ABSTRACT
Through its interaction with the 5′ translation initiation factor eIF4G, poly(A) binding protein (PABP) facilitates the translation of 5′-capped and 3′-poly(A)-tailed mRNAs. Rotavirus mRNAs are capped but not polyadenylated, instead terminating in a 3′ GACC motif that is recognized by the viral protein NSP3, which competes with PABP for eIF4G binding. Upon rotavirus infection, viral, GACC-tailed mRNAs are efficiently translated, while host poly(A)-tailed mRNA translation is, in contrast, severely impaired. To explore the roles of NSP3 in these two opposing events, the translational capabilities of three capped mRNAs, distinguished by either a GACC, a poly(A), or a non-GACC and nonpoly(A) 3′ end, have been monitored after electroporation of cells expressing all rotavirus proteins (infected cells) or only NSP3 (stably or transiently transfected cells). In infected cells, we found that the magnitudes of translation induction (GACC-tailed mRNA) and translation reduction [poly(A)-tailed mRNA] both depended on the rotavirus strain used but that translation reduction not genetically linked to NSP3. In transfected cells, even a small amount of NSP3 was sufficient to dramatically enhance GACC-tailed mRNA translation and, surprisingly, to slightly favor the translation of both poly(A)- and nonpoly(A)-tailed mRNAs, likely by stabilizing the eIF4E-eIF4G interaction. These data suggest that NSP3 is a translational surrogate of the PABP-poly(A) complex; therefore, it cannot by itself be responsible for inhibiting the translation of host poly(A)-tailed mRNAs upon rotavirus infection.
IMPORTANCE To control host cell physiology and to circumvent innate immunity, many viruses have evolved powerful mechanisms aimed at inhibiting host mRNA translation while stimulating translation of their own mRNAs. How rotavirus tackles this challenge is still a matter of debate. Using rotavirus-infected cells, we show that the magnitude of cellular poly(A) mRNA translation differs with respect to rotavirus strains but is not genetically linked to NSP3. Using cells expressing rotavirus NSP3, we show that NSP3 alone not only dramatically enhances rotavirus-like mRNA translation but also enhances poly(A) mRNA translation rather than inhibiting it, likely by stabilizing the eIF4E-eIF4G complex. Thus, the inhibition of cellular polyadenylated mRNA translation during rotavirus infection cannot be attributed solely to NSP3 and is more likely the result of global competition between viral and host mRNAs for the cellular translation machinery.
INTRODUCTION
When delivered into or synthesized by the host cell, viral mRNAs compete with cellular mRNAs already present in the cytoplasm for access to the protein synthesis machinery. Recruitment of the 40S ribosomal subunit onto mRNA (translation initiation) is the rate-limiting and most controlled step of eukaryotic protein biosynthesis; hence, it is highly competitive for both cellular and viral mRNAs. The 5′ cap and 3′ poly(A) tail of most cellular mRNAs are joined by a protein complex containing the cap binding protein eIF4E and the poly(A) binding protein PABP, which are bound together by the scaffold protein eIF4G (1). This complex recruits the preinitiation complex (PIC), which comprises the 40S ribosomal subunit loaded with the methionine initiator tRNA, eIF2, GTP, and several other translation initiation factors (2, 3). PABP is thought to stimulate translation initiation at least in part by promoting cap-to-poly(A) circularization of mRNA (4, 5). This appears to be especially true when mRNAs compete for ribosome binding. In this case, the presence of either structure alone at mRNA extremities is not enough to drive efficient translation, but together they synergize and direct ribosome entry at the 5′ end (6–8).
Rotavirus mRNAs are capped but lack the poly(A) tail required for efficient translation initiation. Instead, each rotavirus mRNA ends with the same 3′ GACC sequence. Despite this apparent handicap, rotavirus mRNAs efficiently compete with cellular mRNAs, and viral proteins are rapidly detectable in infected cells while the synthesis of host proteins is shut off (9).
We and others have shown that the rotavirus nonstructural protein NSP3 is bound to the 3′ end of viral mRNAs during infection (10) and that NSP3 dimers specifically bind the 3′ GACC sequence (11, 12) and eIF4G (9, 13). The simultaneous interactions of NSP3 with the viral mRNA 3′ end and with eIF4G have been shown to enhance the translation of rotavirus-like reporter mRNAs, as does PABP with cellular polyadenylated mRNAs (14, 15).
NSP3 dimers interact with eIF4G at the same position as PABP but with a 10-fold higher affinity (11). In fact, during rotavirus infection, PABP is evicted from eIF4G by NSP3 (9) and relocalizes to the cell nucleus (16). These observations led to the idea that the shutdown of cellular protein synthesis is due to the eviction of PABP and, hence, to the displacement of cellular polyadenylated mRNAs from eIF4G. This model also has been supported by in vivo experiments using recombinant vaccinia virus expressing NSP3 (17), as well as by in vitro translation assays using recombinant NSP3 or its eIF4G-binding domain (6, 15).
The opposite functions of NSP3 in viral versus cellular mRNA translation have been challenged by data obtained in rotavirus-infected cells in which NSP3 levels were downregulated by RNA interference (18). The NSP3 RNA- and eIF4G-binding domains (RNA-BD and 4G-BD, respectively) have been proposed to function independently, with the enhancement of viral mRNA translation due to NSP3 instead resulting from viral mRNA protection by the RNA-BD, whereas 4G-BD is involved in inhibiting host poly(A) mRNA translation (18). In the same vein, the translation of polyadenylated mRNAs introduced directly into the cytoplasm was not inhibited by rotavirus infection, in contrast to mRNAs synthesized in the nucleus, suggesting that a reduction in mRNA nuclear export rather than translation inhibition accounts for the rotavirus-mediated extinction of host protein synthesis (19). However, using an in vivo assay, we recently showed that the NSP3-dependent protection of viral mRNA from degradation was not sufficient to enhance translation and that NSP3 RNA-BD and 4G-BD did not work separately (M. Gratia, unpublished data). Here, using the same in vivo assay in cells infected with either the RF or RRV rotavirus strain or in cells transfected solely with NSP3, we show that both the extent of poly(A) mRNA translation inhibition and the extent of rotavirus-like mRNA translation stimulation vary depending on the rotavirus strain used. We also reveal that, independently of mRNA protection, the translation of reporter mRNAs ending with the rotavirus GACC motif is hugely induced by even low levels of NSP3 expression. Furthermore, we found that low levels of NSP3 similarly enhance the translation of both polyadenylated and nonpolyadenylated mRNAs and favor eIF4E-eIF4G interaction, suggesting that NSP3 alone can substitute for the PABP-poly(A) complex in translation initiation.
MATERIALS AND METHODS
Cells and viruses.
Embryonic rhesus monkey kidney MA104 and baby hamster kidney BSRT7 (20) cells were maintained in Eagle's minimum essential medium (EMEM; Lonza) supplemented with 10% fetal bovine serum, 100 IU/ml penicillin, and 100 μg/ml streptomycin. Human HeLa-S3 cells were obtained from the American Tissue Culture Collection and maintained in Dulbecco's minimum essential medium (Lonza) supplemented with 10% fetal bovine serum (Lonza) and 5 IU/ml penicillin-streptomycin (Lonza) in 5% CO2.
The bovine RF or simian RRV strains of group A rotavirus (RVA) were used to infect MA104 cells. Viral infectivity was determined by plaque assay on MA104 cells as described previously (21). Infections were performed at a multiplicity of infection (MOI) of 10 PFU/cell in EMEM in the presence of trypsin (0.44 μg/ml) and antibiotics but without serum.
Plasmid construction and mutagenesis.
The plasmid for in vitro transcription of the renilla luciferase rotavirus-like reporter mRNA (R-RNA) has been described in detail (Gratia et al., unpublished). Briefly, the DNA template (pT7-RF-Rluc) comprises the following, from 5′ to 3′: the T7 promoter fused to the 5′ untranslated region (UTR) of rotavirus RF gene 11, the renilla luciferase coding sequence, and the 3′ UTR of rotavirus RF gene 6 followed by a BsaI restriction site. The BsaI restriction site is positioned such that runoff transcription of BsaI-linearized plasmids produces RNAs with the canonical GACC rotavirus sequence at the 3′ end. Deletion by site-directed mutagenesis of the last 3 nucleotides of the canonical sequence to generate pT7-RF-Rluc-nona for in vitro synthesis of N-RNA, the addition of 66 adenosines (see below) at the 3′ end of the 3′ UTR to generate pT7-RF-Rluc-p(A) for in vitro synthesis of pA-RNA, and site-directed mutagenesis of the last nucleotides of the canonical 3′ sequence to generate pT7-RF-Rluc-GGCC and pT7-RF-Rluc-GAACC for in vitro synthesis of Nc-RNA have been described previously (Gratia et al., unpublished). The plasmid pEMCV-Fluc for in vitro transcription of the standard RNA (S-RNA) has been described already (Gratia et al., unpublished). S-RNA was obtained by in vitro transcription of the EcoRI-linearized pEMCV-Fluc plasmid.
The construction of the NSP3 cytoplasmic expression vector pT7-IRES NSP3 also has been described (Gratia et al., unpublished); this plasmid is composed of the T7 promoter fused to the encephalomyocarditis virus internal ribosome entry site (EMCV-IRES) cloned upstream of the NSP3 open reading frame (ORF) from either the RF or RRV rotavirus strain. A nonsense codon was introduced via site-directed mutagenesis (22) using PFU DNA polymerase at position 7 of the NSP3 ORF to generate pT7-IRES-NSP3-KO, which was used as a negative control.
The C-terminal half of RF-NSP3, either with (NSP3 150-313; here named cNSP3) or without (NSP3 206-313; here named cΔRX) the RoXaN binding domain, was cloned into the peGFPC1 vector (Clontech) in frame with enhanced green fluorescent protein (eGFP). The eIF4G-binding domain was removed from cNSP3 by the addition of a stop codon at position 240 via site-directed mutagenesis, yielding NSP3 150-239 (here named cΔ4G).
All plasmid constructs were verified by restriction enzyme mapping. PCR fragments were sequenced entirely after cloning into the target plasmids. In the case of site-directed mutagenesis, the whole functional unit (from the T7 promoter to the T7 terminator) was sequenced.
In vitro transcription.
Capped reporter RNAs were synthesized from BsaI-linearized plasmid templates by in vitro transcription using the mMessage mMachine T7 ultra kit (Ambion). S-RNA was synthesized using the T7 MEGAscript kit (Ambion), which does not contain the ARCA cap analog (23). DNA was removed by enzymatic treatment (15 min at 37°C) with RNase-free DNase, and RNAs then were purified on MegaClear (Ambion) silica spin columns to eliminate nonincorporated cap analogue and nucleotides prior to ethanol precipitation and washing with 70% ethanol. Purified RNAs were quantified using a spectrophotometer (NanoDrop), confirmed on denaturing agarose gels, and stored in aliquots at −80°C.
Cell transfection.
DNA was introduced into BSRT7 cells using Lipofectamine 2000 (Life Technologies) and into HeLa cells using ExGen 500 (EuroMedex) according to the manufacturers' instructions.
RNA was introduced into BSRT7 or MA104 cells using a Neon electroporation device (Life Technologies). Cells (106) were trypsinized and suspended in R buffer (Life Technologies) with 50 ng of reporter mRNAs (and 1 μg of standard RNA when indicated). Conditions for optimal electroporation were determined for each cell line; MA104 cells were electroporated by two 30-ms pulses of 1,150 V, and BSRT7 cells were electroporated by two 20-ms pulses of 1,400 V. After electroporation, cells were immediately transferred to complete culture medium and incubated at 37°C. To remove RNA remaining outside the cells, cells were incubated for 15 min at room temperature with 20 μg/ml of RNase A before RNA purification.
MA104 retrotransduction and stable transfection.
The MA104C20bis cell clone (here referred to as C20b) was obtained by retrotransduction. The RF NSP3 coding sequence (amino acids 4 to 365) was amplified using the oligonucleotides 721033newdir (CTATTCATACGTATATTCATA) and 721033newrev (GCCACCATGGAGTCTACACAG) and cloned into the pLenti 6.3 V5 topo vector (Life Technologies) by TA cloning to generate pLenti 6.3 NSP3. Vesicular stomatitis virus (VSV)-pseudotyped lentivirus particles were obtained by cotransfecting 293FT cells with pLenti 6.3 NSP3 and pLP1, pLP2, and pLP/VSVG (Virapower; Life Technologies). Cell culture medium then was used to infect MA104 cells, and cells resistant to blasticidin (1 mg/ml) were selected and cloned. A cell line expressing NSP3 (as detected by indirect immunofluorescence) was subcloned by limited dilution and amplified.
The MA104 cell line constitutively expressing renilla luciferase was obtained by lipofection of the plasmid pcDNA3.1purodsRluc-4xJCV (24), which encodes a destabilized renilla luciferase under the control of the cytomegalovirus (CMV) promoter, into MA104 cells and selection with 10 μg/ml of puromycin for 6 days. Cell clones were obtained by limited dilution and amplified, and a clone expressing a moderate level of renilla was selected.
RNA quantification by reverse transcription-quantitative PCR (RT-qPCR).
Primers for qPCR were selected using OLIGO 7 Primer analysis software (25). The following primers were used: GGAATTATAATGCTTATCTACGTGC and CAGTATTAGGAAACTTCTTGGC for renilla reporters and CAGCCTCAAGATCATCAGCA and TGTGGTCATGAGTCCTTCCA for glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
The PCR efficiency of each primer pair was established by measuring serial dilutions of cDNA from transfected MA104 cells in triplicate. Only threshold cycle (CT) values of <40 were used to calculate the PCR efficiency from the given slope according to the equation PCR efficiency = 10(−1/slope) − 1 × 100. All PCRs displayed an efficiency between 98% and 100%.
RNA was extracted from cells using the NucleoSpin RNA II (Macherey-Nagel) protocol and precipitated using 5 μg of acrylamide (Ambion) as a carrier. RNA was quantified using a NanoDrop spectrophotometer, and 100 ng of total RNA was reverse transcribed by random priming using pdN6 oligonucleotides (10 μM) and Superscript reverse transcriptase (Life Technologies).
qPCR was performed on 5 μl of a 1:5 dilution of reverse-transcribed RNA (1:15 for GAPDH) with MESA green qPCR MasterMix plus (Eurogentec) using an MxPro3000 (Stratagene) apparatus. Primer efficiency was >98%. The results were analyzed according to the 2−ΔΔCT method using GAPDH mRNA as an endogenous reference (26). A calibrator (MA104 cells transfected with renilla and firefly luciferase mRNA) was introduced on each plaque to compare results from plaque to plaque. Three experiments with different in vitro-synthesized RNA reporter preparations were performed in triplicate.
Luciferase activity.
Cells were assayed using the Dual-Glo luciferase assay system (Promega).
Immunoprecipitation and Western blotting.
Immunoprecipitations from HeLa cells were performed as previously described (27). For Western blotting, proteins were separated by SDS-PAGE, transferred to low-fluorescence polyvinylidene difluoride (PVDF) membranes, and probed with a rabbit polyclonal antibody against NSP3 (4-150) (28), anti-GAPDH monoclonal antibody (ab8245; Abcam), rabbit anti-eIF4GI (kindly provided by N. Sonenberg), mouse anti-PABP (10E10; Cell Signaling Technology), mouse anti-eIF4E (9676; Santa Cruz), or rabbit anti-GFP (6556; Abcam). Anti-rabbit and anti-mouse secondary antibodies coupled to Dyelight 800 (Perbio) and IRdye 700 (LI-COR) or coupled to horseradish peroxidase (Pierce) were used. Western blots were visualized either using an OdysseyFC imager and quantified using Image Studio software (LI-COR) or using SuperSignal West Pico chemiluminescent substrate (Pierce) and exposed to Hyper films (GE Healthcare).
Generation and selection of RF-RRV monoreassortants.
Reassortants between the RF and RRV rotaviruses were obtained by coinfecting MA104 cells at an MOI of 5 for each virus. Progeny viruses then were plaque purified and amplified, and their genomic double-stranded RNA (dsRNA) was extracted with TRIzol (Life Technologies) from 0.5 ml of cell culture medium. The dsRNA profiles were determined via polyacrylamide gel electrophoresis and silver staining (silver stain plus kit; Bio-Rad). The parental origins of segments with identical electrophoretic mobilities were further established by RT-PCR using specific primers; genuine monoreassortants were plaque purified twice and amplified.
RESULTS
Translation of R-RNA, N-RNA, and pA-RNA in mock- and rotavirus-infected MA104 cells.
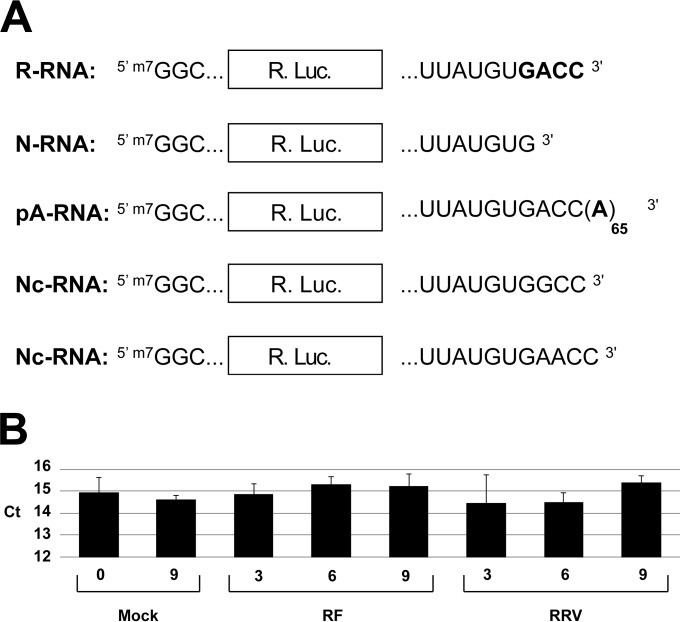
Three types of capped renilla reporters differing only in their extreme 3′-terminal sequences were synthesized in vitro (Fig. 1A). The rotavirus-like mRNA (R-RNA) contains the renilla ORF cloned between the 5′ and 3′ UTRs of rotavirus genes and ends with the canonical 3′ sequence GACC. The nonrotavirus, nonpolyadenylated mRNA (N-RNA) has an identical structure, but due to a deletion of the last three nucleotides, it ends with a UGUG sequence that is not recognized by NSP3 (12). The polyadenylated mRNA (pA-RNA) that mimics a cellular mRNA is identical to R-RNA but ends with a poly(A) sequence containing 65 adenosines.
FIG 1.
Schematic representation of reporter mRNAs and quantification of GAPDH RNA in rotavirus-infected cells. (A) The schematic structures of rotavirus-like mRNA (R-RNA), nonrota and nonpoly(A) RNA (N-RNA), polyadenylated RNA (pA-RNA), and noncanonical rotavirus-like mRNAs (Nc-RNA) are indicated with their 5′ and 3′ ends. The renilla luciferase ORF (Rluc) is boxed and the rotavirus GACC 3′ motif and the poly(A) tail appear in boldface letters. All mRNAs were 5′ capped (m7G). (B) GAPDH RNA quantification in mock- and rotavirus-infected cells. Total mRNA was purified from mock-infected or rotavirus-infected (strain RF or RRV) cells at the indicated times (in hours), and RT-qPCR was performed with GAPDH primers. Data shown represent the mean CT values (± standard errors of the means [SEM]) obtained from three independent experiments performed in triplicate. No significant differences were observed, as determined by a two-tailed Student t test.
R-RNA, N-RNA, and pA-RNA were introduced via electroporation into MA104 cells that were either mock infected or infected for 2.5 h with the RF or RRV rotavirus strain at an MOI of 10. The renilla luciferase activity (Rluc) and the remaining quantities of electroporated reporter RNA were measured in parallel in the same lysates 6 h postelectroporation (8.5 h postinfection).
To establish whether GAPDH cellular mRNA could be used as a stable reference gene over time for RT-qPCR quantification, RNA was purified from mock-, RF-, or RRV-infected cells at different times after infection. As illustrated in Fig. 1B, the GAPDH CT values were not significantly modified up to 9 h after infection with either of the two strains used. This result validates the use of GAPDH as a reference gene for RT-qPCR.
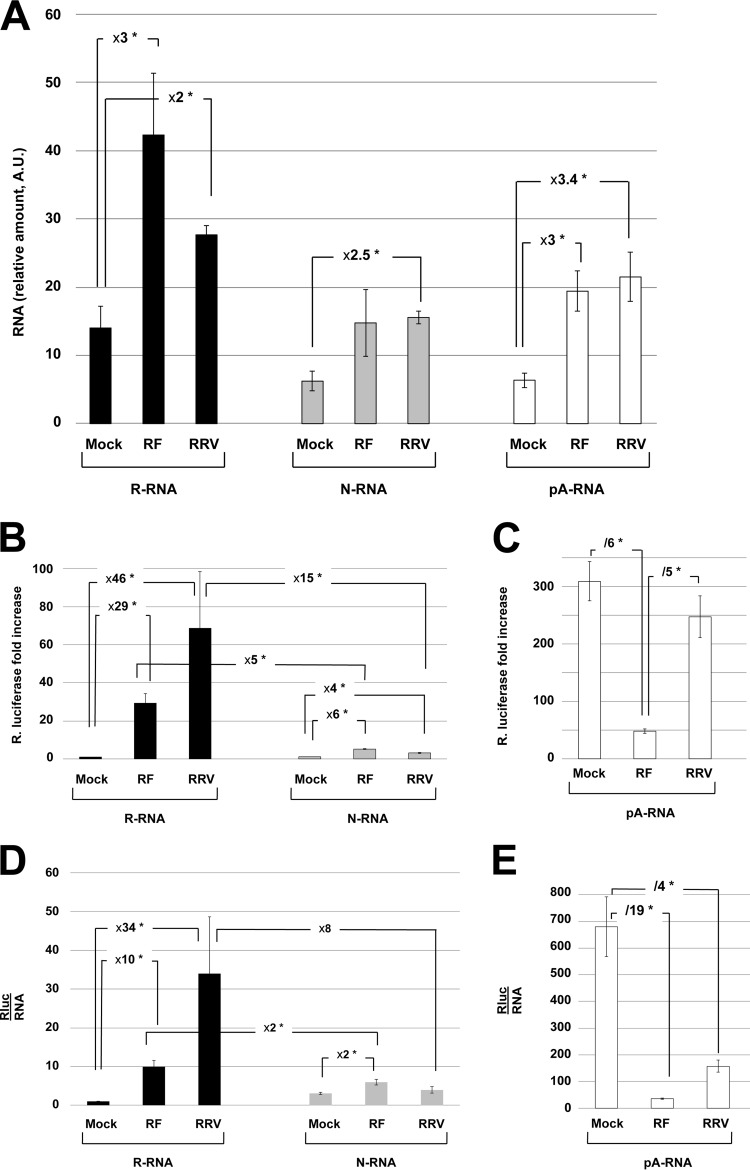
Figure 2A shows the relative amount of the reporter RNAs still present in mock- and rotavirus-infected cells as determined by RT-qPCR. Higher (2- to 3-fold) amounts of reporter RNAs were present in rotavirus-infected cells, but no significant difference was observed between the two strains used. This stabilization of steady-state RNA levels by infection was not more important for pA-RNA or N-RNA than for R-RNA, excluding a role of NSP3 in specifically protecting R-RNA from degradation.
FIG 2.
Quantification and translation of reporter mRNAs introduced into mock- and rotavirus-infected cells. Mock-infected or rotavirus-infected (strain RF or RRV) cells were electroporated with the R-, N-, and pA-RNA reporters and were recovered 6 h after electroporation (8.5 h after infection). The amount of reporter RNA was quantified by RT-qPCR (A), and Rluc activities were measured (B and C). (A) The cellular mRNA encoding GAPDH was used as an internal control, and the amount of reporter RNA relative to GAPDH RNA is presented. A.U., arbitrary units. (B and C) The Rluc activity measured in mock-infected cells electroporated with R-RNA was set to one, but no correction was performed for RNA abundance. (D and E) Rluc activities were normalized to the relative amount of reporter RNA, as measured by RT-qPCR (A), with the ratio (Rluc/RNA) obtained with mock-infected cells electroporated with R-RNA being set to one. Numbers indicate the fold increase/decrease. The means ± SEM for three independent experiments performed in triplicate are shown. Asterisks indicate significant differences (*, P < 0.05) as determined by a two-tailed Student t test.
As the abundance of an mRNA does not always correlate with its translational capability (29, 30), the relative renilla activities measured under these different conditions are presented in Fig. 2B without taking into account the amounts of RNA detected; however, to ease comparison, the Rluc activity obtained with R-RNA electroporated into mock-infected cells was set to 1. In mock-infected cells, the pA-RNA was the only mRNA that was efficiently translated, the R- and N-RNAs being translated with several hundredfold lower efficiency (compare Fig. 2B and C). R-RNA was translated efficiently when cells were infected with either RF or RRV (29-fold and 46-fold stimulation for RF and RRV, respectively, compared to that of mock-infected cells) (Fig. 2B). However, R-RNA was translated 5- to 10-fold less efficiently in rotavirus-infected cells than pA-RNA in mock-infected cells, showing that a GACC sequence at the 3′ end of an mRNA is not as efficient as a poly(A) tail in stimulating translation. A significant increase (4 to 6 times) in the translation of N-RNA upon infection with the RF or RRV strain also was detected. Infection with the RF or RRV rotavirus strain differentially affected pA-RNA translation; a significant (5-fold) decrease was observed when cells were infected with the RF strain, whereas RRV infection did not significantly reduce pA-RNA translation relative to that of mock-infected cells (Fig. 2C).
However, as the electroporated RNAs were 2- to 3-fold more abundant in infected cells (Fig. 2A), when luciferase activities were normalized to RNA abundance, R-RNA translation stimulation by infection still was substantial (10- and 34-fold for RF and RRV, respectively) (Fig. 2D), but pA-RNA translation was strongly diminished in both RF-infected (19-fold) and RRV-infected (4-fold) cells (Fig. 2E). Furthermore, the translation stimulation of R-RNA was greatly reduced compared to that of N-RNA (2- and 8-fold for RF and RRV, respectively; note that the large variations observed with RRV render this difference not statistically significant).
These experiments showed that rotavirus infection did not preferentially stabilize R-RNA and that the extents of rotavirus mRNA translation enhancement and poly(A) mRNA translation inhibition both depended on the rotavirus strain used for infection.
Translation of R-, pA-, and N-mRNAs in MA104 cells expressing NSP3.
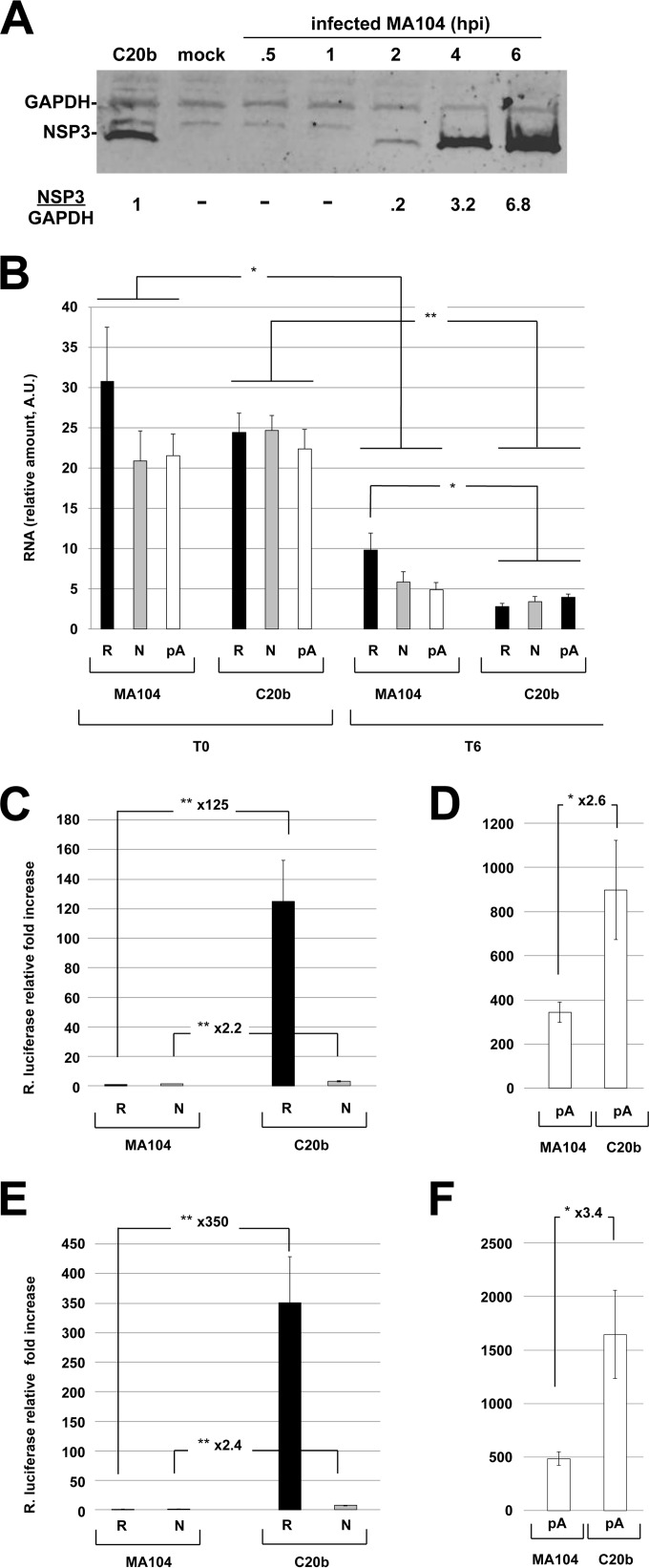
The experiments described above indicated that rotavirus infection increased R-RNA translation but decreased pA-RNA translation, two opposing functions that are attributed to rotavirus NSP3 during infection (15). To determine if these observations were reproducible in cells expressing only NSP3, cells constitutively expressing NSP3-RF were generated. MA104 cells, which are readily infected with rotavirus and can more accurately reflect what occurs in infected cells, were chosen to express RF-NSP3 because this strain strongly inhibits poly(A) mRNA translation (Fig. 2; also see Fig. 4). The established cell line C20b was selected because it expresses a low level of NSP3-RF (less than that of MA104 cells infected with the RF strain at an MOI of 10 for 3 h, as estimated by Western blotting) (Fig. 3A). C20b and parent MA104 cells were transfected with the reporter mRNAs, and RNAs were purified from the transfected cells immediately (time zero [T0]) or 6 h (T6) after electroporation. The amounts of Rluc RNA present at T6 and T0 were quantified by RT-qPCR (Fig. 3B), and Rluc activity was measured at T6 (Fig. 3C to F).
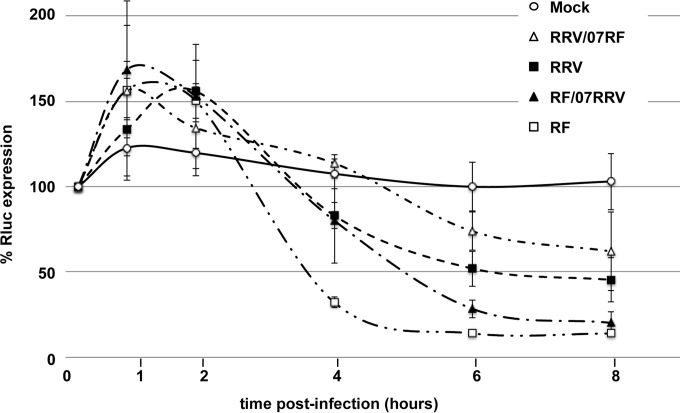
FIG 4.
Kinetics of cellular gene expression after infection with parental (RF and RRV) and monoreassortant (RRV/RF07 and RF/RRV07) strains. Cells constitutively expressing dsRenilla luciferase were mock infected or infected with the indicated parental or reassortant viruses at an MOI of 10 and recovered at the indicated times after infection. Rluc activities were measured, and the level of Rluc measured 20 min after infection was set to 100. The means ± SEM from three independent experiments are shown.
FIG 3.
Quantification and translation of reporter mRNAs in MA104 and C20b cells. (A) Expression of NSP3 in C20b cells. Cell lysates were prepared using identical numbers of C20b and MA104 cells infected with the RF rotavirus strain at an MOI of 10 for the indicated times (hpi, hours postinfection). Viral (NSP3) and cellular (GAPDH) proteins were detected by Western blotting and quantitated using a LI-COR Odyssey Fc fluoroimager. The amounts of NSP3 relative to GAPDH (NSP3/GAPDH) are indicated at the bottom of the figure. The ratio obtained for C20b cells was set to one. (B to F) MA104 or C20b cells were electroporated with the R-, N-, and pA-RNA reporters, total RNA was recovered immediately (T0) or 6 h (T6) after electroporation, and the amount of electroporated reporter RNA was quantified by RT-qPCR. (B) Rluc activities were measured at T6. The amount of reporter RNA relative to GAPDH RNA is presented. Rluc activities were normalized to the relative amounts of reporter RNAs present at T0 (C and D) or at T6 (E and F). The ratio (Rluc/RNA) obtained with mock-infected cells electroporated with R-RNA was set to one. The means ± SEM from three independent experiments performed in triplicate are shown. Numbers indicate the increase/decrease rate between points. Asterisks indicate significant differences (*, P < 0.05; **, P < 0.01) as determined by a two-tailed Student t test.
The amounts of transfected RNA immediately after electroporation (T0) were not significantly different in MA104 and C20b, showing that both cell types were electroporated with similar efficiencies with the three types of mRNAs (Fig. 3B). Six hours after electroporation, all of the reporter mRNAs were diminished in both cell types. Importantly, R-RNA was no more abundant 6 h after electroporation in C20b cells than in MA104 cells (in fact, it was even less abundant), contrary to what might have been expected if NSP3 protected rotavirus mRNAs from degradation.
Figure 3C and D illustrate the luciferase activities measured under each condition; these activities were normalized to the amounts of reporter mRNA present at T0 to take into account the slight, although statistically insignificant, differences in the amount of transfected RNAs (Fig. 3B). The luciferase activity driven by each reporter was enhanced in C20b cells (compared to the parental MA104 cells), albeit to different degrees. The translation of R-RNA was stimulated by 125-fold, whereas the translations of pA-RNA and N-RNA were stimulated by only 2.2- and 2.6-fold, respectively. Thus, if we consider that the C20b cells translated mRNAs twice as well as MA104 cells, then R-RNA still is translated >60-fold better in cells expressing NSP3 than in cells that lack NSP3 expression. This stimulation is even more pronounced than the stimulation of translation of the same R-RNA in RF- or RRV-infected cells versus mock-infected cells (60-fold versus 29- or 46-fold) (Fig. 2C) despite the much lower NSP3 expression in C20b cells (Fig. 3A).
Importantly, the higher (2.2- or 2.6-fold) translation activity of the N- or pA-type RNAs in C20b cells was not related to a higher steady-state RNA level in these cells (Fig. 3B) and more likely reflected an overall increase in translation. Because the amounts of reporter RNAs remaining in MA104 and C20b cells were quite similar, using the amount of RNA measured 6 h after electroporation to normalize reporter expression (Fig. 3E and F) did not modify the conclusions described above.
These experiments showed that even a small amount of NSP3 is sufficient to strongly stimulate the translation of mRNAs ending with GACC. They also reveal that the strong stimulation of R-RNA translation by NSP3 cannot be attributed to NSP3-dependent RNA protection. Furthermore, it appeared that NSP3 per se does not inhibit but rather induces the translation of both poly(A) and nonpoly(A) mRNAs, suggesting that NSP3 alone can substitute for the PABP-poly(A) complex in stimulating translation initiation.
Differences in cellular poly(A) mRNA expression between RF and RRV are not genetically linked to NSP3.
The results described above showed that the extent of pA-RNA translation inhibition differs between the RRV and RF rotavirus strains and that R-RNA translation stimulation but not pA-RNA translation inhibition can be obtained by expressing NSP3 alone.
To establish whether the RF and RRV rotavirus strains also differed in their capacity to inhibit cellular poly(A) mRNA expression, an MA104 cell line constitutively expressing a destabilized renilla luciferase (24) was established. The use of a destabilized luciferase increases the sensitivity and dynamic range of the luciferase reporter assay (31). Luciferase activities then were measured at several time points after cells were infected with the RF or RRV virus. The results are reported in Fig. 4. It can be noted that a slight increase in luciferase activity was observed 1 to 2 h after infection, probably due to a transient inhibition of the proteasome consequential to the activation of the unfolded protein response triggered by rotavirus infection (19, 32). RF required 3.5 h and RRV required 6 h to inhibit 50% of the cellular protein synthesis, and whereas RF reduced expression to 10% of the initial value after 8 h of infection, RRV reduced expression to just below 50% (Fig. 4) during the same period. To determine if these slow or fast phenotypes were linked to the genetic origin of NSP3, reassortants were generated between the RRV and RF viruses. Two monoreassortants were selected and plaque purified twice. The first monoreassortant (RF/07RRV) bears the RRV NSP3 gene (gene 7) but retains the remaining 10 RF genes. Conversely, the second selected monoreassortant (RRV/07RF) bears RF gene 7 but retains the other 10 RRV genes. Interestingly, the different kinetics of protein synthesis inhibition imposed by the two monoreassortants (Fig. 4) were independent of NSP3 origin, as the RF/07RRV reassortant displayed a fast phenotype quite similar to that of RF, whereas the RRV/07RF reassortant displayed a slower phenotype than that of parental RRV. This experiment indicated that the different capacities of the RF and RRV viruses to inhibit cellular poly(A) mRNA translation are not linked only to NSP3.
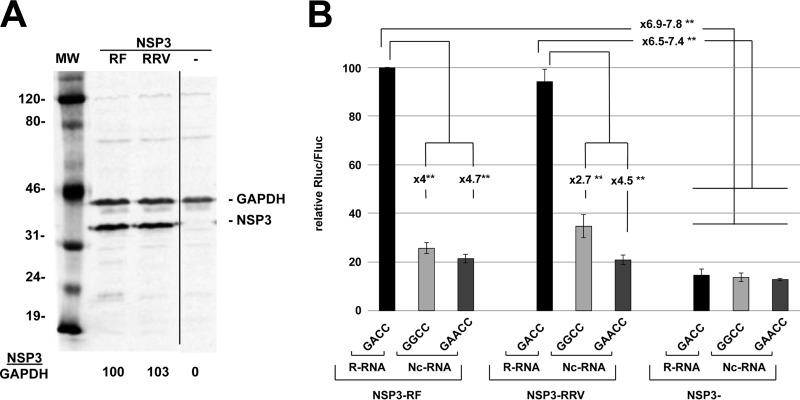
RRV and RF NSP3 stimulate R-RNA translation similarly.
Transfection of R-RNA into cells infected with RF or RRV showed that R-RNA translation enhancement was higher with the former virus (Fig. 2C and E). Therefore, we tested whether such a difference could be attributed to strain-specific NSP3s. To this end, NSP3 from the RF (NSP3-RF) or the RRV (NSP3-RRV) strain was expressed at similar levels in BSRT7 cells (Fig. 5). These cells then were electroporated with three different types of rotavirus-like reporter mRNAs: R-RNA ending with the canonical GACC sequences or Nc-RNAs ending with either a GAACC or a GGCC sequence. The GAACC sequence corresponds to the noncanonical 3′ end found on gene 5 of the SA11 rotavirus strain, with GGCC being found at the 3′ end of genes 5 and 7 of the RRV and SA11 strains, respectively (33). The luciferase activities measured are reported in Fig. 5 and show that NSP3 of either origin stimulated R-RNA translation equally well (7- to 8-fold) compared with translation in the absence of NSP3. Thus, the different translation stimulation of R-RNA observed with the RRV and RF strains (Fig. 2C and E) was not due to the different efficiencies of their cognate NSP3s. Note that a slight (2×) but significant enhancement of Nc-RNA GGCC translation was observed in cells expressing NSP3-RRV versus cells lacking NSP3 (but not versus cells expressing NSP3-RF), suggesting that NSP3-RRV exhibits a recognition specificity for the 3′ end of RNA that differs slightly from that of NSP3-RF. Similar results have been obtained using NSP3 from the SA11 strain (Gratia et al., unpublished).
FIG 5.
Expression of R- and Nc-RNAs in cells expressing NSP3 of RF or RRV origin. (A) NSP3-RF and NSP3-RRV expression in BSRT7 cells. Lysates from BSRT7 cells transfected for 24 h with expression vectors encoding NSP3 from the RF (NSP3-RF) or RRV (NSP3-RRV) strains or killed NSP3-RF (NSP3−) were analyzed by Western blotting with an anti-NSP3 rabbit polyclonal antibody and a mouse monoclonal antibody against the cellular protein GAPDH (used as a loading control). The ratio of NSP3 to GAPDH fluorescence (NSP3/GAPDH) is indicated at the bottom of the figure. MW, molecular weight marker (in thousands). The three lanes are from the same membrane and experiment. (B) Cells expressing RF or RRV NSP3 or killed NSP3 for 24 h were electroporated with R-RNA or Nc-RNAs ending with a GGCC or GAACC and with a standard EMCV-Fluc RNA. Fluc and Rluc activities were measured 6 h after electroporation. The means ± SEM of the renilla-to-firefly luciferase ratio (Rluc/Fluc) from three independent experiments are presented. The Rluc/Fluc ratio obtained with R-RNA electroporated into cells expressing NSP3-RF was set to 100. Numbers indicate the increase/decrease rate between points. Asterisks indicate significant differences (*, P < 0.05; **, P < 0.01) as determined by two-tailed Student t tests.
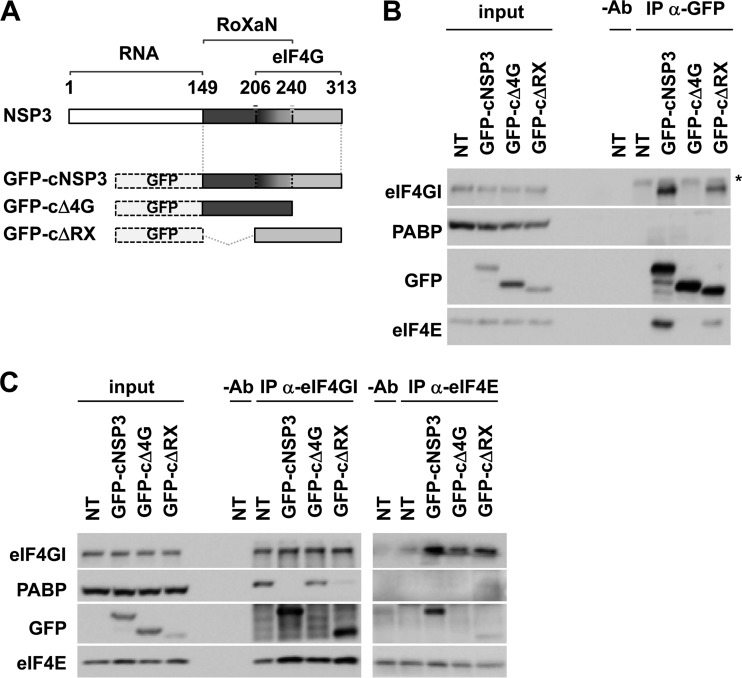
NSP3 binding to eIF4G stabilizes the eIF4E-eIF4G interaction.
We next wished to gain insights into the molecular mechanism underlying the stimulation of poly(A) and nonpoly(A) mRNA translation by NSP3. Because both poly(A) and nonpoly(A) mRNAs are devoid of the 3′ GACC sequence required for specifically binding to the N-terminal RNA-binding domain of NSP3, we anticipated that the RNA-binding domain of NSP3 is not involved. Given that both poly(A) and nonpoly(A) mRNAs are capped, we instead tested the possibility that NSP3 equally stimulated their translation through its interaction with eIF4G and through subsequent changes in eIF4G affinity for the cap-binding protein eIF4E. To this end, three deletion fragments of RF-NSP3 that lack the RNA-binding domain were created and fused to an eGFP tag (Fig. 6A) as follows: one C-terminal fragment (cNSP3) containing both the RoXaN- and eIF4G-binding domains, one shorter fragment lacking the eIF4G-binding domain (cΔ4G), and another shorter fragment lacking the RoXaN-binding domain (cΔRX). The ability of these different fragments to interact with eIF4G first was verified in transiently transfected HeLa cells. As expected, coimmunoprecipitation (co-IP) experiments using anti-GFP antibodies confirmed that NSP3 fragments containing the eIF4G-binding domain (cNSP3 and cΔRX) indeed interacted with eIF4G, whereas the NSP3 fragment devoid of the eIF4G-binding domain (cΔ4G) did not (Fig. 6B). Co-IP experiments performed using anti-eIF4GI antibodies revealed that whereas PABP was evicted from eIF4GI (as expected), interactions between the cNSP3 or cΔRX fragment and eIF4GI were associated with a slight but reproducible increase in the amount of coimmunoprecipitated eIF4E compared to the levels for nontransfected cells (Fig. 6C, left). Similarly, a slight but reproducible increase in the amount of eIF4GI that was coimmunoprecipitated using anti-eIF4E antibodies was observed with cells expressing either of the two NSP3 fragments containing the eIF4G-binding domain (Fig. 6C, right). In contrast, changes in the eIF4E-eIF4GI interaction were not significant when anti-eIF4GI or anti-eIF4E co-IPs were performed with cells expressing the cΔ4G NSP3 fragment devoid of the eIF4G-binding site (Fig. 6B, left and right).
FIG 6.
NSP3 binding to eIF4G stabilizes the eIF4E-eIF4G interaction. (A) Schematic representation of NSP3 fragments. Different fragments of NSP3 (RF strain) containing the RoXaN- and eIF4G-binding sites (cNSP3) or devoid of either the eIF4G- or RoXaN-binding domain (cΔ4G or cΔRX) were fused to GFP. RNA refers to the RNA-binding domain of NSP3, and the numbers refer to the NSP3 amino acid sequence. (B) HeLa cells were transiently transfected with the different GFP-tagged NSP3 fragments, and their relative expression and interaction with eIF4GI (direct interaction), eIF4E (indirect interaction through eIF4G), and PABP (no interaction) were visualized by Western blotting using the indicated antibodies either directly (input) or after immunoprecipitation with anti-GFP antibodies (IP anti-GFP). NT, nontransfected; −Ab, IP without anti-GFP antibodies; *, nonspecific signal. The results are representative of three separate experiments. (C) eIF4E-eIF4GI interaction in HeLa cells transiently transfected with the different GFP-tagged NSP3 fragments was visualized by Western blotting after IP using either anti-eIF4GI (left) or anti-eIF4E (right) antibodies. Coimmunoprecipitated PABP also was visualized. Input, direct Western blotting; NT, nontransfected; −Ab, IP without any antibodies. The results are representative of three separate experiments.
These data indicate that despite the eviction of PABP from eIF4G (and the consequent disruption of the mRNA closed-loop conformation), the interaction of NSP3 with eIF4G somehow enhances the affinity of eIF4G for eIF4E. This effect required neither the RNA-binding domain nor the RoXaN-interacting site of NSP3. This result may at least partially explain how NSP3 stimulates the translation of both poly(A) and nonpoly(A) mRNAs independently of its binding to any mRNA 3′ end.
DISCUSSION
In this work, we have shown that, in living cells, the rotavirus protein NSP3, apart from other viral proteins, is able to enhance the translation of a rotavirus-like mRNA but has no effect on RNA stability. These results are in agreement with results we previously obtained using in vivo and in vitro translation assays (15), as well as with those of Chizhikov and Patton, who used infected cells to show an enhancement of translation when the reporter mRNA ends with a GACC sequence (14).
The role of NSP3 in stimulating rotavirus mRNA translation had been questioned (18). Through the siRNA-mediated downregulation of NSP3 in infected cells, Montero et al. detected via pulse labeling only a transient decrease in de novo viral protein synthesis early after infection. In these experiments, however, NSP3 downregulation was only partial, and it is likely that the remaining NSP3 was sufficient to ensure viral protein synthesis, albeit at a lower rate. Indeed, our data support this idea, as we show that even a very small amount of NSP3 is capable of enhancing rotavirus-like mRNA translation a hundredfold. Thus, NSP3 likely is most important, although at low levels, at the onset of infection, when competition with cellular mRNAs for access to the cell translation machinery is fiercest.
We also showed that the translation capability of R-RNA differs slightly depending on the strain used (RF or RRV), a difference that cannot be attributed to the two different strain-specific NSP3s. This result indicates that other virus-dependent mechanisms are required to modify R-RNA translation efficiency in rotavirus-infected cells. The competition between viral mRNAs and R-RNA for NSP3 likely is one of these mechanisms.
The interaction of NSP3 with eIF4G, which results in the eviction of PABP from translation initiation complexes and its subsequent nuclear relocalization, has been hypothesized to cause the inhibition of host poly(A) mRNA translation following rotavirus infection (9, 13, 15). The reduction of cellular poly(A) mRNA translation induced by the addition of recombinant truncated NSP3 to a rabbit reticulocyte lysate in vitro translation (RRL) assay (6) and the reduction of vaccinia virus gene expression with recombinant vaccinia virus expressing full-length or truncated NSP3 (17) substantiated this hypothesis. Here, we observed a reduction in the translation of pA-RNA introduced into rotavirus-infected cells via electroporation, with the level of translation from one viral strain to another being noticeable in RRV-infected cells only after normalizing to the amount of pA-RNA remaining in the cell. Similar reductions in translation were not observed by Rubio et al. (19) using the RRV rotavirus strain and RNA lipofection even after normalizing to the amount of RNA; it is possible that RNA lipofection distorted RNA quantification, as lipofection has been shown to interfere with RNA half-life, precluding its use in monitoring the expression of reporter RNA (34). The same difference in the capacities of the RF and RRV strains to abate cellular gene expression also was observed in cells constitutively expressing a reporter gene (i.e., expressed as a polyadenylated mRNA synthesized in the nucleus and translated in the cytoplasm); the extent of reduction of cellular mRNA expression was higher and quicker in RF- than in RRV-infected cells. However, the use of monoreassortant strains showed that this variation was not entirely linked to the NSP3 genetic background. Cellular gene expression involves many steps that lie upstream of translation, including transcription, splicing, and nuclear export. Our results suggest that the RF and RRV strains differentially impact one or more of these steps and that the inhibition of translation by NSP3 is not the sole factor involved in shutting down cellular protein synthesis. Another explanation is that the numerous viral mRNA copies present during infection (19, 35) simply titrate away translation initiation factors and render them inaccessible to host poly(A) mRNAs. Thus, the inhibition of cellular polyadenylated mRNA translation during rotavirus infection would be more likely to result from the high level of viral mRNA translation facilitated by NSP3 rather than by the direct exclusion of cellular mRNAs from translation initiation complexes.
Whereas pA-RNA translation was reduced in RF-infected cells, it unexpectedly increased 2.6-fold in cells expressing only NSP3; similarly, a 2.2-fold increase was observed with N-RNA. Thus, in the absence of viral mRNA, NSP3 seems to nonspecifically increase RNA translation. One possibility that could explain such an observation is that the eviction of PABP from eIF4G by NSP3 abolishes the competition between polyadenylated and nonpolyadenylated RNA and renders all RNAs equally competent for translation regardless of their 3′ ends. A similar phenomenon was observed when short poly(A) RNAs were added to an in vitro translation assay using ribosome-depleted RRL (36) or when nonpolyadenylated mRNA degradation was abolished in yeast (37). However, our data showing that NSP3 promotes the eIF4E-eIF4G interaction provides a more direct explanation for this unexpected observation and actually suggests that through its interaction with eIF4G, NSP3, independently of its binding to any 3′ end, can substitute for the PABP-poly(A) complex in facilitating the initiation of capped mRNA translation. Indeed, the binding of PABP to eIF4G has been shown to similarly enhance eIF4E affinity for the cap structure (38, 39), itself further increased by binding of eIF4E to eIF4G (40). However, it remains to be determined whether stabilization of eIF4E-eIF4G interaction in NSP3-expressing cells is directly due to NSP3 binding to eIF4G or whether a more indirect effect, such as liberation of eIF4E from its sequestering proteins (i.e., the eIF4E-binding proteins 1 and 2), is involved.
The results described here, in addition to the study using NSP3 that lacks RNA or eIF4G binding (Gratia et al., unpublished), are totally consistent with the notion that NSP3 behaves as a PABP surrogate for rotavirus mRNA translation through its simultaneous interaction with eIF4G and the viral mRNA GACC 3′ end. However, our results indicate that NSP3 per se cannot explain the inhibition of host poly(A) mRNA translation observed in rotavirus-infected cells. Instead, NSP3 separated from other viral proteins also appears to substitute for PABP in cellular mRNA translation, given that this protein slightly enhances the translation of both poly(A) and nonpoly(A) mRNAs, likely through its interaction with eIF4G and the resulting stabilization of the eIF4E-eIF4G complex.
ACKNOWLEDGMENTS
This work was supported by a grant from Agence Nationale de la Recherche ANR-09-MIEN-04 “Trans-Inf-Rot” to D.P. and S.P. M.G. was supported by a Ph.D. thesis fellowship from Ministère de l'Education Nationale, de l'Enseignement Supérieur et de la Recherche. E.S. was supported by a postdoctoral grant from ANR. P.V., A.C., and D.P. are staff members from the Institut National de la Recherche Agronomique (INRA). M.D. is a staff member from University of Evry Val d'Essones. S.P. is staff member from INSERM.
We thank Cécile Laroche for technical assistance.
Plasmid pcDNA3.1purodsRluc-4xJCV was kindly provided by C. S. Sullivan, The University of Texas at Austin. eIF4G1 antiserum was kindly provided by N. Sonenberg, McGill University (Canada).
REFERENCES
- 1.Hinnebusch AG. 2014. The scanning mechanism of eukaryotic translation initiation. Annu Rev Biochem 83:779–812. doi: 10.1146/annurev-biochem-060713-035802. [DOI] [PubMed] [Google Scholar]
- 2.Jackson RJ, Hellen CU, Pestova TV. 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sonenberg N, Hinnebusch AG. 2009. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wells SE, Hillner PE, Vale RD, Sachs AB. 1998. Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell 2:135–140. doi: 10.1016/S1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- 5.Kuhn U, Wahle E. 2004. Structure and function of poly(A) binding proteins. Biochim Biophys Acta 1678:67–84. doi: 10.1016/j.bbaexp.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 6.Michel YM, Poncet D, Piron M, Kean KM, Borman AM. 2000. Cap-Poly(A) synergy in mammalian cell-free extracts. Investigation of the requirements for poly(A)-mediated stimulation of translation initiation. J Biol Chem 275:32268–32276. doi: 10.1074/jbc.M004304200. [DOI] [PubMed] [Google Scholar]
- 7.Gallie DR. 1998. A tale of two termini: a functional interaction between the termini of an mRNA is a prerequisite for efficient translation initiation. Gene 216:1–11. doi: 10.1016/S0378-1119(98)00318-7. [DOI] [PubMed] [Google Scholar]
- 8.Preiss T, Hentze MW. 1998. Dual function of the messenger RNA cap structure in poly(A)-tail-promoted translation in yeast. Nature 392:516–520. doi: 10.1038/33192. [DOI] [PubMed] [Google Scholar]
- 9.Piron M, Vende P, Cohen J, Poncet D. 1998. Rotavirus RNA-binding protein NSP3 interacts with eIF4GI and evicts the poly(A) binding protein from eIF4F. EMBO J 17:5811–5821. doi: 10.1093/emboj/17.19.5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poncet D, Aponte C, Cohen J. 1993. Rotavirus protein NSP3 (NS34) is bound to the 3′ end consensus sequence of viral mRNAs in infected cells. J Virol 67:3159–3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deo RC, Groft CM, Rajashankar KR, Burley SK. 2002. Recognition of the rotavirus mRNA 3′ consensus by an asymmetric NSP3 homodimer. Cell 108:71–81. doi: 10.1016/S0092-8674(01)00632-8. [DOI] [PubMed] [Google Scholar]
- 12.Poncet D, Laurent S, Cohen J. 1994. Four nucleotides are the minimal requirement for RNA recognition by rotavirus non-structural protein NSP3. EMBO J 13:4165–4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Groft CM, Burley SK. 2002. Recognition of eIF4G by rotavirus NSP3 reveals a basis for mRNA circularization. Mol Cell 9:1273–1283. doi: 10.1016/S1097-2765(02)00555-5. [DOI] [PubMed] [Google Scholar]
- 14.Chizhikov V, Patton JT. 2000. A four-nucleotide translation enhancer in the 3′-terminal consensus sequence of the nonpolyadenylated mRNAs of rotavirus. RNA 6:814–825. doi: 10.1017/S1355838200992264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vende P, Piron M, Castagne N, Poncet D. 2000. Efficient translation of rotavirus mRNA requires simultaneous interaction of NSP3 with the eukaryotic translation initiation factor eIF4G and the mRNA 3′ end. J Virol 74:7064–7071. doi: 10.1128/JVI.74.15.7064-7071.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harb M, Becker MM, Vitour D, Baron CH, Vende P, Brown SC, Bolte S, Arold ST, Poncet D. 2008. Nuclear localization of cytoplasmic poly(A)-binding protein upon rotavirus infection involves the interaction of NSP3 with eIF4G and RoXaN. J Virol 82:11283–11293. doi: 10.1128/JVI.00872-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Padilla-Noriega L, Paniagua O, Guzman-Leon S. 2002. Rotavirus protein NSP3 shuts off host cell protein synthesis. Virology 298:1–7. doi: 10.1006/viro.2002.1477. [DOI] [PubMed] [Google Scholar]
- 18.Montero H, Arias CF, Lopez S. 2006. Rotavirus nonstructural protein NSP3 is not required for viral protein synthesis. J Virol 80:9031–9038. doi: 10.1128/JVI.00437-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rubio RM, Mora SI, Romero P, Arias CF, Lopez S. 2013. Rotavirus prevents the expression of host responses by blocking the nucleocytoplasmic transport of polyadenylated mRNAs. J Virol 87:6336–6345. doi: 10.1128/JVI.00361-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buchholz UJ, Finke S, Conzelmann KK. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J Virol 73:251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poncet D, Cohen J. 1989. A plaque hybridization assay for rotaviruses. J Virol Methods 26:27–33. doi: 10.1016/0166-0934(89)90071-2. [DOI] [PubMed] [Google Scholar]
- 22.Weiner MP, Costa GL, Schoettlin W, Cline J, Mathur E, Bauer JC. 1994. Site-directed mutagenesis of double-stranded DNA by the polymerase chain reaction. Gene 151:119–123. doi: 10.1016/0378-1119(94)90641-6. [DOI] [PubMed] [Google Scholar]
- 23.Rydzik AM, Lukaszewicz M, Zuberek J, Kowalska J, Darzynkiewicz ZM, Darzynkiewicz E, Jemielity J. 2009. Synthetic dinucleotide mRNA cap analogs with tetraphosphate 5′,5′ bridge containing methylenebis (phosphonate) modification. Org Biomol Chem 7:4763–4776. doi: 10.1039/b911347a. [DOI] [PubMed] [Google Scholar]
- 24.Seo GJ, Kincaid RP, Phanaksri T, Burke JM, Pare JM, Cox JE, Hsiang TY, Krug RM, Sullivan CS. 2013. Reciprocal inhibition between intracellular antiviral signaling and the RNAi machinery in mammalian cells. Cell Host Microbe 14:435–445. doi: 10.1016/j.chom.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rychlik W. 2007. OLIGO 7 primer analysis software. Methods Mol Biol 402:35–60. [DOI] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Martineau Y, Wang X, Alain T, Petroulakis E, Shahbazian D, Fabre B, Bousquet-Dubouch MP, Monsarrat B, Pyronnet S, Sonenberg N. 2014. Control of Paip1-eukayrotic translation initiation factor 3 interaction by amino acids through S6 kinase. Mol Cell Biol 34:1046–1053. doi: 10.1128/MCB.01079-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vitour D, Lindenbaum P, Vende P, Becker MM, Poncet D. 2004. RoXaN, a novel cellular protein containing TPR, LD, and zinc finger motifs, forms a ternary complex with eukaryotic initiation factor 4G and rotavirus NSP3. J Virol 78:3851–3862. doi: 10.1128/JVI.78.8.3851-3862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. 2011. Global quantification of mammalian gene expression control. Nature 473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 30.Stevens SG, Brown CM. 2013. In silico estimation of translation efficiency in human cell lines: potential evidence for widespread translational control. PLoS One 8:e57625. doi: 10.1371/journal.pone.0057625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fan F, Wood KV. 2007. Bioluminescent assays for high-throughput screening. Assay Drug Dev Technol 5:127–136. doi: 10.1089/adt.2006.053. [DOI] [PubMed] [Google Scholar]
- 32.Shenkman M, Tolchinsky S, Kondratyev M, Lederkremer GZ. 2007. Transient arrest in proteasomal degradation during inhibition of translation in the unfolded protein response. Biochem J 404:509–516. doi: 10.1042/BJ20061854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kearney K, Chen D, Taraporewala ZF, Vende P, Hoshino Y, Tortorici MA, Barro M, Patton JT. 2004. Cell-line-induced mutation of the rotavirus genome alters expression of an IRF3-interacting protein. EMBO J 23:4072–4081. doi: 10.1038/sj.emboj.7600408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barreau C, Dutertre S, Paillard L, Osborne HB. 2006. Liposome-mediated RNA transfection should be used with caution. RNA 12:1790–1793. doi: 10.1261/rna.191706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson MA, McCrae MA. 1989. Molecular biology of rotaviruses. VIII. Quantitative analysis of regulation of gene expression during virus replication. J Virol 63:2048–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borman AM, Michel YM, Malnou CE, Kean KM. 2002. Free poly(A) stimulates capped mRNA translation in vitro through the eIF4G-poly(A)-binding protein interaction. J Biol Chem 277:36818–36824. doi: 10.1074/jbc.M205065200. [DOI] [PubMed] [Google Scholar]
- 37.Searfoss AM, Wickner RB. 2000. 3′ Poly(A) is dispensable for translation. Proc Natl Acad Sci U S A 97:9133–9137. doi: 10.1073/pnas.97.16.9133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haghighat A, Sonenberg N. 1997. eIF4G dramatically enhances the binding of eIF4E to the mRNA 5′-cap structure. J Biol Chem 272:21677–21680. doi: 10.1074/jbc.272.35.21677. [DOI] [PubMed] [Google Scholar]
- 39.Wei CC, Balasta ML, Ren J, Goss DJ. 1998. Wheat germ poly(A) binding protein enhances the binding affinity of eukaryotic initiation factor 4F and (iso)4F for cap analogues. Biochemistry 37:1910–1916. doi: 10.1021/bi9724570. [DOI] [PubMed] [Google Scholar]
- 40.von Der Haar T, Ball PD, McCarthy JE. 2000. Stabilization of eukaryotic initiation factor 4E binding to the mRNA 5′-cap by domains of eIF4G. J Biol Chem 275:30551–30555. doi: 10.1074/jbc.M004565200. [DOI] [PubMed] [Google Scholar]