Abstract
Erythropoietin (Epo) is produced in the kidney and liver in a hypoxia-inducible manner via the activation of hypoxia-inducible transcription factors (HIFs) to maintain oxygen homeostasis. Accelerating Epo production in hepatocytes is one plausible therapeutic strategy for treating anemia caused by kidney diseases. To elucidate the regulatory mechanisms of hepatic Epo production, we analyzed mouse lines harboring liver-specific deletions of genes encoding HIF-prolyl-hydroxylase isoforms (PHD1, PHD2, and PHD3) that mediate the inactivation of HIF1α and HIF2α under normal oxygen conditions. The loss of all PHD isoforms results in both polycythemia, which is caused by Epo overproduction, and fatty livers. We found that deleting any combination of two PHD isoforms induces polycythemia without steatosis complications, whereas the deletion of a single isoform induces no apparent phenotype. Polycythemia is prevented by the loss of either HIF2α or the hepatocyte-specific Epo gene enhancer (EpoHE). Chromatin analyses show that the histones around EpoHE dissociate from the nucleosome structure after HIF2α activation. HIF2α also induces the expression of HIF3α, which is involved in the attenuation of Epo production. These results demonstrate that the total amount of PHD activity is more important than the specific function of each isoform for hepatic Epo expression regulated by a PHD-HIF2α-EpoHE cascade in vivo.
INTRODUCTION
Under low-oxygen conditions (hypoxia), the number of circulating red blood cells increases to deliver oxygen efficiently into peripheral organs. The red blood cell mass is controlled by the erythroid growth factor erythropoietin (Epo), the majority of which is produced by the kidneys in a hypoxia-dependent manner (1). Therefore, inhabitants of high-altitude areas and patients suffering from chronic respiratory failure with chronic obstructive pulmonary disease often develop polycythemia (erythrocytosis) with elevated levels of Epo in their plasma (2, 3). Epo binds to its receptor (EpoR) on the surfaces of immature erythroid cells and stimulates signaling cascades for proliferation, differentiation, and antiapoptosis (4). Epo or EpoR gene-targeted mouse lines are embryonic lethal, with severe anemia around embryonic day 13, clearly indicating a requirement for Epo signaling in erythropoiesis (5).
Anemia often occurs in patients who suffer from kidney damage (6). We previously reported that renal Epo-producing (REP) cells that are in the interstitial spaces between renal tubules transform into myofibroblastic cells, which are closely associated with renal fibrosis under inflammatory conditions (7–11). Because the transformed REP cells lose their ability to produce Epo even under severely hypoxic conditions, pharmacologically inducing Epo production in the damaged kidneys of anemic patients is difficult. Although REP cells secrete most of the Epo in adult animals, hepatocytes are the primary Epo-producing cells in fetuses (12). The liver maintains its Epo-producing activity throughout adulthood; thus, Epo expression is detectable in the livers of anemic/hypoxic mice (8, 12). However, the level of hepatic Epo production is weak and insufficient to compensate for renal anemia (13). Therefore, pharmacological enhancement of hepatic Epo production is a reasonable strategy for treating anemic patients who have renal diseases (14, 15).
In hepatocytes, Epo transcription, which is the rate-limiting step of Epo production, is controlled by a cis-regulatory element that is proximally downstream of the Epo transcription end site (called Epo hepatic enhancer [EpoHE] in this study). EpoHE contains a binding sequence for hypoxia-inducible transcription factors (HIFs) (16). We previously demonstrated that EpoHE is necessary and sufficient for Epo expression in hepatocytes, whereas renal Epo production is independent of EpoHE (12, 13). Thus, while Epo expression is commonly induced by hypoxic stresses in REP cells and hepatocytes, these two cell types employ different mechanisms of Epo gene regulation.
HIFs are master regulators of hypoxia-inducible gene expression. Each HIF consists of an oxygen-responsive α subunit and a constitutively expressed nuclear β subunit (17). Mammalian genomes contain three genes for HIFα isoforms. HIF1α and HIF2α have similar protein structures, and both exhibit strong transactivities under hypoxic conditions, whereas the function and regulatory mechanisms of HIF3α remain controversial due to the existence of multiple HIF3α splicing variants (18–20). Under normal oxygenation conditions (normoxia), HIFα subunits are consistently synthesized, and their specific proline residues are quickly hydroxylated by HIF-prolyl hydroxylase domain proteins (PHDs) (21, 22). Hydroxylated HIFα is recognized by von Hippel-Lindau protein (pVHL), which is a component of an E3 ubiquitin ligase complex, and degraded by the proteasome (23). Because hypoxia inhibits the enzymatic activities of PHDs, which use oxygen as a substrate, HIF heterodimers have increased activities as transcription factors under hypoxic conditions (24).
Hepatic Epo production is enhanced by the experimental targeting of the HIF pathway using genetically modified mice, small molecules, or small interfering RNA s (siRNAs) (25). For instance, liver-specific pVHL deficiency in mice results in polycythemia with the overexpression of the Epo gene in the liver (26). Because this phenotype depends on HIF2α, it is thought that HIF2α, rather than HIF1α, regulates hypoxia-inducible Epo expression in hepatocytes (26). The contributions of the three PHD isoforms (PHD1, PHD2 and PHD3, which are encoded by the Egln2, Egln1, and Egln3 genes, respectively) to hepatic Epo gene regulation have been investigated using gene-modified mice, and functional redundancy among the isoforms has been demonstrated (27–29). Although these strategies stimulate hepatic Epo production, the constitutive activation of HIFs induces steatosis and ectopic expression of cancer-related genes (25–29). Therefore, establishing a specific strategy for hepatic Epo induction to treat renal anemia is important, and fully understanding the molecular mechanism underlying Epo regulation will be necessary to develop such a strategy.
In this study, we analyzed gene-modified mouse lines lacking PHD isoforms, HIF2α, and/or EpoHE specifically in the liver to elucidate the in vivo signaling pathway between hypoxia sensor PHDs and cis-regulatory elements in the hepatic Epo gene regulatory system. Our results demonstrate that the function of PHDs is gene dose dependent and that HIF2α and EpoHE are necessary for hepatic Epo production. HIF2α functions in both the removal of histones from the region around the Epo gene, which enhances Epo transcription, and the induction of HIF3α expression to attenuate hepatic Epo production. Thus, we propose that a PHD-HIF2α-EpoHE axis regulates hepatic Epo production by controlling HIF3α expression and the chromatin structure of the Epo gene. Our results provide valuable information regarding therapeutic targets that enhance hepatic Epo production for renal-anemia patients.
MATERIALS AND METHODS
Mice.
All mice were maintained at the Institute for Animal Experimentation, Tohoku University Graduate School of Medicine, under the Regulations for Animal Experiments and Related Activities of Tohoku University. A hypoxic box, in which the oxygen concentration was regulated by an oxygen controller (ProOx; BioSpherix) with a nitrogen generator (Nilox; Sanyo Electronic Industries), was used to expose the mice to hypoxic conditions. For acclimation to severe hypoxia (6% O2), the mice were exposed to 10% oxygen for 2 h before the oxygen concentration was reduced to 6%. Mouse lines carrying conditional knockout genes for PHDs (Egln1f/f, Egln2f/f, and Egln3f/f genotypes) and HIF2α (Epas1f/f genotype) were used (30, 31). The in vivo function of the Epo gene hepatic enhancer (EpoHE) was investigated using mice with a deletion of EpoHE (ΔEpoHE mice, EpoΔ3′E/Δ3′E genotype) (9, 12). AlbCre transgenic mice were used to express Cre recombinase exclusively in mature hepatocytes (32). The Epas1f/f and AlbCre mice were purchased from the Jackson Laboratory. Genotyping of genomic DNA from tails was performed by PCR using the primers listed in Table 1.
TABLE 1.
Sequences of the primers used in this study
| Purpose and target | Primer 1 (5′–3′) | Primer 2 (5′–3′) |
|---|---|---|
| Genotyping | ||
| Egln1 (PHD2) | CAAATGGAGATGGAAGATGC | TCAACTCGAGCTGGAAACC |
| Egln2 (PHD1) | TGGGCGCTGCATCACCTGTATCT | ACTGGTGAAGCCTGTAGCCTGTC |
| Egln3 (PHD3) | ATGGCCGCTGTATCACCTGTAT | CCACGTTAACTCTAGAGCCACTGA |
| AlbCre | ACGTTCACCGGCATCAACGT | CTGCATTACCGGTCGATGCA |
| Epas1 (HIF2α) | CAGGCAGCAGGCAGTATGCCTGGCTAATTCCAGTT | CTTCTTCCATCATCTGGGATCTGGGACT |
| EpoHE | CAGGCTCCATTCAAGGC | CCTGCAGTGGACTTTGAAGGC |
| RT-PCR with gel electrophoresis | ||
| PHD1 | ACCGCGCAGCATTCGTG | GGGGCTGGCCATTAGGTAGGTGTA |
| PHD2 | GCGGGAAGCTGGGCAACTAC | CAACCCTCACACCTTTCTCACC |
| PHD3 | CTGCGTGCTGGAGCGAGTCAA | TCATGTGGATTCCTGCGGTCTG |
| IPAS/HIF3α | GAGGGTTTCGTCATGGTACT | TCTTGAAGTTCCTCTTGGTC |
| RT-qPCR with SYBR green reagents for mouse (m) or human (h) mRNA expression | ||
| β-Actin (m, h) | GACAGGATGCAGAAGGAGAT | TTGCTGATCCACATCTGCTG |
| PHD1 (m) | ATGGCTCACGTGGACGCAGTAA | CATTGCCTGGATAACACGCCAC |
| PHD2 (m) | TAAACGGCCGAACGAAAGC | GGGTTATCAACGTGACGGACA |
| PHD3 (m) | CTATGTCAAGGAGCGGTCCAA | GTCCACATGGCGAACATAACC |
| Epo (m) | CATCTGCGACAGTCGAGTTCTG | CACAACCCATCGTGACATTTTC |
| EpoR (m) | GGGCTCCGAAGAACTTCTGTG | ATGACTTTCGTGACTCACCCT |
| Bnip3 (m) | GTTACCCACGAACCCCACTTT | GTGGACAGCAAGGCGAGAAT |
| Vegfa (m) | CTGCTGTAACGATGAAGCCCTG | GCTGTAGGAAGCTCATCTCTCC |
| Fasn (m) | GGCTGCTGTTGGAAGTCAGC | AGTGTTCGTTCCTCGGAGTG |
| Scd1 (m) | TTCTTGCGATACACTCTGGTGC | CGGGATTGAATGTTCTTGTCGT |
| HIF1α (m) | CCTGCACTGAATCAAGAGGTTGC | CCATCAGAAGGACTTGCTGGCT |
| HIF2α (m) | CAATGACAGCTGACAAGGAG | CATAGAAGACCTCCGTCTCC |
| HIF3α (m) | AAGACGCCCTGACCCCCAGG | CCCTCTGCTGGTGAGCGTGC |
| Gpi (m) | CCCTCTTTATAATCGCCTCCA | GAAACCACTCCTTTGCTGTCTC |
| Ldha (m) | GGCACTGACGCAGACAAG | TGATCACCTCGTAGGCACTG |
| Hepcidin (m) | TCTTCTGCATTGGTATCGCA | GAGCAGCACCACCTATCTCC |
| CA9 (h) | CCTTTGCCAGAGTTGACGAG | GACAGCAACTGCTCATAGGC |
| PGK1 (h) | CTAACAAGCTGACGCTGGAC | CTGGTTGTTTGTTATCTGGTTG |
| EPO (h) | TCATCTGTGACAGCCGAGTC | CAAGCTGCAGTGTTCAGCAC |
| qPCR with SYBR green reagents for chromatin analysis | ||
| CA9 promoter | TCTGTGAGTCAGCCTGCTCC | TCCCAGCACACGGTGTGTAC |
| PGK1 promoter | CCCTAAGTCGGGAAGGTTCC | CTGTCCGTCTGCGAGGGTAC |
| EPO promoter | CTCAACCCAGGCGTCCTG | GGGGCTGTTATCTGCATGTG |
| EPO intron1 | CTGTTTGAGCGGGGATTTAGC | TTCCGGGGTCCTTGACAAGT |
| EPO HE | AACCTCCAAATCCCCTGGCTC | CTGTGTGAGACAGCACGTAG |
Reverse transcription and PCR.
Total RNA samples from mouse organs were extracted using Isogen (Nippon Gene). cDNA was synthesized using a SuperScript III system (Invitrogen). To detect both normal and knockout transcripts derived from genes for PHDs simultaneously, cDNAs were amplified with primer pairs that annealed to sequences flanking the deleted exons (Table 1), and the PCR products were subjected to gel electrophoresis, followed by ethidium bromide staining (30). PCR primers that annealed to sequences in exons 3 and 4 of the Hif3a gene were used (Table 1) to detect both IPAS- and HIF3α-specific splicing variants of the Hif3a gene by agarose gel electrophoresis because the IPAS mRNA contains 56-bp sequences from the IPAS-specific exon (exon 4a) that is between exons 3 and 4 (33). Quantitative PCR (qPCR) was performed on cDNA with the primers listed in Table 1, using FastStart SYBR green master mix with a LightCycler 96 system (Roche) or Power SYBR green master mix with a StepOne real-time PCR system (Life Technologies). The expression levels of β-actin mRNA were used as internal controls for the PCR experiments.
Blood analysis.
Peripheral blood (0.2 to 0.3 ml) was collected from the mouse submandibular vein into a 1.5-ml tube containing 5 μl of 0.5 M EDTA. After the blood samples were centrifuged, plasma Epo concentrations were measured using a mouse Epo enzyme-linked immunosorbent assay (ELISA) kit (R&D Systems). To induce anemia by bleeding, 0.3 ml of peripheral blood was withdrawn from the mouse submandibular vein once per day for 3 days, and then the recovery from the anemia was investigated by measuring hematocrit levels in the retro-orbital venous plexus using heparinized microtubes (Drummond). The glucose concentration in blood from tail tips was directly measured using an animal glucometer (LAB Gluco; ForaCare).
Flow cytometry.
Mononucleated cells from the spleen and bone marrow were prepared by isopycnic centrifugation with Histopaque-1083 (Sigma) and were stained with fluorescein isothiocyanate (FITC)-conjugated Ter119 (116215; BioLegend) and phycoerythrin (PE)-conjugated CD71 (113808; BioLegend) antibodies. After washing, the cells were sorted using a FACS Jazz flow cytometer (Becton Dickinson).
Immunoblotting.
Nuclear extracts were obtained from mouse livers using sucrose buffer (0.25 M sucrose in 10 mM HEPES-KOH buffer) and resuspended in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-Cl, 150 mM NaCl, 0.5% sodium deoxycholate, 0.1% SDS, and 1.0% NP-40) supplemented with inhibitors for proteinase (Roche) and proteasome (Proteintech). Whole-cell extracts were directly obtained with RIPA buffer. The cell extracts were incubated with tobacco etch virus (TEV) protease (Promega) at 4°C overnight to remove the HaloTag from HaloTag-HIF3α fusion proteins. Proteins (25 μg) were transferred to nitrocellulose membranes (Bio-Rad) after electrophoresis using 6% or 8% polyacrylamide gels, and the membranes were incubated with primary antibodies against HIF1α (PAB12138; Abnova), HIF2α (C150132; LSBio), HIF3α (HPA041141; Sigma), green fluorescent protein (GFP) (598; MBL), Nup62 (ab96134; Abcam), α-tubulin (T5168; Sigma) or β-tubulin (PA1-21153; Thermo). Horseradish peroxidase-conjugated secondary antibodies against mouse (01803-44; Nacalai Tesque), rabbit (P0448; Dako) or goat (sc-2020; Santa Cruz) immunoglobulins were used with chemiluminescent detection reagents (GE Healthcare) to detect signals with a Chemi-Doc imaging system (Bio-Rad).
Glycogen assays.
To measure the liver glycogen concentration, boiled supernatants from liver homogenates (2.0 mg tissue/ml water) were digested with glucoamylase for 30 min at room temperature. Samples were analyzed using the fluorometric assay protocol in the glycogen assay kit (BioVision). To detect glycogen in liver sections, periodic acid-Schiff (PAS) staining was conducted on paraffin-embedded sections using a PAS staining kit (Muto).
Lipid droplet staining.
Frozen sections (10-μm thickness) of mouse livers that had been fixed in 4% formaldehyde for 4 h were incubated with Nile red solution (100 ng/ml; Wako) and Hoechst 33342 (1.0 μg/ml; Calbiochem) for 5 min (34). After the sections had been washed with PBS, fluorescence signals were detected using the pseudocolor detection system of a BZ9000 microscope (Keyence).
Cell culture and reporter analysis.
Hep3B cells that were derived from a human hepatoma were cultured in high-glucose Dulbecco's modified Eagle medium (DMEM) (Nacalai Tesque) supplemented with 10% fetal bovine serum (Sigma) (35). For hypoxic cell sampling, the cells were incubated and treated in a Sci-tive hypoxia workstation (Ruskinn), in which the air was composed of 1% O2, 5% CO2, and 94% N2.
The plasmid pT81/HRE-luc, which expresses firefly luciferase under the control of HIF-binding sequences from EpoHE, was used as a reporter for EpoHE activity, and pCMX-HIF2αCA was used to overexpress Flag-tagged mouse HIF2α bearing constitutively activated mutations in proline residues (P405A and P530A) (36). We used the pFN21AB6219 plasmid (Kazusa Institute) to express human HIF3α linked to a HaloTag peptide (Promega).
For transient-transfection assays, the cells were cultured to 70% confluence in 12-well plates and transfected with pT81/HRE-luc (300 ng/well), pCMX-HIF2αCA (300 ng/well), and/or pFN21AB6219 (300 or 600 ng/well) using Lipofectamine 2000 (Life Technologies). For internal transfection efficiency controls, 100 ng/well of the reference plasmids pEGFP-N1 (TaKaRa) and pRL-EF (37) were added to all transfection mixtures. The cells were harvested at 48 h after transfection, and the activity of firefly luciferase was measured using a dual luciferase reporter assay system (Promega) after normalization with the activity of Renilla luciferase, which was expressed under the EEF1A1 gene promoter from pRL-EF (37). The cells were observed under a BZ9000 microscope after supplementation of the culture medium with tetramethylrhodamine (TMR)-conjugated HaloTag direct ligand (Promega) to detect HaloTag-HIF3α expression in living cells.
Chemical compounds.
FG4592 (Selleck Chemicals) and acriflavine hydrochloride (Sigma) were dissolved in dimethyl sulfoxide (DMSO) (Nacalai Tesque). HIF2 antagonist 2 {N-(3-chloro-5-fluorophenyl)-4-nitrobenzo[c]1,2,5-oxadiazol-5-amine; CAS no. 1422955-31-4} was synthesized from 3-chloro-5-fluoroaniline (Sigma-Aldrich) and 5-chloro-4-nitrobenzo[c]1,2,5-oxadiazole (Namiki) in accordance with a previously published protocol (38) and resuspended in DMSO after purification.
Chromatin analysis.
Nuclear extracts of Hep3B cells were partially digested with nucleases from an EpiQ chromatin analysis kit (Bio-Rad) to detect the chromatin states of HIF target genes. Using purified genomic DNA from nuclease-digested and undigested samples, qPCR was conducted with the primers listed in Table 1, and the abundance of PCR target sites within the two samples was compared. Because chromatin regions with nucleosome structures (genomic DNA masked by histones) are resistant to nuclease digestion (accessibility), a similar number of target sites were observed in the histone-rich regions in the nuclease-digested and undigested samples (100% protection from nuclease digestion). Nucleosome-free chromatin regions show low levels of protection (39).
Statistics.
The data are presented as means ± standard deviations (SD). P values were calculated using two-tailed, unpaired Student's t tests.
RESULTS
Polycythemia is induced by the loss of two PHD isoforms in the liver.
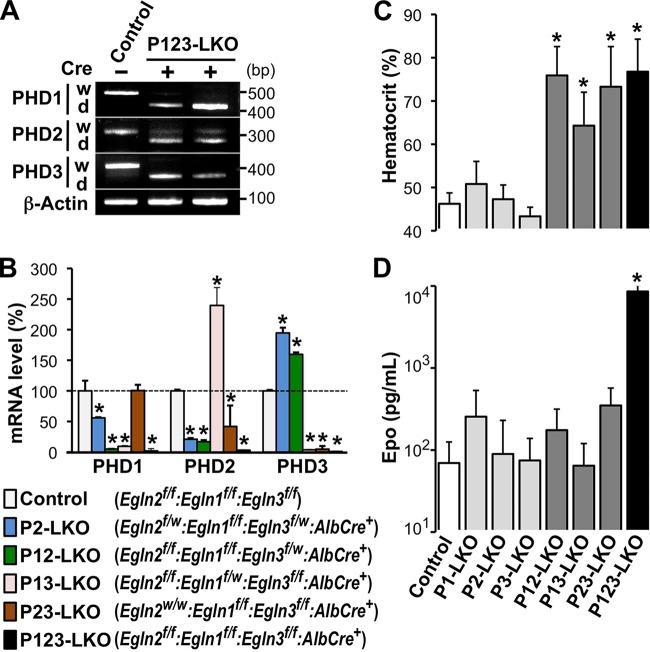
Mice harboring conditional knockout (floxed) genes for PHD1, PHD2, and PHD3 were crossed with AlbCre transgenic mice expressing Cre recombinase in hepatocytes to elucidate the roles of the PHD isoforms in hepatic Epo production (30, 32). Because loxP sequences were inserted at both ends of the exon that encodes the catalytic domain for each PHD in the floxed alleles (f), the PHD transcripts that were produced following Cre-mediated liver-specific knockout (LKO) were shorter than those generated by the wild-type and floxed alleles (30). The shorter transcripts were predominantly detected in the livers of triple-knockout mice (Egln2f/f:Egln3f/f:AlbCre+ genotype; called P123-LKO mice), whereas the control mice, which did not carry the Cre transgene (Egln2f/f:Egln1f/f:Egln3f/f genotype), expressed only transcripts of normal length (Fig. 1A). These data indicated that the expression of functional PHD isoforms was almost eliminated by AlbCre transgene expression in the hepatocytes of P123-LKO mice. Additionally, the levels of mRNA expression for each PHD isoform in the livers of PHD mutant mice were quantitatively measured by reverse transcription-quantitative PCR (RT-qPCR), and the data confirmed the high efficiency of AlbCre-mediated liver-specific gene targeting (Fig. 1B). Interestingly, PHD2 mRNA expression in PHD1/PHD3 double-knockout (P13-LKO) livers was higher than that in the control mice. In addition, PHD3 mRNA expression was increased by the loss of the genes for the other PHD isoforms in livers (Fig. 1B). These data suggest that the expression of the genes for PHD2 and PHD3 is induced by HIFs (40, 41).
FIG 1.
Liver-specific knockout of multiple PHD isoforms causes polycythemia. (A) RT-PCR was performed on livers from P123-LKO (Egln2f/f:Egln1f/f:Egln3f/f:AlbCre+ genotype) or control (Cre− Egln2f/f:Egln1f/f:Egln3f/f genotype) mice. Then, the PCR products were subjected to agarose gel electrophoresis to detect the expression of longer and shorter transcripts from the normal (w) and targeted (d [deletion of the exon encoding the catalytic domain of each PHD isoform]) alleles, respectively. β-Actin was used as an internal control. (B) Average expression levels of PHD isoforms in the livers of the indicated genotype mice were measured by RT-qPCR. The values are averages for 3 to 5 mice in each group, with error bars indicating standard deviations (SD). *, P < 0.01 compared to the expression levels in the control mice, which are set at 100%. (C and D) Average hematocrit (C) and Epo concentration (D) in peripheral blood samples from PHD mutant mice at 8 to 19 weeks after birth. The values are averages for 4 to 12 mice in each group, with error bars indicating SD. *, P < 0.01 compared to the levels in the control mice.
Next, we measured the hematocrit levels (red blood cell mass) in the peripheral blood samples of adult mice (11 to 39 weeks of age) lacking a single PHD isoform in the liver (P1-LKO, P2-LKO, and P3-LKO mice). These mutant mice had no apparent abnormalities, with normal hematocrit levels compared with control mice lacking the Cre transgene (Fig. 1C). A double knockout of PHD isoforms in any combination (P12-LKO, P13-LKO, and P23-LKO mice) resulted in polycythemia, with hematocrit levels ranging from 65% to 85% (Fig. 1C). Mice lacking all three PHD isoforms in the liver (P123-LKO mice) exhibited polycythemia with a severity similar to that observed in the double-knockout mice (Fig. 1C). These data demonstrate that the inhibition of two PHD isoforms in the liver is necessary and sufficient for erythropoietic induction and that the total dose of PHD isoforms in the liver seems to be more important for PHD suppressive activity in erythropoiesis than the specific function of each isoform.
We observed that the Epo concentration in the plasma of P123-LKO mice was significantly higher than that in control mice (Fig. 1D). This result is consistent with the hematocrit data from polycythemic P123-LKO mice. However, plasma Epo levels in double-knockout mice were normal, although the mice suffered from polycythemia (Fig. 1C and D).
Loss of two PHD isoforms induces Epo expression in the liver.
Although the liver-specific double knockout of PHD isoforms was thought to cause polycythemia through Epo oversecretion from the liver, the plasma Epo concentrations in these double-knockout mice were within the normal range (Fig. 1D). Thus, we measured the level of Epo expression in the livers of the PHD mutant mice. Epo expression was increased 10- to 20-fold by the loss of 2 PHD isoforms in the liver (Fig. 2A), while single PHD isoform knockout did not alter the Epo mRNA levels (28, 29). In the triple-knockout mice, the hepatic Epo mRNA level was dramatically (approximately 200-fold) higher than levels in control and double-knockout mice (Fig. 2A). These data demonstrated that PHD isoforms functionally compensate for each other in hepatic Epo production and that any single isoform can partially suppress hepatic Epo expression under normal conditions.
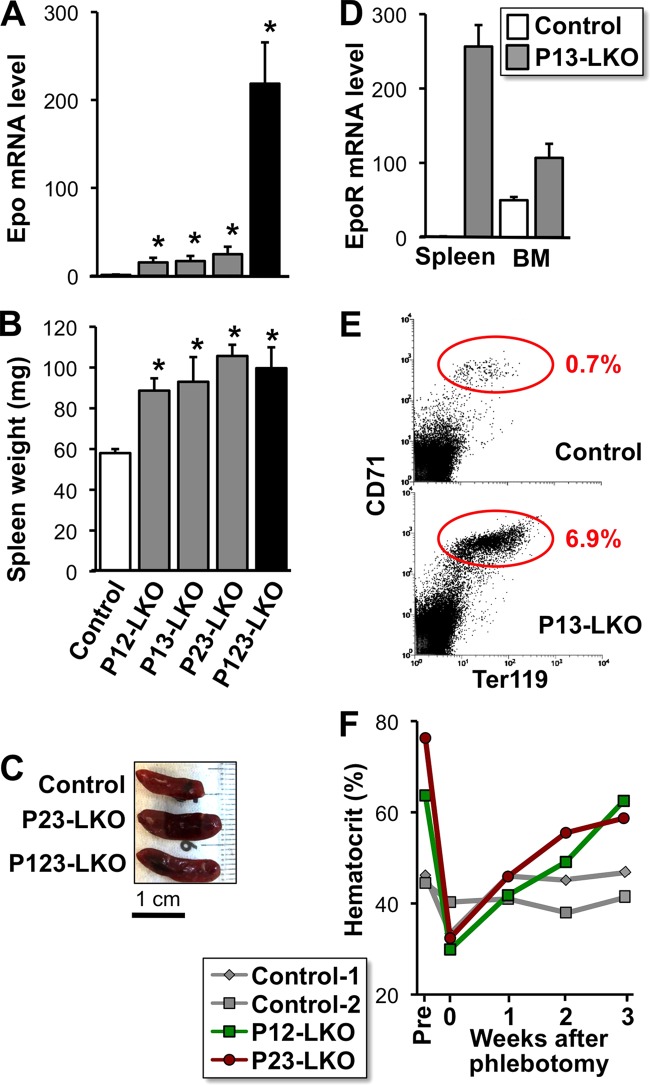
FIG 2.
Overexpression of the Epo gene in livers lacking PHD isoforms. (A) Epo mRNA expression levels in the livers of PHD mutant mice at 8 to 12 weeks of age. The average expression level of control mice was set as 1.0 after normalization to β-actin mRNA levels. The data are means ± SD (n = 3 for each group). *, P < 0.01 compared to the levels in the control mice. (B) Splenic hypertrophy in liver-specific PHD-deficient mice. Wet weights of spleens from the PHD mutant mice (7 to 9 weeks of age) were measured. The values are means and SD (n = 3 for each group). *, P < 0.01 compared to the weights of control mouse spleens. (C) Spleens from the PHD mutant mice were enlarged at 8 weeks of age. (D) EpoR mRNA levels in the spleens and bone marrow of control and P13-LKO mice. The average expression level in control mouse spleens was set as 1.0 after normalization to β-actin mRNA levels. The data are means and SD (n = 3 for each group). (E) Representative data from flow cytometry with CD71 and Ter119 antibodies in mononucleated cells from spleens of control and P13-LKO mice. The percentages of cells found within the circles are shown. (F) Changes in the hematocrit levels of the PHD mutant mice before (Pre) and after phlebotomy. The hematocrit levels in mice at 12 weeks of age were decreased to approximately 30% by phlebotomy. Note that the hematocrit levels of PHD mutant mice returned to polycythemic levels within 3 weeks after phlebotomy.
Next, we predicted that most of the circulating Epo in the double-knockout mice was consumed through the internalization of Epo-EpoR complexes by an expanded population of erythroblastic cells in the spleen after erythropoiesis stimulation (42). As expected, the spleens were significantly enlarged in the PHD double-knockout mice and in the triple-knockout mice (Fig. 2B and C). Splenic EpoR mRNA levels, which were normalized to β-actin mRNA levels, were extremely high in the double-knockout mice compared with control mice (Fig. 2D). EpoR expression was also slightly induced in the bone marrow of liver-specific PHD double-knockout mice (Fig. 2D). Flow cytometry of splenic mononucleated cells demonstrated that the Ter119+ CD71+ erythroblastic cell fraction of the double-knockout mice was approximately 10-fold higher than that in control mice (Fig. 2E) (13, 43). These data suggest that the excess Epo produced by the PHD-deficient livers is absorbed by the expanded erythroblastic cells in spleens. This hypothesis may explain why plasma Epo levels are normal in polycythemic mice lacking two PHD isoforms in their livers.
To test this hypothesis, the hematocrit levels in double-knockout mice were reduced to normal by phlebotomy, and then the postphlebotomy hematocrit levels were evaluated over time. As expected, the mice reverted to their original polycythemic phenotype within 3 weeks postphlebotomy (Fig. 2F), and their plasma Epo levels fell to within the normal range throughout the observation period. These findings suggest that these double-knockout mice constitutively secrete excess Epo from the liver but that most of the Epo is internalized by erythroblasts after initiating the signaling cascade that induces erythropoiesis. Because a large difference in Epo mRNA levels between the double- and triple-knockout livers exists (Fig. 2A), the rate of Epo production in P123-LKO livers likely surpasses the rate of internalization.
Liver-specific loss of all PHD isoforms causes severe steatosis and growth retardation.
Immunoblotting of liver nuclear extracts demonstrated that protein levels of both HIF1α and HIF2α were increased by triple knockout of the PHD isoforms (Fig. 3A), while HIF proteins were undetectable in livers of the double-knockout, single-knockout, and control mice. These data are consistent with previous reports that HIF protein expression is very low or undetectable in PHD double-knockout livers compared with triple-knockout livers (28, 29). Therefore, it is possible that the low-level accumulation of HIF1α and HIF2α is sufficient to induce hepatic Epo gene expression and polycythemia in PHD double-knockout mice. Loss of all 3 PHD isoforms in hepatocytes results in a drastic induction of Epo expression via the hyperactivation of HIFs. Thus, PHD isoforms compensate for each other in the suppression of HIFs in hepatocytes under normal conditions, and no apparent abnormality is caused by loss of one of the three isoforms. The presence of a single functional PHD isoform is insufficient for the oxygen-dependent inactivation of HIFs. HIF1α and/or HIF2α escapes degradation in the hepatocytes and, although present at undetectable levels, induces Epo overexpression in the livers of double-knockout mice with polycythemia.
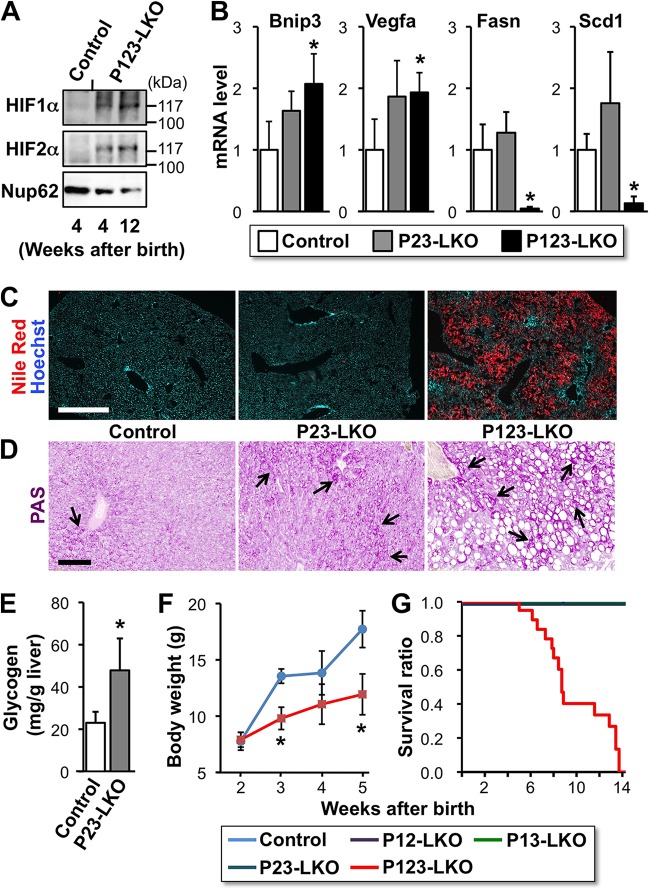
FIG 3.
Disruption of all three PHD isoforms in the liver causes steatosis and growth retardation. (A) Immunoblots for HIF1α and HIF2α in nuclear extracts from the livers of P123-LKO mice at the indicated ages. Nup62 was used as an internal control. (B) Expression levels of Bnip3, Vegfa, Fasn, and Scd1 mRNAs in the livers of PHD mutant mice were analyzed by RT-qPCR. β-Actin was used as an internal control. The expression levels of control mice were set as 1.0. The data are means and SD (n = 3 for each group). (C) Lipid droplets in the livers of the PHD mutant mice at 7 weeks of age were stained with Nile red (red). Hoechst 33342 was used for nuclear staining (blue). (D) Glycogen deposition in the livers of PHD mutant mice at 8 weeks of age was detected by PAS staining (arrows). Bars, 500 μm (C) and 100 μm (D). (E) The glycogen concentration was measured in the livers of the control and P23-LKO mice at 8 weeks of age. The data are means and SD (n = 4 for each group). (F) Changes in body weight between 2 and 5 weeks after birth in P123-LKO and control mice. Error bars indicate SD (n = 3 for each group). (G) Kaplan-Meier survival analysis of the indicated genotype mice (n = 20 for each group). More than half of the P123-LKO mice died by 10 weeks after birth. *, P < 0.01 compared with the control mice (B, E, and F).
Because we showed that Epo expression was induced by HIF activation in mice lacking more than two PHD isoforms in the liver, the expression of other HIF target genes was analyzed in the livers of P23-LKO and P123-LKO mice. The results demonstrated that the expression of Bnip3 and Vegfa mRNA was slightly increased in both P23-LKO and P123-LKO mouse livers compared to control mouse livers (Fig. 3B). These data demonstrated that PHD inactivation causes not only Epo production but also the global induction of HIF target genes, some of which promote mitochondrial autophagy (Bnip3) and angiogenesis (Vegfa).
The livers of P123-LKO mice at 7 weeks of age were large and fatty compared to the livers of mice in other groups. Nile red staining of lipid droplets in the liver sections clearly showed that P123-LKO mice suffered from severe steatosis (Fig. 3C). The double-knockout mice exhibited no lipid droplet accumulation in the liver (Fig. 3C). These data support previous reports showing that the liver-specific activation of the HIF pathway via the genetic inactivation of pVHL causes steatosis (44, 45). The expression of the genes for Fasn and Scd1, both of which are involved in fatty acid synthesis, was strongly suppressed in P123-LKO livers (Fig. 3B). This finding suggests that mitochondrial function is inhibited by HIF activation via Bnip3-mediated mitophagy (46) and that the accumulation of lipids due to decreased fatty acid usage in mitochondria may negatively regulate the expression of fatty acid synthesis genes. Consistent with a previous report that demonstrated increased glycogen concentrations in response to pVHL deficiency in liver (47), the loss of more than 2 PHD isoforms resulted in increased hepatic glycogen deposition (arrows in Fig. 3D). Indeed, the glycogen concentration in P23-LKO mouse livers was significantly higher than that in control mouse livers (Fig. 3E).
P123-LKO mice exhibited growth retardation (Fig. 3F) and a shortened life span of 14 weeks (Fig. 3G). These observations indicate that the universal knockout of all PHD isoforms in livers dramatically increases HIF activity, which causes severe steatosis and lethality. Because the inactivation of two PHD isoforms was sufficient to induce polycythemia without steatosis and lethality, we propose that mild inhibition of PHDs leads to the activation of hepatic Epo production without severe side effects.
HIF2α is a major regulator of hypoxia-inducible Epo gene expression in the liver.
PHD mutant mice with the AlbCre transgene were mated with HIF2α conditional knockout mice to investigate whether HIF1α or HIF2α activates the Epo gene in PHD-deficient hepatocytes (31). The polycythemia exhibited by the double-knockout mice was completely rescued by the loss of the HIF2α gene (Epas1) in hepatocytes (P12H2-LKO, P13H2-LKO, and P23H2-LKO mice in Fig. 4A). The hematocrit levels of HIF2α-deficient P123-LKO (P123H2-LKO) mice were lower than those of P123-LKO mice (Fig. 1C and 4A). Similar to the results of deleting the PHD isoforms in hepatocytes, pVHL deficiency causes Epo overexpression and polycythemia in mice, and this phenotype is blunted by the loss of HIF2α but not of HIF1α (26). Taken together, these data demonstrate that HIF2α, but not HIF1α, predominantly activates the hepatic Epo gene under hypoxic conditions by signaling through PHDs. Because the lethality and severe steatosis in P123-LKO mice were also rescued by HIF2α deficiency, the hyperactivation of HIF2α most likely contributes to these two phenotypes.
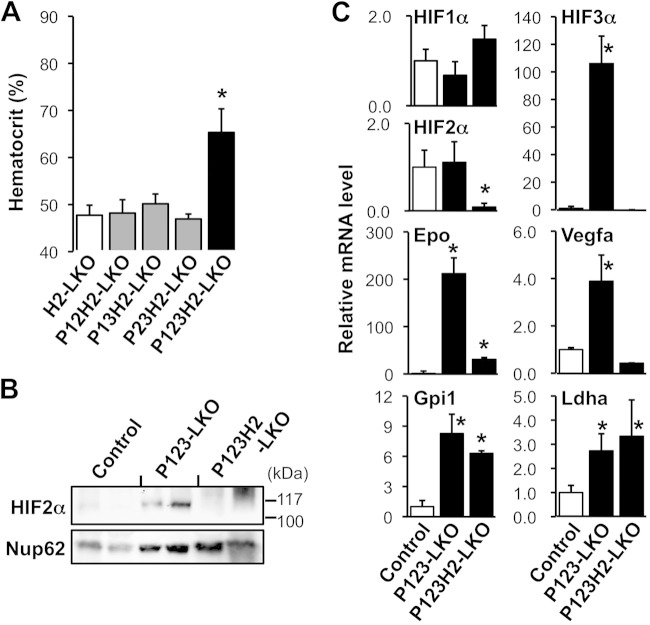
FIG 4.
HIF2α is essential for the development of polycythemia in liver-specific PHD knockout mice. (A) Hematocrit levels in PHD and HIF2α mutant mice at 8 to 16 weeks of age are shown as the means and SD (n = 3 to 5 for each group). *, P < 0.01 compared with control H2-LKO mice. (B) Immunoblots for HIF2α in nuclear extracts from the livers of PHD and HIF2α mutant mice at 4 to 10 weeks of age. Nup62 was used as an internal control. The data from 2 independent samples are shown for each genotype group. (C) mRNA expression levels in the livers of PHD and HIF2α mutant mice were analyzed by RT-qPCR. β-Actin was used as an internal control for RT-qPCR. The expression levels of control mice were set as 1.0. The data are means and SD (n = 3 for each group). *, P < 0.01 compared with the control mice (white bar). Note that the overexpression of Epo mRNA in P123-LKO livers is attenuated by the loss of HIF2α (P123H2-LKO).
Although the immunoblotting (Fig. 4B) and RT-qPCR (Fig. 4C) data showed that HIF2α expression was almost absent in hepatocytes of P123H2-LKO mice, the overexpression of the Epo gene and the polycythemia observed in P123-LKO mice were not completely rescued by HIF2α deletion (Fig. 4A and C). Thus, we suggest that HIF1α accumulation may contribute to the overexpression of Epo mRNA in the triple-knockout livers. The overexpression of the Vegfa gene was also rescued by HIF2α deficiency in P123-LKO livers, while mRNA expression of the glycolytic enzymes (Gpi1 and Ldha) was present at high levels regardless of the HIF2α status (Fig. 4C). These gene expression profiles indicated that HIF2α, rather than HIF1α, upregulates the transcription of the Epo and Vegfa genes in hepatocytes under hypoxic conditions, whereas the glycolytic genes are primarily regulated by HIF1α. Interestingly, the loss of all three PHD isoforms in hepatocytes strongly (100-fold) increased the HIF3α mRNA level; this overexpression was also rescued by HIF2α deficiency (Fig. 4C).
The Epo gene hepatic enhancer is required for Epo overproduction in PHD-deficient livers.
We previously reported that an enhancer sequence that contains a HIF-binding sequence (hypoxia-responsive element [HRE]) and that is proximally downstream of the Epo gene transcriptional endpoint is necessary and sufficient for the expression of the Epo gene in the liver (12). A mouse line (ΔEpoHE) lacking 500 bp of the hepatic enhancer for the Epo gene (EpoHE) presents with fetal anemia due to a loss of Epo production by the fetal liver, which is the major Epo-producing site in fetuses. In contrast, adult ΔEpoHE mice exhibit normal erythropoiesis because adult mice produce most of their Epo in the kidneys, where Epo expression is independent of EpoHE (12). ΔEpoHE mice were crossed with liver-specific PHD-deficient mice to determine whether EpoHE is a cis-regulatory element for the hepatic Epo expression induced by the PHD-HIF2α signaling cascade.
The polycythemia observed in PHD-mutant mice was completely rescued by the loss of EpoHE (Fig. 5A), demonstrating the essential function of EpoHE in hepatic Epo induction by the PHD-HIF2α hypoxia signaling cascade. Indeed, Epo expression in the livers of double (Fig. 5B)- and triple (Fig. 5C)-knockout mice with the EpoHE deficiency fell within the normal range. These data clearly showed that EpoHE is essential for the induction of hypoxia-inducible Epo expression by the PHD-HIF2α signaling cascade.
FIG 5.
The hepatic enhancer for the Epo gene (EpoHE) is necessary for the development of polycythemia in PHD mutant mice. (A) Hematocrit levels in the PHD and EpoHE mutant mice at 6 to 15 weeks of age. The data are means ± SD (n = 3 for each group). (B) Epo mRNA expression levels in the livers of PHD and EpoHE mutant mice at 8 to 15 weeks of age. The expression levels of control mice were set as 1.0 after normalization to β-actin mRNA levels. The data are means ± SD (n = 3 for each group). *, P < 0.01 compared with the levels in the control mice (left bar). (C) Expression levels of Epo, HIF3α, and Scd1 mRNAs in the livers of PHD and EpoHE mutant mice were analyzed by RT-qPCR. β-Actin was used as an internal control. The expression levels of control mice were set as 1.0. The data are means ± SD (n = 3 or 4 for each group). Note that the overexpression of Epo mRNA in P123-LKO livers is completely suppressed by the loss of EpoHE (P123-LKO:ΔEpoHE). (D) Lipid droplets in the livers of PHD mutant mice at 7 weeks after birth were stained with Nile red (red). Hoechst 33342 was used for nuclear staining (blue). (E) Glycogen deposition in the livers of the PHD and EpoHE mutant mice at 8 weeks of age was detected by PAS staining (purple). (F) Glucose concentrations were measured in peripheral blood from the PHD and EpoHE mutant mice at 8 weeks of age. The data are means ± SD (n = 3 for each group). (G) Hepcidin mRNA expression in the livers of PHD and EpoHE mutant mice was analyzed by RT-qPCR. β-Actin was used as an internal control. The expression levels in the control mice were set as 1.0. The data are means ± SD (n = 3 or 4 for each group). Bars, 50 μm (D and E). *, P < 0.01 compared with the control mice (C, F, and G).
HIF3α mRNA levels were unaffected by the loss of EpoHE (Fig. 5C), indicating that EpoHE functions solely in Epo gene regulation. In addition, EpoHE loss did not rescue the severe steatosis (Fig. 5D), the elevated glycogen deposition (Fig. 5E), or the downregulation of Scd1 expression (Fig. 5C) observed in PHD123-LKO livers. Additionally, the lethality of P123-LKO mice was not rescued by the EpoHE deletion. These findings suggested that hepatic steatosis and glycogen deposition are caused by the hyperactivation of HIFs but are not related to Epo overproduction or to polycythemia.
The peripheral blood glucose concentration in the PHD double-knockout mice was decreased compared with that in the control mice, and these reduced blood glucose levels were restored by EpoHE loss (Fig. 5F). This observation indicates that the decreased blood glucose levels are caused by polycythemia but are not related to HIF hyperactivation in liver-specific PHD-deficient mice because erythrocytes actively uptake and utilize blood glucose for energy production (48).
Iron homeostasis is mainly regulated by hepcidin, which inhibits iron usage for erythropoiesis, and hepcidin secretion from livers is downregulated in mice actively undergoing erythropoiesis (49). Hepcidin mRNA levels were dramatically decreased in livers from polycythemic PHD double-knockout mice compared with control mouse livers (Fig. 5G). In nonpolycythemic P23-LKO:ΔEpoHE mice, hepcidin mRNA levels were normal (Fig. 5G). This result is consistent with a previous report that hepcidin expression is suppressed by active erythropoiesis but not by direct effects of Epo or HIFs (49).
HIF2α is involved in nucleosome reorganization around the EPO gene in hepatocytes.
Next, we analyzed the chromatin state of HIF target genes in human hepatoma Hep3B cells that express both HIF1α and HIF2α proteins in a hypoxia-inducible manner to investigate how HIF2α activates transcription of the EPO gene in hepatocytes (Fig. 6A). In Hep3B cells, the expression of EPO and the expression of the well-known HIF targets carbonic anhydrase 9 (CA9) and phosphoglycerate kinase 1 (PGK1) were strongly induced by hypoxia (1% oxygen for 24 h) (Fig. 6B). HIF2α-specific inhibitor (HIF2 antagonist 2) (38) significantly decreased CA9 and EPO mRNA levels under hypoxic conditions, while the PGK1 mRNA level was not affected (Fig. 6B). In contrast, acriflavine, which is an inhibitor for both HIF1α and HIF2α (50), suppressed the expression of all three HIF target genes (Fig. 6B). Because both compounds inhibit the heterodimerization of HIFα and -β subunits (38, 50), these results corroborate the results from P123H2-LKO mice, which suggested that Epo gene expression is primarily activated by HIF2α and not by HIF1α. Indeed, previous studies utilizing RNA interference demonstrated that HIF2α-specific suppression, but not HIF1α-specific suppression, dramatically reduces hypoxia-inducible Epo gene expression in Epo-producing hepatic cell lines (51, 52).
FIG 6.
A HIF2α-specific inhibitor blocks the dissociation of nucleosome structures in the EPO gene under hypoxic conditions in human hepatocytes. (A) Immunoblots for HIF1α and HIF2α in Hep3B human hepatoma cells under normoxic (N) and hypoxic (H24 [1% oxygen for 24 h]) conditions. α-Tubulin was used as an internal control. Both HIF1α and HIF2α proteins accumulated in cells treated with a PHD inhibitor (FG4592) or in cells under hypoxic stimulation, and the protein levels were not altered by supplementation with HIF inhibitors (HIF2 antagonist 2 and acriflavine) under hypoxic conditions. (B) RT-qPCR of CA9, PGK1, and EPO mRNA expression in Hep3B cells cultured with vehicle (v) or HIF inhibitors (HIF2 antagonist 2 and acriflavine) under normoxic (N) or hypoxic (H24) conditions. β-Actin was used as an internal control for RT-qPCR. The expression levels under normal conditions with vehicle supplementation (v/N) were set as 1.0. The data are means and SD from 4 independent experiments. *, P < 0.01 compared with hypoxic samples supplemented with vehicle (v/H24). A HIF2α-specific inhibitor (HIF2 antagonist 2) strongly suppressed hypoxic induction of EPO expression but did not affect the expression of PGK1. (C) Nucleosome occupancy of chromatin loci for the CA9, PGK1, and EPO genes was detected via nuclease accessibility in Hep3B cells cultured with vehicle (v) or HIF inhibitors (HIF2 antagonist 2 and acriflavine) under normoxic (N) or hypoxic (H4 [1% oxygen for 4 h] and H24 [1% oxygen for 24 h]) conditions. The indicated loci were detected by qPCR of nuclease-digested and undigested chromatin from Hep3B cells, and the amount found in undigested samples was set as 100%. The values are means and SD from 4 independent experiments. *, P < 0.01 compared with vehicle-treated samples (v) under each condition. Nuclease accessibility was high in the PGK1 promoter region in all samples, indicating that this region is free from nucleosome structure under both normoxic and hypoxic conditions. The promoter, gene body (intron 1), and hepatic enhancer (HE) of the EPO gene and the CA9 promoter region were well protected from nuclease digestion under normal conditions (v/N), and nuclease accessibility increased under hypoxic conditions. Note that the formation of the nucleosome-free region under hypoxic conditions was repressed by HIF inhibitors.
Next, we used a qPCR-based technique to analyze the changes in nuclease accessibility (sensitivity) to chromatin DNA in Hep3B cells that were exposed to hypoxia (39). Nuclease-sensitive sites are often an indication of transcriptionally active genomic regions, in which histones have dissociated from nucleosomes to open the chromatin for transcription factor binding. Under normal conditions, chromatin DNA was highly protected from nuclease digestion in the promoter, gene body (intron 1) and enhancer (HE, a hepatic enhancer downstream of the transcription end site) regions of the EPO gene (Fig. 6C). Hypoxic stimuli (1% O2 for 4 or 24 h) increased the accessibility of the EPO gene locus (Fig. 6C). The CA9 promoter was similarly affected by hypoxia, whereas the PGK1 promoter remained sensitive to nucleases (Fig. 6C). These data demonstrated that the CA9 and EPO genes in Hep3B cells are covered by histones under normal conditions and that the nucleosome structure is disassembled under hypoxic conditions, facilitating active transcription. The PGK1 promoter is always nucleosome free to enable interactions with transcription factors and to achieve the high level of gene expression.
HIF2 antagonist 2 blocked the hypoxia-induced formation of open chromatin structures in the EPO and CA9 gene loci (gray bars in Fig. 6C). Acriflavine treatment also caused these chromatin regions to become insensitive to nuclease digestion under both 4- and 24-hour hypoxic conditions (Fig. 6C). These data corroborate our gene expression data showing that both inhibitors suppress EPO and CA9 gene expression. These inhibitors did not have any effect on the constitutively open chromatin locus of the PGK1 gene (Fig. 6C). Thus, we concluded that HIF2α is required for the formation of open chromatin structures in the EPO gene locus to induce active transcription under hypoxic conditions in hepatocytes.
HIF3α is induced by hypoxia and attenuates HIF2α-mediated EPO transcription in hepatocytes.
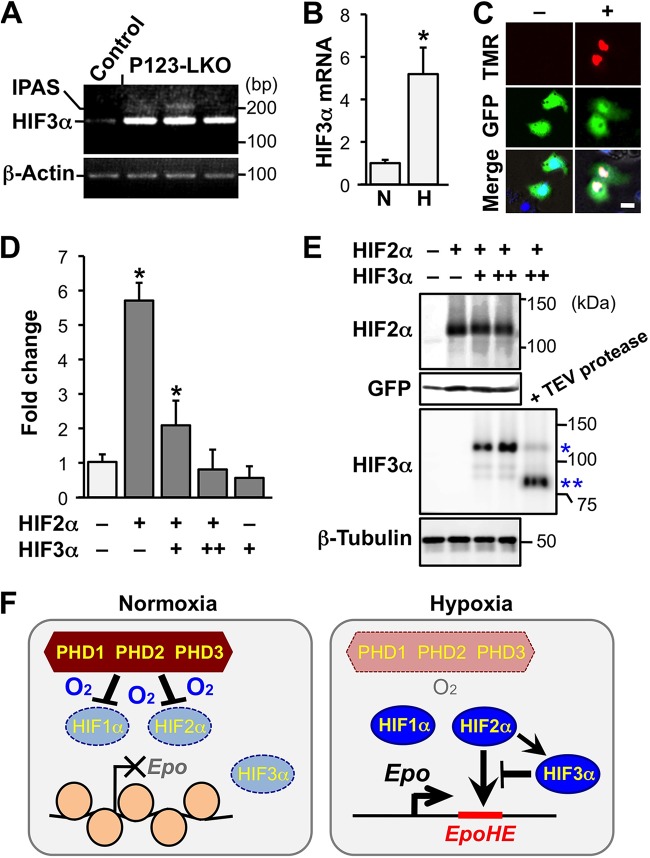
Because we discovered that HIF3α mRNA expression, similar to Epo mRNA expression, is strongly induced by the loss of PHD isoforms in livers in a HIF2α-dependent manner (Fig. 4C and 5C), the role of HIF3α function in hepatic Epo gene regulation was further analyzed. The Hif3a gene in mice mainly generates two splicing isoforms, IPAS and NEPAS, in addition to full-length HIF3α (18, 19). NEPAS is exclusively expressed at embryonic and neonatal stages, whereas IPAS expression is induced by hypoxia in the hearts, lungs, and muscles of adult mice (18). We determined the expression of IPAS-specific transcripts in P123-LKO livers and found that IPAS expression was only weakly induced by PHD deficiency, compared to the strong induction of full-length HIF3α expression caused by PHD deficiency (Fig. 7A). HIF3α mRNA expression in the mouse liver was also stimulated by exposure to hypoxic stress (6% oxygen for 48 h) (Fig. 7B). These data indicated that full-length HIF3α expression is induced by hypoxia through the PHD-HIF pathway in the liver.
FIG 7.
Involvement of HIF3α in the hypoxic regulation of Epo expression. (A) Detection of the IPAS-specific splicing variant of the Hif3a gene by RT-PCR in livers of P123-LKO and control mice. β-Actin was used as an internal control. The 207-bp PCR amplicon derived from the IPAS splicing variant was undetected or detected at extremely low levels by agarose gel electrophoresis of PCR samples compared with the 151-bp amplicon derived from HIF3α-specific splicing variants. (B) Expression levels of HIF3α mRNA in livers of wild-type mice exposed to normal (N) or hypoxic (6% oxygen) air for 48 h were measured by RT-qPCR. The expression levels of normoxic samples were set as 1.0. The data are means and SD from 3 independent mice for each condition. *, P < 0.01 compared with normoxic samples. (C) Overexpression of HIF3α in the nuclei of living Hep3B cells. Hep3B cells were transfected with a control plasmid expressing GFP (pEGFP-N1 [green]) with (+) or without (−) the HaloTag-HIF3α expression plasmid. HaloTag-HIF3α was detected with a HaloTag-TMR direct ligand (red) in Hoechst 33342-stained nuclei (blue [merged images in lower panels]). Bar, 10 μm. (D) Luciferase reporter analysis of EpoHE in Hep3B cells overexpressing HIF2α and/or HIF3α. The HIF2α expression plasmid and/or the HIF3α expression plasmid (+, 300 ng/well; ++, 600 ng/well) were cotransfected with the EpoHE-reporter plasmid. The relative luciferase activity of the control samples (white bar) was set as 1.0 after normalization with internal controls. The values are means and SD from 4 independent experiments. *, P < 0.01 compared with control samples. (E) Expression of HIF2α and HIF3α in cells that were used for the EpoHE reporter assay was confirmed by immunoblotting. The constitutively active HIF2α mutant was detected using anti-HIF2α antibody in whole-cell lysates from the cells used for panel D. Dose-dependent expression of HaloTag-HIF3α (*) was detected using anti-HIF3α antibody. HaloTag-cleaved HIF3α (**) was also detected in a TEV protease-digested sample. GFP and β-tubulin were used as loading controls. (F) A model of hypoxia-inducible Epo gene regulation in hepatocytes. Under normoxic conditions, PHDs suppress the activities of HIF1α and HIF2α via protein degradation, and nucleosome structures on the chromatin silence the Epo gene. In hypoxic cells, HIF1α and HIF2α are activated by the inactivation of PHDs. HIF2α induces Epo transcription via the disassembly of the nucleosome structures. The HIF2α-mediated Epo induction is attenuated by HIF3α, which is also induced by HIF2α.
We analyzed the activity of EpoHE in Hep3B cells that overexpress human full-length HIF3α to investigate the function of HIF3α in EpoHE-mediated gene regulation. We detected the nucleus-specific localization of overexpressed HIF3α, which was conjugated with HaloTag, via staining with a cell-permeable HaloTag ligand in living Hep3B cells (Fig. 7C). The expression of a luciferase reporter driven by EpoHE was significantly increased by HIF2α overexpression, and this induction was suppressed by HIF3α in a dose-dependent manner (Fig. 7D). These data indicated the functional importance of HIF3α in hypoxia-inducible Epo gene regulation. After the removal of the HaloTag by TEV protease, the Western blot data confirmed that the overexpressed HIF3α was the 80-kDa full-length HIF3α (HIF3α1) (20) and that HIF2α protein levels in the constitutively active HIF2α mutant were not affected by HIF3α overexpression (Fig. 7E). Taken together, our results suggest that HIF3α expression in the liver is induced by hypoxia to limit HIF2α-mediated Epo overproduction (Fig. 7F).
DISCUSSION
The REP cells in the renal tubular interstitium are the site of the majority of the Epo production in adult animals (9, 13); the contribution of the liver, which is the other Epo-producing organ, to adult erythropoiesis is extremely small (12). Therefore, damage to the REP cells in chronic kidney diseases causes renal anemia even in patients with an intact liver. Strategies that inhibit PHDs by using small molecules or RNA interference have been investigated in an attempt to develop a treatment that would enhance hepatic Epo production in patients with renal anemia (14, 15, 53). However, PHD inhibition induces several genes, including oncogenes, steatosis-related genes, and the Epo gene, via the activation of both HIF1α and HIF2α (54). In the present study, we investigated the in vivo regulatory mechanisms of hepatic Epo production to identify therapeutic targets that exclusively stimulate Epo production.
The liver-specific knockouts of each PHD isoform exhibited no apparent phenotype, indicating that PHDs have redundant functions in regulating the hypoxia-response pathway (28, 29). The loss of all three genes for PHD isoforms in the liver resulted in Epo overproduction and polycythemia via the upregulation of both HIF1α and HIF2α. However, the strong and constitutive activation of HIFs caused steatosis of the liver and the expression of other HIF target genes (29), and the mice died approximately 10 weeks after birth. We speculate that the development of fatty livers in the P123-LKO mice is related to the HIF-mediated inactivation of mitochondria, which are the organelles that utilize intracellular fatty acids (55). We also showed that these phenotypes depended on neither polycythemia nor Epo overproduction by examining P123-LKO mice lacking EpoHE, which exhibited HIF overexpression and lethal steatosis but not Epo overproduction. The data from the P123-LKO mice suggested that PHD inhibition is responsible for the induction of hepatic Epo production, although universal PHD inhibitors not only induce Epo production but also produce dangerous side effects.
Our data demonstrated that the disruption of two PHD isoforms was sufficient to activate erythropoiesis via Epo gene induction in hepatocytes, without the severe steatosis that was observed in the P123-LKO livers. Surprisingly, the mice lacking two PHD isoforms in any combination exhibited similar polycythemia phenotypes, suggesting that no significant functional differences in hepatic Epo gene regulation exist among the three PHD isoforms. However, we believe that these isoforms may have specific roles in other parts of the hypoxia response pathway because these isoforms have different affinities for HIFs, which differ in protein structure outside their catalytic domains (56).
Although the level of Epo mRNA expression in the double-knockout livers was approximately 10-fold higher than that of the control mice, their plasma Epo concentrations were within the normal range. Epo is internalized and degraded in EpoR-expressing erythroid cells after the transduction of erythropoietic signaling (42). Therefore, our data suggest that most of the Epo produced by the double-knockout livers is absorbed by erythroblastic cells expressing EpoR at a high level, which increase more than 10-fold in number in the spleens of double-knockout mice. The plasma concentration of thrombopoietin (Tpo) depends on the quantity of c-mpl (Tpo receptor)-expressing platelets in peripheral blood (so-called “sponge theory”) (57). Thus, we propose a sponge theory for Epo, in which plasma Epo levels are related to the number of erythroblastic cells in spleens.
Polycythemia in the liver-specific PHD knockout mice was rescued by liver-specific HIF2α deficiency. This result is comparable to that of a previous study, which showed that HIF2α is required for the development of polycythemia in liver-specific knockout of pVHL, which is an E3 ligase of HIFα subunits (26). These findings clearly indicate that HIF2α, rather than HIF1α, is the major transactivator for Epo gene expression. Additionally, we showed that polycythemia was completely rescued by the loss of EpoHE. Taken together, our data suggest that HIF2α binding to EpoHE, which contains an HRE, triggers Epo production in the liver and that EpoHE is the only cis-regulatory element for hypoxia-inducible Epo gene expression in hepatocytes (12). Patients with hepatocellular carcinoma occasionally present both polycythemia and Epo overproduction (58). In these cases, PHDs or other HIF suppression systems might be disrupted in the malignant cells. Recently, hereditary polycythemia has been linked to mutations in the PHD2 and HIF2α genes (59–63), indicating that a PHD-HIF2α pathway regulates EPO gene expression in humans.
Chromatin analyses demonstrated that the genomic DNA around the Epo gene, including the promoter and EpoHE, associates with histones under normal conditions. Under hypoxic conditions, the nucleosome structure is disassembled in a HIF2α-dependent manner to activate Epo transcription (Fig. 7F). Chromatin remodeling factors, such as BRG1, BRM, and SWI/SNF, are implicated in HIF-mediated transcriptional activation (64, 65). Therefore, these factors may be recruited to the Epo gene locus by HIF2α, which binds to EpoHE in hypoxic cells.
We found that HIF3α expression is strongly induced by PHD deficiency in the liver and that this induction requires HIF2α. Some reports have shown that HIF1α, but not HIF2α, activates Hif3a gene expression via binding to its HREs (20, 66). However, we suggest that hypoxia-inducible Hif3a gene expression in normal mouse liver is regulated primarily by HIF2α. Because splicing variants of HIF3α exist, the function of HIF3α in hypoxia-inducible gene regulation is controversial (18, 19, 67, 68). In the present study, we demonstrated that full-length HIF3α is primarily induced in hypoxic livers and that this expression attenuates transcriptional activation driven by the HIF2α-EpoHE axis (Fig. 7F). Since HIF2α deletion did not completely reduce the overexpression of the Epo gene, which was observed in P123-LKO mice, we suggested that HIF1α secondly contributed to the overexpression of Epo mRNA. Alternatively, loss of Hif3a gene induction by HIF2α deletion may be related to the incomplete reduction of the Epo overexpression in the quadruple-knockout livers.
As shown by the dramatic induction of hepatic Epo production following the genetic or pharmacological inactivation of PHDs, PHDs actively inhibit the function of HIFs, even in renal-anemia patients. Therefore, it is plausible to inhibit PHDs in the liver to treat renal anemia. However, mouse genetic studies have demonstrated that the universal inhibition of PHDs strongly induces the activation of HIFs and causes side effects, including steatosis (54, 69). To avoid severe side effects caused by robust inhibition of PHDs, one reasonable strategy suggested by this study is to establish mechanisms to specifically induce HIF2α-EpoHE binding. Because chemical inhibitors usually only partly suppress enzymatic activity and their efficacy can be regulated by dosage, chemical compounds that inhibit PHDs have been tested in clinical trials for renal anemia (14). Additionally, it was recently reported that treating mice with a PHD inhibitor (FG4497) improves metabolic dysfunction, including hepatic steatosis (70). This evidence indicates that renal anemia can be treated with PHD inhibitors.
ACKNOWLEDGMENTS
We thank Atsuko Konuma, Mizuho Tanno, Eriko Naganuma, Aina Fukuda, and Koichiro Kato (Tohoku University). We are also grateful to the Biomedical Research Core and the Centre for Laboratory Animal Research of Tohoku University for technical support.
This work was supported in part by grants-in-aid from MEXT/JSPS KAKENHI (grants 26111002 and 24249015 to M.Y. and 26116702 and 25670157 to N.S.), the Inamori Foundation (N.S.), the Takeda Science Foundation (N.S.), and the Platform for Drug Discovery, Informatics, and Structural Life Science from MEXT, Japan (T.T., T.M., M.Y., and N.S.).
The funders had no role in the design of the study, data collection and analysis, decision to publish or preparation of the manuscript. We have no competing interests to declare.
Y.T. and N.S. designed the study. Y.T., H.S., I.H., X.P., T.S., N.T., and N.S. performed the experiments. K.T., G.-H.F., T.D., T.M., M.Y., and N.S. provided the gene-modified mouse lines. T.T, S.-I.K., and T.M. provided the chemical compounds. Y.T., H.S., and N.S. analyzed the data and constructed the figures. Y.T. and N.S. wrote the manuscript. M.I., T.M., M.Y., and N.S. developed the project.
REFERENCES
- 1.Ebert BL, Bunn HF. 1999. Regulation of the erythropoietin gene. Blood 94:1864–1877. [PubMed] [Google Scholar]
- 2.Milledge JS, Cotes PM. 1985. Serum erythropoietin in humans at high altitude and its relation to plasma renin. J Appl Physiol 59:360–364. [DOI] [PubMed] [Google Scholar]
- 3.Guidet B, Offenstadt G, Boffa G, Najman A, Baillou C, Hatzfeld C, Amstutz P. 1987. Polycythemia in chronic obstructive pulmonary disease. Chest 92:867–870. doi: 10.1378/chest.92.5.867. [DOI] [PubMed] [Google Scholar]
- 4.Franke K, Gassmann M, Wielockx B. 2013. Erythrocytosis: the HIF pathway in control. Blood 122:1122–1128. doi: 10.1182/blood-2013-01-478065. [DOI] [PubMed] [Google Scholar]
- 5.Wu H, Liu X, Jaenisch R, Lodish HF. 1995. Generation of committed erythroid BFU-E and CFU-E progenitors does not require erythropoietin or the erythropoietin receptor. Cell 83:59–67. [DOI] [PubMed] [Google Scholar]
- 6.Babitt JL, Lin HY. 2012. Mechanisms of anemia in CKD. J Am Soc Nephrol 23:1631–1634. doi: 10.1681/ASN.2011111078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suzuki N, Obara N, Yamamoto M. 2007. Use of gene-manipulated mice in the study of erythropoietin gene expression. Methods Enzymol 435:157–177. doi: 10.1016/S0076-6879(07)35009-X. [DOI] [PubMed] [Google Scholar]
- 8.Obara N, Suzuki N, Kim K, Nagasawa T, Imagawa S, Yamamoto M. 2008. Repression via the GATA box is essential for tissue-specific erythropoietin gene expression. Blood 111:5223–5232. doi: 10.1182/blood-2007-10-115857. [DOI] [PubMed] [Google Scholar]
- 9.Pan X, Suzuki N, Hirano I, Yamazaki S, Minegishi N, Yamamoto M. 2011. Isolation and characterization of renal erythropoietin-producing cells from genetically produced anemia mice. PLoS One 6:e25839. doi: 10.1371/journal.pone.0025839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Asada N, Takase M, Nakamura J, Oguchi A, Asada M, Suzuki N, Yamamura K, Nagoshi N, Shibata S, Rao TN, Fehling HJ, Fukatsu A, Minegishi N, Kita T, Kimura T, Okano H, Yamamoto M, Yanagita M. 2011. Dysfunction of fibroblasts of extrarenal origin underlies renal fibrosis and renal anemia in mice. J Clin Invest 121:3981–3990. doi: 10.1172/JCI57301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Souma T, Yamazaki S, Moriguchi T, Suzuki N, Hirano I, Pan X, Minegishi N, Abe M, Kiyomoto H, Ito S, Yamamoto M. 2013. Plasticity of renal erythropoietin-producing cells governs fibrosis. J Am Soc Nephrol 24:1599–1616. doi: 10.1681/ASN.2013010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suzuki N, Obara N, Pan X, Watanabe M, Jishage K, Minegishi N, Yamamoto M. 2011. Specific contribution of the erythropoietin gene 3′ enhancer to hepatic erythropoiesis after late embryonic stages. Mol Cell Biol 31:3896–3905. doi: 10.1128/MCB.05463-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamazaki S, Souma T, Hirano I, Pan X, Minegishi N, Suzuki N, Yamamoto M. 2013. A mouse model of adult-onset anaemia due to erythropoietin deficiency. Nat Commun 4:1950. doi: 10.1038/ncomms2950. [DOI] [PubMed] [Google Scholar]
- 14.Bernhardt WM, Wiesener MS, Scigalla P, Chou J, Schmieder RE, Günzler V, Eckardt KU. 2010. Inhibition of prolyl hydroxylases increases erythropoietin production in ESRD. J Am Soc Nephrol 21:2151–2156. doi: 10.1681/ASN.2010010116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Querbes W, Rogorad RL, Moslehi J, Wong J, Chan AY, Bulgakova E, Kuchimanchi S, Akinc A, Fitzgerald K, Koteliansky Kaelin WG Jr. 2012. Treatment of erythropoietin deficiency in mice with systemically administered siRNA. Blood 120:1916–1922. doi: 10.1182/blood-2012-04-423715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Semenza GL, Koury ST, Nejfelt MK, Gearhart JD, Antonarakis SE. 1991. Cell-type-specific and hypoxia-inducible expression of the human erythropoietin gene in transgenic mice. Proc Natl Acad Sci U S A 88:8725–8729. doi: 10.1073/pnas.88.19.8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lendahl U, Lee KL, Yang H, Poellinger L. 2009. Generating specificity and diversity in the transcriptional response to hypoxia. Nat Rev Genet 10:821–832. doi: 10.1038/nrg2665. [DOI] [PubMed] [Google Scholar]
- 18.Makino Y, Cao R, Svensson K, Bertilsson G, Asman M, Tanaka H, Cao Y, Berkenstam A, Poellinger L. 2001. Inhibitory PAS domain protein is a negative regulator of hypoxia-inducible gene expression. Nature 414:550–554. doi: 10.1038/35107085. [DOI] [PubMed] [Google Scholar]
- 19.Yamashita T, Ohneda O, Nagano M, Iemitsu M, Makino Y, Tanaka H, Miyauchi T, Goto K, Ohneda K, Fujii KY, Poellinger L, Yamamoto M. 2008. Abnormal heart development and lung remodeling in mice lacking the hypoxia-inducible factor-related basic helix-loop-helix PAS protein NEPAS. Mol Cell Biol 28:1285–1297. doi: 10.1128/MCB.01332-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pasanen A, Heikkilä M, Rautavuoma K, Hirsilä M, Kivirikko KI, Myllyharju J. 2010. Hypoxia-inducible factor (HIF)-3alpha is subject to extensive alternative splicing in human tissues and cancer cells and is regulated by HIF-1 but not HIF-2. Int J Biochem Cell Biol 42:1189–1200. doi: 10.1016/j.biocel.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 21.Epstein AC, Gleadle JM, McNeill LA, Hewitson KS, O'Rourke J, Mole DR, Mukherji M, Metzen E, Wilson MI, Dhanda A, Tian YM, Masson N, Hamilton DL, Jaakkola P, Barstead R, Hodgkin J, Maxwell PH, Pugh CW, Schofield CJ, Ratcliffe PJ. 2001. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107:43–54. [DOI] [PubMed] [Google Scholar]
- 22.Ivan M, Haberberger T, Gervasi DC, Michelson KS, Günzler V, Kondo K, Yang H, Sorokina I, Conaway RC, Conaway JW, Kaelin WG Jr. 2002. Biochemical purification and pharmacological inhibition of a mammalian prolyl hydroxylase acting on hypoxia-inducible factor. Proc Natl Acad Sci U S A 99:13459–13464. doi: 10.1073/pnas.192342099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, von Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ, Maxwell PH, Pugh CW, Ratcliffe PJ. 2001. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292:468–472. [DOI] [PubMed] [Google Scholar]
- 24.Miyata T, Suzuki N, van Ypersele de Strihou C. 2013. Diabetic nephropathy: are there new and potentially promising therapies targeting oxygen biology? Kidney Int 84:693–702. doi: 10.1038/ki.2013.74. [DOI] [PubMed] [Google Scholar]
- 25.Haase HV. 2013. Regulation of erythropoiesis by hypoxia-inducible factors. Blood Rev 27:47–53. doi: 10.1016/j.blre.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rankin EB, Biju MP, Liu Q, Unger TL, Rha J, Johnson RS, Simon MC, Keith B, Haase VH. 2007. Hypoxia-inducible factor-2 (HIF-2) regulates hepatic erythropoietin in vivo. J Clin Invest 117:1068–1077. doi: 10.1172/JCI30117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takeda K, Aguila HL, Parikh NS, Li X, Lamothe K, Duan LJ, Takeda H, Lee FS, Fong GH. 2008. Regulation of adult erythropoiesis by prolyl hydroxylase domain proteins. Blood 111:3229–3235. doi: 10.1182/blood-2007-09-114561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minamishima YA, Kaelin WG Jr. 2010. Reactivation of hepatic EPO synthesis in mice after PHD loss. Science 329:407. doi: 10.1126/science.1192811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duan LJ, Takeda K, Fong GH. 2014. Hematological, hepatic, and retinal phenotypes in mice deficient for prolyl hydroxylase domain proteins in the liver. Am J Pathol 184:1240–1250. doi: 10.1016/j.ajpath.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takeda K, Ho VC, Takeda H, Duan LJ, Nagy A, Fong GH. 2006. Placental but not heart defects are associated with elevated hypoxia-inducible factor a levels in mice lacking prolyl hydroxylase domain protein 2. Mol Cell Biol 26:8336–8346. doi: 10.1128/MCB.00425-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gruber M, Hu CJ, Johnson RS, Brown EJ, Keith B, Simon MC. 2007. Acute postnatal ablation of Hif-2alpha results in anemia. Proc Natl Acad Sci U S A 104:2301–2306. doi: 10.1073/pnas.0608382104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Postic C, Shiota M, Niswender KD, Jetton TL, Chen Y, Moates JM, Shelton KD, Lindner J, Cherrington AD, Magnuson MA. 1999. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J Biol Chem 274:305–315. doi: 10.1074/jbc.274.1.305. [DOI] [PubMed] [Google Scholar]
- 33.Makino Y, Kanopka A, Wilson WJ, Tanaka H, Poellinger L. 2002. Inhibitory PAS domain protein (IPAS) is a hypoxia-inducible splicing variant of the hypoxia-inducible factor-3alpha locus. J Biol Chem 277:32405–32408. doi: 10.1074/jbc.C200328200. [DOI] [PubMed] [Google Scholar]
- 34.Greenspan P, Mayer EP, Fowler SD. 1985. Nile red: a selective fluorescent stain for intracellular lipid droplets. J Cell Biol 100:965–973. doi: 10.1083/jcb.100.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goldberg MA, Glass GA, Cunningham JM, Bunn HF. 1987. The regulated expression of erythropoietin by two human hepatoma cell lines. Proc Natl Acad Sci U S A 84:7972–7976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kallio PJ, Wilson WJ, O'Brien S, Makino Y, Poellinger L. 1999. Regulation of the hypoxia-inducible transcription factor 1alpha by the ubiquitin-proteasome pathway. J Biol Chem 274:6519–6525. doi: 10.1074/jbc.274.10.6519. [DOI] [PubMed] [Google Scholar]
- 37.Ohtsuji M, Katsuoka F, Kobayashi A, Aburatani H, Hayes JD, Yamamoto M. 2008. Nrf1 and Nrf2 play distinct roles in activation of antioxidant response element-dependent genes. J Biol Chem 283:33554–33562. doi: 10.1074/jbc.M804597200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scheuermann TH, Li Q, Ma HW, Key J, Zhang L, Chen R, Garcia JA, Naidoo J, Longgood J, Frantz DE, Tambar UK, Gardner KH, Bruick RK. 2013. Allosteric inhibition of hypoxia inducible factor-2 with small molecules. Nat Chem Biol 9:271–276. doi: 10.1038/nchembio.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen P, Zhao J, Wang Y, Wang M, Long H, Liang D, Huang L, Wen Z, Li W, Li X, Feng H, Zhao H, Zhu P, Li M, Wang QF, Li G. 2013. H3.3 actively marks enhancers and primes gene transcription via opening higher-ordered chromatin. Genes Dev 27:2109–2124. doi: 10.1101/gad.222174.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Metzen E, Stiehl DP, Doege K, Marxsen JH, Hellwig-Bürgel T, Jelkmann W. 2005. Regulation of the prolyl hydroxylase domain protein 2 (phd2/egln-1) gene: identification of a functional hypoxia-responsive element. Biochem J 387:711–717. doi: 10.1042/BJ20041736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pescador N, Cuevas Y, Naranjo S, Alcaide M, Villar D, Landázuri MO, Del Peso L. 2005. Identification of a functional hypoxia-responsive element that regulates the expression of the egl nine homologue 3 (egln3/phd2) gene. Biochem J 390:189–197. doi: 10.1042/BJ20042121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gross AW, Lodish HF. 2006. Cellular trafficking and degradation of erythropoietin and novel erythropoiesis stimulating protein (NESP). J Biol Chem 281:2024–2032. doi: 10.1074/jbc.M510493200. [DOI] [PubMed] [Google Scholar]
- 43.Suzuki N, Suwabe N, Ohneda O, Obara N, Imagawa S, Pan X, Motohashi H, Yamamoto M. 2003. Identification and characterization of 2 types of erythroid progenitors that express GATA-1 at distinct levels. Blood 102:3575–3583. doi: 10.1182/blood-2003-04-1154. [DOI] [PubMed] [Google Scholar]
- 44.Haase VH, Glickman JN, Socolovsky M, Jaenisch R. 2001. Vascular tumors in livers with targeted inactivation of the von Hippel-Lindau tumor suppressor. Proc Natl Acad Sci U S A 98:1583–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qu A, Taylor M, Xue X, Matsubara T, Metzger D, Chambon P, Gonzalez FJ, Shah YM. 2011. Hypoxia-inducible transcription factor 2α promotes steatohepatitis through augmenting lipid accumulation, inflammation, and fibrosis. Hepatology 54:472–483. doi: 10.1002/hep.24400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang J, Ney PA. 2009. Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ 16:939–946. doi: 10.1038/cdd.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park SK, Haase VH, Johnson RS. 2007. von Hippel Lindau tumor suppressor regulates hepatic glucose metabolism by controlling expression of glucose transporter 2 and glucose 6-phosphatase. Int J Oncol 30:341–348. doi: 10.3892/ijo.30.2.341. [DOI] [PubMed] [Google Scholar]
- 48.Zhang JZ, Ismail-Beigi F. 1998. Activation of Glut1 glucose transporter in human erythrocytes. Arch Biochem Biophys 356:86–92. doi: 10.1006/abbi.1998.0760. [DOI] [PubMed] [Google Scholar]
- 49.Liu Q, Davidoff O, Niss K, Haase VH. 2012. Hypoxia-inducible factor regulates hepcidin via erythropoietin-induced erythropoiesis. J Clin Invest 122:4635–4644. doi: 10.1172/JCI63924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee K, Zhang H, Qian DZ, Rey S, Liu JO, Semenza GL. 2009. Acriflavine inhibits HIF-1 dimerization, tumor growth, and vascularization. Proc Natl Acad Sci U S A 106:17910–17915. doi: 10.1073/pnas.0909353106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 51.Befani C, Mylonis I, Gkotinakou IM, Georgoulias P, Hu CJ, Simos G, Liakos P. 2013. Cobalt stimulates HIF-1-dependent but inhibits HIF-2-dependent gene expression in liver cancer cells. Int J Biochem Cell Biol 45:2359–2368. doi: 10.1016/j.biocel.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Warnecke C, Zaborowska Z, Kurreck J, Erdmann VA, Frei U, Wiesener M, Eckardt KU. 2004. Differentiating the functional role of hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha (EPAS-1) by the use of RNA interference: erythropoietin is a HIF-2alpha target gene in Hep3B and Kelly cells. FASEB J 18:1462–1464. doi: 10.1096/fj.04-1640fje. [DOI] [PubMed] [Google Scholar]
- 53.Vachal P, Miao S, Pierce JM, Guiadeen D, Colandrea VJ, Wyvratt MJ, Salowe SP, Sonatore LM, Milligan JA, Hajdu R, Gollapudi A, Keohane CA, Lingham RB, Mandala SM, DeMartino JA, Tong X, Wolff M, Steinhuebel D, Kieczykowski GR, Fleitz FJ, Chapman K, Athanasopoulos J, Adam G, Akyuz CD, Jena DK, Lusen JW, Meng J, Stein BD, Xia L, Sherer EC, Hale JJ. 2012. 1,3,8-Triazaspiro[4.5]decane-2,4-diones as efficacious pan-inhibitors of hypoxia-inducible factor prolyl hydroxylase 1-3 (HIF PHD1-3) for the treatment of anemia. J Med Chem 55:2945–2959. doi: 10.1021/jm201542d. [DOI] [PubMed] [Google Scholar]
- 54.Rankin EB, Rha J, Selak MA, Unger TL, Keith B, Liu Q, Haase VH. 2009. Hypoxia-inducible factor 2 regulates hepatic lipid metabolism. Mol Cell Biol 29:4527–4538. doi: 10.1128/MCB.00200-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Semenza GL. 2011. Regulation of metabolism by hypoxia-inducible factor 1. Cold Spring Harbor Symp Quant Biol 76:347–353. doi: 10.1101/sqb.2011.76.010678. [DOI] [PubMed] [Google Scholar]
- 56.Pappalardi MB, Mcnulty DE, Martin JD, Fisher KE, Jiang Y, Burns MC, Zhao H, Thau HO, Sweitzer S, Schwartz Annan B RS, Copeland RA, Tummino PJ, Luo L. 2011. Biochemical characterization of human HIF hydroxylases using HIF protein substrates that contain all three hydroxylation sites. Biochem J 436:363–369. doi: 10.1042/BJ20101201. [DOI] [PubMed] [Google Scholar]
- 57.Fielder PJ, Gurney AL, Stefanich E, Marian M, Moore MW, Carver-Moore K, de Sauvage FJ. 1996. Regulation of thrombopoietin levels by c-mpl-mediated binding to platelets. Blood 87:2154–2161. [PubMed] [Google Scholar]
- 58.Sakisaka S, Watanabe M, Tateishi H, Harada M, Shakado S, Mimura Y, Gondo K, Yoshitake M, Noguchi K, Hino T. 1993. Erythropoietin production in hepatocellular carcinoma cells associated with polycythemia: immunohistochemical evidence. Hepatology 18:1357–1362. [PubMed] [Google Scholar]
- 59.Percy MJ, Zhao Q, Flores A, Harrison C, Lappin TR, Maxwell PH, McMullin MF, Lee FS. 2006. A family with erythrocytosis establishes a role for prolyl hydroxylase domain protein 2 in oxygen homeostasis. Proc Natl Acad Sci U S A 103:654–659. doi: 10.1073/pnas.0508423103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ladroue C, Hoogewijs D, Gad S, Carcenac R, Storti F, Barrois M, Gimenez-Rogueplo AP, Leporrier M, Casadevall N, Hermine O. 2012. Distinct deregulation of the hypoxia inducible factor by PHD2 mutants identified in germline DNA of patients with polycythemia. Haematologicaae 97:9–14. doi: 10.3324/haematol.2011.044644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Albiero E, Ruggeri M, Fortuna S, Finotto S, Bernardi M, Madeo D, Rodeghiero F. 2012. Isolated erythrocytosis: study of 67 patients and identification of three novel germ-line mutations in the prolyl hydroxylase domain protein 2 (PHD2) gene. Haematologicaae 97:123–127. doi: 10.3324/haematol.2010.039545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Percy MJ, Chung YJ, Harrison C, Mercieca J, Hoffbrand AV, Dinardo CL, Santos PC, Fonseca GH, Gualandro SF, Pereira AC, Lappin TR, McMullin MF, Lee FS. 2012. Two new mutations in the HIF2A gene associated with erythrocytosis. Am J Hematol 87:439–442. doi: 10.1002/ajh.23123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Perrotta S, Stiehl DP, Punzo F, Scianguetta S, Borriello A, Bencivenga D, Casale M, Nobili B, Fasoli S, Balduzzi A. 2013. Congenital erythrocytosis associated with gain-of-function HIF2A gene mutations and erythropoietin levels in the normal range. Haematologicaae 98:1624–1632. doi: 10.3324/haematol.2013.088369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang F, Zhang R, Beischlag TV, Muchardt C, Yaniv M, Hankinson O. 2004. Roles of Brahma and Brahma/SWI2-related gene 1 in hypoxic induction of the erythropoietin gene. J Biol Chem 279:46733–46741. doi: 10.1074/jbc.M409002200. [DOI] [PubMed] [Google Scholar]
- 65.Sena JA, Wang L, Hu CJ. 2013. BRG1 and BRM chromatin-remodeling complexes regulate the hypoxia response by acting as coactivators for a subset of hypoxia-inducible transcription factor target genes. Mol Cell Biol 33:3849–3863. doi: 10.1128/MCB.00731-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Makino Y, Uenishi R, Okamoto K, Isoe T, Hosono O, Tanaka H, Kanopka A, Poellinger L, Haneda M, Morimoto C. 2007. Transcriptional up-regulation of inhibitory PAS domain protein gene expression by hypoxia-inducible factor 1 (HIF-1): a negative feedback regulatory circuit in HIF-1-mediated signaling in hypoxic cells. J Biol Chem 282:14073–14082. doi: 10.1074/jbc.M700732200. [DOI] [PubMed] [Google Scholar]
- 67.Tanaka T, Wiesener M, Bernhardt W, Eckardt KU, Warnecke C. 2009. The human HIF (hypoxia-inducible factor)-3alpha gene is a HIF-1 target gene and may modulate hypoxic gene induction. Biochem J 424:143–151. doi: 10.1042/BJ20090120. [DOI] [PubMed] [Google Scholar]
- 68.Maynard MA, Qi H, Chung J, Lee EH, Kondo Y, Hara S, Conaway RC, Conaway JW, Ohh M. 2003. Multiple splice variants of the human HIF-3 alpha locus are targets of the von Hippel-Lindau E3 ubiquitin ligase complex. J Biol Chem 278:11032–11040. doi: 10.1074/jbc.M208681200. [DOI] [PubMed] [Google Scholar]
- 69.Minamishima YA, Moslehi J, Padera RF, Bronson RT, Liao R, Kaelin WG Jr. 2009. A feedback loop involving the Phd2 prolyl hydroxylase tunes the mammalian hypoxic response in vivo. Mol Cell Biol 29:5729–5741. doi: 10.1128/MCB.00331-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rahtu-Korpela L, Karsikas S, Hörkkö S, Blanco Sequeiros R, Lammentausta E, Mäkelä KA, Herzig KH, Walkinshaw G, Kivirikko KI, Myllyharju J, Serpi R, Koivunen P. 2014. HIF prolyl 4-hydroxylase-2 inhibition improves glucose and lipid metabolism and protects against obesity and metabolic dysfunction. Diabetes 63:3324–3333. doi: 10.2337/db14-0472. [DOI] [PubMed] [Google Scholar]