ABSTRACT
Polydnaviruses form a group of unconventional double-stranded DNA (dsDNA) viruses transmitted by endoparasitic wasps during egg laying into caterpillar hosts, where viral gene expression is essential to immature wasp survival. A copy of the viral genome is present in wasp chromosomes, thus ensuring vertical transmission. Polydnaviruses comprise two taxa, Bracovirus and Ichnovirus, shown to have distinct viral ancestors whose genomes were “captured” by ancestral wasps. While evidence indicates that bracoviruses derive from a nudivirus ancestor, the identity of the ichnovirus progenitor remains unknown. In addition, ichnoviruses are found in two ichneumonid wasp subfamilies, Campopleginae and Banchinae, where they constitute morphologically and genomically different virus types. To address the question of whether these two ichnovirus subgroups have distinct ancestors, we used genomic, proteomic, and transcriptomic analyses to characterize particle proteins of the banchine Glypta fumiferanae ichnovirus and the genes encoding them. Several proteins were found to be homologous to those identified earlier for campoplegine ichnoviruses while the corresponding genes were located in clusters of the wasp genome similar to those observed previously in a campoplegine wasp. However, for the first time in a polydnavirus system, these clusters also revealed sequences encoding enzymes presumed to form the replicative machinery of the progenitor virus and observed to be overexpressed in the virogenic tissue. Homology searches pointed to nucleocytoplasmic large DNA viruses as the likely source of these genes. These data, along with an analysis of the chromosomal form of five viral genome segments, provide clear evidence for the relatedness of the banchine and campoplegine ichnovirus ancestors.
IMPORTANCE Recent work indicates that the two recognized polydnavirus taxa, Bracovirus and Ichnovirus, are derived from distinct viruses whose genomes integrated into the genomes of ancestral wasps. However, the identity of the ichnovirus ancestor is unknown, and questions remain regarding the possibility that the two described ichnovirus subgroups, banchine and campoplegine ichnoviruses, have distinct origins. Our study provides unequivocal evidence that these two ichnovirus types are derived from related viral progenitors. This suggests that morphological and genomic differences observed between the ichnovirus lineages, including features unique to banchine ichnovirus genome segments, result from evolutionary divergence either before or after their endogenization. Strikingly, analysis of selected wasp genomic regions revealed genes presumed to be part of the replicative machinery of the progenitor virus, shedding new light on the likely identity of this virus. Finally, these genes could well play a role in ichnovirus replication as they were overexpressed in the virogenic tissue.
INTRODUCTION
Wasps that lay their eggs within the body cavity of caterpillars spend their larval life as endoparasites. Feeding on caterpillar tissues culminates in host death and egression of the wasp larva, which then spins a cocoon and undergoes metamorphosis. Within the caterpillar, the wasp egg and larva must either evade or abrogate the host immune response, which would otherwise neutralize the parasitic invader through encasement within a sheath of hemocytes, a process known as encapsulation. In addition, the wasp often needs to alter the development of its host to suit its own development. To help them overcome these challenges, some parasitic wasps have domesticated a virus, which they introduce into their host during egg laying (“stinging”). These unusual mutualistic viruses, known as polydnaviruses (PDVs; family Polydnaviridae), replicate exclusively in wasp ovaries, where their segmented double-stranded DNA (dsDNA) circular genomes are generated from linear DNA copies present in the wasp chromosomes (referred to here as proviral DNA). Although no viral replication takes place in the parasitized caterpillar, viral gene expression causes alterations in host physiology, thereby enabling parasite survival and growth. Thus, successful development of the parasite ensures vertical transmission of the integrated viral genome to the next wasp generation (reviewed in reference 1).
PDVs have so far been observed in members of two wasp families, the Ichneumonidae and the Braconidae. Both morphological and genomic differences distinguish the viruses found in these two wasp taxa; as a consequence, they are considered as forming two separate taxonomic entities referred to as ichnoviruses (IVs) and bracoviruses (BVs). Within the Braconidae, BVs appear to be confined to a group of seven subfamilies that form a complex of >20,000 species, referred to as the microgastroid complex (2–4). All BV representatives examined to date display virions containing one or several cylindrical nucleocapsids enveloped by a single-unit membrane generated within the nuclei of calyx epithelial cells. These cells are located in the lateral oviducts near their junction with ovarioles. BV virions are released in the lumen of the oviduct by calyx cell lysis (5–7). Evidence has recently been presented that BV core genes derive from a nudivirus (formally classified as a baculovirus lineage [8, 9]) whose genome became integrated into the genome of an ancestor of all extant microgastroid wasps ∼100 million years ago. Nudivirus-related genes encoding BV structural proteins were found in a microgastroid wasp, in a region of its genome distinct from that harboring the proviral sequences that are packaged in BV particles (10). In support of this evolutionary scenario, related nudiviral sequences were identified in several other microgastroid wasps, either from genomic DNA or mRNA (10–12).
Within the Ichneumonidae, IVs have so far been observed in only two subfamilies, the Campopleginae and the Banchinae, which are separated by several subfamilies according to a recently published ichneumonid phylogeny (13). To date, most studies on IVs have focused on the former group, with only two banchine IVs having been characterized thus far: Glypta fumiferanae ichnovirus (GfIV) (14, 15) and Apophua simplicipes ichnovirus (AsIV) (16). Interestingly, the virus particles of the two IV groups display strikingly different morphologies: whereas those of campoplegine IVs harbor singly enveloped lenticular nucleocapsids, each banchine IV virion contains several, smaller, rod-shaped nucleocapsids; however, both display a two-unit-membrane envelope, with the outer membrane acquired by budding through the plasma membrane of calyx epithelial cells (Fig. 1). In addition, the packaged genomes of these two IV groups display major differences in terms of gene content, degree of genome segmentation and coding density. Strikingly, banchine IV genomes share more genes with BVs than with their campoplegine counterparts, featuring primarily protein tyrosine phosphatases (PTPs), ankyrins, and a family unique to banchine IVs, the NTPase-like proteins (14–16). Proteomic and genomic analyses of the wasp Hyposoter didymator and its ichnovirus, HdIV, have revealed that campoplegine IVs did not originate from a nudivirus (17). However, their likely viral ancestor has not yet been identified; while bearing a clear viral signature (absence of introns and high coding density), the clusters of genes that encode HdIV particle proteins (termed Hd-IVSPERs, for H. didymator ichnovirus structural protein encoding regions) appear to be derived from a currently unknown virus taxon. Thus, observed genomic and morphological differences between campoplegine and banchine IVs, coupled with the apparent absence of PDVs in the wasp lineages that separate the Campopleginae from the Banchinae (15), have raised the question of whether these two IV types have a common ancestor or whether the association between Banchinae and their PDVs represents a third example of virus domestication during parasitic wasp evolution.
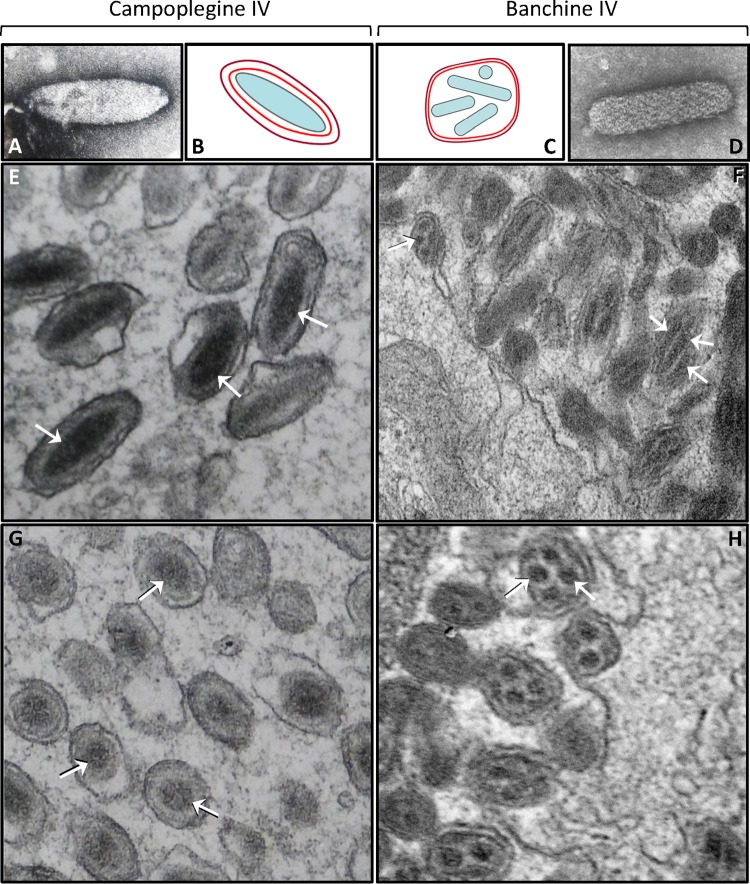
FIG 1.
Examples of campoplegine and banchine IV virions and nucleocapsids. Negatively stained nucleocapsids of Hyposoter exiguae IV (A) and Glypta fumiferanae IV (D) are shown. Diagrams show typical campoplegine (B) and banchine (C) IV virion structures; inner and outer membranes are shown in red and purple, respectively. Longitudinal (E and F) and cross (G, H) sections of representative Hyposoter fugitivus (Campopleginae) and A. simplicipes (Banchinae) virions, as seen in the lumen of the oviduct. Arrows indicate positively stained nucleocapsids; the arrow on the left side of panel F points to a particle budding from the apical surface of a calyx epithelial cell.
In an effort to shed light on this controversial issue, we conducted a proteomic analysis of GfIV virion structural proteins and characterized the wasp genomic regions encoding these proteins. In parallel, we isolated and sequenced several proviral forms of packaged GfIV genome segments in order to examine their organization and compare their putative modes of excision/circularization with those described for campoplegine IVs and BVs. Our analyses indicate that several GfIV virion structural proteins are homologous to those of campoplegine IVs and that the putative genome segment excision mechanism producing GfIV circles resembles that proposed for campoplegine IVs. In addition, for the first time in a polydnavirus system, analysis of the wasp genomic regions harboring the genes encoding virion structural proteins revealed coding sequences presumed to be part of the replicative machinery of the progenitor virus; these genes are overexpressed in the lateral oviducts and could thus play a role in the replication of the GfIV packaged genome.
MATERIALS AND METHODS
Insects.
Glypta fumiferanae wasps were obtained from Choristoneura fumiferana host larvae collected in natural stands of balsam fir and white spruce near Baie-Comeau (49°13′N, 68°09′W), Canada, during the 2010-2012 field seasons. Species identification was confirmed by G. Pelletier (Natural Resources Canada, Quebec City, Canada).
Construction of ovarian cDNA library and sequencing.
Total RNA was extracted from a pool of five ovary pairs dissected from G. fumiferanae adult females a few days after emergence, using a QIAshredder and RNeasy minikit (Qiagen) according to the manufacturer's instructions. A total of 500 ng of RNA was used to generate a cDNA library, as described previously (17, 18). About 2,500 clones were selected and sequenced from both ends at Genoscope (Centre National de Séquençage, Institut de Génomique, Evry, France).
Proteomics analyses. (i) Virus preparation and gel electrophoresis.
Ovaries of six G. fumiferanae wasps were dissected in phosphate-buffered saline (PBS) and gently teased open to release the calyx fluid. This fluid was collected with a pulled pipette, partially purified by passage through a 0.45-μm-pore-size filter, and centrifuged at 13,793 × g. The resulting virus pellet was resuspended in 20 μl of PBS and boiled with 5 μl of 5× sample buffer (0.22 M Tris-HCl, pH 6.8, 49% glycerol, 5.5% SDS, 0.005% bromophenol blue, 0.25 M dithiothreitol [DTT]). The virus preparation (10 μl) was fractionated on a 10% SDS-PAGE gel, followed by silver staining using standard procedures. Protein bands were excised from the gel and frozen at −80°C.
(ii) In-gel digestion.
Excised bands were digested using an automatic digestion robot (ProGest; Genomic Solutions) as described previously (19), with minor modifications. Briefly, the gel bands were reduced with dithiothreitol, alkylated with iodoacetamide, and then digested with trypsin. Peptides were extracted from the gel bands with 50% acetonitrile–5% formic acid (JT Baker, Phillipsburg, NJ), dried by vacuum centrifugation (SPD SpeedVac; Thermo Electron Corp., Waltham, MA), and resuspended in 30 μl of 5% acetonitrile–0.5% formic acid in high-performance liquid chromatography (HPLC)-grade water.
(iii) HPLC-MS/MS.
HPLC-tandem mass spectrometry (HPLC-MS/MS) was performed using a NanoFlow LC Packings (Amsterdam, Netherlands) system interfaced to a hybrid triple quadrupole linear ion trap (Qtrap) mass spectrometer (Applied Biosystems, Foster City, CA). Samples were injected onto an Onyx monolithic C18 capillary column (0.1 by 150 mm; Phenomenex, Torrance, CA), at a flow rate of 1.0 μl/min. Spectra were acquired using the information-dependent acquisition mode.
(iv) Data analysis.
Protein identification was performed using the Trans-Proteomic Pipeline (20). Database searching was done using the X!Tandem search engine. Peptide validation was performed using PeptideProphet, with a 0.01 false discovery rate (FDR) setting. A custom database (i.e., the ovarian expressed sequence tag [EST] library described above) was used for peptide mapping.
Fosmid library construction, screening, and sequencing.
For fosmid library construction, DNA was prepared from G. fumiferanae larvae (of undetermined sex) obtained by parasitization of prediapause second-instar C. fumiferana larvae. A pool of 99 G. fumiferanae larvae was used for DNA extraction, using Genomic-tips and a blood and cell culture midi-kit (Qiagen), according to the manufacturer's instructions. A total of 5 μg of high-molecular size (40 to 50 kb) genomic DNA was cloned into the pCC1FOS vector using a CopyControl Fosmid Library Production kit (Epicentre), according to the manufacturer's instructions. After the titers of the packaged fosmid library were determined, 70,000 clones (representing three to four times genome coverage) were used to infect EPI300-T1 cells plated onto 20, 150- by 15-mm, petri dishes (3,500 clones/plate). For each plate, colonies were resuspended in 1.5 ml of LB medium supplemented with 20% glycerol and conserved as the fractionated amplified library at −80°C.
Primer pairs were designed specifically against target sequences (see GenBank entries KP706795 to KP706803 for primer sequences) and used to perform a PCR screen of serial dilutions of the library. DNA used for amplification was prepared from a 30-μl aliquot of a 100-μl overnight culture inoculated with the number of CFU needed for a given dilution (the remaining 70 μl was kept at 4°C to be used as starting material for subsequent dilution of positive fractions). The 30-μl aliquot was first centrifuged for 1 min at maximum speed, the medium was removed, and the bacterial pellet was resuspended in 80 μl of double-distilled H2O (ddH2O). The cells were then heated at 99°C for 5 min and centrifuged for 2 min at maximum speed to pellet the cell debris. A total of 5 μl of the supernatant was directly used for PCR amplification in PCR buffer (1× final concentration) supplemented with 0.33 mM each deoxynucleoside triphosphate (dNTP), 0.25 μM each primer, and 1 unit of Taq DNA polymerase in a 30-μl reaction volume. The amplification was performed for 35 cycles consisting of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min, with a final extension step at 72°C for 5 min. Five microliters per reaction was analyzed by 1% agarose gel electrophoresis to detect positive fractions.
An initial PCR screen was carried out on each of the 20 fractions of the amplified library (3,500 CFU/fraction). Positive fractions, for a given primer pair, were diluted and divided into 10 fractions of 500 CFU each for a second PCR screen. Then, positive 500-CFU pools were further diluted and divided into eight fractions of 100 CFU each for a third PCR screen. Lastly, the positive 100-CFU fractions were plated, and single colonies were transferred to each well of a 96-well plate. After overnight incubation, row- and column-specific pools (based on the plate grid layout) were prepared by combining 5 μl of culture from each of the 8 wells in a given column (12 pools) and each of the 12 wells in a given row (8 pools), where each pool was screened by PCR. Wells containing positive clones were identified from plate coordinates following the plotting of row-versus column-specific positives.
Fosmid DNA from positive clones was extracted using a FosmidMax DNA purification kit (Epicentre) and 1 ml of a DNA pool containing equimolar amounts of 16 distinct clones prepared at a final concentration of 10 ng/μl for Roche 454 pyrosequencing, conducted at the Institut de Biologie Intégrative et des Systèmes (IBIS), Université Laval, Québec, Canada. Contigs were assembled using Newbler, version 2.8 (Roche), and analyzed using SeqManPro (Lasergene DNAStar software package). To fill remaining gaps, we used a genome-walking strategy and Sanger sequencing of PCR amplicons generated from purified fosmid clones. Complete sequences were obtained by ordering/aligning contigs and Sanger sequences using BioEdit (21).
Quantitative PCR (qPCR) transcriptional analysis.
Ovaries were dissected from newly emerged and 5- to 10-day-old cold-anesthetized virgin G. fumiferanae females in a drop of PBS, using a pair of fine forceps. The ovarioles and the lateral oviducts (the latter comprise the calyx region where virogenesis takes place) were teased apart so that the two tissues could be processed independently. RNA was extracted from tissue pooled from two to six females, using a Qiagen RNeasy minikit, with on-column DNase treatment. Each RNA sample was eluted twice with 30 μl of water. RNA was quantified using a NanoDrop 1000 spectrophotometer (Thermo Scientific). Prior to cDNA synthesis, RNA was DNase treated again, using 2 units of Invitrogen DNase I. cDNA was synthesized from 7 μl of RNA, using 100 U of Invitrogen Superscript II reverse transcriptase for each reaction volume. All cDNA samples were diluted to 5 ng/μl using 10 mM Tris, pH 8.0. Because the values obtained for transcript abundance in newly emerged females were very similar to those measured in older ones, we pooled the data across all ages examined.
Primers were designed using the Oligo Explorer program (Gene Link, Hawthorne, NY). Criteria for primer selection included a thermal denaturation midpoint temperature (Tm) of 60°C and predicted amplicon size ranging between 85 and 165 bp. Quantitative PCR was performed using an Applied Biosystems 7500 Fast Real Time PCR machine. BrightWhite real-time PCR plates (96-well; Primerdesign, United Kingdom) were used with Applied Biosystems MicroAmp Optical Adhesive Film. Fifty PCR cycles of 95°C for 15 s, 60°C for 30 s, and 65°C for 90 s were performed using a Qiagen Quantitect SYBR green PCR kit. Two technical replicates were amplified for each sample. Quantification was initially done using 2.5 ng of cDNA per well; however, some samples showed baseline drift, requiring further dilution to 100 pg or less to eliminate the drift. Transcript levels were measured for 24 target genes (see GenBank entries KP706795 to KP706799 for primer sequences) and two housekeeping genes (sequences were obtained from the ovarian EST library; primer sequences: β-tubulin1-F, GCTTCCGTACCCTTAAATTATCCA; β-tubulin1-R, GCGCAGATCGGCGTTCAAT; EF1g-F, CGTTTGGAGAAAATGAGAAAGGCAG; EF1g-R, CTTAGAGTGAAAGCGAGGTCTTGA). To confirm the specificity of reaction, a melting curve analysis was carried out at the end of the PCR run. Linear regression of efficiency (LRE) analysis developed for modeling qPCR amplification (22–25) was used to determine absolute quantities of target molecules. Lambda genomic DNA was used as a quantitative standard.
Nucleotide sequence accession numbers.
All G. fumiferanae sequences reported here have been deposited in the GenBank database under accession numbers KP706795 to KP706803.
RESULTS
Proteomic analysis shows that banchine and campoplegine IVs share virion structural proteins.
Electrophoretic fractionation of a filter-purified preparation of GfIV virions revealed a complex mixture of proteins of various mobilities (Fig. 2). Six of the most prominent and well-resolved bands were excised from the gel and submitted to LC-MS/MS analysis, using a G. fumiferanae ovarian EST library as a reference to identify the proteins. Proteins 1 to 5 could be unequivocally identified using this approach (i.e., each had a matching EST). Whereas proteins 1, 2, 3, and 4 were found to be homologs of virion structural proteins identified in HdIV (Table 1), protein 5 displayed significant similarity to the baculoviral protein p26 (26) (i.e., presence of a baculo_p26/pfam04766 domain; E value of 6E−32 for p26 of Agrotis segetum nucleopolyhedrovirus [AsNPV]). Protein 6 contained two peptide fragments identical to fragments identified in protein 4, but a sequence matching the size of the predicted protein could not be found in the ovarian EST library, suggesting that it may be a truncated version of protein 4 or a homolog thereof with no EST match.
FIG 2.
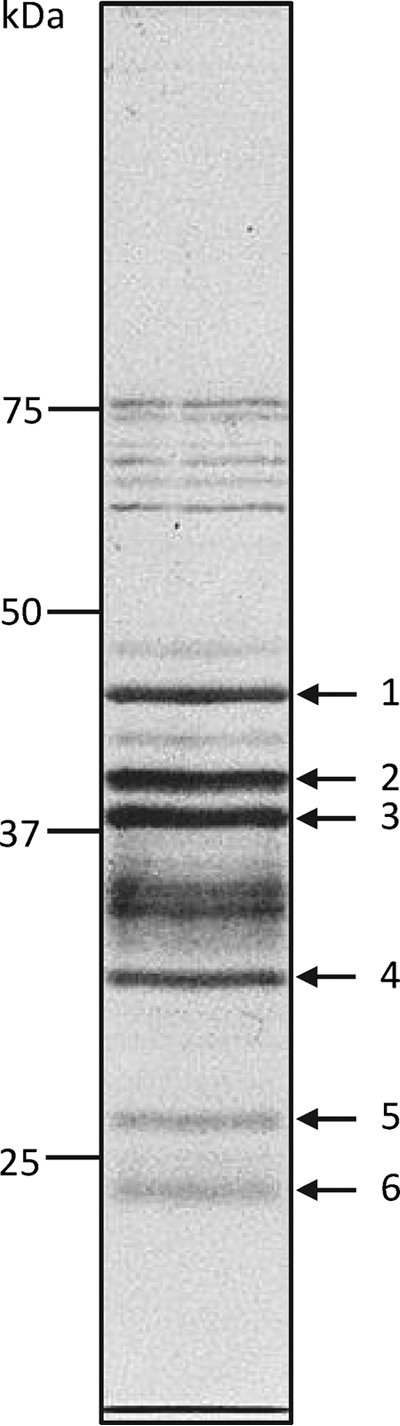
SDS-PAGE fractionation of GfIV virion proteins. Molecular mass markers (kDa) are shown on the left. The protein bands (1 to 6) that were excised from the gel for proteomics analysis are shown on the right; the remaining bands were not submitted to proteomics analysis.
TABLE 1.
Glypta fumiferanae homologs of Hyposoter didymator IVSPER genes, as determined by blastp searches
| G. fumiferanae CDS | Protein no.a | Match in EST library | H. didymator CDS | GenBank accession no. | E value |
|---|---|---|---|---|---|
| Gf-IVSP1La | 4 | IVSP1-2 | ADI40464 | 2E−20 | |
| Gf-IVSP1Lb | IVSP1-2 | ADI40464 | 7E−32 | ||
| Gf-IVSP2La | IVSP2-1 | ADI40459 | 4E−59 | ||
| Gf-IVSP2Lb | Yes | IVSP2-1 | ADI40459 | 5E−49 | |
| Gf-IVSP3La | IVSP3-1 | ADI40465 | 1E−138 | ||
| Gf-IVSP3Lb | Yes | IVSP3-1 | ADI40465 | 2E−116 | |
| Gf-IVSP4L | 1 | IVSP4-2 | ADI40484 | 1E−114 | |
| Gf-NLa | N-2 | ADI40491 | 6E−79 | ||
| Gf-NLb | N-2 | ADI40491 | 2E−80 | ||
| Gf-P12L | P12-like 1 | CAR31591 | NDb | ||
| Gf-P53L | P53-like 1 | CAR31590 | 2E−26 | ||
| Gf-U1L | Yes | U1 | ADI40452 | 3E−47 | |
| Gf-U3L | U3 | ADI40455 | 2E−29 | ||
| Gf-U4L | U4 | ADI40456 | 4E−13 | ||
| Gf-U5L | U5 | ADI40458 | 3E−38 | ||
| Gf-U6L | Yes | U6 | ADI40462 | 2E−88 | |
| Gf-U8L | U8 | ADI40466 | 7.6 | ||
| Gf-U9L | U9 | ADI40467 | 0.25 | ||
| Gf-U13L | Yesc | U13 | ADI40471 | 2E−05 | |
| Gf-U15L | U15 | ADI40477 | 1E−89 | ||
| Gf-U16L | U16 | ADI40479 | 7E−177 | ||
| Gf-U17L | Yesc | U17 | ADI40480 | 1E−05 | |
| Gf-U19L | U19 | ADI40483 | 3E−109 | ||
| Gf-U22L | U22 | ADI40487 | 7E−30 | ||
| Gf-U23La | 3 | Yes | U23 | ADI40488 | 5E−79 |
| Gf-U23Lb | 2 | Yes | U23 | ADI40488 | 4E−64 |
| Gf-U24L | U24 | ADI40490 | 8E−97 |
Numbering is according to Fig. 2.
Although the blastp algorithm found no detectable (ND) similarity between Gf-P12L and H. didymator (Hd)-P12-like 1, it did identify C. sonorensis P12 (AAD01200) as a homolog (E value = 5E−06), which is a homolog of Hd-P12-like 1 (E value = 6E−07).
These are on the same EST.
G. fumiferanae fosmid clones contain genomic regions related to H. didymator IVSPERs.
To characterize the G. fumiferanae genomic regions containing the genes coding for the proteins identified by proteomic analysis, we constructed a fosmid library that was then screened by PCR using primer pairs designed against ESTs matching the proteins identified above as well as against additional HdIV-related sequences found in the G. fumiferanae ovarian EST library; positive clones were then sequenced.
We detected 27 G. fumiferanae coding DNA sequences (CDSs) homologous to coding regions identified earlier in H. didymator IVSPERs (17) (Table 1 and Fig. 3). These CDSs were contained within three genomic regions referred to here as Gf-IVSPER1, -2, and -3 (Fig. 3); whereas Gf-IVSPER1 and -2 displayed a relatively high density of coding sequences, all devoid of introns, putative CDSs were more sparsely distributed in Gf-IVSPER3, which contained only three sequences related to H. didymator IVSPER genes. Interestingly, none of these three genomic regions contained CDSs that could be clearly identified as coding for insect proteins (typically containing several exons) although the deduced product of (single-exon) U44 on Gf-IVSPER3 (Fig. 3) had an uncharacterized hymenopteran protein as its closest homolog (E value, 1E−22). This lack of typical wasp genes suggests that the Gf-IVSPERs we identified may extend beyond the clones sequenced here (40 to 45 kb).
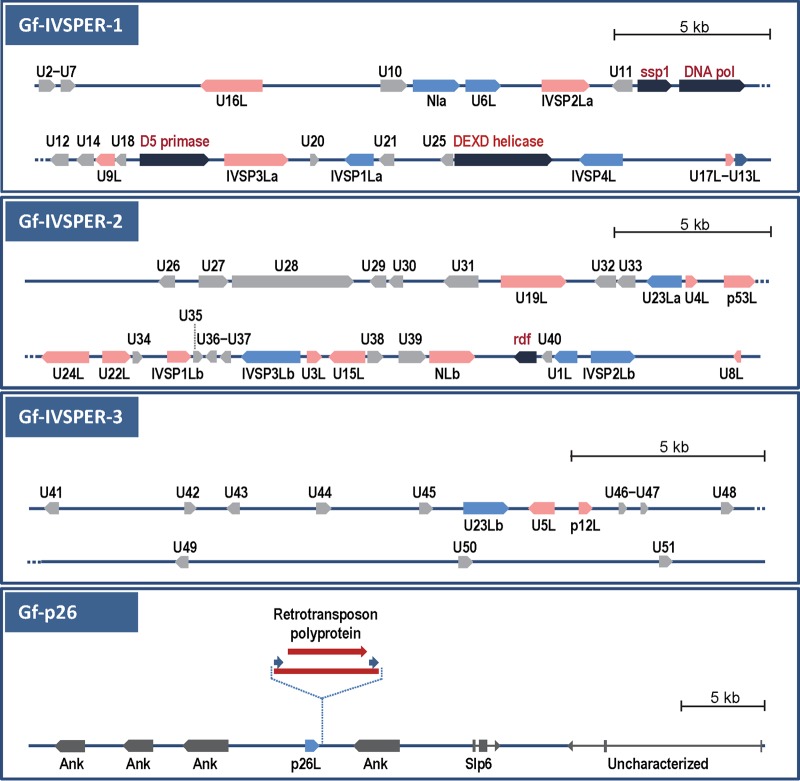
FIG 3.
Schematic representation of G. fumiferanae genomic regions that were sequenced and annotated following PCR screening of a G. fumiferanae fosmid library using primers targeting putative GfIV structural proteins identified through proteomic/EST library analysis. Each region shown here represents individual or overlapping fosmid clones. The top three panels show genomic regions that contain CDSs displaying homology to CDSs identified in H. didymator IVSPERs (17); these regions are designated Gf-IVSPER1, Gf-IVSPER2, and Gf-IVSPER3. Black arrows, replicative machinery genes from a putative NCLDV ancestor; pink and blue arrows, CDSs that display significant similarity to known genes identified in H. didymator IVSPERs (blue arrows designate CDSs that were targeted for PCR screening of the fosmid library); gray arrows, unknown proteins apparently unique to Gf-IVSPERs. The names of CDSs that have homologs in H. didymator contain an “L” (“like”) at the end (e.g., U16L is a homolog of H. didymator U16). A hyphen between two CDS names (e.g., U2-U7) indicates that both were found on a single transcript in the EST library. As a rule, only CDSs encoding ≥100 amino acids are shown here; however, smaller ones that had a match in the EST library or in H. didymator were also included. Among U proteins, some displayed significant similarity to one another: U10 and U39 (7E−56), U14 and U26 (9E−38), U29 and U31 (5E−57), U30 and U31 (2E−37), U32 and U33 (3E−29), and U23La and U23Lb (3E−54). The bottom panel shows the genomic region that contains the baculoviral p26-like CDS; none of the neighboring sequences display similarity to H. didymator IVSPER genes, and all appear to be insect genes: Gf-slp6 and Gf-uncharacterized have introns (3 and 2, respectively), and the closest homologs of the Ank repeats are from the wasp Nasonia vitripennis, with E values ranging from E−70 to E−154. Two p26-positive fosmid clones were selected for sequencing, and one of them contained a retrotransposon (red arrow) immediately downstream from the p26L CDS.
These Gf-IVSPERs contained homologs of most H. didymator IVSPER genes (recognized by the presence of an “L” at the end of the names), including the IVSP1, -2, -3, and -4 gene families, the N family, the p12 and p53 virion structural proteins identified earlier in Campoletis sonorensis (27, 28), and 16 unidentified (U) genes, two of which appear to be abundant GfIV virion structural proteins (proteins 2 and 3 in Fig. 2, designated G. fumiferanae U23Lb [Gf-U23Lb] and Gf-U23La, respectively, in Table 1). Comparison of G. fumiferanae and H. didymator IVSPER-encoded proteins indicated that, with a few exceptions, orthologous pairs displayed highly significant E values (14 protein pairs had E values ranging between E−50 and E−177) (Table 1), clearly establishing their relatedness. Little or no synteny was detected between Hd- and Gf-IVSPERs, except for three CDSs on Gf-IVSPER2 (U23La, p53L, and U24L) that were in the same order and orientation as their homologs in Hd-IVSPER3 (Fig. 3) (17). The Gf-IVSPERs contained 26 additional putative CDSs for which no H. didymator homologs could be detected in available databases and whose products (named with a U prefix) displayed very limited similarity to other known proteins.
It is noteworthy that the G. fumiferanae homolog of the baculoviral p26 protein (Fig. 2, protein 5) was absent from the three Gf-IVSPERs described above but was found on a distinct fosmid clone, in the midst of CDSs encoding insect proteins, including ankyrin repeat-containing proteins (Fig. 3).
Gf-IVSPERs contain genes from a viral replicative machinery.
Most interestingly, the Gf-IVSPER1 also contained three specific CDSs belonging to a set of genes conserved among DNA viruses known as nucleocytoplasmic large DNA viruses (NCLDVs). These genes encode proteins containing conserved domains, namely, a DNA polymerase, a D5 primase-helicase, and a DEXD helicase (Fig. 3, black arrows in Gf-IVSPER1 diagram). The D5-primase-helicase-like CDS was unequivocally identified as a viral gene in a blastp search (i.e., all matches were viral), with the closest homolog recognized as a member of the Iridoviridae (Table 2). This protein is related to the NTPase-like proteins identified earlier in the GfIV encapsidated genome (14) but displayed very limited similarity to these enzymes (data not shown). Although the closest homologs of the proteins encoded by the DNA-polymerase (Pol)- and DEXD-helicase-like CDSs were not viral, according to blastp searches, their closest viral homologs were both from the Phycodnaviridae (Table 2), another NCLDV family. Two other CDSs, a RING finger domain gene (rdf), on Gf-IVSPER2, and a sentrin-specific peptidase (ssp1), on Gf-IVSPER1, are also suspected of being of viral origin, given that their closest viral homologs were also found to be phycodnaviral sequences, albeit at a low level of significance (Table 2).
TABLE 2.
Identification of closest homologs to putative GfIV core genes in both the full NCBI NR and virus databasesa
| CDS name | Domain IDb | Domain E value | ID in full NR database |
ID in virus database |
||||
|---|---|---|---|---|---|---|---|---|
| Best hit (taxon) | GenBank accession no. | E value | Best hit (family) | GenBank accession no. | E value | |||
| DNA Pol | POLBc SF | 7.0E−18 | Bacteria | WP_048203595 | 9E−12 | Phycodnaviridae | AGE53060 | 3E−09 |
| D5 primase-helicase | Primase C-term F | 5.5E−06 | Virus | YP_009046717 | 1E−10 | Iridoviridae | YP_009046717 | 2E−12 |
| DEXDc helicase | DEAD-like helicase SF | 4.0E−07 | Fungus | XP_001727986 | 0.004 | Phycodnaviridae | AGE55615 | 0.023 |
| RDF | RING SF | 1.2E−03 | Mammal | XP_008827864 | 5E−04 | Phycodnaviridae | AFK66233 | 0.013 |
| SSP1 | Peptidase C48 SF | 2.6E−08 | Fungus | ESA17555 | 6E−06 | Phycodnaviridae | AAR26952 | 0.54 |
Databases were accessed in April 2015. NR, nonredundant.
ID, identification; SF, superfamily; F, family.
The results of the above analyses raised questions as to whether the products of the other Gf-IVSPER CDSs may show distant phylogenetic relationships with proteins from NCLDVs or other dsDNA viruses. This was shown not to be the case since blastp analyses against the NCBI virus database, using these other Gf-IVSPER CDSs as queries, yielded only very marginal matches distributed among >12 virus taxa (data not shown).
Gf-IVSPER genes are specifically transcribed in the GfIV-producing tissue.
To determine whether CDSs identified in the IVSPER and Gf-p26 genomic regions (Fig. 3) were specifically expressed in the tissue where GfIV particles are produced (the calyx region of the lateral oviduct), lateral oviducts and ovarioles were dissected from adult females. Each tissue was then processed independently for RNA extraction and qPCR quantification of transcripts using gene-specific primers. On average, transcript abundance for the 24 genes examined was 164 times higher (range, 104 to 228 times) in the lateral oviduct than in the ovarioles (Fig. 4).
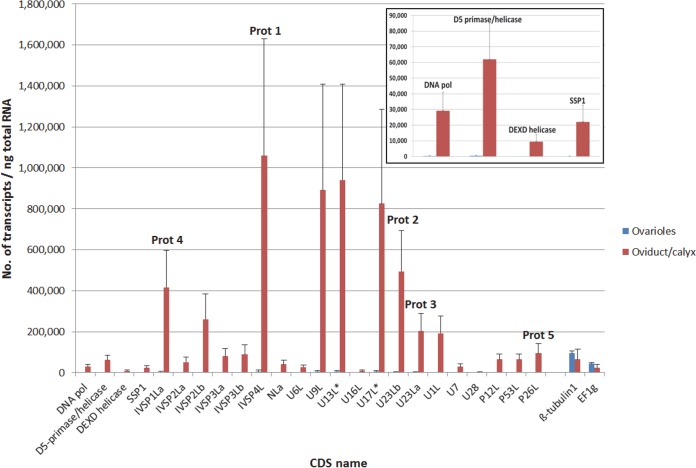
FIG 4.
qPCR assessment of transcript abundance for a subset of genes, shown in Fig. 3, in G. fumiferanae ovarioles and lateral oviducts, which contain the calyx region where GfIV virions are produced. Values presented here are the means + standard deviations of four biological replicates, each containing tissue dissected from two to six individual female wasps of various ages. Absolute transcript amounts were assessed using the LRE analyzer (22–24) and normalized against values obtained for two housekeeping genes (β-tubulin 1 and EF1g; absolute transcript amounts for these two genes are shown at the right end of the histogram) using the geNorm algorithm (48). Five transcripts that correspond to proteins identified through proteomic analysis (Fig. 2) are labeled Prot 1 to Prot 5. CDSs U13L* and U17L* were found on a single transcript in the EST library. The inset shows transcript abundance for four of the five genes presumed to be part of the replicative machinery of the progenitor virus, shown on a different scale.
Transcripts corresponding to proteins 1 to 4 identified on the gel (Fig. 2) were among the most abundant in lateral oviducts, with levels above 200,000 copies/ng total RNA. Other transcripts that were very abundant included those corresponding to Gf-IVSP2Lb, Gf-U9L, Gf-13L, and Gf-17L. The last two appeared to be bicistronic (i.e., the two CDSs were found on a single EST) (Fig. 3); accordingly, levels measured using primer pairs targeting each of these two CDSs generated similar values (Fig. 4). Protein 5 (p26L), whose coding region was found outside the three IVSPERs described above, was clearly overexpressed in the lateral oviducts. Interestingly, four of the putative replicative machinery genes identified in Gf-IVSPER1 were also overexpressed in lateral oviducts (average of 128-fold relative to ovarioles) although their absolute transcript levels were lower than those of the other genes examined (Fig. 4, inset); this observation is consistent with the hypothesis of a role in DNA replication, which requires smaller amounts of proteins than for the production of virion structural components.
Banchine IV genome segments share an ∼260-bp region.
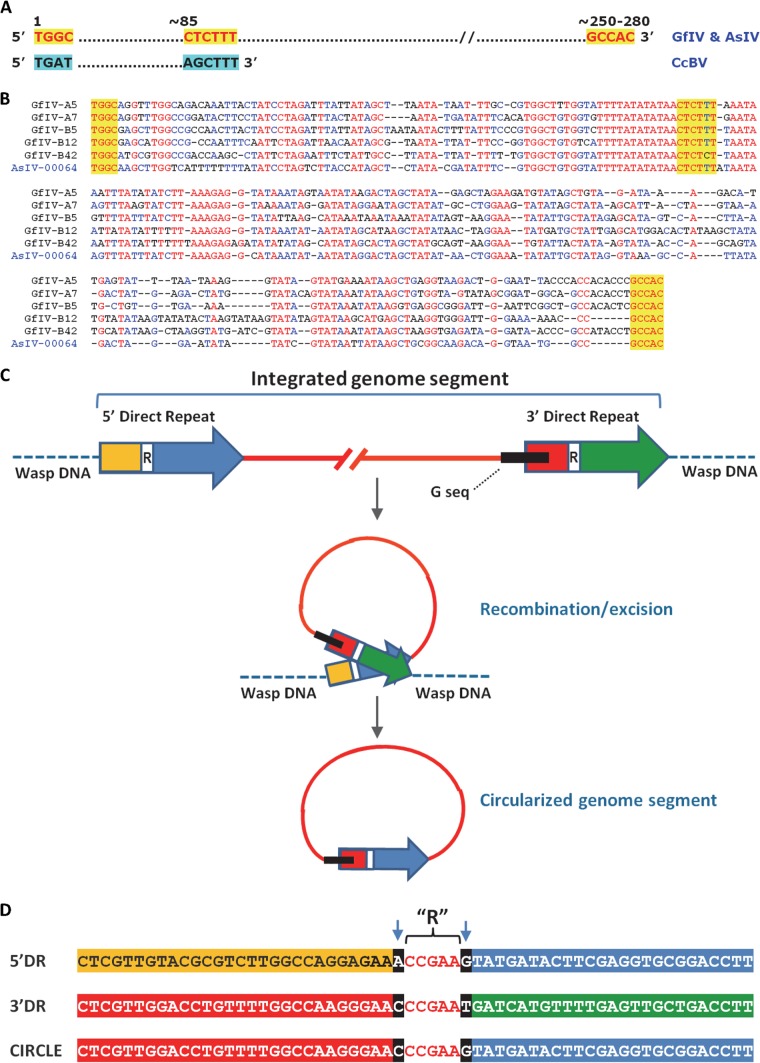
In an effort to determine if the GfIV and AsIV genome segments reported in earlier studies (14, 16) displayed sequence features unique to banchine IVs, we undertook their further characterization through blastn analyses. This endeavor led to the identification of a conserved region of ∼260 bp present on all banchine IV circles analyzed. This sequence, referred to here as the G sequence, was present as a single copy in each segment, except for segment E1 (the largest), which contained two copies. To our knowledge, similar conserved regions have not been found among campoplegine IV genome segments, but a portion of the GfIV/AsIV ∼260-bp conserved region displayed similarity with the region upstream of the BV direct repeat junction ([DRJ] AGCTTT) (29–31), raising questions about their origin and their possible involvement as DRJs in processing of proviral segments after amplification, as hypothesized for BVs (32, 33) (Fig. 5A and B).
FIG 5.
G sequences and direct repeats within chromosomal copies of GfIV genome segments and their proposed role in genome segment excision/circularization. (A) Three recognizable motifs (shaded in yellow) within an ∼250- to 280-bp region (G sequence) conserved among packaged GfIV and AsIV genome segments are shown; below are shown BV motifs resembling those found in GfIV and AsIV, including the DRJ motif, AGCTTT. (B) Alignment of the G sequences of the five GfIV genome segments examined here. A sample from AsIV is also included for comparison. Sequences shaded in yellow are those highlighted in panel A. (C) Schematic representation of the direct repeats (DRs) and associated G sequence found in proviral homologs of GfIV genome segments, along with a model for their involvement in segment excision/circularization. The DRs are represented by an arrow at each end of the integrated segment. Each DR contains a recombination region (indicated by an R and white box) whose position varies among segments (Table 3). The G sequence is represented by a black box. For genome segment excision, the DRs come in close proximity to allow recombination through the recombination regions; following recombination and excision, the circle contains a single, hybrid DR whose 3′ and 5′ ends come from the 5′ DR and the 3′ DR of the proviral genome segment, respectively. The exact role of the G sequence in this process, if any, is unclear at this stage. (D) Identification of the R region, using genome segment A7 as an example. The full 5′ and 3′ DRs are 313 and 312 bp long, respectively (Table 3); only their central portion is shown here. The R region was identified by aligning the two DRs and the homologous portion of the corresponding circle. The R region displayed perfect sequence identity among the three aligned sequences, while sequence variation was observed on either side of it (arrows). This sequence variation enabled identification of DR regions retained in the circle following recombination (the color scheme is the same as that shown in panel C).
Sequences of junctions between GfIV genome segments and other wasp DNA segments are variable.
To address the above question, we next examined chromosomal sequences located at the junctions between proviral sequences and flanking nonpackaged wasp DNA. PCR screening of the fosmid library with primer pairs targeting specific packaged GfIV circles led to the identification of positive clones bearing proviral homologs of five genome segments in total. None of the proviral sequences harbored IVSPER-like genes, and none appeared to be part of a dense cluster of proviral segments, as observed for subsets of BV proviral DNAs (12, 31, 34), with three observed on distinct ∼40- to 60-kb clones and two found on the same clone but separated by ∼20 kb of wasp DNA. All five proviral segments contained the same sets of CDSs as those identified earlier in the encapsidated DNA circles (14, 16).
Unlike what has been observed for BV genome segments, where a common DRJ sequence is present at the chromosomal wasp-virus junction in all cases (31), the sequences we found at the wasp-viral junctions of G. fumiferanae proviral segments differed from one segment to another, and these junctions were generally separated from the G sequence by ∼100 to 200 bp (Table 3), indicating that the G sequence does not act as a DRJ. However, in the five GfIV proviral segments examined, the G sequence was always positioned near the same end although the distance between this sequence and the end of the viral genome segment varied slightly (Fig. 5C), suggesting that the G sequence might, nonetheless, be involved in the excision/circularization mechanism.
TABLE 3.
Features of direct repeats within five GfIV proviral genome segments
| Proviral genome segment | 5′ DR size (bp) | 3′ DR size (bp) | Recombination regiona | G sequence size (bp) | G-R distance (bp)b |
|---|---|---|---|---|---|
| A5 | 15 | 15c | AGCT | 248 | 173 |
| A7 | 313 | 312 | CCGAA | 251 | 77 |
| B5 | 198 | 197 | CCATTTTGAGATTGT | 252 | 93 |
| B12 | 194 | 199 | ATGT | 270 | 227 |
| B42 | 248 | 235 | GGTATCTACAAC | 268 | 115 |
Region near the center of DRs and common to both DRs within the proviral copy of a given genome segment. Comparison of 5′ and 3′ DRs shows sequence variation on each side of the recombination region (R) (Fig. 5C and D).
Distance between the C residue of the GCCAC motif at the end of the G sequence (Fig. 5A and B) and the middle of the recombination motif.
The A5 proviral sequence was incomplete at its 3′ end; the size of the 3′ DR was thus estimated by reconstituting the missing portion from the sequence of circle A5.
A thorough analysis and comparison of the five GfIV proviral segments led to the identification of segment-specific terminal, duplicated regions in the same orientation and to the development of a model for the involvement of these direct repeats (DRs) in genome segment excision and circularization (Fig. 5C). The 5′ and 3′ DRs of each proviral segment showed ∼80% conservation relative to one another but displayed no significant similarity to DRs of other segments. In addition, we found no DR-like sequences other than the two terminal ones in any of the genome segments. The G sequence either overlapped the 3′ DR (segments A7, B5, and B42) or was located adjacent to it (segments A5 and B12) (Table 3). Within each direct repeat, we identified a region (R) that, to all appearances, is involved in performing recombination during excision and circularization of the genome segments; the R sequence (Fig. 5C, white boxes, and D) and its position within the direct repeats varied from one genome segment to another (Table 3). The 3′ portion of the repeats upstream of R and the 5′ end downstream of R are represented in different colors in Fig. 5C.
A comparison of the five genome segments and their proviral homologs led us to propose the following scenario for genome segment excision and circularization: (i) the direct repeats come in close proximity to allow recombination through the R sequences (Table 3); (ii) following recombination and excision, the DNA circle contains a single, hybrid direct repeat whose 3′ end comes from the 5′ direct repeat of the proviral sequence, while its 5′ end comes from the 3′ direct repeat (Fig. 5C). The exact role of the G sequence in this process, if any, remains unclear at this stage.
DISCUSSION
Do banchine and campoplegine IVs have a common origin?
The present study shows very clearly that the genome of G. fumiferanae contains specific regions rich in single-exon genes, many of which are homologous to genes identified earlier in the H. didymator wasp genome, within regions referred to as IVSPERs (Fig. 3 and Table 1) (17). In the case of the latter species, proteomic analysis of virion structural proteins showed that at least 26 out of 40 Hd-IVSPER genes code for proteins found in HdIV particles (17, 35). Conspicuous among these were members of the IVSP1 and IVSP4 families, as well as the U23 protein, homologs of which were identified here as abundant GfIV structural components (Fig. 2; Table 1). Given that multiple Gf-IVSPER genes are overexpressed in G. fumiferanae lateral oviducts (Fig. 4), where virogenesis takes place, an exhaustive proteomic analysis of GfIV virion proteins would likely yield several additional structural proteins common to the two viruses. Thus, on the basis of similarities in the virion structural protein machinery of GfIV and HdIV, it is reasonable to conclude that banchine and campoplegine IVs have either the same ancestor (a conventional virus) or two closely related ones. However, it is not possible at this time to determine whether the two IV lineages arose from a unique event of viral genome integration into the genome of an ancestor of both wasp subfamilies, in a process similar to that proposed for all BV-bearing wasps (10), or whether they are the result of two distinct integration events involving very similar viral entities (Fig. 6). Given that the 10 genes within the nudiviral clusters of two microgastrine BV-carrying wasps showed perfect synteny (12), the near absence of synteny observed between the IVSPERs of G. fumiferanae and H. didymator could be interpreted as pointing to distinct ancestors/integration events. On the other hand, the microsynteny detected in a small region of Gf-IVSPER2 could provide arguments for the alternative hypothesis, particularly in view of the greater phylogenetic distance between banchine and campoplegine wasps than among species of the Microgastrinae subfamily (14). The presence of key NCLDV viral genes in Gf-IVSPERs and their absence from those of H. didymator could likewise be viewed as suggesting that the two IV lineages arose independently. However, such genes, in H. didymator, could simply have escaped detection in earlier analyses (17). Lastly, although banchine genome segments display features not seen in their campoplegine counterparts, the mode of proviral segment excision appears similar for the two IV lineages (see detailed discussion below). As already pointed out, the Banchinae are phylogenetically separated from the Campopleginae by several subfamilies for which the presence of polydnaviruses has not been reported to date (15). Sequencing the genomes of wasps belonging to these subfamilies would likely shed some light on this issue. For example, should the genomes of species within these intervening subfamilies be found to be devoid of traces of IVSPERs, it may indicate that the two IV lineages are the products of two independent viral genome integration events. Certainly, there exists a precedent for the independent acquisition of closely related viral genomes by distinct groups of insects: a microgastroid wasp (i.e., BVs) (10) and a planthopper (36) both captured a nudivirus genome, indicating that distinct integration events of related viral genomes in different insect species are possible.
FIG 6.
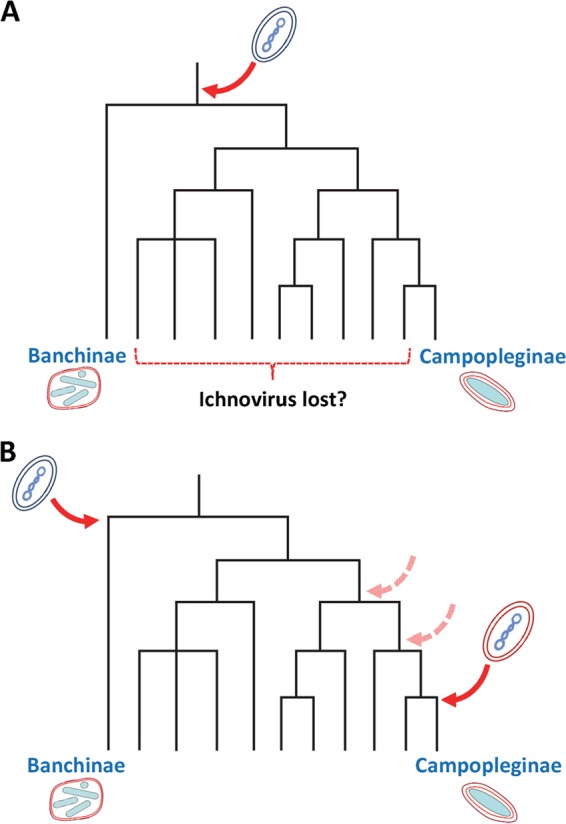
Two hypothetical evolutionary scenarios to explain the homology between banchine and campoplegine IVSPERs (15). (A) Single IV ancestor. The dsDNA genome of a putative NCLDV integrated (red arrow) into the genome of an ancestor of banchine and campoplegine wasps; following genome integration, IVs of banchine and campoplegine wasps took different evolutionary courses, resulting in virions with distinct morphological and genomic features. To date, no PDV has been observed in wasps belonging to the intervening subfamilies; if this absence were to be confirmed, the present scenario would imply a loss of the capacity to produce PDV particles in these subfamilies although traces of IVSPERs could remain in their genomes. (B) Independent acquisition of closely related IV ancestors. Here, two separate integration events involving two very similar but distinct putative NCLDVs took place in ancestors of banchine and campoplegine wasps. Note that in this scenario, the integration event depicted on the right-hand side could have taken place in a more basal ancestor (dashed arrows).
Interestingly, the coding sequence of one of the proteins identified as a GfIV particle component through proteomic analysis, a homolog of the baculoviral p26 (Fig. 2, protein 5), was not found within the Gf-IVSPERs. Rather, it was detected in a distinct region of the G. fumiferanae genome that contained typical insect CDSs (Fig. 3) but yet was overexpressed in lateral oviducts (Fig. 4). Given that the other Gf-IVSPER-encoded proteins display no striking homology with baculovirus proteins, p26 is likely the result of a horizontal gene transfer event that bears no connection with the integration of the putative viral progenitor of GfIV. Interestingly, proteomic analysis of HdIV particle proteins did not reveal the presence of a p26 homolog (17), pointing to another potential difference between campoplegine and banchine IVs, although this feature could also be unique to GfIV. The role of p26 in baculovirus replication is not entirely clear, but it appears to be required, along with p10 and p74, for optimal occlusion of virions (37). Thus, the significance of the presence of a p26-like protein in GfIV particles is unknown. Similarly, the mechanism through which its expression is coordinated with that of Gf-IVSPER genes needs to be examined. In H. didymator, the IVSPERs were observed to be amplified in calyx cells during virogenesis, a phenomenon that likely contributed to the overexpression of Hd-IVSPER genes in this tissue (17). Whether the Gf-IVSPER and Gf-p26 genomic regions are similarly amplified during GfIV particle production is currently unknown. In Microplitis demolitor, some of the nudivirus-like genes involved in BV virion production are likewise located well outside the main nudiviral cluster (12) and are clearly overexpressed in ovaries (11), where a nudivirus-related RNA polymerase has been shown to provide transcriptional specificity through promoter recognition (10, 12, 33).
Genes of the viral replicative machinery: functions and origins.
The present study provides the first example of CDSs related to replication-associated genes of viral origin found in wasp DNA within a cluster of genes involved in polydnavirus particle production. A sequence encoding a putative nudivirus-like helicase was also found in the genome of the BV-carrying wasp M. demolitor, but this gene was located outside the main nudiviral cluster (12). Whether the products of the G. fumiferanae genes referred to here are active enzymes that play a role in GfIV replication is not known at this point. Certainly, the four that we examined in our transcriptomic analysis (DNA Pol, D5 primase-helicase, DEXD helicase, and ssp1) are all significantly overexpressed in lateral oviducts (Fig. 4), suggesting that they may play a role in virogenesis. In support of this hypothesis, the products of these genes are of appropriate size in comparison with their respective viral homologs (i.e., absence of stop codons, which could have been introduced following a loss of function), and the relevant domain was identified in each of them (Table 2), albeit at a modest level of significance. In addition, a blast search against the Protein Data Bank (PDB blast) indicated that the DNA Pol sequence identified here contains the motifs corresponding to the palm, thumb, and exonuclease domains of DNA Pol, required to accommodate the DNA molecule. An RNA interference (RNAi) strategy targeting these genes before the onset of GfIV replication might give clues as to their role, if any, in particle production.
Our discovery of replication-associated genes provided new elements to address the issue of what the IV progenitor might be. Given that homologs of IV particle proteins could not be found in public databases, presumably because members of the ancestral virus taxon have not yet been submitted to genomic analyses, or else constitute an extinct group, we did not expect the core GfIV genes identified here to point to an unequivocal ancestral candidate. However, blastp searches using the protein products of these genes as queries suggested a relatedness to proteins encoded by nucleocytoplasmic large DNA viruses (NCLDVs), which form a monophyletic group encompassing seven families: Poxviridae, Asfarviridae, Iridoviridae, Ascoviridae, Phycodnaviridae, Mimiviridae, and the new family Marseilleviridae (38–40). Thus, although not clearly related to any of these families, the IV ancestor may be expected to belong to an as yet undescribed NCLDV family. Efforts to shed additional light on the relatedness of Gf-IVSPER replicative enzymes to their dsDNA viral homologs through phylogenetic analyses have so far proven inconclusive (i.e., phylogenetic positioning of Gf-IVSPER proteins highly sensitive to analytical parameters [data not shown]), likely as a result of the limited number of homologous sites between Gf-IVSPER proteins and their respective homologs in other taxa. Interestingly, a recent review on the evolution of viruses of eukaryotes tentatively placed the polydnaviruses, along with other dsDNA viruses of arthropods, as highly derived offshoots of the NCLDVs (41).
In view of the limited phylogenetic signal obtained for the five sequences examined here (Table 2), the fact that the best blastp matches were not always from viruses was not completely unexpected as many viral genes have been acquired from prokaryotic or eukaryotic hosts. Central to our conclusion that these CDSs are of viral origin, however, are the following observations: (i) each of the five CDSs contained a single exon; (ii) one of them (the D5 primase/helicase) generated only viral (NCLDV) hits when used as a query in a blastp search (top hit, Iridoviridae); (iii) the best viral hits for the remaining four CDSs were all from the Phycodnaviridae, a family that displays a relatively close phylogenetic relationship to the Iridoviridae (42); and (iv) four of these sequences were found in relatively close proximity to one another, within a genomic region (Gf-IVSPER-1) (Fig. 3) presumed to result from the endogenization of the progenitor viral genome. The observation that the best match for the D5 primase-helicase was not a phycodnaviral sequence does not contradict the hypothesis of a common origin for these genes as the D5 primase-helicase in phycodnaviruses has apparently been displaced by a bacteriophage homolog (38).
On the basis of a striking similarity discovered between a family of protein-coding genes found in the encapsidated genome of GfIV, the NTPase-like family (14), and an ascoviral gene, it has been suggested that IVs are derived from an ascoviral ancestor (43). However, the data presented here as well as results presented earlier for campoplegine IVs (17) tend to rule out this scenario, given that none of the IV virion structural proteins bear an ascoviral signature. The alternative hypothesis, that banchine IVs acquired the NTPase-like gene from an ascovirus through a horizontal gene transfer event (15), is further supported by the present work. Indeed, while the D5-primase-helicase-like protein identified in this study and the GfIV NTPase-like proteins are members of the same enzyme family, they display marginal similarity to one another, whereas GfIV NTPase-like proteins and their ascoviral homologs show a high degree of similarity (15, 16). It should be pointed out, however, that with the Ascoviridae being one of the families forming the monophyletic NCLDV group, it is not unreasonable to expect the IV ancestor to share at least some features with this group of viruses.
Proviral copies of GfIV genome segments display both shared and unique features relative to campoplegine IVs.
In an effort to further characterize differences and similarities between banchine and campoplegine IVs, we examined wasp proviral homologs of a sample of GfIV genome segments. Although only two campoplegine IV sequences of this type have been described in the literature (44, 45), comparison of these with the data presented here suggests that the two IV lineages use similar excision/circularization mechanisms while displaying some differences relative to BVs (29–31). Judging from the available data, proviral copies of genome segments in both IV groups contain direct repeats (DRs) at their 5′ and 3′ ends. These DRs show a high degree of similarity to one another within a segment but, unlike what has been shown for BVs, differ both in sequence and size among segments, and DRs appear to be involved in a mechanism of recombination leading to segment excision and circularization (Fig. 5C), involving as yet undescribed recombinases.
However, packaged GfIV genome segments share an ∼260 bp G sequence that has not been found in campoplegine IV circles. This G sequence is also present in segments of the only other banchine IV characterized to date, AsIV (16), and is located just upstream of the 3′ DR in GfIV proviral homologs (Fig. 5C). Its function is currently unknown, but we speculate that it may play a role in the integration of GfIV genome segments into the genome of the lepidopteran host, as shown for the HIM motif in M. demolitor BV (46). Indirect evidence that GfIV genome segments can integrate into the lepidopteran host genome has already been presented and is based on the presence, in the Choristoneura fumiferana genome, of a sequence that is clearly derived from a GfIV genome segment or a homolog thereof (15). However, the integrated sequence found in the C. fumiferana genome is limited to a portion of the CDS identified in the corresponding GfIV circle, with no trace left of the other portion of the segment. Alternative hypotheses include the provision of a recognition signal for genome segment encapsidation, through an as yet uncharacterized mechanism, and a role in the process of DNA recombination described above although what this role would be is not immediately apparent from existing knowledge of site-specific recombination systems (47).
In conclusion, the present work provides solid evidence that banchine and campoplegine IVs have closely related viral ancestors. Strikingly, the IVSPERs characterized here also contained coding sequences believed to be part of the replicative machinery of the progenitor virus. Whether the genomes of IV-bearing campoplegine wasps contain the same machinery needs to be determined as homologous sequences were not found within the three IVSPERs characterized in H. didymator (17). The factors responsible for the genomic and morphological differences observed between banchine and campoplegine IVs remain unknown but are likely the result of evolutionary divergence between the wasp lineages. Distinct events of viral genome acquisition by ancestors of banchine and campoplegine wasps could also account, at least in part, for these differences. A thorough assessment of the latter hypothesis will require extensive genomic analysis of wasp species belonging to ichneumonid subfamilies on branches separating Campopleginae from Banchinae in the phylogenetic tree, a research avenue that should prove most rewarding.
ACKNOWLEDGMENTS
This work was supported by Discovery grants from the Natural Sciences and Engineering Research Council of Canada to M. Cusson (171350-2012) and D. Stoltz (171215-2008), Genoscope (France), the Cluster de Recherche en Infectiologie de la Region Center (France), and the Canadian Forest Service.
We thank G. Pelletier (Canadian Forest Service, Quebec City, Canada) for confirming the identification of field-collected G. fumiferanae wasps. We are grateful to H. Maaroufi, J. Laroche, and R. Shi (Université Laval) for assistance with bioinformatics analyses. We also thank three anonymous reviewers for constructive comments on the original version of the manuscript.
REFERENCES
- 1.Strand MR. 2010. Polydnaviruses, p 216–241. In Asgari S, Johnson KN (ed), Insect Virology Caister Academic Press, Norwich. [Google Scholar]
- 2.Murphy N, Banks JC, Whitfield JB, Austin AD. 2008. Phylogeny of the parasitic microgastroid subfamilies (Hymenoptera: Braconidae) based on sequence data from seven genes, with an improved time estimate of the origin of the lineage. Mol Phylogenet Evol 47:378–395. doi: 10.1016/j.ympev.2008.01.022. [DOI] [PubMed] [Google Scholar]
- 3.Whitfield JB. 2002. Estimating the age of the polydnavirus/braconid wasp symbiosis. Proc Natl Acad Sci U S A 99:7508–7513. doi: 10.1073/pnas.112067199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whitfield JB, O'Connor JM. 2012. Molecular systematics of wasp and polydnavirus genomes and their coevolution, p 89–97. In Beckage NE, Drezen J-M (ed), Parasitoid viruses: symbionts and pathogens. Elsevier, Inc., London, United Kingdom. [Google Scholar]
- 5.Stoltz DB, Vinson SB, MacKinnon EA. 1976. Baculovirus-like particles in the reproductive tracts of female parasitoid wasps. Can J Microbiol 22:1013–1023. doi: 10.1139/m76-148. [DOI] [PubMed] [Google Scholar]
- 6.de Buron I, Beckage NE. 1992. Characterization of a polydnavirus (PDV) and virus-like filamentous particle (VLFP) in the braconid wasp Cotesia congregata (Hymenoptera: Braconidae). J Invertebr Pathol 59:315–327. doi: 10.1016/0022-2011(92)90139-U. [DOI] [Google Scholar]
- 7.Wyler T, Lanzrein B. 2003. Ovary development and polydnavirus morphogenesis in the parasitic wasp Chelonus inanitus. II. Ultrastructural analysis of calyx cell development, virion formation and release. J Gen Virol 84:1151–1163. [DOI] [PubMed] [Google Scholar]
- 8.Wang YJ, Burand JP, Jehle JA. 2007. Nudivirus genomics: diversity and classification. Virol Sin 22:128–136. doi: 10.1007/s12250-007-0014-3. [DOI] [Google Scholar]
- 9.Bézier A, Thézé J, Gavory F, Gaillard J, Poulain J, Drezen J-M, Herniou EA. 2015. The genome of the nucleopolyhedrosis causing virus from Tipula oleracea (ToNV) sheds new light into the Nudiviridae family. J Virol 89:3008–3025. doi: 10.1128/JVI.02884-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bézier A, Annaheim M, Herbiniere J, Wetterwald C, Gyapay G, Bernard-Samain S, Wincker P, Roditi I, Heller M, Belghazi M, Pfister-Wilhem R, Periquet G, Dupuy C, Huguet E, Volkoff AN, Lanzrein B, Drezen JM. 2009. Polydnaviruses of braconid wasps derive from an ancestral nudivirus. Science 323:926–930. doi: 10.1126/science.1166788. [DOI] [PubMed] [Google Scholar]
- 11.Burke GR, Strand MR. 2012. Deep sequencing identifies viral and wasp genes with potential roles in replication of Microplitis demolitor bracovirus. J Virol 86:3293–3306. doi: 10.1128/JVI.06434-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burke GR, Walden KK, Whitfield JB, Robertson HM, Strand MR. 2014. Widespread genome reorganization of an obligate virus mutualist. PLoS Genet 10:e1004660. doi: 10.1371/journal.pgen.1004660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quicke DLJ, Laurenne NM, Fitton MG, Broad GR. 2009. A thousand and one wasps: a 28S rDNA and morphological phylogeny of the Ichneumonidae (Insecta: Hymenoptera) with an investigation into alignment parameter space and elision. J Nat Hist 43:1305–1421. doi: 10.1080/00222930902807783. [DOI] [Google Scholar]
- 14.Lapointe R, Tanaka K, Barney WE, Whitfield JB, Banks JC, Béliveau C, Stoltz D, Webb BA, Cusson M. 2007. Genomic and morphological features of a banchine polydnavirus: comparison with bracoviruses and ichnoviruses. J Virol 81:6491–6501. doi: 10.1128/JVI.02702-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cusson M, Stoltz D, Lapointe R, Béliveau C, Nisole A, Volkoff AN, Drezen J-M, Maaroufi H, Levesque RC. 2012. Genomics of banchine ichnoviruses: insights into their relationship to bracoviruses and campoplegine ichnoviruses, p 79–87. In Beckage NE, Drezen J-M (ed), Parasitoid viruses: symbionts and pathogens. Elsevier, Inc., London, United Kingdom. [Google Scholar]
- 16.Djoumad A, Stoltz D, Béliveau C, Boyle B, Kuhn L, Cusson M. 2013. Ultrastructural and genomic characterization of a second banchine polydnavirus confirms the existence of shared features within this ichnovirus lineage. J Gen Virol 94:1888–1895. doi: 10.1099/vir.0.052506-0. [DOI] [PubMed] [Google Scholar]
- 17.Volkoff AN, Jouan V, Urbach S, Samain S, Bergoin M, Wincker P, Demettre E, Cousserans F, Provost B, Coulibaly F, Legeai F, Béliveau C, Cusson M, Gyapay G, Drezen JM. 2010. Analysis of virion structural components reveals vestiges of the ancestral ichnovirus genome. PLoS Pathog 6:e1000923. doi: 10.1371/journal.ppat.1000923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matz MV. 2003. Amplification of representative cDNA pools from microscopic amounts of animal tissue. Methods Mol Biol 221:103–116. [DOI] [PubMed] [Google Scholar]
- 19.Shevchenko A, Tomas H, Havli J, Olsen JV, Mann M. 2006. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1:2856–2860. [DOI] [PubMed] [Google Scholar]
- 20.Deutsch EW, Mendoza L, Shteynberg D, Farrah T, Lam H, Tasman N, Sun Z, Nilsson E, Pratt B, Prazen B, Eng JK, Martin DB, Nesvizhskii AI, Aebersold R. 2010. A guided tour of the trans-proteomic pipeline. Proteomics 10:1150–1159. doi: 10.1002/pmic.200900375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98. [Google Scholar]
- 22.Rutledge RG, Stewart D. 2008. Critical evaluation of methods used to determine amplification efficiency refutes the exponential character of real-time PCR. BMC Mol Biol 9:96. doi: 10.1186/1471-2199-9-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rutledge RG, Stewart D. 2008. A kinetic-based sigmoidal model for the polymerase chain reaction and its application to high-capacity absolute quantitative real-time PCR. BMC Biotechnol 8:47. doi: 10.1186/1472-6750-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rutledge RG, Stewart D. 2010. Assessing the performance capabilities of LRE-based assays for absolute quantitative real-time PCR. PLoS One 5:e9731. doi: 10.1371/journal.pone.0009731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rasoolizadeh A, Béliveau C, Stewart D, Cloutier C, Cusson M. 2009. Tranosema rostrale ichnovirus repeat element genes display distinct transcriptional patterns in caterpillar and wasp hosts. J Gen Virol 90:1505–1514. doi: 10.1099/vir.0.008664-0. [DOI] [PubMed] [Google Scholar]
- 26.Bicknell JN, Leisy DJ, Rohrmann GF, Beaudreau GS. 1987. Comparison of the p26 gene region of two baculoviruses. Virology 161:589–592. doi: 10.1016/0042-6822(87)90155-3. [DOI] [PubMed] [Google Scholar]
- 27.Deng L, Webb BA. 1999. Cloning and expression of a gene encoding a Campoletis sonorensis polydnavirus structural protein. Arch Insect Biochem Physiol 40:30–40. doi:. [DOI] [PubMed] [Google Scholar]
- 28.Deng L, Stoltz DB, Webb BA. 2000. A gene encoding a polydnavirus structural polypeptide is not encapsidated. Virology 269:440–450. doi: 10.1006/viro.2000.0248. [DOI] [PubMed] [Google Scholar]
- 29.Savary S, Beckage N, Tan F, Periquet G, Drezen JM. 1997. Excision of the polydnavirus chromosomal integrated EP1 sequence of the parasitoid wasp Cotesia congregata (Braconidae, Microgastinae) at potential recombinase binding sites. J Gen Virol 78:3125–3134. [DOI] [PubMed] [Google Scholar]
- 30.Dupuy C, Gundersen-Rindal D, Cusson M. 2012. Genomics and replication of polydnaviruses, p 47–61. In Beckage NE, Drezen J-M (ed), parasitoid viruses: symbionts and pathogens. Elsevier, Inc., London, United Kingdom. [Google Scholar]
- 31.Bézier A, Louis F, Jancek S, Periquet G, Thézé J, Gyapay G, Musset K, Lesobre J, Lenoble P, Dupuy C, Gundersen-Rindal D, Herniou EA, Drezen J-M. 2013. Functional endogenous viral elements in the genome of the parasitoid wasp Cotesia congregata: insights into the evolutionary dynamics of bracoviruses. Philos Trans R Soc B Biol Sci 368:20130047. doi: 10.1098/rstb.2013.0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Louis F, Bézier A, Periquet G, Ferras C, Drezen JM, Dupuy C. 2013. The bracovirus genome of the parasitoid wasp Cotesia congregata is amplified within 13 replication units, including sequences not packaged in the particles. J Virol 87:9649–9660. doi: 10.1128/JVI.00886-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burke GR, Thomas SA, Eum JH, Strand MR. 2013. Mutualistic polydnaviruses share essential replication gene functions with pathogenic ancestors. PLoS Pathog 9:e1003348. doi: 10.1371/journal.ppat.1003348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Desjardins CA, Gundersen-Rindal DE, Hostetler JB, Tallon LJ, Fuester RW, Schatz MC, Pedroni MJ, Fadrosh DW, Haas BJ, Toms BS, Chen D, Nene V. 2007. Structure and evolution of a proviral locus of Glyptapanteles indiensis bracovirus. BMC Microbiol 7:61. doi: 10.1186/1471-2180-7-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Volkoff AN, Drezen JM, Cusson M, Webb BA. 2012. The organization of genes encoding ichnovirus structural proteins, p 33–45. In Beckage NE, Drezen J-M (ed), Parasitoid viruses: symbionts and pathogens. Elsevier, Inc., London, United Kingdom. [Google Scholar]
- 36.Cheng RL, Xi Y, Lou YH, Wang Z, Xu JY, Xu HJ, Zhang CX. 2014. Brown planthopper nudivirus DNA integrated in its host genome. J Virol 88:5310–5308. doi: 10.1128/JVI.03166-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang L, Salem TZ, Campbell DJ, Turney CM, Kumar CM, Cheng XW. 2009. Characterization of a virion occlusion-defective Autographa californica multiple nucleopolyhedrovirus mutant lacking the p26, p10 and p74 genes. J Gen Virol 90:1641–1648. doi: 10.1099/vir.0.010397-0. [DOI] [PubMed] [Google Scholar]
- 38.Yutin N, Koonin EV. 2012. Hidden evolutionary complexity of nucleo-cytoplasmic large DNA viruses of eukaryotes. Virol J 9:161. doi: 10.1186/1743-422X-9-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Colson P, Pagnier I, Yoosuf N, Fournous G, La Scola B, Raoult D. 2013. “Marseilleviridae,” a new family of giant viruses infecting amoebae. Arch Virol 158:915–920. doi: 10.1007/s00705-012-1537-y. [DOI] [PubMed] [Google Scholar]
- 40.Colson P, De Lamballerie X, Yutin N, Asgari S, Bigot Y, Bideshi DK, Cheng XW, Federici BA, Van Etten JL, Koonin EV, La Scola B, Raoult D. 2013. “Megavirales,” a proposed new order for eukaryotic nucleocytoplasmic large DNA viruses. Arch Virol 158:2517–2521. doi: 10.1007/s00705-013-1768-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koonin EV, Dolja VV, Krupovic M. 2015. Origins and evolution of viruses of eukaryotes: the ultimate modularity. Virology 479-480C:2–25. doi: 10.1016/j.virol.2015.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boyer M, Yutin N, Pagnier I, Barrassi L, Fournous G, Espinosa L, Robert C, Azza S, Sun S, Rossmann MG, Suzan-Monti M, La Scola B, Koonin EV, Raoult D. 2009. Giant Marseillevirus highlights the role of amoebae as a melting pot in emergence of chimeric microorganisms. Proc Natl Acad Sci U S A 106:21848–21853. doi: 10.1073/pnas.0911354106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bigot Y, Samain S, Auge-Gouillou C, Federici BA. 2008. Molecular evidence for the evolution of ichnoviruses from ascoviruses by symbiogenesis. BMC Evol Biol 8:253. doi: 10.1186/1471-2148-8-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cui L, Webb BA. 1997. Homologous sequences in the Campoletis sonorensis polydnavirus genome are implicated in replication and nesting of the W segment family. J Virol 71:8504–8513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rattanadechakul W, Webb BA. 2003. Characterization of Campoletis sonorensis ichnovirus unique segment B and excision locus structure. J Insect Physiol 49:523–532. doi: 10.1016/S0022-1910(03)00053-2. [DOI] [PubMed] [Google Scholar]
- 46.Beck MH, Zhang S, Bitra K, Burke GR, Strand MR. 2011. The encapsidated genome of Microplitis demolitor bracovirus integrates into the host Pseudoplusia includens. J Virol 85:11685–11696. doi: 10.1128/JVI.05726-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Turan S, Bode J. 2011. Site-specific recombinases: from tag-and-target- to tag-and-exchange-based genomic modifications. FASEB J 25:4088–4107. doi: 10.1096/fj.11-186940. [DOI] [PubMed] [Google Scholar]
- 48.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:research0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]