ABSTRACT
Viruses exploit molecules on the target membrane as receptors for attachment and entry into host cells. Thus, receptor expression patterns can define viral tissue tropism and might to some extent predict the susceptibility of a host to a particular virus. Previously, others and we have shown that respiratory pathogens of the genus Gammacoronavirus, including chicken infectious bronchitis virus (IBV), require specific α2,3-linked sialylated glycans for attachment and entry. Here, we studied determinants of binding of enterotropic avian gammacoronaviruses, including turkey coronavirus (TCoV), guineafowl coronavirus (GfCoV), and quail coronavirus (QCoV), which are evolutionarily distant from respiratory avian coronaviruses based on the viral attachment protein spike (S1). We profiled the binding of recombinantly expressed S1 proteins of TCoV, GfCoV, and QCoV to tissues of their respective hosts. Protein histochemistry showed that the tissue binding specificity of S1 proteins of turkey, quail, and guineafowl CoVs was limited to intestinal tissues of each particular host, in accordance with the reported pathogenicity of these viruses in vivo. Glycan array analyses revealed that, in contrast to the S1 protein of IBV, S1 proteins of enteric gammacoronaviruses recognize a unique set of nonsialylated type 2 poly-N-acetyl-lactosamines. Lectin histochemistry as well as tissue binding patterns of TCoV S1 further indicated that these complex N-glycans are prominently expressed on the intestinal tract of various avian species. In conclusion, our data demonstrate not only that enteric gammacoronaviruses recognize a novel glycan receptor but also that enterotropism may be correlated with the high specificity of spike proteins for such glycans expressed in the intestines of the avian host.
IMPORTANCE Avian coronaviruses are economically important viruses for the poultry industry. While infectious bronchitis virus (IBV), a respiratory pathogen of chickens, is rather well known, other viruses of the genus Gammacoronavirus, including those causing enteric disease, are hardly studied. In turkey, guineafowl, and quail, coronaviruses have been reported to be the major causative agent of enteric diseases. Specifically, turkey coronavirus outbreaks have been reported in North America, Europe, and Australia for several decades. Recently, a gammacoronavirus was isolated from guineafowl with fulminating disease. To date, it is not clear why these avian coronaviruses are enteropathogenic, whereas other closely related avian coronaviruses like IBV cause respiratory disease. A comprehensive understanding of the tropism and pathogenicity of these viruses explained by their receptor specificity and receptor expression on tissues was therefore needed. Here, we identify a novel glycan receptor for enteric avian coronaviruses, which will further support the development of vaccines.
INTRODUCTION
Entry of a virus into a host cell is defined by the interaction of a virus particle with its specific receptors on the target membrane. By exploiting these host cell receptors, viruses initiate attachment to, fusion with, and entry into cells. A plethora of viruses, including influenza virus, adenovirus, reovirus, and rotavirus, use glycans on host cell surfaces as receptors (1). Unlike protein receptors, which are differentially expressed in tissues and host species, glycans are distributed universally on many different cell surfaces. Recently, others (2–4) and we (5) showed that viral attachment proteins of avian coronaviruses (CoVs), in particular infectious bronchitis virus (IBV) and IBV-like viruses causing respiratory disease, interact with tissue-specific sialic acid structures.
Avian coronaviruses infect both domesticated and wild birds. IBV and turkey coronavirus (TCoV) belong to the genus Gammacoronavirus within the order Nidovirales. They are important pathogens of chickens and turkeys, respectively, and have huge economic implications for the poultry industry. While IBV is a respiratory pathogen, TCoV causes gastrointestinal disease resulting in growth retardation or mortality of entire flocks. Enterotropism has also been observed for some IBV serotypes; however, all IBV strains infect primarily the respiratory system, resulting in mild to severe inflammation of the nasal and tracheal epithelia (6, 7). In contrast, TCoV infects primarily the intestine, thereby inducing severe enteritis, and appears to possess strict tropism for the intestinal epithelium (8, 9). Recently, guineafowl coronavirus (GfCoV) and quail coronavirus (QCoV) were added to the enterotropic group of avian gammacoronaviruses as a cause of enteritis in guineafowl (8, 10) and quails (11), respectively.
For avian gammacoronaviruses the spike glycoprotein in the viral envelope acts as the viral attachment protein and is thus described as a determinant of cell tropism (12) and tissue tropism (13). The spike protein contains two subunits, S1 and S2; while S1 mediates virus attachment, S2 initiates the fusion and internalization of the virus with the host cell. Previously, by comparing tissue binding patterns of the S1 domains of IBV serotypes with different degrees of virulence, we showed that the in vivo tropism and pathogenicity of these viruses are in agreement with the patterns of binding of S1 to respiratory tissues (5). Similar observations were reported for binding of the S1 domains of IBV-like viruses, including those of pigeon and partridge CoVs (14). In particular, binding of S1 of respiratory gammacoronaviruses requires α2,3-linked sialic acids on host tissues; the specific involvement of sialic acid type 1 lactosamines was demonstrated for IBV strain M41 S1 (5, 13). In contrast to these respiratory viruses, hardly anything is known about virus-host interactions that determine the in vivo tropism and pathogenicity of enterotropic gammacoronaviruses, including the receptor specificity of turkey, guineafowl, and quail CoVs.
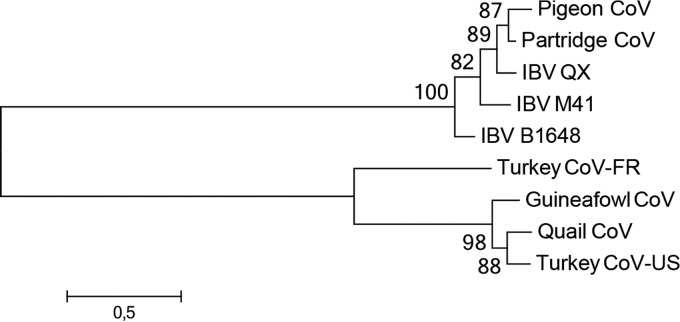
Genomic analysis indicates that IBV, TCoV-US (isolate ATCC from the United States), and GfCoV are closely related, demonstrating 84.7 to 88.7% nucleic acid sequence identity over the full lengths of their genomes. The S1 domain of the spike gene, however, is highly variable, with the amino acid sequences of IBV and TCoV-US sharing <25% sequence identity. Phylogenetic analysis of the S1 gene shows grouping of IBV and IBV-like viruses on the one hand and TCoV-US and GfCoV on the other hand (Fig. 1). QCoV clusters with the latter viruses, but further analysis is hampered by the lack of additional sequence data.
FIG 1.
Phylogenetic analysis of the S1 proteins of selected avian gammacoronaviruses. Amino acid sequences were aligned with ClustalX under default settings. The unrooted maximum likelihood tree was constructed by using MEGA6.06 with the best-fit substitution model with complete deletion (WAG+G+F model) and 100 bootstrap replicates (values indicated at the nodes). Pigeon CoV, pigeon coronavirus strain PSH050513 (GenBank accession number AAZ85066.1); Partridge CoV, partridge coronavirus strain GD/S14/2003 (accession number AAT70772.1); IBV QX, IBV strain QX (accession number AAF19629.1); IBV M41, IBV strain Mass 41 (accession number AAW33786.1); IBV B1648, IBV Belgian isolate B1648 (accession number CAA60684.1); Turkey CoV-FR, turkey coronavirus isolate FR070341j from France (accession number ADK88938.1); Guineafowl CoV, guineafowl coronavirus strain GfCoV/FR/2011 (accession number CCN27367.1); Quail CoV, quail coronavirus isolate quail/Italy/Elvia/2005 (accession number ABO45243.1); Turkey CoV-US, turkey coronavirus isolate ATCC from the United States (accession number ABW75138.1).
Here, we investigated the receptor specificity of avian coronaviruses known to cause enteritis in poultry. By analyzing the tissue and glycan specificities of the S1 domains of TCoV-US, GfCoV, and QCoV, we revealed that avian coronaviruses that cause enteritis use a novel glycan receptor for binding of the spike to tissues. In particular, they recognize nonsialylated lactosamine repeats (poly-N-acetyllactosamine [poly-LacNAc]) on complex-type N-glycans. Analysis of tissues revealed that poly-LacNAc was expressed predominantly on the intestinal epithelium of not only the respective host but also various other avian species. Taken together, we reveal that enteric gammacoronaviruses recognize a novel glycan receptor on target intestinal tissues of birds. In addition, the observed restricted expression patterns of these receptors in the intestine may contribute to the marked difference between the tissue tropisms of these avian coronaviruses from chickens and those from other poultry species such as turkey, guineafowl, and quail.
MATERIALS AND METHODS
Animal experiments.
Respiratory tissues of specific-pathogen-free (SPF) chickens (Gallus gallus) intranasally inoculated with 105 50% egg infectious doses (EID50) of IBV strain M41 (virus stocks were grown in embryonated eggs and kindly provided by the Animal Health Service, Deventer, The Netherlands) at 7 days of age were obtained from an experiment performed with the permission of the Dutch Ethic Committee at the Department of Pathobiology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands, according to local animal welfare regulations. Birds were humanely sacrificed at 1, 2, 3, and 7 days postinfection (dpi), followed by necropsy.
Intestinal tissues of SPF turkeys (Meleagris gallopavo) experimentally inoculated orally at 1 day of age with 104.7 EID50 of TCoV-US (isolate IN P4/1.1; kind gift of C. C. Wu and Y. M. Saif) (15, 16) were obtained from an experiment performed at the French Agency for Food, Environmental and Occupational Heath Safety (Anses), Ploufragan-Plouzane Laboratory, Ploufragan, France, with the approval of the French national committee for ethics in animal research. Virus stocks were prepared in turkey eggs as previously described (17). Birds were humanely sacrificed at 3, 6, 14, 21, and 50 dpi, after which necropsy was performed.
Histology and immunohistochemistry.
Tissues from experimentally infected chickens and turkeys were fixed in 4% formalin and subsequently dehydrated, embedded in paraffin, and prepared as 3- to 4-μm sections on coated glass slides (Klinipath, The Netherlands). Hematoxylin and eosin (H&E) staining and immunohistochemistry were performed on serial sections. H&E-stained sections were analyzed for histopathological changes by a certified pathologist. For immunohistochemistry, antigen retrieval was performed by boiling sections in Tris-EDTA (pH 9.0) (preheated for 10 min) for 10 min at 1,100 kW in a microwave. After washing in phosphate-buffered saline (PBS)–0.1% Tween, primary antibody against S2 of IBV (18, 19) or nucleocapsid of TCoV (20) was applied onto tissues from chicken and turkey at a 1:100 dilution. Sections was incubated for 2 h at room temperature and washed again in PBS (pH 7.4)–0.1% Tween before application of polyclonal mouse antibody (Envision) at a dilution of 1:1. Finally, 3-amino-9-ethylcarbazole (AEC; Sigma-Aldrich) was applied as the substrate to visualize the signal.
Genes and expression vectors.
S1-encoding sequences were obtained from GenBank (National Center for Biotechnology Information). GenBank accession numbers for S1-encoding sequences of TCoV-US, GfCoV, and QCoV are ABW75138.1, CCN27367, and ABO45243, respectively. Codon-optimized sequences for both S1 genes in pUC57 with upstream NheI and downstream PacI were obtained from GenScript and cloned into the pCD5 expression vector by restriction digestion, as previously described (5). The pCD5 expression vector containing S1 of IBV M41 was previously described (5) and used as a control.
Expression and purification of proteins.
Recombinant S1 proteins were expressed and purified as described previously by Ambepitiya Wickramasinghe et al. (5). Briefly, pCD5 vectors containing the S1 domains of TCoV-US, GfCoV, and GfCoV were transfected into human embryo kidney cells (HEK293T), and cell culture supernatants were harvested after 7 days. S1 proteins were purified by adding a 50% Strep-Tactin Sepharose suspension (IBA GmbH) to the mixture and analyzed by SDS-PAGE and Western blotting.
Protein histochemistry with S1.
Protein histochemistry was performed on tissue slides from our previously developed single- or multispecies tissue microarray (TMA) (14). For the purpose of this study, we selected TMAs containing respiratory and intestinal tissues of chickens and turkeys. When the indicated S1 proteins were precomplexed with Strep-Tactin horseradish peroxidase (HRPO) for 30 min on ice and subsequently mixed with polyacrylic acid (PAA)-3′ LacNAc type 1 (Neu5Acα2,3Galβ1,3GlcNAcβ-OCH2CH2CH2NH-polyacrylamide) or PAA-diLacNAc (Galβ1,4GlcNAcβ1,3Galβ1-GlcNAcβ-OCH2CH2CH2NH-polyacrylamide) (both from Lectinity Holding Inc., Russia) in a molar ratio of 1:2 (S1:PAA neoconjugates). S1:Strep-Tactin HRPO:PAA conjugate complexes were incubated on ice for another 30 min and subsequently applied onto tissue sections. The next day, sections were washed with PBS–0.1% Tween, and AEC was applied (5). When indicated, the tissues were treated with Arthrobacter ureafaciens neuraminidase (Roche, USA) at 1 mU/100 μl in PBS (pH 5) overnight at 37°C.
Glycan array.
Glycan arrays for S1 proteins were performed by using Consortium for Functional Glycomics (CFG) version 5.2 arrays. As described above, S1 proteins were precomplexed with anti-Strep-tag antibody and Alexa 647-linked anti-mouse IgG (4:2:1 molar ratio) prior to incubation for 15 min on ice in 100 μl PBS containing 0.1% Tween and were incubated on the array surface in a humidified chamber for 90 min. Slides were subsequently washed by successive rinses with PBS–0.1% Tween, PBS, and deionized H2O. Washed arrays were dried by centrifugation and immediately scanned for fluorescein isothiocyanate (FITC) signals on a Perkin-Elmer ProScanArray Express confocal microarray scanner. The fluorescent signal intensity was measured by using Imagene (Biodiscovery), and the mean intensity minus the mean background was calculated and graphed by using MS Excel. For each glycan, the mean signal intensity was calculated from 6 replicates spots. The highest and lowest signals of the 6 replicates were removed, and the remaining 4 replicates were used to calculate the mean signal, standard deviation (SD), and standard error measurement (SEM). A list of glycans on the microarray is included and can be found on the website of the Consortium for Functional Glycomics (http://www.functionalglycomics.org/).
ELISA-like binding assay for S1 proteins.
Binding affinities were compared by using enzyme-linked immunosorbent assay (ELISA)-like binding assays, performed similarly to the glycan array analyses, with minor modifications. Here, the total volume of the precomplexing mixture was 20 μl, the mixture was incubated for 15 min on ice, and samples were subsequently diluted 1:1 in a 384-well plate. Next, 8 μl was transferred onto microwell slides containing 48 wells. Each well was imprinted with diLacNAc, α2,3-linked sialylated diLacNAc, and α2,6-linked sialylated diLacNAc structures with and without PAA conjugation, all in 6 replicates.
Lectin histochemistry.
Lectin histochemistry was performed to detect poly-LacNAc structures and complex-type N-glycan cores. To this end, either Erythrina cristagalli lectin (ECA; Vector Laboratories) or Phaseolus vulgaris agglutinin (PHA; Vector Laboratories) was diluted in PBS to a concentration of 2 μg/ml and applied onto multispecies TMAs containing intestinal tissues of 10 avian species, including turkey, quail, guineafowl, Canada goose, graylag goose, mallard duck, teal, pigeon, partridge, and pheasant. After incubation overnight at 4°C, the slides were washed in PBS, and an avidin-biotin complex (ABC kit; Vector Laboratories) was applied for 30 min to visualize the signal.
RESULTS
Tissue tropism of avian gammacoronaviruses IBV and TCoV-US in vivo.
While most gammacoronaviruses, including prototype IBV strain M41 in chickens, induce primarily respiratory disease, other viruses of this genus, like TCoV, are known to be enteropathogenic. To correlate the clinical manifestations of these diseases with the histopathological changes and virus replication in target organs, we performed two in vivo experiments, in which SPF chickens and turkeys were infected with IBV M41 and TCoV-US, respectively. Infected chickens showed clinical signs of respiratory disease, such as mild dyspnea, sneezing, and serous exudate from the nostrils, from 4 dpi; infected turkeys showed signs of gastrointestinal disease with diarrhea, starting at 3 dpi. While chickens did not show any signs of gastrointestinal disease during the course of infection, the infected turkeys did not display any notorious signs of respiratory illness.
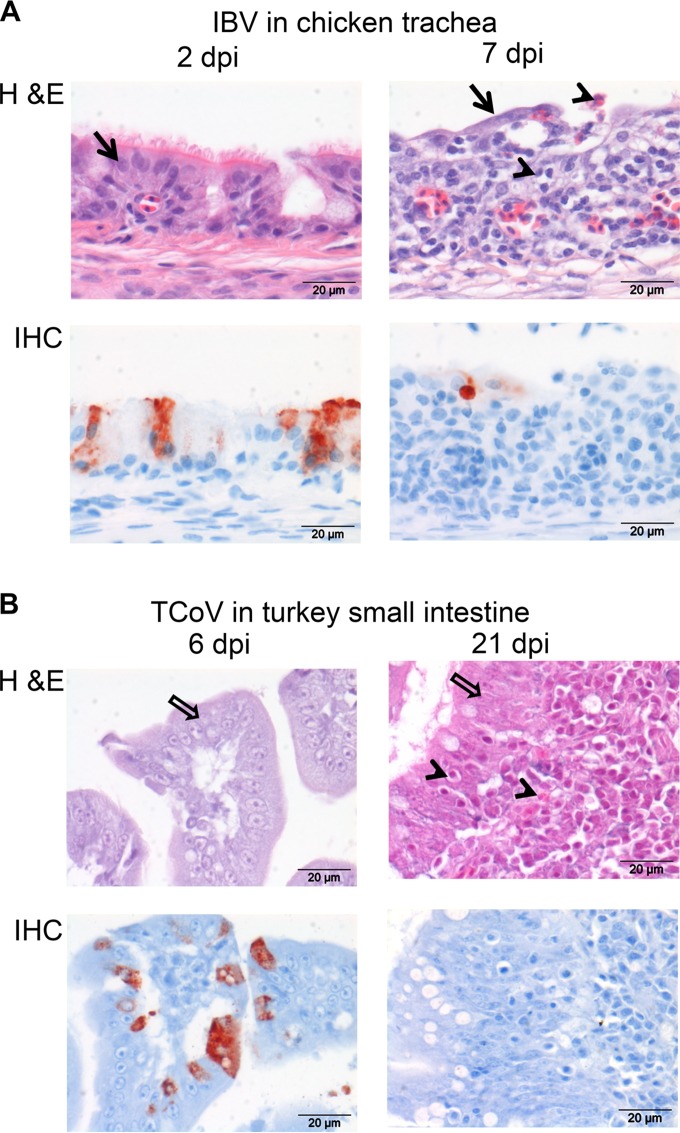
In the chicken trachea, significant histomorphological changes were detected from day 3 postinfection onwards, comprising a loss of normal columnar ciliated epithelium and mucous glands, attenuation, and squamous-like changes of the epithelium. Infiltration of the mucosa and submucosa by marked numbers of lymphocytes and macrophages and fewer plasma cells was also observed. In addition, mucosal transmigration and intraluminal accumulation of moderate numbers of heterophils and marked hyperemia of submucosal capillaries were detected (Fig. 2A). While intestinal sections of infected chickens did not show any significant histomorphological changes, obvious changes in the small intestine of the turkeys were seen from day 13 postinfection onwards, including multifocal marked pseudostratification of the epithelium, infiltration of the epithelium by small numbers of mainly lymphocytes, formation of lymphoid aggregates in the lamina propria and submucosa, and multifocally scattered infiltration of moderate numbers of plasma cells and heterophils (Fig. 2B).
FIG 2.
Histopathological and immunohistochemical findings in infected tissues of chicken and turkey. (A) Tracheas of chickens infected with IBV M41 were stained with H&E or with an antibody against the S2 protein of IBV by performing immunohistochemistry (IHC). (B) Small intestines of turkeys infected with TCoV were stained with H&E or with an antibody against the nucleocapsid protein of TCoV. dpi, days postinfection. Epithelium of chicken trachea (black arrows), inflammatory cells (arrowheads), and epithelium of turkey intestine (white arrows) are indicated.
To investigate the presence of viral antigen in infected tissues, immunohistochemistry was performed by using monoclonal antibodies against the S2 domain of the spike protein of IBV (Fig. 2A) and the nucleocapsid protein of TCoV (Fig. 2B). The IBV S2 protein was detected in the ciliated epithelium of the trachea from days 2 to 7 postinfection. At 2 dpi, many morphologically unchanged epithelial cells contained large amounts of the S2 protein, whereas at 7 dpi, when histopathological changes were most significant, only a few cells containing the viral antigen remained (Fig. 2A). The presence of TCoV nucleocapsid proteins was detected from days 6 to 13 postinfection in the enterocytes of the turkey small intestine (Fig. 2B). No viral proteins were detected by day 21 postinfection, the time point at which the small intestine showed the most remarkable histopathological changes. In conclusion, both viruses infected the epithelial cells of the target organs, defined by clinical symptoms, but the course of infection of TCoV appeared to be longer than that of IBV. Moreover, histopathological changes and the presence of viral proteins confirmed that unlike IBV infection, TCoV infection is localized in the intestine, as previously described (9, 10, 21).
Enterotropic coronavirus spike binding is not sialic acid dependent.
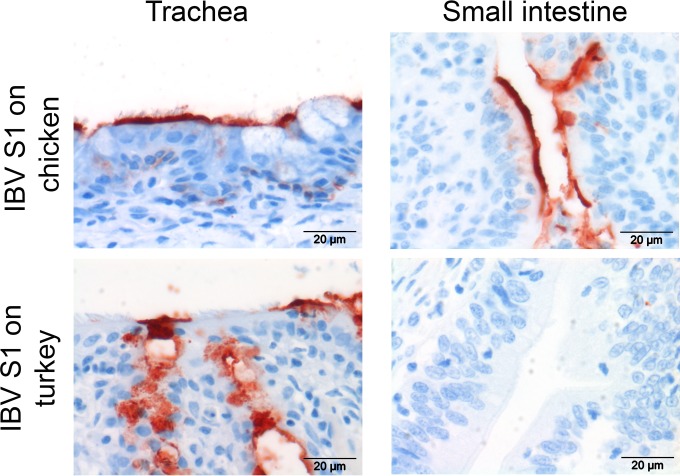
Binding of S1 of IBV and IBV-like CoVs (14) to host tissues is sialic acid dependent. In particular, IBV M41 uses sialic acids on type 1 lactosamine (Neu5Acα2,3Galβ1,3GcNAc) to determine its tissue-specific binding (5). To reveal whether turkey tissues express the previously identified avian coronavirus receptor, a recombinant IBV S1 protein was applied onto turkey tissues, and protein histochemistry was performed (5). IBV S1 bound to tracheal and intestinal epithelia of chicken, as observed previously (5), as well as to trachea of turkey (Fig. 3). No binding to the intestinal epithelium of turkey was observed, indicating that the turkey intestine does not express IBV-specific sialic acid-containing glycans. Moreover, this result suggests that gastrointestinal avian coronaviruses might use a different receptor for attachment and entry into the epithelial cells of the intestine.
FIG 3.
Protein histochemistry of IBV M41 S1 on chicken and turkey tissues. Protein histochemistry was performed by applying recombinant IBV M41 S1 (5) onto trachea and small intestines of chicken and turkey.
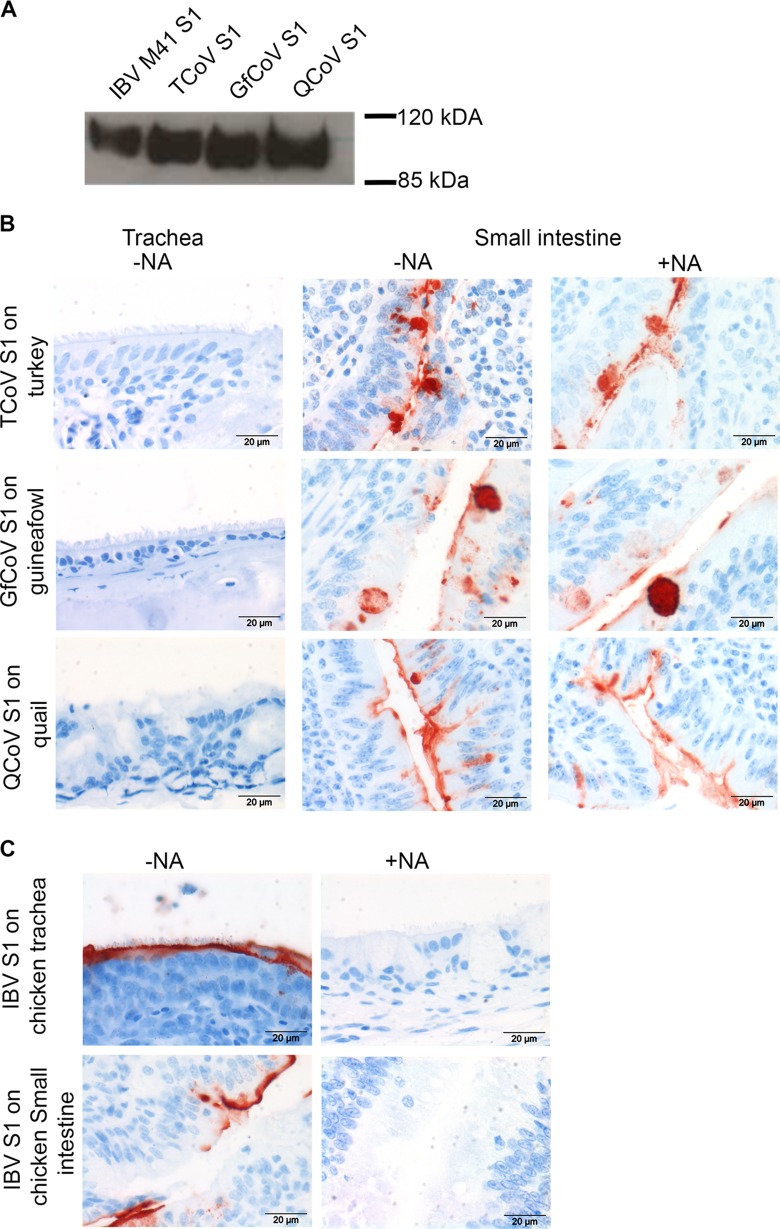
As tissue-specific binding of S1 proteins can provide insights into the receptor usage of the respective viruses, we next expressed S1 of TCoV-US in our mammalian expression system, in a manner similar to that for IBV S1 (5). In parallel, we produced S1 proteins of two other avian coronaviruses with reported gastrointestinal tract tropism, namely, guineafowl (8) and quail (11) CoVs. Analysis of the proteins by SDS-PAGE followed by Western blotting confirmed the production of the S1 proteins of TCoV, GfCoV, and QCoV, migrating at ∼110 kDa (Fig. 4A). Protein histochemistry was performed by applying these S1 proteins to TMAs containing gastrointestinal systems of turkey, guineafowl, and quail. All three S1 proteins attached strongly to the brush border of epithelial cells and goblet cells of the small (Fig. 4B) and large (not shown) intestines of the respective hosts, and no binding to the tracheas was observed (Fig. 4B). Hence, the profile of binding of the spike protein of TCoV to the intestinal tissue was comparable to that for the in vivo replication sites. In addition, the tissue binding patterns of S1 proteins of GfCoV and QCoV were comparable to those of S1 of TCoV and correlated to their in vivo tissue tropism (8, 11). To determine if the tissue binding of these S1 proteins was independent of the presence of sialylated glycans, we applied S1 proteins of TCoV, GfCoV, and QCoV onto the intestinal tissue of the respective host after treatment of the tissue with neuraminidase. The S1 protein of enterotropic CoV still had the ability to bind to desialylated tissues (Fig. 4B), in contrast to what was observed for IBV (5) (Fig. 4C). These results serve to confirm the above-described observation that TCoV, GfCoV, and QCoV bind to host cells via a different host receptor than that used by IBV M41.
FIG 4.
Expression and tissue binding specificities of S1 proteins. (A) Recombinantly expressed S1 domains of chicken IBV-M41 (IBV S1), turkey coronavirus (TCoV S1), guineafowl coronavirus (GfCoV S1), and quail coronavirus (QCoV S1) were analyzed by SDS-PAGE and Western blotting. (B) Enteric CoV S1 proteins were applied onto respiratory and intestinal tissues of their respective hosts. S1 was applied onto both sialylated (−NA) and desialylated (+NA) tissues. (C) IBV S1 was applied onto sialylated (−NA) and desialylated (+NA) chicken tracheas as a control. Desialylation was performed by treating tissues with bacterial neuraminidase (NA), as described in Materials and Methods. Results are representative of data from three independent experiments.
Novel glycan binding specificity of TCoV-US, GfCoV, and QCoV spike proteins.
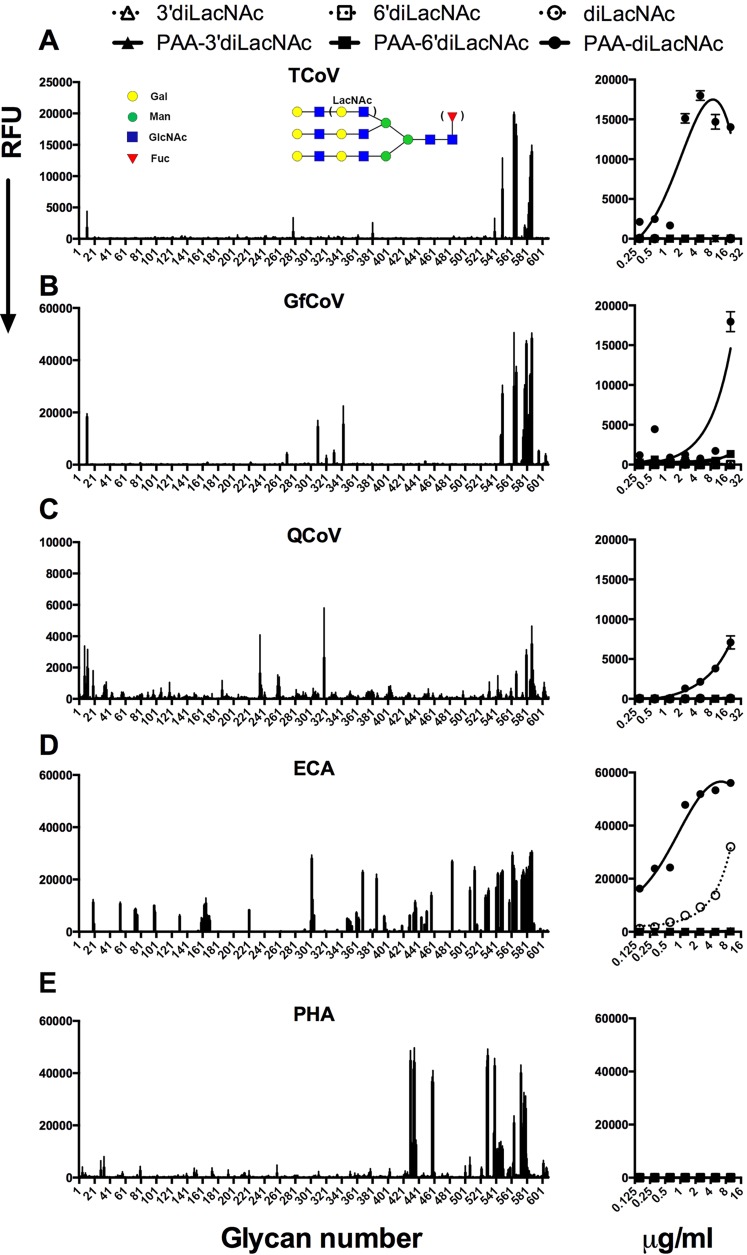
To reveal whether gastrointestinal avian coronaviruses can exploit other, nonsialylated glycans for attachment, we analyzed the binding of S1 proteins of TCoV-US, GfCoV, and QCoV on Consortium for Functional Glycomics (CFG) version 5.2 arrays (as described in Materials and Methods). The results showed that TCoV S1 recognized nonsialylated type 2 poly-LacNAc structures (Fig. 5; see also Table SA1 in the supplemental material). Strikingly, TCoV S1 bound to these glycans only if the core structure was either a bi- or triantennary N-glycan (as exemplified in Fig. 5A, left). To confirm binding and to determine the affinity of TCoV S1 for these structures, we printed diLacNAc, α2,3-linked sialylated diLacNAc, and α2,6-linked sialylated diLacNAc structures with and without conjugation to PAA in an ELISA-like glycan binding setup. PAA is commonly used to coat 96-well plates for solid-phase ELISAs, as its multivalent structure increases the binding affinity of the viral proteins (22). Here, we observed specific binding of TCoV S1 to nonsialylated PAA-diLacNAc (Fig. 5A, right), confirming that TCoV S1 did not bind to sialic acids, in contrast to IBV. The GfCoV and QCoV S1 proteins showed very similar glycan binding specificities (Fig. 5B and C, left; see also Table SA1 in the supplemental material). Interestingly, however, GfCoV S1 also recognized several N-glycans with α2,6-linked sialic acids, while QCoV S1 additionally recognized N-glycans with α2,3-linked sialic acids in the glycan array. The ELISA-like glycan binding assay, however, confirmed specificity only for LacNAc structures and not for sialylated structures (Fig. 5B and C, right). Furthermore, the binding affinity for PAA-diLacNAc appeared to be slightly higher for TCoV S1 than for GfCoV and QCoV S1 proteins (Fig. 5, right). The biological relevance of this is as yet unclear.
FIG 5.
Glycan binding specificity of avian coronavirus spike proteins. Shown are glycan binding specificities of S1 proteins of TCoV (A), GfCoV (B), QCoV d (C) and the plant lectins Erythrina cristagalli lectin (ECA) (D) and Phaseolus vulgaris agglutinin (PHA) (E). A representative example of the bound glycans for TCoV is depicted schematically. The left panels show the results of the glycan array; bar graphs were plotted in Prism and represent the averaged mean signal minus the background for each glycan sample, and error bars are the SEM values (from six replicates). The right panel shows the results of an ELISA-like assay in which the affinity of binding to sialylated and nonsialylated diLacNAc printed with and without conjugation to PAA was measured. RFU, relative fluorescence units.
Erythrina cristagalli lectin is known to bind specifically to terminal galactose residues on type 2 LacNAc structures (23, 24) and was therefore used as a control in our assays. On the glycan array, we observed a more promiscuous binding pattern than that for S1 proteins, as this lectin is not N-glycan dependent (Fig. 5D, left) (25). In the ELISA-like assay, we observed a significantly higher binding avidity of Erythrina cristagalli lectin for LacNAc structures than that of the avian coronavirus spike proteins, as it was also able to bind non-PAA-conjugated LacNAc (Fig. 5D, right). Finally, to study N-glycan dependency, we used Phaseolus vulgaris agglutinin (24, 25), which indeed bound the same N-glycans in the glycan array (Fig. 5E, left). However, in contrast to ECA, PHA could not bind to LacNAc structures in the ELISA-like assay (Fig. 5E, right). Thus, while the more promiscuous lectin ECA can be used to profile galactose residues on type 2 LacNAc structures, enteric coronavirus S1 proteins show a more strict specificity for the LacNAc N-glycans.
Glycan binding specificity of TCoV-US, GfCoV, and QCoV spike proteins on tissues.
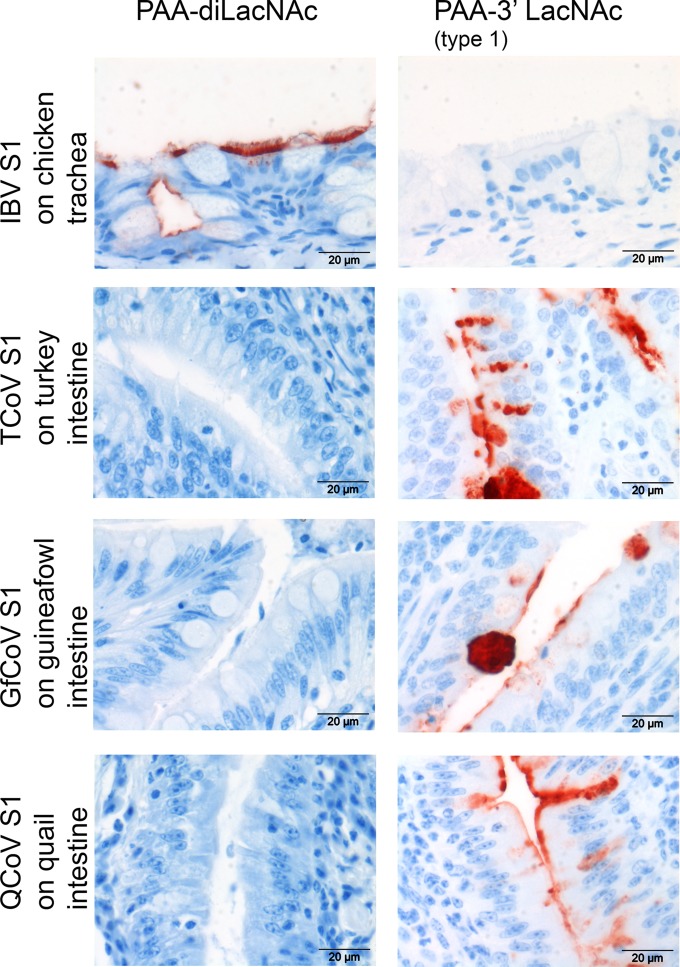
Next, we analyzed the biological relevance of the glycan binding specificities of the spike proteins of TCoV-US, GfCoV, and QCoV on tissues. Therefore, we mixed the S1 proteins with synthetic glycopolymers containing diLacNAc (Galβ1,4GlcNAcβ1,3Galβ1,4GlcNAc) prior to application of the mixture onto tissues. Binding of all three S1 proteins to intestinal tissues from their respective hosts was completely blocked in the presence of PAA-diLacNAc, while this compound could not block the binding of IBV S1 (Fig. 6, left). These results indicate that specific intestinal binding of the TCoV, GfCoV, and QCoV S1 proteins is dependent on the availability of LacNAc on the tissues rather than on sialylated glycans. Reciprocally, the binding of S1 proteins of TCoV, GfCoV, and QCoV could not be blocked with the sialylglycopolymer containing Neu5Acα2,3Galβ1,3GlcNAc (PAA-3′ LacNAc) (Fig. 6, right), while this glycan blocked IBV S1 binding to chicken trachea. These data confirm the relevance of these glycans for determining the receptor specificity of CoVs causing gastrointestinal disease.
FIG 6.
Blocking of binding of S1 to tissues with PAA-linked glycans. S1 proteins were mixed with PAA-diLacNAc or PAA-3′ LacNAc type 1 (Neu5Acα2,3Galβ1,3GlcNAc) prior to applying them onto tissues. Representative pictures from 4 independent experiments are shown.
Additionally, we aimed to reveal the biological relevance of the PAA-glycans in avian coronavirus infection in embryonated eggs, as continuous cell lines supporting IBV M41 and TCoV infection are not available. Infection of chicken eggs inoculated via the allantoic cavity with IBV M41 preincubated with PAA-Neu5Acα2,3Galβ1,3GlcNAc resulted in a 10-fold reduction in the abundance of viral genomes (from allantoic fluid harvested at 12 h postinfection), compared to inoculation with PBS- and PAA-diLacNAc-pretreated virus (data not shown). For TCoV, infection can be achieved via inoculation of 104 EID50 into the amniotic cavity of embryonated turkey eggs (17) and harvesting of intestines at 4 dpi. Initial experiments to block TCoV infection by using PAA-diLacNAc unfortunately were not successful, likely because the conditions for blocking with glycans require minimizing the virus dose, which could result in 100% infected eggs detected at earlier time points after inoculation. Thus, while we demonstrated that Neu5Acα2,3Galβ1,3GlcNAc is required for infection by the avian coronavirus IBV, the role of diLacNAc in TCoV infection in a complex system such as that of eggs awaits further study.
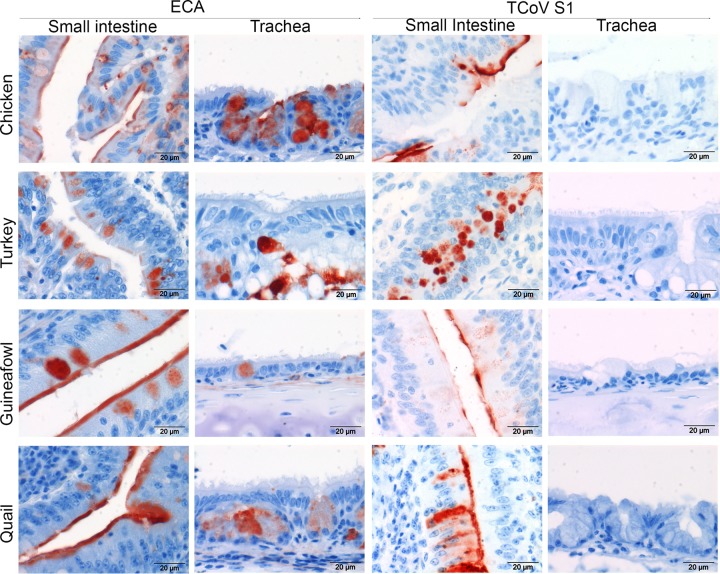
Erythrina cristagalli lectin appeared to have a somewhat broader glycan preference than the viral attachment proteins of avian coronaviruses (Fig. 7; see also Table SA1 in the supplemental material). To compare the specificities of ECA with the recognition of viral receptors on host tissues, we applied ECA onto tissues from chicken, turkey, guineafowl, and quail. Interestingly, ECA stained the intestine and the trachea of all four species; however, strong staining was observed for the epithelium of the small intestine (Fig. 7, left) compared to that for goblet cells of the trachea (Fig. 7, right) of each host. In the intestine, the staining of ECA was comparable to that of TCoV S1 (Fig. 7), while no staining of the tracheas was observed (Fig. 7). Thus, the observed differences in the glycan specificities of ECA and the CoV S1 proteins are reflected by broader binding of ECA to tissues.
FIG 7.
Lectin and protein histochemistry for respiratory and intestinal tissues. ECA or TCoV S1 was applied onto TMAs containing trachea and small intestinal tissues of chicken, turkey, guineafowl, and quail.
LacNAc receptors are widely distributed across avian species.
Finally, we investigated whether this novel receptor is present across avian species. To this end, lectin and spike histochemistry using ECA and TCoV-US S1, respectively, was performed on the multispecies TMA containing intestinal tissues of various other avian species, including Canada goose, graylag goose, mallard duck, teal, partridge, pigeon, pheasant, and chicken. ECA histochemistry revealed that, except for mallard duck, LacNAc was prominently distributed across the intestines (Table 1) of all the other selected birds. Similar results were obtained for TCoV S1, suggesting that the availability of N-glycans with nonsialylated poly-LacNAc may predispose these avian species to infection by gammacoronaviruses that require poly-LacNAc for attachment. However, while ECA staining was prominent in respiratory tissues of birds, only very low or no affinity of TCoV S1 for respiratory tissues was observed (Fig. 7), indicating that TCoV S1 recognizes a specific set of LacNAc-containing N-glycans, which may be expressed uniquely in the intestines of these birds. The biological relevance of the expression of the viral receptor for TCoV in other birds is further supported by the observation that TCoV has the ability to infect not only turkeys but also chickens under experimental conditions (21).
TABLE 1.
Distribution of enterotropic gammacoronavirus receptors in intestines of avian speciesa
| Avian species | Presence of poly-LacNAc in intestine determined by ECA histochemistry | Presence of receptor in intestine determined by TCoV S1 histochemistry |
|---|---|---|
| Chicken | + | + |
| Turkey | + | + |
| Guineafowl | + | + |
| Quail | + | + |
| Canada goose | + | + |
| Graylag goose | + | + |
| Mallard duck | − | − |
| Partridge | + | + |
| Pheasant | + | + |
| Teal | + | + |
| Pigeon | + | + |
ECA and TCoV S1 were applied onto TMAs containing intestinal tissues of 11 avian species. −, absence of poly-LacNAc or the receptor; +, presence of poly-LacNAc or the receptor.
DISCUSSION
Here, we revealed a novel receptor for coronaviruses that cause gastrointestinal tract disease in poultry. In particular, the spike viral attachment proteins of turkey, guineafowl, and quail coronaviruses required nonsialylated type 2 poly-LacNAc structures on N-glycan cores for binding. This was in marked contrast to the sialic acid-dependent binding of avian coronaviruses that induce primarily respiratory tract disease, including IBV and IBV-like viruses from partridge and pigeon (14). The resemblances of the tissue binding profiles of the S1 proteins of enteric CoVs with their in vivo tropism and the expression of poly-LacNAc-containing N-glycans in the small intestines of various avian species suggest that tropism of these coronaviruses is defined by the restricted expression patterns of this novel avian coronavirus receptor in target tissues.
The binding of S1 proteins of enteric and respiratory CoVs to the intestinal epithelium (this study) and the tracheal and lung epithelia (5), respectively, was in agreement with the reported in vivo pathotypes of avian coronaviruses. Inoculation of turkey CoV into turkey poults resulted in marked macroscopic and microscopic changes in the intestine but not in other organs, including respiratory tissues (21). Similarly, both guineafowl and quail CoVs were reported to produce gastrointestinally related clinical signs, matching the binding of the S1 protein to these tissues. Our data indicate that the spike proteins contribute largely to determining the in vivo tissue tropism of a particular avian coronavirus. This is in line with data from previous reports in which the cell culture-adapted IBV Beaudette strain gained tropism for trachea of chickens only by replacing the S1 domain of the respiratory tropic IBV strains (26, 27). In this respect, it is interesting to see that phylogenetic trees based on the S1 gene (Fig. 1) clearly separate respiratory from gastrointestinal avian coronaviruses. Remarkably, the spike protein of a TCoV isolate from France (TCoV-FR) (16) had only 40% sequence identity to that of the TCoV-US strain. This diversity was reflected by the prominent binding of this S1 protein to the epithelium of the bursa of Fabricius and the only rather mild staining of the small intestine of turkey (data not shown). As the TCoV-FR S1 protein did not show affinity for diLacNAc and did not recognize any particular glycan on the glycan array, further studies are needed to elucidate whether this virus uses yet another receptor for binding, and to determine its tropism, in turkeys.
In our study, we observed that tissue binding of S1 proteins of turkey, guineafowl, and quail coronaviruses was dependent on the availability of nonsialylated poly-LacNAc-containing N-glycans. Compared to the wide distribution of α2,3-linked sialic acids across avian tissues and hosts (14), poly-LacNAc was detected conspicuously in the epithelial cells of the intestines of various birds. As poly-LacNAc-containing N-glycans appear to act as specific receptors for enteric CoVs, we speculate that the restricted expression of LacNAc can account for spatially restricted intestinal disease, like enteritis. In this line, the broader distribution of α2,3-linked sialic acids across organ systems can explain why respiratory avian coronaviruses often show tropism for other organs (6, 7). Spatial and temporal expression of glycans exploited by viruses as receptors might thus define tissue tropism and, ultimately, disease. In this respect, it is of interest to note that poly-LacNAcs are described to be an age-specific receptor for human rotavirus (28). As they are expressed predominantly in saliva of infants, neonates and young children appear to be more susceptible to diarrhea caused by human rotavirus (28). As TCoV-induced enteritis in turkeys is more frequently described for young animals (9), it might well be that LacNAc expression in bird is also age dependent. Whether the observed presence of LacNAc in the intestines of various other bird species, based on binding of ECA and TCoV S1, truly predicts the susceptibility of these species to these, or other, enterotropic coronaviruses remains to be determined.
Lectin histochemistry was used previously to profile viral glycan receptor expression on tissues (5, 29, 30). Here, we used Erythrina cristagalli lectin to identify the terminal LacNAc structures, but not the core carbohydrate portion, of the LacNAc-containing N-glycans (23). With this, we revealed that such LacNAc structures are expressed not only in the intestinal tracts but also in the respiratory systems of several bird species. Other plant lectins, for example, Phaseolus vulgaris agglutinin, identify N-glycans rather than terminal LacNAcs (24). By using PHA, however, no affinity for diLacNAc in the ELISA-like assay (Fig. 5) or PHA staining of the intestinal epithelia of chicken, turkey, guineafowl, and quail was detected (data not shown). Compared to these lectins, S1 proteins of enteric CoVs recognize nonsialylated poly-LacNAc-containing N-glycans with high specificity (Fig. 5; see also Table SA1 in the supplemental material), and we detected this attachment factor only in the intestine of birds. This strict specificity of specific viral lectins points toward the preferred use of viral attachment proteins over lectins for profiling glycans in other cells and tissues.
In our ELISA-like assay, we determined that the affinity of TCoV S1 for LacNAc was higher than that of the GfCoV and QCoV S1 proteins. The reason for this difference in avidity is as yet unclear; it might be linked to the virulence and the persistence of these viruses in the field. Indeed, for influenza virus, it has been suggested that high avidity for glycoconjugates might prevent the release of viruses from decoy receptor-expressing cells (31–33), thereby reducing virus infection. Although avian coronaviruses do not contain a virus-releasing enzyme like influenza virus, it might well be that they evolved a mechanism based on high avidity as well as high specificity for glycans to regulate infection. In this respect, it is of interest to note the phenotypic differences between these viruses: while TCoV causes enteritis with rather low mortality rates (up to 10% [34]), the mortality rates for GfCoV and QCoV infections are much higher, up to 100% (8, 11). Whether the higher avidity of S1 of TCoV for PAA-diLacNAc actually resulted in the observed lower virulence remains to be seen.
In conclusion, we elucidated the receptor specificities of the S1 viral attachment proteins of TCoV, GfCoV, and QCoV. The preference of these viruses for nonsialylated poly-LacNAc-containing glycans and their particular expression on tissues might predispose organ systems and hosts to infection by coronaviruses that cause gastrointestinal disease in poultry.
Supplementary Material
ACKNOWLEDGMENTS
R.P.D.V. and M.H.V. are financially supported by NWO Rubicon and VENI and by NWO MEERVOUD, respectively. This work was funded in part by National Institutes of Health grant AI099274 to J.C.P. Several glycans used for binding assays were provided by the Consortium for Functional Glycomics (http://www.functionalglycomics.org/), funded by NIGMS grant GM62116 to J.C.P.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00745-15.
REFERENCES
- 1.Stencel-Baerenwald JE, Reiss K, Reiter DM, Stehle T, Dermody TS. 2014. The sweet spot: defining virus-sialic acid interactions. Nat Rev Microbiol 12:739–749. doi: 10.1038/nrmicro3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Winter C, Schwegmann-Wessels C, Cavanagh D, Neumann U, Herrler G. 2006. Sialic acid is a receptor determinant for infection of cells by avian infectious bronchitis virus. J Gen Virol 87:1209–1216. doi: 10.1099/vir.0.81651-0. [DOI] [PubMed] [Google Scholar]
- 3.Winter C, Herrler G, Neumann U. 2008. Infection of the tracheal epithelium by infectious bronchitis virus is sialic acid dependent. Microbes Infect 10:367–373. doi: 10.1016/j.micinf.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shahwan K, Hesse M, Mork AK, Herrler G, Winter C. 2013. Sialic acid binding properties of soluble coronavirus spike (S1) proteins: differences between infectious bronchitis virus and transmissible gastroenteritis virus. Viruses 5:1924–1933. doi: 10.3390/v5081924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ambepitiya Wickramasinghe IN, de Vries RP, Gröne A, de Haan CA, Verheije MH. 2011. Binding of avian coronavirus spike proteins to host factors reflects virus tropism and pathogenicity. J Virol 85:8903–8912. doi: 10.1128/JVI.05112-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cavanagh D. 2007. Coronavirus avian infectious bronchitis virus. Vet Res 38:281–297. doi: 10.1051/vetres:2006055. [DOI] [PubMed] [Google Scholar]
- 7.Cavanagh D. 2005. Coronaviruses in poultry and other birds. Avian Pathol 34:439–448. doi: 10.1080/03079450500367682. [DOI] [PubMed] [Google Scholar]
- 8.Liais E, Croville G, Mariette J, Delverdier M, Lucas MN, Klopp C, Lluch J, Donnadieu C, Guy JS, Corrand L, Ducatez MF, Guerin JL. 2014. Novel avian coronavirus and fulminating disease in guinea fowl, France. Emerg Infect Dis 20:105–108. doi: 10.3201/eid2001.130774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guy JS. 2008. Turkey coronavirus enteritis, p 330–338. In Saif YM, Fadly AM, Glisson JR, MacDougald LR, Nolan LK, Swayne DE (ed), Diseases of poultry, 12th ed Blackwell Publishing, Oxford, United Kingdom. [Google Scholar]
- 10.Guy JS. 2000. Turkey coronavirus is more closely related to avian infectious bronchitis virus than to mammalian coronaviruses: a review. Avian Pathol 29:207–212. doi: 10.1080/03079450050045459. [DOI] [PubMed] [Google Scholar]
- 11.Circella E, Camarda A, Martella V, Bruni G, Lavazza A, Buonavoglia C. 2007. Coronavirus associated with an enteric syndrome on a quail farm. Avian Pathol 36:251–258. doi: 10.1080/03079450701344738. [DOI] [PubMed] [Google Scholar]
- 12.Casais R, Dove B, Cavanagh D, Britton P. 2003. Recombinant avian infectious bronchitis virus expressing a heterologous spike gene demonstrates that the spike protein is a determinant of cell tropism. J Virol 77:9084–9089. doi: 10.1128/JVI.77.16.9084-9089.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ambepitiya Wickramasinghe IN, van Beurden SJ, Weerts EA, Verheije MH. 2014. The avian coronavirus spike protein. Virus Res 194:37–48. doi: 10.1016/j.virusres.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ambepitiya Wickramasinghe IN, de Vries RP, Eggert AM, Wandee N, de Haan CA, Gröne A, Verheije MH. 2015. Host tissue and glycan binding specificities of avian viral attachment proteins using novel avian tissue microarrays. PLoS One 10:e0128893. doi: 10.1371/journal.pone.0128893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cao J, Wu CC, Lin TL. 2008. Complete nucleotide sequence of polyprotein gene 1 and genome organization of turkey coronavirus. Virus Res 136:43–49. doi: 10.1016/j.virusres.2008.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maurel S, Toquin D, Briand FX, Queguiner M, Allee C, Bertin J, Ravillion L, Retaux C, Turblin V, Morvan H, Eterradossi N. 2011. First full-length sequences of the S gene of European isolates reveal further diversity among turkey coronaviruses. Avian Pathol 40:179–189. doi: 10.1080/03079457.2011.551936. [DOI] [PubMed] [Google Scholar]
- 17.Guionie O, Courtillon C, Allee C, Maurel S, Queguiner M, Eterradossi N. 2013. An experimental study of the survival of turkey coronavirus at room temperature and +4 degrees C. Avian Pathol 42:248–252. doi: 10.1080/03079457.2013.779364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koch G, Hartog L, Kant A, van Roozelaar DJ. 1990. Antigenic domains on the peplomer protein of avian infectious bronchitis virus: correlation with biological functions. J Gen Virol 71(Part 9):1929–1935. [DOI] [PubMed] [Google Scholar]
- 19.Koch G, Hartog L, Kant A, van Roozelaar DJ, de Boer GF. 1986. Antigenic differentiation of avian bronchitis virus variant strains employing monoclonal antibodies, p 128–139. In McFerran JB, McNulty MS (ed), Acute virus infections of poultry, vol 37 Springer Science+Business Media BV, Dordrecht, The Netherlands. [Google Scholar]
- 20.Breslin JJ, Smith LG, Barnes HJ, Guy JS. 2000. Comparison of virus isolation, immunohistochemistry, and reverse transcriptase-polymerase chain reaction procedures for detection of turkey coronavirus. Avian Dis 44:624–631. doi: 10.2307/1593102. [DOI] [PubMed] [Google Scholar]
- 21.Gomes DE, Hirata KY, Saheki K, Rosa AC, Luvizotto MC, Cardoso TC. 2010. Pathology and tissue distribution of turkey coronavirus in experimentally infected chicks and turkey poults. J Comp Pathol 143:8–13. doi: 10.1016/j.jcpa.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu W, Air GM. 2004. Binding of influenza viruses to sialic acids: reassortant viruses with A/NWS/33 hemagglutinin bind to alpha2,8-linked sialic acid. Virology 325:340–350. doi: 10.1016/j.virol.2004.05.013. [DOI] [PubMed] [Google Scholar]
- 23.Vierbuchen M, Uhlenbruck G, Ortmann M, Dufhues G, Fischer R. 1988. Occurrence and distribution of glycoconjugates in human tissues as detected by the Erythrina cristagalli lectin. J Histochem Cytochem 36:367–376. doi: 10.1177/36.4.3346539. [DOI] [PubMed] [Google Scholar]
- 24.Loris R, Hamelryck T, Bouckaert J, Wyns L. 1998. Legume lectin structure. Biochim Biophys Acta 1383:9–36. doi: 10.1016/S0167-4838(97)00182-9. [DOI] [PubMed] [Google Scholar]
- 25.Wang L, Cummings RD, Smith DF, Huflejt M, Campbell CT, Gildersleeve JC, Gerlach JQ, Kilcoyne M, Joshi L, Serna S, Reichardt NC, Parera Pera N, Pieters RJ, Eng W, Mahal LK. 2014. Cross-platform comparison of glycan microarray formats. Glycobiology 24:507–517. doi: 10.1093/glycob/cwu019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei YQ, Guo HC, Dong H, Wang HM, Xu J, Sun DH, Fang SG, Cai XP, Liu DX, Sun SQ. 2014. Development and characterization of a recombinant infectious bronchitis virus expressing the ectodomain region of S1 gene of H120 strain. Appl Microbiol Biotechnol 98:1727–1735. doi: 10.1007/s00253-013-5352-5. [DOI] [PubMed] [Google Scholar]
- 27.Hodgson T, Casais R, Dove B, Britton P, Cavanagh D. 2004. Recombinant infectious bronchitis coronavirus Beaudette with the spike protein gene of the pathogenic M41 strain remains attenuated but induces protective immunity. J Virol 78:13804–13811. doi: 10.1128/JVI.78.24.13804-13811.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y, Huang P, Jiang B, Tan M, Morrow AL, Jiang X. 2013. Poly-LacNAc as an age-specific ligand for rotavirus P[11] in neonates and infants. PLoS One 8:e78113. doi: 10.1371/journal.pone.0078113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Costa T, Chaves AJ, Valle R, Darji A, van Riel D, Kuiken T, Majo N, Ramis A. 2012. Distribution patterns of influenza virus receptors and viral attachment patterns in the respiratory and intestinal tracts of seven avian species. Vet Res 43:28. doi: 10.1186/1297-9716-43-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kimble B, Nieto GR, Perez DR. 2010. Characterization of influenza virus sialic acid receptors in minor poultry species. Virol J 7:365. doi: 10.1186/1743-422X-7-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Munier S, Larcher T, Cormier-Aline F, Soubieux D, Su B, Guigand L, Labrosse B, Cherel Y, Quere P, Marc D, Naffakh N. 2010. A genetically engineered waterfowl influenza virus with a deletion in the stalk of the neuraminidase has increased virulence for chickens. J Virol 84:940–952. doi: 10.1128/JVI.01581-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schultze B, Cavanagh D, Herrler G. 1992. Neuraminidase treatment of avian infectious bronchitis coronavirus reveals a hemagglutinating activity that is dependent on sialic acid-containing receptors on erythrocytes. Virology 189:792–794. doi: 10.1016/0042-6822(92)90608-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wagner R, Matrosovich M, Klenk HD. 2002. Functional balance between haemagglutinin and neuraminidase in influenza virus infections. Rev Med Virol 12:159–166. doi: 10.1002/rmv.352. [DOI] [PubMed] [Google Scholar]
- 34.Gomaa MH, Yoo D, Ojkic D, Barta JR. 2009. Infection with a pathogenic turkey coronavirus isolate negatively affects growth performance and intestinal morphology of young turkey poults in Canada. Avian Pathol 38:279–286. doi: 10.1080/03079450903055389. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.