Abstract
Probiotic microorganisms are ingested as food or supplements and impart positive health benefits to consumers. Previous studies have indicated that probiotics transiently reside in the gastrointestinal tract and, in addition to modulating commensal species diversity, increase the expression of genes for carbohydrate metabolism in resident commensal bacterial species. In this study, it is demonstrated that the human gut commensal species Bacteroides thetaiotaomicron efficiently metabolizes fructan exopolysaccharide (EPS) synthesized by probiotic Lactobacillus reuteri strain 121 while only partially degrading reuteran and isomalto/malto-polysaccharide (IMMP) α-glucan EPS polymers. B. thetaiotaomicron metabolized these EPS molecules via the activation of enzymes and transport systems encoded by dedicated polysaccharide utilization loci specific for β-fructans and α-glucans. Reduced metabolism of reuteran and IMMP α-glucan EPS molecules may be due to reduced substrate binding by components of the starch utilization system (sus). This study reveals that microbial EPS substrates activate genes for carbohydrate metabolism in B. thetaiotaomicron and suggests that microbially derived carbohydrates provide a carbohydrate-rich reservoir for B. thetaiotaomicron nutrient acquisition in the gastrointestinal tract.
INTRODUCTION
Trillions of bacteria inhabit the gut, imparting symbiotic effects that benefit the overall health and well-being of individuals (1). While the human genome has approximately 30,000 genes, the human microbiome contributes an additional 3 to 9 million gene products (termed the gut metagenome) that contribute to functionalities in human lifestyle (2). One prominent example is the human utilization of carbohydrates. Carbohydrates provide a substantial proportion of the daily energy intake of an individual. However, humans have the ability to metabolize only very few dietary carbohydrate compounds, such as lactose, starch, and sucrose, by enzymes either present in saliva (amylases) or anchored to the epithelial wall of the small intestine with glycosylphosphatidylinositol (GPI) anchors (such as invertases and lactases). All other carbohydrates that traverse the gastrointestinal (GI) tract have the potential to be metabolized by commensal gut bacteria, where it is estimated that gut microbiota provide an additional 30,000 enzymes which facilitate the breakdown of carbohydrates in the gut (3).
Approximately 30% of resident human gut microbiota are from the Bacteriodetes phylum. These Gram-negative, obligate anaerobes are well armed with a repertoire of carbohydrate-degrading enzymes for harvesting carbohydrate nutritional resources. One species, Bacteroides thetaiotaomicron, devotes approximately 18% of its genome to carbohydrate foraging (4). Discrete polysaccharide utilization loci (PULs) within its genome are upregulated in response to carbohydrates dependent on the source. Most carbohydrate sources for B. thetaiotaomicron nutrient acquisition originate from dietary sources (such as nondigestible plant polysaccharides) or from the mucosal layer lining the gastrointestinal tract (mucins and mucopolysaccharides) (5). The ability of B. thetaiotaomicron to efficiently utilize a wide range of carbohydrates makes this organism well suited to inhabit the dynamic environment of the gastrointestinal tract.
Probiotic bacteria can be ingested without harm to an individual and provide a positive health benefit to the consumer (6). These strains most commonly are consumed by ingesting fermented milk products (FMPs) or are found at therapeutic levels by ingesting dried bacterial strains in capsular form. Ingesting probiotic bacteria and FMPs is correlated with creating balanced gut homeostasis by altering gut microbial composition, altering bacterial metabolic activity, and producing metabolites that directly affect host intestinal epithelium (7). Therefore, consuming probiotics is one strategy used to treat microbiome-associated diseases (8, 9).
Lactic acid bacteria (LAB) are a diverse group of probiotic microorganisms that reside in the upper GI tract and have immunomodulatory properties (10). Additionally, they are among the most abundantly consumed bacteria, as they are commonly used in food production for meats, beverages, pickling agents, and FMPs, producing flavor and textural features while reducing food spoilage. LAB produce lactic acid from fermentation of carbohydrates, which leads to acidification of the surrounding environment, and synthesize extensive exopolysaccharides (EPS) that promote their adhesion and growth in the gastrointestinal tract (11) and offer protection from pathogenic bacteria (12). The most widely known example is EPS from Streptococcus mutans that synthesizes a sticky biofilm called mutan, which aids in its colonization of the oral cavity (13). S. mutans and the majority of other LAB genera, such as Leuconostoc and Lactobacillus, synthesize β-fructan and α-glucan EPS polymers, which protect the bacteria from immune detection and aid in attachment of bacteria to the mucosal lining of the intestine (14, 15). These homopolysaccharide EPS molecules are derived from dietary sucrose and are synthesized mainly in the oral cavity and small intestine, where there is an abundance of dietary sucrose present. LAB employ cell surface-associated transglycosidases using sucrose as the donor substrate to synthesize these extensive β-fructan and α-glucan EPS polymers (16, 17).
Several studies have investigated the effects of probiotics on altering gut microbial composition and how this may contribute to the health of an individual (7, 18–21). One area of particular interest is the observation that ingesting probiotic bacteria alters the gene expression profile and metabolic output of commensal bacteria, and the changes in metabolic output correlate with health benefits in an individual (20). Research by McNulty et al. showed that monozygotic twins and gnotobiotic mice administered common probiotic strains from FMPs exhibited little change in bacterial composition over time; however, gene expression profiles in commensal bacterial strains were significantly altered (20). Most significant increases were observed for genes involved in carbohydrate metabolism and transport, specifically starch and sucrose metabolism pathways, including levanases (EC 3.2.1.65), pectinesterases (EC 3.1.1.11), and cellobiose phosphorylase (EC 2.4.1.20). The researchers could not establish what exactly caused the effect of increased carbohydrate metabolism, but they determined that the effect was rapid and consistent during probiotic supplementation.
As probiotic LAB strains transit the GI tract, commensal microbiota will be exposed to LAB-synthesized carbohydrate-based EPS molecules which would be available for fermentation. If EPS is a fermentable substrate, its presence would cause an upregulation of genes required for metabolism of these carbohydrate compounds. Therefore, the hypothesis in this study posits that human gut commensal bacteria have the ability to degrade LAB EPS by activating and producing genome-encoded carbohydrate-active enzymes (CAZymes) and transport systems with activity directed toward β-fructan and α-glucan EPS substrates, specifically genes encoding levanases and starch-metabolizing CAZymes, leading to the metabolism of LAB EPS molecules by gut commensal microbes.
To test this hypothesis, different preparations of EPS derived from Lactobacillus reuteri 121 (termed LrEPS and isomalto/malto-polysaccharide [IMMP]) were synthesized and purified. Purified EPS was provided as a sole carbon source to the human saccharolytic bacterial symbiont Bacteroides thetaiotaomicron to see whether bacteria can metabolize LAB EPS products and, furthermore, what molecular mechanisms are involved in EPS breakdown. Results show that B. thetaiotaomicron completely metabolized LAB-derived β-fructan EPS, as would be predicted from previous work on selective levan metabolism by this species (22), and partially degraded α-glucan EPS. Furthermore, EPS degradation was associated with the production of enzymes and transport proteins encoded by polysaccharide utilization loci (PULs) in the B. thetaiotaomicron genome, namely, PULs for fructan and starch catabolism (22–24). Although L. reuteri α-glucan EPS triggered the production of starch and dextran PUL-associated components, B. thetaiotaomicron was unable to completely degrade α-glucan EPS polymers. To our knowledge, this is the first study to demonstrate that B. thetaiotaomicron activates PULs in response to LAB-derived EPS molecules. These results provide a plausible explanation for the observed upregulation of carbohydrate metabolism genes in commensal gut bacterial species in the presence of probiotic bacteria. Furthermore, these findings suggest a mechanism for EPS metabolism by members of gut microbial species that may transiently or persistently colonize the gut.
MATERIALS AND METHODS
Bacterial strains, media, and reagents.
Bacteroides thetaiotaomicron VPI-5482 was purchased from the ATCC. A B. thetaiotaomicron starch deletion mutant (BtΔ3702) and β-fructan deletion mutant (BtΔ1760) were provided by E. C. Martens. Lactobacillus reuteri strain 121 and L. reuteri strain 35-5 were from TNO Quality of Life, Zeist, The Netherlands. All medium components were purchased from Sigma (Zwijndrecht, Netherlands) or as mentioned.
Purification of EPS components.
L. reuteri EPS was purified as described in reference 25 using the following procedure. L. reuteri 121 and the 35-5 mutant were grown on MRS agar containing either 10% sucrose (to obtain levan plus reuteran EPS) or 10% maltodextrin (to obtain IMMP) and incubated anaerobically in anaerobic jars (Oxoid) containing a GasPak (BD) for 48 h. After growth, plates were scraped of colonies and resuspended in deionized water. Tubes were stirred for 2 h at room temperature to release EPS into solution. After 2 h, cells were pelleted and supernatants were collected, to which 3 volumes of ice-cold 100% ethanol were added and stirred overnight at 4°C to allow precipitation of EPS polysaccharides. The precipitated material was pelleted, dried, and dissolved in deionized water. Ten percent trichloroacetic acid (TCA) was added to each preparation to precipitate out any contaminating proteins. After 1 h of incubation with 10% TCA at room temperature, precipitated material was pelleted and supernatants containing EPS were dialyzed into deionized water for 3 days with 3,500-molecular-weight-cutoff (MWCO) tubing (Thermo Scientific). After dialysis, polysaccharides were freeze-dried and stored under desiccating conditions until further use.
NMR analysis.
Nuclear magnetic resonance (NMR) was carried out as described previously (25). Samples were exchanged twice with 200 μl of 99.9 atom% D2O (Cambridge Isotope Laboratories Ltd., United Kingdom) with intermediate lyophilization and finally dissolved in 650 μl of 99.9 atom% D2O containing 25 ppm acetone as an internal standard (δ1H 2.225). One-dimensional 600-MHz 1H NMR spectra were recorded on a Varian Inova 600 spectrometer (NMR Department, University of Groningen, The Netherlands), collecting 32 transients of 16,000 complex data points, using a 5,000-Hz spectral width. The residual HOD (chemical shift of H2O) peak was suppressed using a WET1D suppression pulse. All spectra were processed using MestReNova 5.3 (Mestrelabs Research SL, Santiago de Compostella, Spain), using Whittaker Smoother baseline correction.
B. thetaiotaomicron growth experiments.
B. thetaiotaomicron growth was carried out as described previously (26). Briefly, overnight cultures of B. thetaiotaomicron were grown at 37°C under anaerobic conditions. The following day, 1 ml of B. thetaiotaomicron overnight culture was prepared in a carbon-limited minimally defined medium of 100 mM KH2PO4 (pH 7.2), 15 mM NaCl, 8.5 mM (NH4)2SO4, 4 mM l-cysteine, 1.9 μM hematin, 200 μM l-histidine, 100 nM MgCl2, 1.4 nM FeSO4 · 7 H2O, 50 μM CaCl2, 1 μg/ml vitamin K3, 5 ng/ml vitamin B12, and individual carbon sources (0.5%, wt/vol). Growth curves were obtained by incubating microtiter plates at 37°C, and the optical density at 600 nm (OD600) was recorded at 30-min intervals.
To obtain culture supernatants for analysis of carbohydrate activity, B. thetaiotaomicron cells were prepared in the same carbon-limited minimal media as that for the microtiter plates. One milliliter of bacterial cell suspension was added to 1 ml of 10 mg/ml carbohydrate solution (final concentration of 5 mg/ml) in a sterilized glass test tube with a metal cap, placed in an anaerobic jar (Oxoid) with a GasPak (BD), and placed in a 37°C incubator for 48 h.
After growth, cultures of B. thetaiotaomicron in carbohydrate-defined media were centrifuged to obtain supernatants which subsequently were filtered through a 0.2-μm-pore-size syringe filter (Millipore). One hundred μl of culture supernatant was added to 100 μl of a 10 mg/ml solution of carbohydrate (35-5 EPS reuteran, wild-type LrEPS, or IMMP) in deionized water and incubated at 37°C overnight to allow the enzyme reactions to proceed.
qPCR of PUL genes.
Isolation of RNA, synthesis of cDNA, and quantitative PCR (qPCR) measurements were carried out as described previously (27). Values reported are the fold difference compared to that of the glucose control RNA and were normalized against 16S gene expression. Primers used for quantification of PUL activation were directed against susC-like genes for each designated PUL and are listed in Table S1 in the supplemental material.
TLC.
Thin-layer chromatography (TLC) of carbohydrate products was completed as described in the literature (28) Briefly, 2 μl of each reaction mixture was spotted, dried, and then subsequently spotted again on a silica gel 60 plate (Millipore). The solvent system used was 3:1:1 isopropanol, ethylacetate, and deionized water, and plates were run in a TLC jar for approximately 4 to 5 h. Plates were removed from the jar and dried, and spots were visualized by staining with 20% sulfuric acid plus 5% orcinol in methanol and heated at 110°C for half an hour. Plates were scanned and figures prepared using Adobe Photoshop. For enzymatic degradation of reuteran, 5 U of each enzyme was added to 100 μl of 1% reuteran solution in phosphate-buffered saline (PBS) and incubated for 1 h at appropriate temperatures. Products were observed by TLC as mentioned above.
HPAEC-PAD.
The oligosaccharides produced by B. thetaiotaomicron culture supernatants were analyzed by high-pH anion-exchange chromatography on a Dionex DX500 work station equipped with an ED40 pulsed amperometric detection system (HPAEC-PAD). The oligosaccharides were separated on a CarboPac PA-1 column (250 by 5 mm; Dionex) by using a linear gradient of 10 to 240 mM sodium acetate in 100 mM NaOH (1 ml/min).
Proteomic analysis of culture supernatants.
The proteins in the supernatants were precipitated using cold trichloroacetic acid and washed 3 times with ice-cold acetone. Proteins were redissolved in 50 mM ammonium bicarbonate (pH 8), chemically reduced by adding 5 μl of 5 mM tris(2-carboxyethyl)phosphine, and alkylated by adding 5 μl of 100 mM iodoacetamide at room temperature for 30 min. To incorporate proteins into a gel directly in the Eppendorf vial, 18.5 μl of acrylamide-bisacrylamide solution (40%, vol/vol, 29:1), 2.5 μl of 10% (wt/vol) ammonium persulfate, and 1 μl of 100% N,N,N′,N′-tetramethylethylenediamine (TEMED) was applied to the protein solution and polymerized in 30 min. The resulting gel was cut into small pieces and washed several times with 1 ml of 50 mM ammonium bicarbonate containing 50% (vol/vol) acetonitrile. The gel samples were further dehydrated with 100% acetonitrile. Proteolytic digestion then was performed with trypsin in 50 mM ammonium bicarbonate with incubation overnight at 37°C. The peptide mixture was acidified by adding 5 μl formic acid and extracted. Samples were analyzed by nano-liquid chromatography-tandem mass spectrometry (nano-LC-MS/MS) on an Ultimate 3000 system (Dionex, Amsterdam, The Netherlands) interfaced on-line with an LTQ-Orbitrap-XL mass spectrometer (Thermo Fisher Scientific, San Jose, CA). Peptide mixtures were loaded onto a trapping microcolumn (inner diameter [i.d.], 5 mm by 300 μm) packed with C18 PepMAP100 5-μm particles (Dionex) in 0.1% formic acid at a flow rate of 20 μl/min. After loading and washing for 5 min, peptides were back-flush eluted onto a nanocolumn (15 cm by 75 μm i.d.) packed with C18 PepMAP100 3-μm particles (Dionex). The following mobile-phase gradient was delivered at a flow rate of 300 nl/min: 2 to 50% solvent B for 60 min, 50 to 90% B for 7 min, 90% B for 10 min, and back to 2% B for 5 min. Solvent A was H2O-acetonitrile (100:0, vol/vol) with 0.1% formic acid, and solvent B was H2O-acetonitrile (0:100, vol/vol) with 0.1% formic acid.
Peptides were infused into the mass spectrometer via a dynamic nanospray probe (Thermo Electron Corp.) with a stainless steel emitter (Proxeon, Odense, Denmark). The typical spray voltage was 1.8 kV with no sheath and auxiliary gas flow; the ion transfer tube temperature was 200°C. The mass spectrometer was operated in data-dependent mode. The automated gain control (AGC) was set to 5 × 105 charges and 1 × 104 charges for MS/MS on the linear ion trap analyzer. The data-dependent acquisition (DDA) cycle consisted of the survey scan within m/z 300 to 1,300 on the Orbitrap analyzer with target mass resolution of 60,000 (full width at half maximum [FWHM] at m/z 400), followed by MS/MS fragmentation of the five most intense precursor ions under the relative collision energy of 35% in the linear trap. Singly charged ions were excluded from MS/MS experiments, and the m/z values of fragmented precursor ions were dynamically excluded for a further 60 s. The ion selection threshold for triggering MS/MS experiments was set to 500 counts.
The software PEAKS Studio (version 7) was applied to the spectra generated by the LTQ Orbitrap XL to search against either the protein sequence database UniProtKB/Trembl of the UniProt Knowledgebase (UniProtKB), limited to protein sequences of Bacteroides thetaiotaomicron, or the Swiss-Prot database. Searching for the fixed-modification carbamidomethylation of cysteine and the variable posttranslational modifications, oxidation of methionine was done with a maximum of 5 posttranslational modifications per peptide at a parent mass error tolerance of 10 ppm. The false discovery rate was set at 0.1%.
SusD binding activity assay.
SusD protein was purified as described previously (26). SusD was buffer exchanged into 100 mM sodium bicarbonate buffer and labeled with fluorescein isothiocyanate (FITC). Labeled SusD was isolated and buffer exchanged into 20 mM Tris, pH 8.0, 150 mM NaCl, 5 mM CaCl2 (BB) buffer solution and concentrated with an Amicon Centricon device with a 10,000-MWCO membrane to a concentration of ∼20 mg/ml. The control protein, Thermotoga maritima CBM41 (TmCBM41), was prepared as previously described (29) and FITC labeled. Membranes for carbohydrate macroarrays were prepared in-house based on previously published procedures (29). Briefly, 1 μl of a 1% (wt/vol) carbohydrate solution was spotted onto a nitrocellulose membrane grid and allowed to dry for 1 h at room temperature. After macroarrays were prepared, membranes were blocked in BB plus 0.5% Tween 20 (BBT) and 1% bovine serum albumin (BSA) for 1 h at room temperature. After blocking, membranes were washed 3× and placed in BB plus 200 μg of corresponding protein at a final concentration of 40 μg/ml and incubated overnight at 4°C with continuous agitation. The following day, membranes were washed 3× with BBT and visualized on a Chemidoc with fluorescein filters (Thermo Scientific).
For testing binding to insoluble starch, 10 mg of insoluble wheat starch was washed 3× with BBT and resuspended in 1 ml of BB. Two hundred micrograms of FITC-labeled SusD or TmCBM41 was added to individual tubes and incubated at room temperature for 1 h with continuous agitation. After 1 h, Eppendorf tubes containing starch and bound protein were centrifuged to pellet the starch. The starch pellet was thoroughly washed 3× with BBT. It was resuspended in a 100-μl final volume of BB, and 5 μl of the suspension was spotted on a nitrocellulose membrane, dried, and visualized.
SusG activity.
Enzymatic reactions were carried out as described previously (28). Reuteran, IMMP, amylopectin, and insoluble amylose were prepared at a concentration of 10 mg/ml in 20 mM Tris, pH 8.0, 150 mM NaCl. Reactions were carried out in 20-μl volumes, where 2 μl (approximately 50 μg) of SusG was added to 18 μl of each carbohydrate solution. Reaction mixtures were incubated at 37°C for 4 h and analyzed by TLC as described above.
RESULTS
B. thetaiotaomicron metabolized L. reuteri β-fructan exopolysaccharides.
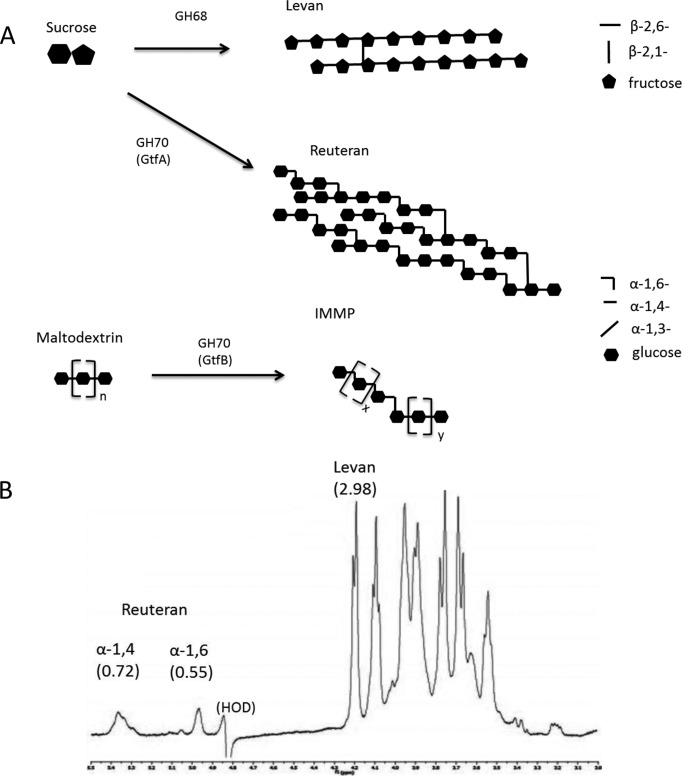
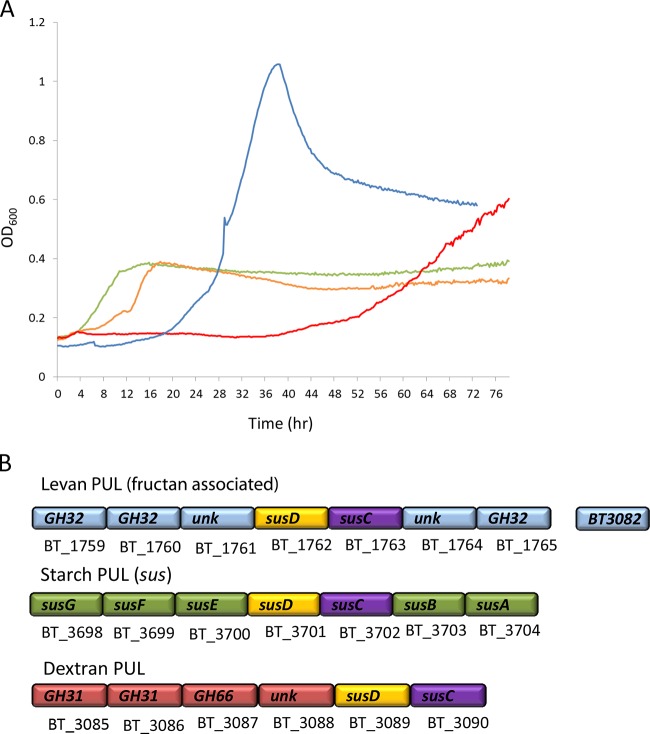
EPS was purified from the probiotic strain Lactobacillus reuteri 121, which synthesizes a thick EPS layer composed of the fructan polymer levan (β-2,6-fructan) and the α-glucan polymer reuteran from a sucrose donor substrate and IMMP (isomalto/malto-polysaccharides) from maltodextrins (Fig. 1A). L. reuteri was chosen as our model LAB strain, as it is an endogenous mammalian gut strain more adapted to human colonization than other LAB species involved in food preparation, and much is known on the structure of EPS produced by this strain (25, 30, 31). Additionally, it is used as a probiotic supplement, such as treatment of Helicobacter pylori infections and infant ailments such as colic (32, 33). We observed that wild-type L. reuteri grown in the presence of sucrose synthesized both levan and reuteran EPS molecules at a ratio of approximately 2.3:1, as calculated by integration of the area under the peak corresponding to the anomeric center for the monosaccharide (Fig. 1B), and produced a pure IMMP α-glucan EPS polymer when grown on maltodextrins (Fig. 1A) (31, 34). Thus, three forms of L. reuteri EPS were used for this study: LrEPS from L. reuteri 121 grown on sucrose, which consists of both levan and reuteran, reuteran only, which was produced by the mutant L. reuteri 121 strain 35-5 lacking the levansucrase enzyme grown on sucrose (25, 35), and IMMP from wild-type L. reuteri 121 grown on maltodextrins (31). B. thetaiotaomicron grown in the presence of 5 mg/ml LrEPS demonstrated only moderate growth on LrEPS (Fig. 2A, green line) compared to a 5 mg/ml glucose standard (Fig. 2A, blue line), suggesting that B. thetaiotaomicron was using only a portion of the EPS substrate. Because B. thetaiotaomicron contains dedicated PULs in its genome for the metabolism of β-fructans (β-2,6-linked fructose) (22), starch (sus) (α-1,4-linked glucose) (23), and dextran (α1,6-linked glucose) (Fig. 2B), we suspected that these PULs contribute to the observed bacterial growth. To determine the contribution of the fructan-associated and sus PULs to the degradation of LrEPS, B. thetaiotaomicron fructan-associated PUL deletion mutant BtΔ1763 and sus deletion mutant BtΔ3702 were grown in the presence of LrEPS. BtΔ3702, which lacks the ability to degrade starch, showed a slightly delayed growth profile but reached wild-type growth levels after 16 h (Fig. 2A, yellow line). BtΔ1760, which lacks the ability to grow on levan, exhibited no growth on LrEPS after 16 h, while after 60 h it begins to show a slow increase in cell density (Fig. 2A, red line), implying that levan is the preferred EPS substrate contributing to bacterial growth. To further verify these results, quantification of sus, dextran-associated, and fructan-associated PUL activation was determined by qPCR to analyze expression levels of mRNA extracted from wild-type B. thetaiotaomicron grown in the presence of LrEPS. The level of the fructan-associated PUL was increased 19.2-fold ± 3.93-fold compared to the level for glucose, whereas there was a slight decrease in expression levels of sus and dextran-associated PUL (Table 1). Together, these results demonstrate that B. thetaiotaomicron activates the β-fructan PUL (BT1759-BT1765) for metabolism of LrEPS when a combination of levan and reuteran is present.
FIG 1.
(A) Graphical representation of β-fructan levan and α-glucans reuteran and IMMP exopolysaccharides produced by Lactobacillus reuteri 121 via transglycosylation reactions using glycoside hydrolase family 68 (GH68) or GH70 enzymes (www.cazy.org [39]). (B) NMR analysis of LrEPS products indicating the abundance of monosaccharides present as ratios of each peak representing the anomeric carbon present.
FIG 2.
(A) Growth of B. thetaiotaomicron VPI-5482 and mutants derived on Lactobacillus reuteri EPS. Wild type, green; BtΔ3702 (sus mutant), yellow; BtΔ1763 (fructan-associated PUL mutant), red; and glucose control, blue. B. thetaiotaomicron and deletion mutants were grown under anaerobic conditions at 37°C in minimal growth media supplemented with wild-type LrEPS (levan plus reuteran) or glucose (control) at 5 mg/ml final concentration. Only one curve was carried out for each sample due to the limited availability of substrate, and results were verified by qPCR. (B) Graphical representation of B. thetaiotaomicron PULs mentioned in this study.
TABLE 1.
Major proteins identified from proteomic analysis of material from culture supernatants of B. thetaiotaomicrona
| Protein category | Proteins identified withb: |
||
|---|---|---|---|
| LrEPS | Reuteran | IMMP | |
| PUL-associated carbohydrate-active enzymes and binding proteins | Endo-2,6-β-fructanase GH32 BT1760 (fructan associated), 2,6-β-fructofuranosidase GH32 BT1759 (fructan associated), 2,6-β-fructofuranosidase GH32 BT3082 (fructan associated), outer membrane protein SusE BT3700 (sus), outer membrane protein SusF BT3699 (sus), and α-glucosidase GH97 BT2620 | SusG amylase GH13 BT3698 (sus), SusB α-glucosidase GH97 BT3703 (sus), outer membrane protein SusF BT3699 (sus), and outer membrane protein SusE BT3700 (sus) | Endodextranase BT3087 (dextran associated), α-glucosidase BT3086 (dextran associated), SusE-F-like binding protein BT3088 (dextran associated), SusB α-glucosidase GH97 BT3703 (sus), SusA pullulanase BT3704 (sus), and α-glucosidase GH97 BT2620 |
| Non-PUL-associated carbohydrate-active enzymes | α-Amylase GH57 BT4305 and 4,6-glucanotransferase BT4303 | α-Glucosidase GH97 BT4581, α-amylase GH57 BT4305, and 4,6-glucanotransferase BT4303 | |
Proteins were identified from carbon-limited experiments with 5 mg/ml LrEPS, reuteran, or IMMP. PUL systems were identified based on the presence of both members of SusC-D pairs (values in parentheses are from qPCR results): the BT1762/BT1763 (fructan-associated) pair was present on LrEPS (19.2 ± 3.93) but not on reuteran or IMMP, the BT3089/BT3090 (dextran-associated) pair was present on IMMP but not on reuteran or LrEPS (0.54 ± 0.12), and the BT3701/BT3702 (sus) pair was present on LrEPS (0.49 ± 0.10), reuteran, and IMPP.
Proteins and enzymes are identified and compared to the glucose controls (a full list can be found in the supplemental material) and correlated with their associated PULs, which are identified in parentheses (4).
B. thetaiotaomicron partially metabolized reuteran and IMMP EPS.
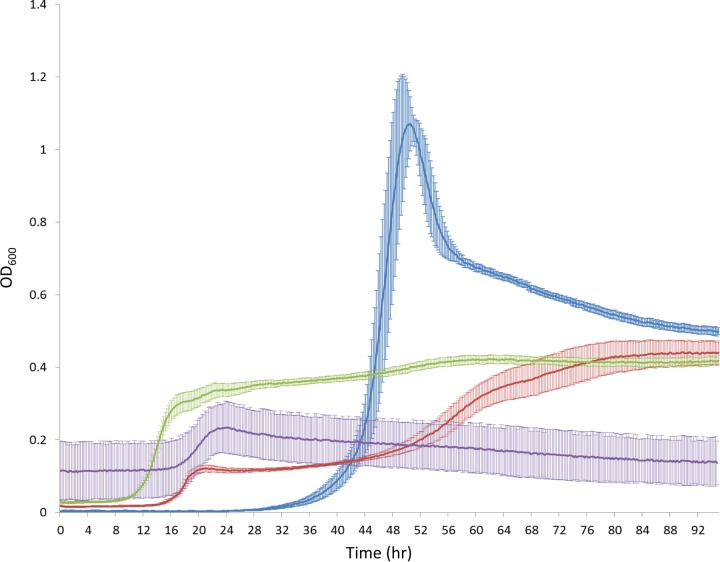
Since B. thetaiotaomicron is well equipped with PULs for the degradation of starch and dextran, it was surprising to see that reuteran from the LrEPS mixture was not being metabolized. Therefore, independent B. thetaiotaomicron culture experiments were performed to test for bacterial growth on the α-glucan EPS substrates reuteran and IMMP separately. Compared to B. thetaiotaomicron cultures of LrEPS (2.3:1 levan-reuteran) (Fig. 3, green line), reuteran substrate showed a biphasic growth profile with a slight increase in growth after 16 h and an increase up to an OD600 of 0.4 after 52 h (Fig. 3, red line), while IMMP as a substrate showed very little increase in growth after 72 h (change in OD600 of 0.12 ± 0.09) (Fig. 3, purple line). Although B. thetaiotaomicron can effectively degrade starch, a polymer of amylose (α-1,4-glucan) and amylopectin (α-1,4-α-1,6-mixed glucan), it was difficult for B. thetaiotaomicron to metabolize α-glucan EPS substrates containing the same types of glucan linkages found in starch.
FIG 3.
Growth of B. thetaiotaomicron VPI-5482 on individual α-glucan-based EPS components reuteran (red) and IMMP (purple) compared to LrEPS (green) and glucose (blue) controls. The lines indicate averages from six growth experiments, and error bars indicate the standard deviations for these experiments.
The identification and activity of B. thetaiotaomicron CAZymes induced by LrEPS, reuteran, and IMMP.
Previous observations from the seminal work of the Anderson and Salyers laboratory on starch degradation by B. thetaiotaomicron showed that PUL-activated CAZymes can be found in culture supernatants (23). Therefore, to further characterize the activity of CAZymes produced by B. thetaiotaomicron in the presence of LrEPS, fresh B. thetaiotaomicron culture supernatants from whole LrEPS, reuteran, and IMMP growth experiments were incubated with EPS molecules, and the products of EPS degradation by CAZymes present in culture supernatants were analyzed by thin-layer chromatography and HPAEC-PAD. Enzymes present in culture supernatants from wild-type LrEPS cultures completely degraded levan into its monosaccharide component, fructose, and small amounts of fructose disaccharide (Fig. 4 red line; also see the inset, lane 1) but showed no activity on reuteran (Fig. 4, inset, lane 2). Culture supernatants then were analyzed for the presence of PUL-encoded proteins using proteomics (see Materials and Methods). Results from proteomic analysis of LrEPS culture supernatants showed that almost all extracellular enzymatic components of the fructan-associated PUL were present (Table 1 and Fig. 2B) (see File S1 in the supplemental material for a full listing of proteins identified in culture supernatants), which accounts for the observed degradation of levan in LrEPS.
FIG 4.
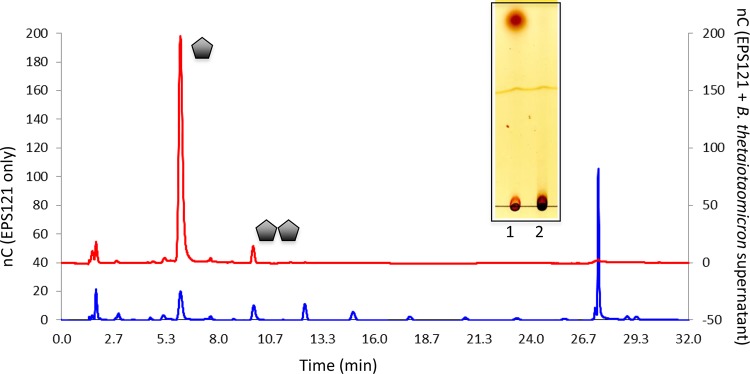
Thin-layer chromatography analysis and HPAEC-PAD profile of L. reuteri 121 β-fructan (levan) EPS degradation. The graph shows the levan (blue line) control and products from levan hydrolysis by B. thetaiotaomicron culture supernatants after growth on levan (red line). The inset is the corresponding TLC profile from enzymatic activity of CAZymes present in B. thetaiotaomicron culture supernatants from growth on wild-type LrEPS activity on LrEPS (lane 1) and reuteran (lane 2). The y axes show electric charges (in nanocoulombs [nC]).
Culture supernatants from B. thetaiotaomicron growth on reuteran and IMMP also were tested for enzymatic activity on pure reuteran or IMMP substrates. The activity of reuteran culture supernatants shows that small amounts of glucose and maltose were produced (Fig. 5, lane 1), which is not observed in the glucose control supernatant (Fig. 5, lane 3), while IMMP culture supernatant activity on IMMP showed the same general product profile (Fig. 5, lane 2). Proteomic analysis of reuteran culture supernatants identified several components from the sus starch utilization PUL (Table 1 and Fig. 2B), including the transporter component SusD and extracellular amylase SusG, suggesting that the partial growth phenotype exhibited by B. thetaiotaomicron on reuteran is due to sus PUL-associated enzymes. Culture supernatants from IMMP growth revealed that enzymes from the sus operon were present, whereas additional enzymes from the dextran-associated PUL BT3085-BT3090 also were present (Table 1 and Fig. 2B; also see File S1 in the supplemental material for a full listing). Additional enzymes found in the IMMP supernatant include endodextranase BT3087 (GH66) and α-glucosidase II BT3086 (GH31) (Table 1). These results suggest that sus and dextran-associated PUL-encoded α-glucan-specific CAZymes are produced by B. thetaiotaomicron in the presence of LAB α-glucan EPS, implying that other factors, such as substrate recognition or enzymatic activity, prevent the full degradation and metabolism of these substrates.
FIG 5.
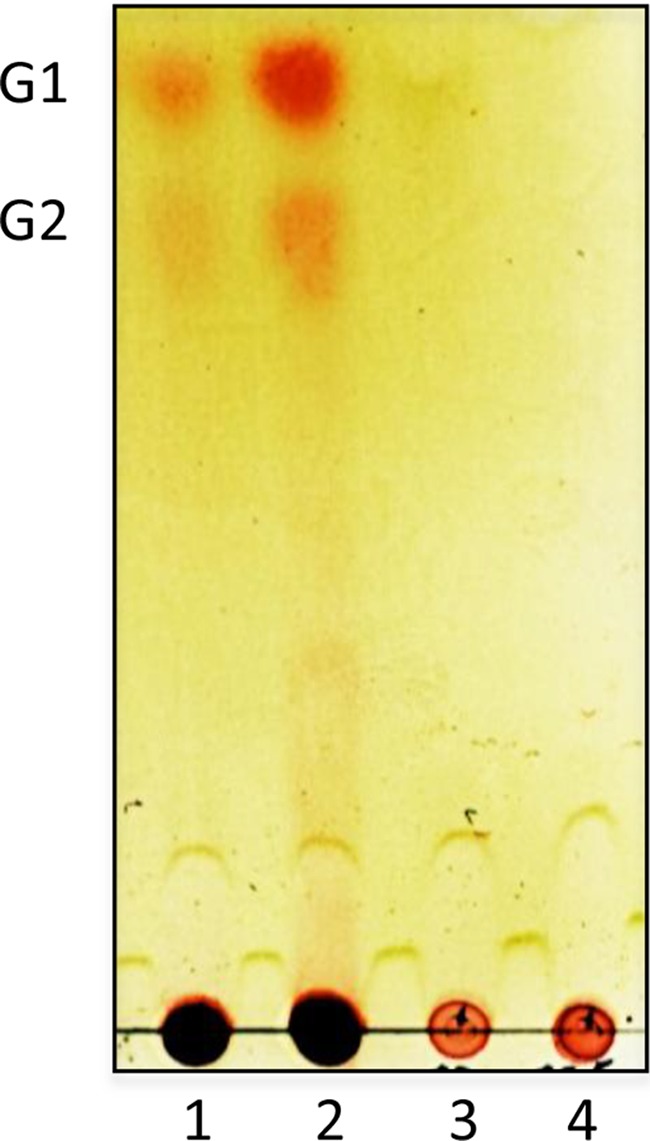
Thin-layer chromatography analysis of the products of degradation on reuteran and IMMP incubated with culture supernatants (CS) of B. thetaiotaomicron grown in the presence of either substrate. Lane 1, reuteran and reuteran CS; lane 2, IMMP and IMMP CS; lane 3, reuteran and glucose CS; lane 4, IMMP and glucose CS. Culture supernatants were incubated with EPS substrate for 1 h before being spotted on a TLC plate.
Role of sus-encoded transport component SusD and surface-associated α-amylase SusG on α-glucan EPS metabolism.
The extracellular α-amylase SusG and transport component SusD were identified in B. thetaiotaomicron culture supernatant grown in reuteran only (Table 1). To further elucidate the effects of sus system components on reuteran digestion by B. thetaiotaomicron, purified SusG and SusD were tested for their activity on reuteran. SusG is the surface-associated α-amylase from the sus system involved in the breakdown of starch into smaller glucooligosaccharides (28). SusG was tested for its degradation properties on reuteran, where characteristic high activity was seen with amylopectin (Fig. 6, lane 4), with only limited activity observed on reuteran, producing relatively small amounts of glucose and maltose (Fig. 6, lanes 2 and 3). The SusG product profile was similar to the profile observed from activity of the B. thetaiotaomicron culture supernatants from reuteran growth experiments containing SusG enzyme (Fig. 5); therefore, it is likely that SusG contributes to partial growth of B. thetaiotaomicron on reuteran. However, this suggests the enzyme itself is not able to sufficiently hydrolyze the polymer. Reuteran resembles starch in its composition and α-1,4- and α-1,6-glucosidic linkages, except that alternating α-1,4 and α-1,6 linkages are much more complex in reuteran than in starch, making it difficult for SusG to access the substrate (Fig. 1). Thus, reduced enzymatic activity of SusG on reuteran would indeed partially contribute to reduced bacterial growth on this substrate.
FIG 6.
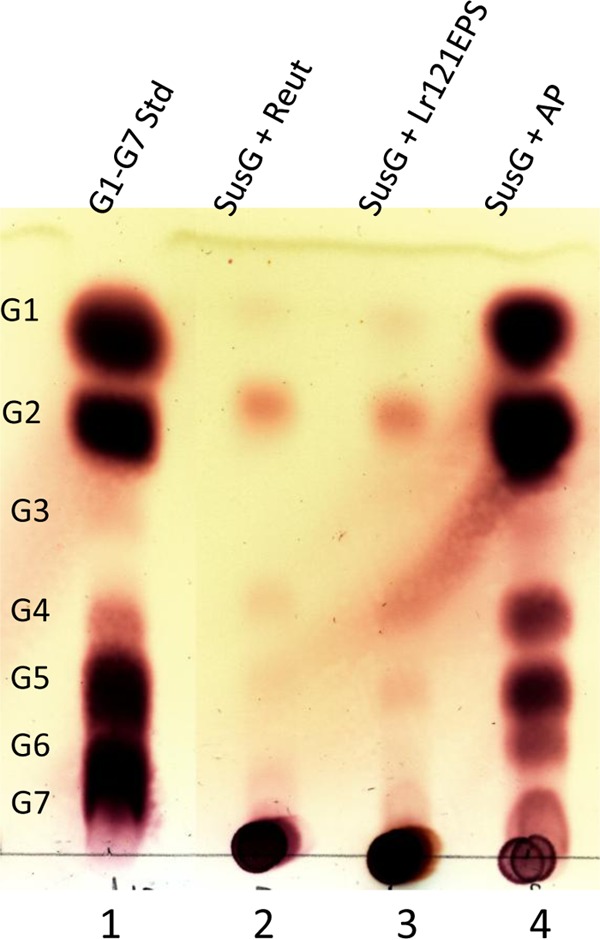
Thin-layer chromatography of reuteran hydrolysis by SusG. All reaction mixtures contain 10 mg/ml polysaccharide and 22 μg SusG in 20 mM HEPES, pH 7.0, 150 mM NaCl, 5 mM CaCl2. Reaction products were sampled after 4 h of incubation at 37°C. Lane 1, G1 to G7 maltooligosaccharide standards (Std); lane 2, reuteran (Reut); lane 3, Lr121EPS; lane 4, amylopectin (AP).
To further investigate which CAZymes are able to degrade reuteran, several commercially available α-glucan-specific CAZymes were tested for their activity on reuteran. Enzymes with specific activity on α-1,6-linkages, such as pullulanases (endo-α-1,6-glucosidases, GH13 subfamily 14) and isoamylases, were able to degrade reuteran most efficiently (Fig. 7A). B. thetaiotaomicron has two pullulanase genes in its genome, BT1663 and BT4689, both predicted to be surface enzymes; however, neither pullulanase is associated with a specific PUL, and neither enzyme was identified in the proteomics analysis of reuteran culture supernatants. Therefore, attempts were made to induce the expression of these pullulanases by adding equal amounts of isomaltose, maltose, or panose, in addition to reuteran, to the B. thetaiotaomicron growth media. The addition of these sugars did not yield any further activity on reuteran by culture supernatants (Fig. 7B), while strong growth was observed only in the cultures containing additional maltose. In the TLC analysis it is apparent that B. thetaiotaomicron is not able to transport either isomaltose (6-O-α-d-glucopyranosyl–d-glucose) or panose [O-α-d-glucopyranosyl-(1→6)-O-α-d-glucopyranosyl-(1→4)-d-glucose] glucooligosaccharides, since these sugars still are present in culture supernatants. The addition of pullulan components did not promote further activation of α-glucan-specific CAZymes with activity on reuteran. Although B. thetaiotaomicron has pullulanases encoded by its genome, it is not known what induces expression of pullulanases or other non-PUL-associated CAZymes with activity on α-glucans in B. thetaiotaomicron. These results demonstrate that reuteran is partially resistant to SusG activity but has the potential to be degraded by B. thetaiotaomicron genome-encoded pullulanases; however, these enzymes are not produced by the bacterium in the presence of reuteran or mixed-linkage glucooligosaccharides.
FIG 7.
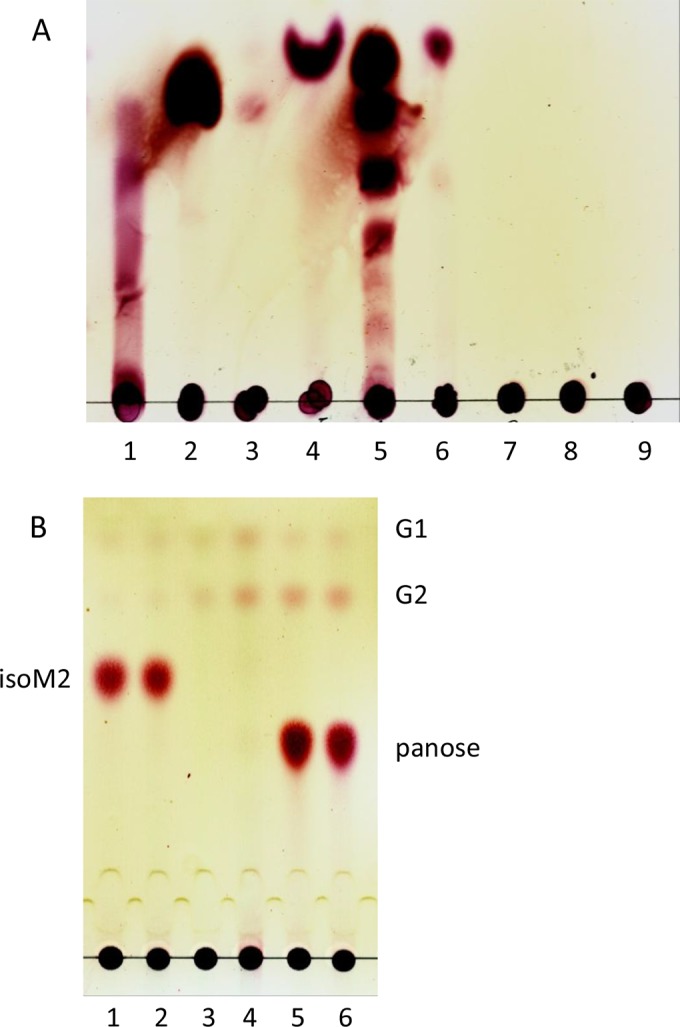
(A) Thin-layer chromatography analysis of the products of reuteran digestion by various α-glucan-degrading enzymes. Lane 1, Pseudomonas species isoamylase (EC 3.2.1.68); lane 2, B. licheniformis α-amylase (EC 3.2.1.1) (the dark spot is sucrose from enzyme storage solution); lane 3, Microbacterium aurum amylase A1 (EC3.2.1.1); lane 4, Aspergillus niger amyloglucosidase (EC 3.2.1.3); lane 5, Bacillus acidopullulyticus pullulanase (EC 3.2.1.41); lane 6, L. reuteri GTFA (EC 2.4.1.5); lane 7, GTF180; lane 8, Microbacterium aurum amylase B; lane 9, untreated reuteran control. (B) Activity of culture supernatants from B. thetaiotaomicron grown on reuteran with the addition of dextran- or pullulan-derived oligosaccharides isomaltose, maltose, and panose. Lane 1, reuteran plus isomaltose supernatant only; lane 2, reuteran plus isomaltose supernatant with reuteran; lane 3, reuteran plus maltose supernatant only; lane 4, reuteran plus maltose supernatant with reuteran; lane 5, reuteran plus panose culture supernatant only; lane 6, reuteran plus panose culture supernatant with reuteran. The addition of small oligosaccharides did not increase the ability of B. thetaiotaomicron to degrade reuteran. Results also indicate that isomaltose and panose are not transported by B. thetaiotaomicron, since they remain in the culture supernatant.
SusD forms an essential component of the SusC/D transport system and is essential for the metabolism of starch by B. thetaiotaomicron (26). SusD also was found to be present in reuteran culture supernatants; therefore, its production is activated by the presence of reuteran (Table 1). To further characterize SusD interactions with reuteran and IMMP, carbohydrate macroarray analysis was performed with purified and fluorescently labeled SusD on a range of soluble α-glucans. SusD was not able to bind reuteran or IMMP (Fig. 8), which also was confirmed by isothermal titration calorimetry (not shown). However, SusD also did not interact with any soluble glucans tested, including soluble potato starch, amylopectin, and pullulan (Fig. 8), while the appropriate control protein TmCBM41 showed the expected binding pattern (Fig. 8). SusD recognizes amylose helices in starch, and due to its crystallinity, amylose exists as an insoluble polymer. To see whether solubility of the substrate, and not the experimental setup, was the problem, pulldown assays were performed using insoluble starch as a substrate for SusD binding. In this case it was observed that SusD efficiently bound to insoluble wheat starch, suggesting that SusD is able to interact only with crystalline amylose. Thus, it appears that SusD is limited to interacting with insoluble α-1,4-linked glucose polymers, such as amylose, and is not able to interact with soluble α-glucans, including soluble starch molecules, reuteran, and IMMP. Since the interaction of SusD with its cognate PUL substrate is an important step in substrate-targeted degradation by B. thetaiotaomicron, it is likely that the lack of SusD binding with reuteran and IMMP prevents utilization of this polymer. The results of SusG activity and SusD binding experiments suggest it is a combined effect of poor SusD interaction and poor SusG enzymatic activity on IMMP and reuteran substrates that makes reuteran plus IMMP a metabolically poorer growth substrate for B. thetaiotaomicron than β-fructan EPS.
FIG 8.
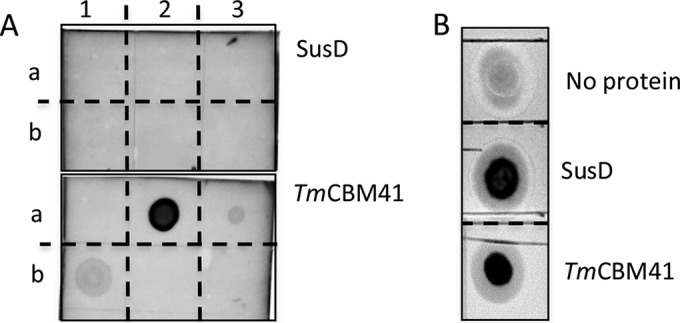
(A) Carbohydrate macroarray analysis of SusD binding to soluble α-glucans. A1, 1% pullulan; A2, 1% soluble potato starch; A3, 1% amylopectin; B1, 1% reuteran; B2, 1% IMMP; B3, 1% dextran. (B) Pulldown assay of SusD with insoluble wheat starch. TmCBM41 is a control for α-glucan binding activity and displays expected binding patterns (29). FITC-labeled SusD and TmCBM41 binding were carried out in 20 mM Tris, 150 mM NaCl, 5 mM CaCl2, and 0.05% Tween 20 at room temperature for 1 h. Binding was visualized at 520 nm.
DISCUSSION
This study demonstrates that B. thetaiotaomicron, a human gut commensal organism, is able to actively target and metabolize microbially synthesized carbohydrates, such as EPS produced by probiotic LAB strains, and achieves this by activating dedicated PUL systems to target EPS molecules. β-Fructan EPS metabolism was most prominent in the experiments where levan EPS activated the fructan-associated PUL, and cell surface levanase enzymes were able to fully metabolize levan to fructose monosaccharide, following previously established data showing that B. thetaiotaomicron degrades levan (β-2,6-fructan) but cannot degrade inulin (β-2,1-fructan) (22). Reuteran and IMMP trigger the expression of sus and dextran-associated PUL components, respectively (Table 1), as observed in proteomic analysis of culture supernatants. Reuteran and IMMP partial degradation may be linked to two factors. First, reuteran and IMMP are unable to be recognized by the transport component SusD, which is shown to be essential for growth on starch (Fig. 8A) (24, 26). Koropatkin et al. have shown that mutants of B. thetaiotaomicron lacking SusD are not able to grow on starch, showing that SusD recognition is essential for substrate utilization by B. thetaiotaomicron (26). SusD is closely associated with the transmembrane transporter SusC and facilitates transport of larger (molecules greater than 5 glucose units [>DP5]) maltooligosaccharides into the cell. Without an interaction between SusD and reuteran or IMMP, full metabolism of glucan EPS substrates would not be achieved. Second, the sus PUL-encoded CAZyme SusG encounters difficulties in degrading reuteran. Reuteran has a much more complex structure than starch, as it contains a much larger proportion of alternating α-1,4- and α-1,6-glycosidic linkages (Fig. 1A). Although reuteran and IMMP activate the production of starch- and dextran-degrading enzymes, it is most likely the unique architecture of LAB α-glucan EPS polymers that causes inaccessibility issues for α-amylase SusG and also for SusD to bind to the substrate. The complexity of the substrate also may be why there is an apparent lack of growth on IMMP, a dextran-like molecule containing large stretches of α-1,6-linked glucose residues. As yet there are no studies on dextran metabolism by B. thetaiotaomicron arising from dextran-associated PUL activation, making it difficult to speculate why IMMP was not degraded by B. thetaiotaomicron. Other studies on LAB α-glucan-based EPS molecules have shown that large α-glucan polymers synthesized by purified glucansucrases from lactic acid bacteria are fermented in fecal inocula (34, 36). Thus, even though B. thetaiotaomicron cannot fully metabolize these polymers, there still may be cross-feeding that occurs between bacterial taxa in the gut environment.
EPS degradation by B. thetaiotaomicron may confer two main advantages to the bacterium. First, in addition to dietary or mammalian carbohydrate sources, microbial sources also would provide an abundant and continuous nutritional source for B. thetaiotaomicron which would further add to its success as an established human gut symbiont. Second, by digesting EPS from competing gut microbiota in the intestinal tract, B. thetaiotaomicron would leave the competition vulnerable to clearance from the gastrointestinal tract, as the competition lacking EPS would lose its ability to efficiently adhere to the intestinal cell wall, while removal of the protective outer EPS layer would expose competing bacteria to cells of the innate immune system that reside in the intestinal mucosal layer.
Overall, our results describe a novel role for PUL activation in B. thetaiotaomicron directed at metabolism of LAB-derived EPS molecules. Activation of PULs in response to microbially derived carbohydrates has, to our knowledge, not been described before. EPS molecules are differentially metabolized, with levan being completely consumed, reuteran being partially consumed in the absence of levan, and very little IMMP consumed. If we consider the observations by McNulty et al. (20), who demonstrated that genes for levan and starch carbohydrate metabolism are upregulated in commensal bacteria in the presence of probiotic species, the results of this study using the model glycolytic bacterium B. thetaiotaomicron provide evidence for the first time that increased activation of genes for carbohydrate metabolism in commensal species is connected to the metabolism of EPS molecules produced by probiotic LAB strains. Other studies indicate EPS from Bifidobacterium as fermentable substrates for intestinal bacteria (37, 38), suggesting a more widespread mechanism of EPS metabolism by human gut commensal bacteria occurring in the gastrointestinal tract.
Conclusion.
B. thetaiotaomicron activates distinct polysaccharide utilization loci to degrade microbially synthesized polysaccharides, describing a novel role for PULs within Bacteroides. L. reuteri EPS molecules were differentially metabolized in the order of levan, reuteran, and IMMP. This phenomenon provides an explanation as to why genes for carbohydrate metabolism, specifically levanases and enzymes for starch metabolism, are significantly upregulated by commensal bacteria during probiotic supplementation (20). Bacterial EPS is likely an important alternative energy source for commensal bacterial species residing in the gut and may play an important role in how microbial communities are shaped in our gastrointestinal tract. Future research is aimed at further studying how EPS, and other microbially derived carbohydrates, are metabolized by B. thetaiotaomicron via the action of genome-based PULs and assessing the importance of microbially derived carbohydrates associated with transient and persistent microbial colonizers of the gut.
Supplementary Material
ACKNOWLEDGMENTS
We thank Nicole Koropatkin for providing plasmids and protein for SusD and SusG, Vincent Valk for providing Microbacterium aurum α-amylase A and B, Xiangfeng Meng for providing L. reuteri GTFA enzyme, Sander van Leeuwen and Justyna Dobruchowska for assistance with NMR analysis, Marcel de Vries and Hjalmar Permentier (Mass Spectrometry Facility, ERIBA, University of Groningen) for assistance with proteomics, and Alisdair Boraston for helpful discussions.
E.C.M. is supported by NIH grant GM099513. L.D. acknowledges support from the TKI Agri&Food program as coordinated by the Carbohydrate Competence Center (CCC3 and CCC-ABC; www.cccresearch.nl). A.L.V.B. is the recipient of a Veni grant from the Netherlands Organization for Scientific Research (NWO).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00149-15.
REFERENCES
- 1.Koropatkin NM, Cameron EA, Martens EC. 2012. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol 10:323–335. doi: 10.1038/nrmicro2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto J-M, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Li S, Jian M, Zhou Y, Li Y, Zhang X, Li S, Qin N, Yang H, Wang J, Brunak S, Doré J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, Bork P, Ehrlich SD, Wang J. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cantarel BL, Lombard V, Henrissat B. 2012. Complex carbohydrate utilization by the healthy human microbiome. PLoS One 7:e28742. doi: 10.1371/journal.pone.0028742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martens EC, Koropatkin NM, Smith TJ, Gordon JI. 2009. Complex glycan catabolism by the human gut microbiota: the Bacteroidetes Sus-like paradigm. J Biol Chem 284:24673–24677. doi: 10.1074/jbc.R109.022848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sonnenburg JL, Xu J, Leip DD, Chen C-H, Westover BP, Weatherford J, Buhler JD, Gordon JI. 2005. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 6.Szajewska H, Gyrczuk E, Horvath A. 2013. Lactobacillus reuteri DSM 17938 for the management of infantile colic in breastfed infants: a randomized, double-blind, placebo-controlled trial. J Pediatr 162:257–262. doi: 10.1016/j.jpeds.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 7.Veiga P, Pons N, Agrawal A, Oozeer R, Guyonnet D, Faurie J, van Hylckama Vlieg JE, Houghton LA, Whorwell PJ, Ehrlich SD, Kennedy SP. 2014. Changes of the human gut microbiome induced by a fermented milk product. Sci Rep 4:1–9. doi: 10.1038/srep06328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerritsen J, Smidt H, Rijkers GT, de Vos WM. 2011. Intestinal microbiota in human health and disease: the impact of probiotics. Genes Nutr 6:209–240. doi: 10.1007/s12263-011-0229-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Roock S, van Elk M, Hoekstra MO, Prakken BJ, Rijkers GT, de Kleer IM. 2011. Gut derived lactic acid bacteria induce strain specific CD4(+) T cell responses in human PBMC. Clin Nutr 30:845–851. doi: 10.1016/j.clnu.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 10.Van Baarlen P, Wells JM, Kleerebezem M. 2013. Regulation of intestinal homeostasis and immunity with probiotic lactobacilli. Trends Immunol 34:208–215. doi: 10.1016/j.it.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 11.Roberts IS. 1996. The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu Rev Microbiol 50:285–315. doi: 10.1146/annurev.micro.50.1.285. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y, Gänzle MG, Schwab C. 2010. Exopolysaccharide synthesized by Lactobacillus reuteri decreases the ability of enterotoxigenic Escherichia coli to bind to porcine erythrocytes. Appl Environ Microbiol 76:4863–4866. doi: 10.1128/AEM.03137-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klein MI, Duarte S, Xiao J, Mitra S, Foster TH, Koo H. 2009. Structural and molecular basis of the role of starch and sucrose in Streptococcus mutans biofilm development. Appl Environ Microbiol 75:837–841. doi: 10.1128/AEM.01299-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leemhuis H, Pijning T, Dobruchowska JM, van Leeuwen SS, Kralj S, Dijkstra BW, Dijkhuizen L. 2013. Glucansucrases: three-dimensional structures, reactions, mechanism, α-glucan analysis and their implications in biotechnology and food applications. J Biotechnol 163:250–272. doi: 10.1016/j.jbiotec.2012.06.037. [DOI] [PubMed] [Google Scholar]
- 15.Horn N, Wegmann U, Dertli E, Mulholland F, Collins SR, Waldron KW, Bongaerts RJ, Mayer MJ, Narbad A. 2013. Spontaneous mutation reveals influence of exopolysaccharide on Lactobacillus johnsonii surface characteristics. PLoS One 8:e59957. doi: 10.1371/journal.pone.0059957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ozimek LK, Kralj S, van der Maarel MJEC, Dijkhuizen L. 2006. The levansucrase and inulosucrase enzymes of Lactobacillus reuteri 121 catalyse processive and non-processive transglycosylation reactions. Microbiology 152:1187–1196. doi: 10.1099/mic.0.28484-0. [DOI] [PubMed] [Google Scholar]
- 17.Van Hijum SAFT, Kralj S, Ozimek LK, Dijkhuizen L, van Geel-Schutten IGH. 2006. Structure-function relationships of glucansucrase and fructansucrase enzymes from lactic acid bacteria. Microbiol Mol Biol Rev 70:157–176. doi: 10.1128/MMBR.70.1.157-176.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duncan SH, Flint HJ. 2013. Probiotics and prebiotics and health in ageing populations. Maturitas 75:44–50. doi: 10.1016/j.maturitas.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 19.Arthur JC, Gharaibeh RZ, Uronis JM, Perez-Chanona E, Sha W, Tomkovich S, Mühlbauer M, Fodor A, Jobin C. 2013. VSL#3 probiotic modifies mucosal microbial composition but does not reduce colitis-associated colorectal cancer. Sci Rep 3:2868. doi: 10.1038/srep02868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McNulty NP, Yatsunenko T, Hsiao A, Faith JJ, Muegge BD, Goodman AL, Henrissat B, Oozeer R, Cools-Portier S, Gobert G, Chervaux C, Knights D, Lozupone CA, Knight R, Duncan AE, Bain JR, Muehlbauer MJ, Newgard CB, Heath AC, Gordon JI. 2011. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Sci Transl Med 3:106ra106. doi: 10.1126/scitranslmed.3002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uyeno Y, Sekiguchi Y, Kamagata Y. 2008. Impact of consumption of probiotic lactobacilli-containing yogurt on microbial composition in human feces. Int J Food Microbiol 122:16–22. doi: 10.1016/j.ijfoodmicro.2007.11.042. [DOI] [PubMed] [Google Scholar]
- 22.Sonnenburg ED, Zheng H, Joglekar P, Higginbottom SK, Firbank SJ, Bolam DN, Sonnenburg JL. 2010. Specificity of polysaccharide use in intestinal bacteroides species determines diet-induced microbiota alterations. Cell 141:1241–1252. doi: 10.1016/j.cell.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anderson KL, Salyers AA. 1989. Biochemical evidence that starch breakdown by Bacteroides thetaiotaomicron involves outer membrane starch-binding sites and periplasmic starch-degrading enzymes. J Bacteriol 171:3192–3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cameron EA, Kwiatkowski KJ, Lee B, Hamaker BR, Koropatkin NM, Martens C. 2014. Multifunctional nutrient-binding proteins adapt human symbiotic bacteria for glycan competition in the gut by separately promoting enhanced sensing and catalysis. mBio 5:e01441-14. doi: 10.1128/mBio.01441-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Leeuwen SS, Kralj S, van Geel-Schutten IH, Gerwig GJ, Dijkhuizen L, Kamerling JP. 2008. Structural analysis of the alpha-d-glucan (EPS35-5) produced by the Lactobacillus reuteri strain 35-5 glucansucrase GTFA enzyme. Carbohydr Res 343:1251–1265. doi: 10.1016/j.carres.2008.01.044. [DOI] [PubMed] [Google Scholar]
- 26.Koropatkin NM, Martens EC, Gordon JI, Smith TJ. 2008. Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices. Structure 16:1105–1115. doi: 10.1016/j.str.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rogers TE, Pudlo N, Koropatkin NM, Bell JSK, Moya Balasch M, Jasker K, Martens EC. 2013. Dynamic responses of Bacteroides thetaiotaomicron during growth on glycan mixtures. Mol Microbiol 88:876–890. doi: 10.1111/mmi.12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koropatkin NM, Smith TJ. 2010. SusG: a unique cell-membrane-associated alpha-amylase from a prominent human gut symbiont targets complex starch molecules. Structure 18:200–215. doi: 10.1016/j.str.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 29.Lammerts van Bueren A, Finn R, Ausio J, Boraston AB. 2004. A-glucan recognition by a new family of carbohydrate-binding modules found primarily in bacterial pathogens. Biochemistry 43:15633–15642. doi: 10.1021/bi048215z. [DOI] [PubMed] [Google Scholar]
- 30.Van Hijum SAFT, Szalowska E, van der Maarel MJEC, Dijkhuizen L. 2004. Biochemical and molecular characterization of a levansucrase from Lactobacillus reuteri. Microbiology 150:621–630. doi: 10.1099/mic.0.26671-0. [DOI] [PubMed] [Google Scholar]
- 31.Dobruchowska JM, Gerwig GJ, Kralj S, Grijpstra P, Leemhuis H, Dijkhuizen L, Kamerling JP. 2012. Structural characterization of linear isomalto-/malto-oligomer products synthesized by the novel GTFB 4,6-α-glucanotransferase enzyme from Lactobacillus reuteri 121. Glycobiology 22:517–528. doi: 10.1093/glycob/cwr167. [DOI] [PubMed] [Google Scholar]
- 32.Savino F, Pelle E, Palumeri E, Oggero R, Miniero R. 2007. Lactobacillus reuteri (American Type Culture Collection strain 55730) versus simethicone in the treatment of infantile colic: a prospective randomized study. Pediatrics 119:e124–e130. doi: 10.1542/peds.2006-1222. [DOI] [PubMed] [Google Scholar]
- 33.Lionetti E, Miniello VL, Castellaneta SP, Magistá A, Mde Canio A, Maurogiovanni G, Ierardi E, Cavallo L, Francavilla R. 2006. Lactobacillus reuteri therapy to reduce side-effects during anti-Helicobacter pylori treatment in children: a randomized placebo controlled trial. Aliment Pharmacol Ther 24:1461–1468. doi: 10.1111/j.1365-2036.2006.03145.x. [DOI] [PubMed] [Google Scholar]
- 34.Leemhuis H, Dobruchowska JM, Ebbelaar M, Faber F, Buwalda PL, van der Maarel MJEC, Kamerling JP, Dijkhuizen L. 2014. Isomalto/malto-polysaccharide, a novel soluble dietary fiber made via enzymatic conversion of starch. J Agric Food Chem 62:12034–12044. doi: 10.1021/jf503970a. [DOI] [PubMed] [Google Scholar]
- 35.Van Geel-Schutten GH, Faber EJ, Smit E, Bonting K, Smith MR, Brink BTEN, Kamerling JP, Vliegenthart JFG, Dijkhuizen L. 1999. Biochemical and structural characterization of the glucan and fructan exopolysaccharides synthesized by the Lactobacillus reuteri wild-type strain and by mutant strains. Appl Environ Microbiol 65:3008–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hernandez-Hernandez O, Côté GL, Kolida S, Rastall R, Sanz ML. 2011. In vitro fermentation of alternansucrase raffinose-derived oligosaccharides by human gut bacteria. J Agric Food Chem 59:10901–10906. doi: 10.1021/jf202466s. [DOI] [PubMed] [Google Scholar]
- 37.Salazar N, Gueimonde M, Hernández-Barranco AM, Ruas-Madiedo P, de los Reyes-Gavilán CG. 2008. Exopolysaccharides produced by intestinal Bifidobacterium strains act as fermentable substrates for human intestinal bacteria. Appl Environ Microbiol 74:4737–4745. doi: 10.1128/AEM.00325-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rios-Covian D, Arboleya S, Hernandez-Barranco AM, Alvarez-Buylla JR, Ruas-Madiedo P, Gueimonde M, de los Reyes-Gavilan CG. 2013. Interactions between Bifidobacterium and Bacteroides species in cofermentations are affected by carbon sources, including exopolysaccharides produced by bifidobacteria. Appl Environ Microbiol 79:7518–7524. doi: 10.1128/AEM.02545-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:490–495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.