Abstract
H7N9 was a cause of significant global health concern due to its severe infection and approximately 35% mortality in humans. By screening a Fab antibody phage library derived from patients who recovered from H7N9 infections, we characterized two human monoclonal antibodies (HuMAbs), HNIgGD5 and HNIgGH8. The epitope of these two antibodies was dependent on two residues in the receptor binding site at positions V186 and L226 of the hemagglutinin glycoprotein. Both antibodies possessed high neutralizing activity.
TEXT
In the spring of 2013, there was an outbreak of a novel influenza A virus, H7N9, in China (1). Influenza A viruses are enveloped RNA viruses in the family Orthomyxoviridae. They possess eight negative-sense genomic segments and are classified into subtypes based on the two surface glycoproteins, hemagglutinin (HA) and neuraminidase (NA). HA is a trimer of identical subunits. Each HA monomer contains a receptor binding site (RBS) in its globular head, involved in virus-receptor recognition and viral invasion into cells. NA is a mushroom-shaped tetramer of identical subunits. During replication, NA removes sialic acid from cellular glycoproteins and glycolipids and from both of the virus glycoproteins and thus enables the virus to be released from the host cell. So far, 16 HA subtypes (H1 to H16) and 9 NA subtypes (N1 to N9) have been identified from avian natural reservoirs. However, this is the first time that an H7N9 virus has been infectious and lethal to humans. Within a few months, the H7N9 virus had caused 137 cases of infection, including 45 fatalities (2). Direct contact with infected poultry was responsible for most of the human cases (1, 3). Genetic analysis also indicated that both HA and NA of the H7N9 influenza virus were closely related to that from ducks in China (HA) and from wild birds and ducks from China and Korea (NA) (1, 4).
Amantadine and rimantadine have been widely utilized as anti-influenza drugs. Unfortunately, neither was effective against H7N9 infection. Recent studies revealed that H7N9 could be inhibited by the NA inhibitors oseltamivir and zanamivir (5). Unfortunately, an R292K mutation and an R152K mutation (N9 numbering) that conferred resistance to both inhibitors were detected (5, 6). Neutralizing antibodies could prevent influenza A virus infection, especially in sporadic human infections (7). However, the only neutralizing antibody reported to react with H7 was less effective against this recent H7N9 virus (8).
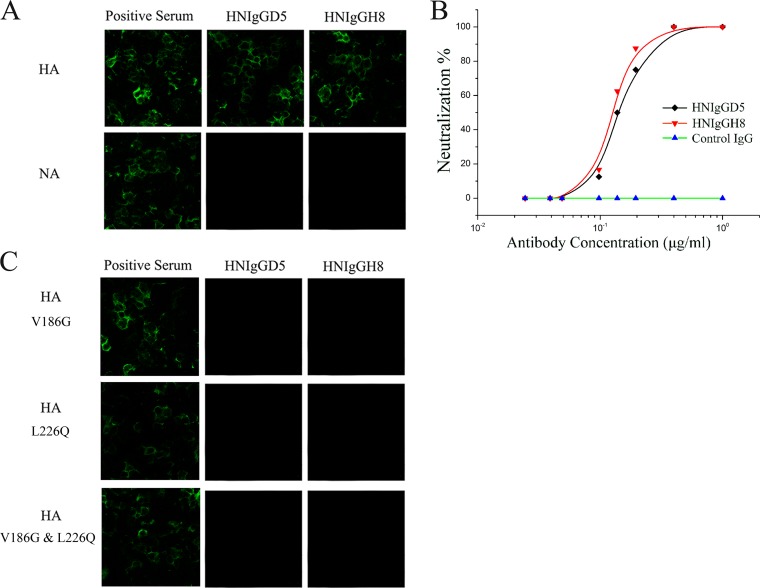
To screen for human neutralizing antibodies against novel H7N9 virus, we established two Fab phage display libraries for λ and κ light chains from the peripheral blood mononuclear cells from three recovered patients 3 to 4 weeks after hospitalization, who were infected with H7N9 (A/Anhui/1/2013 [here termed H7N9-AH1]). After three rounds of panning with purified H7N9 virions, 816 positive Fab clones and 18 unique clones were identified. Among these 18 Fabs, 6 derive from the VH3 family, and 12 derive from the VH4 family. The light chains of these Fabs are derived from the lambda gene family Vλ1 and the kappa gene families Vκ1, Vκ2, and Vκ3. All 18 clones demonstrated reactivity with purified virions or viral antigens, while 6 showed potent in vitro neutralizing activity. We then converted these 6 Fabs into full-length IgG1 molecules for further study. The results for HNIgGD5 and HNIgGH8 (derived from H7N9Fab13 and H7N9Fab14) are reported here. Both Fab clones possessed unique VH4 and Vκ1 sequences (Table 1). There are four amino acid substitutions between the two Fabs, with one located in the CDR3 region in the light chain. The immunospecificity for HNIgGD5 and HNIgGH8 was further tested by immunofluorescence assay (IFA). Both reacted with HA protein but not NA (Fig. 1A). A hemagglutination inhibition (HI) assay was then performed. HNIgGD5 and HNIgGH8 showed equally strong HI activity, with the HI titer as low as 0.8 μg/ml. Next, their neutralizing activity against live H7N9 virus was determined on MDCK cells as described before (9). As shown in Fig. 1B, both HNIgGD5 and HNIgGH8 substantially neutralized H7N9 virus on MDCK cells in a dose-dependent manner. To determine the epitope regions recognized by the antibodies, escape mutants were selected as described previously (9, 10). After five passages, variants were confirmed by their similar levels of growth in the presence and absence of the antibody and lack of inhibition in HI tests. The entire HA gene was then sequenced. As expected, amino acid substitutions were detected. Interestingly, the substitutions occurred at identical positions, V186G or L226Q (H3 numbering), both located in the RBS. It has been reported that these two amino acids had significant roles in human H7 HA binding with human receptor (11, 12). To verify this result, we constructed three HA mutants: each had one or both of the two amino acids substituted. The constructs were transiently expressed in HEK293T cells and further detected by IFA. As shown in Fig. 1C, either V186G or L226Q abolished binding of HA with the antibodies. These results together demonstrated that the epitope of HNIgGD5 and HNIgGH8 was dependent on two residues at positions V186 and L226 of human H7N9 HA. The findings are also in agreement with previous findings that antibodies recognizing the globular head had strong HI activity (13). Considering the crucial roles for V186 and L226 in human H7 HA binding with the human receptor (11, 12), it was plausible that the antibodies HNIgGD5 and HNIgGH8 bound with the HA protein through the RBS and thus interfered with its recognition and interaction with the human cellular receptor.
TABLE 1.
Amino acid sequences of variable regions in the H and L chains of H7N9 virus-specific Fabs
| Fab | VRa | Sequence of CDR in VRb: |
||
|---|---|---|---|---|
| CDR1 | CDR2 | CDR3 | ||
| H7N9Fab13 | VH | SGGYYWS | YIYYSGSTDYNPSLKS | GSTGDRHYYYYGMDV |
| VL | RASQSISSYLN | AASSLQS | QQSYSTPPT | |
| H7N9Fab14 | VH | SGGYYWS | YIYYSGSTDYNPSLKS | GSTGDRHYYYYGMDV |
| VL | RASQSISSYLN | AASSLQS | QQSYSTPST | |
VR, variable region; VH, and VL, V gene segments of the heavy and light chains, respectively.
CDR, complementarity-determining region.
FIG 1.
HNIgGD5 and HNIgGH8 are able to neutralize H7N9 virus. (A) The binding activity of the antibodies to HA and NA proteins were determined by IFA. (B) Neutralizing activities of HNIgGD5 and HNIgGH8 against H7N9 virus were tested on MDCK cells. An irrelevant human IgG was utilized as a control. (C) Epitope mapping of the antibodies. The selected amino acids in H7N9 HA were converted to the residues indicated. The mutated HA protein was then transiently expressed in cells and detected by IFA.
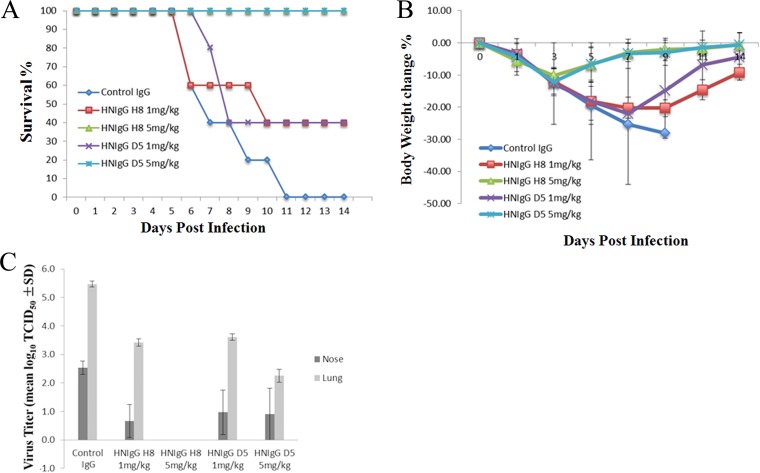
The in vivo therapeutic efficacies for both antibodies were tested in BALB/c mice. Ten mice per group were intraperitoneally injected with 1 or 5 mg/kg of purified human monoclonal antibodies (HuMAbs) 24 h before the mice were challenged intranasally with 50 μl of a 5× 50% lethal dose (LD50) mouse infectious dose of H7N9 virus. Another 10 mice immunized with an irrelevant human IgG were also infected as controls. Mice were observed daily for signs of disease and mortality for up to 14 days. As shown in Fig. 2A, animals that received an irrelevant control antibody succumbed to infection within 5 to 11 days after viral challenge. In contrast, 40% of the mice that received 1 mg/kg body weight HNIgGD5 or HNIgGH8 survived, and at 5 mg/kg, the antibodies conferred 100% protection from lethality by H7N9 in the infected mice. In addition, mice immunized with an irrelevant control antibody rapidly lost weight, compared to mice receiving HNIgGD5 or HNIgGH8, which gradually regained body weight at approximately 5 days postinfection (dpi) (Fig. 2B). To verify that the protection in the infected mice was due to the inhibition of viral proliferation by the antibodies, titers of H7N9 virus in the nose and lungs were determined. As shown in Fig. 2C, mice that received an irrelevant human IgG had high titers of virus in both the nose and lungs at 5 dpi. However, after passive immunization with either HNIgGD5 or HNIgGH8, viral proliferation was noticeably inhibited, and virus was difficult to detect in mice treated with 5 mg/kg HNIgGH8 (Fig. 2C).
FIG 2.
In vivo therapeutic efficacies of HNIgGD5 and HNIgGH8 in mice. Mice received HNIgGD5 or HNIgGH8 24 h prior to H7N9 infection and were monitored daily for 14 days. (A) The survival rate is presented. (B) The average body weight change of the mice is shown. (C) Virus titers from nose and lung tissue were determined at 5 dpi. Virus titers were substantially reduced in the HuMAb-treated groups (P < 0.05, t test).
A total of 454 H7N9 cases resulting in at least 171 deaths were reported by September 2014, and most cases were antigenically similar to the H7N9-AH1 isolate (14). In this study, we characterized two HuMAbs by screening a Fab antibody phage library derived from patients who recovered from H7N9 infection. These two antibodies showed high neutralizing activity against H7N9-AH1 in vitro and in vivo. At present, the neuraminidase inhibitors have still been the main therapeutic antiviral agents utilized for treatment of human infections with H7N9. Unfortunately, appearance of escape mutants was monitored (5, 6). In this regard, HNIgGD5 and HNIgGH8, attacking the virus directly toward the HA protein, thus provide an alternative choice for H7N9 treatment. The antibodies were of human origin, were originally generated in response to the natural infectious H7N9 virus, and had protected the infected individual, also powerfully supporting this rationale. Epitope identification demonstrated that two human receptor preferable residues at V186 and L226 were essential for antigen recognition. The essential roles played by the two residues in receptor binding limit their variation and consequent antigenic drift. Even if escape occurred, the antibodies would be suitable to be utilized as adjuncts with other established methods. At last, considering the possibility for antigen drift caused by widespread presence of H7N9 influenza virus in natural reservoirs and the continuing zoonotic introductions into humans, more HuMAbs, especially antibodies targeting distinct or more conserved epitopes, are still needed.
ACKNOWLEDGMENTS
This study was supported by PUMC Youth Fund and the Fundamental Research Funds for the Central Universities (33320140031), the National Natural Science Foundation of China (no. 31300154), the Chinese Ministry of Science and Technology (KJYJ-2013-01-01-01), and the Chinese National Major S & T Project (2013ZX10004-101). We thank the Shanghai and Beijing Centers for Disease Control and Prevention for providing the samples.
REFERENCES
- 1.Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, Xu X, Lu H, Zhu W, Gao Z, Xiang N, Shen Y, He Z, Gu Y, Zhang Z, Yang Y, Zhao X, Zhou L, Li X, Zou S, Zhang Y, Yang L, Guo J, Dong J, Li Q, Dong L, Zhu Y, Bai T, Wang S, Hao P, Yang W, Han J, Yu H, Li D, Gao GF, Wu G, Wang Y, Yuan Z, Shu Y. 2013. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med 368:1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- 2.Salathe M, Freifeld CC, Mekaru SR, Tomasulo AF, Brownstein JS. 2013. Influenza A (H7N9) and the importance of digital epidemiology. N Engl J Med 369:401–404. doi: 10.1056/NEJMp1307752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen Y, Liang W, Yang S, Wu N, Gao H, Sheng J, Yao H, Wo J, Fang Q, Cui D, Li Y, Yao X, Zhang Y, Wu H, Zheng S, Diao H, Xia S, Chan KH, Tsoi HW, Teng JL, Song W, Wang P, Lau SY, Zheng M, Chan JF, To KK, Chen H, Li L, Yuen KY. 2013. Human infections with the emerging avian influenza A H7N9 virus from wet market poultry: clinical analysis and characterisation of viral genome. Lancet 381:1916–1925. doi: 10.1016/S0140-6736(13)60903-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lam TT, Wang J, Shen Y, Zhou B, Duan L, Cheung CL, Ma C, Lycett SJ, Leung CY, Chen X, Li L, Hong W, Chai Y, Zhou L, Liang H, Ou Z, Liu Y, Farooqui A, Kelvin DJ, Poon LL, Smith DK, Pybus OG, Leung GM, Shu Y, Webster RG, Webby RJ, Peiris JS, Rambaut A, Zhu H, Guan Y. 2013. The genesis and source of the H7N9 influenza viruses causing human infections in China. Nature 502:241–244. doi: 10.1038/nature12515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu Y, Lu S, Song Z, Wang W, Hao P, Li J, Zhang X, Yen HL, Shi B, Li T, Guan W, Xu L, Liu Y, Wang S, Tian D, Zhu Z, He J, Huang K, Chen H, Zheng L, Li X, Ping J, Kang B, Xi X, Zha L, Li Y, Zhang Z, Peiris M, Yuan Z. 2013. Association between adverse clinical outcome in human disease caused by novel influenza A H7N9 virus and sustained viral shedding and emergence of antiviral resistance. Lancet 381:2273–2279. doi: 10.1016/S0140-6736(13)61125-3. [DOI] [PubMed] [Google Scholar]
- 6.Zurcher T, Yates PJ, Daly J, Sahasrabudhe A, Walters M, Dash L, Tisdale M, McKimm-Breschkin JL. 2006. Mutations conferring zanamivir resistance in human influenza virus N2 neuraminidases compromise virus fitness and are not stably maintained in vitro. J Antimicrob Chemother 58:723–732. doi: 10.1093/jac/dkl321. [DOI] [PubMed] [Google Scholar]
- 7.Traggiai E, Becker S, Subbarao K, Kolesnikova L, Uematsu Y, Gismondo MR, Murphy BR, Rappuoli R, Lanzavecchia A. 2004. An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nat Med 10:871–875. doi: 10.1038/nm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tharakaraman K, Subramanian V, Cain D, Sasisekharan V, Sasisekharan R. 2014. Broadly neutralizing influenza hemagglutinin stem-specific antibody CR8020 targets residues that are prone to escape due to host selection pressure. Cell Host Microbe 15:644–651. doi: 10.1016/j.chom.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Z, Wang J, Bao L, Guo L, Zhang W, Xue Y, Zhou H, Xiao Y, Wu F, Deng Y, Qin C, Jin Q. 2015. Human monoclonal antibodies targeting the haemagglutinin glycoprotein can neutralize H7N9 influenza virus. Nat Commun 6:6714. doi: 10.1038/ncomms7714. [DOI] [PubMed] [Google Scholar]
- 10.Yasugi M, Kubota-Koketsu R, Yamashita A, Kawashita N, Du A, Sasaki T, Nishimura M, Misaki R, Kuhara M, Boonsathorn N, Fujiyama K, Okuno Y, Nakaya T, Ikuta K. 2013. Human monoclonal antibodies broadly neutralizing against influenza B virus. PLoS Pathog 9:e1003150. doi: 10.1371/journal.ppat.1003150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi Y, Zhang W, Wang F, Qi J, Wu Y, Song H, Gao F, Bi Y, Zhang Y, Fan Z, Qin C, Sun H, Liu J, Haywood J, Liu W, Gong W, Wang D, Shu Y, Wang Y, Yan J, Gao GF. 2013. Structures and receptor binding of hemagglutinins from human-infecting H7N9 influenza viruses. Science 342:243–247. doi: 10.1126/science.1242917. [DOI] [PubMed] [Google Scholar]
- 12.Xiong X, Martin SR, Haire LF, Wharton SA, Daniels RS, Bennett MS, McCauley JW, Collins PJ, Walker PA, Skehel JJ, Gamblin SJ. 2013. Receptor binding by an H7N9 influenza virus from humans. Nature 499:496–499. doi: 10.1038/nature12372. [DOI] [PubMed] [Google Scholar]
- 13.Okuno Y, Isegawa Y, Sasao F, Ueda S. 1993. A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J Virol 67:2552–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wkly Epidemiol Rec. 2014. Antigenic and genetic characteristics of zoonotic influenza viruses and development of candidate vaccine viruses for pandemic preparedness. Wkly Epidemiol Rec 89:457–464. [PubMed] [Google Scholar]