Abstract
A pilot-scale field experiment demonstrated that a one-time amendment of emulsified vegetable oil (EVO) reduced groundwater U(VI) concentrations for 1 year in a fast-flowing aquifer. However, little is known about how EVO amendment stimulates the functional gene composition, structure, and dynamics of groundwater microbial communities toward prolonged U(VI) reduction. In this study, we hypothesized that EVO amendment would shift the functional gene composition and structure of groundwater microbial communities and stimulate key functional genes/groups involved in EVO biodegradation and reduction of electron acceptors in the aquifer. To test these hypotheses, groundwater microbial communities after EVO amendment were analyzed using a comprehensive functional gene microarray. Our results showed that EVO amendment stimulated sequential shifts in the functional composition and structure of groundwater microbial communities. Particularly, the relative abundance of key functional genes/groups involved in EVO biodegradation and the reduction of NO3−, Mn(IV), Fe(III), U(VI), and SO42− significantly increased, especially during the active U(VI) reduction period. The relative abundance for some of these key functional genes/groups remained elevated over 9 months. Montel tests suggested that the dynamics in the abundance, composition, and structure of these key functional genes/groups were significantly correlated with groundwater concentrations of acetate, NO3−, Mn(II), Fe(II), U(VI), and SO42−. Our results suggest that EVO amendment stimulated dynamic succession of key functional microbial communities. This study improves our understanding of the composition, structure, and function changes needed for groundwater microbial communities to sustain a long-term U(VI) reduction.
INTRODUCTION
Mining, ore processing, and weapons and fuel production have left many sites around the world contaminated with uranium (U) (1). Groundwater contamination in particular is a critical public concern because the transport of highly soluble and toxic U(VI) within groundwater threatens drinking water resources. Bioreduction of U(VI) to insoluble U(IV) has been recognized as an effective approach to immobilize U in situ (2). However, while microorganisms capable of U(VI) reduction are present in aquifers, U(VI) still persists because of the lack of available electron donors and, in some cases, the presence of excess competing electron acceptors (e.g., NO3−) (3). Injection of a substrate that would provide electron donors is essential to stimulate indigenous microbial communities toward U(VI) reduction (4, 5).
Some U.S. Department of Energy (DOE) sites, for example, the Oak Ridge Integrated Field Research Challenge (ORIFRC) and the Old Rifle uranium mill tailing remedial action sites, are contaminated with U(VI). To remediate these sites, various fast-degrading substrates (e.g., acetate, ethanol, and lactate) have been used. The substrate injection stimulated microbial populations important to U(VI) reduction, resulting in distinct microbial communities whose functions were dependent upon the choice of substrate (6–13). However, the use of these fast-degrading, simple substrates has several drawbacks. First, it requires frequent (continuous, daily, or weekly) injections to maintain U(VI) reduction because after substrate is consumed, the reduced U(IV) could be reoxidized (14, 15). Thus, high, long-term operating costs could be incurred. Second, these fast-degrading substrates could cause overgrowth of microorganisms near injection wells, resulting in biomass clogging, which could then affect substrate delivery into contaminated plumes (16). Slowly degrading, complex substrates are promising alternatives because prolonged reducing conditions could be maintained with one-time injection. Such substrates have been successfully used for bioremediation of contaminated aquifers. For example, a one-time amendment of glycerol polylactate decreased groundwater Cr(VI) concentrations for >3.5 years (17), and perchlorate degradation was maintained for >2 years after a single oil emulsion injection (18). The prolonged effectiveness of these complex substrates is largely due to their high energy density, slow biodegradation, and retarded flow in groundwater systems (19), allowing a slow release of electron donors and carbon (C) sources to sustain bioremediation.
At the ORIFRC site, emulsified vegetable oil (EVO) was injected within 2 h into a fast-flowing aquifer within a uranium-bearing contaminant plume. The amendment reduced the groundwater U(VI) concentrations for 1 year (20, 21), and the phylogenetic structure of groundwater microbial communities was altered (22). Another study reported functional responses of groundwater microbial communities (e.g., increases of some enzymes involved in EVO degradation and denitrification), but the study captured only the initial responses (i.e., 4 days postamendment) using a sample collected from a downgradient well (23). In this study, the experimental design and EVO amendment were the same as the general survey of groundwater microbial communities using pyrosequencing of 16S rRNA genes (22). However, this study used a comprehensive functional gene microarray (GeoChip3.0) to further analyze the groundwater microbial communities from the aspect of functional genes. Particularly, we examined how key functional genes respond to EVO amendment, and how such responses are linked to key geochemical variables related to U(VI) reduction. Groundwater samples were collected from one upgradient and seven downgradient wells at different time points after EVO amendment (see Fig. S1 in the supplemental material). We hypothesized that (i) EVO amendment would alter the functional gene composition and structure of groundwater microbial communities, (ii) EVO amendment would stimulate key functional genes/groups involved in EVO biodegradation and reduction of electron acceptors in the aquifer, and (iii) the changes in the functional microbial communities would promote prolonged U(VI) reduction, primarily through a long-term supply of electron donors and stimulation of microbial processes involved in electron acceptor reduction.
MATERIALS AND METHODS
Site description, EVO amendment, and sampling.
A detailed description of the experimental site, design, and sampling was given previously (20–22) and is provided in the supplemental material. Briefly, this study was conducted in Area 2 of the ORIFRC (www.esd.ornl.gov/orifrc/). The groundwater flows from an upgradient zone across a control well (W8) and three injection wells and then passes through the downgradient zone installed with seven monitoring wells (W1 to W7) (see Fig. S1 in the supplemental material). Groundwater took 10 h to flow through the experimental plot. Prior to the experiment (8 December 2008), the groundwater flow pattern was characterized by injecting a potassium bromide solution (450 mg/liter, 3,400 liters) into the three injection wells. Peak bromide concentrations were then mapped as an indicator of hydraulic connection among the wells. Dissolved oxygen was near zero, although oxygen can infiltrate into the upper vadose zone from the atmosphere. The composition of EVO (SRS; Terra Systems, Wilmington, DE) was (wt/wt) 60% vegetable oil, 6% food grade surfactants, 0.3% yeast extract, 0.05% (NH4)3PO4, and the reminder water. An EVO emulsion (680 liters of EVO diluted to 3,400 liters with site groundwater) was evenly injected into the three injection wells over a 2-h time period on 9 February 2009. Groundwater samples were collected by pumping from W1 to W8 before injection and at 4, 17, 31, 80, 140, and 269 days after the injection. For microbial community analysis, groundwater (1 liter) was filtered on site with sterile 8-μm filters to remove large particles, followed by filtering with 0.2-μm filters to collect biomass. The filters were immediately frozen, shipped on dry ice to the laboratory, and stored at −80°C until DNA extraction.
Analytic methods.
Groundwater samples for metal analysis (10 ml) were filtered via a 0.3-μm filter, acidified with 0.05 ml of concentrated nitric acid, and then stored at 4°C until analysis. Details for all analytic methods are described in the supplemental material. Briefly, anions (acetate, NO3−, NO2−, Cl−, and SO42−) were analyzed with an ion chromatograph equipped with an IonPac AS-14 analytical column and an AG-14 guard column (DX-120; Dionex, Sunnyvale, CA) (14, 15). Aqueous Fe(II), total Fe, and sulfide were measured colorimetrically using a HACH DR 2000 spectrophotometer (Hach Chemical, Loveland, CO).
DNA extraction, GeoChip hybridization, and statistical analyses.
The community DNA was extracted using a freeze-grinding method (24) and quantified with PicoGreen (25) (Quant-It PicoGreen kit; Invitrogen, Carlsbad, CA). DNA (20 ng) was amplified using whole-community genome amplification with a TempliPhi kit (GE Healthcare, Piscataway, NJ), and 3.0 μg amplified DNA was hybridized to the GeoChip 3.0 as previously described (26, 27). The hybridized GeoChip was scanned, and the signal intensity for each spot was determined using ImaGene 6.0 (Biodiscovery, El Segundo, CA). GeoChip data are available at http://ieg.ou.edu/. Various statistical approaches, including two-tailed t test, clustering analysis, Mantel tests, analysis of similarity (ANOSIM), permutational multivariate analysis of variance (Adonis), and the multiresponse permutation procedure (MRPP), were used to analyze various data sets as previously described (28).
RESULTS
Changes in key geochemical variables after EVO amendment.
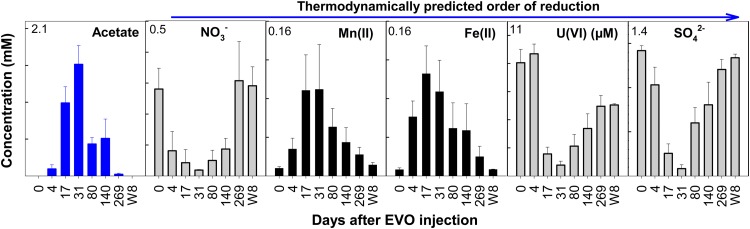
Before EVO amendment (day 0), groundwater in the experiment plot contained a considerable amount of soluble electron acceptors (0.35 mM NO3− and 1.2 mM SO42−) but not acetate or reduced products (NO2− and H2S; NH4+ was not measured), and concentrations of Fe(II) and Mn(II) were also low, indicating that bioreduction was limited in the absence of electron donors and/or other nutrients in the oligotrophic environment (Fig. 1). After EVO amendment, substantial acetate production, presumably from biodegradation of EVO (e.g., glycerol, long-chain fatty acids [LCFAs]), was observed in the seven downgradient wells (W1 to W7) (see Fig. S2 in the supplemental material). Acetate was detected at day 4, increased rapidly to ∼1.5 mM by day 31, and remained at ∼0.5 mM from day 80 to day 140 (Fig. 1). Sequential reduction of NO3−, Mn(IV), Fe(III), U(VI), and SO42− was also detected. NO3− decreased within 4 days, and then Fe(II) and Mn(II) concentrations increased, indicating Fe(III) and Mn(IV) reduction. A decline in U(VI) was observed later, almost concurrently with a substantial decrease in SO42−. U(VI) decreased from ∼10 μM to 1 μM within 24 days, and a U(VI) concentration lower than that in an upgradient control well (W8) was maintained for ∼140 days in W1 to W7 and for 269 days in W1, W3, and W5 (see Fig. S2 in the supplemental material). U(VI) reduction to U(IV) has been confirmed (21).
FIG 1.
Changes of average groundwater concentrations of acetate (blue), soluble electron acceptors [NO3−, U(VI), and SO42−] (gray), and reduced products [Fe(II) and Mn(II)] (black) in the seven downgradient wells (W1 to W7) after EVO amendment. Data detected at the same time points in a upgradient control well (W8) were also included for comparison. All data are presented as mean ± standard error (SE) of measurements in the seven downgradient wells (W1 to W7) at each time point and mean ± SE of seven measurements in W8 over time. The electron acceptors are presented in an order of thermodynamically predicted reduction preference. For y axes, the unit is μM for U(VI) and mM for the remainder, and the maximum label for each variable is shown. Detailed changes of these variables in each well are shown in Fig. S2 in the supplemental material.
Three complementary nonparametric multivariate statistical tests (MRPP, ANOSIM, and Adonis) of groundwater concentrations of acetate, NO3−, Fe(II), Mn(II), U(VI), and SO42− suggested that significant (P < 0.05 or 0.01) EVO biodegradation and reduction of multiple electron acceptors occurred in the downgradient zone after EVO amendment (see Table S1 in the supplemental material). In contrast, no significant electron acceptor reduction was observed in the upgradient control W8 (Fig. 1; see Fig. S2 in the supplemental material).
Shifts of overall groundwater microbial communities.
To examine whether the overall functional composition and structure of groundwater microbial communities changed after EVO amendment, a few statistical analyses of all detected functional genes were performed. Clustering analysis showed that groupings of the 56 samples were largely consistent with the observed differences in EVO biodegradation and electron acceptor reduction (see Fig. S3 in the supplemental material). First, the communities during the active redox period (days 4 to 140 after EVO amendment) (Fig. 1; see Table S1 in the supplemental material) were separated from the day 0 (before EVO amendment) and W8 (upgradient control) samples. Further, most of the day 4 to 17 samples were grouped together and were separated from the day 80 to 140 samples. By day 269, the communities were closer to those of day 0 samples, indicating sequential shifts of functional microbial communities after EVO amendment. MRPP, ANOSIM, and Adonis tests indicated that the functional composition and structure of groundwater microbial communities were significantly (P < 0.05 or 0.01) altered after EVO amendment (days 4 to 269) (see Table S1 in the supplemental material).
Dynamics of key functional genes/groups involved in important microbial processes.
To determine how the groundwater microbial community changed after EVO amendment, the relative abundance of various functional genes was examined, which was calculated by dividing the total signal intensity of detected all probes within each gene family or group by the total signal intensity of all probes detected. In this study, we focused on genes involved in EVO degradation and electron acceptor reduction.
(i) C cycling genes.
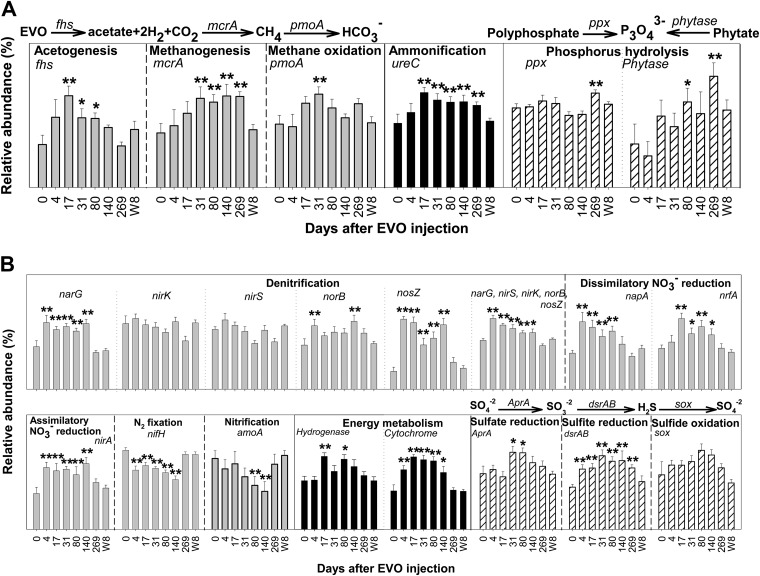
Some key genes involved in the final steps of EVO biodegradation and electron donor (e.g., acetate or H2) production were enriched after EVO amendment. These genes included fhs (encoding formyltetrahydrofolate synthetase), which is involved in acetogenesis, mcrA (encoding methyl coenzyme M reductase), which is involved in methanogenesis, and pmoA (encoding particulate methane monooxygenase), which is involved in methane oxidation (Fig. 2A). The increase in acetogenesis genes (fhs) was observed at 17 to 80 days after EVO amendment, while a significant increase in methanogenesis genes (mcrA) was not detected until 31 to 269 days. Also, the abundance of genes involved in the degradation of organic C (e.g., the glucoamylase, mannanase, and cadherin genes for starch, hemicellulose, and cellulose degradation), nitrogen (N) (e.g., ureC for ammonification), and phosphorus (P) (the phytase gene for phytate hydrolysis) compounds significantly (P < 0.05) increased after EVO amendment (Fig. 2A; see Fig. S4a in the supplemental material). In contrast, no significant increase was observed in the abundance of genes involved in CO2 fixation (e.g., aclB encoding ATP citrate lyase, the CODH gene encoding carbon monoxide dehydrogenase, pcc encoding propionyl coenzyme A [propionyl-CoA] carboxylase, and the RUBISCO gene encoding ribulose-l,5-bisphosphate carboxylase/oxygenase) (see Fig. S4b in the supplemental material).
FIG 2.
Changes in the average relative abundance of key genes involved in degradation of organic carbon (dark gray), nitrogen (black), and phosphorus (white) compounds (A) and in nitrogen (gray) and sulfur (white) cycling and energy metabolism (black) (B) in the seven downgradient wells (W1 to W7) after EVO amendment. Because the total abundance varies for each gene depending on probe number on the array, y axis scales for gene abundance are not shown. The significance (**, P < 0.05; *<0.10) of differences between each time point and day 0 was tested using the Student t test, which applies to Fig. S4, S8, and S9 in the supplemental material. Data detected at the same time points in a upgradient control well (W8) were also included for comparison. All data are presented as mean ± SE of measurements in the seven downgradient wells (W1 to W7) at each time point and mean±SE of seven measurements in W8 over time. The relative abundance was calculated by dividing the total signal intensity of detected individual gene sequences for each gene or gene group by the total signal intensity of all genes detected on the GeoChip. The processes catalyzed by these genes are shown, and processes catalyzed by the N cycling genes and gene description are shown in Fig. S6 in the supplemental material. Other genes: fhs, encoding formyltetrahydrofolate synthetase responsible for acetogenesis; mcrA, encoding methyl coenzyme M reductase responsible for methanogenesis; pmoA, encoding particulate methane monooxygenase responsible for methane oxidation; ppx, encoding exopolyphosphatase responsible for polyphosphate hydrolysis; phytase gene, encoding phytate phosphohydrolase responsible for phytate hydrolysis; aprA, encoding adenosine-5′-phosphosulfate reductase responsible for dissimilatory sulfate reduction; dsrAB, encoding dissimilatory sulfite reductase responsible for dissimilatory sulfite reduction; sox, encoding a sulfur-oxidizing enzyme for H2S reoxidation. More details (e.g., the protein identification numbers and derived microorganisms) for the genes involved in acetogenesis (fhs), methanogenesis (mcrA), methane oxidation (pmoA), dissimilatory nitrate reduction (napA and nrfA), dissimilatory sulfite reduction (dsrAB), and hydrogenase are shown in Fig. S5, S7, S8, and S11 in the supplemental material.
To further examine the enriched fhs, mcrA, and pmoA genes, clustering analysis of those genes detected at day 0 (before EVO amendment) and at 17 days postamendment was performed. The day 17 samples were clustered together and were well separated from day 0 samples. A considerable variation of functional gene distribution was observed between these two time points for the downgradient wells, while changes in the upgradient control W8 were small. Although some genes were common in all samples (see Fig. S5a in the supplemental material), many genes were stimulated and detected in most downgradient wells only after EVO amendment. The fhs genes from known species were enriched, including Syntrophomonas wolfei, able to degrade LCFAs, Alkaliphilus metalliredigens, reported to grow on short-chain fatty acids (SCFAs) and Fe(III) or Cr(VI), and Clostridium spp., known to produce acetate and H2 (29). Enriched methanogenesis genes included mcrA, similar to known methanogens (e.g., Methanococcus and Methanobacterium), but most were from uncultured archaea (see Fig. S5b in the supplemental material), while most of the stimulated methane oxidation genes (pmoA) were from uncultured bacteria (see Fig. S5c in the supplemental material).
(ii) N cycling genes.
The relative abundance of various N cycling genes was assessed (Fig. 2B; see Fig. S6 in the supplemental material). The results suggested that most (75%) of the genes involved in nitrate reduction processes were significantly (P < 0.05) enriched after EVO amendment. For example, narG and napA (encoding nitrate reductase) are involved in the reduction of nitrate to nitrite, nrfA (encoding the c-type cytochrome nitrite reductase) involves dissimilatory nitrite reduction and nirA (encoding nitrite reductase) involves assimilatory nitrite reduction, and nosZ (encoding nitrous oxide reductase) is involved in the reduction of N2O to N2. The relative abundance for most of these genes peaked at day 4 and remained elevated until day 140 (Fig. 2B). In contrast, the relative abundance of genes involved in nitrification (e.g., amoA) and N2 fixation (nifH) decreased after EVO amendment (Fig. 2B; see Fig. S6 in the supplemental material).
Further analysis suggested that most (∼90%) of the enriched denitrification genes were derived from uncultured bacteria. Pseudomonas spp. (e.g., P. stutzeri) and Dechloromonas spp. were among the enriched denitrifiers. Interestingly, most of the increased dissimilatory nitrate reduction genes were derived from known bacteria, including Fe(III)-reducing bacteria (FeRB) (e.g., Geobacter uraniireducens and Geobacter bemidjiensis), sulfate-reducing bacteria (SRB) (e.g., Desulfovibrio desulfuricans), and bacteria (e.g., Desulfitobacterium hafniense) capable of sulfite reduction (see Fig. S7 in the supplemental material). These bacteria are known to be capable of U(VI) reduction.
(iii) Sulfur cycling genes.
Sulfate (∼1.2 mM) in the groundwater is another significant electron acceptor, and SRB are an important functional group frequently detected in U(VI)-contaminated aquifers during bioremediation (10). The relative abundance of dsrAB genes (encoding dissimilatory sulfite reductase), which are involved in sulfate reduction, significantly (P < 0.05) increased after 4 days and peaked at day 31 (Fig. 2B), suggesting a stimulation of overall SRB community. More specifically, although a large portion (80%) of the enriched genes were from uncultured SRB, analysis of dsrAB genes derived from known species suggested that EVO amendment significantly (P < 0.05) stimulated some genera (e.g., Desulfovibrio and Desulfotomaculum) with members known to be capable of U(VI) reduction. Before EVO amendment, a limited number (13) of dsrAB genes from known SRB were detected, and only one was from Clostridium, which has members known to be capable of U(VI) reduction (30, 31) (see Fig. S8a in the supplemental material). The number of detected dsrAB genes increased ≥4-fold after EVO amendment, and a large portion (39%) of these enriched genes were from Desulfovibrio and Desulfotomaculum, both of which have members known to be capable of U(VI) reduction. Further analysis suggested that the significant increase in dsrAB genes derived from these two genera occurred during the active U(VI) reduction period (days 4 to 140) (see Fig. S8b in the supplemental material). The relative abundance of aprA, encoding adenosine-5′-phosphosulfate reductase, which is involved in the reduction of sulfate to sulfite, also increased after EVO amendment (Fig. 2B).
(iv) Genes involved in energy metabolism.
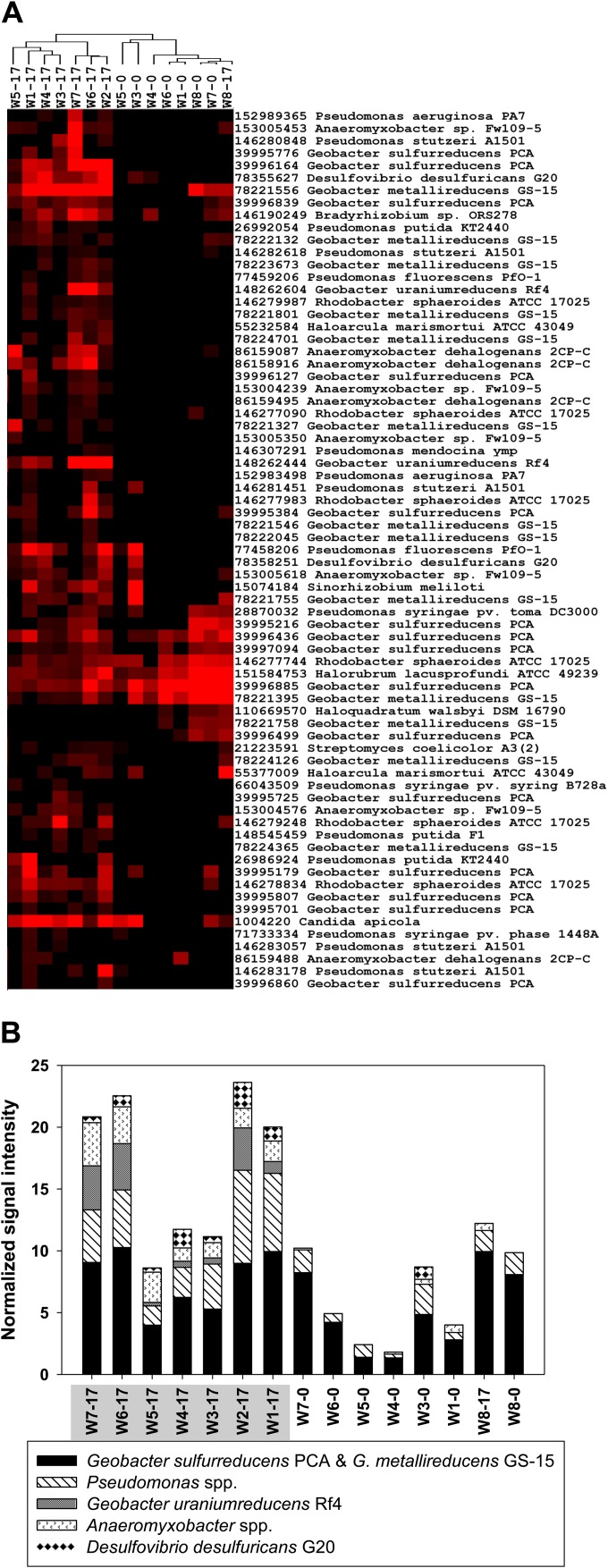
Previous studies with FeRB and SRB (e.g., Geobacter and Desulfovibrio) have demonstrated that cytochromes c3 are involved in U(VI) reduction (32–35). GeoChip 3.0 contains 364 probes specifically designed to detect cytochrome c3 genes from important U(VI)-reducing Geobacter, Desulfovibrio, and Anaeromyxobacter species (e.g., G. uraniumreducens Rf4). Analysis of the 190 detected cytochrome c3 genes indicated that EVO amendment significantly (P < 0.05) increased the relative abundance, diversity, and detected number of cytochrome c3 genes and changed the composition and structure of cytochrome-containing communities (Fig. 2B). Before EVO amendment, only 11 cytochrome c3 genes were detected, with those from G. sulfurreducens PCA and G. metallireducens GS-15 predominating (Fig. 3). After EVO amendment, while cytochrome c3 genes from G. sulfurreducens PCA and G. metallireducens GS-15 continued to be abundant, those from other species appeared and were significantly (P < 0.05) enriched in all downgradient wells (W1 to W7). At day 17, the abundance of cytochrome c3 genes from G. uraniumreducens Rf4, Anaeromyxobacter (A. dehalogenans 2CP-C and Anaeromyxobacter sp. strain Fw109-5), and Desulfovibrio desulfuricans G20, which were rarely detected before EVO amendment, accounted for up to 31%, 12%, and 16% of the cytochrome-containing community, respectively (see Fig. S9 in the supplemental material). The elevated levels of cytochrome c3 genes derived from these species were maintained (see Fig. S10 in the supplemental material), and by day 269, the composition and structure of cytochrome-containing communities were still different from those before EVO amendment (P = 0.017, 0.008, and 0.013 for the MRPP, ANOSIM, and Adonis tests, respectively).
FIG 3.
(A) Two distinct major clusters, with the right cluster showing cytochrome c3 genes detected before EVO amendment (day 0) and in the upgra dient control well (W8) and the left cluster showing enrichment of the genes in the seven downgradient wells (W1 to W7) 17 days after the amendment. In the sample identification, the number following the dash is 0 for day 0 samples (we lost the day 0 sample from W2) and is 17 for day 17 samples. Results were generated in Cluster3.0 and visualized using TreeView. Black indicates signal intensities below background, while red indicates signal intensities above background and brighter red indicates higher signal intensities. (B) Signal intensity of cytochrome c3 genes derived from U(VI)-reducing species. The samples highlighted in gray represent the enriched cluster (left). Anaeromyxobacter spp. include A. dehalogenans 2CP-C and Anaeromyxobacter sp. Fw109-5. Pseudomonas spp. include primarily P. putida KT2440, P. stutzeri A1501, P. syringae, P. fluorescens, and P. aeruginosa PA7. The time-series dynamics and SE and P values are shown in Fig. S9 and S10 in the supplemental material. Other detected cytochrome-containing genera include Rhodobacter, Haloarcula, Sinorhizobium, Halorubrum, and Candida.
Hydrogenases are known to be involved in electron transport from H2 to cytochromes c3 and then to U(VI) (33). Our results showed that hydrogenase genes were also stimulated after EVO amendment (Fig. 2B). Particularly, the genes from SRB (e.g., Desulfovibrio desulfuricans and Desulfitobacterium spp.) and FeRB (Geobacter uraniumreducens Rf4 and Anaeromyxobacter sp. Fw109-5) were enriched (see Fig. S11 in the supplemental material).
(v) Genes for metal resistance and organic contaminant degradation.
The Oak Ridge site examined in this study is contaminated with various metals [e.g., U(VI), Al, Cr, and Zn] and organic contaminants (e.g., tetrachloroethylene and trichloroethylene), though at low (0.2 to 15 μM) concentrations (22, 36). It is expected that indigenous groundwater microbial communities have developed resistance/degradation to these contaminants and that EVO amendment would stimulate such populations. Clustering analyses comparing genes detected in the day 0 and day 17 samples indicated that ∼55% of the metal resistance genes were detected primarily after EVO amendment (see Fig. S12a and c in the supplemental material). About 14% of these enriched genes were from Geobacter, Anaeromyxobacter, Desulfovibrio, Desulfotomaculum, Desulfitobacterium, and Pseudomonas. Similarly, ∼62% of the organic contaminant degradation genes were detected primarily after EVO amendment (see Fig. S12b and d in the supplemental material). Some of these enriched genes were also from Geobacter, Desulfovibrio, Desulfitobacterium, and Pseudomonas. U(VI)-reducing species affiliated with these genera have been frequently detected at this site during bioremediation (10, 37, 38). The number and abundance of metal resistance genes [e.g., efflux transporters for Cr(VI) (chrA) and Zn (czcA or czcD and zntA)] and organic contaminant degradation genes (e.g., the toluene dioxygenase gene for trichloroethylene degradation) derived from these genera significantly (P < 0.001) increased after EVO amendment.
Geochemical variables governing microbial community shifts.
Mantel tests were performed to identify correlations between changes in the microbial community composition and structure after EVO amendment and geochemical variables. Although the shifts in the functional composition and structure of overall community were not significantly (P = 0.378) correlated with 19 geochemical variables [pH, specific conductivity, Cl, Ag, Al, Ba, Ca, Cr, Ga, K, Mg, Sr, Zn, acetate, NO3−, Fe(II), Mn(II), U(VI), and SO42−], a marginal correlation (P = 0.098) was observed between the overall community and 6 key geochemical variables representing EVO degradation and electron acceptor reduction [acetate, NO3−, Fe(II), Mn(II), U(VI), and SO42−] (data not shown). More specifically, the changes of 13 key functional genes involved in EVO degradation (fhs for acetogenesis, mcrA for methanogenesis, and pmoA and mmoX for methane oxidation) and electron acceptor reduction (narG, nirS or nirK, norB, and nosZ for denitrification, napA and nrfA for dissimilatory nitrate reduction, dsrAB for sulfate reduction, and the cytochrome c3 gene for energy metabolism) were significantly (P = 0.048) correlated with these key geochemical variables (data not shown). Among these key geochemical variables, the correlations were significant with nitrate (P = 0.047), Fe(II) (P = 0.050), and sulfate (P = 0.006) (Table 1), three major electron acceptors in the aquifer.
TABLE 1.
Correlations between key functional genes and groundwater concentrations of acetate, Fe(II), U(VI), NO3−, SO42−, and Mn(II) revealed by the Mantel testa
| Gene category | Genes | No. of detected sequences | Correlation with groundwater concn ofb: |
|||||
|---|---|---|---|---|---|---|---|---|
| Acetate | Fe(II) | U(VI) | NO3− | SO42− | Mn(II) | |||
| 7 key processesc | 13 key genese | 1,566 | 0.068 | 0.050 | 0.182 | 0.047 | 0.006 | 0.153 |
| EVO degradationd | fhs, mcrA, pmoA, mmoX | 154 | 0.048 | 0.148 | 0.391 | 0.216 | 0.020 | 0.207 |
| Denitrification | narG, nirS or nirK, norB, nosZ | 647 | 0.268 | 0.174 | 0.459 | 0.048 | 0.122 | 0.366 |
| Dissimilatory NO3− reduction | napA, nrfA | 102 | 0.150 | 0.006 | 0.302 | 0.048 | 0.015 | 0.088 |
| Sulfate reduction | dsrAB | 473 | 0.334 | 0.100 | 0.077 | 0.006 | 0.008 | 0.157 |
| Energy metabolism | Cytochrome c3 gene | 190 | 0.068 | 0.017 | 0.048 | 0.413 | 0.028 | 0.017 |
The signal intensities of functional genes from 56 samples were used as the first matrix and the chemical concentrations were used as the second matrix. Mantel tests were performed using R package Vegan.
Boldface, P < 0.1.
Acetogenesis, methanogenesis, methane oxidation, denitrification, dissimilatory nitrate reduction, metal reduction, and sulfate reduction.
Acetogenesis, methanogenesis, and methane oxidation.
fhs, mcrA, pmoA, mmoX, cytochrome c3 gene, narG, nirS or nirK, norB, nosZ, napA, nrfA, and dsrAB.
Additional Mantel tests were performed to assess whether changes in the composition and structure of 13 key functional genes individually or as smaller groups were correlated with the 6 key geochemical variables. The changes in genes involved in EVO degradation (fhs, mcrA, pmoA, and mmoX) were significantly correlated with groundwater acetate (P = 0.048) and SO42− (P = 0.020) concentrations. Also, there were significant correlations between denitrification genes (narG, nirS or nirK, norB, and nosZ) and NO3− (P = 0.048) concentrations, between dissimilatory nitrate reduction genes (napA and nrfA) and NO3− (P = 0.048), Fe(II) (P = 0.006), and SO42− (P = 0.015) concentrations, between sulfate reduction genes (dsrAB) and SO42− (P = 0.008) and NO3− (P = 0.006) concentrations, and between cytochrome c3 genes and Fe(II) (P = 0.017), U(VI) (P = 0.048), SO42− (P = 0.028), and Mn(II) (P = 0.017) concentrations. The relatively poor correlation of cytochrome c3 genes with acetate concentrations (P = 0.068) was likely due to the presence of other important electron donors (such as H2) in this system. Regression analysis also indicated significant correlations between denitrification gene abundance and NO3− concentrations (r = 0.787, P = 0.02) and between cytochrome c3 gene abundance and Fe(II) (r = 0.876, P = 0.020) and U(VI) (r = 0.776, P = 0.023) concentrations.
DISCUSSION
Knowledge of the composition, structure, and activities of microbial communities represents the baseline information necessary to understand how remediation treatments alter communities and what molecular markers signal bioremediation success and reoxidation, which are important for monitoring bioremediation efforts as well as natural attenuation. One of the big challenges in bioremediation is the uncertainty in biotransformation rates inferred from chemical data and relating biogeochemical processes to associated microbial populations. It is expected that molecular data from microbial communities could help to reduce such uncertainty and provide insights into strategies that could be used to sustain the desired microbial populations. Using comprehensive GeoChip technology, this study examined the responses of groundwater microbial communities to EVO amendment and linked the community structure to geochemical variables. Our results indicated that the functional composition and structure of groundwater microbial communities was altered after EVO amendment, and the relative abundance of key functional genes/groups involved in EVO degradation and NO3−, Mn(IV), Fe(III), U(VI), and SO42− reduction (e.g., acetogenesis, methanogenesis, denitrification, dissimilatory nitrate reduction, energy metabolism, and sulfate reduction) significantly increased, with highest abundance during the active U(VI) reduction period, and declined with U(IV) reoxidation. The dynamics of these key functional genes were significantly correlated with groundwater acetate, NO3−, Fe(II), Mn(II), U(VI), and SO42− concentrations. This study improves our understanding of key functional genes and the diversity in response to substrate amendment.
The availability of C substrates is one of the most important factors affecting microbial community composition and structure. The effect could be particularly significant in U(VI)-contaminated groundwater like that at the Oak Ridge site, where electron donors and C sources are limited and multiple electron acceptors are present (6). Previous studies demonstrated that injection of fast-degrading, simple substrates (e.g., acetate or ethanol) stimulated distinct microbial communities (7, 9, 12, 13). In this study, a slowly degrading, complex substrate, EVO, was used. Based on a conceptual model (22), complete biodegradation of EVO requires multiple steps. After injection, EVO is first hydrolyzed into glycerol and LCFAs [e.g., linoleic acid (C18H32O2) or oleic acid (C18H34O2)], which are then biodegraded to H2,CO2, acetate, and propionate. Subsequently, these biodegraded intermediates/products could serve as electron donors and C sources and stimulate reduction of electron acceptors in the aquifer and finally methanogenesis. Geochemical analysis detected an increase of acetate and methane in the treatment zone and sequential reduction of NO3−, Mn(IV), Fe(III), U(VI), and SO42− (21). Proteomic analysis confirmed the enrichment of some key enzymes involved in EVO degradation and denitrification shortly (4 days) after EVO amendment (23). Since all of these microbial processes involve an array of functional genes/groups in the groundwater microbial community, sequential shifts in the overall functional composition and structure are expected, which are in general agreement with 16S rRNA gene-based analysis (22).
We further hypothesized that EVO amendment would stimulate a variety of key genes/groups in the groundwater microbial community, particularly those genes/groups involved in EVO biodegradation and electron acceptor reduction. It has been reported that some key enzymes (e.g., flavoproteins and thiolase) involved in EVO degradation were abundant in one of the downgradient wells early (4 days) after EVO amendment and that Desulforegula spp. could be key degraders (22, 23). However, limited information is available about the temporal dynamics of key genes involved in substrate degradation, which is important for continued electron donor production and U(VI) reduction. By analyzing 56 samples, we observed increased abundance of key genes, including fhs (acetogenesis), mcrA (methanogenesis), and pmoA (methane oxidation), and significant correlations of these genes with EVO degradation, e.g., acetate and sulfate concentrations. In addition, genes involved in the degradation of organic C, N, and P compounds increased and could be involved in degradation of microbial biomass. Microbial biomass in the groundwater increased >100-fold after EVO amendment, and this biomass may decay and could be an important C and energy source at later stages of organic amendment (17, 22). Overall, our data demonstrated a stimulation of various microbial processes involved in EVO degradation and biomass decomposition.
EVO amendment appeared to stimulate key genes/groups involved in the sequential reduction of NO3−, Mn(IV), Fe(III), U(VI), and SO42−, which typically coexist in contaminated aquifers. Competition for electrons among some of these electron acceptors has been reported. With ethanol injections, NO3− was reduced before Fe(III) and U(VI) (4). With acetate injections, the abundance of cytochrome c3 genes decreased while dsrAB genes increased when the dominant electron-accepting process shifted from Fe(III)-reducing to sulfate-reducing conditions (9). Similar results were observed shortly (4 days) after EVO amendment. Some key denitrification enzymes (e.g., NosZ) were highly abundant, but cytochrome c3 abundance was still fairly low (23). In this study, we observed increased abundance of a diverse array of key functional genes involved in reduction of multiple electron acceptors. Also, the peak in abundance for most of these key genes generally followed the observed reduction sequence of their corresponding electron acceptor: day 4 for nitrate (e.g., narG, norB, nosZ, napA, and nirA), day 17 for metals [Fe(III), Mn(IV), and U(VI)] (hydrogenase and cytochrome c3 genes), and day 31 for SO42− (aprA and dsrAB). Further, the dynamics in the composition and structure of these key genes were significantly correlated with groundwater concentrations of the corresponding electron acceptors, indicating a stimulation of these electron-accepting processes after EVO amendment.
Finally, we anticipated that shifts in the community functional composition and structure would enhance the ecosystem's ability to reduce U(VI) long-term. We expected that EVO amendment provides a longstanding supply of electron donors and C sources and subsequently stimulates the reduction of multiple electron acceptors [NO3−, Mn(IV), Fe(III), U(VI), and SO42−]. Previous studies indicated that enrichment of nitrate-reducing bacteria (NRB), FeRB, and SRB may prompt long-term U(VI) reduction. For example, with frequent ethanol injections, U(VI) reduction was maintained for >2 years with the enrichment of Geobacter, Anaeromyxobacter, Desulfovibrio, and Desulfotomaculum (37, 39, 40). It was suggested that after Fe(III) and Mn(IV) were depleted, Anaeromyxobacter, Desulfovibrio, and Desulfotomaculum could continue to grow by using the nitrate and/or sulfate that continuously entered the system via groundwater flow and sustain U(VI) reduction (7, 41–43). After EVO amendment, an increase in 16S rRNA gene sequences of Geobacter and Desulfovibrio was also observed (22). In addition to 16S rRNA genes, some functional genes can also be used as biomarkers to track the associated microbial populations. For example, the highly conserved dsrAB genes have been used for in-depth analysis of various SRB communities (44–48), and cytochromes c3 and hydrogenases are involved in U(VI) reduction (32, 33, 35). In this study, analysis of multiple functional genes [e.g., genes for denitrification (narG, norB, and nosZ), dissimilatory nitrate reduction (napA and nifA), sulfate reduction (dsrAB), energy metabolism (cytochrome c3 and hydrogenase genes), organic contaminant degradation, and metal resistance] suggested that diverse NRB, FeRB, and SRB were enriched after EVO amendment, including various species of Geobacter, Anaeromyxobacter, Desulfovibrio, and Desulfotomaculum. Further, the enriched functional genes derived from these microorganisms were diverse, suggesting that they have the potential to reduce/grow on multiple electron acceptors [e.g., NO3−, Fe(III), Mn(IV), U(VI), and SO42−), degrade organic contaminants, and be resistant to various toxic metals. These results suggest that the enriched NRB, FeRB, and SRB could be phylogenetically diverse, functionally versatile, and ecologically robust in the aquifer and play important roles in prolonging U(VI) reduction. Such observations could provide a mechanistic explanation for the predominance of NRB, FeRB, and SRB in various metal-reducing environments (49, 50).
In summary, this study examined the response of functional microbial communities to a slowly degrading, complex substrate, EVO, and its impacts on prolonged U(VI) reduction. Our results showed that EVO amendment stimulated sequential shifts in the functional composition and structure of groundwater microbial communities. Our results also demonstrated that key functional genes/groups involved in EVO biodegradation and reduction of multiple electron acceptors were greatly stimulated, and the dynamics of these key functional genes were significantly correlated with groundwater acetate, NO3−, Fe(II), Mn(II), U(VI), and SO42− concentrations. To the best of our knowledge, this is the first study to examine the time-series dynamics of functional microbial communities important to U(VI) reduction after substrate amendment. The results from this study improve our understanding of the diversity of functional genes/groups stimulated toward prolonged U(VI) reduction. While additional studies of sediment microbial communities are needed, the knowledge gained from examining the groundwater microbial community will help us further improve bioremediation designs for long-term bioremediation of various contaminated aquifers (e.g., heavy metal reduction, dechlorination of solvents, and nitrate removal).
Supplementary Material
ACKNOWLEDGMENTS
We thank Janna Phillips, Jennifer Earles, Kenneth Lowe, Tonia Mehlhorn, and Xiangping Yin for field sampling.
The field sampling was supported by the Subsurface Biogeochemical Research Program under contract no. DE-FG02-07ER64398, and the microbial community analysis was supported by ENIGMA-Ecosystems and Networks Integrated with Genes and Molecular Assemblies under contract no. DE-AC02-05CH11231 through the Office of Science, Office of Biological and Environmental Research, of the U.S. Department of Energy.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00043-15.
REFERENCES
- 1.Abdelouas A, Lutze W, Nuttall HE. 1999. Uranium contamination in the subsurface: characterization and remediation. Rev Mineral D 38:433–473. [Google Scholar]
- 2.Lovley DR, Phillips EJP, Gorby YA, Landa ER. 1991. Microbial reduction of uranium. Nature 350:413–416. doi: 10.1038/350413a0. [DOI] [Google Scholar]
- 3.Abdelouas A, Lu YM, Lutze W, Nuttall HE. 1998. Reduction of U(VI) to U(IV) by indigenous bacteria in contaminated ground water. J Contam Hydrol 35:217–233. doi: 10.1016/S0169-7722(98)00134-X. [DOI] [Google Scholar]
- 4.Istok JD, Senko JM, Krumholz LR, Watson D, Bogle MA, Peacock A, Chang YJ, White DC. 2004. In situ bioreduction of technetium and uranium in a nitrate-contaminated aquifer. Environ Sci Technol 38:468–475. doi: 10.1021/es034639p. [DOI] [PubMed] [Google Scholar]
- 5.Wall JD, Krumholz LR. 2006. Uranium reduction. Annu Rev Microbiol 60:149–166. doi: 10.1146/annurev.micro.59.030804.121357. [DOI] [PubMed] [Google Scholar]
- 6.Akob DM, Mills HJ, Gihring TM, Kerkhof L, Stucki JW, Anastacio AS, Chin KJ, Kusel K, Palumbo AV, Watson DB, Kostka JE. 2008. Functional diversity and electron donor dependence of microbial populations capable of U(VI) reduction in radionuclide-contaminated subsurface sediments. Appl Environ Microbiol 74:3159–3170. doi: 10.1128/AEM.02881-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anderson RT, Vrionis HA, Ortiz-Bernad I, Resch CT, Long PE, Dayvault R, Karp K, Marutzky S, Metzler DR, Peacock A, White DC, Lowe M, Lovley DR. 2003. Stimulating the in situ activity of Geobacter species to remove uranium from the groundwater of a uranium-contaminated aquifer. Appl Environ Microbiol 69:5884–5891. doi: 10.1128/AEM.69.10.5884-5891.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Handley KM, VerBerkmoes NC, Steefel CI, Williams KH, Sharon I, Miller CS, Frischkorn KR, Chourey K, Thomas BC, Shah MB, Long PE, Hettich RL, Banfield JF. 2013. Biostimulation induces syntrophic interactions that impact C, S and N cycling in a sediment microbial community. ISME J 7:800–816. doi: 10.1038/ismej.2012.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang YT, Van Nostrand JD, N′Guessan LA, Peacock AD, Deng Y, Long PE, Resch CT, Wu LY, He ZL, Li GH, Hazen TC, Lovley DR, Zhou JZ. 2012. Microbial functional gene diversity with a shift of subsurface redox conditions during in situ uranium reduction. Appl Environ Microbiol 78:2966–2972. doi: 10.1128/AEM.06528-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cardenas E, Wu WM, Leigh MB, Carley J, Carroll S, Gentry T, Luo J, Watson D, Gu BH, Ginder-Vogel M, Kitanidis PK, Jardine PM, Zhou JZ, Criddle CS, Marsh TL, Tiedje JM. 2010. Significant association between sulfate-reducing bacteria and uranium-reducing microbial communities as revealed by a combined massively parallel sequencing-indicator species approach. Appl Environ Microbiol 76:6778–6786. doi: 10.1128/AEM.01097-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Nostrand JD, Wu L, Wu WM, Huang Z, Gentry TJ, Deng Y, Carley J, Carroll S, He Z, Gu B, Luo J, Criddle CS, Watson DB, Jardine PM, Marsh TL, Tiedje JM, Hazen TC, Zhou J. 2011. Dynamics of microbial community composition and function during in situ bioremediation of a uranium-contaminated aquifer. Appl Environ Microbiol 77:3860–3869. doi: 10.1128/AEM.01981-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Nostrand JD, Wu WM, Wu LY, Deng Y, Carley J, Carroll S, He ZL, Gu BH, Luo J, Criddle CS, Watson DB, Jardine PM, Marsh TL, Tiedje JM, Hazen TC, Zhou JZ. 2009. GeoChip-based analysis of functional microbial communities during the reoxidation of a bioreduced uranium-contaminated aquifer. Environ Microbiol 11:2611–2626. doi: 10.1111/j.1462-2920.2009.01986.x. [DOI] [PubMed] [Google Scholar]
- 13.Xu M, Wu WM, Wu L, He Z, Van Nostrand JD, Deng Y, Luo J, Carley J, Ginder-Vogel M, Gentry TJ, Gu B, Watson D, Jardine PM, Marsh TL, Tiedje JM, Hazen T, Criddle CS, Zhou J. 2010. Responses of microbial community functional structures to pilot-scale uranium in situ bioremediation. ISME J 4:1060–1070. doi: 10.1038/ismej.2010.31. [DOI] [PubMed] [Google Scholar]
- 14.Wu WM, Carley J, Green SJ, Luo J, Kelly SD, Van Nostrand J, Lowe K, Mehlhorn T, Carroll S, Boonchayanant B, Lofller FE, Watson D, Kemner KM, Zhou JZ, Kitanidis PK, Kostka JE, Jardine PM, Criddle CS. 2010. Effects of nitrate on the stability of uranium in a bioreduced region of the subsurface. Environ Sci Technol 44:5104–5111. doi: 10.1021/es1000837. [DOI] [PubMed] [Google Scholar]
- 15.Wu WM, Carley J, Luo J, Ginder-Vogel MA, Cardenas E, Leigh MB, Hwang CC, Kelly SD, Ruan CM, Wu LY, Van Nostrand J, Gentry T, Lowe K, Mehlhorn T, Carroll S, Luo WS, Fields MW, Gu BH, Watson D, Kemner KM, Marsh T, Tiedje J, Zhou JZ, Fendorf S, Kitanidis PK, Jardine PM, Criddle CS. 2007. In situ bioreduction of uranium(VI) to submicromolar levels and reoxidation by dissolved oxygen. Environ Sci Technol 41:5716–5723. doi: 10.1021/es062657b. [DOI] [PubMed] [Google Scholar]
- 16.Wu WM, Carley J, Gentry T, Ginder-Vogel MA, Fienen M, Mehlhorn T, Yan H, Caroll S, Pace MN, Nyman J, Luo J, Gentile ME, Fields MW, Hickey RF, Gu BH, Watson D, Cirpka OA, Zhou JZ, Fendorf S, Kitanidis PK, Jardine PM, Criddle CS. 2006. Pilot-scale in situ bioremedation of uranium in a highly contaminated aquifer. 2. Reduction of U(VI) and geochemical control of U(VI) bioavailability. Environ Sci Technol 40:3986–3995. doi: 10.1021/es051960u. [DOI] [PubMed] [Google Scholar]
- 17.Faybishenko B, Hazen TC, Long PE, Brodie EL, Conrad ME, Hubbard SS, Christensen JN, Joyner D, Borglin SE, Chakraborty R, Williams KH, Peterson JE, Chen JS, Brown ST, Tokunaga TK, Wan JM, Firestone M, Newcomer DR, Resch CT, Cantrell KJ, Willett A, Koenigsberg S. 2008. In situ long-term reductive bioimmobilization of Cr(VI) in groundwater using hydrogen release compound. Environ Sci Technol 42:8478–8485. doi: 10.1021/es801383r. [DOI] [PubMed] [Google Scholar]
- 18.Borden RC. 2007. Concurrent bioremediation of perchlorate and 1,1,1-trichloroethane in an emulsified oil barrier. J Contam Hydrol 94:13–33. doi: 10.1016/j.jconhyd.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 19.Borden RC. 2006. Evaluation of slow release substrates for anaerobic bioremediation. Bioremed J 10:59–69. doi: 10.1080/10889860600835492. [DOI] [Google Scholar]
- 20.Tang G, Watson DB, Wu WM, Schadt CW, Parker JC, Brooks SC. 2013. U(VI) bioreduction with emulsified vegetable oil as the electron donor—model application to a field test. Environ Sci Technol 47:3218–3225. doi: 10.1021/es304643h. [DOI] [PubMed] [Google Scholar]
- 21.Watson DB, Wu WM, Mehlhorn T, Tang G, Earles J, Lowe K, Gihring TM, Zhang G, Phillips J, Boyanov MI, Spalding BP, Schadt C, Kemner KM, Criddle CS, Jardine PM, Brooks SC. 2013. In situ bioremediation of uranium with emulsified vegetable oil as the electron donor. Environ Sci Technol 47:6440–6448. doi: 10.1021/es3033555. [DOI] [PubMed] [Google Scholar]
- 22.Gihring TM, Zhang GX, Brandt CC, Brooks SC, Campbell JH, Carroll S, Criddle CS, Green SJ, Jardine P, Kostka JE, Lowe K, Mehlhorn TL, Overholt W, Watson DB, Yang ZM, Wu WM, Schadt CW. 2011. A limited microbial consortium is responsible for extended bioreduction of uranium in a contaminated aquifer. Appl Environ Microbiol 77:5955–5965. doi: 10.1128/AEM.00220-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chourey K, Nissen S, Vishnivetskaya T, Shah M, Pfiffner S, Hettich RL, Loffler FE. 2013. Environmental proteomics reveals early microbial community responses to biostimulation at a uranium- and nitrate-contaminated site. Proteomics 13:2921–2930. doi: 10.1002/pmic.201300155. [DOI] [PubMed] [Google Scholar]
- 24.Zhou JZ, Bruns MA, Tiedje JM. 1996. DNA recovery from soils of diverse composition. Appl Environ Microbiol 62:316–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahn SJ, Costa J, Emanuel JR. 1996. PicoGreen quantitation of DNA: effective evaluation of samples pre- or post-PCR. Nucleic Acids Res 24:2623–2625. doi: 10.1093/nar/24.13.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He Z, Deng Y, Van Nostrand JD, Tu Q, Xu M, Hemme CL, Li X, Wu L, Gentry TJ, Yin Y, Liebich J, Hazen TC, Zhou J. 2010. GeoChip 3.0 as a high-throughput tool for analyzing microbial community composition, structure and functional activity. ISME J 4:1167–1179. doi: 10.1038/ismej.2010.46. [DOI] [PubMed] [Google Scholar]
- 27.Wu L, Liu X, Schadt CW, Zhou J. 2006. Microarray-based analysis of subnanogram quantities of microbial community DNAs by using whole-community genome amplification. Appl Environ Microbiol 72:4931–4941. doi: 10.1128/AEM.02738-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He ZL, Xu MY, Deng Y, Kang SH, Kellogg L, Wu LY, Van Nostrand JD, Hobbie SE, Reich PB, Zhou JZ. 2010. Metagenomic analysis reveals a marked divergence in the structure of belowground microbial communities at elevated CO2. Ecol Lett 13:564–575. doi: 10.1111/j.1461-0248.2010.01453.x. [DOI] [PubMed] [Google Scholar]
- 29.Ye Q, Roh Y, Carroll SL, Blair B, Zhou J, Zhang CL, Fields MW. 2004. Alkaline anaerobic respiration: isolation and characterization of a novel alkaliphilic and metal-reducing bacterium. Appl Environ Microbiol 70:5595–5602. doi: 10.1128/AEM.70.9.5595-5602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Francis AJ, Dodge CJ, Lu F, Halada GP, Clayton CR. 1994. XPS and XANES studies of uranium reduction by Clostridium sp. Environ Sci Technol 28:636–639. doi: 10.1021/es00053a016. [DOI] [PubMed] [Google Scholar]
- 31.Gao WM, Francis AJ. 2008. Reduction of uranium(VI) to uranium(IV) by clostridia. Appl Environ Microbiol 74:4580–4584. doi: 10.1128/AEM.00239-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elias DA, Suflita JM, McInerney MJ, Krumholz LR. 2004. Periplasmic cytochrome c(3) of Desulfovibrio vulgaris is directly involved in H-2-mediated metal but not sulfate reduction. Appl Environ Microbiol 70:413–420. doi: 10.1128/AEM.70.1.413-420.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lovley DR, Widman PK, Woodward JC, Phillips EJP. 1993. Reduction of uranium by cytochrome-C(3) of Desulfovibrio vulgaris. Appl Environ Microbiol 59:3572–3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Payne RB, Gentry DA, Rapp-Giles BJ, Casalot L, Wall JD. 2002. Uranium reduction by Desulfovibrio desulfuricans strain G20 and a cytochrome c3 mutant. Appl Environ Microbiol 68:3129–3132. doi: 10.1128/AEM.68.6.3129-3132.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shelobolina ES, Coppi MV, Korenevsky AA, DiDonato LN, Sullivan SA, Konishi H, Xu HF, Leang C, Butler JE, Kim BC, Lovley DR. 2007. Importance of c-type cytochromes for U(VI) reduction by Geobacter sulfurreducens. BMC Microbiol 7:16. doi: 10.1186/1471-2180-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu WM, Carley J, Fienen M, Mehlhorn T, Lowe K, Nyman J, Luo J, Gentile ME, Rajan R, Wagner D, Hickey RF, Gu B, Watson D, Cirpka OA, Kitanidis PK, Jardine PM, Criddle CS. 2006. Pilot-scale in situ bioremediation of uranium in a highly contaminated aquifer. 1. Conditioning of a treatment zone. Environ Sci Technol 40:3978–3985. doi: 10.1021/es051954y. [DOI] [PubMed] [Google Scholar]
- 37.Cardenas E, Wu WM, Leigh MB, Carley J, Carroll S, Gentry T, Luo J, Watson D, Gu B, Ginder-Vogel M, Kitanidis PK, Jardine PM, Zhou J, Criddle CS, Marsh TL, Tiedje JA. 2008. Microbial communities in contaminated sediments, associated with bioremediation of uranium to submicromolar levels. Appl Environ Microbiol 74:3718–3729. doi: 10.1128/AEM.02308-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas SH, Padilla-Crespo E, Jardine PM, Sanford RA, Loffler FE. 2009. Diversity and distribution of Anaeromyxobacter strains in a uranium-contaminated subsurface environment with a nonuniform groundwater flow. Appl Environ Microbiol 75:3679–3687. doi: 10.1128/AEM.02473-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hwang CC, Wu WM, Gentry TJ, Carley J, Corbin GA, Carroll SL, Watson DB, Jardine PM, Zhou JZ, Criddle CS, Fields MW. 2009. Bacterial community succession during in situ uranium bioremediation: spatial similarities along controlled flow paths. ISME J 3:47–64. doi: 10.1038/ismej.2008.77. [DOI] [PubMed] [Google Scholar]
- 40.North NN, Dollhopf SL, Petrie L, Istok JD, Balkwill DL, Kostka JE. 2004. Change in bacterial community structure during in situ biostimulation of subsurface sediment cocontaminated with uranium and nitrate. Appl Environ Microbiol 70:4911–4920. doi: 10.1128/AEM.70.8.4911-4920.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lovley DR, Phillips EJP. 1992. Reduction of uranium by Desulfovibrio-Desulfuricans. Appl Environ Microbiol 58:850–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tebo BM, Obraztsova AY. 1998. Sulfate-reducing bacterium grows with Cr(VI), U(VI), Mn(IV), and Fe(III) as electron acceptors. FEMS Microbiol Lett 162:193–198. doi: 10.1111/j.1574-6968.1998.tb12998.x. [DOI] [Google Scholar]
- 43.Wu Q, Sanford RA, Loffler FE. 2006. Uranium(VI) reduction by Anaeromyxobacter dehalogenans strain 2CP-C. Appl Environ Microbiol 72:3608–3614. doi: 10.1128/AEM.72.5.3608-3614.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chang YJ, Peacock AD, Long PE, Stephen JR, McKinley JP, Macnaughton SJ, Hussain AK, Saxton AM, White DC. 2001. Diversity and characterization of sulfate-reducing bacteria in groundwater at a uranium mill tailings site. Appl Environ Microbiol 67:3149–3160. doi: 10.1128/AEM.67.7.3149-3160.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karr EA, Sattley WM, Rice MR, Jung DO, Madigan MT, Achenbach LA. 2005. Diversity and distribution of sulfate-reducing bacteria in permanently frozen Lake Fryxell, McMurdo Dry Valleys, Antarctica. Appl Environ Microbiol 71:6353–6359. doi: 10.1128/AEM.71.10.6353-6359.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu XD, Bagwell CE, Wu LY, Devol AH, Zhou JH. 2003. Molecular diversity of sulfate-reducing bacteria from two different continental margin habitats. Appl Environ Microbiol 69:6073–6081. doi: 10.1128/AEM.69.10.6073-6081.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miletto M, Williams KH, N′Guessan AL, Lovley DR. 2011. Molecular analysis of the metabolic rates of discrete subsurface populations of sulfate reducers. Appl Environ Microbiol 77:6502–6509. doi: 10.1128/AEM.00576-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perez-Jimenez JR, Kerkhof LJ. 2005. Phylogeography of sulfate-reducing bacteria among disturbed sediments, disclosed by analysis of the dissimilatory sulfite reductase genes (dsrAB). Appl Environ Microbiol 71:1004–1011. doi: 10.1128/AEM.71.2.1004-1011.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holmes DE, Nevin KP, Lovley DR. 2004. Comparison of 16S rRNA, nifD, recA, gyrB, rpoB and fusA genes within the family Geobacteraceae fam. nov. Int J Syst Evol Micr 54:1591–1599. doi: 10.1099/ijs.0.02958-0. [DOI] [PubMed] [Google Scholar]
- 50.Holmes DE, O'Neil RA, Vrionis HA, N′Guessan LA, Ortiz-Bernad I, Larrahondo MJ, Adams LA, Ward JA, Nicoll JS, Nevin KP, Chavan MA, Johnson JP, Long PE, Lovley DR. 2007. Subsurface clade of Geobacteraceae that predominates in a diversity of Fe(III)-reducing subsurface environments. ISME J 1:663–677. doi: 10.1038/ismej.2007.85. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.