ABSTRACT
The small hydrophobic (SH) gene of respiratory syncytial virus (RSV), a major cause of infant hospitalization, encodes a viroporin of unknown function. SH gene knockout virus (RSV ΔSH) is partially attenuated in vivo, but not in vitro, suggesting that the SH protein may have an immunomodulatory role. RSV ΔSH has been tested as a live attenuated vaccine in humans and cattle, and here we demonstrate that it protected against viral rechallenge in mice. We compared the immune response to infection with RSV wild type and RSV ΔSH in vivo using BALB/c mice and in vitro using epithelial cells, neutrophils, and macrophages. Strikingly, the interleukin-1β (IL-1β) response to RSV ΔSH infection was greater than to wild-type RSV, in spite of a decreased viral load, and when IL-1β was blocked in vivo, the viral load returned to wild-type levels. A significantly greater IL-1β response to RSV ΔSH was also detected in vitro, with higher-magnitude responses in neutrophils and macrophages than in epithelial cells. Depleting macrophages (with clodronate liposome) and neutrophils (with anti-Ly6G/1A8) demonstrated the contribution of these cells to the IL-1β response in vivo, the first demonstration of neutrophilic IL-1β production in response to viral lung infection. In this study, we describe an increased IL-1β response to RSV ΔSH, which may explain the attenuation in vivo and supports targeting the SH gene in live attenuated vaccines.
IMPORTANCE There is a pressing need for a vaccine for respiratory syncytial virus (RSV). A number of live attenuated RSV vaccine strains have been developed in which the small hydrophobic (SH) gene has been deleted, even though the function of the SH protein is unknown. The structure of the SH protein has recently been solved, showing it is a pore-forming protein (viroporin). Here, we demonstrate that the IL-1β response to RSV ΔSH is greater in spite of a lower viral load, which contributes to the attenuation in vivo. This potentially suggests a novel method by which viruses can evade the host response. As all Pneumovirinae and some Paramyxovirinae carry similar SH genes, this new understanding may also enable the development of live attenuated vaccines for both RSV and other members of the Paramyxoviridae.
INTRODUCTION
Respiratory syncytial virus (RSV) is the most significant cause of bronchiolitis and pneumonia in infants for which there is no vaccine (1). Recent advances in the understanding of the infant immune response to vaccination suggest that a live attenuated vaccine given in infancy may be the most effective approach to prevent RSV infection (2), potentially in combination with maternal immunization using recombinant F protein (3). This is supported by the successful introduction of live attenuated influenza vaccine to the childhood vaccination schedule (4) in conjunction with immunization during pregnancy with the trivalent inactivated vaccine (5). One issue with live attenuated RSV vaccines has been balancing immunogenicity and safety (6). Two approaches are used to develop live attenuated vaccines: biological derivation of strains, usually by multiple passages, often at lower temperatures to mimic the upper respiratory tract, and targeted gene deletion by reverse genetics.
To most effectively attenuate a virus by reverse genetics, understanding the proteins it encodes is required. In the current generation of live attenuated RSV vaccines, genes encoding the nonstructural protein 2 (NS2) and the small hydrophobic protein (SH) have been targeted. NS2 acts as an inhibitor of the type I interferon response (7), modulating NF-κB (7), but the function of the SH protein is currently unknown. In silico analysis suggests that a transmembrane pentamer is the most energetically favorable conformation of SH (8). The formation of SH protein pentamers has been confirmed by cryo-electron microscopy (cryo-EM) studies (9), and SH pentamers enable the passage of ions and small molecules (9–11). When transfected into HEK293 cells, SH is located at the plasma membrane (10), and during RSV infection, SH is located at the Golgi complex (12). These studies suggest that the SH protein belongs to the family of viroporins (13); however, the role of the pore encoded by the SH gene is unknown.
Recombinant RSV that does not express the SH protein is partially attenuated in vivo (14), but not in vitro (15). This suggests it may play a role in modulating the immune response, with earlier studies showing that RSV SH inhibits tumor necrosis factor (TNF) signaling (16). In a recent study, recombinant bovine RSV (bRSV) with an SH gene deletion was attenuated (17), inducing increased levels of the cytokines interleukin-1β (IL-1β) and TNF. Viral pore proteins have been proposed to modulate the inflammasome, a multiprotein pattern recognition complex that catalyzes the cleavage of the proforms of IL-1β and IL-18 into their active forms via caspase 1 (18).
We observed that recombinant RSV lacking the SH gene was attenuated but protective against subsequent virus challenge. We propose that the attenuation observed in vivo following deletion of the RSV SH gene is due to its effect on the host immune response. To test this, we compared the response to infection with wild-type (WT) RSV (strain A2) and its derivative in which the SH gene had been deleted (RSV ΔSH) in vitro and in vivo. We observed that RSV ΔSH induced significantly higher levels of IL-1β, especially from macrophages and neutrophils. These studies demonstrate increased inflammation due to the virus with SH deleted, which may contribute to its attenuation.
(Part of this information was presented at the Gordon Research Conference, Biology of Acute Respiratory Infection, Barga, Italy, February 2014.)
MATERIALS AND METHODS
Virus.
RSV strain A2 was used as the WT and compared to an SH gene deletion recombinant on an A2 background (19). Infectious stocks of virus were grown using the human laryngeal carcinoma cell line HEp-2. The viral titer was calculated by an immunoplaque assay using biotinylated goat anti-RSV polyclonal antibody (AbD Serotec, Oxford, United Kingdom) to detect plaques. Prior to in vitro and in vivo studies, stocks were screened for lipopolysaccharide (LPS) contamination. Virus was inactivated by UV irradiation at 1.3 × 105 μJ/cm2 for 15 min on ice in a CX-2000 cross-linker (UVP, Cambridge, United Kingdom).
Animals.
Female BALB/c mice were obtained from Harlan Scientific (Brook House, United Kingdom) and used at 6 to 8 weeks of age. All procedures undertaken were approved by the local animal welfare and ethical review board and performed by personal licensees under the appropriate project license. Experiments were carried out in accordance with the Animals (Scientific Procedures) Act of 1986. The mice were infected with 2.5 × 105 PFU of virus or medium alone in a 100-μl volume intranasally while under isoflurane anesthesia. The animals were weighed prior to RSV challenge and daily thereafter. Where described, neutrophils were depleted with 0.5 mg anti-Ly6G (clone 1A8; BioXCell), and IL-1β was blocked with 0.5 mg anti-murine IL-1β (mIL-1β) (clone B122; BioXCell) in vivo in 500 μl delivered intraperitoneally on days −1 and +1 of primary RSV infection. For macrophage depletion, mice were treated with 100 μl of clodronate liposome (CL) suspension (Boehringer GmbH, Mannheim, Germany) or control empty liposomes (PL) intranasally. After infection, bronchoalveolar lavage (BAL) fluids, lung tissues, and serum samples were harvested as described previously (20).
RSV load.
The viral load in vivo was assessed by extracting RNA from frozen lung tissue disrupted in a TissueLyzer (Qiagen, Manchester, United Kingdom) using TRIzol extraction and then converting it into cDNA. Quantitative real-time (RT)-PCR was carried out using bulk viral RNA for the RSV L gene and mRNA, using 900 nM forward primer (5′-GAACTCAGTGTAGGTAGAATGTTTGCA-3′), 300 nM reverse primer (5′-TTCAGCTATCATTTTCTCTGCCAAT-3′), and 100 nM probe (5′-6-carboxyfluorescein [FAM]-TTTGAACCTGTCTGAACAT-6-carboxytetramethylrhodamine [TAMRA]-3′) on a Stratagene Mx3005p (Agilent Technologies, Santa Clara, CA, USA). The L-specific RNA copy number was determined using an RSV L gene standard.
Cytokine quantification.
IL-1β was measured in samples by human or mouse duoset enzyme-linked immunosorbent assay (ELISA) according to the manufacturer's instructions (R&D Systems, Oxford, United Kingdom). IL-6 and CXCL1/KC were measured by Luminex (Bio-Rad, Hemel Hempstead, United Kingdom).
Flow cytometric analysis.
Live cells were suspended in Fc block (anti-CD16/32; BD) in phosphate-buffered saline (PBS)-1% bovine serum albumin (BSA) and stained with surface antibodies (panel 1, RSV M2 82-90 pentamer R-PE [Proimmune, Oxford, United Kingdom[, CD3-fluorescein isothiocyanate [FITC] [BD, Oxford, United Kingdom], CD4-allophycocyanin [APC] [BD], CD8-APC Alexa 75 (Invitrogen, Paisley, United Kingdom), and CD19-eFluor 450 [eBioscience, Hatfield, United Kingdom]; panel 2, CD11c-FITC [BD], CD80-APC [eBioscience], major histocompatibility complex class II [MHC-II]-efluor450 [eBioscience], F4/80-phycoerythrin [PE]-Cy7 [eBioscience], and Ly6G-BV605 [BD]). For intracellular IL-1β staining, after surface staining, cells were fixed and permeabilized with Cytofix/Cytoperm (BD) and stained with IL-1β (BD). Analysis was performed on an LSR Fortessa flow cytometer (BD). Fluorescence minus one (FMO) controls were used for surface stains, and an IgG1-PE isotype control was used for IL-1β intracellular staining.
Antigen-specific ELISA.
A quantitative assay (adapted from reference 21) was used to determine serum antibody levels. Plates were coated with 1 μg/ml RSV antigen and blocked with PBS-1% BSA. Bound IgG was detected with horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (AbD Serotec, Kidlington, United Kingdom). A dilution series of recombinant murine IgG was used as a standard to quantify RSV-specific antibodies. 3,3′,5,5′-Tetramethylbenzidine (TMB) with H2SO4 to quench the reaction was used to detect the response, and optical densities were read at 450 nm.
In vitro cells.
HEp-2 cells were grown in Dulbecco's modified Eagle's medium (DMEM) (Sigma) with 10% fetal calf serum (FCS), l-glutamine, and penicillin-streptomycin. THP-1 cells (a macrophage-like cell line) were grown in RPMI with 10% FCS, l-glutamine, and penicillin-streptomycin. The THP-1 cells were differentiated into phagocyte-type cells by seeding into new tissue culture plates, supplementing with 20 ng/ml phorbol myristate acetate (PMA), and incubating for 24 h before resting for a further 24 h (adapted from reference 22). For PBMC, mixed donor pools from three NC24 leukocyte clones were sourced from the NHS Blood and Transplant Unit (Colindale, United Kingdom). RosetteSep CD8 depletion cocktail (StemCell Technologies, Cambridge, United Kingdom) was used to prepare CD8-depleted PBMC populations. Cells were dispensed into tissue culture plates at the required density following the addition of 10 U/ml IL-2. For primary neutrophils, neutrophils were separated from fresh blood collected from healthy donors with written consent according to local research committee guidelines. A modified Percoll method was used (23). The neutrophils were separated on a 70% over 60% Percoll solution and centrifuged at 500 × g for 35 min at 22°C, with acceleration/braking at 2 m/s2. This gave a population that was >90% neutrophils, as confirmed by differential staining.
In vitro viral infection.
Cells were seeded into 24-well plates at the following densities: HEp-2, 5 × 105; THP-1, 5 × 105; PBMC, 5 × 106; neutrophils, 5 × 106. The cells were incubated for a minimum of 24 h at 37°C, 5% CO2 before infection with RSV ΔSH or RSV WT at a multiplicity of infection (MOI) of 0.5. At the time points specified in the results, supernatants were collected and subjected to cytokine analysis by ELISA.
Fluorescence microscopy and imaging.
The HEp-2 and THP-1 cell lines and primary neutrophils were infected with green fluorescent protein (GFP)-tagged RSV (24) at an MOI of 0.1 24 or 48 h prior to imaging. Images were captured using a Nikon Eclipse TE2000 inverted microscope attached to a Nikon digital camera (DXM 1200F), with 20× magnification. Blue (400- to 446-nm) fluorescence filters were used to detect GFP.
Statistical analysis.
The calculations described in the figure legends were performed using Prism 6 (GraphPad Software Inc., La Jolla, CA, USA).
RESULTS
RSV ΔSH infection is protective against subsequent viral challenge.
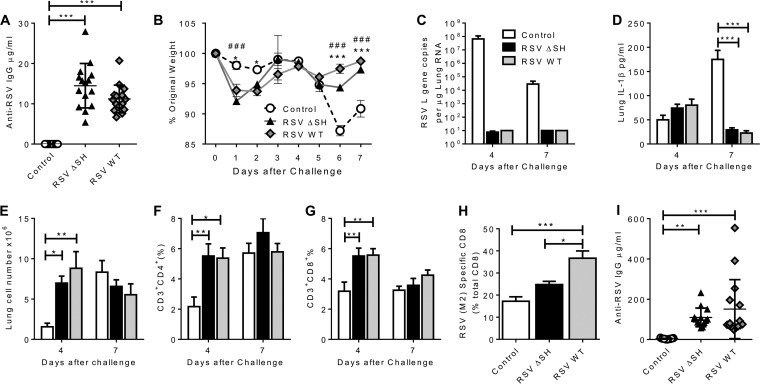
SH gene deletion has been proposed as a possible attenuating mutation for the generation of vaccine strains of RSV. To confirm that RSV ΔSH protected against RSV challenge, mice were infected intranasally with either RSV WT or RSV ΔSH at 2.5 × 105 PFU in a 100-μl volume or with medium alone as a control and then 28 days later challenged intranasally with 2.5 × 105 PFU RSV WT in 100 μl. Sera were collected prior to challenge infection, and RSV WT and ΔSH infection generated an RSV-specific IgG response, with the response to ΔSH slightly, but not significantly, greater than that to RSV WT (Fig. 1A). Mice that had primary infection with either ΔSH or WT were protected against disease following subsequent RSV challenge; on days 6 and 7 after RSV challenge infection, control-treated mice lost significantly more weight (P < 0.001) (Fig. 1B). Interestingly, as we have previously observed (25), there was acute weight loss on days 1 and 2 in the primed groups. There was no detectable viral load in either primed group, but RSV L gene-specific RNA was detectable in the control group on both days 4 and 7 after RSV infection (Fig. 1C). There was no difference in the IL-1β level in the lungs (Fig. 1D) on day 4 after infection, but the control group had significantly more IL-1β in the lungs on day 7. The primed groups recruited more cells to the lungs (Fig. 1E) on day 4, with a greater proportion of CD4+ and CD8+ T cells on day 4. On day 7 after infection, the mice previously infected with RSV WT had significantly more RSV-specific CD8 cells (Fig. 1H). Both previously infected groups had significantly higher anti-RSV IgG responses than the control-treated mice on day 7 and an increase from the level prior to challenge, indicating a memory response to RSV (Fig. 1I). We thus confirmed that RSV ΔSH can induce a protective immune response.
FIG 1.
RSV ΔSH is protective against RSV infection. Mice were infected intranasally with 2.5 × 105 PFU of either RSV WT (gray symbols) or RSV ΔSH (black symbols) or control treated (white symbols), and 4 weeks later, all the mice were challenged with RSV WT. (A) Anti-RSV IgG was measured by ELISA 1 day before RSV challenge. (B to E) Weight change (B), lung viral load (C), lung IL-1B (D), and lung cell numbers (E) were measured after secondary infection. (F to H) Lung CD4+ (F) and CD8+ (G) T cells on day 4 and day 7 and lung RSV-specific CD8 T cells on day 7 (H) were measured by flow cytometry. (I) Anti-RSV IgG was measured by ELISA on day 7 after infection. (B to H) The points represent individual animals. The data represent means ± standard errors of the mean (SEM) (n = 5 animals). *, P < 0.05; **, P < 0.01; and ***, P < 0.001 comparing WT RSV and the control; #, P < 0.05; and ###, P < 0.001 comparing RSV ΔSH and the control, measured by multiple t tests with Holm-Sidak correction (B) or analysis of variance (ANOVA) (A and C to I).
RSV lacking the SH gene is attenuated in vivo but induces a greater IL-1β response.
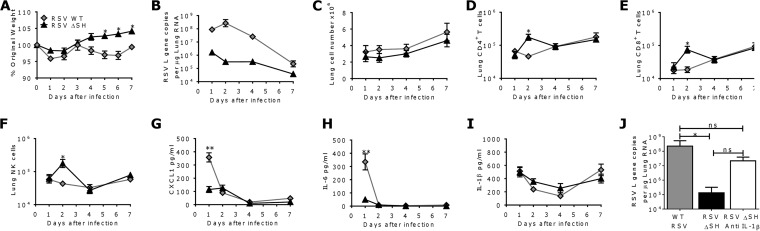
To assess the role of the SH protein during RSV infection, BALB/c mice were infected with 2.5 × 105 PFU RSV WT or RSV ΔSH (19). The ΔSH virus was attenuated; mice infected with RSV WT lost significantly more weight than mice infected with RSV ΔSH on days 5 to 7 after infection (P < 0.05) (Fig. 2A), and markedly less RSV RNA was detected in the RSV ΔSH groups than in RSV WT-infected animals (Fig. 2B). There was no difference in the total cell counts recovered from the lungs (Fig. 2C). However, there was significantly greater recruitment of CD4+ (P < 0.05) (Fig. 2D), CD8+ (P < 0.05) (Fig. 2E), and DX5+ NK (P < 0.05) (Fig. 2F) cells in the lungs of mice infected with RSV ΔSH on day 2 after infection. Lung CXCL1 (P < 0.001) (Fig. 2G) and IL-6 (P < 0.001) (Fig. 2H) levels were significantly greater in RSV WT-infected mice on day 1 after infection. IL-1β levels were slightly, but not significantly, higher in mice infected with RSV ΔSH on days 2 and 4 after infection (Fig. 2I). This was striking, because the viral load was so much lower in the RSV ΔSH-infected mice, suggesting that the IL-1β response was modulated by the SH gene. IL-1β was blocked during RSV ΔSH infection to determine its contribution to the attenuated phenotype of RSV ΔSH. Mice were treated with anti-IL-1β on days −1 and +1 of infection, and the viral load was measured on day 4 after infection. RSV ΔSH was significantly attenuated compared to the wild-type virus, but anti-IL-1β treatment restored the ΔSH viral load to wild-type levels (Fig. 2J).
FIG 2.
RSV ΔSH is attenuated in vivo but induces a greater IL-1β response than the wild type. Mice were infected intranasally with RSV WT or RSV ΔSH. (A to C) Weight loss (A), lung viral load (B), and lung cell numbers (C) were measured after infection. (D to F) Lung CD4+ T (D), CD8+ T (E), and DX5+ NK (F) cells were measured by flow cytometry. (G to I) CXCL1 (G), IL-6 (H), and IL-1β (I) were measured in lung supernatants by ELISA. (J) Mice were treated with anti-IL-1β prior to infection with RSV ΔSH, and the viral load was measured on day 4 after infection. The points represent means ± SEM (n > 5 mice). *, P < 0.05; **, P < 0.01; calculated by multiple t tests with Holm-Sidak correction.
RSV lacking the SH gene has growth kinetics similar to those of the wild-type virus but induces a greater IL-1β response in vitro.
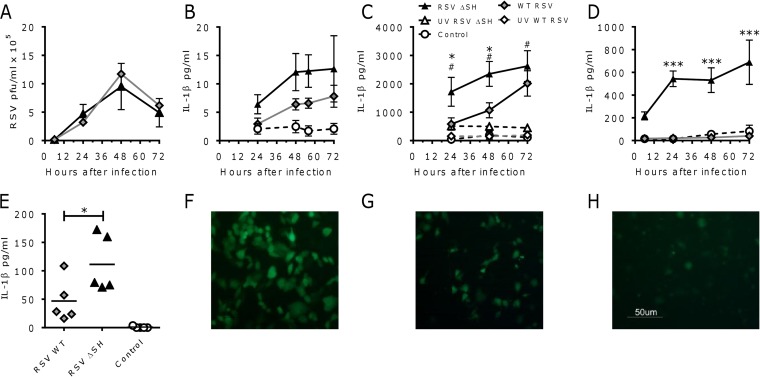
To dissect the effect of SH gene deletion on the IL-1β response, a range of cell types were infected in vitro. HEp-2 cells were infected at an MOI of 0.5 with RSV WT or RSV ΔSH, and supernatants were collected at various time points after infection. RSV ΔSH has been observed to replicate to titers similar to those of wild-type virus in vitro, and we observed a similar effect in HEp-2 cells (Fig. 3A). The levels of IL-1β induced by viral infection of HEp-2 cells were low, but RSV ΔSH induced more IL-1β than RSV WT (Fig. 3B). We have previously observed that macrophages contribute to the inflammatory response to RSV infection (26) and wished to compare the IL-1β responses in these cells. THP-1 cells and CD8-depleted, adherent PBMC were cultured with RSV WT or RSV ΔSH. RSV WT induced an IL-1β response that was greater than that of control-treated cells, at a level similar to that observed in previous studies (Fig. 3C and D) (27). RSV ΔSH induced a significantly greater IL-1β response than RSV WT in both THP-1 cells (P < 0.05) (Fig. 3C) and PBMC (P < 0.05) (Fig. 3D). To determine whether viral replication or virus-associated pathogen-associated molecular patterns (PAMPs) were driving the IL-1β response, the IL-1β responses in THP-1 cells exposed to live or UV-inactivated virus were compared. Inactivation of the virus was confirmed by in vitro plaque assay (data not shown). Live RSV induced a significantly greater IL-1β response than UV-inactivated virus with both the ΔSH and WT viruses, indicating that the virus needs to replicate to induce cytokine release in macrophages. Since neutrophils are also a significant source of inflammatory cytokines in the lungs after infection, IL-1β levels were measured at 24 h after RSV infection of primary human neutrophils. Infection of neutrophils with RSV ΔSH led to a significantly greater IL-1β response than RSV WT (P < 0.05) (Fig. 3E). From these studies, we speculate that the SH protein of RSV antagonizes the IL-1β response.
FIG 3.
Recombinant RSV lacking the SH gene induces a greater IL-1β response than the wild type in vitro. (A) HEp-2 cells were infected with RSV WT or RSV ΔSH (MOI = 0.5), and the viral load was assessed by plaque assay. (B to E) Supernatants were collected and analyzed for IL-1β levels by ELISA following infection of HEp-2 cells (B), THP-1 cells (C), PBMC (D), and neutrophils (E). (F to H) HEp-2 (F) and THP-1 (G) cells were infected with RSV GFP (MOI = 0.1), and primary neutrophils (H) were infected with RSV (MOI = 0.25) and imaged at 24 h after infection. The points represent means ± SEM (n = 3 repeats) of HEp-2, PBMC, and THP-1 cells and 3 individual PBMC and neutrophil donors. *, P < 0.05, and ***, P < 0.001 between RSV ΔSH and RSV WT; #, P < 0.05 between ΔSH and UV-inactivated ΔSH; calculated by two-way ANOVA (A to D) or ANOVA (E). The images are representative of 2 studies.
IL-1β release following exposure to RSV may be in response to intracellular infection of the cell (in cis) or in response to extracellular danger signals (in trans). To understand the responses in different cell types, we determined whether RSV infects these cell types. Cells were cultured with GFP-expressing RSV and imaged by fluorescence microscopy. Green fluorescence was detectable in HEp-2 (Fig. 3F) and THP-1 (Fig. 3G) cells after infection with RSV, indicating that there is transcription of virus-encoded RNA. More green HEp-2 cells were detectable than THP-1 cells for an equivalent MOI of RSV. Very limited green fluorescence was observed in primary neutrophils at 24 h after infection, even with a higher MOI of RSV, suggesting they do not support viral replication and are therefore responding to extracellular virus, though further studies are required to confirm this (Fig. 3H).
IL-1β is produced in response to RSV by macrophages and neutrophils in vivo.
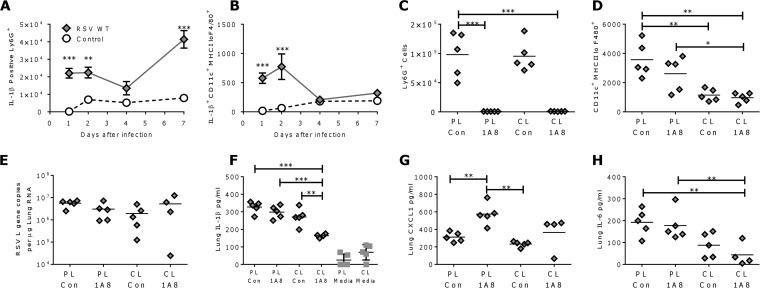
Having observed differences in the amounts of IL-1β produced by different cell types in vitro, we wished to determine which cell types produce IL-1β in vivo. Cells were isolated from the lungs after RSV WT infection or mock infection with medium alone, and IL-1β-positive cells were identified by intracellular cytokine staining in the absence of stimulation. The number of IL-1β+ Ly6G+ cells (neutrophils) (Fig. 4A) was significantly greater in the infected group than in the uninfected group. Interestingly, the percentage of Ly6G cells positive for IL-1β increased over the course of infection. Detectable expression of IL-1β by CD11c+ F480+ MHC-II low cells (alveolar macrophages [28]) (Fig. 4B) was highest on days 1 and 2 after infection but returned to control levels by day 4 after infection. No lymphocytes were observed to be IL-1β positive. A similar profile of IL-1β-positive cells was observed following infection with RSV ΔSH.
FIG 4.
IL-1β is produced by neutrophils and macrophages in vivo. (A and B) Mice were infected with RSV WT or control treated intranasally. Expression of IL-1β by Ly6G+ neutrophils (A) and CD11c+ F480+ MHC-II low alveolar macrophages (B) was measured by flow cytometry at various time points after infection. (C to H) Mice were treated with anti-Ly6G-depleting antibody (1A8) or control antibody (Con) intraperitoneally and CL or PL intranasally prior to RSV WT infection. Neutrophil (C) and alveolar macrophage (D) numbers were analyzed by flow cytometry and the RSV L gene (E) by RT-PCR, and lung IL-1β (F), CXCL1 (G), and IL-6 (H) were measured on day 1 after infection. The points represent means ± SEM (n = 5 mice) (A and B) and individual animals; the bars represent means (n = 5) (C to H). *, P < 0.05; **, P < 0.01; ***, P < 0.001; calculated by multiple t tests with Holm-Sidak correction (A and B) or ANOVA (C to H). Gray squares, media-alone groups.
Cell-specific depletion was used to determine whether a specific cell type was the major contributor to lung IL-1β after infection. The effects on acute RSV infection (day 1 after infection) of alveolar macrophage depletion (by CL treatment) or neutrophil depletion (using anti Ly6G [clone 1A8] antibody) were compared. 1A8 treatment led to a significant reduction in the number of neutrophils detectable in the lungs (Fig. 4C) and BAL fluid (data not shown). CL treatment led to a significant decrease in the percentage of CD11c+ F480+ MHC-II low cells (Fig. 4D). Cell depletion had no effect on the viral load at day 1 after infection (Fig. 4E). In spite of the high percentage of IL-1β-positive neutrophils detected, neutrophil depletion by 1A8 treatment alone had no effect on IL-1β (Fig. 4F). Interestingly, more CXCL1 was detected in the lungs of these mice (Fig. 4G), and there was no effect on IL-6 levels (Fig. 4H). CL treatment alone significantly reduced the level of IL-6 but had no effect on lung IL-1β or CXCL1 levels. Mice that received both clodronate and 1A8 treatments had significantly reduced IL-1β, suggesting that either there is some redundancy in the cells that produce IL-1β or these cells act cooperatively. CL treatment alone without RSV infection did not significantly increase IL-1β production in the lungs compared to control PL treatment alone (Fig. 4F). We therefore conclude that IL-1β is produced by a number of different cellular sources, including neutrophils and CL-sensitive cells, presumably alveolar macrophages.
DISCUSSION
One rationale for understanding the function of the SH gene is to target it for deletion in live attenuated RSV vaccines. Here, we demonstrate enhanced inflammation after infection with RSV lacking the SH gene, particularly in the IL-1β pathway. In the current study, we demonstrated that RSV ΔSH protected against viral challenge in the mouse model. A recent study has demonstrated that a bRSV ΔSH vaccine is highly protective in infant calves, even in the presence of maternal antibody (29), and a live attenuated RSV ΔSH vaccine with additional point mutations (Medi-559) has been shown to be safe and immunogenic in clinical trials in children (30). Data from chimpanzee studies (31), which showed moderate attenuation, suggest SH alone would be insufficient for a safe live vaccine, though it has been shown to be highly effective in cattle (29). While addition of ΔSH to other temperature-sensitive vaccines had a marginal effect (32, 33) on attenuation, we speculate that SH has an immunomodulatory function and that its deletion as part of a live attenuated RSV vaccine design would be beneficial.
The observation that deletion of RSV SH leads to an increase in IL-1β is supported by studies in bRSV, where a ΔSH vaccine strain induced higher levels of IL-1β in the lungs of infected cattle (17). In the current study, blockade of IL-1β prior to infection increased the viral load, supporting the idea that SH might enable immunomodulation. Interestingly, bRSV ΔSH also induced greater levels of apoptosis, which can be associated with inflammasome activation. In contrast, a study with different live attenuated RSV vaccines showed that the most heavily attenuated vaccine (rA2cp248/404/1030/ΔSH) induced lower responses in nasal wash samples to a number of cytokines than other live attenuated vaccines (6). However, the virus tested was highly attenuated, and the reduced cytokine response was likely a consequence of strong attenuation of replication related to the point mutations in the backbone of the virus rather than the SH deletion. A recent study by Triantafilou et al. proposed that the RSV SH protein activates the inflammasome (34). Unlike the current study, the virus used by Triantafilou et al. had the SH gene replaced with GFP. We believe that there are flaws in the published study by Triantafillou et al. that warrant further investigation: figures showing the relative growth of the two viruses are not included in the study, the study was performed at only a single time point in a single cell line using a high MOI of virus, and there was no statistical analysis comparing the responses between the viruses. In contrast to the published study, we have carefully dissected the inflammasome response to RSV infection, with and without the SH protein, both in vivo and in vitro, performing multiple time course studies using primary cells, cell lines, and a mouse model.
We observed that both neutrophils and clodronate-sensitive alveolar macrophages contribute to the lung IL-1β response (in vivo and in vitro). We propose a model where the alveolar macrophages act as sentinels for infection, initiating the immune response, and the neutrophils then amplify the signal. Of note, in our hands, epithelial cells and macrophages supported transcription of the viral genome, but neutrophils did not. While we (26, 35) and other groups (36, 37) have explored the role of alveolar macrophages in initiating the immune response to RSV infection by cytokine production, the role of neutrophils in RSV infection is not known. Studies have shown IL-9 (38), TNF (39), and chemokine (40) production by neutrophils in response to RSV. Neutrophils are a source of IL-1β in lung injury models (41), but this is the first study that has demonstrated IL-1β production by neutrophils in response to respiratory viral infection. Airway neutrophilia is observed in patients with bronchiolitis (42), and an increase in neutrophils is detected in blood prior to the influx of CD8 T cells (43). We have previously observed persistent airway neutrophilia following regulatory T cell depletion (44), suggesting they are associated with disease, but other studies have demonstrated that neutrophils can have a protective (45) or antiviral (46) role. While RSV RNA and protein have been observed in airway neutrophils from infected infants (47), the data generated using GFP-expressing virus suggest that macrophages, but not neutrophils, can support viral replication.
We speculate that SH is an immune evasion protein and that this mechanism of immune evasion may be conserved across other viruses. SH genes are found in all members of the subfamily Pneumovirinae and some of the Paramyxovirinae, including mumps virus (48) and human parainfluenza virus 5 (hPIV5) (49). The SH proteins from human metapneumovirus (hMPV) (50), bRSV (51), hPIV5 (49), and mumps virus (52) are nonessential for in vitro growth, and an immunomodulatory function has been described for the SH proteins from PIV5 (49), bRSV (17), and hMPV (53), which also encodes a viroporin (54). We hypothesize that the pore structure of the SH protein (10) facilitates this immunomodulatory function, counteracting the activation of NLRP3 by ionic changes induced by RSV infection (27). However, not all viroporins inhibit inflammasome function; for example, the M2 protein of influenza A virus is a viroporin that has been demonstrated to activate the inflammasome (55). The differences between RSV SH and influenza virus M2 may be due to the differences in structure (13). Of note, another viroporin with a structure similar to that of RSV SH, influenza B virus NB, has a similar, limited effect on the replication of influenza virus in vitro (56). Alternatively, SH has been shown to form a complex with the G protein (57), which has also been shown to have an immunomodulatory function (58), and the complex is potentially important; more research is required to determine the exact mechanism of action. In addition to depletion in live vaccines, targeting the SH protein may open up novel antiviral therapies, and a recent study has demonstrated that its function as an ion pore can be blocked with a small-molecule inhibitor, pyronin B (11). In the current study, we show that deletion of the SH gene changes the immune response on infection, increasing IL-1β responses by an as yet undetermined mechanism, and we believe that further research on this aspect of SH gene function is important in the development of an RSV vaccine.
ACKNOWLEDGMENTS
We thank Peter L. Collins (NIH) for providing the ΔSH and GFP viruses; Joanna Dreux, Sajad Bhat, Bhavika Parmanand, and Ephram Roresa for initial studies not included in the manuscript; and Fiona Culley and Robin Shattock (Imperial College London) for advice on the manuscript.
We have no commercial conflicts of interest in this study.
The research leading to these results has received support from the Innovative Medicines Initiative Joint Undertaking under grant agreement no. 115308 Biovacsafe, the resources of which are composed of financial contributions from the European Union's Seventh Framework Program (FP7/2007-2013) and EFPIA members' in-kind contributions. This work was supported by the European Community's European Seventh Framework Program ADITEC (HEALTH-F4-2011-18 280873).
REFERENCES
- 1.Tregoning JS, Schwarze J. 2010. Respiratory viral infections in infants: causes, clinical symptoms, virology, and immunology. Clin Microbiol Rev 23:74–98. doi: 10.1128/CMR.00032-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sande CJ, Mutunga MN, Okiro EA, Medley GF, Cane PA, Nokes DJ. 2013. Kinetics of the neutralizing antibody response to respiratory syncytial virus infections in a birth cohort. J Med Virol 85:2020–2025. doi: 10.1002/jmv.23696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Graham BS. 2014. Protecting the family to protect the child: vaccination strategy guided by RSV transmission dynamics. J Infect Dis 209:1679–1681. doi: 10.1093/infdis/jiu075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prutsky GJ, Domecq JP, Elraiyah T, Prokop LJ, Murad MH. 2014. Assessing the evidence: live attenuated influenza vaccine in children younger than 2 years. A systematic review. Pediatr Infect Dis J 33:e106–e115. doi: 10.1097/INF.0000000000000200. [DOI] [PubMed] [Google Scholar]
- 5.Steinhoff MC, MacDonald N, Pfeifer D, Muglia LJ. 2014. Influenza vaccine in pregnancy: policy and research strategies. Lancet 383:1611–1613. doi: 10.1016/S0140-6736(14)60583-3. [DOI] [PubMed] [Google Scholar]
- 6.Karron RA, Buchholz UJ, Collins PL. 2013. Live-attenuated respiratory syncytial virus vaccines. Curr Top Microbiol Immunol 372:259–284. doi: 10.1007/978-3-642-38919-1_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spann KM, Tran KC, Collins PL. 2005. Effects of nonstructural proteins NS1 and NS2 of human respiratory syncytial virus on interferon regulatory factor 3, NF-kappaB, and proinflammatory cytokines. J Virol 79:5353–5362. doi: 10.1128/JVI.79.9.5353-5362.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gan SW, Ng L, Lin X, Gong X, Torres J. 2008. Structure and ion channel activity of the human respiratory syncytial virus (hRSV) small hydrophobic protein transmembrane domain. Protein Sci 17:813–820. doi: 10.1110/ps.073366208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carter SD, Dent KC, Atkins E, Foster TL, Verow M, Gorny P, Harris M, Hiscox JA, Ranson NA, Griffin S, Barr JN. 2010. Direct visualization of the small hydrophobic protein of human respiratory syncytial virus reveals the structural basis for membrane permeability. FEBS Lett 584:2786–2790. doi: 10.1016/j.febslet.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gan SW, Tan E, Lin X, Yu D, Wang J, Tan GM, Vararattanavech A, Yeo CY, Soon CH, Soong TW, Pervushin K, Torres J. 2012. The small hydrophobic protein of the human respiratory syncytial virus forms pentameric ion channels. J Biol Chem 287:24671–24689. doi: 10.1074/jbc.M111.332791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y, To J, Verdia-Baguena C, Dossena S, Surya W, Huang M, Paulmichl M, Liu DX, Aguilella VM, Torres J. 2014. Inhibition of the human respiratory syncytial virus small hydrophobic protein and structural variations in a bicelle environment. J Virol 88:11899–11914. doi: 10.1128/JVI.00839-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rixon HW, Brown G, Aitken J, McDonald T, Graham S, Sugrue RJ. 2004. The small hydrophobic (SH) protein accumulates within lipid-raft structures of the Golgi complex during respiratory syncytial virus infection. J Gen Virol 85:1153–1165. doi: 10.1099/vir.0.19769-0. [DOI] [PubMed] [Google Scholar]
- 13.Nieva JL, Madan V, Carrasco L. 2012. Viroporins: structure and biological functions. Nat Rev Microbiol 10:563–574. doi: 10.1038/nrmicro2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin H, Zhou H, Cheng X, Tang R, Munoz M, Nguyen N. 2000. Recombinant respiratory syncytial viruses with deletions in the NS1, NS2, SH, and M2-2 genes are attenuated in vitro and in vivo. Virology 273:210–218. doi: 10.1006/viro.2000.0393. [DOI] [PubMed] [Google Scholar]
- 15.Bukreyev A, Whitehead SS, Murphy BR, Collins PL. 1997. Recombinant respiratory syncytial virus from which the entire SH gene has been deleted grows efficiently in cell culture and exhibits site-specific attenuation in the respiratory tract of the mouse. J Virol 71:8973–8982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuentes S, Tran KC, Luthra P, Teng MN, He B. 2007. Function of the respiratory syncytial virus small hydrophobic protein. J Virol 81:8361–8366. doi: 10.1128/JVI.02717-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taylor G, Wyld S, Valarcher JF, Guzman E, Thom M, Widdison S, Buchholz UJ. 2014. Recombinant bovine respiratory syncytial virus with deletion of the SH gene induces increased apoptosis and pro-inflammatory cytokines in vitro, and is attenuated and induces protective immunity in calves. J Gen Virol 95:1244–1254. doi: 10.1099/vir.0.064931-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamazaki T, Ichinohe T. 2014. Inflammasomes in antiviral immunity: clues for influenza vaccine development. Clin Exp Vaccine Res 3:5–11. doi: 10.7774/cevr.2014.3.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bukreyev A, Camargo E, Collins PL. 1996. Recovery of infectious respiratory syncytial virus expressing an additional, foreign gene. J Virol 70:6634–6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tregoning JS, Yamaguchi Y, Wang B, Mihm D, Harker JA, Bushell ES, Zheng M, Liao G, Peltz G, Openshaw PJ. 2010. Genetic susceptibility to the delayed sequelae of neonatal respiratory syncytial virus infection is MHC dependent. J Immunol 185:5384–5391. doi: 10.4049/jimmunol.1001594. [DOI] [PubMed] [Google Scholar]
- 21.Donnelly L, Curran RM, Tregoning JS, McKay PF, Cole T, Morrow RJ, Kett VL, Andrews GP, Woolfson AD, Malcolm RK, Shattock RJ. 2011. Intravaginal immunization using the recombinant HIV-1 clade-C trimeric envelope glycoprotein CN54gp140 formulated within lyophilized solid dosage forms. Vaccine 29:4512–4520. doi: 10.1016/j.vaccine.2011.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park EK, Jung HS, Yang HI, Yoo MC, Kim C, Kim KS. 2007. Optimized THP-1 differentiation is required for the detection of responses to weak stimuli. Inflamm Res 56:45–50. doi: 10.1007/s00011-007-6115-5. [DOI] [PubMed] [Google Scholar]
- 23.Eggleton P. 1998. Separation of cells using free flow electrophoresis, p 213–251. In Fisher DF, Gillian E, Rickwood D (ed), Cell separation: a practical approach. Oxford University Press, Oxford, United Kingdom. [Google Scholar]
- 24.Zhang L, Peeples ME, Boucher RC, Collins PL, Pickles RJ. 2002. Respiratory syncytial virus infection of human airway epithelial cells is polarized, specific to ciliated cells, and without obvious cytopathology. J Virol 76:5654–5666. doi: 10.1128/JVI.76.11.5654-5666.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harker JA, Godlee A, Wahlsten JL, Lee DC, Thorne LG, Sawant D, Tregoning JS, Caspi RR, Bukreyev A, Collins PL, Openshaw PJ. 2010. Interleukin 18 coexpression during respiratory syncytial virus infection results in enhanced disease mediated by natural killer cells. J Virol 84:4073–4082. doi: 10.1128/JVI.02014-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pribul PK, Harker J, Wang B, Wang H, Tregoning JS, Schwarze J, Openshaw PJ. 2008. Alveolar macrophages are a major determinant of early responses to viral lung infection but do not influence subsequent disease development. J Virol 82:4441–4448. doi: 10.1128/JVI.02541-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Segovia J, Sabbah A, Mgbemena V, Tsai SY, Chang TH, Berton MT, Morris IR, Allen IC, Ting JP, Bose S. 2012. TLR2/MyD88/NF-kappaB pathway, reactive oxygen species, potassium efflux activates NLRP3/ASC inflammasome during respiratory syncytial virus infection. PLoS One 7:e29695. doi: 10.1371/journal.pone.0029695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guilliams M, De Kleer I, Henri S, Post S, Vanhoutte L, De Prijck S, Deswarte K, Malissen B, Hammad H, Lambrecht BN. 2013. Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF. J Exp Med 210:1977–1992. doi: 10.1084/jem.20131199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blodorn K, Hagglund S, Fix J, Dubuquoy C, Makabi-Panzu B, Thom M, Karlsson P, Roque JL, Karlstam E, Pringle J, Eleouet JF, Riffault S, Taylor G, Valarcher JF. 2014. Vaccine safety and efficacy evaluation of a recombinant bovine respiratory syncytial virus (BRSV) with deletion of the SH gene and subunit vaccines based on recombinant human RSV proteins: N-nanorings, P and M2-1, in calves with maternal antibodies. PLoS One 9:e100392. doi: 10.1371/journal.pone.0100392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malkin E, Yogev R, Abughali N, Sliman J, Wang CK, Zuo F, Yang CF, Eickhoff M, Esser MT, Tang RS, Dubovsky F. 2013. Safety and immunogenicity of a live attenuated RSV vaccine in healthy RSV-seronegative children 5 to 24 months of age. PLoS One 8:e77104. doi: 10.1371/journal.pone.0077104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whitehead SS, Bukreyev A, Teng MN, Firestone CY, St Claire M, Elkins WR, Collins PL, Murphy BR. 1999. Recombinant respiratory syncytial virus bearing a deletion of either the NS2 or SH gene is attenuated in chimpanzees. J Virol 73:3438–3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wright PF, Karron RA, Belshe RB, Thompson J, Crowe JE Jr, Boyce TG, Halburnt LL, Reed GW, Whitehead SS, Anderson EL, Wittek AE, Casey R, Eichelberger M, Thumar B, Randolph VB, Udem SA, Chanock RM, Murphy BR. 2000. Evaluation of a live, cold-passaged, temperature-sensitive, respiratory syncytial virus vaccine candidate in infancy. J Infect Dis 182:1331–1342. doi: 10.1086/315859. [DOI] [PubMed] [Google Scholar]
- 33.Karron RA, Wright PF, Belshe RB, Thumar B, Casey R, Newman F, Polack FP, Randolph VB, Deatly A, Hackell J, Gruber W, Murphy BR, Collins PL. 2005. Identification of a recombinant live attenuated respiratory syncytial virus vaccine candidate that is highly attenuated in infants. J Infect Dis 191:1093–1104. doi: 10.1086/427813. [DOI] [PubMed] [Google Scholar]
- 34.Triantafilou K, Kar S, Vakakis E, Kotecha S, Triantafilou M. 2013. Human respiratory syncytial virus viroporin SH: a viral recognition pathway used by the host to signal inflammasome activation. Thorax 68:66–75. doi: 10.1136/thoraxjnl-2012-202182. [DOI] [PubMed] [Google Scholar]
- 35.Bukreyev A, Yang L, Collins PL. 2012. The secreted G protein of human respiratory syncytial virus antagonizes antibody-mediated restriction of replication involving macrophages and complement. J Virol 86:10880–10884. doi: 10.1128/JVI.01162-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kolli D, Gupta MR, Sbrana E, Velayutham TS, Hong C, Casola A, Garofalo RP. 2014. Alveolar macrophages contribute to the pathogenesis of hMPV infection while protecting against RSV infection. Am J Respir Cell Mol Biol 51:502–515. doi: 10.1165/rcmb.2013-0414OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goritzka M, Makris S, Kausar F, Durant LR, Pereira C, Kumagai Y, Culley FJ, Mack M, Akira S, Johansson C. 2015. Alveolar macrophage-derived type I interferons orchestrate innate immunity to RSV through recruitment of antiviral monocytes. J Exp Med 212:699–714. doi: 10.1084/jem.20140825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McNamara PS, Flanagan BF, Baldwin LM, Newland P, Hart CA, Smyth RL. 2004. Interleukin 9 production in the lungs of infants with severe respiratory syncytial virus bronchiolitis. Lancet 363:1031–1037. doi: 10.1016/S0140-6736(04)15838-8. [DOI] [PubMed] [Google Scholar]
- 39.Stokes KL, Currier MG, Sakamoto K, Lee S, Collins PL, Plemper RK, Moore ML. 2013. The respiratory syncytial virus fusion protein and neutrophils mediate the airway mucin response to pathogenic respiratory syncytial virus infection. J Virol 87:10070–10082. doi: 10.1128/JVI.01347-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jaovisidha P, Peeples ME, Brees AA, Carpenter LR, Moy JN. 1999. Respiratory syncytial virus stimulates neutrophil degranulation and chemokine release. J Immunol 163:2816–2820. [PubMed] [Google Scholar]
- 41.Grailer JJ, Canning BA, Kalbitz M, Haggadone MD, Dhond RM, Andjelkovic AV, Zetoune FS, Ward PA. 2014. Critical Role for the NLRP3 Inflammasome during Acute Lung Injury. J Immunol 192:5974–5983. doi: 10.4049/jimmunol.1400368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Everard ML, Swarbrick A, Wrightham M, McIntyre J, Dunkley C, James PD, Sewell HF, Milner AD. 1994. Analysis of cells obtained by bronchial lavage of infants with respiratory syncytial virus infection. Arch Dis Child 71:428–432. doi: 10.1136/adc.71.5.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lukens MV, van de Pol AC, Coenjaerts FE, Jansen NJ, Kamp VM, Kimpen JL, Rossen JW, Ulfman LH, Tacke CE, Viveen MC, Koenderman L, Wolfs TF, van Bleek GM. 2010. A systemic neutrophil response precedes robust CD8(+) T-cell activation during natural respiratory syncytial virus infection in infants. J Virol 84:2374–2383. doi: 10.1128/JVI.01807-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee DC, Harker JA, Tregoning JS, Atabani SF, Johansson C, Schwarze J, Openshaw PJ. 2010. CD25+ natural regulatory T cells are critical in limiting innate and adaptive immunity and resolving disease following respiratory syncytial virus infection. J Virol 84:8790–8798. doi: 10.1128/JVI.00796-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tate MD, Deng YM, Jones JE, Anderson GP, Brooks AG, Reading PC. 2009. Neutrophils ameliorate lung injury and the development of severe disease during influenza infection. J Immunol 183:7441–7450. doi: 10.4049/jimmunol.0902497. [DOI] [PubMed] [Google Scholar]
- 46.Stacey MA, Marsden M, Pham NT, Clare S, Dolton G, Stack G, Jones E, Klenerman P, Gallimore AM, Taylor PR, Snelgrove RJ, Lawley TD, Dougan G, Benedict CA, Jones SA, Wilkinson GW, Humphreys IR. 2014. Neutrophils recruited by IL-22 in peripheral tissues function as TRAIL-dependent antiviral effectors against MCMV. Cell Host Microbe 15:471–483. doi: 10.1016/j.chom.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Halfhide CP, Flanagan BF, Brearey SP, Hunt JA, Fonceca AM, McNamara PS, Howarth D, Edwards S, Smyth RL. 2011. Respiratory syncytial virus binds and undergoes transcription in neutrophils from the blood and airways of infants with severe bronchiolitis. J Infect Dis 204:451–458. doi: 10.1093/infdis/jir280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cui A, Myers R, Xu W, Jin L. 2009. Analysis of the genetic variability of the mumps SH gene in viruses circulating in the UK between 1996 and 2005. Infect Genet Evol 9:71–80. doi: 10.1016/j.meegid.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 49.He B, Lin GY, Durbin JE, Durbin RK, Lamb RA. 2001. The SH integral membrane protein of the paramyxovirus simian virus 5 is required to block apoptosis in MDBK cells. J Virol 75:4068–4079. doi: 10.1128/JVI.75.9.4068-4079.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de Graaf M, Herfst S, Aarbiou J, Burgers PC, Zaaraoui-Boutahar F, Bijl M, van Ijcken W, Schrauwen EJ, Osterhaus AD, Luider TM, Scholte BJ, Fouchier RA, Andeweg AC. 2013. Small hydrophobic protein of human metapneumovirus does not affect virus replication and host gene expression in vitro. PLoS One 8:e58572. doi: 10.1371/journal.pone.0058572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karger A, Schmidt U, Buchholz UJ. 2001. Recombinant bovine respiratory syncytial virus with deletions of the G or SH genes: G and F proteins bind heparin. J Gen Virol 82:631–640. [DOI] [PubMed] [Google Scholar]
- 52.Wilson RL, Fuentes SM, Wang P, Taddeo EC, Klatt A, Henderson AJ, He B. 2006. Function of small hydrophobic proteins of paramyxovirus. J Virol 80:1700–1709. doi: 10.1128/JVI.80.4.1700-1709.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bao X, Kolli D, Liu T, Shan Y, Garofalo RP, Casola A. 2008. Human metapneumovirus small hydrophobic protein inhibits NF-kappaB transcriptional activity. J Virol 82:8224–8229. doi: 10.1128/JVI.02584-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Masante C, El Najjar F, Chang A, Jones A, Moncman CL, Dutch RE. 2014. The human metapneumovirus small hydrophobic protein has properties consistent with those of a viroporin and can modulate viral fusogenic activity. J Virol 88:6423–6433. doi: 10.1128/JVI.02848-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ichinohe T, Pang IK, Iwasaki A. 2010. Influenza virus activates inflammasomes via its intracellular M2 ion channel. Nat Immunol 11:404–410. doi: 10.1038/ni.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hatta M, Kawaoka Y. 2003. The NB protein of influenza B virus is not necessary for virus replication in vitro. J Virol 77:6050–6054. doi: 10.1128/JVI.77.10.6050-6054.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Low KW, Tan T, Ng K, Tan BH, Sugrue RJ. 2008. The RSV F and G glycoproteins interact to form a complex on the surface of infected cells. Biochem Biophys Res Commun 366:308–313. doi: 10.1016/j.bbrc.2007.11.042. [DOI] [PubMed] [Google Scholar]
- 58.Polack FP, Irusta PM, Hoffman SJ, Schiatti MP, Melendi GA, Delgado MF, Laham FR, Thumar B, Hendry RM, Melero JA, Karron RA, Collins PL, Kleeberger SR. 2005. The cysteine-rich region of respiratory syncytial virus attachment protein inhibits innate immunity elicited by the virus and endotoxin. Proc Natl Acad Sci U S A 102:8996–9001. doi: 10.1073/pnas.0409478102. [DOI] [PMC free article] [PubMed] [Google Scholar]