Abstract
Toll-like receptors (TLRs) induce inflammation and tissue repair through multiple signaling pathways. The Nrf2 pathway plays a key role in defending against the tissue damage incurred by microbial infection or inflammation-associated diseases. The critical event that mediates TLR-induced Nrf2 activation is still poorly understood. In this study, we found that lipopolysaccharide (LPS) and other Toll-like receptor (TLR) agonists activate Nrf2 signaling and the activation is due to the reduction of Keap1, the key Nrf2 inhibitor. TLR signaling-induced Keap1 reduction promoted Nrf2 translocation from the cytoplasm to the nucleus, where it activated transcription of its target genes. TLR agonists modulated Keap1 at the protein posttranslation level through autophagy. TLR signaling increased the expression of autophagy protein p62 and LC3-II and induced their association with Keap1 in the autophagosome-like structures. We also characterized the interaction between p62 and Keap1 and found that p62 is indispensable for TLR-mediated Keap1 reduction: TLR signaling had no effect on Keap1 if cells lacked p62 or if cells expressed a mutant Keap1 that could not interact with p62. Our study indicates that p62-mediated Keap1 degradation through autophagy represents a critical linkage for TLR signaling regulation of the major defense network, the Nrf2 signaling pathway.
INTRODUCTION
Toll-like receptors (TLRs) act as the first line of host defense against microbial infection and play a pivotal role in both innate and adaptive immunity (1). TLRs recognize the molecular patterns from both invading pathogens and endogenous inflammatory stimuli and subsequently activate distinct intracellular pathways that lead to the inflammatory response (2). A deregulated inflammatory response can cause cell damage and tissue injuries, which are associated with many inflammation-related disorders. On the other hand, the host has developed multiple mechanisms to counteract the excessive inflammatory response and to repair the deleterious tissue damage. Mounting evidence suggests that the nuclear factor-erythroid 2-related factor 2 (Nrf2) signaling pathway is an important component in host anti-inflammation defense.
Nrf2 is a key transcription factor that mainly regulates cellular defenses against oxidative stress and electrophilic insults (3, 4). Under resting conditions, Nrf2 is sequestered in the cytoplasm by Kelch-like ECH-associated protein 1 (Keap1) and removed by Keap1-mediated ubiquitination and proteasomal degradation. When cells are exposed to oxidative stress or other noxious attacks, Nrf2 is released from Keap1 and translocated into the nucleus, where it binds to the promoters of many genes whose products have cellular defensive functions (5–8). The Nrf2 signaling pathway is also activated during inflammation and negatively regulates inflammatory responses. Inflammation can induce the expression of Nrf2 downstream genes (9), and the Nrf2 pathway cross talks with NF-κB and other inflammatory signaling pathways to repress the inflammatory response (10).
Autophagy is a bulk degradation system by which cytoplasmic materials are engulfed by double-membrane vesicles, known as autophagosomes, and delivered to lysosomes for degradation (11). Autophagy is also a selective process involved in the degradation of unnecessary or toxic structure proteins, organelles, and intracellular pathogens (12). The autophagic protein p62 acts as a cargo receptor for ubiquitinated substrates. The association of p62 with the selective substrates forms an autophagic complex which fuses with the lysosome and leads to substrate degradation (13). Recent research suggests that autophagy is a part of the innate immune responses. Toll-like receptors trigger the activation of selective autophagy and Nrf2 signaling to eliminate invading microbes and to promote tissue repairs (14, 15). Although many studies show that TLR signaling, autophagy, and Nrf2 signaling are all activated during inflammation, little is known about how the interplays of these innate immune components are regulated.
In the present study, we sought to determine the key event that leads to Nrf2 pathway activation during TLR signaling. Our results suggest that p62-mediated Keap1 degradation is a critical switch that leads to TLR signaling-induced Nrf2 activation.
MATERIALS AND METHODS
Antibodies and regents.
Keap1, Nrf2, C-myc, glutathione S-transferase (GST), lamin B, and rabbit anti-goat IgG–horseradish peroxidase (HRP) antibodies were from Santa Cruz Biotech, USA; SQSTM1/p62 antibody was from Abcam, USA; rabbit anti-heme oxygenase 1 (anti-HO-1) antibody was from StressGen, USA; β-actin, goat anti-rabbit IgG–HRP, and goat anti-mouse IgG–HRP antibodies were from Sunshine Bio, China; and DyLight 594-conjugated AffiniPure donkey anti-goat IgG and DyLight 649-conjugated AffiniPure donkey anti-rabbit IgG were from Jackson ImmunoResearch, USA. Anti-LC3 antibody, lipopolysaccharide (LPS), poly(I·C), cycloheximide (CHX), MG132, staurosporine, chloroquine (CQ), and 3-methylamphetamine (3-MA) were from Sigma, USA.
Cell culture and transfection.
RAW cells were maintained in Dulbecco modified Eagle medium (DMEM; Gibco, USA) supplemented with newborn calf serum (10%,vol/vol; Gibco, USA). HEK293 cells were grown in DMEM supplemented with 10% fetal bovine serum (FBS). All cells were cultured in a 5% CO2 incubator at 37°C.
A cell viability assay was performed with a cell counting kit (CCK-8 kit; Beyotime, China) following the manufacturer's instruction.
Animals.
Nrf2−/− mice were a generous gift from Masayuki Yamamoto. Wild-type (WT) control mice were generated from Nrf2+/− heterozygous mice in-house. All experimental protocols were approved by the Institutional Animal Care Committee of the Nanjing University Medical School and were conducted in accordance with the guidelines of the Nanjing University Medical School. Mice were fed a regular sterile chow diet, provided water ad libitum, and housed under controlled conditions (25 ± 2°C, 12-h light and 12-h dark periods). After the experiments, the mice were euthanized by CO2 inhalation. Mouse tissue was removed by surgical procedures and frozen at −80°C until further analysis.
Plasmid constructs.
The HO-1 promoter was amplified by PCR from RAW cell genome DNA using the primers 5′-GGAAGATCTCTGCAGAGCCCCACTGGAG-3′ and 5′-CCCAAGCTTGGAACAGCAACGCTGT-3′. The PCR product was then inserted into the pGL3 vector at the HindIII and BglII sites. The wild-type Keap1 expression plasmid (WT-Keap1-myc) was constructed by insertion of Keap1 full-length cDNA, which was generated by reverse transcription-PCR (RT-PCR), at the BamHI/HindIII sites of pcDNA3.1 that was premodified with 3 copies of the myc tag. Two truncated Keap1 expression plasmids, myc-tagged Keap1 containing the N terminus from amino acids (aa) 1 to 314 and lacking Keap1 aa 315 to 424 (K1-N-myc) and myc-tagged Keap1 containing the C terminus from aa 314 to 624 and lacking aa 1 to 314 (K1-C-myc), were constructed in the same vector used to construct WT-Keap1-myc. The mutant Keap1 expression plasmid mKeap1-myc with three arginines replaced by alanines (R380A, R415A, and R483A) was constructed by use of a site-directed mutagenesis kit (TaKaRa, Japan). All the constructs were confirmed by sequencing. Plasmid shRNA-p62 carrying p62-specific short hairpin RNA (shRNA) was constructed by inserting a short oligonucleotide (the underlined sequence covering positions 624 to 642 of p62 in the sequence of the shRNA-p62-F oligonucleotide presented below) at the BamHI and HindIII sites of the GV62 vector. The sequence of oligonucleotide shRNA-p62-F was GATCCGCTGAAACATGGACACTTTTTCAAGAGAAAAGTGTCCATGTTTCAGCTTTTTTGGAAA, and that of oligonucleotide shRNA-p62-R was AGCTTTTCCAAAAAAGCTGAAACATGGACACTTTTCTCTTGAAAAAGTGTCCATGTTTCAGCG. A nonspecific oligonucleotide was used to construct a control plasmid.
Luciferase reporter assays.
HEK293 cells grown on 24-well plates were transfected with plasmids and treated with LPS for the periods of time indicated below. Transfection was performed with the Lipofectamine 2000 reagent according to the manufacturer's instructions. Luciferase activities were measured using a luciferase reporter assay system (Promega, USA).
RT-PCR.
Cell total RNA was extracted using the TRIzol reagent (Invitrogen, USA) according to the manufacturer's instruction. The cDNA was generated with a Transcript first-strand cDNA synthesis kit (Vazyme, China) according to the manufacturer's instructions. PCR was performed using the follow primers: GAPDH F (AACGACCCCTTCATTGAC), GAPDH R (TCCACGACATACTCAGCA), Keap1F (CACACTAGAGGATCACACCAAG), and Keap1R (CCGTGTAGGCGAACTCAATAA). The PCR products were resolved on a 2% agarose gel and visualized under UV light.
Immunofluorescence staining.
Cells were grown on glass coverslips in a 12-well chamber and treated with LPS. After washing with ice-cold phosphate-buffered saline (PBS), the cells were fixed with 4% paraformaldehyde and then permeabilized in PBS containing 2% Triton X-100. The cells were then incubated with anti-LC3 or anti-Keap1 antibody and DyLight 649-conjugated AffiniPure donkey anti-rabbit IgG or DyLight 594-conjugated AffiniPure donkey anti-goat IgG secondary antibodies. The stained cells were observed under a fluorescence confocal microscope.
Western blotting and coimmunoprecipitation.
Western blotting and coimmunoprecipitation assays were performed essentially as described before (16) with the antibodies indicated below at a predetermined dilution.
Statistics.
Quantitative data were expressed as the mean ± standard deviation (SD). Statistical significance was analyzed by one-way analysis of variance, and a P value of <0.05 was considered statistically significant.
RESULTS
TLR agonists activate the Nrf2 pathway in RAW cells.
To test the effect of TLR signaling on Nrf2 pathway activation, we first treated RAW cells with different doses of LPS and assayed for expression of the Nrf2 target HO-1 by Western blotting. LPS induced HO-1 expression in a dose-dependent manner, with marked induction occurring at 0.5 μg/ml (Fig. 1A). We then tested the time course of LPS induction of HO-1 and another Nrf2 target, GST. LPS induced HO-1 and GST expression in 8 to 16 h (Fig. 1B). Similarly, we tested the TLR2 and TLR3 agonists peptidoglycan (PGN) and poly(I·C), respectively, for their capacity to induce Nrf2 downstream protein expression. PGN and poly(I·C) also induced HO-1 expression in a dose-dependent manner (Fig. 1C and D). To confirm that TLR-activated Nrf2 activation occurs in real inflammation, we injected LPS into mice peritoneally and assayed for HO-1 from mouse kidney. LPS induced HO-1 expression as well (Fig. 1E). To determine if LPS induced Nrf2 target protein expression through Nrf2 activation, we assayed the effect of LPS treatment on Nrf2 protein levels. LPS enhancements of Nrf2 were also time and dose dependent (Fig. 1F and G). Since Nrf2 nuclear translocation is a necessary step for Nrf2 activation, we then tested the nucleus Nrf2 protein upon LPS treatment. LPS caused Nrf2 accumulation in the nucleus in 3 h (Fig. 1H). Finally, to confirm that LPS activation of Nrf2 increased the transactivation ability of Nrf2, we tested the effects of LPS on an HO-1 promoter reporter plasmid that contained an Nrf2 binding motif. LPS treatment caused a significant increase in luciferase activity in a dose-dependent fashion (Fig. 1I). In addition, we also tested the viability of RAW cells (Fig. 1J) and HEK293 cells (data not shown) under conditions with LPS at microgram-per-milliliter concentrations. We treated the cells with LPS at up to 20 μg/ml for 24 h and did not observe obvious cell death. Taken together, these results indicate that TLR signaling can indeed activate the Nrf2 pathway and cause Nrf2 downstream gene induction.
FIG 1.
TLR signaling activates the Nrf2 pathway. (A) RAW cells were treated with increasing doses of LPS for 16 h. HO-1 was assayed by Western blotting. (B) RAW cells were treated with 1 μg/ml of LPS for various times. HO-1 and GST were assayed by Western blotting. (C) RAW cells were treated with various doses of PGN for 16 h. HO-1 was assayed by Western blotting. (D) RAW cells were treated with various doses of poly(I·C) for 16 h. HO-1 was assayed by Western blotting. (E) WT mice were treated with LPS (10 μg/kg of body weight) by intraperitoneal injection for 24 h. Kidneys from two mice in each group were removed, and HO-1 was assayed by Western blotting. Con., control mice. (F and G) Time-dependent (F) and dose-dependent (G) study of Nrf2 protein levels. RAW cells were treated with 1 μg/ml LPS for different times or with different doses of LPS for 16 h, and the Nrf2 protein level was assayed by Western blotting. (H) RAW cells were treated with 0.1 and 1 μg/ml LPS for 3 h. Nuclear Nrf2 (N. Nfr2) was assayed by Western blotting. (I) Luciferase assay. HEK293 cells were transfected with the HO-1 promoter reporter plasmid pHO-1p/Luc, followed by treatment with increasing doses of LPS for 16 h. Cell lysates were assayed for luciferase activity. The experiments were repeated at least three times, and representative results are shown. *, P < 0.05 compared to the results for the control. (J) Cell viability assay. RAW cells were treated with various doses of LPS for 24 h. A cell counting kit (CCK-8) was used to measure the living cells. The assay was performed with cells in triplicate.
TLR signaling reduces the Keap1 protein level.
To identify the responsive molecule that mediates Nrf2 activation by TLR signaling, we examined Keap1, the critical inhibitor of Nrf2. As seen in Fig. 2A and B, LPS stimulated a marked decrease in the level of the Keap1 protein in a time- and dose-dependent manner. We also tested the TLR2 agonist PGN and the TLR3 agonist poly(I·C) for their induction of Keap1 reduction. Both the PGN and poly(I·C) treatments caused a decrease in Keap1 levels in a dose-dependent manner (Fig. 2C and D). Similarly, the bacterium Escherichia coli decreased Keap1 expression and increased HO-1 expression (Fig. 2E). Mice that received an LPS injection also showed marked reductions in Keap1 levels in the kidney (Fig. 2F) and liver (not shown). These results suggest that Keap1 reduction and subsequent Nrf2 activation occur when cells are exposed to TLR agonists as well as during in vivo inflammation.
FIG 2.
TLR signaling reduces the Keap1 protein level. (A and B) RAW cells were treated with LPS (1 μg/ml) for various times (A) or with different doses of LPS for 16 h (B). Keap1 was assayed by Western blotting. (C to E) RAW cells were treated with various doses of PGN (C), poly(I·C) (D), or E. coli (E). Keap1 was assayed by Western blotting. (F) A WT mouse was treated with LPS (10 μg/kg of body weight) by intraperitoneal injection for 24 h. The mouse kidneys were removed, and Keap1 was assayed by Western blotting.
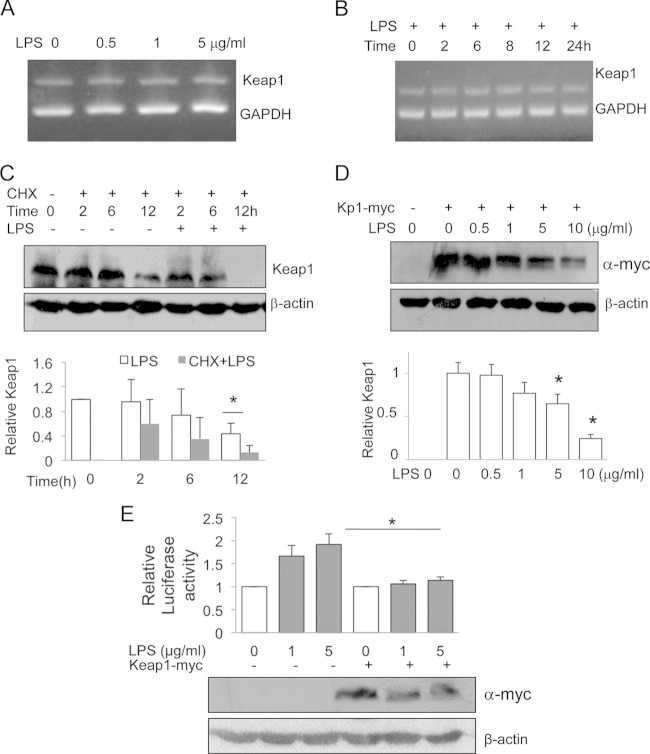
LPS-induced Keap1 protein reduction occurs at the Keap1 posttranscriptional level.
To dissect the steps in which TLR represses Keap1 along the Keap1 biosynthesis pathway, we measured Keap1 mRNA levels by RT-PCR. We used a range of LPS doses (0.05 to 5 μg/ml) (Fig. 3A) and treatment times (2 to 24 h) (Fig. 3B) and found that LPS did not affect Keap1 mRNA levels, excluding the possibility of regulation at the transcriptional or posttranscriptional level. We then tested whether LPS regulated Keap1 at the translational or posttranslational level. We first treated cells with cycloheximide (CHX) to block protein synthesis and then measured Keap1 levels in the presence or absence of LPS over time. Keap1 levels underwent a gradual decrease, with a 50% reduction occurring by 12 h in the presence of CHX alone. However, the additional LPS treatment led to a reduction in the level of the Keap1 protein by more than 90% by 12 h (Fig. 3C), suggesting that LPS induced the Keap1 reduction through a posttranslational mechanism.
FIG 3.
LPS induces Keap1 degradation. (A and B) RAW cells were treated with different doses of LPS for 16 h (A) or with 1 μg/ml of LPS for various times (B). Keap1 and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNAs were simultaneously analyzed by RT-PCR in a single PCR and visualized by agarose gel electrophoresis. (C) RAW cells were pretreated with CHX (0.5 μg/ml) alone or in the presence of LPS (200 ng/ml) for various times. Keap1 protein was examined by Western blotting (top), and Keap1 levels were quantified (bottom). The experiments were repeated at least three times, and representative results are shown. *, P < 0.05. (D) HEK293 cells were transfected with a plasmid carrying WT-Keap1-myc (Kp1-myc) and treated with various amounts of LPS for another 16 h. The cell lysates were subjected to Western blotting analysis using anti-myc antibody (top), and Keap1 levels were quantified (bottom). (E) HEK293 cells were cotransfected with WT-Keap1-myc and an HO-1 promoter reporter. Then, the cells were treated with LPS for 16 h and the luciferase activities were measured (top). The expression of exogenous Keap1-myc was monitored by Western blotting (bottom). The experiments were repeated at least three times, and representative results are shown. *, P < 0.05.
To confirm that LPS induced the Keap1 reduction through a posttranslational modification, we constructed a myc-tagged Keap1 expression plasmid and transfected it into HEK293 cells. We treated the cells with different doses of LPS and measured the amount of exogenously expressed Keap1 protein. LPS treatment also decreased myc-tagged Keap1 levels in a dose-dependent manner (Fig. 3D), indicating that LPS can indeed induce the Keap1 reduction at the protein level. To further determine whether the reduced amount of Keap1 accounts for LPS-induced Nrf2 pathway activation, we overexpressed Keap1 and found that Keap1 overexpression significantly inhibited LPS-induced HO-1 promoter luciferase activity (Fig. 3E), suggesting that the LPS-induced Keap1 reduction plays a key role in controlling the Nrf2 pathway signaling.
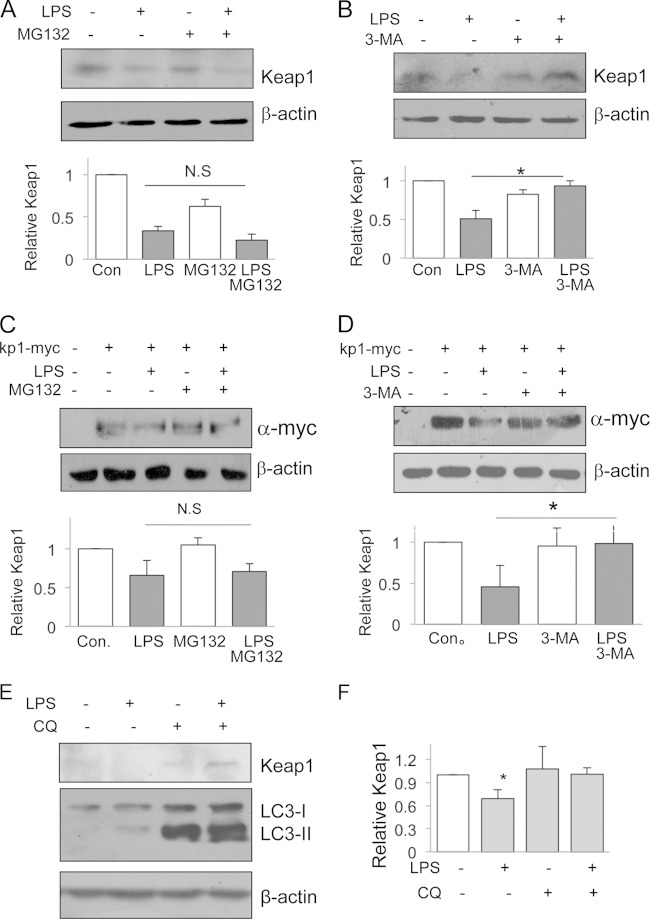
LPS-induced Keap1 degradation is mediated by autophagy.
The ubiquitin-proteasomal pathway and autophagy-lysosomal pathway are two major pathways for protein degradation (17). To test if the LPS-induced Keap1 reduction was mediated through the ubiquitination pathway, we treated cells with LPS and/or MG132, an ubiquitination-proteasome inhibitor. MG132 treatment did not significantly affect the LPS-induced Keap1 reduction (Fig. 4A). When cells were treated with LPS and/or 3-MA, an autophagy inhibitor, the LPS-induced Keap1 protein reduction was blocked (Fig. 4B). To confirm these results, we also tested exogenous Keap1-myc protein for its responsiveness to the inhibitors. Similarly, MG132 had no effect on the LPS-induced Keap1-myc protein reduction (Fig. 4C), but 3-MA clearly abolished the LPS-induced Keap1-myc protein reduction (Fig. 4D). Since 3-MA affects many cellular processes, in addition to its antiautophagy activity, we also used an autophagosome-lysosome inhibitor, chloroquine (CQ), to block autophagy. Cells treated with CQ showed no Keap1 degradation in response to LPS treatment (Fig. 4E and F). However, LPS caused an increase in LC3-II levels in the presence of CQ. Taken together, these results suggest that the LPS-induced Keap1 protein reduction occurs through an autophagy degradation pathway.
FIG 4.
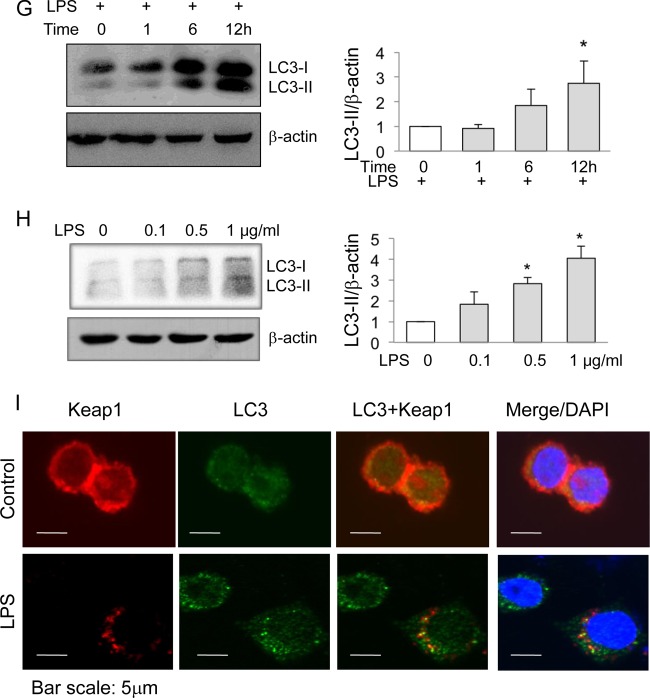
Autophagy mediates LPS-induced Keap1 degradation. (A and B) RAW cells were treated with MG132 (1 μM) (A) or 3-MA (1 mM) (B) for 1 h, and then LPS (200 ng/ml) was added to some cells for 12 h. Expression of endogenous Keap1 protein was examined by Western blotting (top). Keap1 quantities are shown at the bottom. *, P < 0.05; N.S, no significant difference. (C and D) HEK293 cells were first transfected with plasmids encoding myc-tagged Keap1 and then treated with MG132 (2.5 μM) (C) or 3-MA (1 mM) (D) for 1 h in the presence or absence of LPS (200 ng/ml) for an additional 12 h. Cell lysates were analyzed for exogenous Keap1 expression by Western blotting with an anti-myc antibody (top). Keap1 quantities are shown at the bottom. The experiments were repeated at least three times, and representative results are shown. *, P < 0.05; N.S, no significant difference. (E and F) RAW cells were treated with LPS (200 ng/ml) or CQ (25 μM), or both, for 12 h. Keap1 and LC3 were analyzed by Western blotting (E), and Keap1 levels were quantified (F). (G) RAW cells were treated with LPS (200 ng/ml) for various times, and LC3 was analyzed by Western blotting (left). The ratio of LC3-II to β-actin was calculated and plotted (right). *, P < 0.05 compared to the results for the control. (H) RAW cells were treated with various doses of LPS for 12 h, and LC3 was analyzed by Western blotting (left). The ratio of LC3-II to β-actin was calculated and plotted (right). *, P < 0.05 compared to the results for the control. (I) RAW cells were treated with LPS (200 ng/ml) for 12 h, and then LC3 and Keap1 proteins were examined by immunostaining. DAPI (4′,6-diamidino-2-phenylindole) stain was applied as a nuclear marker.
To search for additional evidence that the autophagy pathway is involved in LPS-induced Keap1 degradation, we tested LC3-I and its activation. LC3-I is an autophagosomal membrane protein. Upon autophagy pathway activation, LC3-I is cleaved to produce LC3-II. The amount of LC3-II represents the number of autophagosomes and is a solid indication of autophagy pathway activation (18). As expected, treatment of RAW cells with LPS not only caused an increase in the LC3-I level but also increased the LC3-II level in a time-dependent (Fig. 4G) and dose-dependent (Fig. 4H) manner. To further explore the possibility that the LPS-induced Keap1 reduction was related to autophagy activation, we performed immunofluorescent staining of LC3 and Keap1 in RAW cells. LPS treatment caused a redistribution of LC3 from a diffuse staining pattern to a punctate, autophagosome-like pattern. The merged results show that Keap1 and LC3 colocalized in such punctate structures (Fig. 4I). Collectively, these results indicate that LPS-induced autophagy activation leads to the Keap1 reduction.
p62 is indispensable for LPS-induced Keap1 degradation.
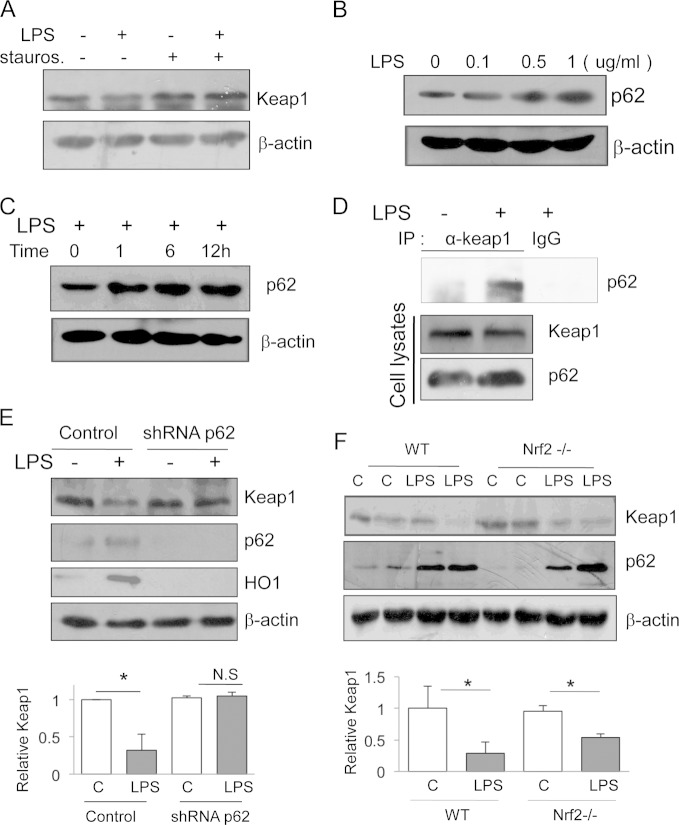
A previous study reported that LPS can activate Nrf2 through protein kinase C (PKC)-mediated Nrf2 phosphorylation (19), but it was unclear if Keap1 was also affected or Nrf2 phosphorylation preceded Keap1 degradation. To assess the role of PKC in LPS-induced Keap1 degradation, we tested a PKC inhibitor, staurosporine. Staurosporine reversed the LPS-induced Keap1 degradation (Fig. 5A), indicating that PKC acts upstream of Keap1 degradation and Nrf2 phosphorylation.
FIG 5.
p62 mediates LPS-induced Keap1 degradation. (A) RAW cells were treated with LPS in the presence or absence of the PKC inhibitor staurosporine (stauros.; 15 nM) or LPS (200 ng/ml) for 16 h. Keap1 was assayed by Western blotting. (B and C) RAW cells were treated with different doses of LPS for 12 h (B) or with 200 ng/ml LPS for various times (C). p62 was assayed by Western blotting. (D) RAW cells were treated with LPS (200 ng/ml) for 3 h. The cell lysates were subjected to coimmunoprecipitation with anti-Keap1 or an isoform control antibody. The level of p62 was assayed by Western blotting. Nonimmunoprecipitated cell lysates were assayed for p62 and Keap1 as controls. IP, immunoprecipitation. (E) RAW cells were transfected with shRNA specific for p62 or a nonspecific control shRNA for 36 h and then treated with LPS (200 ng/ml) for 12 h. Keap1, p62, and HO-1 proteins were assayed by Western blotting (top). The results of quantification analysis are also shown (bottom). The experiments were repeated at least three times, and representative results are shown. *, P < 0.05; N.S, no significant difference. (F) Nrf2−/− and WT mice were treated with LPS (10 μg/kg of body weight) by injection for 24 h, and then Keap1 and p62 from mouse kidney were analyzed by Western blotting (top). C, control. The results of quantification analysis are also shown (bottom). The experiments were repeated at least three times, and representative results are shown. *, P < 0.05.
To search for a direct mediator related to autophagy, we focused on p62 since previous studies showed that increased amounts of p62 due to autophagy deficiency promote the p62/Keap1 association and contribute to Nrf2 pathway activation (20, 21). LPS treatment increased the p62 protein level in a dose-dependent (Fig. 5B) and time-dependent (Fig. 5C) manner. A coimmunoprecipitation assay demonstrated that LPS induced the p62 association with Keap1 (Fig. 5D), suggesting that p62 mediated LPS-induced Keap1 degradation. To determine the role of p62 in Keap1 degradation, we used a p62-specific interfering RNA strategy to knock down p62 and test the effect of LPS on the Keap1 reduction. The results showed that knockdown of p62 prevented LPS-induced Keap1 degradation (Fig. 5E), supporting the idea that LPS induced Keap1 degradation through the p62-mediated autophagy pathway.
The promoter of p62 contains a functional Nrf2 binding site, and p62 has been reported to be an Nrf2 target (22); therefore, it is possible that the elevated p62 protein level from LPS stimulation happens due to LPS-induced Nrf2 activation. To answer this critical question, we compared the p62 and Keap1 levels in the livers of wild-type and Nrf2-knockout mice (Nrf2−/−). A lack of Nrf2 did not prevent LPS-induced p62 induction or Keap1 reduction (Fig. 5F), suggesting that LPS can induce p62 accumulation and Keap1 degradation independently of Nrf2 pathway activation.
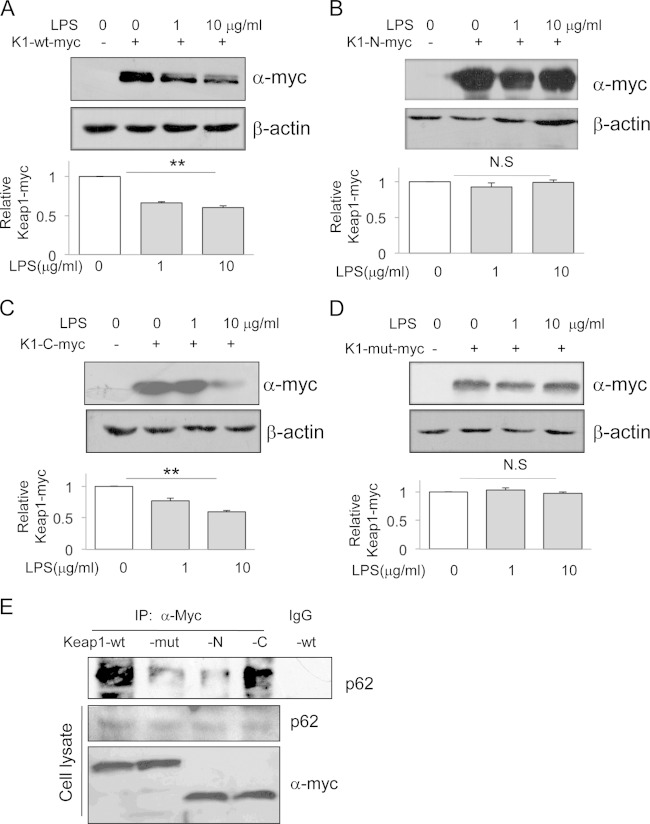
Three arginines in Keap1 are crucial for LPS-induced Keap1 degradation.
Keap1 contains five functional domains, and the Kelch domain, located on the C-terminal part of Keap1, involves its binding with Nrf2 (7). In order to determine the critical Keap1 domain that mediates LPS-induced Keap1 degradation, we constructed two additional truncation mutants that express the Keap1 N terminus (aa 1 to 314; K1-N-myc) or the C terminus part (aa 314 to 624; K1-C-myc). We tested wild-type and two truncated Keap1 proteins for their responsiveness to LPS. LPS reduced the full-length Keap1, as expected (Fig. 6A). LPS also reduced the Keap1 C-terminal fragment to an extent similar to that for wild-type Keap1 (Fig. 6C). However, LPS failed to reduce the Keap1 N-terminal fragment (Fig. 6B), suggesting that the Keap1 C-terminal part contains a responsive element that mediates LPS-induced Keap1 degradation.
FIG 6.
Arginines R380, R415, and R483 on Keap1 are crucial for LPS-induced Keap1 degradation. HEK293 cells were transfected with plasmids carrying myc-tagged wild-type Keap1 (K1-wt-myc) (A), the Keap1 N-terminal fragment from aa 1 to 314 (K1-N-myc) (B), the Keap1 C-terminal fragment from aa 314 to 624 (K1-C-myc) (C), or a mutant Keap1 in which the three arginines at positions 380, 415, and 483 were replaced with alanines (K1-mut-myc) (D). After transfection for 12 h, the cells were treated with 1 or 10 μg/ml LPS for an additional 12 h. The myc-tagged Keap1 proteins were assayed by Western blotting with an anti-Myc antibody (top). The results of quantification analysis are also shown (bottom). The experiments were repeated at least three times, and representative results are shown, *, P < 0.05; N.S, no significant difference. (E) Coimmunoprecipitation and Western blotting assays. HEK293 cells were transfected with a plasmid carrying K1-wt-myc, K1-N-myc, K1-C-myc, or K1-mut-myc. After 18 h, cells were treated with 1 μg/ml LPS for an additional 4 h. The cell lysates were precipitated with an anti-myc antibody, and immunoprecipitates were analyzed by Western blotting with an anti-p62 antibody. As loading and transfection controls, cell lysates were also analyzed by Western blotting with anti-p62 and anti-myc antibodies. The experiments were repeated at least three times, and representative results are shown.
There are three arginines (R380, R415, and R483) in the Keap1 C-terminal part that are necessary for Keap1 and Nrf2 interactions (23). These three arginines were also essential for Keap1 binding to p62 (24), implying that p62 may be competing with Nrf2 for Keap1 binding. To test the role of these three arginines in LPS/p62-induced Keap1 degradation, we constructed an additional plasmid carrying mutated Keap1 in which the three arginines were replaced by alanines (K1-mut-myc). We then transfected this plasmid into HEK293 cells and tested LPS-induced Keap1 degradation. LPS did not reduce the mutated Keap1 protein (Fig. 6D). We also performed a coimmunoprecipitation assay to verify the association between p62 and the various Keap1 proteins. As expected, p62 was associated with wild-type Keap1 and the C-terminal fragment of Keap1, but the associations between p62 and the N-terminal fragment or the Keap1 protein with three arginine mutations were barely detectable (Fig. 6E). Taken together, these data indicate that the three arginines located in the C terminus of Keap1 are essential for LPS-induced Keap1 degradation and that the degradation is mediated by p62.
DISCUSSION
The major findings of our study are that TLR signaling can activate autophagy through the induction of p62 and LC3 and that the autophagy-associated Keap1 degradation is a critical trigger that leads to activation of the Nrf2 pathway. It is well-known that TLR signaling activates the Nrf2 antioxidative stress pathway and that TLR signaling also induces selective autophagy (25–27). During autophagy deficiency, aberrant p62 accumulation could target Keap1 and cause Nrf2 pathway activation, which would lead to liver injury (20, 24). It has also been reported that Keap1 is an ubiquitinated protein but that its degradation is independent of 26S proteasome-mediated degradation (28); instead, Keap1 was constitutively degraded through autophagy even under the basal and native/unmodified conditions (29). However, the role of autophagy in the process of TLR activation of Nrf2 has not been fully revealed. Our study expands on these previous studies and provides direct evidence that TLR signaling controls the host innate response induced by Nrf2 activation through autophagic Keap1 degradation. It also establishes that autophagy is an important link between TLR signaling and Nrf2 activity during inflammation.
Recent studies have suggested that TLR signaling is linked to the induction of autophagy and at least two selective autophagy processes are associated with TLR signaling: autophagic lysis of intracellular microbes (called xenophagy) and autophagic clearance of TLR-induced aggregation of signaling proteins (called aggrephagy) (12). In regard to xenophagy, one of the central questions is how TLR-mediated substrates induce selective autophagy machinery and promote phagophore assembly. Now it is generally accepted that autophagy cargo receptor p62 plays a critical role in this process. The accumulation of ubiquitinated cellular signaling molecules can be recognized by cargo receptor p62, which leads to targeted phagophore assembly and subsequent cargo degradation (13, 30). Since p62 is an Nrf2 downstream target and Nrf2 activation increases p62 expression (22, 27), it is possible that p62-associated autophagy activation is a result of TLR4-induced Nrf2 activation. To address this, we compared the LPS-induced Keap1 levels in the kidneys of Nrf2-knockout and wild-type mice. Our results showed that injection of LPS still caused increases in p62 levels and Keap1 degradation in the absence of Nrf2, suggesting that TLR signaling can activate autophagy independently of Nrf2. It is worth noting that in our assay system Keap1 degradation occurred after the initial accumulation of Nrf2 upon LPS stimulation. Previous studies showed that Nrf2 phosphorylation controls its activity at the early stage (19, 27, 31). We speculate that TLR signaling-induced Nrf2 activation probably involves an early Nrf2 phosphorylation-associated activation and a later Keap1 degradation-triggered Nrf2 activation. The latter might play a critical role in maintaining Nrf2 pathway activation for an extended period of time, which is beneficial for tissue repair since autophagic p62-induced Nrf2 pathway activation inhibits the TLR signaling-induced deregulation of inflammatory responses (25).
It is well established that Keap1 regulates the Nrf2 signaling pathway, but the regulation of Keap1 itself is less well documented. Recent studies showed that Keap1 is subjected to posttranscription regulation: its mRNA level was affected by microRNA 200a through its 3′ untranslated region (32, 33). Another study reported that Keap1 overexpression in HepG2 cells inhibits Nrf2 activity (34). Genetic knockdown of Keap1 was also found to enhance Nrf2 activity and decrease fasting-induced steatosis and oxidative stress (35, 36). These studies suggest that the level of endogenous Keap1 is important in the proper control of Nrf2 activity. Our results add new evidence that reduction of the Keap1 protein level directly affects Nrf2 activity. We demonstrated that the LPS-induced Keap1 reduction occurs at the protein level, since Keap1 mRNA levels remained constant in the presence of LPS treatment (Fig. 3A and B). We further showed that LPS also causes a reduction in the amount of exogenously expressed Keap1 in a manner similar to that for endogenous Keap1 (Fig. 3D), indicating that regulation of the Keap1 protein level constitutes an important step in TLR signaling-induced Nrf2 activation.
Ubiquitination and autophagy are two major degradation pathways that remove cellular and microbial components to maintain cell homeostasis (37). We have presented several lines of evidence indicating that autophagy mediates the inhibitory effect of LPS on the Keap1 protein. First, a proteasome inhibitor, MG132, had no effect, but 3-MA and a specific autophagy inhibitor, CQ, abolished the LPS-induced Keap1 protein reduction. Second, LPS treatment increased LC3 and LC3-II cleavage, which correlated with Keap1 and LC3 colocalization and redistribution in punctate autophagosome-like structures. Finally, LPS enhanced p62 protein expression and its association with Keap1. All these pieces of evidence support the notion that autophagy-mediated Keap1 causes Nrf2 activation.
Keap1 contains multiple highly reactive cysteine residues, located in the intervening region between BTB (broad complex, tram-track, bric-a-brac) and Kelch repeat domains, which function as sensors for electrophilic compounds and oxidants. Upon stimulation of cells with reactive oxygen species, the reactive cysteine residues undergo oxidation and form an intramolecular disulfide bond (38, 39). It is generally accepted that the modification of critical cysteines Cys257, Cys273, and Cys288 causes a conformational change of Keap1 and leads to the release of Nrf2 from Keap1 and subsequent Nrf2 nuclear translocation and activation (40, 41). Due to the different properties of oxidants and inflammation, we surmise that TLR signaling might induce Keap1 to release Nrf2 through a different mechanism. Indeed, we found that three arginines (Arg380, Arg415, and Arg483) on Keap1 are required for the p62 and Keap1 interaction induced by LPS since replacement of the three arginines with alanines abolished the LPS-induced Keap1 protein reduction (Fig. 6D). Our results do not exclude the possibility that the critical cysteines on Keap1 are also involved in LPS-induced Nrf2 activation but present a clear direction for future study to explore the role of these three cysteines in this process.
Autophagy protein p62 itself is subjected to degradation by autophagy (42). Mice with deficient autophagy exhibit elevated p62 protein levels in their livers, and the level of the p62 protein is generally considered to correlate inversely with autophagy activity (20, 43). However, during inflammation, p62 can be transcriptionally upregulated and its enhanced expression can be correlated with increased autophagy activity (11, 27). Indeed, our study has provided direct evidence demonstrating that LPS treatment induces autophagy by enhancing p62 and LC3 protein levels and increasing the level of association of p62 with Keap1 as well as subsequent Keap1 degradation. LPS did not induce Keap1 degradation when autophagy was blocked by 3-MA or chloroquine. Therefore, our results support the suggestion that TLR agonist-induced p62 correlates with increased autophagy. We further tested the role of p62 in Keap1 degradation and Nrf2 activation by a p62-knockdown assay. LPS treatment did not cause Keap1 degradation when p62 was absent (Fig. 5E), indicating that LPS-induced Keap1 degradation depends on intact p62 and autophagy is a major regulator of Keap1 degradation.
To our knowledge, this is the first study showing how TLR agonists regulate the Nrf2 signaling pathway through p62-mediated Keap1 degradation. Our study provides another example of aggrephagy in which selective autophagy affects a key signaling molecule, Keap1, and regulates the antistress and anti-inflammation responses.
ACKNOWLEDGMENTS
This work was supported by research grants from the National Nature Science Foundation of China (81070940, 81271301, and 81470940) to W. Cao.
We declare no competing interests.
REFERENCES
- 1.Akira S. 2003. Toll-like receptor signaling. J Biol Chem 278:38105–38108. doi: 10.1074/jbc.R300028200. [DOI] [PubMed] [Google Scholar]
- 2.Sabroe I, Parker LC, Dower SK, Whyte MK. 2008. The role of TLR activation in inflammation. J Pathol 214:126–135. doi: 10.1002/path.2264. [DOI] [PubMed] [Google Scholar]
- 3.Kobayashi M, Yamamoto M. 2006. Nrf2-Keap1 regulation of cellular defense mechanisms against electrophiles and reactive oxygen species. Adv Enzyme Regul 46:113–140. doi: 10.1016/j.advenzreg.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 4.Takaya K, Suzuki T, Motohashi H, Onodera K, Satomi S, Kensler TW, Yamamoto M. 2012. Validation of the multiple sensor mechanism of the Keap1-Nrf2 system. Free Radic Biol Med 53:817–827. doi: 10.1016/j.freeradbiomed.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rangasamy T, Guo J, Mitzner WA, Roman J, Singh A, Fryer AD, Yamamoto M, Kensler TW, Tuder RM, Georas SN, Biswal S. 2005. Disruption of Nrf2 enhances susceptibility to severe airway inflammation and asthma in mice. J Exp Med 202:47–59. doi: 10.1084/jem.20050538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Velichkova M, Hasson T. 2005. Keap1 regulates the oxidation-sensitive shuttling of Nrf2 into and out of the nucleus via a Crm1-dependent nuclear export mechanism. Mol Cell Biol 25:4501–4513. doi: 10.1128/MCB.25.11.4501-4513.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, Yamamoto M. 1999. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev 13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nguyen T, Yang CS, Pickett CB. 2004. The pathways and molecular mechanisms regulating Nrf2 activation in response to chemical stress. Free Radic Biol Med 37:433–441. doi: 10.1016/j.freeradbiomed.2004.04.033. [DOI] [PubMed] [Google Scholar]
- 9.Rushworth SA, MacEwan DJ. 2008. HO-1 underlies resistance of AML cells to TNF-induced apoptosis. Blood 111:3793–3801. doi: 10.1182/blood-2007-07-104042. [DOI] [PubMed] [Google Scholar]
- 10.Yang Y, Cai X, Yang J, Sun X, Hu C, Yan Z, Xu X, Lu W, Wang X, Cao P. 2014. Chemoprevention of dietary digitoflavone on colitis-associated colon tumorigenesis through inducing Nrf2 signaling pathway and inhibition of inflammation. Mol Cancer 13:48. doi: 10.1186/1476-4598-13-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujita K, Srinivasula SM. 2011. TLR4-mediated autophagy in macrophages is a p62-dependent type of selective autophagy of aggresome-like induced structures (ALIS). Autophagy 7:552–554. doi: 10.4161/auto.7.5.15101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johansen T, Lamark T. 2011. Selective autophagy mediated by autophagic adapter proteins. Autophagy 7:279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lippai M, Low P. 2014. The role of the selective adaptor p62 and ubiquitin-like proteins in autophagy. Biomed Res Int 2014:832704. doi: 10.1155/2014/832704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Birgisdottir AB, Lamark T, Johansen T. 2013. The LIR motif—crucial for selective autophagy. J Cell Sci 126(Pt 15):3237–3247. doi: 10.1242/jcs.126128. [DOI] [PubMed] [Google Scholar]
- 15.Vijayan V, Baumgart-Vogt E, Naidu S, Qian G, Immenschuh S. 2011. Bruton's tyrosine kinase is required for TLR-dependent heme oxygenase-1 gene activation via Nrf2 in macrophages. J Immunol 187:817–827. doi: 10.4049/jimmunol.1003631. [DOI] [PubMed] [Google Scholar]
- 16.Jeong Y, Du R, Zhu X, Yin S, Wang J, Cui H, Cao W, Lowenstein CJ. 2014. Histone deacetylase isoforms regulate innate immune responses by deacetylating mitogen-activated protein kinase phosphatase-1. J Leukoc Biol 95:651–659. doi: 10.1189/jlb.1013565. [DOI] [PubMed] [Google Scholar]
- 17.Wang H, Khor TO, Saw CLL, Lin W, Wu T, Huang Y, Kong A-NT. 2010. Role of Nrf2 in suppressing LPS-induced inflammation in mouse peritoneal macrophages by polyunsaturated fatty acids docosahexaenoic acid and eicosapentaenoic acid. Mol Pharm 7:2185–2193. doi: 10.1021/mp100199m. [DOI] [PubMed] [Google Scholar]
- 18.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rushworth SA, Chen XL, Mackman N, Ogborne RM, O'Connell MA. 2005. Lipopolysaccharide-induced heme oxygenase-1 expression in human monocytic cells is mediated via Nrf2 and protein kinase C. J Immunol 175:4408–4415. doi: 10.4049/jimmunol.175.7.4408. [DOI] [PubMed] [Google Scholar]
- 20.Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI, Kim M, Nishito Y, Iemura S, Natsume T, Ueno T, Kominami E, Motohashi H, Tanaka K, Yamamoto M. 2010. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol 12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 21.Hayashi K, Dan K, Goto F, Tshuchihashi N, Nomura Y, Fujioka M, Kanzaki S, Ogawa K. 2015. The autophagy pathway maintained signaling crosstalk with the Keap1-Nrf2 system through p62 in auditory cells under oxidative stress. Cell Signal 27:382–393. doi: 10.1016/j.cellsig.2014.11.024. [DOI] [PubMed] [Google Scholar]
- 22.Jain A, Lamark T, Sjottem E, Larsen KB, Awuh JA, Overvatn A, McMahon M, Hayes JD, Johansen T. 2010. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J Biol Chem 285:22576–22591. doi: 10.1074/jbc.M110.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Padmanabhan B, Tong KI, Ohta T, Nakamura Y, Scharlock M, Ohtsuji M, Kang MI, Kobayashi A, Yokoyama S, Yamamoto M. 2006. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol Cell 21:689–700. doi: 10.1016/j.molcel.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 24.Lau A, Wang XJ, Zhao F, Villeneuve NF, Wu T, Jiang T, Sun Z, White E, Zhang DD. 2010. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol Cell Biol 30:3275–3285. doi: 10.1128/MCB.00248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee HM, Shin DM, Yuk JM, Shi G, Choi DK, Lee SH, Huang SM, Kim JM, Kim CD, Lee JH, Jo EK. 2011. Autophagy negatively regulates keratinocyte inflammatory responses via scaffolding protein p62/SQSTM1. J Immunol 186:1248–1258. doi: 10.4049/jimmunol.1001954. [DOI] [PubMed] [Google Scholar]
- 26.Kobayashi A, Kang MI, Watai Y, Tong KI, Shibata T, Uchida K, Yamamoto M. 2006. Oxidative and electrophilic stresses activate Nrf2 through inhibition of ubiquitination activity of Keap1. Mol Cell Biol 26:221–229. doi: 10.1128/MCB.26.1.221-229.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujita K, Maeda D, Xiao Q, Srinivasula SM. 2011. Nrf2-mediated induction of p62 controls Toll-like receptor-4-driven aggresome-like induced structure formation and autophagic degradation. Proc Natl Acad Sci U S A 108:1427–1432. doi: 10.1073/pnas.1014156108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang DD, Lo SC, Sun Z, Habib GM, Lieberman MW, Hannink M. 2005. Ubiquitination of Keap1, a BTB-Kelch substrate adaptor protein for Cul3, targets Keap1 for degradation by a proteasome-independent pathway. J Biol Chem 280:30091–30099. doi: 10.1074/jbc.M501279200. [DOI] [PubMed] [Google Scholar]
- 29.Taguchi K, Fujikawa N, Komatsu M, Ishii T, Unno M, Akaike T, Motohashi H, Yamamoto M. 2012. Keap1 degradation by autophagy for the maintenance of redox homeostasis. Proc Natl Acad Sci U S A 109:13561–13566. doi: 10.1073/pnas.1121572109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kirkin V, McEwan DG, Novak I, Dikic I. 2009. A role for ubiquitin in selective autophagy. Mol Cell 34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 31.Jain AK, Mahajan S, Jaiswal AK. 2008. Phosphorylation and dephosphorylation of tyrosine 141 regulate stability and degradation of INrf2: a novel mechanism in Nrf2 activation. J Biol Chem 283:17712–17720. doi: 10.1074/jbc.M709854200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Eades G, Yang M, Yao Y, Zhang Y, Zhou Q. 2011. miR-200a regulates Nrf2 activation by targeting Keap1 mRNA in breast cancer cells. J Biol Chem 286:40725–40733. doi: 10.1074/jbc.M111.275495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang JJ, Tao H, Hu W, Liu LP, Shi KH, Deng ZY, Li J. 2014. MicroRNA-200a controls Nrf2 activation by target Keap1 in hepatic stellate cell proliferation and fibrosis. Cell Signal 26:2381–2389. doi: 10.1016/j.cellsig.2014.07.016. [DOI] [PubMed] [Google Scholar]
- 34.Yueh MF, Tukey RH. 2007. Nrf2-Keap1 signaling pathway regulates human UGT1A1 expression in vitro and in transgenic UGT1 mice. J Biol Chem 282:8749–8758. doi: 10.1074/jbc.M610790200. [DOI] [PubMed] [Google Scholar]
- 35.Xu J, Donepudi AC, Moscovitz JE, Slitt AL. 2013. Keap1-knockdown decreases fasting-induced fatty liver via altered lipid metabolism and decreased fatty acid mobilization from adipose tissue. PLoS One 8:e79841. doi: 10.1371/journal.pone.0079841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williamson TP, Johnson DA, Johnson JA. 2012. Activation of the Nrf2-ARE pathway by siRNA knockdown of Keap1 reduces oxidative stress and provides partial protection from MPTP-mediated neurotoxicity. Neurotoxicology 33:272–279. doi: 10.1016/j.neuro.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shaid S, Brandts CH, Serve H, Dikic I. 2013. Ubiquitination and selective autophagy. Cell Death Differ 20:21–30. doi: 10.1038/cdd.2012.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu C, Eggler AL, Mesecar AD, van Breemen RB. 2011. Modification of Keap1 cysteine residues by sulforaphane. Chem Res Toxicol 24:515–521. doi: 10.1021/tx100389r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sekhar KR, Rachakonda G, Freeman ML. 2010. Cysteine-based regulation of the CUL3 adaptor protein Keap1. Toxicol Appl Pharmacol 244:21–26. doi: 10.1016/j.taap.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamamoto T, Suzuki T, Kobayashi A, Wakabayashi J, Maher J, Motohashi H, Yamamoto M. 2008. Physiological significance of reactive cysteine residues of Keap1 in determining Nrf2 activity. Mol Cell Biol 28:2758–2770. doi: 10.1128/MCB.01704-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang DD, Hannink M. 2003. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol Cell Biol 23:8137–8151. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ichimura Y, Komatsu M. 2010. Selective degradation of p62 by autophagy. Semin Immunopathol 32:431–436. doi: 10.1007/s00281-010-0220-1. [DOI] [PubMed] [Google Scholar]
- 43.Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata J, Ezaki J, Murata S, Hamazaki J, Nishito Y, Iemura S, Natsume T, Yanagawa T, Uwayama J, Warabi E, Yoshida H, Ishii T, Kobayashi A, Yamamoto M, Yue Z, Uchiyama Y, Kominami E, Tanaka K. 2007. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]