Abstract
The human gut is inhabited by thousands of microbial species, most of which are still uncharacterized. Gut microbes have adapted to each other's presence as well as to the host and engage in complex cross feeding. Constraint-based modeling has been successfully applied to predicting microbe-microbe interactions, such as commensalism, mutualism, and competition. Here, we apply a constraint-based approach to model pairwise interactions between 11 representative gut microbes. Microbe-microbe interactions were computationally modeled in conjunction with human small intestinal enterocytes, and the microbe pairs were subjected to three diets with various levels of carbohydrate, fat, and protein in normoxic or anoxic environments. Each microbe engaged in species-specific commensal, parasitic, mutualistic, or competitive interactions. For instance, Streptococcus thermophilus efficiently outcompeted microbes with which it was paired, in agreement with the domination of streptococci in the small intestinal microbiota. Under anoxic conditions, the probiotic organism Lactobacillus plantarum displayed mutualistic behavior toward six other species, which, surprisingly, were almost entirely abolished under normoxic conditions. This finding suggests that the anoxic conditions in the large intestine drive mutualistic cross feeding, leading to the evolvement of an ecosystem more complex than that of the small intestinal microbiota. Moreover, we predict that the presence of the small intestinal enterocyte induces competition over host-derived nutrients. The presented framework can readily be expanded to a larger gut microbial community. This modeling approach will be of great value for subsequent studies aiming to predict conditions favoring desirable microbes or suppressing pathogens.
INTRODUCTION
The human intestine is inhabited by a complex ecosystem consisting of thousands of microbial species. Its collective genome (the microbiome) contains more than 150 times as many genes as our own genome (1). The community of gut microbes has coevolved with humans and has adapted to the competitive conditions in the intestine. Gut microbes differ in their metabolic potential to exploit various environmental conditions and to persist in the intestine (2). They respond differently to the availability of dietary nutrients (3). Moreover, the gut microbiota has to contend with a steep oxygen gradient, which ranges from a partial O2 pressure of approximately 80 mm Hg to nearly anoxic conditions (4). The diverse physiology of the small intestine creates a wide range of local oxygen microenvironments that favor particular groups of bacteria (4). Furthermore, metabolic interaction patterns between microbes affect microbial growth. Generally, six types of species-species interactions can be distinguished (5) (Fig. 1a). In the case of neutralism, two organisms do not depend on each other for growth. If shared resources become scarce, this condition leads to competition (5). In the gut, competition over fermentable carbohydrates typically arises (6). If only one species is negatively affected by the competition, this is known as amensalism (5). Commensalism is observed when one organism (the giver) provides something to another (the consumer) while not benefitting itself. In such a case, the microbe grows on the waste products of the other microbe (5). If the giver pays a fitness price for the interaction, this interaction can be considered parasitism or predation (5). Finally, an interaction in which both organisms convey mutual benefit to each other is considered mutualism, cooperation, or syntrophy (5).
FIG 1.
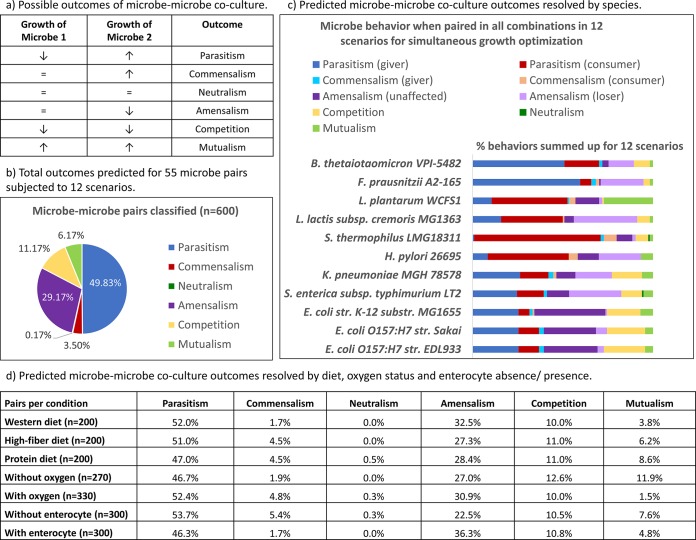
Overview of cogrowth outcomes predicted for the 11 microbes paired in all possible combinations. (a) Description of all possible outcomes of coculturing of two microbes; (b) outcomes predicted for 55 microbe pairs subjected to 12 scenarios (600 cogrowth predictions in total); (c) depiction of the 600 pairs from panel b resolved at the species level; (d) depiction of the 600 pairs from panel b resolved by diet, oxygen status, and enterocyte presence or absence.
Established tools for studying microbe-microbe interactions include in vitro model communities that contain selected representative species (6–8). A typical example of mutualistic cross feeding in the gut is the interaction between Roseburia intestinalis, which converts xylan and acetate to butyrate and hydrogen, and Blautia hydrogenotrophica, which consumes hydrogen and provides acetate to R. intestinalis in return (8). An example of commensalism is the interaction between Bifidobacterium adolescentis and Eubacterium hallii. Unlike the latter organism, B. adolescentis is able to use starch and fructooligosaccharides. Its metabolic products, acetate and lactate, are then utilized by E. hallii (7). Another established experimental model is gnotobiotic animal models, which are germfree animals colonized with selected representative microbes. For example, a gnotobiotic mouse model revealed the interplay of the Bacteroidetes representative Bacteroides thetaiotaomicron and the Firmicutes Eubacterium rectale in the mouse gut (9). Other gnotobiotic mouse models investigated the interaction between B. thetaiotaomicron and the Archaea representative Methanobrevibacter smithii (10) and between B. thetaiotaomicron and two acetogens (11). M. smithii used the formate produced by B. thetaiotaomicron for methanogenesis (10). The acetogen Blautia hydrogenotrophica removed the hydrogen produced by B. thetaiotaomicron, thereby enabling the latter to reoxidize NADH (11). However, due to the complexity of the gut ecosystem, in vitro and gnotobiotic mouse models are limited in scope. In silico models, which can resolve complex interactions at the molecular level, are useful tools that can complement in vitro and in vivo models (12).
Computational models can successfully predict interspecies interactions, such as competition and mutualism. For example, the reverse ecology approach investigates the genome-scale metabolic network of an organism under the assumption that selection pressure from the environment and the presence of other species is reflected in its metabolism (13). In this approach, a seed set, which is the set of compounds that the metabolic network can extract from its environment, is determined. For example, intracellular parasites have smaller seed sets than free-living bacteria (13). Recently, the seed set approach was applied to 154 gut microbes and their competitive potential was predicted (14). Interspecies competition correlated with co-occurrence, indicating that habitat filtering drives microbiome assembly (14). Another method that can successfully predict microbe-microbe interactions in silico is constraint-based modeling (15–21). While constraint-based models operate under the steady-state assumption and do not account for kinetic parameters, their advantage is that the predictions are based on manually curated, genome-scale networks at the biochemical level and are therefore mechanistic. Mechanisms behind a community's behavior can be proposed in silico and subsequently validated experimentally (16). For example, in an in silico bioremediation model, added acetate and Fe(III) were predicted to be major factors influencing competition, which led to the proposal of a long-term bioremediation strategy (21, 22). The applications of constraint-based multispecies modeling to the complex ecosystem residing in the human gut are particularly promising (16). For instance, constraint-based models of an ex-germfree mouse colonized with a single gut symbiont (23) and of a human gut containing up to 11 microbes, including commensals, probiotics, and pathogens, in different combinations have been constructed and analyzed (24). In the latter study, the model community of 11 microbes was shown to significantly affect the human metabolism and to increase the secretion flux of host body fluid metabolites by up to 100 times (24). In the present study, the pairwise interactions between 11 microbes were systematically investigated while imposing various environmental constraints on the pairs. The microbe pairs were subjected to three different dietary regimes and both anoxic and oxic conditions. Moreover, a cell type-specific reconstruction of the human small intestinal enterocyte (25) was included in the modeling framework, making it possible to model microbe-microbe interactions with and without the human host as the background.
MATERIALS AND METHODS
Definition of scenarios.
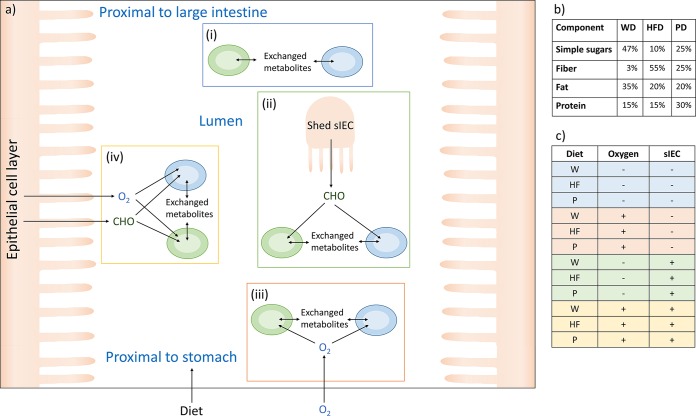
To simulate different regions of the small intestine, scenarios with and without the host and with varied oxygen and nutrient availabilities that represented the range of microenvironments in the small intestine due to its oxygen gradient and anatomy were defined (4) (Fig. 2a). These scenarios were oversimplifications made for modeling purposes, but they allowed us to evaluate certain parameters that define environmental niches found in the human gut. In vivo, an oxygen gradient (4) rather than a binary switch between the total absence/presence of oxygen occurs.
FIG 2.
Overview of study design. (a) Diagram of the modeling framework. The four main scenarios are depicted: (i) no oxygen, without an enterocyte, representing the near anoxic conditions at the midpoint of the lumen (4); (ii) no oxygen, with an enterocyte, representing a scenario in which the microbes were exposed to sloughed epithelial cells (4) but were under anoxic conditions because they were far from the epithelial cell layer; (iii) with oxygen, without an enterocyte, representing the midpoint of the lumen in the duodenum, where some oxygen swallowed with the food is present (60); (iv) with oxygen, with an enterocyte, simulating the growth of bacteria near the epithelial cell layer with oxygen dissipating from the enterocyte (4). (b) Compositions of the simulated dietary regimes. (c) Table of the 12 scenarios that the 55 microbe pairs were subjected to. W and WD, Western diet; HF and HFD, high-fiber diet; P and PD, protein diet; CHO, carbohydrates derived from the enterocyte; sIEC, small intestine enterocyte.
Assembly of the host and a representative microbial community.
We used 11 manually curated, published gut microbe reconstructions (23, 26–33) that we had refined previously (24). The reconstructed strains are listed in Table S1 in the supplemental material, and the reconstructions are shown in spreadsheet format in Table S2 in the supplemental material. To simulate a small intestine environment, a manually curated and validated model of the small intestinal enterocyte, hs_sIEC611 (25), was used. Host and microbes were joined through a separate compartment (u) simulating the intestinal lumen. This compartment served as an inlet for nutrients derived from the simulated diet and the enterocyte; it allowed metabolite exchange between the microbes and provided an outlet for fermentation end products (Fig. 2). The 11 microbes were paired in every possible combination. To simulate baseline conditions or controls, the corresponding microbe-microbe models were used, and one microbe was silenced by constraining the fluxes through all of the reactions to zero (lower bound [lb] = upper bound [ub] = 0 mmol/gDW/h, where gDW is the number of grams [dry weight]). The microbe-microbe models and single-microbe models were then subjected to four basic scenarios, which were defined as follows (Fig. 2). (i) The microbe models were used separately (n = 10) and joined pairwise in all combinations (n = 45) while simulating anoxic conditions. (ii) The microbe models were used separately (n = 10) and joined pairwise with hs_sIEC611 in all combinations (n = 45) while simulating anoxic conditions. (iii) The microbe models were used separately (n = 11) and joined pairwise in all combinations (n = 55) while allowing oxygen uptake. (iv) The microbe models were used separately (n = 11) and joined pairwise with hs_sIEC611 in all combinations (n = 55) while allowing oxygen uptake.
Pairs including Helicobacter pylori, a microaerophilic bacterium, were not subjected to scenarios (i) and (ii). Each basic scenario was performed for three diets varying in carbohydrate, fat, and protein contents (Fig. 2b), resulting in 12 scenarios (Fig. 2c). In total, 600 species-species interactions and 126 single-microbe models were simulated. The growth rate of the enterocyte was set to a maintenance rate by constraining the lower and upper bounds on the biomass objective function of hs_sIEC611 (lb = ub = 0.01 mmol/gDW/h). The lower bounds on the extracellular oxygen exchange reaction were set to −10 mmol/gDW/h for scenarios (iii) and (iv). The lower and upper bounds on the luminal oxygen exchange of the enterocyte were fixed at zero. All models are available in Matlab format at www.thielelab.eu.
Diet definition.
To investigate the impact of diet on microbe-microbe interactions, three diets that differed in their carbohydrate, fat, and protein contents were defined (Fig. 2). The Western diet was high in simple sugar and fat content and low in fiber content. In contrast, the high-fiber diet was high in fiber content but low in simple sugar and fat content. The protein diet had twice the protein content of the other diets, and it had a low fat content with a moderate simple sugar and fiber content. The constraints applied to simulate the three diets are listed in Table S3 in the supplemental material.
In silico analysis.
For all of the simulations, the methods implemented in the COBRA toolbox (34) were used within Matlab (Mathworks, Inc., Natick, MA, USA). Tomopt (Tomlab, Inc., Seattle, WA, USA) was used as a linear programming solver for the flux balance analysis (35). For the flux variability analysis (36), ILOG CPLEX (IBM) was used as a linear programming solver.
Pareto optimality analysis.
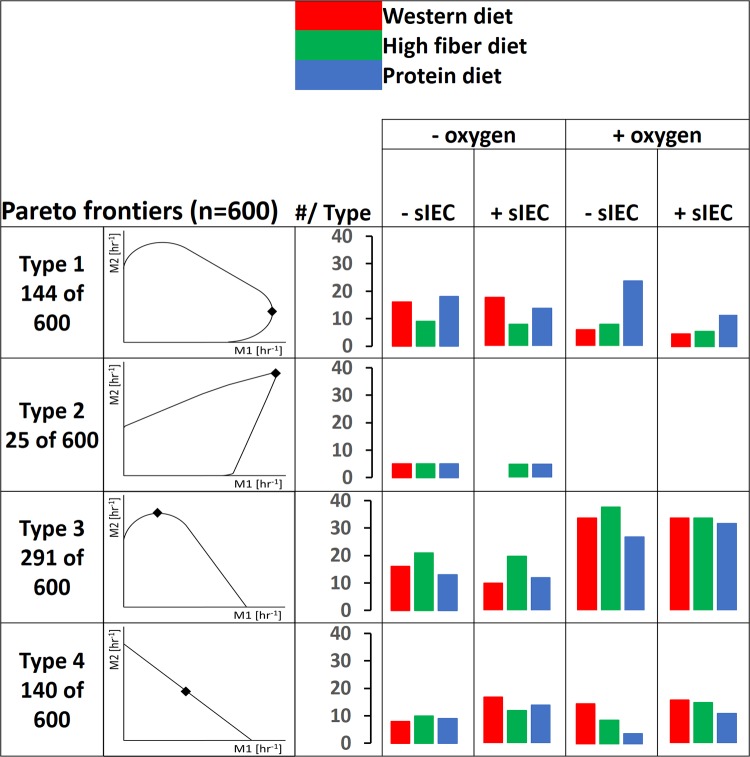
Pareto optimality analysis was performed as described previously (23). Briefly, the fluxes through the biomass objective functions of each microbe were fixed at different intervals while optimizing the flux through the other biomass objective function. Hence, the trade-offs in biomass production for each pair were computed. Each Pareto frontier contained all of the possible interactions between both microbes when optimizing for biomass production. Pareto optimality analysis was performed for all 12 defined scenarios (Fig. 2). Each microbe was included in 114 Pareto frontiers, with the exception of the microaerophile Helicobacter pylori, which participated in only 60 Pareto frontiers. In total, 600 Pareto frontiers were computed (see Fig. S1 in the supplemental material).
Computation of optimal total biomass production for microbe-microbe pairs.
The combined maximal growth was optimized by simultaneously maximizing the biomass objective functions of both microbes. As a baseline condition, growth rates for the microbes alone were computed for the 12 scenarios (Fig. 2). The growth rate of each microbe in each pair for each scenario was compared with the growth rate achieved by the microbe alone. Eight microbe behaviors could be distinguished, and these were grouped into six basic cogrowth outcomes (Fig. 1a; Table 1).
TABLE 1.
Description of nine possible outcomes of cogrowth per microbea
| Description | Type |
|---|---|
| The microbe grows slower in the presence of the other microbe, whereas the other microbe grows faster. | Parasitism |
| The microbe grows faster in the presence of the other microbe, whereas the other microbe grows more slowly. | Parasitism |
| The microbe's growth is not affected, whereas the other microbe grows faster. | Commensalism |
| The microbe grows faster in the presence of the other microbe, whereas the other microbe's growth is not affected. | Commensalism |
| Neither microbe affects the growth of its partner significantly. | Neutralism |
| The microbe grows more slowly in the presence of the other microbe, whereas the other microbe's growth is not affected. | Amensalism |
| The microbe's growth is not affected, whereas the other microbe grows more slowly. | Amensalism |
| Both microbes grow more slowly when they are grown together than when they are grown separately. | Competition |
| Both microbes achieve a significantly higher growth rate when they are grown together than when they are grown separately. | Mutualism |
A difference in growth rate of at least 10% compared with the growth rate of the microbe grown separately was considered significant.
Identification of obligate metabolic exchanges.
Obligate metabolic exchanges were defined as metabolite shuttles between microbe-microbe pairs that occurred in every alternative steady-state solution when simulating simultaneous growth. To compute alternative steady-state solutions, flux variability analysis (36, 37) was used. Metabolites exchanged between microbe pairs were determined by inspecting the internal exchange reactions in the model (Fig. 2a). To identify metabolite pairs that were shuttled between microbes while being interconverted, the pathway utilization in the microbe models was inspected for each transported metabolite.
Identification of metabolic products supplied by the enterocyte.
The internal exchanges of the enterocyte and the microbe pairs in scenarios (ii) and (iv) were inspected to identify valuable nutrients provided by the enterocyte. This condition was fulfilled when the enterocyte secreted the nutrient in every alternative solution and at least one microbe consumed it at a corresponding uptake rate.
RESULTS
In this study, the potential of pairs of gut microbes to engage in mutualistic, commensal, and competitive interactions was systematically investigated. A computational framework that could predict interspecies behavior under various environmental conditions, such as dietary input, oxygen availability, and exposure to the host intestinal cell layer, was developed. The results revealed a variety of trade-offs within the microbe pairs that depended on both the metabolic potential of each microbe and the imposed environmental conditions. Maximizing the total growth of each pair revealed that giver-consumer interactions were the most frequent. Moreover, a substantial percentage of mutualistic interactions involving Lactobacillus plantarum were predicted, but these were abolished in the presence of oxygen. Both known and nonintuitive metabolic cross feedings in a small intestinal model community of 11 microbes were proposed.
Representativeness of the model gut microbiota community.
The model gut microbiota community consisted of 11 strains belonging to nine species. The 11 microbes account for three of the four main phyla in the human gut microbiome (see Table S1 in the supplemental material), and their metabolic functions are representative of those detected in the gut microbiomes of 124 human volunteers (1, 24). Moreover, they contained metabolically similar species, such as Escherichia coli MG1655 and Klebsiella pneumoniae, but they also had metabolically distant representatives (24), the presence of which has been proposed to be important for collaboration in a community (38). All metabolic reconstructions used in this study were constructed in a manual curation effort (23, 26–33) and account for not only the genomic annotation but also the known biochemical and physiological traits of their target organisms. The modeling framework can thus be expected to accurately capture the metabolic capabilities of the 11 representative gut microbes. As a result, the 11 microbes captured distinct interaction patterns that are typical of those detected in microbial communities (Fig. 1b and c). All of the included species and strains, except Faecalibacterium prausnitzii and K. pneumoniae, have been detected in the small intestine microbiome (39); therefore, our model community is also representative of the small intestine microbiota. Species display strain-specific differences, which were reflected by the observed differences in the behavior of commensal and pathogenic E. coli strains (Fig. 1c). It can be expected that the use of different strains for the other species would also result in different outcomes.
Trade-offs in biomass production between microbe pairs.
Pareto optimality analysis was performed for each of the 12 scenarios (see Fig. S1 in the supplemental material). Four types of Pareto frontiers could be distinguished (Fig. 3). In the type 1 interaction (24% of the pairs), both of the microbes benefitted from the other microbe at low growth rates of the latter (increase in growth rate, at least 10%). Higher growth rates led to increasing competition for growth-limiting nutrients (Fig. 3). This type of interaction has been previously observed in a model of the mouse and its gut symbiont B. thetaiotaomicron (23). In the rare type 2 interaction (4% of pairs), which was observed only for pairs including L. plantarum (see Fig. S1 in the supplemental material), mutual benefit was observed even at the highest possible growth rates, and the microbes never entered a phase of competition. Metabolic exchange between the two microbes was obligatory to achieve the highest possible growth rates (Fig. 3), which may be considered syntrophy. In the type 3 interaction, which was observed in the highest number of pairs (49%), only one microbe benefitted at low growth rates of the other microbe (Fig. 3). At higher growth rates of the benefitting microbe, this interaction can be described as parasitism, as it came at the expense of the giving microbe (Fig. 3). Finally, in the type 4 interaction, competition was observed throughout the Pareto frontier, as neither microbe benefitted the other (Fig. 3; 23% of pairs). Notably, the same species fell into different types of interactions depending on the scenario and depending on which microbe it was paired with. For example, E. coli MG1655 was able to engage in all four types of interactions (see Fig. S1 in the supplemental material). Moreover, the same pair could display various behaviors depending on the scenario. One example is the pair of L. plantarum and K. pneumoniae, which displayed a type 1 interaction in three scenarios, a type 2 interaction in five scenarios, a type 3 interaction in three scenarios, and a type 4 interaction in one scenario (see Fig. S1.20 and S1.68 in the supplemental material). As can be expected, the computed Pareto frontiers reflected not only the microbe's metabolic potential but also the nutrient regime, oxygen availability, and enterocyte presence or absence. For instance, type 2 interactions were computed only in the absence of oxygen (Fig. 3; see also Table S4 in the supplemental material). These results revealed a complex interplay of microbe-microbe interactions and strong influences of dietary nutrients, host-derived nutrients, and oxygen on the possible trade-offs for each microbe pair.
FIG 3.
The four types of Pareto frontiers computed when predicting microbe-microbe growth trade-offs and the number of pairs falling into each type for all models, resolved by scenario. M1, microbe 1; M2, microbe 2. Points on the Pareto frontiers where total growth is maximal are indicated by diamonds. The locations of these points differ for the individual microbe-microbe plots (and are shown in detail in Fig. S1 in the supplemental material). Note that the figure displays extreme cases for types 3 and 4, in which the growth benefit is 0%.
Microbe-microbe interactions enabling maximal biomass production.
The Pareto frontier contains an infinite number of steady-state flux solutions, each representing an optimal trade-off between the growth rates of the two microbes. However, many of these solutions may not be attained in vivo, as they result in low biomass production overall or significantly prioritize one of the two microbes. Under the assumption that a coculture of two bacteria would result in the highest possible bacterial density, the simultaneous growth of each pair in the 12 scenarios, which corresponded to the point of the Pareto frontier resulting in a combined optimal biomass production, was optimized. For type 1, type 2, and type 3 interactions, this point was located at the saddle points of the frontiers where the incline changed (see Fig. S1 in the supplemental material). To characterize the type of behavior observed at this point, the growth rate achieved for each microbe in each pair was compared with the growth rate achieved by the microbe alone (Fig. 2). An increase or a decrease in the growth rate under the pairwise condition of at least 10% compared with that of each microbe alone was considered a significant difference. The analysis was performed in the presence and absence of the enterocyte model. The outcome for each microbe could be grouped into nine types (Table 1). The most commonly observed interaction was a giver-consumer interaction that was detrimental to the giver (parasitism). This interaction accounted for 50% of all pairs in the present study (Fig. 1b). This result agrees with the predictions of Freilich et al. (19), who also found give-take relationships to be the most common when computing pairwise interactions in an in silico community. The least frequent interaction, neutralism, occurred in only one pair (0.17%) (Fig. 1b). This indicates that most of the microbes in our study utilized similar nutrients, causing competitive interactions to be more frequent (11%). Because all 11 microbes are gut inhabitants and likely use carbon sources typically found in the human diet, such an outcome was expected. Often, only one microbe was affected by the competitive interaction, indicating that the microbes differed in their ability to cope with limited nutrient intake. Commensalism was rarely observed (4%). Finally, the microbes in a small subset of pairs (6%) were able to complement each other's metabolisms to mutual benefit (mutualism).
Microbe-microbe interactions are species specific.
To identify species-specific differences in the commensal, mutualistic, and competitive interaction profiles due to the distinct metabolisms of the microbes, we resolved the computed interactions for each microbe (Fig. 1c). Each microbe was capable of acting as a giver and a consumer in at least one pair and scenario, though the percentage of interactions in which it participated varied greatly. For instance, B. thetaiotaomicron and F. prausnitzii were givers in the majority of their interactions (Fig. 1c; see also Table S5 in the supplemental material). In agreement with B. thetaiotaomicron's predicted role as a giver (only 21% of its interactions benefitted the microbe itself), Bacteroides representatives, such as B. thetaiotaomicron, are considered primary degraders, in contrast to secondary fermenters, such as streptococci and lactobacilli (40). F. prausnitzii benefitted from only 10% of its interactions (Fig. 1c). This finding indicates that F. prausnitzii is easily outcompeted in the presence of facultative anaerobes and may partially explain why this microbe is not detected in the small intestine (39).
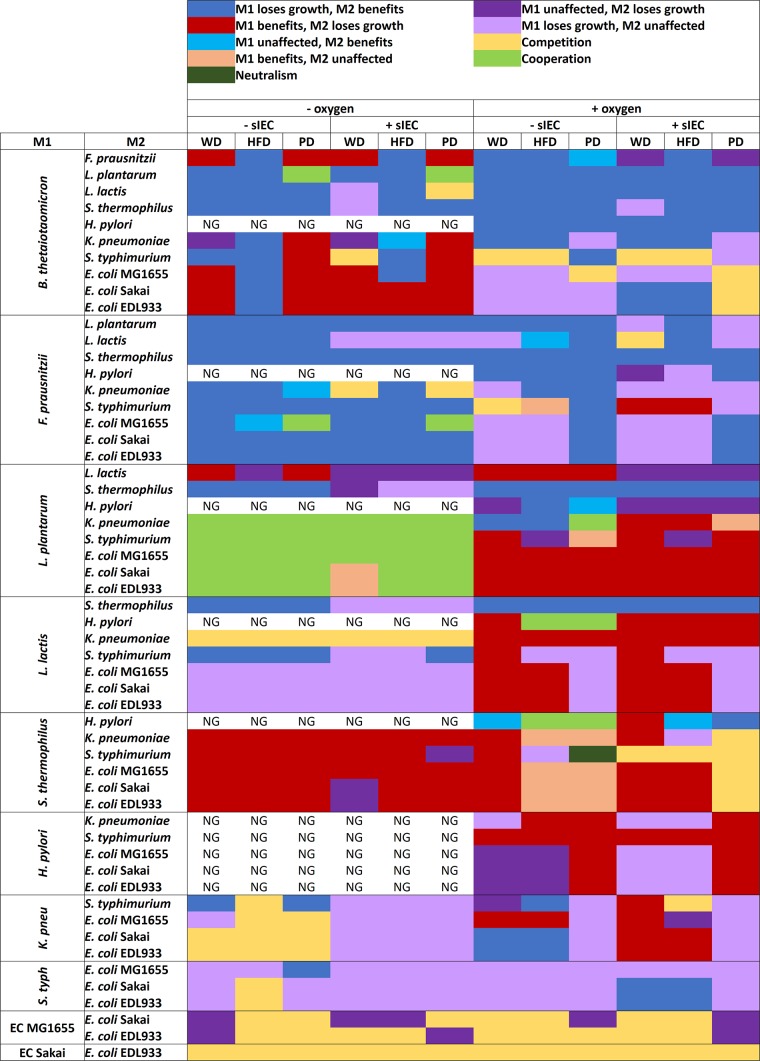
A surprisingly high number of mutualistic interactions was predicted for L. plantarum (Fig. 1c). In fact, 31 of the 37 (84%) predicted mutualistic pairs included this bacterium (Fig. 4). In contrast, Streptococcus thermophilus, Lactococcus lactis, and H. pylori were mainly consumers and as such received one-sided benefits from other microbes. S. thermophilus showed a particularly strong ability to benefit from other microbes, acting as a consumer in 77% of its interactions (Fig. 1c). In only 4% of pairs, S. thermophilus played a benefactor role (giver or mutualism) (Fig. 1c), indicating that the microbe would benefit greatly from the modeled microbial community to the detriment of the other microbes. Consistently, S. thermophilus grew poorly by itself, but in the presence of other microbes it achieved the second highest growth rates overall after L. plantarum (see Table S5 in the supplemental material). Our results suggest that S. thermophilus was poorly adapted to grow individually under certain conditions (especially on the high-fiber diet), but it was able to use metabolic end products and simple sugars liberated by other microbes efficiently. K. pneumoniae, Salmonella enterica subsp. Typhimurium, and the E. coli strains engaged in significant competitive interactions (Fig. 1c), in agreement with the fact that proteobacteria compete over similar carbon sources (41). In fact, 56 to 58% of the interactions predicted for the E. coli strains were competitive (Fig. 1c), and the E. coli strains were almost exclusively competitive when paired among themselves (Fig. 4). In more than half of the competitive interactions that they participated in, the E. coli strains were unaffected, but the competing microbe had a decreased growth rate (Fig. 1c). This result suggests that E. coli can cope well with competition for dietary nutrients. In agreement with this observation, E. coli strains are commonly detected in the small intestine (39). One possible explanation for these differences in pairwise interactions is metabolic distance, which was previously determined for the 11 microbes (24). However, the metabolic distance for the 11 microbes correlated poorly with giver-consumer interactions and competitive interactions (see the Supplemental Results section in the supplemental material).
FIG 4.
Overview of the interactions predicted for all pairs in 12 scenarios. M1, microbe 1; M2, microbe 2; sIEC, small intestine enterocyte; WD, Western diet; HFD, high-fiber diet; PD, protein diet; NG, no growth due to H. pylori being unable to grow without oxygen; K. pneu, K. pneumoniae; S. typh, S. enterica subsp. Typhimurium; EC, E. coli.
Environmental constraints affect the predicted microbe behavior.
The availability of nutrients (e.g., carbohydrates) and oxygen is well-known to influence the gut microbiota (3, 42). The models were used to interrogate how microbe-microbe interactions differed in the defined 12 scenarios that placed the 55 species-species pairs in different metabolic environments (Fig. 2). Microbe-microbe interactions for the three diets were generally comparable, except that fewer giver-consumer pairs and more mutualistic pairs were predicted on the protein diet (Fig. 1d). Resolving the microbe behavior by diet on the species level, however, revealed significant species-specific differences (see Fig. S5a to c in the supplemental material). For instance, B. thetaiotaomicron provided benefits to the other microbes in significantly more pairs on the high-fiber diet (see Fig. S5b in the supplemental material). These relationships were due to the microbe's well-known fiber-degrading capabilities (43), which enabled it to provide otherwise inaccessible plant polysaccharides to the other microbes. Accordingly, the plant polysaccharide content was highest in the high-fiber diet (see Table S3 in the supplemental material). In particular, the presence of B. thetaiotaomicron increased the growth rate of S. thermophilus by up to 31-fold, demonstrating that it liberated simple sugars from fiber in the diet that were usable by S. thermophilus (see Table S5 in the supplemental material).
An anoxic environment is mutualism inducing.
The predicted microbe-microbe interactions with and without oxygen differed significantly (Fig. 1d and 4). Allowing oxygen uptake caused a decrease in mutually beneficial interactions and an increase in giver-consumer interactions (Fig. 1d and 4). In fact, all but one of the mutualistic interactions observed without oxygen were abolished in the presence of oxygen (Fig. 4; see also Fig. S6a and b in the supplemental material). Of the five cases of mutualistic interactions predicted in the presence of oxygen, four involved the strict microaerophile H. pylori (Fig. 4). As can be expected, all of the microbes except Lactococcus lactis showed growth rate increases of at least 10% and a growth rate up to 11-fold higher in the presence of oxygen than in the absence of oxygen (see Table S5 in the supplemental material). These results highlight that in the presence of oxygen, most microbes were able to efficiently extract energy from the supplied dietary nutrients and did not rely on metabolites secreted by other microbes. In the absence of oxygen, however, the microbes were forced to cooperate to achieve optimal growth by exchanging metabolites with each other. Accordingly, mutualistic pairs switched to parasitic giver-consumer interactions in the presence of oxygen (Fig. 4).
Obligate metabolic exchanges reoccur in microbe-microbe pairs.
To identify the mechanisms behind the observed oxygen-dependent microbe-microbe interactions, the pathway usage in the computed alternative solutions was inspected for all of the pairs in the 12 scenarios. Obligate metabolic interactions, which we defined as cross-feeding cycles occurring in every alternative solution and therefore required for optimal growth, were identified. Certain obligate metabolic interactions reoccurred in multiple pairs and/or scenarios (see Table S6 in the supplemental material). These interactions included well-known cross feedings occurring in the gut microbiota, such as the conversion of acetate to butyrate (44) and of ethanol to acetaldehyde (45), as well as nonintuitive exchanges, such as threonine/glycine interconversion (see Table S6 in the supplemental material). Up to three of these interactions were observed in each pair and scenario (see Fig. S3 in the supplemental material); however, there was no clear correlation between the sum of the cross-feeding interactions and the pair's ability to engage in mutual benefit. For example, the pair of L. lactis and K. pneumoniae displayed 24 metabolic interactions in total in the 12 scenarios yet showed no commensal or mutualistic behavior (see Fig. S3 in the supplemental material). Furthermore, the highest computed number of cross-feeding behaviors (three) was observed only for the pair of F. prausnitzii and E. coli MG1655 in 7 out of 12 scenarios, but only two of these cases resulted in mutual benefit (see Fig. S3 in the supplemental material). In conclusion, the number of reoccurring exchanges per pair alone could not entirely explain the behavior of the pairs.
The co-occurrence of metabolic exchanges involving NAD+/NADH interconversion drives mutualism.
To identify the exchange co-occurrence patterns that induced mutualism, the above-described types of computed exchange strategies were plotted by interaction (see Fig. S4 in the supplemental material). As was expected, mutualistic pairs displayed more metabolic exchanges on average than giver-consumer or competitive pairs (1.97 exchanges on average per pair compared with 0.67 to 0.84 exchanges per pair for giver-consumer pairs and 0.25 to 0.30 exchanges per pair for competitive pairs) (see Fig. S4 in the supplemental material). Mutualistic behavior was linked to exchange co-occurrences, with on average 1.1 exchange strategies co-occurring per mutualistic pair (see Table S7 in the supplemental material). In 29 of the 37 mutualistic pairs, all of which included L. plantarum, pyruvate/d-lactate and acetaldehyde/ethanol exchanges co-occurred, indicating that pairwise reoccurring metabolic exchanges were linked to mutualism. Both exchanges involved the interconversion between NAD+ and NADH, which is required to maintain the flux through glycolysis (Fig. 5a). These predictions indicate that the mutually beneficial behavior in many pairs involving L. plantarum was due to an improved ability to maintain the redox balance via d-lactate dehydrogenase and alcohol dehydrogenase (Fig. 5a). When oxygen uptake was allowed, the bacteria instead regenerated NAD+ from NADH via cytochrome oxidase and NADH dehydrogenase, eliminating the need to cooperate to maintain their NAD+/NADH balance (Fig. 5b). In conclusion, certain pairs were able to optimize the redox balance to each other's benefit by complementing each other's metabolism. These pairs typically involved L. plantarum and a metabolically distant partner (B. thetaiotaomicron or a representative of the gammaproteobacteria) (Fig. 4), suggesting that parallel pathways in bacteria with otherwise distinct metabolisms were mutualism inducing.
FIG 5.
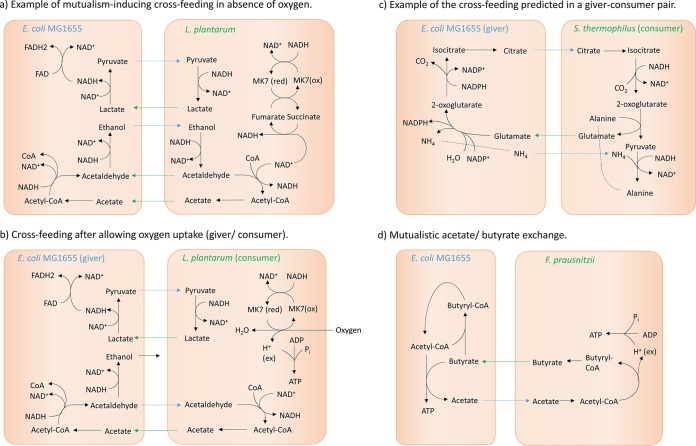
Simplified depiction of the cross feeding observed in selected cases of microbe-microbe interaction. (a) Example of a mutualistic pair under anoxic conditions, in which the microbes benefit from each other through pyruvate/d-lactate exchange and acetaldehyde/ethanol exchange. MK7(red), menaquinol 7; MK7(ox), menaquinone 7; FAD, flavin adenine dinucleotide. (b) Altered behavior observed in the pair from panel a after allowing oxygen uptake, which abolishes the mutualistic behavior. (ex), extracellular. (c) Example of the cross feeding predicted for a typical giver-consumer pair. (d) Depiction of the specific cross feeding between F. prausnitzii, which converts acetate to butyrate, and E. coli MG1655, which utilizes butyrate as a carbon source and produces acetate.
We then examined exchange co-occurrences that benefit only one partner in pairs (giver-consumer pairs). The most frequent exchange co-occurrence in parasitic giver-consumer pairs was fumarate/succinate and glutamate/citrate exchange (co-occurring in 26 out of 299 pairs, or 9%) (see Table S7 in the supplemental material). In all 26 pairs, F. prausnitzii acted as a giver to the benefit of a representative of the gammaproteobacteria. Fumarate/succinate exchange enabled the representative of the gammaproteobacteria to regenerate NAD+ via fumarate reductase and a quinone-dependent NADH dehydrogenase (Fig. 5c). Glutamate/citrate exchange caused citrate transfer from giver to consumer, thereby enabling the consumer to convert 2 units of NADH to NAD+ (Fig. 5c). Finally, we predicted that acetate/butyrate exchange was specific for the pair of F. prausnitzii and E. coli MG1655 (Fig. 5d). The latter utilized the butyrate produced by F. prausnitzii as a carbon source, thereby gaining a significant growth advantage in seven scenarios (Fig. 4; see also Table S5 in the supplemental material). In two scenarios (protein diet without oxygen), this strategy caused the usually one-sided giver-consumer interaction between F. prausnitzii and E. coli MG1655 to switch to mutualism (Fig. 4). Thus, under certain nutrient regimes, E. coli MG1655 was able to assist F. prausnitzii through acetate production and in return benefitted from F. prausnitzii's waste product, butyrate. In contrast to commensal E. coli, the O157:H7 (enterohemorrhagic E. coli [EHEC]) strains were unable to utilize butyrate due to the lack of acetyl coenzyme A (acetyl-CoA):butyrate-CoA transferase (encoded by the genome of strain MG1655; NCBI gene identifiers b2221 and b2222).
In summary, we predicted that the co-occurrences of specific metabolite exchange strategies could explain most cases of mutualism as well as specific cases of giver-consumer interactions. These co-occurring metabolite strategies involved reactions that maintained the NAD+/NADH balance. In mutualistic pairs, both microbes were able to regenerate NAD+ from NADH through cross feeding. In one-sided giver-consumer interactions (parasitism), only one microbe was able to exploit metabolite exchange to maintain its NAD+/NADH balance. Competing pairs were unable to balance the availability of redox equivalents, causing the forced cogrowth to be a burden on one or both microbes. In the specific pair of F. prausnitzii and E. coli MG1655, cross feeding of acetate and butyrate was able to induce mutualism only under anoxic, high-protein-intake conditions (Fig. 4).
The small intestinal enterocyte induces competition between microbes through secretion of carbon sources.
It is well-known that intestinal microbes feed on host-derived carbohydrates (43, 46). Therefore, the host background can be expected to influence the pairwise interactions. In the presence of the enterocyte, the number of competitive interactions affecting only one microbe (amensalism) was higher. The number of competitive interactions affecting both microbes was comparable with and without the presence of the enterocyte (Fig. 1d). Species losing more frequently in amensalism-type interactions in the presence of the enterocyte included F. prausnitzii, L. lactis, H. pylori, and K. pneumoniae. In contrast, L. plantarum, S. thermophilus, and E. coli MG1655 were rarely outcompeted in the presence of the enterocyte (see Fig. S7a and b in the supplemental material). When joined separately with the enterocyte, the growth rates of all of the microbes increased by at least 10% and up to 13-fold under most nutrient conditions, demonstrating that the microbes can utilize enterocyte-derived nutrients (see Table S5 in the supplemental material). Thus, exposing the pairs to the enterocyte opened up a source of energy and carbon apart from the simulated diet that can explain the predicted differences in interactions induced by the enterocyte.
To identify these enterocyte-derived energy and carbon sources, the metabolites exchanged between the enterocyte and each microbe pair were inspected (see Table S8 in the supplemental material). It must be noted that the simulation setup is not entirely realistic because it assumes that the enterocyte is working for the microbes' benefit. However, it still provides valuable insight into the abilities of the 11 microbes to utilize enterocyte-derived nutrients to their advantage. The enterocyte provided significant glucose to the microbes by performing gluconeogenesis, which is accounted for in hs_sIEC611 (25). Glucose was partially derived from the starch fraction of the diet, which can be degraded only by the human host (46) and by B. thetaiotaomicron (43). Thus, the enterocyte provided extra glucose, in addition to the dietary glucose, to the microbes (see Table S8a in the supplemental material), resulting in competition over the glucose provided by the enterocyte. L. lactis and K. pneumoniae were frequently deprived of glucose when paired with another microbe (see Table S8a in the supplemental material), explaining why they were largely losing in amensalism-type interactions in the presence of the enterocyte. In contrast, S. thermophilus consistently consumed glucose at a high uptake rate, depriving the microbes it was paired with of this carbon source (see Table S8a in the supplemental material). This result indicates that S. thermophilus was able to outcompete other bacteria through its efficient use of glycolysis. Another carbon source derived from the enterocyte was glycerol, which was, however, not produced in every enterocyte-associated scenario. Glycerol could not be consumed by B. thetaiotaomicron, F. prausnitzii, S. thermophilus, or H. pylori (see Table S8b in the supplemental material) but was consumed at high rates by K. pneumoniae, S. Typhimurium, and the E. coli strains, and thus, it constituted another source of competition in pairings with these bacteria (Fig. 4; see also Table S8b). This finding, rather than metabolic closeness, may explain why competition was the dominant interaction predicted for pairs of Gammaproteobacteria (Fig. 4). In conclusion, the enterocyte induced competition by secreting glucose and, to a lesser extent, by secreting glycerol.
DISCUSSION
The pairwise interactions of 11 gut microbes spanning three phyla, including commensals, probiotics, opportunistic pathogens, and pathogens, were systematically investigated under various environmental constraints. Our main findings were as follows: (i) the trade-off between the growth rates in the microbe pairs varied significantly depending on the participating microbes and was influenced by the nutrient environment; (ii) the potential to engage in giver-consumer, mutualistic, or competitive interactions was species specific; (iii) allowing oxygen uptake abolished most mutualistic interactions between microbes; (iv) the NAD+/NADH balance drove mutualism in pairs involving L. plantarum; and (v) the enterocyte induced competition by secreting a carbon source that could be used by the microbes. In summary, both the distinct metabolisms of the 11 microbes and the imposed environmental constraints significantly influenced the pairwise interaction patterns and resulted in a variety of cogrowth outcomes.
Both the participating microbes and the nutrient environment were predicted to alter the trade-offs between two microbes. In fact, many microbe pairs produced drastically different trade-offs depending on the scenario (see Fig. S1 in the supplemental material). The in silico prediction of potential outcomes of gut microbial coculture has useful applications. One of the defense mechanisms against pathogens is exclusion through competition. For instance, commensal proteobacteria can outcompete closely related pathogens, such as S. Typhimurium (41). Using metabolic modeling, gut microbes could be screened for species that are particularly efficient at outcompeting persistent pathogens, such as Clostridium difficile. Such predictions could subsequently be experimentally validated. In addition, mutualistic microbe-microbe interactions could be predicted in silico. For example, mutualistic partners that improve the growth of poorly growing beneficial microbes could be identified. Multistrain and multispecies probiotics have, in some cases, been found to be more effective than monostrain probiotics; however, the development of such probiotic mixtures is expensive (47). The presented metabolic modeling framework, which can incorporate any reconstructed species, could predict optimal combinations of probiotics to promote community survival in the gut. Moreover, nutrient conditions supporting the optimal growth of microbe pairs could be predicted. Although the presented framework currently limits Pareto optimality analysis to two biomass functions, it is possible to compute the trade-offs for three or more competing objectives using constraint-based methods (48), which may lead to novel insights into multispecies interactions.
Singular cogrowth outcomes were computed by maximizing for total biomass production, which corresponds to maximal community growth as the objective function. A downside of this method is the inherent assumption that one species would sacrifice its own growth to increase the growth rate of another species (49). Another alternative would be fixing the growth rates of the included microbes at experimentally determined ratios (49). This alternative was unsuitable for the present study because there are no experimental cogrowth data available for the simulated pairs under the given conditions. Species-specific differences in microbial growth rates between the three diets were predicted, in agreement with the fact that diet affects the gut microbiota (3). For instance, L. plantarum consistently showed higher growth rates on the Western diet than on the high-fiber diet, except when paired with the fiber degrader B. thetaiotaomicron (see Table S5 in the supplemental material). Consistently, the persistence of L. plantarum WCFS1 was 10 to 100 times higher in mice fed a Western diet than in animals fed a low-fat chow (50). Moreover, E. coli MG1655, but not the two E. coli O157:H7 strains or any of the other species, could use butyrate as a carbon source. These predictions confirm the results of Monk et al., who found that commensal E. coli strains but not EHEC strains exploit butyrate as a carbon source (51). Butyrate utilization by commensal E. coli strains may be an adaptation to the presence of butyrate producers, such as F. prausnitzii.
All but one of the predicted mutualistic interactions under anoxic conditions were abolished when oxygen uptake was allowed (Fig. 4). The colonic microbiota is made up mainly of strict anaerobes (52), whereas the small intestinal microbiota is dominated by facultative anaerobes, such as streptococci and E. coli (39), due to the higher oxygen partial pressure. Notably, the colonic microbiota is also more diverse and complex than the small intestinal microbiota (39). We speculate that the lack of oxygen availability in the large intestine may have forced the members of the large intestinal microbiota to coevolve to maintain their redox balance and a positive energy balance, thus leading to an ecosystem that is more complex and cooperative than that of the small intestinal microbiota.
Pyruvate/d-lactate interconversion and acetaldehyde/ethanol interconversion induced mutualism by enabling the microbes to balance their NAD+/NADH levels (Fig. 5a; see also Table S7 in the supplemental material). Allowing oxygen uptake instead permitted the microbes to regenerate NAD+ via the electron transport chain (Fig. 5a and b). Ethanol and acetaldehyde are known products of the small intestinal microbiota that can disrupt intestinal barrier integrity (53). Moreover, ethanol consumption can promote small intestine overgrowth and increase the proteobacterial population (53). Our model proposes a mutualism-inducing effect of ethanol conversion to acetaldehyde for pairs including L. plantarum and representatives of the proteobacteria, which may have implications for the proteobacterium-promoting effect of ethanol consumption.
Exposing the microbe pairs to the small intestinal enterocyte mainly induced one-sided competition over readily available carbon sources (glucose, glycerol; see Table S8 in the supplemental material). The clear winner in the competition for glucose was S. thermophilus (see Table S8a in the supplemental material). Consistently, streptococci dominate the small intestinal microbiota through the efficient and rapid uptake of simple carbohydrates (39), and carbohydrate metabolism is essential for the colonization of the intestine by S. thermophilus (54). The losers included L. lactis and K. pneumoniae. L. lactis cannot establish itself permanently in the conventional microbiota (55), and K. pneumoniae has not been detected in samples from the human small intestine (39). Their limited ability to compete over host-derived carbohydrates may partly explain their poor adaptation to the gut ecosystem. It is not clear whether the enterocyte would readily provide microbes with glucose and glycerol in vivo. Glycerol can be transported into the lumen via facilitated transport, and the glucose uniport between the lumen and the cytosol is reversible in the small intestine cell model (25), indicating that export of these metabolites is possible in principle. Furthermore, it can be expected that enterocyte-derived nutrients regularly become available through cell lysis and that small intestine microbes have adapted to utilize these host-derived carbohydrates.
In summary, the modeling framework presented here allowed the systematic investigation of microbe-microbe interactions. A collection of just 11 microbes was sufficient to predict all possible outcomes of microbe-microbe coculture (Fig. 1). Constraint-based modeling successfully predicted the effects of imposing various nutrient environments and subjecting the microbe pairs to the presence of the host. As more metabolic reconstructions of gut microbes become available, future efforts will make it possible to systematically investigate the metabolic cross talk in more representative synthetic gut microbe communities. We expect that more extensive community modeling will provide valuable insight into cross feeding among the gut microbiota. For instance, oxygen has been proposed to play a role in microbial dysbiosis in inflammatory bowel diseases, causing a shift from oxygen-sensitive anaerobes, such as F. prausnitzii, to facultative anaerobes, such as E. coli (56). Such changes in gut communities caused by various environmental constraints could be modeled in silico. The present study also provides a framework for studying the effects of the host on microbe-microbe interactions. An in vitro model capturing the microbiota and an enterocyte layer has recently been established (57). Such in vitro data could be put into context using our in silico modeling approach. Similarly, microbe-microbe interactions in gnotobiotic animal models could be predicted in silico and subsequently experimentally validated. For example, germfree mice were colonized with random consortia of cultured bacteria (58). The outcomes of such costly and time-intensive experiments could be predicted in advance in silico. Moreover, using a similar setup, other environments, such as ocean, soil communities, or entirely synthetic communities, could be modeled to learn about the dynamics and the basic biology behind microbe-microbe interactions. For instance, germfree Drosophila melanogaster flies were colonized with pairwise combinations of five main gut bacteria (59). The experiment revealed that certain combinations of two microbes were sufficient for the development of a conventional host phenotype (59). The mechanisms behind such microbial interspecies interactions and their effects on the host could be predicted in silico. The in silico framework presented here can readily incorporate any reconstructed host or microbe, allowing it to be adapted to any microbial ecosystem of interest.
Supplementary Material
ACKNOWLEDGMENT
This work has been funded by an ATTRACT program grant to I.T. (FNR/A12/01) from the Luxembourg National Research Fund (FNR).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00101-15.
REFERENCES
- 1.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Li S, Jian M, Zhou Y, Li Y, Zhang X, Li S, Qin N, Yang H, Wang J, Brunak S, Dore J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, et al. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S. 2012. Host-gut microbiota metabolic interactions. Science 336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 3.Scott KP, Gratz SW, Sheridan PO, Flint HJ, Duncan SH. 2013. The influence of diet on the gut microbiota. Pharmacol Res 69:52–60. doi: 10.1016/j.phrs.2012.10.020. [DOI] [PubMed] [Google Scholar]
- 4.Espey MG. 2013. Role of oxygen gradients in shaping redox relationships between the human intestine and its microbiota. Free Radic Biol Med 55:130–140. doi: 10.1016/j.freeradbiomed.2012.10.554. [DOI] [PubMed] [Google Scholar]
- 5.Grosskopf T, Soyer OS. 2014. Synthetic microbial communities. Curr Opin Microbiol 18:72–77. doi: 10.1016/j.mib.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flint HJ, Duncan SH, Scott KP, Louis P. 2007. Interactions and competition within the microbial community of the human colon: links between diet and health. Environ Microbiol 9:1101–1111. doi: 10.1111/j.1462-2920.2007.01281.x. [DOI] [PubMed] [Google Scholar]
- 7.Belenguer A, Duncan SH, Calder AG, Holtrop G, Louis P, Lobley GE, Flint HJ. 2006. Two routes of metabolic cross-feeding between Bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl Environ Microbiol 72:3593–3599. doi: 10.1128/AEM.72.5.3593-3599.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chassard C, Bernalier-Donadille A. 2006. H2 and acetate transfers during xylan fermentation between a butyrate-producing xylanolytic species and hydrogenotrophic microorganisms from the human gut. FEMS Microbiol Lett 254:116–122. doi: 10.1111/j.1574-6968.2005.00016.x. [DOI] [PubMed] [Google Scholar]
- 9.Mahowald MA, Rey FE, Seedorf H, Turnbaugh PJ, Fulton RS, Wollam A, Shah N, Wang CY, Magrini V, Wilson RK, Cantarel BL, Coutinho PM, Henrissat B, Crock LW, Russell A, Verberkmoes NC, Hettich RL, Gordon JI. 2009. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc Natl Acad Sci U S A 106:5859–5864. doi: 10.1073/pnas.0901529106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Samuel BS, Gordon JI. 2006. A humanized gnotobiotic mouse model of host-archaeal-bacterial mutualism. Proc Natl Acad Sci U S A 103:10011–10016. doi: 10.1073/pnas.0602187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rey FE, Faith JJ, Bain J, Muehlbauer MJ, Stevens RD, Newgard CB, Gordon JI. 2010. Dissecting the in vivo metabolic potential of two human gut acetogens. J Biol Chem 285:22082–22090. doi: 10.1074/jbc.M110.117713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fritz JV, Desai MS, Shah P, Schneider JG, Wilmes P. 2013. From meta-omics to causality: experimental models for human microbiome research. Microbiome 1:14. doi: 10.1186/2049-2618-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borenstein E, Kupiec M, Feldman MW, Ruppin E. 2008. Large-scale reconstruction and phylogenetic analysis of metabolic environments. Proc Natl Acad Sci U S A 105:14482–14487. doi: 10.1073/pnas.0806162105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levy R, Borenstein E. 2013. Metabolic modeling of species interaction in the human microbiome elucidates community-level assembly rules. Proc Natl Acad Sci U S A 110:12804–12809. doi: 10.1073/pnas.1300926110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zengler K, Palsson BO. 2012. A road map for the development of community systems (CoSy) biology. Nat Rev Microbiol 10:366–372. doi: 10.1038/nrmicro2763. [DOI] [PubMed] [Google Scholar]
- 16.Thiele I, Heinken A, Fleming RM. 2013. A systems biology approach to studying the role of microbes in human health. Curr Opin Biotechnol 24:4–12. doi: 10.1016/j.copbio.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 17.Klitgord N, Segre D. 2010. Environments that induce synthetic microbial ecosystems. PLoS Comput Biol 6:e1001002. doi: 10.1371/journal.pcbi.1001002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wintermute EH, Silver PA. 2010. Emergent cooperation in microbial metabolism. Mol Syst Biol 6:407. doi: 10.1038/msb.2010.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freilich S, Zarecki R, Eilam O, Segal ES, Henry CS, Kupiec M, Gophna U, Sharan R, Ruppin E. 2011. Competitive and cooperative metabolic interactions in bacterial communities. Nat Commun 2:589. doi: 10.1038/ncomms1597. [DOI] [PubMed] [Google Scholar]
- 20.Zomorrodi AR, Maranas CD. 2012. OptCom: a multi-level optimization framework for the metabolic modeling and analysis of microbial communities. PLoS Comput Biol 8:e1002363. doi: 10.1371/journal.pcbi.1002363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhuang K, Izallalen M, Mouser P, Richter H, Risso C, Mahadevan R, Lovley DR. 2011. Genome-scale dynamic modeling of the competition between Rhodoferax and Geobacter in anoxic subsurface environments. ISME J 5:305–316. doi: 10.1038/ismej.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhuang K, Ma E, Lovley DR, Mahadevan R. 2012. The design of long-term effective uranium bioremediation strategy using a community metabolic model. Biotechnol Bioeng 109:2475–2483. doi: 10.1002/bit.24528. [DOI] [PubMed] [Google Scholar]
- 23.Heinken A, Sahoo S, Fleming RM, Thiele I. 2013. Systems-level characterization of a host-microbe metabolic symbiosis in the mammalian gut. Gut Microbes 4:28–40. doi: 10.4161/gmic.22370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heinken A, Thiele I. 2015. Systematic prediction of health-relevant human-microbial co-metabolism through a computational framework. Gut Microbes 6:120–130. doi: 10.1080/19490976.2015.1023494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sahoo S, Thiele I. 2013. Predicting the impact of diet and enzymopathies on human small intestinal epithelial cells. Hum Mol Genet 22:2705–2722. doi: 10.1093/hmg/ddt119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teusink B, Wiersma A, Molenaar D, Francke C, de Vos WM, Siezen RJ, Smid EJ. 2006. Analysis of growth of Lactobacillus plantarum WCFS1 on a complex medium using a genome-scale metabolic model. J Biol Chem 281:40041–40048. doi: 10.1074/jbc.M606263200. [DOI] [PubMed] [Google Scholar]
- 27.Thiele I, Vo TD, Price ND, Palsson BO. 2005. Expanded metabolic reconstruction of Helicobacter pylori (iIT341 GSM/GPR): an in silico genome-scale characterization of single- and double-deletion mutants. J Bacteriol 187:5818–5830. doi: 10.1128/JB.187.16.5818-5830.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thiele I, Hyduke DR, Steeb B, Fankam G, Allen DK, Bazzani S, Charusanti P, Chen FC, Fleming RM, Hsiung CA, De Keersmaecker SC, Liao YC, Marchal K, Mo ML, Ozdemir E, Raghunathan A, Reed JL, Shin SI, Sigurbjornsdottir S, Steinmann J, Sudarsan S, Swainston N, Thijs IM, Zengler K, Palsson BO, Adkins JN, Bumann D. 2011. A community effort towards a knowledge-base and mathematical model of the human pathogen Salmonella Typhimurium LT2. BMC Syst Biol 5:8. doi: 10.1186/1752-0509-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pastink MI, Teusink B, Hols P, Visser S, de Vos WM, Hugenholtz J. 2009. Genome-scale model of Streptococcus thermophilus LMG18311 for metabolic comparison of lactic acid bacteria. Appl Environ Microbiol 75:3627–3633. doi: 10.1128/AEM.00138-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baumler DJ, Peplinski RG, Reed JL, Glasner JD, Perna NT. 2011. The evolution of metabolic networks of E. coli. BMC Syst Biol 5:182. doi: 10.1186/1752-0509-5-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liao YC, Huang TW, Chen FC, Charusanti P, Hong JS, Chang HY, Tsai SF, Palsson BO, Hsiung CA. 2011. An experimentally validated genome-scale metabolic reconstruction of Klebsiella pneumoniae MGH 78578, iYL1228. J Bacteriol 193:1710–1717. doi: 10.1128/JB.01218-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heinken A, Khan MT, Paglia G, Rodionov DA, Harmsen HJ, Thiele I. 2014. A functional metabolic map of Faecalibacterium prausnitzii, a beneficial human gut microbe. J Bacteriol 196:3289–3302. doi: 10.1128/JB.01780-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Flahaut NA, Wiersma A, van de Bunt B, Martens DE, Schaap PJ, Sijtsma L, Dos Santos VA, de Vos WM. 2013. Genome-scale metabolic model for Lactococcus lactis MG1363 and its application to the analysis of flavor formation. Appl Microbiol Biotechnol 97:8729–8739. doi: 10.1007/s00253-013-5140-2. [DOI] [PubMed] [Google Scholar]
- 34.Schellenberger J, Que R, Fleming RM, Thiele I, Orth JD, Feist AM, Zielinski DC, Bordbar A, Lewis NE, Rahmanian S, Kang J, Hyduke DR, Palsson BO. 2011. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA toolbox v2.0. Nat Protoc 6:1290–1307. doi: 10.1038/nprot.2011.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Orth JD, Thiele I, Palsson BO. 2010. What is flux balance analysis? Nat Biotechnol 28:245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gudmundsson S, Thiele I. 2010. Computationally efficient flux variability analysis. BMC Bioinformatics 11:489. doi: 10.1186/1471-2105-11-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thiele I, Fleming RM, Bordbar A, Schellenberger J, Palsson BO. 2010. Functional characterization of alternate optimal solutions of Escherichia coli's transcriptional and translational machinery. Biophys J 98:2072–2081. doi: 10.1016/j.bpj.2010.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mazumdar V, Amar S, Segre D. 2013. Metabolic proximity in the order of colonization of a microbial community. PLoS One 8:e77617. doi: 10.1371/journal.pone.0077617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zoetendal EG, Raes J, van den Bogert B, Arumugam M, Booijink CC, Troost FJ, Bork P, Wels M, de Vos WM, Kleerebezem M. 2012. The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbohydrates. ISME J 6:1415–1426. doi: 10.1038/ismej.2011.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Macfarlane GT, Macfarlane S. 2012. Bacteria, colonic fermentation, and gastrointestinal health. J AOAC Int 95:50–60. doi: 10.5740/jaoacint.SGE_Macfarlane. [DOI] [PubMed] [Google Scholar]
- 41.Spees AM, Lopez CA, Kingsbury DD, Winter SE, Baumler AJ. 2013. Colonization resistance: battle of the bugs or ménage à trois with the host? PLoS Pathog 9:e1003730. doi: 10.1371/journal.ppat.1003730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marteyn B, Scorza FB, Sansonetti PJ, Tang C. 2011. Breathing life into pathogens: the influence of oxygen on bacterial virulence and host responses in the gastrointestinal tract. Cell Microbiol 13:171–176. doi: 10.1111/j.1462-5822.2010.01549.x. [DOI] [PubMed] [Google Scholar]
- 43.Martens EC, Lowe EC, Chiang H, Pudlo NA, Wu M, McNulty NP, Abbott DW, Henrissat B, Gilbert HJ, Bolam DN, Gordon JI. 2011. Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol 9:e1001221. doi: 10.1371/journal.pbio.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duncan SH, Barcenilla A, Stewart CS, Pryde SE, Flint HJ. 2002. Acetate utilization and butyryl coenzyme A (CoA):acetate-CoA transferase in butyrate-producing bacteria from the human large intestine. Appl Environ Microbiol 68:5186–5190. doi: 10.1128/AEM.68.10.5186-5190.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Elamin EE, Masclee AA, Dekker J, Jonkers DM. 2013. Ethanol metabolism and its effects on the intestinal epithelial barrier. Nutr Rev 71:483–499. doi: 10.1111/nure.12027. [DOI] [PubMed] [Google Scholar]
- 46.Hooper LV, Midtvedt T, Gordon JI. 2002. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr 22:283–307. doi: 10.1146/annurev.nutr.22.011602.092259. [DOI] [PubMed] [Google Scholar]
- 47.Timmerman HM, Koning CJ, Mulder L, Rombouts FM, Beynen AC. 2004. Monostrain, multistrain and multispecies probiotics—a comparison of functionality and efficacy. Int J Food Microbiol 96:219–233. doi: 10.1016/j.ijfoodmicro.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 48.Schuetz R, Zamboni N, Zampieri M, Heinemann M, Sauer U. 2012. Multidimensional optimality of microbial metabolism. Science 336:601–604. doi: 10.1126/science.1216882. [DOI] [PubMed] [Google Scholar]
- 49.Manor O, Levy R, Borenstein E. 2014. Mapping the inner workings of the microbiome: genomic- and metagenomic-based study of metabolism and metabolic interactions in the human microbiome. Cell Metab 20:742–752. doi: 10.1016/j.cmet.2014.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tachon S, Lee B, Marco ML. 2014. Diet alters probiotic Lactobacillus persistence and function in the intestine. Environ Microbiol 16:2915–2926. doi: 10.1111/1462-2920.12297. [DOI] [PubMed] [Google Scholar]
- 51.Monk JM, Charusanti P, Aziz RK, Lerman JA, Premyodhin N, Orth JD, Feist AM, Palsson BO. 2013. Genome-scale metabolic reconstructions of multiple Escherichia coli strains highlight strain-specific adaptations to nutritional environments. Proc Natl Acad Sci U S A 110:20338–20343. doi: 10.1073/pnas.1307797110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. 2005. Diversity of the human intestinal microbial flora. Science 308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Joyce SA, Gahan CG. 2014. The gut microbiota and the metabolic health of the host. Curr Opin Gastroenterol 30:120–127. doi: 10.1097/MOG.0000000000000039. [DOI] [PubMed] [Google Scholar]
- 54.Thomas M, Wrzosek L, Ben-Yahia L, Noordine ML, Gitton C, Chevret D, Langella P, Mayeur C, Cherbuy C, Rul F. 2011. Carbohydrate metabolism is essential for the colonization of Streptococcus thermophilus in the digestive tract of gnotobiotic rats. PLoS One 6:e28789. doi: 10.1371/journal.pone.0028789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roy K, Meyrand M, Corthier G, Monnet V, Mistou MY. 2008. Proteomic investigation of the adaptation of Lactococcus lactis to the mouse digestive tract. Proteomics 8:1661–1676. doi: 10.1002/pmic.200700698. [DOI] [PubMed] [Google Scholar]
- 56.Rigottier-Gois L. 2013. Dysbiosis in inflammatory bowel diseases: the oxygen hypothesis. ISME J 7:1256–1261. doi: 10.1038/ismej.2013.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marzorati M, Vanhoecke B, De Ryck T, Sadaghian Sadabad M, Pinheiro I, Possemiers S, Van den Abbeele P, Derycke L, Bracke M, Pieters J, Hennebel T, Harmsen HJ, Verstraete W, Van de Wiele T. 2014. The HMI module: a new tool to study the host-microbiota interaction in the human gastrointestinal tract in vitro. BMC Microbiol 14:133. doi: 10.1186/1471-2180-14-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Faith JJ, Ahern PP, Ridaura VK, Cheng J, Gordon JI. 2014. Identifying gut microbe-host phenotype relationships using combinatorial communities in gnotobiotic mice. Sci Transl Med 6:220ra211. doi: 10.1126/scitranslmed.3008051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Newell PD, Douglas AE. 2014. Interspecies interactions determine the impact of the gut microbiota on nutrient allocation in Drosophila melanogaster. Appl Environ Microbiol 80:788–796. doi: 10.1128/AEM.02742-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.He G, Shankar RA, Chzhan M, Samouilov A, Kuppusamy P, Zweier JL. 1999. Noninvasive measurement of anatomic structure and intraluminal oxygenation in the gastrointestinal tract of living mice with spatial and spectral EPR imaging. Proc Natl Acad Sci U S A 96:4586–4591. doi: 10.1073/pnas.96.8.4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.