Abstract
Epstein-Barr-related herpesviruses, or lymphocryptoviruses (LCV), naturally infect humans and nonhuman primates (NHP), but their host range is not well characterized. Using LCV and B cells from multiple species of Hominidae and Cercopithecidae, we show that LCV can immortalize B cells from some nonnative species but that growth transformation is restricted to B cells from their own family of hominoids or Old World NHP, suggesting a high degree of LCV adaptation to their natural primate host.
TEXT
Herpesviruses in the lymphocryptovirus (LCV) genus are found naturally only in hominoids (human, great apes, and gibbons) and Old and New World nonhuman primates (NHP), suggesting that they evolved relatively recently. LCVs infecting hominoids and Old World NHP have well-conserved viral genomes and biological properties, including the ability to immortalize B cells from the natural host in vitro (1, 2). Epstein-Barr virus (EBV), i.e., human LCV, can also immortalize B cells derived from New World NHP, leading to the assumption that LCV might have the ability to immortalize B cells from other NHP species (3, 4). However, a clear model for the host range of LCV-induced B cell immortalization has not emerged from the literature.
While EBV has been reported to immortalize B cells from chimpanzees, it was reported to be unable to immortalize cells from baboons or macaques (5–8). LCV from cynomolgus macaques (cyLCV) and rhesus macaques (rhLCV) could transform B cells from both macaque species but not human cells (8, 9). These data suggest a model of a restricted host range in which LCV can immortalize B cells from their own and closely related primate species but not cells from more distant species.
This model is challenged by reports that LCV from chimpanzees (chLCV) and baboons (baLCV) could immortalize B cells from humans, baboons, and different macaque species, suggesting no restriction of LCV host range among hominoids and Old World NHP (6, 7). However, the cell line used as the source of chLCV in those studies was obtained not by spontaneous outgrowth from chimpanzee peripheral blood mononuclear cells (PBMC) but by infecting baboon PBMC with throat swab material from chimpanzees (5, 6, 10). Virtually all domesticated Old World NHP raised in conventional groups are persistently infected with their natural LCV. Thus, the cell line derived from baboon PBMC could theoretically have been infected with either chLCV or endogenous baLCV, and techniques to distinguish the two were not available at the time at which those studies were performed. In our previous studies, we obtained immortalized human B cell lines after exposing human PBMC to baLCV, but in all cases the cell lines contained both baLCV and endogenous EBV; i.e., baLCV required EBV coinfection for human B cell immortalization (8). Those studies demonstrated the potential problem posed by persistent B cell infection with endogenous LCV and the importance of confirming the molecular signature, or identity, of the transforming virus in immortalized cells.
Unfortunately, the cell lines putatively immortalized with chLCV are no longer readily available for analyses to determine whether they produced chLCV or baLCV or both. For this report, we derived new transforming LCV isolates from a chimpanzee (m5305) and a cynomolgus macaque (m4409) and defined molecular signatures for each. This investigation focused on testing the host range models for hominoid and Old World LCV using these new LCV isolates in addition to well-characterized hominoid (EBV) and Old World NHP LCV (baLCV and rhLCV) isolates.
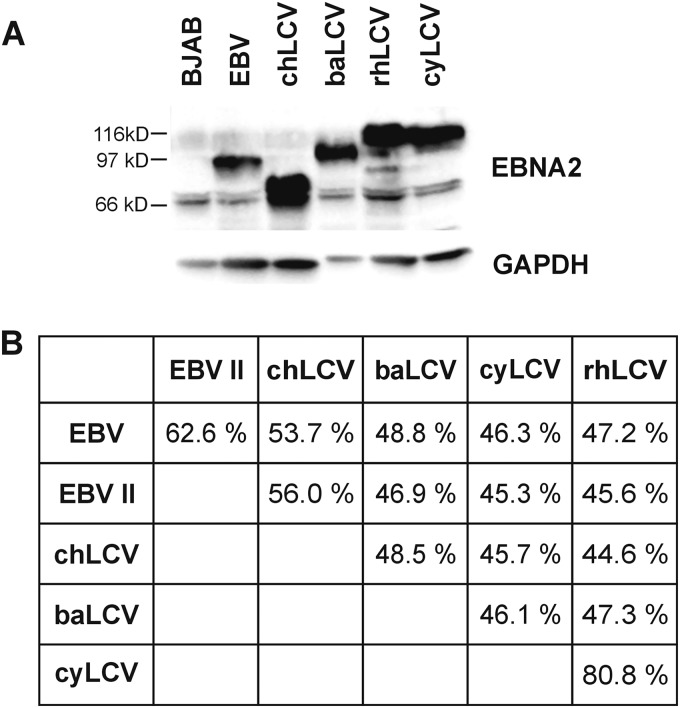
The m5305 chLCV and m4409 cyLCV isolates were derived by spontaneous outgrowth of virus-infected lymphoma cells from a chimpanzee and a cynomolgus macaque cultured in vitro. LCV infection was confirmed by detection of EBNA2 orthologues in both cell lines. Immunoblotting showed that the chLCV EBNA2 protein (Fig. 1A; lane 3) migrates with a molecular mass lower than those of its orthologues from EBV (lane 2), baLCV (lane 4), or rhLCV (lane 5), whereas cyEBNA2 (lane 6) is comparable to rhEBNA2 with respect to its molecular mass. Additional PE2-reactive bands with lower molecular mass in lysates from chLCL and rhLCL were observed occasionally and at various degrees of intensity consistent with a protein degradation product. Expression levels of full-length EBNA2 proteins detected by the PE2 monoclonal antibody in B cell lines infected with the respective LCVs were comparable.
FIG 1.
Comparison of EBNA2 proteins from hominoid and Old World lymphocryptoviruses. (A) EBNA2 protein expression in B cell lines (105 cells per lane) harboring EBV (B95-8 strain), chLCV (m5305), baLCV (S594), rhLCV (LCL8664), or cyLCV (m4409) detected by immunoblotting with PE2 monoclonal antibody. (B) EBNA2 amino acid similarity of hominoid and Old World LCV. EBV type II is represented by the AG876 strain.
The EBNA2 open reading frames (ORFs) from chLCV and cyLCV were PCR amplified and sequenced. ChEBNA2 was most closely related to type I and II EBNA2 sequences (53.7% and 56% amino acid similarity; Fig. 1B). CyEBNA2 was closely related to rhEBNA2 (80.8% amino acid similarity) and more distantly related to EBV types I and II EBNA2 (46.3% and 45.3%). Since we used a baLCV strain (S594; isolated from Papio hamadryas) different from that used for the published baEBNA2 sequence (BA65; isolated from Papio cynocephalus), we sequenced baEBNA2 of S594 and found only four nucleotide and two amino acid changes compared to BA65 baEBNA2 (Fig. 2) (6, 8, 11–13). The stronger EBNA2 similarity of chLCV with EBV and of cyLCV with rhLCV confirmed the hominoid and Old World NHP origins of m5305 chLCV and m4409 cyLCV, respectively. Furthermore, strain-specific differences in the EBNA2 sequence provide a unique molecular signature to distinguish each LCV species.
FIG 2.
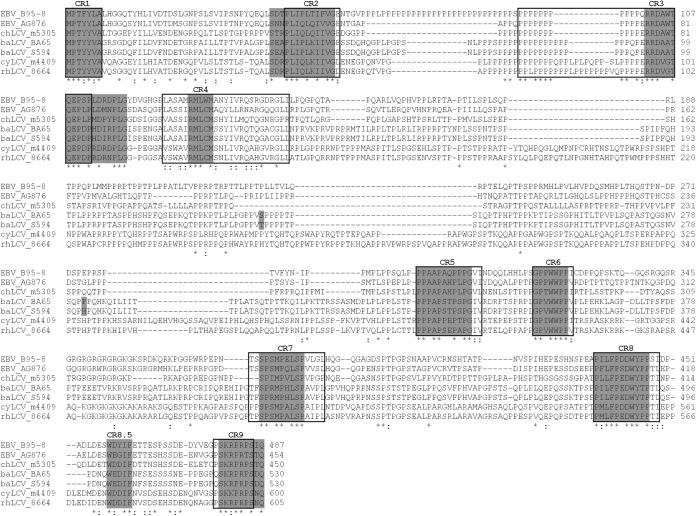
Amino acid alignment of all available hominoid and Old World LCV EBNA2 sequences. Amino acid sequences from EBV type I (B95-8) and type II (AG876), chLCV (m5305), baLCV (BA65 and S594), cyLCV (m4409), and rhLCV (LCL8664) EBNA2 were aligned using the ClustalW2 program. Conserved regions defined by Peng et al. (14) are highlighted as open boxes, and new CR borders proposed in this work are highlighted as gray boxes. The newly identified CR between CR8 and CR9 was named CR8.5 due to its location. Amino acid differences between baEBNA2 of BA65 and S594 strains are highlighted by a gray background.
The new chEBNA2, baEBNA2 (S594), and cyEBNA2 sequences also provide additional insight into the nine conserved regions (CR) in EBNA2 originally defined by Ling et al. (13) on the basis of alignment of baEBNA2 (BA65) with EBV type I and II EBNA2s and later with rhEBNA2 (13, 14). Figure 2 indicates that CR4, which started with the lowest percentage of identical residues (30%), is even less conserved, as shown by alignment with our expanded repertoire of EBNA2s (14). Among the 25 amino acids that originally formed CR4, only 5 are well conserved among all LCV. In contrast, the expanded alignment confirms and provides a better definition for the strongly conserved amino acids in the remaining CRs. The alignment also identifies one new cluster of residues (CR8.5) in between CR8 and CR9 that are highly conserved in the carboxy terminus important for EBNA2 transactivating activity.
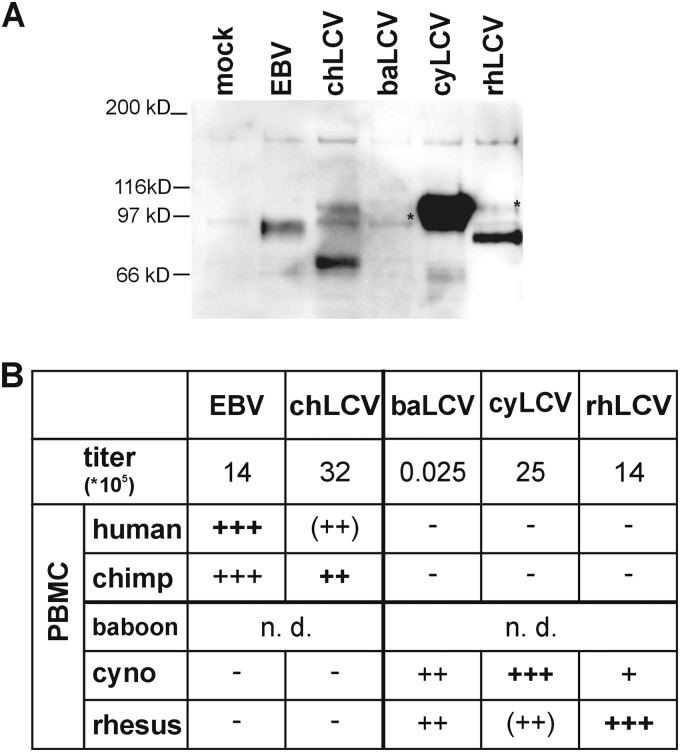
Virus stocks were produced from cell-free supernatants of cell lines infected with EBV (B95-8), chLCV (m5305), baLCV (S594), cyLCV (m4409), and rhLCV (LCL8664) and induced for lytic replication. The relative amounts of viral DNA in the supernatants determined by PCR (Fig. 3B) were comparable (14 × 105 to 32 × 105 DNA units) except for the amount of baLCV DNA, which was approximately 3 logs lower than the amounts seen with the other viral supernatants.
FIG 3.
LCV-induced immortalization of B cells from different primate host species. (A) Confirmation of infectious virus in cell-free supernatants by infection of Louckes cells and detection of EBNA2 expression. Cell-free LCV supernatants were produced from cell lines induced for lytic replication by culturing in media supplemented with 12-O-tetradecanoylphorbol-13-acetate (TPA) and butyrate. Louckes cells were infected with virus stocks and harvested 48 h after infection. EBNA2 was immunoprecipitated from cell lysates using PE2 monoclonal antibody. Immunoprecipitates were subjected to SDS-polyacrylamide gel electrophoresis followed by immunoblot detection of EBNA2 with the PE2 monoclonal antibody conjugated with horseradish peroxidase. Asterisks indicate full-length EBNA2 protein bands. (B) Virus titer and B cell immortalization. The relative amounts of LCV in each virus stock were quantitated by endpoint limiting dilution PCR analysis of a highly conserved sequence in the DNA polymerase gene. Equal volumes of virus stock containing the total amounts of virus shown were used for infection of 107 PBMC of human, chimpanzee, cynomolgus macaque (cyno), or rhesus macaque origin. Infections of baboon PBMC were not done (n.d.). Cells were cultured in RPMI media with 10% fetal calf serum and 0.5 μg/ml cyclosporine in microtiter plates with 200,000 cells/well for 6 weeks, and the number of wells with lymphoblastoid growth was determined (−, no positive wells; +, 1 to 20 positive wells/50 total wells; ++, 21 to 44; +++, 45 to 50). Scores in parentheses indicate wells where outgrowth was visibly present but cells were difficult to expand. Medium control infections were carried out in parallel for all PBMC infections, and no proliferation was observed at 6 weeks in any control infections. Nonnative virus-cell combinations were tested for the presence of the inoculating virus by EBNA2 PCR and sequencing.
The relative amounts of infectious virus in the stocks were tested by acute infection of the EBV-negative human B lymphoma cell line Louckes and detection of EBNA2 after 48 h (Fig. 3A). EBNA2 expression was readily detected after EBV, chLCV, cyLCV, and rhLCV infection. EBNA2 expression was detected in Louckes cells after infection with each LCV, indicating there was no absolute block to entry of different LCV species into human B cells. The rank order of the relative levels of EBNA2 expression in Louckes cells was similar to, but did not exactly match, the rank order of DNA titers measured by PCR, which may be related to slight differences in the infectious titer versus viral DNA, efficiency of human B cell infection, or expression levels of different LCV EBNA2s in human cells. The low level of baEBNA2 expression in baLCV-infected Louckes cells was consistent with the much lower viral DNA titer in that stock. Thus, EBNA2 expression after acute infection of Louckes cells confirmed the presence of infectious EBV, chLCV, baLCV, cyLCV, and rhLCV in the virus stocks and the ability of all LCVs to infect human B cells.
The same virus stocks were then tested for immortalization of human, chimpanzee, cynomolgus macaque, and rhesus macaque PBMC. Infected PBMC were cultured in the presence of cyclosporine for 4 to 6 weeks and scored visually for immortalization by counting the number of wells with proliferating lymphoblastoid cells. Since we used the same viral stocks with all species of B cells and since every B cell preparation was efficiently immortalized by the autologous LCV, we controlled for the quality of both the viruses and the cells used. EBV efficiently induced proliferation of both human and chimpanzee PBMC, with cell outgrowth in a high percentage of wells (Fig. 3B). Cells could be readily expanded from both EBV-infected human PBMC and EBV-infected chimpanzee PBMC, suggesting no preference of EBV for either hominoid B cell type. EBV induced no proliferation of cynomolgus macaque or rhesus macaque PBMC. chLCV readily induced proliferation of both chimpanzee and human PBMC, with comparable numbers of positive microtiter wells for the two hominoid species. chLCV-infected human PBMC proliferated robustly in microtiter plates but were difficult to expand beyond the microtiter plates, suggesting a stronger growth advantage in the native hominoid B cell. chLCV did not induce proliferation of PBMC from either Old World NHP macaque species.
rhLCV and cyLCV both induced robust proliferation of their native PBMC and had weaker proliferative activity against PBMC from the heterologous macaque species. cyLCV infection of rhesus macaque PBMC induced proliferation in several microtiter wells, but the cells were difficult to expand. In the case of rhLCV infection of cynomolgous macaque PBMC, only a single well displayed proliferation, but we were able to expand this into an immortalized cell line that had growth kinetics similar to that of the rhLCV-infected rhesus macaque cell lines. baLCV infection resulted in high and comparable numbers of positive wells with PBMC from the two macaque species despite the low viral titers, and the cell lines were readily expandable from both macaque species. rhLCV, baLCV, and cyLCV all failed to induce any proliferation of human or chimpanzee PBMC.
EBNA2 was characterized from cell lines derived from infection with nonnative LCV in order to rule out the possibility of spontaneous outgrowth of cells infected with native LCV. PCR primers capable of amplifying all EBNA2 species were used to amplify and sequence DNA from independent cell lines of chimpanzee PBMC immortalized with EBV (n = 6), PBMC from cynomolgus (n = 6) and rhesus (n = 6) macaques immortalized with baLCV, and cynomolgus macaque PBMC immortalized with rhLCV (n = 1). In all cases, only the inoculated virus species but no endogenous virus was detected, confirming the ability of LCV to immortalize B cells across species.
Our experiments support the model that hominoid and Old World LCV have a relative, but not absolute, host range restriction. EBV and baLCV provide the most clear-cut results, with the ability to robustly immortalize B cells from the natural host and from at least some closely related members within the same primate family, i.e., Hominoidae or Old World NHP (Cercopithecidae). Results obtained with chLCV, cyLCV, and rhLCV were consistent with this model, showing robust proliferation of cells in microtiter wells from closely, but not from more distantly, related species even though the cell growth was not robust enough to establish cell lines under standard tissue culture conditions.
Since viral entry does not appear to be a barrier to LCV infection of B cells across primate species, the most likely explanation for the block to B cell immortalization is that of differences in expression or function of the LCV latent-infection genes in B cells from another species. Given the close evolutionary relationship between hominoids and Old World NHP, one would predict that the transcriptional regulation and cell signaling pathways important for LCV-induced B cell immortalization, e.g., those represented by NF-κB and NOTCH, are highly conserved. Indeed, in all instances tested to date, latent infection proteins from baLCV and rhLCV are interchangeable with their EBV orthologues in functional assays, e.g., genome maintenance by EBNA1, transcriptional activation by EBNA2, or NF-κB activation by LMP1, although not always with the same level of activity (14–16). Thus, it is surprising that LCVs show such a strong preference for the autologous or closely related B cell. This suggests that one or more cellular pathways essential for LCV-induced B cell immortalization are significantly different among primate hosts, requiring evolution of LCV specific for a given primate species. Identifying the differences in virus-host interaction that restrict LCV host range provides an opportunity to elucidate novel pathways critical for EBV-induced growth transformation. A first step in that direction would be to isolate the viral protein with restricted function. This could be tested by replacing EBV proteins with, for example, their rhLCV orthologues and asking whether this would rescue the defect of EBV-induced immortalization of rhesus macaque B cells.
Nucleotide sequence accession numbers.
EBNA2 sequences from cyLCV (m4409), chLCV (m5305), and baLCV (S594) were deposited in GenBank under accession numbers KT072774, KT072775, and KT072776, respectively.
ACKNOWLEDGMENTS
This work was supported by grants from the US Public Health Service (grants CA68051 and AI114557). Rhesus macaque PBMC were provided with support from the New England Primate Research Center base grant (OD0111103). Chimpanzee PBMC were provided with support from the Yerkes National Primate Research Center (OD011132).
We thank Tatsuo Kawai at Massachusetts General Hospital, Harvard Medical School, for providing specimens of cynomolgus macaque lymphoma tissue and PBMC.
REFERENCES
- 1.Rivailler P, Jiang H, Cho YG, Quink C, Wang F. 2002. Complete nucleotide sequence of the rhesus lymphocryptovirus: genetic validation for an Epstein-Barr virus animal model. J Virol 76:421–426. doi: 10.1128/JVI.76.1.421-426.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moghaddam A, Rosenzweig M, Lee-Parritz D, Annis B, Johnson RP, Wang F. 1997. An animal model for acute and persistent Epstein-Barr virus infection. Science 276:2030–2033. doi: 10.1126/science.276.5321.2030. [DOI] [PubMed] [Google Scholar]
- 3.Falk L, Wolfe L, Deinhardt F, Paciga J, Dombos L, Klein G, Henle W, Henle G. 1974. Epstein-Barr virus: transformation of non-human primate lymphocytes in vitro. Int J Cancer 13:363–376. doi: 10.1002/ijc.2910130312. [DOI] [PubMed] [Google Scholar]
- 4.Miller G, Shope T, Lisco H, Stitt D, Lipman M. 1972. Epstein-Barr virus: transformation, cytopathic changes, and viral antigens in squirrel monkey and marmoset leukocytes. Proc Natl Acad Sci U S A 69:383–387. doi: 10.1073/pnas.69.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerber P, Pritchett RF, Kieff ED. 1976. Antigens and DNA of a chimpanzee agent related to Epstein-Barr virus. J Virol 19:1090–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerber P, Kalter SS, Schidlovsky G, Peterson WD Jr, Daniel MD. 1977. Biologic and antigenic characteristics of Epstein-Barr virus-related herpesviruses of chimpanzees and baboons. Int J Cancer 20:448–459. doi: 10.1002/ijc.2910200318. [DOI] [PubMed] [Google Scholar]
- 7.Rabin H, Neubauer RH, Hopkins RF III, Nonoyama M. 1978. Further characterization of a herpesvirus-positive orang-utan cell line and comparative aspects of in vitro transformation with lymphotropic old world primate herpesviruses. Int J Cancer 21:762–767. doi: 10.1002/ijc.2910210614. [DOI] [PubMed] [Google Scholar]
- 8.Moghaddam A, Koch J, Annis B, Wang F. 1998. Infection of human B lymphocytes with lymphocryptoviruses related to Epstein-Barr virus. J Virol 72:3205–3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heberling RL, Bieber CP, Kalter SS. 1981. Establishment of a lymphoblastoid cell line from a lymphomous cynomolgus monkey, p 385–386. In Yohn DS, Blakeslee JR (ed), Advances in comparative leukemia research. Elsevier, Amsterdam, the Netherlands. [Google Scholar]
- 10.Landon JC, Ellis LB, Zeve VH, Fabrizio DP. 1968. Herpes-type virus in cultured leukocytes from chimpanzees. J Natl Cancer Inst 40:181–192. [PubMed] [Google Scholar]
- 11.Rabin H, Neubauer RH, Hopkins RF III, Dzhikidze EK, Shevtsova ZV, Lapin BA. 1977. Transforming activity and antigenicity of an Epstein-Barr-like virus from lymphoblastoid cell lines of baboons with lymphoid disease. Intervirology 8:240–249. doi: 10.1159/000148899. [DOI] [PubMed] [Google Scholar]
- 12.Falk LA, Henle G, Henle W, Deinhardt F, Schudel A. 1977. Transformation of lymphocytes by Herpesvirus papio. Int J Cancer 20:219–226. doi: 10.1002/ijc.2910200209. [DOI] [PubMed] [Google Scholar]
- 13.Ling PD, Ryon JJ, Hayward SD. 1993. EBNA-2 of herpesvirus papio diverges significantly from the type A and type B EBNA-2 proteins of Epstein-Barr virus but retains an efficient transactivation domain with a conserved hydrophobic motif. J Virol 67:2990–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peng R, Gordadze AV, Fuentes Panana EM, Wang F, Zong J, Hayward GS, Tan J, Ling PD. 2000. Sequence and functional analysis of EBNA-LP and EBNA2 proteins from nonhuman primate lymphocryptoviruses. J Virol 74:379–389. doi: 10.1128/JVI.74.1.379-389.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yates JL, Camiolo SM, Ali S, Ying A. 1996. Comparison of the EBNA1 proteins of Epstein-Barr virus and herpesvirus papio in sequence and function. Virology 222:1–13. doi: 10.1006/viro.1996.0392. [DOI] [PubMed] [Google Scholar]
- 16.Franken M, Devergne O, Rosenzweig M, Annis B, Kieff E, Wang F. 1996. Comparative analysis identifies conserved tumor necrosis factor receptor-associated factor 3 binding sites in the human and simian Epstein-Barr virus oncogene LMP1. J Virol 70:7819–7826. [DOI] [PMC free article] [PubMed] [Google Scholar]