, JR4, and N5-i5 all engage the same residues of the α1-helix for binding that include the highly conserved Gln103, Glu106, and Asp107 residues of gp120. MAb 2.2c does not contact the α1-helix and focuses its binding almost entirely onto the α0-helix and layer 1. The N60-i3, JR4, and N5-i5 contacts to the α1-helix are mediated exclusively by arginines of their heavy chains (Arg99 in CDR H3, Arg30 and Arg31 in CDR H1, and Arg55 in CDR H2 of N60-i3, JR4, and N5-i5, respectively) coordinating the Asn103-Glu106-Asp107 triad through an invariant salt bridge and network of H bonds (Fig. 5) (53). Thus, to reach the α1-helix through the heavy chain and target their cognate epitopes, N60-i3, JR4, and N5-i5 must approach the gp120 antigen at similar angles and contact gp120 antigen via similar variable domain contact surfaces. Although there are differences in the modes of attachment among N60-i3, JR4, and N5-i5, defined by the exact orientation of the contact surfaces of the heavy and light chain variable regions (VH and VL, respectively) on gp120, the heavy chain contributions are all in very close proximity (Fig. 6C). Using the conserved framework of the VH domain with the gp120 center of mass as the origin, MAbs N60-i3 and JR4 rotate in their complex by 8.7°and 9.3°, respectively, relative to the VH domain of N5-i5. In contrast, the VH domain of 2.2c rotates 25.5° relative to the N5-i5-defined orientation. Furthermore, although the exact position of the target cell membrane is unknown, based on the model for cell fusion as shown in Fig. 6C, N60-i3, JR4, and N5-i5 approach the gp120 antigen at an angle 16.2° or more farther from the target cell membrane to bind their epitopes than 2.2c. We showed previously that the N5-i5 epitope is more accessible on the target cell surface than the 2.2c epitope, resulting in effective gp120-CD4 complexes cross-linking and potent Fc-mediated effector function. Indeed, although SPR studies show that there are essentially no differences in KD values between the MAbs N60-i3, JR4, and N5-i5 and MAb 2.2c for binding to monomeric gp120-CD4 complexes (FLSCs) (see Fig. S1 in the supplemental material) (53), when tested by ELISA, MAb 2.2c can be easily cross-competed by N60-i3, JR4, or N5-i5 (data not shown). This indicates that the N60-i3/JR4/N5-i5 epitope is more accessible for antibody cross-linking in the ELISA format and that its engagement results in a more stable epitope-paratope complex. Furthermore, experiments with hybrid variants with interconverted CH2 domain orientations indicated that the mode of attachment, as defined by the relative orientations of its light and heavy chains bound to gp120 antigen, contribute to the relative impotency of 2.2c in ADCC assays. It did not affect the ADCC potency of N5-i5 (53). MAbs N60-i3 and JR4, similar to N5-i5, attach their heavy chains to the α1-helix, but the positions and gp120 binding contacts of their light chains differ noticeably (Fig. 6C). This suggests that the epitope footprint and the precise epitope targeting determine the ADCC potency for N60-i3 and JR4. By directing their heavy chains to the α1-helix, N60-i3, JR4, and N5-i5 are not only more accessible for antibody cross-linking on the target cell but also position their CH2 domains optimally for effective Fc receptor interaction within the immune complex. Analysis of the residues subject to somatic mutation from the germ line sequences revealed that N60-i3, JR4, and N5-i5 were selected to have arginines at positions Arg99, Arg30/Arg31, and Arg55, respectively, enabling them to interact with the Asn103-Glu106-Asp107 triad of the α1-helix. Thus, N60-i3, JR4, and N5-i5 seem to have been selected to specifically target the α1-helix and recognize an epitope that encompasses structures in two inner-domain mobile layers and utilize cooperative binding in the α0- and α1-helices to link the two layers into one binding unit.
DISCUSSION
In conclusion, our findings indicate that the potent ADCC to the cluster A region focuses on a highly conserved epitope surface involving the α0- and α1-helices of the inner domains of the C1 and C2 regions of gp120, respectively. The cluster A region is buried and not accessible for antibodies in native and soluble CD4-triggered HIV-1 Env trimers and becomes exposed within a viral spike only upon binding to the cell surface form of CD4, where it is readily accessible for antibody avidity interactions and effective antigen cross-linking (47, 48, 53). Furthermore, these studies confirm our previous observation that precise epitope targeting—a combination of both the epitope footprint and the mode of antibody attachment—plays a major role in determining the potency of ADCC. Cluster A MAbs capable of potent Fc-mediated effector function cross-link the epitopes on the target cell surface by attaching their heavy chains to the α1-helix of gp120. This mode of binding allows positioning of CH2 domains for more effective Fc receptor interaction.
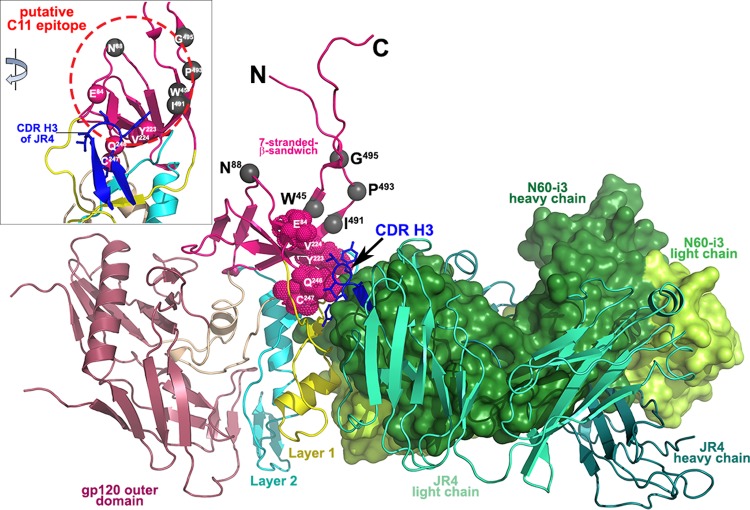
Epitopes in the cluster A region may be restricted to the surface-engaging residues of layers 1 and 2 only (N60-i3 epitope) and include also involvement of residues of the seven-stranded β-sandwich (JR4 epitope). MAb N60-i3, similar to MAb N5-i5, competes in the binding to the Env antigen only with A32 Fab; thus, its epitope represents the A32-like epitope in the cluster A region. In contrast, the Fab of rhesus macaque MAb JR4 competes entirely for A32 Fab binding and partially for C11 Fab binding in ELISA, FCS-FRET, and SPR competition assays. This indicates that the JR4 epitope footprint on gp120 antigen involves elements of both the A32- and C11-binding surfaces and represents a mixed A32-C11 epitope of the cluster A region. MAb JR4 was isolated from SHIV-infected rhesus macaques, but we have shown previously that antibodies of similar A32-C11 mixed specificity are also induced in HIV-1-infected individuals (41). This indicates that the A32-C11 mixed specificity could be induced in both nonhuman primates and humans following HIV-1 and SHIV infection, respectively. Since the exact epitope footprint of MAb C11 is not known, our studies allow us for the first time to define a putative contact region of MAb C11 with gp120. As shown in Fig. 7, the gp120 residues previously shown by mutagenesis to be involved in MAb C11 binding (68, 71) mapped to the seven-stranded β-sandwich in the gp41-interactive region (PDB accession number 3JWD) (72), with N60-i3 and JR4 Fabs bound as in their CD4-triggered gp120 complexes. As previously indicated, both N60-i3 and JR4 bind to gp120 in largely overlapping regions with only the protruding region of the CDR H3 of JR4 attaching to the seven-stranded β-sandwich (Fig. 7). We propose that the CDR H3 of JR4 bound to the seven-stranded β-sandwich interferes with MAb C11 binding in this area. The residues shown previously by mutagenesis to decrease C11 binding to gp120 map to this region. Thus, the putative C11 epitope involves residues of the seven-stranded β-sandwich and maps immediately adjacent to the A32-like epitope surface (Fig. 7, inset). This is also in agreement with our FCS-FRET measurements showing a distance of 79.8 Å between C11 and N60-i3 Fabs bound to FLSC.
FIG 7.
Putative binding site of MAb C11. The crystal structures of N60-i3 Fab-gp12093TH057 coree-M48 and JR4 Fab-gp12093TH057 coree-M48U1 are superimposed onto the HIV-1 gp120 with the gp41-interactive region (PDB code 3JWD) (72). Only the Fabs of complexes are shown, and a molecular surface is displayed over the N60-i3 Fab. The CDR H3 of JR4 predicted to block MAb C11 binding is shown in blue. The residue contacts of CDR H3 on the seven-stranded β-sandwich are shown as red spheres. Residues shown by mutagenesis studies to affect binding of MAb C11 to gp120 Env are shown as gray spheres. The inset shows a 45° rotation of the assembly where only gp120 and the CDR H3 of JR4 are shown, and the putative C11 epitope is encircled in red.
Supplementary Material
ACKNOWLEDGMENTS
Support for this work was provided by a grant from The Bill and Melinda Gates Foundation (OPP1033109 to G.K.L.), by National Institute of Allergy and Infectious Diseases (NIAID)/NIH grants R01 AI-084830 and R01 AI-087181 (to G.K.L), K25-AI087968 (to K.R.), and 5K23AI084580-04 (to M.M.S.), and by the Intramural Research Program of the VRC, National Institute of Allergy and Infectious Diseases, NIH. Crystallographic data were collected at the Stanford Synchrotron Radiation Lightsource (SSRL), a Directorate of the SLAC National Accelerator Laboratory and an Office of Science User Facility operated for the U.S. Department of Energy Office of Science by Stanford University. The SSRL Structural Molecular Biology Program is supported by the U.S. Department of Energy Office of Biological and Environmental Research, by the National Institutes of Health (NIH) National Center for Research Resources, Biomedical Technology Program (P41RR001209), and by the National Institute of General Medical Sciences.
We thank Daniel A. Bonsor for assistance with the ITC experiment and Christine Obreicht for outstanding technical support in protein expression.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01232-15.
REFERENCES
- 1.Su B, Moog C. 2014. Which antibody functions are important for an HIV vaccine? Front Immunol 5:289. doi: 10.3389/fimmu.2014.00289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Excler JL, Ake J, Robb ML, Kim JH, Plotkin SA. 2014. Nonneutralizing functional antibodies: a new “old” paradigm for HIV vaccines. Clin Vaccine Immunol 21:1023–1036. doi: 10.1128/CVI.00230-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forthal D, Hope TJ, Alter G. 2013. New paradigms for functional HIV-specific nonneutralizing antibodies. Curr Opin HIV AIDS 8:393–401. doi: 10.1097/COH.0b013e328363d486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewis GK. 2014. Role of Fc-mediated antibody function in protective immunity against HIV-1. Immunology 142:46–57. doi: 10.1111/imm.12232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parren PW, Marx PA, Hessell AJ, Luckay A, Harouse J, Cheng-Mayer C, Moore JP, Burton DR. 2001. Antibody protects macaques against vaginal challenge with a pathogenic R5 simian/human immunodeficiency virus at serum levels giving complete neutralization in vitro. J Virol 75:8340–8347. doi: 10.1128/JVI.75.17.8340-8347.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baba TW, Liska V, Hofmann-Lehmann R, Vlasak J, Xu W, Ayehunie S, Cavacini LA, Posner MR, Katinger H, Stiegler G, Bernacky BJ, Rizvi TA, Schmidt R, Hill LR, Keeling ME, Lu Y, Wright JE, Chou TC, Ruprecht RM. 2000. Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian-human immunodeficiency virus infection. Nat Med 6:200–206. doi: 10.1038/72309. [DOI] [PubMed] [Google Scholar]
- 7.Mascola JR, Lewis MG, Stiegler G, Harris D, VanCott TC, Hayes D, Louder MK, Brown CR, Sapan CV, Frankel SS, Lu Y, Robb ML, Katinger H, Birx DL. 1999. Protection of macaques against pathogenic simian/human immunodeficiency virus 89.6PD by passive transfer of neutralizing antibodies. J Virol 73:4009–4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walker LM, Burton DR. 2010. Rational antibody-based HIV-1 vaccine design: current approaches and future directions. Curr Opin Immunol 22:358–366. doi: 10.1016/j.coi.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Overbaugh J, Morris L. 2012. The antibody response against HIV-1. Cold Spring Harb Perspect Med 2:a007039. doi: 10.1101/cshperspect.a007039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, Benenson M, Gurunathan S, Tartaglia J, McNeil JG, Francis DP, Stablein D, Birx DL, Chunsuttiwat S, Khamboonruang C, Thongcharoen P, Robb ML, Michael NL, Kunasol P, Kim JH, Investigators M-T. 2009. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med 361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 11.Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans DT, Montefiori DC, Karnasuta C, Sutthent R, Liao HX, DeVico AL, Lewis GK, Williams C, Pinter A, Fong Y, Janes H, DeCamp A, Huang Y, Rao M, Billings E, Karasavvas N, Robb ML, Ngauy V, de Souza MS, Paris R, Ferrari G, Bailer RT, Soderberg KA, Andrews C, Berman PW, Frahm N, De Rosa SC, Alpert MD, Yates NL, Shen X, Koup RA, Pitisuttithum P, Kaewkungwal J, Nitayaphan S, Rerks-Ngarm S, Michael NL, Kim JH. 2012. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med 366:1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yates NL, Liao HX, Fong Y, Decamp A, Vandergrift NA, Williams WT, Alam SM, Ferrari G, Yang ZY, Seaton KE, Berman PW, Alpert MD, Evans DT, O'Connell RJ, Francis D, Sinangil F, Lee C, Nitayaphan S, Rerks-Ngarm S, Kaewkungwal J, Pitisuttithum P, Tartaglia J, Pinter A, Zolla-Pazner S, Gilbert PB, Nabel GJ, Michael NL, Kim JH, Montefiori DC, Haynes BF, Tomaras GD. 2014. Vaccine-induced Env V1-V2 IgG3 correlates with lower HIV-1 infection risk and declines soon after vaccination. Sci Transl Med 6:228ra39. doi: 10.1126/scitranslmed.3007730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chung AW, Ghebremichael M, Robinson H, Brown E, Choi I, Lane S, Dugast AS, Schoen MK, Rolland M, Suscovich TJ, Mahan AE, Liao L, Streeck H, Andrews C, Rerks-Ngarm S, Nitayaphan S, de Souza MS, Kaewkungwal J, Pitisuttithum P, Francis D, Michael NL, Kim JH, Bailey-Kellogg C, Ackerman ME, Alter G. 2014. Polyfunctional Fc-effector profiles mediated by IgG subclass selection distinguish RV144 and VAX003 vaccines. Sci Transl Med 6:228ra38. doi: 10.1126/scitranslmed.3007736. [DOI] [PubMed] [Google Scholar]
- 14.Xiao P, Patterson LJ, Kuate S, Brocca-Cofano E, Thomas MA, Venzon D, Zhao J, DiPasquale J, Fenizia C, Lee EM, Kalisz I, Kalyanaraman VS, Pal R, Montefiori D, Keele BF, Robert-Guroff M. 2012. Replicating adenovirus-simian immunodeficiency virus (SIV) recombinant priming and envelope protein boosting elicits localized, mucosal IgA immunity in rhesus macaques correlated with delayed acquisition following a repeated low-dose rectal SIVmac251 challenge. J Virol 86:4644–4657. doi: 10.1128/JVI.06812-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alpert MD, Harvey JD, Lauer WA, Reeves RK, Piatak M Jr, Carville A, Mansfield KG, Lifson JD, Li W, Desrosiers RC, Johnson RP, Evans DT. 2012. ADCC develops over time during persistent infection with live-attenuated SIV and Is associated with complete protection against SIVmac251 challenge. PLoS Pathog 8:e1002890. doi: 10.1371/journal.ppat.1002890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forthal DN, Gilbert PB, Landucci G, Phan T. 2007. Recombinant gp120 vaccine-induced antibodies inhibit clinical strains of HIV-1 in the presence of Fc receptor-bearing effector cells and correlate inversely with HIV infection rate. J Immunol 178:6596–6603. doi: 10.4049/jimmunol.178.10.6596. [DOI] [PubMed] [Google Scholar]
- 17.Fouts TR, Bagley K, Prado IJ, Bobb KL, Schwartz JA, Xu R, Zagursky RJ, Egan MA, Eldridge JH, LaBranche CC, Montefiori DC, Le Buanec H, Zagury D, Pal R, Pavlakis GN, Felber BK, Franchini G, Gordon S, Vaccari M, Lewis GK, DeVico AL, Gallo RC. 2015. Balance of cellular and humoral immunity determines the level of protection by HIV vaccines in rhesus macaque models of HIV infection. Proc Natl Acad Sci U S A 112:E992–E999. doi: 10.1073/pnas.1423669112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Rompay KK, Berardi CJ, Dillard-Telm S, Tarara RP, Canfield DR, Valverde CR, Montefiori DC, Cole KS, Montelaro RC, Miller CJ. 1998. Passive immunization of newborn rhesus macaques prevents oral simian immunodeficiency virus infection. J Infect Dis 177:1247–1259. doi: 10.1086/515270. [DOI] [PubMed] [Google Scholar]
- 19.Forthal DN, Landucci G, Cole KS, Marthas M, Becerra JC, Van Rompay K. 2006. Rhesus macaque polyclonal and monoclonal antibodies inhibit simian immunodeficiency virus in the presence of human or autologous rhesus effector cells. J Virol 80:9217–9225. doi: 10.1128/JVI.02746-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mabuka J, Nduati R, Odem-Davis K, Peterson D, Overbaugh J. 2012. HIV-specific antibodies capable of ADCC are common in breastmilk and are associated with reduced risk of transmission in women with high viral loads. PLoS Pathog 8:e1002739. doi: 10.1371/journal.ppat.1002739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Milligan C, Richardson BA, John-Stewart G, Nduati R, Overbaugh J. 2015. Passively acquired antibody-dependent cellular cytotoxicity (ADCC) activity in HIV-infected infants is associated with reduced mortality. Cell Host Microbe 17:500–506. doi: 10.1016/j.chom.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bournazos S, Klein F, Pietzsch J, Seaman MS, Nussenzweig MC, Ravetch JV. 2014. Broadly neutralizing anti-HIV-1 antibodies require Fc effector functions for in vivo activity. Cell 158:1243–1253. doi: 10.1016/j.cell.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiang SH, Doka N, Choudhary RK, Sodroski J, Robinson JE. 2002. Characterization of CD4-induced epitopes on the HIV type 1 gp120 envelope glycoprotein recognized by neutralizing human monoclonal antibodies. AIDS Res Hum Retroviruses 18:1207–1217. doi: 10.1089/08892220260387959. [DOI] [PubMed] [Google Scholar]
- 25.Huang CC, Lam SN, Acharya P, Tang M, Xiang SH, Hussan SS, Stanfield RL, Robinson J, Sodroski J, Wilson IA, Wyatt R, Bewley CA, Kwong PD. 2007. Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4. Science 317:1930–1934. doi: 10.1126/science.1145373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang CC, Tang M, Zhang MY, Majeed S, Montabana E, Stanfield RL, Dimitrov DS, Korber B, Sodroski J, Wilson IA, Wyatt R, Kwong PD. 2005. Structure of a V3-containing HIV-1 gp120 core. Science 310:1025–1028. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Kwon YD, Scheid JF, Shi W, Xu L, Yang Y, Zhu J, Nussenzweig MC, Sodroski J, Shapiro L, Nabel GJ, Mascola JR, Kwong PD. 2010. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cardoso RM, Zwick MB, Stanfield RL, Kunert R, Binley JM, Katinger H, Burton DR, Wilson IA. 2005. Broadly neutralizing anti-HIV antibody 4E10 recognizes a helical conformation of a highly conserved fusion-associated motif in gp41. Immunity 22:163–173. doi: 10.1016/j.immuni.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 29.Zwick MB, Jensen R, Church S, Wang M, Stiegler G, Kunert R, Katinger H, Burton DR. 2005. Anti-human immunodeficiency virus type 1 (HIV-1) antibodies 2F5 and 4E10 require surprisingly few crucial residues in the membrane-proximal external region of glycoprotein gp41 to neutralize HIV-1. J Virol 79:1252–1261. doi: 10.1128/JVI.79.2.1252-1261.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brunel FM, Zwick MB, Cardoso RM, Nelson JD, Wilson IA, Burton DR, Dawson PE. 2006. Structure-function analysis of the epitope for 4E10, a broadly neutralizing human immunodeficiency virus type 1 antibody. J Virol 80:1680–1687. doi: 10.1128/JVI.80.4.1680-1687.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saphire EO, Parren PW, Pantophlet R, Zwick MB, Morris GM, Rudd PM, Dwek RA, Stanfield RL, Burton DR, Wilson IA. 2001. Crystal structure of a neutralizing human IGG against HIV-1: a template for vaccine design. Science 293:1155–1159. doi: 10.1126/science.1061692. [DOI] [PubMed] [Google Scholar]
- 32.Diskin R, Marcovecchio PM, Bjorkman PJ. 2010. Structure of a clade C HIV-1 gp120 bound to CD4 and CD4-induced antibody reveals anti-CD4 polyreactivity. Nat Struct Mol Biol 17:608–613. doi: 10.1038/nsmb.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang X, Burke V, Totrov M, Williams C, Cardozo T, Gorny MK, Zolla-Pazner S, Kong XP. 2010. Conserved structural elements in the V3 crown of HIV-1 gp120. Nat Struct Mol Biol 17:955–961. doi: 10.1038/nsmb.1861. [DOI] [PubMed] [Google Scholar]
- 34.Huang CC, Venturi M, Majeed S, Moore MJ, Phogat S, Zhang MY, Dimitrov DS, Hendrickson WA, Robinson J, Sodroski J, Wyatt R, Choe H, Farzan M, Kwong PD. 2004. Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120. Proc Natl Acad Sci U S A 101:2706–2711. doi: 10.1073/pnas.0308527100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koup RA, Robinson JE, Nguyen QV, Pikora CA, Blais B, Roskey A, Panicali D, Sullivan JL. 1991. Antibody-dependent cell-mediated cytotoxicity directed by a human monoclonal antibody reactive with gp120 of HIV-1. AIDS 5:1309–1314. doi: 10.1097/00002030-199111000-00004. [DOI] [PubMed] [Google Scholar]
- 36.Alsmadi O, Tilley SA. 1998. Antibody-dependent cellular cytotoxicity directed against cells expressing human immunodeficiency virus type 1 envelope of primary or laboratory-adapted strains by human and chimpanzee monoclonal antibodies of different epitope specificities. J Virol 72:286–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lewis GK, Guan Y, Kamin-Lewis R, Sajadi M, Pazgier M, Devico AL. 2014. Epitope target structures of Fc-mediated effector function during HIV-1 acquisition. Curr Opin HIV AIDS 9:263–270. doi: 10.1097/COH.0000000000000055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pollara J, Bonsignori M, Moody MA, Pazgier M, Haynes BF, Ferrari G. 2013. Epitope specificity of human immunodeficiency virus-1 antibody dependent cellular cytotoxicity (ADCC) responses. Curr HIV Res 11:378–387. doi: 10.2174/1570162X113116660059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferrari G, Pollara J, Kozink D, Harms T, Drinker M, Freel S, Moody MA, Alam SM, Tomaras GD, Ochsenbauer C, Kappes JC, Shaw GM, Hoxie JA, Robinson JE, Haynes BF. 2011. An HIV-1 gp120 envelope human monoclonal antibody that recognizes a C1 conformational epitope mediates potent antibody-dependent cellular cytotoxicity (ADCC) activity and defines a common ADCC epitope in human HIV-1 serum. J Virol 85:7029–7036. doi: 10.1128/JVI.00171-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ampol S, Pattanapanyasat K, Sutthent R, Permpikul P, Kantakamalakul W. 2012. Comprehensive investigation of common antibody-dependent cell-mediated cytotoxicity antibody epitopes of HIV-1 CRF01_AE gp120. AIDS Res Hum Retroviruses 28:1250–1258. doi: 10.1089/aid.2011.0346. [DOI] [PubMed] [Google Scholar]
- 41.Guan Y, Pazgier M, Sajadi MM, Kamin-Lewis R, Al-Darmarki S, Flinko R, Lovo E, Wu X, Robinson JE, Seaman MS, Fouts TR, Gallo RC, DeVico AL, Lewis GK. 2013. Diverse specificity and effector function among human antibodies to HIV-1 envelope glycoprotein epitopes exposed by CD4 binding. Proc Natl Acad Sci U S A 110:E69–E78. doi: 10.1073/pnas.1217609110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Veillette M, Desormeaux A, Medjahed H, Gharsallah NE, Coutu M, Baalwa J, Guan Y, Lewis G, Ferrari G, Hahn BH, Haynes BF, Robinson JE, Kaufmann DE, Bonsignori M, Sodroski J, Finzi A. 2014. Interaction with cellular CD4 exposes HIV-1 envelope epitopes targeted by antibody-dependent cell-mediated cytotoxicity. J Virol 88:2633–2644. doi: 10.1128/JVI.03230-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bonsignori M, Pollara J, Moody MA, Alpert MD, Chen X, Hwang KK, Gilbert PB, Huang Y, Gurley TC, Kozink DM, Marshall DJ, Whitesides JF, Tsao CY, Kaewkungwal J, Nitayaphan S, Pitisuttithum P, Rerks-Ngarm S, Kim JH, Michael NL, Tomaras GD, Montefiori DC, Lewis GK, DeVico A, Evans DT, Ferrari G, Liao HX, Haynes BF. 2012. Antibody-dependent cellular cytotoxicity-mediating antibodies from an HIV-1 vaccine efficacy trial target multiple epitopes and preferentially use the VH1 gene family. J Virol 86:11521–11532. doi: 10.1128/JVI.01023-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pollara J, Bonsignori M, Moody MA, Liu P, Alam SM, Hwang KK, Gurley TC, Kozink DM, Armand LC, Marshall DJ, Whitesides JF, Kaewkungwal J, Nitayaphan S, Pitisuttithum P, Rerks-Ngarm S, Robb ML, O'Connell RJ, Kim JH, Michael NL, Montefiori DC, Tomaras GD, Liao HX, Haynes BF, Ferrari G. 2014. HIV-1 vaccine-induced C1 and V2 Env-specific antibodies synergize for increased antiviral activities. J Virol 88:7715–7726. doi: 10.1128/JVI.00156-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tomaras GD, Ferrari G, Shen X, Alam SM, Liao HX, Pollara J, Bonsignori M, Moody MA, Fong Y, Chen X, Poling B, Nicholson CO, Zhang R, Lu X, Parks R, Kaewkungwal J, Nitayaphan S, Pitisuttithum P, Rerks-Ngarm S, Gilbert PB, Kim JH, Michael NL, Montefiori DC, Haynes BF. 2013. Vaccine-induced plasma IgA specific for the C1 region of the HIV-1 envelope blocks binding and effector function of IgG. Proc Natl Acad Sci U S A 110:9019–9024. doi: 10.1073/pnas.1301456110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Finnegan CM, Berg W, Lewis GK, DeVico AL. 2001. Antigenic properties of the human immunodeficiency virus envelope during cell-cell fusion. J Virol 75:11096–11105. doi: 10.1128/JVI.75.22.11096-11105.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ray K, Mengistu M, Yu L, Lewis GK, Lakowicz JR, DeVico AL. 2014. Antigenic properties of the HIV envelope on virions in solution. J Virol 88:1795–1808. doi: 10.1128/JVI.03048-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mengistu M, Ray K, Lewis GK, DeVico AL. 2015. Antigenic properties of the human immunodeficiency virus envelope glycoprotein gp120 on virions bound to target cells. PLoS Pathog 11:e1004772. doi: 10.1371/journal.ppat.1004772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robinson JE, Elliott DH, Martin EA, Micken K, Rosenberg ES. 2005. High frequencies of antibody responses to CD4 induced epitopes in HIV infected patients started on HAART during acute infection. Hum Antibodies 14:115–121. [PubMed] [Google Scholar]
- 50.Veillette M, Coutu M, Richard J, Batraville LA, Dagher O, Bernard N, Tremblay C, Kaufmann DE, Roger M, Finzi A. 2015. The HIV-1 gp120 CD4-bound conformation is preferentially targeted by antibody-dependent cellular cytotoxicity-mediating antibodies in sera from HIV-1-infected individuals. J Virol 89:545–551. doi: 10.1128/JVI.02868-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chung AW, Isitman G, Navis M, Kramski M, Center RJ, Kent SJ, Stratov I. 2011. Immune escape from HIV-specific antibody-dependent cellular cytotoxicity (ADCC) pressure. Proc Natl Acad Sci U S A 108:7505–7510. doi: 10.1073/pnas.1016048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pham TN, Lukhele S, Hajjar F, Routy JP, Cohen EA. 2014. HIV Nef and Vpu protect HIV-infected CD4+ T cells from antibody-mediated cell lysis through down-modulation of CD4 and BST2. Retrovirology 11:15. doi: 10.1186/1742-4690-11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Acharya P, Tolbert WD, Gohain N, Wu X, Yu L, Liu T, Huang W, Huang CC, Kwon YD, Louder RK, Luongo TS, McLellan JS, Pancera M, Yang Y, Zhang B, Flinko R, Foulke JS Jr, Sajadi MM, Kamin-Lewis R, Robinson JE, Martin L, Kwong PD, Guan Y, DeVico AL, Lewis GK, Pazgier M. 2014. Structural definition of an antibody-dependent cellular cytotoxicity response implicated in reduced risk for HIV-1 infection. J Virol 88:12895–12906. doi: 10.1128/JVI.02194-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Acharya P, Luongo TS, Louder MK, McKee K, Yang Y, Kwon YD, Mascola JR, Kessler P, Martin L, Kwong PD. 2013. Structural basis for highly effective HIV-1 neutralization by CD4-mimetic miniproteins revealed by 1.5 Å cocrystal structure of gp120 and M48U1. Structure 21:1018–1029. doi: 10.1016/j.str.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van Herrewege Y, Morellato L, Descours A, Aerts L, Michiels J, Heyndrickx L, Martin L, Vanham G. 2008. CD4 mimetic miniproteins: potent anti-HIV compounds with promising activity as microbicides. J Antimicrob Chemother 61:818–826. doi: 10.1093/jac/dkn042. [DOI] [PubMed] [Google Scholar]
- 56.Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276:307–326. [DOI] [PubMed] [Google Scholar]
- 57.McCoy AJ. 2007. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr 63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.NCCP. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 59.Murshudov GN, Vagin AA, Dodson EJ. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 60.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58:1948–1954. doi: 10.1107/S0907444902016657. [DOI] [PubMed] [Google Scholar]
- 61.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 62.Chen VB, Arendall WB III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Krissinel E, Henrick K. 2007. Inference of macromolecular assemblies from crystalline state. J Mol Biol 372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 64.Tina KG, Bhadra R, Srinivasan N. 2007. PIC: protein interactions calculator. Nucleic Acids Res 35:W473–W476. doi: 10.1093/nar/gkm423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schuler B, Lipman EA, Eaton WA. 2002. Probing the free-energy surface for protein folding with single-molecule fluorescence spectroscopy. Nature 419:743–747. doi: 10.1038/nature01060. [DOI] [PubMed] [Google Scholar]
- 66.Fouts TR, Tuskan R, Godfrey K, Reitz M, Hone D, Lewis GK, DeVico AL. 2000. Expression and characterization of a single-chain polypeptide analogue of the human immunodeficiency virus type 1 gp120-CD4 receptor complex. J Virol 74:11427–11436. doi: 10.1128/JVI.74.24.11427-11436.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gomez-Roman VR, Florese RH, Patterson LJ, Peng B, Venzon D, Aldrich K, Robert-Guroff M. 2006. A simplified method for the rapid fluorometric assessment of antibody-dependent cell-mediated cytotoxicity. J Immunol Methods 308:53–67. doi: 10.1016/j.jim.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 68.Moore JP, Willey RL, Lewis GK, Robinson J, Sodroski J. 1994. Immunological evidence for interactions between the first, second, and fifth conserved domains of the gp120 surface glycoprotein of human immunodeficiency virus type 1. J Virol 68:6836–6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Säfsten P. 2009. Methods in molecular biology: epitope mapping by surface plasmon resonance, p 67–76. In Reineke U, Schutkowski M (ed), Epitope mapping protocols, 2nd ed Humana Press, Hatfield, Hertfordshire, United Kingdom. [DOI] [PubMed] [Google Scholar]
- 70.LaLonde JM, Kwon YD, Jones DM, Sun AW, Courter JR, Soeta T, Kobayashi T, Princiotto AM, Wu X, Schon A, Freire E, Kwong PD, Mascola JR, Sodroski J, Madani N, Smith AB III. 2012. Structure-based design, synthesis, and characterization of dual hotspot small-molecule HIV-1 entry inhibitors. J Med Chem 55:4382–4396. doi: 10.1021/jm300265j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Finzi A, Xiang SH, Pacheco B, Wang L, Haight J, Kassa A, Danek B, Pancera M, Kwong PD, Sodroski J. 2010. Topological layers in the HIV-1 gp120 inner domain regulate gp41 interaction and CD4-triggered conformational transitions. Mol Cell 37:656–667. doi: 10.1016/j.molcel.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pancera M, Majeed S, Ban YE, Chen L, Huang CC, Kong L, Kwon YD, Stuckey J, Zhou T, Robinson JE, Schief WR, Sodroski J, Wyatt R, Kwong PD. 2010. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc Natl Acad Sci U S A 107:1166–1171. doi: 10.1073/pnas.0911004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brunger AT. 1992. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.