Abstract
Limited uptake is one of the bottlenecks for l-arabinose fermentation from lignocellulosic hydrolysates in engineered Saccharomyces cerevisiae. This study characterized two novel l-arabinose transporters, LAT-1 from Neurospora crassa and MtLAT-1 from Myceliophthora thermophila. Although the two proteins share high identity (about 83%), they display different substrate specificities. Sugar transport assays using the S. cerevisiae strain EBY.VW4000 indicated that LAT-1 accepts a broad substrate spectrum. In contrast, MtLAT-1 appeared much more specific for l-arabinose. Determination of the kinetic properties of both transporters revealed that the Km values of LAT-1 and MtLAT-1 for l-arabinose were 58.12 ± 4.06 mM and 29.39 ± 3.60 mM, respectively, with corresponding Vmax values of 116.7 ± 3.0 mmol/h/g dry cell weight (DCW) and 10.29 ± 0.35 mmol/h/g DCW, respectively. In addition, both transporters were found to use a proton-coupled symport mechanism and showed only partial inhibition by d-glucose during l-arabinose uptake. Moreover, LAT-1 and MtLAT-1 were expressed in the S. cerevisiae strain BSW2AP containing an l-arabinose metabolic pathway. Both recombinant strains exhibited much faster l-arabinose utilization, greater biomass accumulation, and higher ethanol production than the control strain. In conclusion, because of higher maximum velocities and reduced inhibition by d-glucose, the genes for the two characterized transporters are promising targets for improved l-arabinose utilization and fermentation in S. cerevisiae.
INTRODUCTION
Biorefining of lignocellulosic biomass has attracted considerable attention in recent years because of its abundance, sustainability and potential environmental benefits (1, 2). The main sugars in hydrolysates from currently used feedstocks are a mixture of d-glucose, d-xylose, and l-arabinose (3). For a cost-effective conversion of biomass into fuels or chemicals, the fermentative organisms are required to utilize all three sugars efficiently. Even though many organisms are able to natively convert these substrates, the most commonly selected microbe is Saccharomyces cerevisiae (baker's yeast) because of its high ethanol productivity and high tolerance for inhibitors present in biomass hydrolysates, as well as the fact that it is amenable to genetic engineering (4–6).
Wild-type S. cerevisiae cannot utilize the two pentose sugars d-xylose and l-arabinose. Thus, improving import and the intracellular pentose utilization efficiency is very critical, and intensive efforts have been made to do so by yeast metabolic engineering (7–13). Sugar uptake is the initial step for its utilization, and therefore, efficient molecular transport is a prerequisite to achieve enhanced fermentation rates in the presence of a working pentose metabolism pathway. Baker's yeast is able to absorb pentoses through its native hexose transporters, such as Hxt5 and Hxt7, and the galactose transporter Gal2 (14–16). However, these transporters show higher affinity for hexoses, and thus, it was suggested that d-glucose impairs the simultaneous utilization of pentoses mainly by inhibition of pentose uptake. Therefore, heterologous expression of pentose transporters can alleviate this inhibition and lead to improved pentose utilization and cofermentation rates of hexoses and pentoses (17, 18). Recently, multiple novel transporters from pentose-assimilating microorganisms have been discovered (19–23). In addition, it was shown that pentose utilization could be improved by increasing its uptake in S. cerevisiae via heterologous expression of glucose/xylose facilitator 1 (Gxf1) and glucose/xylose symporter 1 (Gxs1) from Candida intermedia (24) or by overexpression of the endogenous Gal2 (25). Alternative approaches using transporter engineering and directed evolution found that their substrate specificity could be rewired, and d-glucose inhibition-free d-xylose transporters were engineered through modification of protein motifs (26, 27).
l-Arabinose is the second most abundant sugar in hemicellulose; however, because d-xylose is considered the most representative pentose sugar, l-arabinose has so far attracted much less attention, and its use in biorefining has been less well studied. Although l-arabinose and d-xylose are both pentoses and their catabolic pathways are interconnected, we found that their presence elicited substantially different responses on the transcriptional level in our previous study using Neurospora crassa (28). Moreover, most of the known pentose transporters were shown to be able to differentiate between d-xylose and l-arabinose. For example, Gxf1 and Gxs1 can transport d-xylose but not l-arabinose, while the d-xylose/l-arabinose transporter (XAT-1) transports l-arabinose much better than d-xylose (16, 20, 24). To date, only a few l-arabinose-specific transporters have been reported, including two transporters for l-arabinose cloned from Ambrosiozyma monospora in 2011 (21). Shortly thereafter, another three l-arabinose transporters were identified and partially characterized: AraT from Scheffersomyces stiptis, Stp2 from Arabidopsis thaliana, and LAT-1 (NCU02188) from N. crassa (22, 23). We found that lat-1 is the most strongly induced sugar transporter gene in N. crassa on l-arabinose and is coexpressed with lad-1, encoding the l-arabinitol 4-dehydrogenase. The corresponding deletion strain was shown to be impaired in l-arabinose uptake, intracellular accumulation, and signaling. Moreover, the lat-1 deletion strain in N. crassa displayed an obvious growth defect on arabinan and impaired l-arabinose consumption (23, 28). Together, these results strongly suggested that lat-1 encodes a bona fide l-arabinose transporter. However, no detailed kinetic properties of this protein have yet been reported, and knowing these properties is important to improve our knowledge concerning the mechanism of transport specification and substrate recognition, as well as to determine the possibility of its application to S. cerevisiae metabolic engineering.
In this study, we report the comprehensive characterization of LAT-1, as well as its ortholog in the related thermophilic fungus Myceliophthora thermophila (syn. Sporotrichum thermophile, in the same order Sordariales with N. crassa) (29). Both l-arabinose transporters were found to use a proton-coupled symport mechanism but to differ in their substrate specificities. Moreover, they exert a greater maximum velocity (Vmax) and are less inhibited by d-glucose than all other transporters reported so far. The characteristics of these two novel l-arabinose transporters investigated in the present study demonstrate that both genes should be good future candidates for metabolic engineering to improve l-arabinose fermentation in microbes, such as S. cerevisiae, that are unable to naturally use l-arabinose in biorefining.
MATERIALS AND METHODS
Strains and culture conditions.
S. cerevisiae EBY.VW4000 (30), a gift from Eckhard Boles (Goethe Universität Frankfurt am Main), was cultured in YPM medium (10 g/liter yeast extract, 20 g/liter peptone, and 20 g/liter maltose). Recombinant EBY.VW400 strains were grown in synthetic complete (SC) medium (6.7 g/liter yeast nitrogen base and 2 g/liter amino acid dropout mix) supplemented with amino acids lacking uracil (SC−Ura) (in order to maintain transporter-expressing plasmids) and various carbon sources as described previously (31).
S. cerevisiae BSW2AP (25), a gift from Xiaoming Bao, Shandong University, was grown in SC medium supplemented with amino acids lacking leucine and uracil (SC−Leu−Ura) and 20 g/liter l-arabinose. The medium was supplemented with 400 μg/ml Geneticin (G418; Amresco, Solon, OH, USA) when required to maintain the plasmid with the KanMX4 marker. All yeast strains were cultured at 30°C and 200 rpm for aerobic growth.
For serial dilution growth assays, a single colony was precultured to exponential phase with an optical density at 600 nm (OD600) of 1 to 2. The cells were washed twice with sterile water and resuspended in sterile water with an OD600 of 3. The cells were serially 10-fold diluted in multiple steps, and aliquots of 2 μl were spotted on agar plates with various carbon sources.
Escherichia coli DH5α was used for gene cloning and manipulation. Strains were cultivated in lysogeny broth (LB) medium with 100 μg/ml ampicillin for plasmid selection (32).
For transcription analysis, N. crassa (FGSC 2489), obtained from the Fungal Genetics Stock Center (FGSC), was cultured on Vogel's minimal medium supplemented with 2% sucrose for 10 days at 28°C with constant light, and conidia were inoculated into 100 ml of liquid medium (1× Vogel's salt with 2% d-glucose or 2% l-arabinose) at 106 conidia per ml in a 250-ml Erlenmeyer flask. Two replicate flasks of each culture were grown. After growth for 16 to 26 h at 25°C with shaking at 200 rpm under constant light, mycelia were harvested by vacuum filtration with Whatman filter paper, frozen immediately in liquid nitrogen, and stored at −80°C for RNA extraction.
M. thermophila ATCC 42464, obtained from the American Type Culture Collection (ATCC), was cultured on Vogel's minimal medium supplemented with 2% sucrose for 15 days at 45°C, and conidia were inoculated into 100 ml of liquid medium (1× Vogel's salt with 2% d-glucose or 2% l-arabinose) at 2.5 × 105 conidia per ml in a 250-ml Erlenmeyer flask. Two replicate flasks were inoculated for each culture. After growth for 16 h at 45°C with shaking at 150 rpm under constant light, mycelia were harvested by vacuum filtration with Whatman filter paper, frozen immediately in liquid nitrogen, and stored at −80°C for RNA extraction.
d-Glucose, l-arabinose, maltose, d-fructose, d-mannose, d-galactose, lactose, and d-sorbose were purchased from Sigma-Aldrich (St. Louis, MO, USA) with a purity of more than 99%, and sucrose was obtained from Amresco with a purity of more than 99%.
RNA extraction.
Total RNA from frozen samples was isolated using zirconia/silica beads (0.5-mm diameter; Biospec, Bartlesville, OK, USA) and a MiniBeadbeater (Biospec) with 1 ml TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA, USA) according to the manufacturer's protocol. To eliminate genomic DNA contamination, an additional cleanup with DNase was performed using the Qiagen (Hilden, Germany) RNeasy minikit as described in the manufacture's RNA cleanup introduction. A NanoDrop (Thermo Scientific, South Logan, UT, USA) and agarose gel electrophoresis were used to check the RNA concentration and integrity (33). RNAs with A260/A280 values between 2.0 and 2.2, A260/A230 values between 2.0 and 2.4, and no visible degradation on agarose gel electrophoresis were used.
Quantitative real-time PCR.
Quantitative real-time PCR was performed using a CFX96 real-time PCR detection system (Bio-Rad, Hercules, CA, USA) with reagents from the One-step qPCR kit (Toyobo, Osaka, Japan). The PCR mixture (20 μl), with three replicates, included approximately 75 ng template RNA, 0.4 μM primers, and 10 μl RNA-direct SYBR green Real-time PCR master mix, according to the manufacturer's instructions. The relative transcript levels of putative l-arabinose transporters in N. crassa and M. thermophila on l-arabinose were calculated by the 2−ΔΔCT method, with expression on d-glucose as the control and the expression level of actin (NCU04173 and MYCTH_2303024 in N. crassa and M. thermophila, respectively) as an internal standard.
Plasmid and strain construction.
RNA samples from N. crassa and M. thermophila grown on l-arabinose were reverse transcribed into cDNA with a RevertAid First Strand cDNA synthesis kit (Thermo Scientific, Waltham, MA, USA) according to the instructions and used as the template for amplification of genes encoding putative l-arabinose transporters.
For analysis of the localization and transport kinetics of the l-arabinose transporters in recombinant EBY.VW4000 strains, the plasmid p426PGK with the PGK promoter, green fluorescent protein (GFP) open reading frame, and CYC terminator was used as the shuttle vector, which was constructed from the 2μ plasmid pRS426 according to the method described previously (34). The gene encoding LAT-1 (NCU02188) was amplified from N. crassa and inserted between the BamHI and EcoRI sites (respective underlined sequences) using the primers CGCGGATCCATGGGGCTCGGGCTTAAGCTAC and CGGAATTCAACCTTCTCATGCTCATGCACCTC, resulting in plasmid p426LATgfp. The MtLAT-1-encoding gene (MYCTH_95427) amplified from M. thermophila was inserted between the BamHI and EcoRI sites using the primers CGCGGATCCATGAAGCTGCCCACGATTTAC and CCGGAATTCAACCTTCTCCTGCTCGCCGAC, resulting in plasmid p426MtLATgfp.
For expression of transporters in strain BWS2AP, the plasmid p426 with the PGK promoter and CYC terminator was used as the shuttle vector, which was constructed from the 2μ plasmid pRS426 as described previously (34), and the plasmid p426kanmx4 with KanMX4 as the selective marker was constructed. KanMX4 cloned from pUG8 was inserted into p426 between the NdeI and ApaI sites to replace the URA3 fragment using the primers GGGAATTCCATATGGATCTGTTTAGCTTGCCTCGTC and ATGGGCCCCGACACTGGATGGCGGCGTTAG, resulting in plasmid p426kanmx4. Subsequently, the genes encoding LAT-1 and MtLAT-1 were inserted into the shuttle plasmid p426kanmx4, behind the PGK promoter, with the primers CGCGGATCCATGGGGCTCGGGCTTAAGCTAC/CGGAATTCCTAAACCTTCTCATGCTCATGCAC and CGCGGATCCATGAAGCTGCCCACGATTTAC/CCGGAATTCTTAAACCTTCTCCTGCTCGCCGAC, respectively. All restriction enzymes were obtained from Thermo Scientific.
All the target genes cloned into shuttle vectors were sequenced to verify the authenticity of the plasmid constructs. Yeast transformations were performed as described previously (32).
GFP fluorescence and confocal fluorescence microscopy.
The recombinant EBY.VW4000 strains expressing sugar transporters tagged with GFP were streaked from 15% glycerol stocks onto SC−Ura plates with 20 g/liter maltose. A single clone was inoculated into SC−Ura liquid medium supplemented with 20 g/liter maltose and grown to exponential phase with an OD600 of 1 to 2. Cells were collected, washed twice with sterile water, and then resuspended in sterile water. Ten microliters of culture was spotted on a cover glass for confocal microscopy using a Leica TCS SP5 II microscope with a 100× 1.4-numerical-aperture (NA) oil immersion objective. The images were processed and analyzed with ImageJ (version 1.47).
l-Arabinose transport kinetics.
Initial l-arabinose uptake rates were determined using a modification of the method described previously (19, 35). Cell cultures at exponential phase were collected, washed twice with 100 mM Tris-citrate buffer at pH 5.0, and then resuspended to an OD600 of 40. An aliquot (100 μl) of yeast cells was preincubated at 30°C for 5 min and then mixed with 42 μl of a sugar solution containing l-[1-3H]arabinose (American Radiolabeled Chemicals Inc., St. Louis, MO, USA) at various concentrations (1 mM, 5 mM, 10 mM, 50 mM, 200 mM, 400 mM, and 600 mM) and incubated for 2 min at 30°C. One milliliter of ice-cold water was added to stop the reaction. After centrifugation (10,000 rpm; 1 min), the cells were washed three times with ice-cold water, mixed with 1 ml of 1 M NaOH, and then transferred to a 5-ml scintillation cocktail (PerkinElmer, Waltham, MA, USA). The radioactivity was measured using a scintillation counter (Tri-Carb 2910TR Liquid Scintillation Analyzer; PerkinElmer). A 300-μl aliquot of each yeast strain was dried at 80°C for determination of the dry cell weight (DCW). The results shown are the averages of at least two independent experiments. The Km and maximum velocity (Vmax) were determined as described previously (19).
Uncoupling assays.
To determine the transport type of LAT-1 and MtLAT-1, the dependence of the l-arabinose transport on the plasma membrane proton gradient was monitored by a previously described method (36) with minor modifications. Strains were prepared in the same way as for the kinetics assay described above. Fifty-microliter aliquots of yeast suspensions were mixed with various amounts of carbonyl cyanide m-chlorophenylhydrazone (CCCP) (Sigma-Aldrich), resulting in final CCCP concentrations of 0 mM, 0.5 mM, 1.0 mM, 25 mM, and 50 mM, and incubated for 10 min at 30°C. After incubation, 50 μl of a sugar solution containing l-[1-3H]arabinose at a concentration of 10 mM was added and incubated for 2 min at 30°C, and the transport rate was determined as for the kinetic assay described above. All assays were performed in biological triplicates.
d-Glucose inhibition assay.
Yeast cells were prepared as for the methods described above. Fifty-microliter aliquots of cell cultures were mixed with 50 μl of d-glucose solutions at different concentrations (0 mM, 30 mM, 75 mM, 150 mM, and 300 mM) and incubated at 30°C for 5 min. Subsequently, 50 μl of a sugar solution containing l-[1-3H]arabinose at 300 mM was added and incubated for 2 min at 30°C. The transport rate was determined as described above. The experiments were performed in independent triplicates.
Fermentation experiment.
A single colony from a fresh agar plate was inoculated into 10 ml synthetic complete medium supplemented with 400 μg/ml G418 with 15 g/liter l-arabinose and 5 g/liter d-glucose as carbon sources. Precultured cells were harvested at late exponential phase, washed with sterile water, and then inoculated into 30 ml synthetic complete medium in 250-ml Erlenmeyer flasks under aerobic conditions. Oxygen-limited fermentation was performed on 50-ml synthetic medium in 100-ml serum vials sealed with rubber caps. l-Arabinose (2%) served as the sole carbon source, and 400 μg/ml G418 was added to maintain the plasmid for all fermentations. The initial cell densities of all fermentation experiments were adjusted to an OD600 of 1.0. Under aerobic or oxygen-limited conditions, cells were cultivated in an orbital shaker at 200 rpm at 30°C, and samples were taken at intervals. All assays were carried out in biological duplicates for each investigated strain.
Analytical methods.
Cell growth was monitored by measuring the absorbance at 600 nm using a UV-1800 spectrophotometer (Shimadzu, Kyoto, Japan). The residual l-arabinose concentration and ethanol production were determined by high-performance liquid chromatography (HPLC) with an instrument (e2695; Waters, Manchester, United Kingdom) equipped with a Waters 2414 refractive index detector and an Aminex HPX-87H column (Bio-Rad). The column was eluted at a temperature of 63°C, and 5 mM H2SO4 was used as a mobile phase with a constant flow rate of 0.6 ml/min. Data analysis was performed using a Waters e2695 separation module.
Protein sequence alignment.
l-Arabinose transporters were aligned by the PRALINE multiple-sequence alignment server (37), and the predicted result was visualized with ALINE software (38).
RESULTS
Cloning and heterologous expression in S. cerevisiae of two fungal l-arabinose transporters.
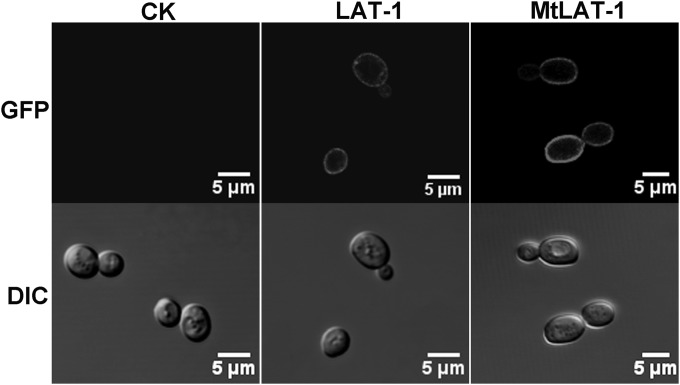
Our previous transcriptomic analysis of N. crassa identified the major facilitator superfamily (MFS) transporter gene NCU02188, named lat-1, as one of the most strongly induced genes on l-arabinose (23). Interestingly, we found that its ortholog (MYCTH_95427; MtLAT-1) in M. thermophila, a thermophilic filamentous fungus that can use l-arabinose even more efficiently than N. crassa (unpublished data), was also strongly induced by l-arabinose (see Fig. S1 in the supplemental material). Because the two proteins are highly homologous (about 83% identity based on the predicted amino acid sequence; see Fig. S2 and S3 in the supplemental material), we assumed the molecular function and biochemical properties would be comparable. To characterize the two putative l-arabinose transporters in more detail, we cloned the corresponding genes and introduced them into the S. cerevisiae null-hexose-transporter strain EBY.VW4000 (30). The transporters fused to GFP displayed correct plasma membrane localization in the recombinant overexpression strains, as visualized by confocal microscopy (Fig. 1).
FIG 1.
Confocal fluorescence imaging of recombinant EBY.VW4000 strains expressing LAT-1 and MtLAT-1, demonstrating correct localization at the yeast plasma membrane. CK, the EBY.VW4000 strain without LAT-1 or MtLAT-1 used as a negative control. DIC, differential interference contrast.
The substrate specificities of LAT-1 and MtLAT-1 differ considerably.
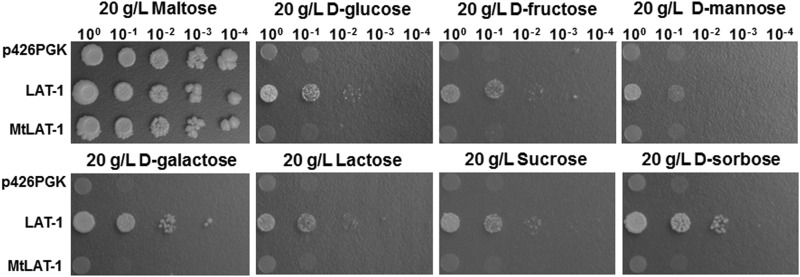
To determine the substrate spectra of LAT-1 and MtLAT-1, we performed growth assays of the recombinant EBY.VW4000 yeast strains on various carbon sources. To this end, precultured strains in serial dilutions were spotted on agar plates with various mono- and disaccharides as carbon sources at a concentration of 20 g/liter. Due to the absence of any hexose transporter, the EBY.VW4000 strain harboring only the empty plasmid control did not grow on any tested carbon sources except d-maltose, as expected (Fig. 2). Conversely, LAT-1, expressed under the control of the strong PGK promoter, supported slow growth on 20 g/liter concentrations of d-glucose, d-fructose, d-mannose, d-galactose, lactose, sucrose, and d-sorbose (Fig. 2). In addition, the cells expressing LAT-1 showed transport capacity for l-arabinose in an isotope assay (data not shown). These results suggested that under our conditions, LAT-1 was not an l-arabinose-specific transporter but a multifunctional transporter with a potentially broad substrate spectrum. In contrast to LAT-1, the EBY.VW4000 transformant expressing MtLAT-1 showed a growth pattern similar to that of the empty-plasmid control with no capacity for growth on all tested sugars except d-maltose, while MtLAT-1-mediated l-arabinose uptake was observable in an isotope assay (data not shown). These results suggested that MtLAT-1 was considerably more l-arabinose specific. Considering that M. thermophila is naturally efficient at using l-arabinose, the specificity of MtLAT-1 for this substrate would make it an excellent target for metabolic engineering in S. cerevisiae and would make M. thermophila a good fungal candidate for rapid industrial l-arabinose fermentation.
FIG 2.
Growth properties of recombinant EBY.VW4000 expressing LAT-1 and MtLAT-1. Cells were spotted in serial dilutions on synthetic complete medium agar plates with various carbon sources at the indicated concentrations. Cells harboring the empty vector p426PGK served as a negative control. The cells were grown at 30°C for 5 days.
LAT-1 and MtLAT-1 display a low-affinity–high-capacity type of transport kinetics.
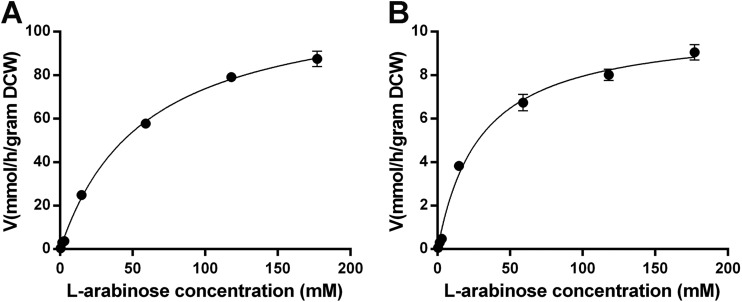
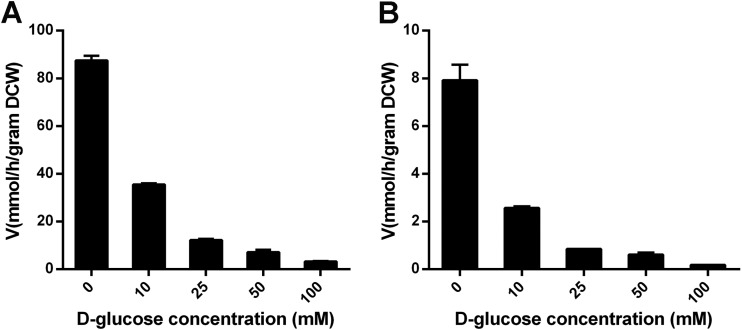
To get a better understanding of the biochemical properties of both MFS transporters, the kinetic parameters were determined by measuring initial uptake rates. The recombinant EBY.VW4000 strains were incubated with l-arabinose at various concentrations using a radiolabeled tracer in 2-min experiments. We found that the Km values of LAT-1 and MtLAT-1 for l-arabinose uptake were 58.12 ± 4.06 mM and 29.39 ± 3.60 mM, respectively, with corresponding Vmax values of 116.7 ± 3.0 mmol/h/g DCW and 10.29 ± 0.35 mmol/h/g DCW, respectively (Fig. 3). Compared with previously characterized proteins, the affinity of the two fungal transporters for l-arabinose is therefore similar to that of Gal2 but lower than that measured for AraT and Stp2 (22). However, the Vmax values are much higher than those of any l-arabinose transporters reported so far, particularly for LAT-1 from N. crassa, which displayed a Vmax 3 orders of magnitude higher (Table 1), indicating it is possible to improve the speed of l-arabinose uptake by heterologous expression of LAT-1 and MtLAT-1 in microbes that have no or very low capability for natural l-arabinose uptake, such as S. cerevisiae.
FIG 3.
Kinetic characterization of LAT-1 (A) and MtLAT-1 (B) for l-arabinose uptake. The initial rate of sugar transport was determined as a function of the sugar concentration. The transport rate was normalized by yeast DCW. The background values determined with cells containing the empty vector were subtracted. The error bars indicate the standard deviations of the results of at least two independent experiments.
TABLE 1.
Kinetic parameters of known l-arabinose transporters
| Organism | Gene product | Km (mM) | Vmax (mmol/h/g DCW)a | Reference |
|---|---|---|---|---|
| S. cerevisiae | Gal2 | 57 ± 11 | 0.132 ± 0.0156 | 22 |
| S. stipitis | AraT | 3.8 ± 1.7 | 0.024 ± 0.0036 | 22 |
| A. thaliana | Stp2 | 4.5 ± 2.2 | 0.036 ± 0.0048 | 22 |
| N. crassa | LAT-1 | 58.12 ± 4.06 | 116.7 ± 3.0 | This study |
| M. thermophila | MtLAT-1 | 29.39 ± 3.60 | 10.29 ± 0.35 | This study |
Vmax from the references was recalculated to match the dimension mmol/h/g DCW.
l-Arabinose uptake by LAT-1 and MtLAT-1 shows characteristics of a proton symport type.
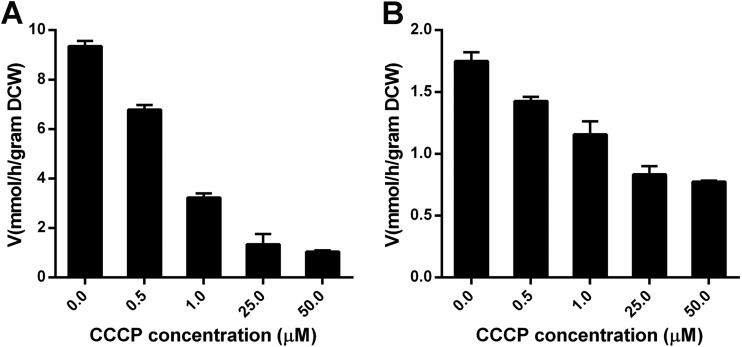
We subsequently subjected the recombinant EBY.VW4000 yeast strains to sensitivity assays for CCCP, a standard method to investigate whether the MFS protein energy requirement is either the symporter or facilitator type. CCCP, a chemical inhibitor of oxidative phosphorylation, is capable of dissipating the proton gradient over the plasma membrane. Therefore, transport rates in the presence of CCCP indicate whether the uptake of a specific substrate is dependent on a proton gradient (symporter) or not (facilitator).
The recombinant EBY.VW4000 strains overexpressing LAT-1 and MtLAT-1 were mixed with different concentrations of CCCP, and the initial uptake rates of l-arabinose were measured (Fig. 4). With increasing CCCP concentrations, transport rates for l-arabinose were reduced in both strains, indicating that both LAT-1 and MtLAT-1 are symporters, and the transport is dependent on an intact plasma membrane proton gradient. Intriguingly, l-arabinose rates of uptake of both transporters did not decrease considerably below about 0.8 mmol/h/g DCW at CCCP concentrations exceeding 25 μM. The precise reason is not known, but one possibility is that the two l-arabinose symporters might have a basal l-arabinose transport capability without a proton gradient, such as a separate facilitator-like mode.
FIG 4.
Sensitivity of l-arabinose transport by LAT-1 (A) and MtLAT-1 (B) to the proton uncoupler CCCP. S. cerevisiae strains expressing sugar transporters were incubated with the indicated concentration of CCCP for 10 min at 30°C. Subsequently, the rate at which the cells imported 5 mM l-arabinose was measured. The error bars indicate the standard deviations of the results of duplicate assays.
l-Arabinose uptake by LAT-1 and MtLAT-1 is affected but not completely inhibited by d-glucose.
In particular for their industrial applicability, the degree of inhibition by d-glucose is a very important characteristic for newly discovered transporters. To investigate the inhibition potential of LAT-1 and MtLAT-1, initial uptake rates at a final concentration of 100 mM l-arabinose were recorded in the presence of increasing concentrations of d-glucose. The results, depicted in Fig. 5, demonstrate that the l-arabinose transport activity of LAT-1 and MtLAT-1 decreased with increasing concentrations of d-glucose. The transport rate in both transporters was reduced to about 40% when 10 mM d-glucose was added, while activity of AraT and Stp2 was previously shown to be completely inhibited under these concentrations (22). Additionally, l-arabinose uptake activity by LAT-1 and MtLAT-1 was still detectable at 10-fold-higher concentrations (100 mM d-glucose). These results suggest that the l-arabinose transporters identified here are much less severely inhibited by d-glucose.
FIG 5.
Inhibitory effect of d-glucose on l-arabinose uptake of LAT-1 (A) and MtLAT-1 (B). The initial transport rate of 100 mM l-arabinose by both transporters was monitored at various d-glucose concentrations, as indicated. The error bars indicate the standard deviations of the results of biological duplicates.
Overexpression of LAT-1 and MtLAT-1 improves l-arabinose utilization in S. cerevisiae.
To test whether the high transport velocities of the two transporters can efficiently help to improve l-arabinose fermentation, we investigated cell growth and l-arabinose consumption rates by overexpression of LAT-1 and MtLAT-1 in the previously created S. cerevisiae strain BSW2AP (25). This strain harbors the l-arabinose utilization pathway genes araA, araB, and araD from Lactobacillus plantarum. Precultured cells containing LAT-1 or MtLAT-1 were initially grown in synthetic complete medium containing 15 g/liter l-arabinose and 5 g/liter d-glucose. The BSW2AP strain harboring an empty vector (p426kanmx4) was used as the control. After being washed with sterile water, cells in late exponential phase were inoculated into synthetic medium containing 20 g/liter l-arabinose and grown under aerobic conditions. As shown in Fig. 6, the cells with empty vector grew very slowly on 20 g/liter l-arabinose and consumed only minor amounts under aerobic conditions. However, the LAT-1- and MtLAT-1-overexpressing strains supported 2- to 3-fold-stronger growth (Fig. 6A), consistent with concurrently higher l-arabinose consumption rates (Fig. 6B). Compared with MtLAT-1, LAT-1 enabled even faster growth and l-arabinose utilization.
FIG 6.
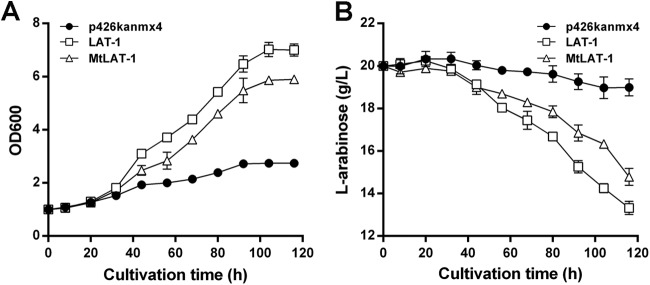
Growth and sugar utilization of BSW2AP strains expressing l-arabinose transporters under aerobic conditions. Precultured cells expressing LAT-1 or MtLAT-1 or harboring the empty vector p426kanmx4 (as a control) were inoculated into synthetic complete medium containing 20 g/liter l-arabinose and 400 μg/ml G418 to an initial optical density of 1.0 at 600 nm. The results shown are average values of two independent experiments.
We further tested the effect of heterologously expressed LAT-1 or MtLAT-1 on ethanol fermentation rates from l-arabinose in BSW2AP cells. Compared with empty-vector-harboring control cells under oxygen-limited conditions, the levels of sugar utilization and the ethanol production in strains expressing LAT-1 or MtLAT-1 were considerably higher (Fig. 7). Although the total titer of ethanol was still very low, it can surely be improved by downstream optimization of the l-arabinose utilization pathway in the future.
FIG 7.
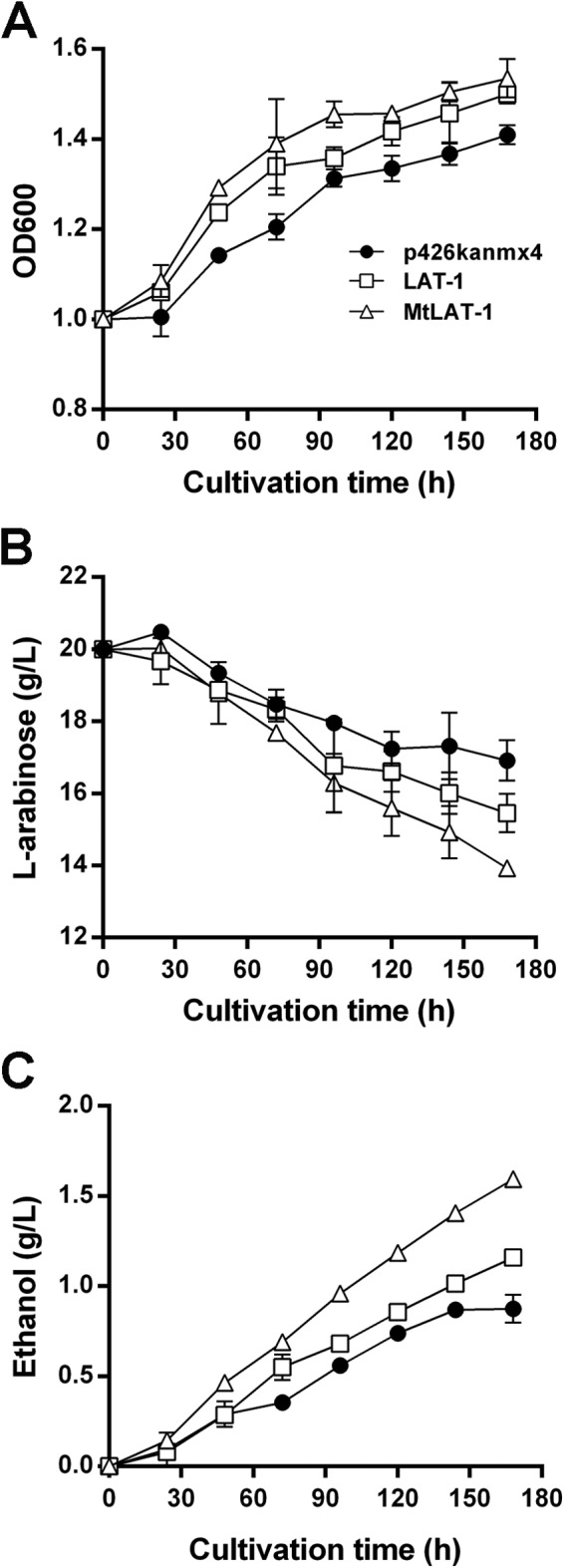
Profiles of l-arabinose fermentation by recombinant S. cerevisiae strains under oxygen-limited conditions. Fermentation experiments were performed on synthetic complete medium containing 20 g/liter l-arabinose and 400 μg/ml G418 with an initial OD600 of 1.0 under oxygen-limited conditions. The error bars show the standard derivations of the results of two independent experiments. BSW2AP strains were expressing either LAT-1 or MtLAT-1 or harboring an empty vector (as the control).
DISCUSSION
For an economically feasible biorefining process, it will be crucial to completely utilize the sugars derived from biomass, including l-arabinose. However, l-arabinose fermentation in recombinant S. cerevisiae is far from efficient, and uptake of the sugar is still a limiting factor. Although S. cerevisiae can take up l-arabinose via its hexose transporters, such as Gal2, this process is carried out with low affinity and is strongly inhibited by d-glucose (17, 18). Recently, several novel l-arabinose-specific transporters have been identified (19–21, 23), and the transporting kinetics of two of them (AraT and Stp2) were characterized (22). However, the transporters discovered so far do not allow efficient l-arabinose fermentation. Therefore, the identification and detailed characterization of additional transporters is of paramount importance for the field.
Filamentous fungi with the capability for lignocellulosic depolymerization are a promising reservoir of industrial enzymes and transporters. Several sugar transporters from lignocellulose-degrading fungi have already contributed to the metabolic engineering of S. cerevisiae for efficient fermentation of sustainable feedstock, such as the cellodextrin transporters (CDT-1 and CDT-2) and a xylose transporter (An25) from N. crassa (19, 34). Here, we cloned and characterized two l-arabinose transporters from filamentous fungi: LAT-1 from N. crassa and MtLAT-1 from M. thermophila. When expressed in the hexose-transporter-less S. cerevisiae strain EBY.VW4000, both recombinant strains were able to take up l-arabinose with low affinity and high capacity, even though the two transporters showed distinct characteristics, including the l-arabinose uptake rates and their substrate spectra. Since both l-arabinose transporter constructs in the recombinant strains displayed similar transcript levels (data not shown), this suggests that the observed differences were not caused by gene expression levels. Whether some differences might exist at the protein level is worth checking in the future. However, we consider it more likely that the markedly different lifestyles of the two fungi that the two transporters genes were cloned from caused the divergence of the gene products. For example, the optimal temperature for the thermophile M. thermophila is 40 to 50°C, whereas it is 25 to 30°C for the mesophilic N. crassa. We also noticed that the speed of l-arabinose utilization by M. thermophila is at least twice that of N. crassa, which might be due to the better specificity of MtLAT-1 that we observed. The observed difference in substrate preference is interesting, considering that the two transporters are highly homologous (sharing about 83% identity) and use the same type of energy requirement (proton symport). In the future, it will be worthwhile to test what motif or amino acid residue(s) could be the determining factor for this substrate specificity based on a comparative analysis of the two proteins and using techniques such as reciprocal amino acid point mutations.
The transport velocity of LAT-1 was found to be 3 orders of magnitude higher than that of any of the three previously identified l-arabinose transporters (Table 1) (19). Theoretically, the sugar consumption rate could also be increased by heterologous expression of the transporters with higher uptake velocity. A direct relationship between sugar transport kinetics and its consumption rate has been shown (39), implying a potential for improved l-arabinose consumption by overexpressing both transporters. Moreover, both transporters also displayed less competitive inhibition by d-glucose. Regarding the mechanism of d-glucose inhibition, it was reported recently that amino acid N376 of Gal2 is critical for this process. A mutation of N376 to phenylalanine (N376F) prevented d-glucose from entering the binding pocket and thereby relieved competitive inhibition (26). Based on sequence alignments (see Fig. S4 in the supplemental material), the corresponding amino acid residue of LAT-1 and MtLAT-1 has evolved naturally into phenylalanine, and this amino acid change might be the reason for the reduced potential for inhibition of LAT-1 and MtLAT-1 by d-glucose. Consistent with this hypothesis, the same positions in AraT and Stp2, which are completely inhibited by 10 mM d-glucose, still encode asparagine (22).
Finally, when we tested the beneficial properties of LAT-1 and MtLAT-1 in an aerobic fermentation experiment by introducing the two transporters into the l-arabinose-catabolizing yeast strain BSW2AP (25), both strains outperformed the parental strain and achieved higher growth rates and had better ethanol production capability. However, we noticed that the total amount of ethanol production in the recombinant strain was still very low, suggesting that containing a good transporter is not enough for efficient l-arabinose fermentation. This seems analogous to xylose fermentation in yeast, where many modifications of the intracellular utilization pathway and even extensive adaptive evolution were needed to reach good titers (40, 41). Consistently, the advantage of d-xylose-specific transporter overexpression was not observed for xylose fermentation because of a deficient d-xylose utilization pathway (21). Moreover, it was shown that the effect of increased transport performance is a function of the strain background and the substrate concentration (39). Therefore, cooptimization of the downstream metabolic flux for l-arabinose is required in the future.
In conclusion, we characterized in detail two l-arabinose transporters, LAT-1 and MtLAT-1, from the two cellulolytic fungi N. crassa and M. thermophila. Both proteins are symporter-type transporters and exhibited good Vmax values and lower d-glucose inhibition during l-arabinose uptake than any other l-arabinose transporter analyzed so far. The genes for the two transporters characterized here should be promising target genes for the elucidation of the working mechanism of MFS transporters and for improving l-arabinose fermentation in engineered microbes, such as S. cerevisiae and industrial fungi for bio-based chemical production in the future.
Supplementary Material
ACKNOWLEDGMENTS
We thank Eckhard Boles for providing the S. cerevisiae EBY.VW4000 strain, and we thank Xiaoming Bao for providing S. cerevisiae BSW2AP. We also thank all members of the Tian laboratory, especially Qian Liu, Liangcai Lin, and Feiyu Fan, for critical comments and technical assistance.
This work was supported by funding from the National Natural Science Foundation of China (31171207 and 31471186) and the 973 Program of China (2011CB707403 and 2011CBA00803).
We declare that we have no competing interests.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00165-15.
REFERENCES
- 1.Lynd LR, Laser MS, Brandsby D, Dale BE, Davison B, Hamilton R, Himmel M, Keller M, McMillan JD, Sheehan J, Wyman CE. 2008. How biotech can transform biofuels. Nat Biotechnol 26:169–172. doi: 10.1038/nbt0208-169. [DOI] [PubMed] [Google Scholar]
- 2.Rubin EM. 2008. Genomics of cellulosic biofuels. Nature 454:841–845. doi: 10.1038/nature07190. [DOI] [PubMed] [Google Scholar]
- 3.Hendriks ATWM, Zeeman G. 2009. Pretreatments to enhance the digestibility of lignocellulosic biomass. Bioresour Technol 100:10–18. doi: 10.1016/j.biortech.2008.05.027. [DOI] [PubMed] [Google Scholar]
- 4.Olsson L, Hahn-Hägerdal B. 1993. Fermentative performance of bacteria and yeasts in lignocellulose hydrolysates. Process Biochem 28:249–257. doi: 10.1016/0032-9592(93)80041-E. [DOI] [Google Scholar]
- 5.van Maris AJ, Abbott DA, Bellissimi E, van den Brink J, Kuyper M, Luttik MA, Wisselink HW, Scheffers WA, van Dijken JP, Pronk JT. 2006. Alcoholic fermentation of carbon sources in biomass hydrolysates by Saccharomyces cerevisiae: current status. Antonie Van Leeuwenhoek 90:391–418. doi: 10.1007/s10482-006-9085-7. [DOI] [PubMed] [Google Scholar]
- 6.Bajpai P, Sharma A, Raghuram N, Bajpai PK. 1988. Rapid production of ethanol in high-concentration by immobilized cells of Saccharomyces cerevisiae through soya flour supplementation. Biotechnol Lett 10:217–220. doi: 10.1007/BF01134833. [DOI] [Google Scholar]
- 7.Ha SJ, Galazka JM, Kim SR, Choi JH, Yang X, Seo JH, Glass NL, Cate JH, Jin YS. 2011. Engineered Saccharomyces cerevisiae capable of simultaneous cellobiose and xylose fermentation. Proc Natl Acad Sci U S A 108:504–509. doi: 10.1073/pnas.1010456108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee SM, Jellison T, Alper HS. 2014. Systematic and evolutionary engineering of a xylose isomerase-based pathway in Saccharomyces cerevisiae for efficient conversion yields. Biotechnol Biofuels 7:122. doi: 10.1186/s13068-014-0122-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Latimer LN, Lee ME, Medina-Cleghorn D, Kohnz RA, Nomura DK, Dueber JE. 2014. Employing a combinatorial expression approach to characterize xylose utilization in Saccharomyces cerevisiae. Metab Eng 25:20–29. doi: 10.1016/j.ymben.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 10.Cao L, Tang X, Zhang X, Zhang J, Tian X, Wang J, Xiong M, Xiao W. 2014. Two-stage transcriptional reprogramming in Saccharomyces cerevisiae for optimizing ethanol production from xylose. Metab Eng 24:150–159. doi: 10.1016/j.ymben.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 11.Matsushika A, Sawayama S. 2008. Efficient bioethanol production from xylose by recombinant Saccharomyces cerevisiae requires high activity of xylose reductase and moderate xylulokinase activity. J Biosci Bioeng 106:306–309. doi: 10.1263/jbb.106.306. [DOI] [PubMed] [Google Scholar]
- 12.Richard P, Putkonen M, Väänänen R, Londesborough J, Penttilä M. 2002. The missing link in the fungal l-arabinose catabolic pathway, identification of the l-xylulose reductase gene. Biochemistry 41:6432–6437. doi: 10.1021/bi025529i. [DOI] [PubMed] [Google Scholar]
- 13.Jojima T, Omumasaba CA, Inui M, Yukawa H. 2010. Sugar transporters in efficient utilization of mixed sugar substrates: current knowledge and outlook. Appl Microbiol Biotechnol 85:471–480. doi: 10.1007/s00253-009-2292-1. [DOI] [PubMed] [Google Scholar]
- 14.Becker J, Boles E. 2003. A modified Saccharomyces cerevisiae strain that consumes l-arabinose and produces ethanol. Appl Environ Microbiol 69:4144–4150. doi: 10.1128/AEM.69.7.4144-4150.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Madhavan A, Tamalampudi S, Srivastava A, Fukuda H, Bisaria VS, Kondo A. 2009. Alcoholic fermentation of xylose and mixed sugars using recombinant Saccharomyces cerevisiae engineered for xylose utilization. Appl Microbiol Biotechnol 82:1037–1047. doi: 10.1007/s00253-008-1818-2. [DOI] [PubMed] [Google Scholar]
- 16.Hamacher T, Becker J, Gárdonyi M, Hahn-Hägerdal B, Boles E. 2002. Characterization of the xylose-transporting properties of yeast hexose transporters and their influence on xylose utilization. Microbiology 148:2783–2788. [DOI] [PubMed] [Google Scholar]
- 17.Kotter P, Ciriacy M. 1993. Xylose fermentation by Saccharomyces cerevisiae. Appl Microbiol Biotechnol 38:776–783. doi: 10.1007/BF00167144. [DOI] [Google Scholar]
- 18.Subtil T, Boles E. 2012. Competition between pentoses and glucose during uptake and catabolism in recombinant Saccharomyces cerevisiae. Biotechnol Biofuels 5:14. doi: 10.1186/1754-6834-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du J, Li S, Zhao H. 2010. Discovery and characterization of novel d-xylose-specific transporters from Neurospora crassa and Pichia stipitis. Mol Biosyst 6:2150–2156. doi: 10.1039/c0mb00007h. [DOI] [PubMed] [Google Scholar]
- 20.Runquist D, Fonseca C, Radstrom P, Spencer-Martins I, Hahn-Hägerdal B. 2009. Expression of the Gxf1 transporter from Candida intermedia improves fermentation performance in recombinant xylose-utilizing Saccharomyces cerevisiae. Appl Microbiol Biotechnol 82:123–130. doi: 10.1007/s00253-008-1773-y. [DOI] [PubMed] [Google Scholar]
- 21.Verho R, Penttila M, Richard P. 2011. Cloning of two genes (LAT1,2) encoding specific l-arabinose transporters of the l-arabinose fermenting yeast Ambrosiozyma monospora. Appl Biochem Biotechnol 164:604–611. doi: 10.1007/s12010-011-9161-y. [DOI] [PubMed] [Google Scholar]
- 22.Subtil T, Boles E. 2011. Improving l-arabinose utilization of pentose fermenting Saccharomyces cerevisiae cells by heterologous expression of l-arabinose transporting sugar transporters. Biotechnol Biofuels 4:38. doi: 10.1186/1754-6834-4-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benz JP, Chau BH, Zheng D, Bauer S, Glass NL, Somerville CR. 2014. A comparative systems analysis of polysaccharide-elicited responses in Neurospora crassa reveals carbon source-specific cellular adaptations. Mol Microbiol 91:275–299. doi: 10.1111/mmi.12459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leandro MJ, Spencer-Martins I, Goncalves P. 2008. The expression in Saccharomyces cerevisiae of a glucose/xylose symporter from Candida intermedia is affected by the presence of a glucose/xylose facilitator. Microbiology 154:1646–1655. doi: 10.1099/mic.0.2007/015511-0. [DOI] [PubMed] [Google Scholar]
- 25.Wang C, Shen Y, Zhang Y, Suo F, Hou J, Bao X. 2013. Improvement of l-arabinose fermentation by modifying the metabolic pathway and transport in Saccharomyces cerevisiae. Biomed Res Int 2013:461204. doi: 10.1155/2013/461204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farwick A, Bruder S, Schadeweg V, Oreb M, Boles E. 2014. Engineering of yeast hexose transporters to transport d-xylose without inhibition by d-glucose. Proc Natl Acad Sci U S A 111:5159–5164. doi: 10.1073/pnas.1323464111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Young EM, Tong A, Bui H, Spofford C, Alper HS. 2014. Rewiring yeast sugar transporter preference through modifying a conserved protein motif. Proc Natl Acad Sci U S A 111:131–136. doi: 10.1073/pnas.1311970111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J, Lin L, Li H, Tian C, Ma Y. 2014. Transcriptional comparison of the filamentous fungus Neurospora crassa growing on three major monosaccharides d-glucose, d-xylose and l-arabinose. Biotechnol Biofuels 7:31. doi: 10.1186/1754-6834-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Visser H, Joosten V, Punt PJ, Gusakov AV, Olson PT, Joosten R, Bartels J, Visser J, Sinitsyn AP, Emalfarb MA, Verdoes JC, Wery J. 2011. Development of a mature fungal technology and production platform for industrial enzymes based on a Myceliophthora thermophila isolate, previously known as Chrysosporium lucknowense C1. Ind Biotechnol 7:10. doi: 10.1089/ind.2011.7.214. [DOI] [Google Scholar]
- 30.Wieczorke R, Krampe S, Weierstall T, Freidel K, Hollenberg CP, Boles E. 1999. Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae. FEBS Lett 464:123–128. doi: 10.1016/S0014-5793(99)01698-1. [DOI] [PubMed] [Google Scholar]
- 31.Zimmermann FK. 1975. Procedures used in the induction of mitotic recombination and mutation in the yeast Saccharomyces cerevisiae. Mutat Res 31:71–86. doi: 10.1016/0165-1161(75)90069-2. [DOI] [PubMed] [Google Scholar]
- 32.Gietz RD, Woods RA. 2002. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol 350:87–96. doi: 10.1016/S0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- 33.Tian C, Beeson WT, Iavarone AT, Sun J, Marletta MA, Cate JH, Glass NL. 2009. Systems analysis of plant cell wall degradation by the model filamentous fungus Neurospora crassa. Proc Natl Acad Sci U S A 106:22157–22162. doi: 10.1073/pnas.0906810106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Galazka JM, Tian C, Beeson WT, Martinez B, Glass NL, Cate JH. 2010. Cellodextrin transport in yeast for improved biofuel production. Science 330:84–86. doi: 10.1126/science.1192838. [DOI] [PubMed] [Google Scholar]
- 35.Bisson LF, Fraenkel DG. 1983. Involvement of kinases in glucose and fructose uptake by Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 80:1730–1734. doi: 10.1073/pnas.80.6.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim H, Lee WH, Galazka JM, Cate JH, Jin YS. 2014. Analysis of cellodextrin transporters from Neurospora crassa in Saccharomyces cerevisiae for cellobiose fermentation. Appl Microbiol Biotechnol 98:1087–1094. doi: 10.1007/s00253-013-5339-2. [DOI] [PubMed] [Google Scholar]
- 37.Simossis VA, Heringa J. 2005. PRALINE: a multiple sequence alignment toolbox that integrates homology-extended and secondary structure information. Nucleic Acids Res 33:W289–W294. doi: 10.1093/nar/gki390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bond CS, Schuttelkopf AW. 2009. ALINE: a WYSIWYG protein-sequence alignment editor for publication-quality alignments. Acta Crystallogr D 65:510–512. doi: 10.1107/S0907444909007835. [DOI] [PubMed] [Google Scholar]
- 39.Runquist D, Hahn-Hagerdal B, Radstrom P. 2010. Comparison of heterologous xylose transporters in recombinant Saccharomyces cerevisiae. Biotechnol Biofuels 3:5. doi: 10.1186/1754-6834-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zuo Q, Zhao XQ, Xiong L, Liu HJ, Xu YH, Hu SY, Ma ZY, Zhu QW, Bai FW. 2013. Fine-tuning of xylose metabolism in genetically engineered Saccharomyces cerevisiae by scattered integration of xylose assimilation genes. Biochem Biophys Res Commun 440:241–244. doi: 10.1016/j.bbrc.2013.09.046. [DOI] [PubMed] [Google Scholar]
- 41.Wisselink HW, Toirkens MJ, del Rosario F, Berriel M, Winkler AA, van Dijken JP, Pronk JT, van Maris AJ. 2007. Engineering of Saccharomyces cerevisiae for efficient anaerobic alcoholic fermentation of l-arabinose. Appl Environ Microbiol 73:4881–4891. doi: 10.1128/AEM.00177-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.