ABSTRACT
The airways of patients with cystic fibrosis are colonized with diverse bacterial communities that change dynamically during pediatric years and early adulthood. Staphylococcus aureus is the most prevalent pathogen during early childhood, but during late teens and early adulthood, a shift in microbial composition occurs leading to Pseudomonas aeruginosa community predominance in ∼50% of adults. We developed a robust dual-bacterial in vitro coculture system of P. aeruginosa and S. aureus on monolayers of human bronchial epithelial cells homozygous for the ΔF508 cystic fibrosis transmembrane conductance regulator (CFTR) mutation to better model the mechanisms of this interaction. We show that P. aeruginosa drives the S. aureus expression profile from that of aerobic respiration to fermentation. This shift is dependent on the production of both 2-heptyl-4-hydroxyquinoline N-oxide (HQNO) and siderophores by P. aeruginosa. Furthermore, S. aureus-produced lactate is a carbon source that P. aeruginosa preferentially consumes over medium-supplied glucose. We find that initially S. aureus and P. aeruginosa coexist; however, over extended coculture P. aeruginosa reduces S. aureus viability, also in an HQNO- and P. aeruginosa siderophore-dependent manner. Interestingly, S. aureus small-colony-variant (SCV) genetic mutant strains, which have defects in their electron transport chain, experience reduced killing by P. aeruginosa compared to their wild-type parent strains; thus, SCVs may provide a mechanism for persistence of S. aureus in the presence of P. aeruginosa. We propose that the mechanism of P. aeruginosa-mediated killing of S. aureus is multifactorial, requiring HQNO and P. aeruginosa siderophores as well as additional genetic, environmental, and nutritional factors.
IMPORTANCE In individuals with cystic fibrosis, Staphylococcus aureus is the primary respiratory pathogen during childhood. During adulthood, Pseudomonas aeruginosa predominates and correlates with worse patient outcome. The mechanism(s) by which P. aeruginosa outcompetes or kills S. aureus is not well understood. We describe an in vitro dual-bacterial species coculture system on cystic fibrosis-derived airway cells, which models interactions relevant to patients with cystic fibrosis. Further, we show that molecules produced by P. aeruginosa additively induce a transition of S. aureus metabolism from aerobic respiration to fermentation and eventually lead to loss of S. aureus viability. Elucidating the molecular mechanisms of P. aeruginosa community predominance can provide new therapeutic targets and approaches to impede this microbial community transition and subsequent patient worsening.
INTRODUCTION
Complex polymicrobial communities colonize the airways of cystic fibrosis (CF) patients within the first month of life (1). Culture-independent studies have revealed the simultaneous presence of numerous bacterial taxa, fungi, and viruses in respiratory samples from CF patients at all stages of life (2–8). This abundance of microbes colonizing the respiratory tract, and particularly the lower airways, is facilitated by thick airway mucus and deficient mucociliary clearance that result from mutation of the cystic fibrosis transmembrane conductance regulator (CFTR) gene (9–11). These polymicrobial airway communities are diverse and dynamic, especially in young patients (12). During the first decade of life, bacterial diversity generally increases in patient samples. As patients age, diversity decreases until adulthood, when lower airway communities often become stable and resilient to significant perturbation by treatment (7, 12–15). In recent years, our understanding of the complexities of these polymicrobial communities has improved by characterizing patient samples using deep-sequencing technologies; however, how these communities impact interbacterial interactions, the immune response, antibiotic efficacy, and overall patient health is not well understood.
Culture-independent studies have emphasized the relevance of several emerging pathogens, like Streptococcus milleri group species and Mycobacterium abscessus, as well as nonpathogenic community members (16–19). Despite the identification of these emerging pathogens, Staphylococcus aureus and Pseudomonas aeruginosa remain two of the most relevant bacterial pathogens for CF patients. S. aureus is the most prevalent pathogen detected in pediatric patients clinically (20). The presence of S. aureus is correlated with worse lung function and lower forced expiratory volume (FEV), poorer nutrition, and increased inflammation in children (21, 22). Further, strains of S. aureus may persist in patients through adaptation of numerous virulence factors, including capsule, hemolysis, biofilm formation, and antibiotic resistance (particularly methicillin resistance) (23) and through the formation of small-colony variants (SCVs) (24). P. aeruginosa is detected in patients of all ages but is typically most prevalent and abundant in adult patients (20). Similarly to S. aureus, P. aeruginosa is associated with decreased FEV (25) and increased inflammation in children (26). Further, early colonization with P. aeruginosa is correlated with increased exacerbation and morbidity (27, 28). P. aeruginosa has a repertoire of virulence factors (including proteases, rhamnolipids, phospholipase C, hemolysin, and others) that may aid early-stage colonization. During chronic P. aeruginosa infection, expression of such virulence factors subsequently decreases over time to promote persistence (29). During long-term infections, hypermutators and adaptations like mucoidy and antibiotic resistance, as well as changes in gene expression, aid P. aeruginosa's ability to chronically colonize patient airways (30–34).
While S. aureus is well recognized as the predominant CF pathogen throughout childhood, P. aeruginosa is arguably the second most relevant pathogen in CF children. In some patients, P. aeruginosa may be the first pathogen isolated in infants and is detected in about 25 to 40% of young children by culture-based methods (12, 20, 35). Early in life, P. aeruginosa acquisition is associated with increased isolation of methicillin-resistant S. aureus (28). Interestingly, during late adolescence and adulthood the frequency of S. aureus in CF patients steeply declines and this microbe is displaced by P. aeruginosa (20). P. aeruginosa is detected in 80 to 90% of adults and becomes the predominant organism in the sputum of at least half of patients (3, 12, 20).
This striking negative clinical correlation between S. aureus and P. aeruginosa during teenage years and young adulthood has driven several in vitro and in vivo studies geared toward characterizing the interbacterial interactions of these two organisms (36–44). These previous studies have found that P. aeruginosa secretes various antistaphylococcal products and proteases, such as LasA, that can cause both biofilm dispersion and cell lysis of S. aureus (42, 45). Further, P. aeruginosa produces several inhibitors of S. aureus respiration, including hydrogen cyanide, quinoline N-oxides, and the phenazine pyocyanin (41, 46–48). P. aeruginosa has also been described to benefit from coculture with S. aureus by using this Gram-positive organism as an iron source (43) and a cue to produce extracellular virulence factors against both prokaryotic neighbors and the host (49). In response to the hostile environment, S. aureus may adapt to P. aeruginosa exoproducts, particularly in the presence of 2-heptyl-4-hydroxyquinoline N-oxide (HQNO), through increased biofilm formation (41) or increased frequency of SCVs (50, 51).
Despite significant efforts to characterize P. aeruginosa and S. aureus in CF patients, the molecular mechanism of their interaction and the role of the host (52) in driving the dynamics of the transition between P. aeruginosa and S. aureus during teenage years are not well understood. Studies of these two organisms, to date, have characterized isolated interactions using various in vitro or in vivo models. Here, we use a single in vitro model that can be adapted to study interactions on epithelial cells or plastic, during coculture in both the planktonic and biofilm phases. Using this system, we describe a mechanism by which P. aeruginosa drives S. aureus toward a lactic acid fermentative lifestyle through the action of HQNO and P. aeruginosa-produced siderophores early in coculture, eventually leading to killing of S. aureus and predominance of P. aeruginosa in the community.
MATERIALS AND METHODS
Strains and culture conditions.
Overnight cultures were inoculated with a single colony of P. aeruginosa or S. aureus into 5 ml lysogeny broth (LB) or tryptic soy broth (TSB), respectively, and grown for ∼12 h at 37°C with aeration on a roller drum. Methicillin-resistant S. aureus strains were grown in TSB plus 16 μg/ml oxacillin under these same conditions. SCV strains were grown statically for 20 h. A complete list of strains used throughout this study is provided in Table S1 in the supplemental material.
Coculture assays. (i) Coculture assay on CFBE monolayers.
Coculture assays were performed as previously described (53), with indicated adaptations to accommodate multiple bacterial species, and are depicted in Fig. 1A. Liquid cultures of P. aeruginosa and S. aureus were individually centrifuged (10,000 × g, 3 min) and washed in minimal essential medium plus 2 mM l-glutamine (MEM + l-Gln). Washed bacterial cells were resuspended in MEM + l-Gln to an optical density at 600 nm (OD600) of 0.1. For coculture samples, 250 μl of 0.1-OD600 P. aeruginosa and 250 μl of 0.1-OD600 S. aureus were added to triplicate wells of monolayers of CF bronchial epithelial (CFBE) airway cells in a 24-well plate. CFBE monolayers were grown as previously described (54). For monoculture controls, 250 μl of 0.1-OD600 P. aeruginosa or 250 μl of 0.1-OD600 S. aureus and 250 μl of MEM + l-Gln were added to triplicate wells. Target inputs were 0.5 × 107 to 1 × 107 CFU per well for each species in a 24-well plate. CFBE coculture plates were incubated at 37°C with 5% CO2 for 1 h, at which point any unattached bacterial cells were removed and 500 μl of MEM + l-Gln + 0.4% l-arginine was added to each well and incubated for an additional 5 h. After this additional 5-h incubation, the planktonic cells were removed and 500 μl of fresh MEM + l-Gln + 0.4% l-arginine was once again replaced in each well. The established coculture biofilm was incubated for an additional 16 h, followed by viable cell counts. For planktonic CFU enumeration, culture supernatant was 10-fold serially diluted in phosphate-buffered saline (PBS) and plated on Pseudomonas isolation agar or mannitol salt agar, for P. aeruginosa and S. aureus selective growth, respectively. S. aureus SCV strains were selected on tryptic soy agar plates grown in an anaerobic chamber for 3 days. After removal of culture supernatants, the biofilm fraction was removed by treatment with 250 μl of 0.1% Triton X-100 in PBS, with gentle shaking for 15 min. Biofilm bacteria were then scraped and vortexed in the plate for 2 min (covered with aluminum tape). Biofilm fractions were then serially diluted and plated as described for the planktonic fraction.
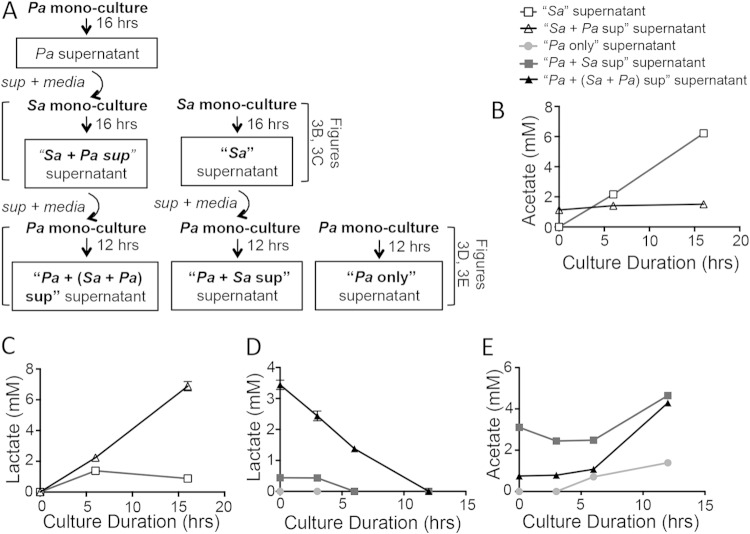
FIG 1.
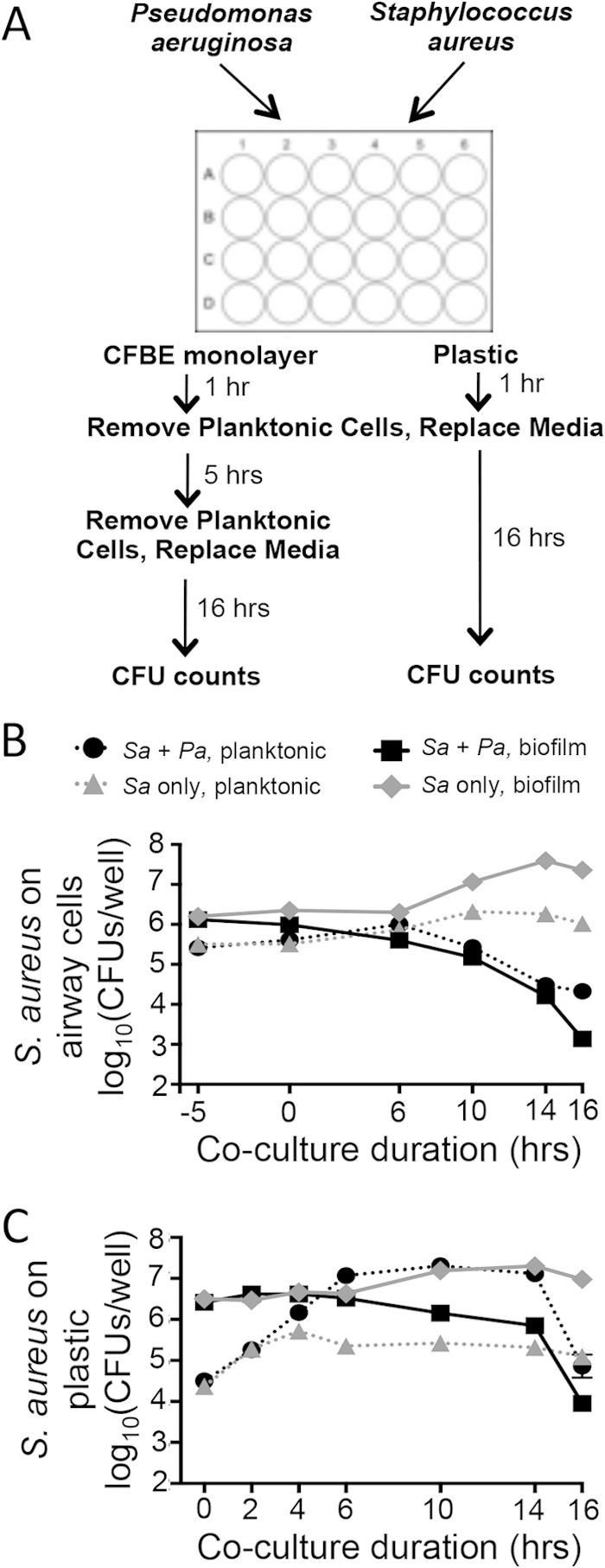
In vitro P. aeruginosa-S. aureus coculture assays on CFBE monolayers or plastic model early-stage coexistence and late-stage S. aureus killing by P. aeruginosa. (A) Assay design displaying P. aeruginosa-S. aureus coculture assays on CFBE monolayers and a plastic substrate. (B and C) Time course analyses were performed for S. aureus 8325-4 in planktonic and biofilm fractions. S. aureus viability was measured as log10(CFU/well) in a 24-well plate either in coculture with P. aeruginosa PA14 or as a monoculture. The key for panels B and C is shown at the top of panel B. Assays were performed in the CFBE coculture (B) or plastic coculture (C). Error bars indicate standard errors of the means from a representative triplicate time course assay. Sa, S. aureus; Pa, P. aeruginosa.
(ii) Coculture assay on plastic substrate.
In the plastic coculture assay, in which the bacterial biofilms are grown on a plastic substrate rather than on CFBE cells, bacterial culture preparation and inoculation were performed as described above. Cultures were inoculated directly into tissue culture-treated 24-well plastic plates. Unattached cells were removed after 1 h, and 500 μl MEM + l-Gln was added to each well. The cultures were then incubated for an additional 16 h, followed by CFU counts performed as described for the coculture assay on CFBE cells. For iron supplementation assays, bacterial cells were inoculated in MEM + l-Gln on plastic substrate. After 1 h, unattached cells were removed and MEM + l-Gln + 8 μM FeCl3 was added to the well. Cocultures then proceeded as described above.
RNA isolation.
Coculture assay mixtures for RNA analysis were prepared and inoculated as described in the CFBE and plastic assays and incubated for 6 total hours. After 6 h, planktonic cells and supernatant were removed and surface-attached biofilms were treated with RNAlater. Treated samples were scraped from the substrate using a cell scraper and collected. Samples were subsequently pelleted (16,000 × g, 5 min), and bacterial cells were lysed for 30 min at 37°C in 0.25 μg/μl lysostaphin and 2.5 μg/μl lysozyme in Tris-EDTA (TE) buffer. Total RNA was isolated using TRIzol and the Direct-Zol RNA miniprep kit (Zymo Research), followed by Turbo DNA-free DNase (Life Technologies) treatment, per the manufacturer's recommendation.
RNA-seq.
One microgram of total RNA was treated for rRNA and tRNA removal before sequencing. Single-read transcriptome sequencing (RNA-seq) was performed on the HiSeq platform at the Helmholtz Centre for Infection Research (Braunschweig, Germany). Raw RNA-seq reads were processed using the CLC Genomics Workbench. Processing was performed using default parameter settings set by the manufacturer. For each sample, sequences were trimmed and mapped to the P. aeruginosa UCBPP-PA14 (P. aeruginosa PA14) and S. aureus 8325 reference genomes using the RNA-seq analysis tool. Mapped sequences were further analyzed using R-project and the Bioconductor package edgeR (55). Read count data sets were filtered to remove low-expression reads prior to RNA expression analysis, with a cutoff of at least 15 counts, to reduce bias from library size differences and to address the fact that there is little power to detect differential expression of very low detection reads. Only a single replicate was performed for each sample; therefore, a conservative cutoff of 10-fold increase or decrease in gene expression was applied to identify genes of interest.
Reverse transcription and qRT-PCR.
cDNA was synthesized from total RNA for each coculture or monoculture using the Invitrogen Superscript III first-strand synthesis system according to the manufacturer's protocol. RNAs from plastic-grown coculture biofilm samples were synthesized using random hexamers and ∼100 ng total RNA. cDNA for CFBE-grown cocultures was synthesized using gene-specific primers (GSP) (see Table S2 in the supplemental material) in a single reaction to decrease nonspecific detection from host RNA. Briefly, total RNA, 10 mM deoxynucleoside triphosphates (dNTPs), and primers (50 ng/μl random hexamers or 2 μM [each] GSP) were incubated for 5 min at 65°C in a 10-μl total volume. Following this incubation, 10 μl of cDNA synthesis mix was added for a final 20-μl reaction mixture of 1× reaction buffer, 5 mM MgCl2, 0.01 M dithiothreitol (DTT), 2 U/μl RNaseOUT, and 10 U/μl Superscript III reverse transcriptase. Reaction mixtures were incubated at 25°C for 10 min, 50°C for 50 min, 85°C for 5 min, and 4°C for 5 min. Afterward, 1 μl RNase H was added to each reaction mixture and incubated at 37°C for 20 min. Quantitative real-time PCR (qRT-PCR) was performed with three technical replicates for each sample. S. aureus-specific primers were designed for each gene of interest and are specified in Table S2 in the supplemental material. For qRT-PCR, 1 μl of cDNA was diluted in a 10-μl reaction mixture with 0.25 μM forward and reverse primers and 1× iQ Sybr green Supermix (Bio-Rad). The qRT-PCR was as follows: 95°C for 3 min and then 95°C for 30 s, annealing temperature (Tanneal) for 30 s, and 72°C for 20 s, for 40 cycles. cDNA to each gene of interest was quantified based on cycle threshold (CT) compared to a standard curve of purified P. aeruginosa PA14 rplU DNA and normalized from sample to sample based on S. aureus rpoB quantification (56). S. aureus gyrB was used as a second normalization control for initial experiments and showed results consistent with those of rpoB; therefore, a single gene, rpoB, was used for later assays and is reported here.
Quantification of metabolite accumulation and utilization.
Sequential monocultures of P. aeruginosa PA14 and S. aureus 8325-4 were treated with culture supernatant containing exoproducts and metabolites from previous cultures, as depicted in Fig. 3A. Six-well tissue culture-treated plates were inoculated with 2 ml 0.05-OD600 P. aeruginosa PA14 or S. aureus 8325-4 in MEM + l-Gln + 0.5 g/liter yeast extract. In many samples, lactate, acetate, and ethanol were produced at levels near the limit of detection; therefore, yeast extract (0.5 g/liter) was added to amplify production of these metabolites above background. Cultures were incubated at 37°C and 5% CO2. Similarly to the plastic-grown coculture assay, supernatant and unattached cells were removed after 1 h and replaced with 2 ml fresh MEM + l-Gln + 0.5 g/liter yeast extract or 1 ml fresh medium plus 1 ml spent culture supernatant from the appropriate 16-h monoculture. A 16-h monoculture of P. aeruginosa PA14 was followed by a 16-h monoculture of S. aureus 8325-4 and, finally, a 12-h monoculture of P. aeruginosa PA14. Culture supernatants were collected at indicated time points throughout the 16-h or 12-h monocultures to examine the kinetics of metabolite accumulation and consumption. Lactate, ethanol, acetate, formate, and glucose accumulation and/or utilization from culture supernatants was analyzed by high-performance liquid chromatography (HPLC) as previously described (57). Briefly, 400 μl culture supernatant was filtered using 0.22-μm nylon centrifugal filters and acidified with 10 μl of 10% sulfuric acid sample. Samples were analyzed on a Bio-Rad HPX-87H column.
FIG 3.
P. aeruginosa exoproducts induce lactate production by S. aureus, which is subsequently consumed by P. aeruginosa. (A) Schematic of the sequential P. aeruginosa PA14 and S. aureus 8325-4 monoculture and spent supernatant treatment protocol used to analyze metabolite accumulation and utilization. Metabolites were quantified by HPLC from clarified supernatants collected throughout the assay at the indicated time points (B to E). The key above panel B applies to the graphs in panels B to E. (B and C) Accumulation of acetate (B) or lactate (C) by S. aureus monoculture in the presence (white triangles) or absence (white squares) of P. aeruginosa culture supernatants. (D and E) Accumulation/utilization of lactate (D) or acetate (E) by P. aeruginosa monocultures in the absence of S. aureus culture supernatants (circles), the presence of S. aureus supernatants when S. aureus was grown in fresh MEM (black squares), or the presence of S. aureus supernatants when S. aureus was grown in MEM plus spent P. aeruginosa supernatants (black triangles). Sa, S. aureus; Pa, P. aeruginosa.
Statistical analysis.
Differences in survival, bacterial load, or growth of monoculture versus cocultures of S. aureus or P. aeruginosa viabilities were compared for log10 normalized values using Student's t tests with the Holm-Šidák method for correcting for multiple comparisons or analysis of variance (ANOVA) with Dunnett's or Šidák's multiple-comparison posttest, as specified. Normalized RNA expression, quantified by qRT-PCR, was compared using the Friedman rank test and Dunn's posttest.
RESULTS
P. aeruginosa coexists with and then kills S. aureus in late-stage coculture on airway cells and plastic.
Clinical culture data and non-culture-based microbial analyses from both pediatric and adult populations show dynamic community structure earlier in life, particularly pertaining to the evolving predominance of P. aeruginosa versus S. aureus (1, 7, 20, 58). To better understand the nature of these community dynamics and interbacterial interactions, we adapted the previously described in vitro airway cell-P. aeruginosa coculture assay, using human bronchial epithelial cells (CFBE41o−) homozygous for ΔF508 CFTR (referred to as CFBE here) (53), to include a dual-bacterial system (Fig. 1A). As depicted in Fig. 1A, during the in vitro CFBE coculture assay, P. aeruginosa PA14 and S. aureus 8325-4 are coinoculated onto a monolayer of CFBE cells. Unattached bacterial cells are removed after the first hour. A dual-species biofilm is allowed to form for 5 additional hours on the airway epithelium. After the initial 6 h, the medium is again replaced and the established biofilm coculture is incubated for an additional 16 h.
Over the course of 22 h, we observed that the population of P. aeruginosa PA14 is not impacted by the presence or absence of S. aureus 8325-4, in either the planktonic or the biofilm phase (see Fig. S1A in the supplemental material). In contrast, we observed a dynamic interaction between P. aeruginosa PA14 and S. aureus 8325-4 in regard to the S. aureus 8325-4 population. The kinetics of the CFBE-inoculated coculture assay displayed an extended period of coexistence, followed by late-stage killing of S. aureus 8325-4 by P. aeruginosa PA14 (Fig. 1B). It is important to note that we interpret the reduced population of S. aureus as attributable to the killing of this microbe as we are measuring viable counts of each microbe. A rapid decline in the S. aureus 8325-4 population occurs between 10 h and 16 h of coincubation with P. aeruginosa PA14. This S. aureus 8325-4-killing phenotype by P. aeruginosa PA14 occurs on airway cells but does not require the presence of CFBE cells, as a simplified coculture assay performed on plastic displays similar late-stage interaction kinetics (Fig. 1A and C). On both the CFBE and plastic substrates, S. aureus 8325-4 killing is observed in both the biofilm and planktonic phases (Fig. 1B and C). We will focus on S. aureus biofilm results here, but it should be noted that the reported phenotypes are consistent between the planktonic populations and the biofilm-grown bacteria.
The coculture assay performed on plastic also displays a strikingly enhanced planktonic S. aureus 8325-4 population in the presence of P. aeruginosa PA14 between 4 h and 6 h (Fig. 1C). The increased planktonic S. aureus 8325-4 population may be due, at least in part, to a biofilm dispersion phenotype in the presence of P. aeruginosa PA14. This conclusion is indicated by the decreased biofilm population of S. aureus 8325-4 and a concomitant increase in the planktonic S. aureus 8325-4 population at 6 h (see Fig. S2 in the supplemental material). Less dramatic biofilm dispersion similarly occurs on CFBE monolayers prior to the second medium change in the airway cell coculture system. This S. aureus biofilm dispersion phenomenon was previously reported by Park et al. to be mediated by P. aeruginosa proteases (42). While the described S. aureus biofilm dispersion phenotype is consistent with the observations in our coculture model on plastic, other mechanisms leading to an increased planktonic population are possible. Additionally, similarly to the CFBE coculture model, when bacteria are coincubated on plastic, the P. aeruginosa PA14 population is not differentially impacted by the presence or absence of S. aureus 8325-4 (see Fig. S1B in the supplemental material).
Killing of S. aureus by P. aeruginosa is not specific to the S. aureus 8325-4–P. aeruginosa PA14 strain combination. There is, however, variation in the degree or kinetics of S. aureus killing by P. aeruginosa strains (see Fig. S3A in the supplemental material). Two other tested laboratory strains of P. aeruginosa (PAO1 and FRD1) killed S. aureus 8325-4, but less dramatically than P. aeruginosa PA14. S. aureus 8325-4 killing by seven clinical isolates of P. aeruginosa ranged from no killing (no difference in viable cell count from S. aureus 8325-4 monoculture) to killing that resulted in S. aureus 8325-4 viable cell counts below the limit of detection in our coculture assay (<2 log10[CFU/well]). Notably, P. aeruginosa strains with lower biofilm populations at the end of the assay, likely due to decreased initial attachment and/or reduced growth/survival despite similar starting inocula, were unable to kill S. aureus 8325-4 (SMC1585, SMC5450, and SMC5451 [see Fig. S3B in the supplemental material]), indicating a dose-dependent relationship between P. aeruginosa population and S. aureus killing. However, end-of-assay P. aeruginosa population was not the only factor contributing to this variation, as demonstrated in the coculture with SMC1587, which developed a P. aeruginosa biofilm population comparable to that of P. aeruginosa PA14 at the end of assay but showed no killing of S. aureus 8325-4 (see Fig. S3 in the supplemental material). Importantly, both mucoid and nonmucoid strains of P. aeruginosa can reduce S. aureus 8325-4 populations (see Fig. S3A in the supplemental material). This finding was confirmed through equivalent S. aureus 8325-4 killing by the parent strain FRD1 (mucoid) and its transposon mutant derivative FRD1 algT::Tn501, which is nonmucoid due to the mutation in the alginate biosynthetic genes.
Furthermore, killing of S. aureus by P. aeruginosa PA14 in our coculture model is not specific to S. aureus 8325-4. All S. aureus laboratory strains tested (8325-4, Newman, MW2, and USA300) and clinical isolates of S. aureus (including both methicillin-sensitive and methicillin-resistant strains) were killed by P. aeruginosa PA14 (see Fig. S4 in the supplemental material). Susceptibility to killing did vary, but all strains experienced at least a 20-fold decrease in population when in coculture with P. aeruginosa PA14 compared to monoculture.
Taken together, our data show that multiple strains of P. aeruginosa, including mucoid and nonmucoid strains, are capable of killing a variety of S. aureus laboratory and clinical isolates, either in a biofilm or planktonically, indicating a general mechanism of interaction between these organisms. Furthermore, while the kinetics of the interaction shows some variation, the killing of S. aureus by P. aeruginosa does not require the presence of CFBE cells. We took advantage of the latter conclusion in many of the subsequent experiments presented below.
Coculture with P. aeruginosa upregulates S. aureus fermentation pathway gene expression.
To better understand the molecular nature of P. aeruginosa-S. aureus interactions, an unbiased approach to examine changes in gene expression was employed. RNA-seq analysis was performed on single samples of 6-h biofilms from the plastic coculture assay, as follows: (i) P. aeruginosa PA14 monoculture, (ii) S. aureus 8325-4 monoculture, and (iii) P. aeruginosa PA14 plus S. aureus 8325-4 coculture. P. aeruginosa PA14 and S. aureus 8325-4 gene read assignments for monocultures and cocultures are provided in Tables S3 and S4 in the supplemental material, respectively. We utilized the RNA-seq results as a hypothesis-generating approach; therefore, a conservative cutoff of 10-fold change or greater was used to identify differentially expressed genes of potential interest for further investigation.
P. aeruginosa PA14 gene expression was minimally impacted by the presence of S. aureus 8325-4 compared to growth of P. aeruginosa PA14 alone (see Fig. S5 in the supplemental material). Only one gene, PA_10470, displayed a greater-than-10-fold change in coculture compared to monoculture. PA_10470, which encodes a major facilitator superfamily (MFS) transporter, was downregulated 15-fold in the presence of S. aureus 8325-4.
In contrast, coculture of P. aeruginosa PA14 with S. aureus 8325-4 greatly impacted S. aureus 8325-4 gene expression compared to an S. aureus monoculture (Fig. 2A). A summary of genes with >10-fold change in expression (increased or decreased) in the presence of P. aeruginosa PA14 compared to S. aureus 8325-4 alone is provided in Table S5 in the supplemental material. The four most differentially upregulated S. aureus 8325-4 genes in coculture with P. aeruginosa PA14 compared to S. aureus 8325-4 monoculture were pyruvate formate-lyase 1-activating enzyme (pflA, SAOUHSC_00188), formate acetyltransferase (pflB, SAOUHSC_00187), l-lactate dehydrogenase (ldh, SAOUHSC_00206), and alcohol dehydrogenase (adh, SAOUHSC_00113), all of which are associated with fermentation pathways in S. aureus (Fig. 2A).
FIG 2.
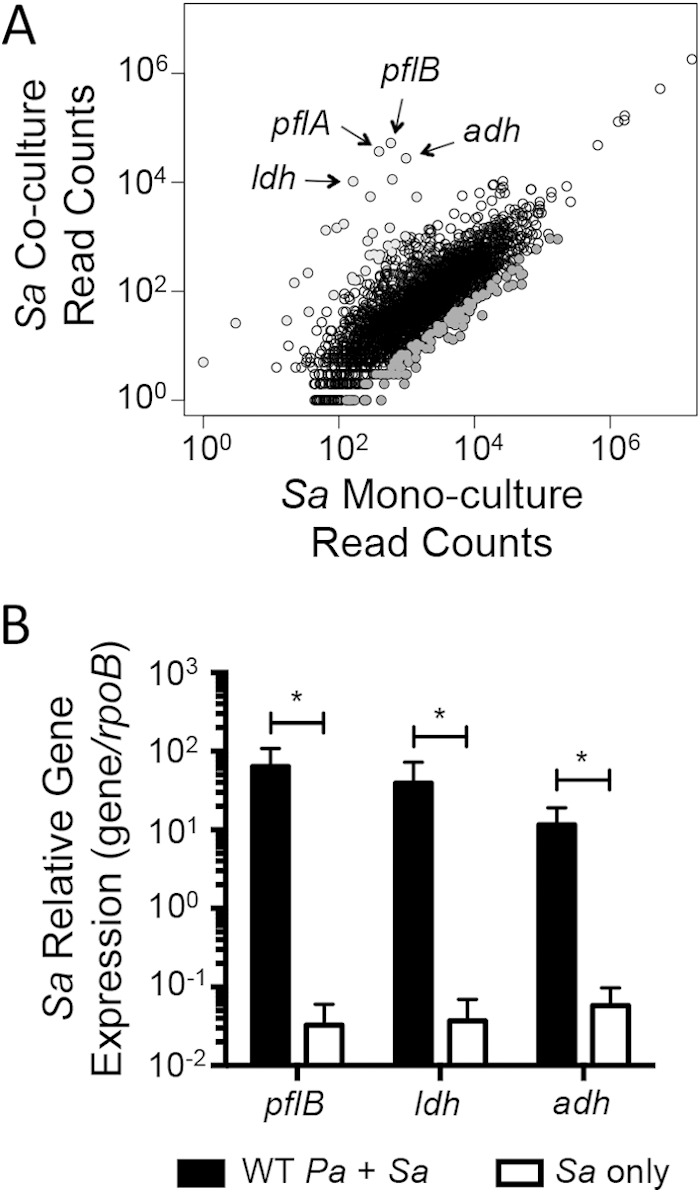
S. aureus fermentation pathway gene expression is upregulated in the presence of P. aeruginosa. S. aureus 8325-4 gene expression was compared in coculture with P. aeruginosa PA14 versus S. aureus monoculture. Total RNA was isolated from 6-h biofilm fractions on plastic. (A) Filtered RNA-seq reads corresponding to all expressed genes by S. aureus in coculture and monoculture. Highly differentially expressed genes in coculture with P. aeruginosa compared to S. aureus monoculture are indicated (light gray, >10-fold-increased expression in coculture; dark gray, >10-fold-decreased expression in coculture). Key fermentation pathway genes are indicated. (B) Relative expression of marker genes from formate (pflB, SAOUHSC_00187), lactate (ldh, SAOUHSC_00206), and ethanol (adh, SAOUHSC_00113) fermentation pathways. S. aureus gene expression in coculture with P. aeruginosa is compared with that in S. aureus monoculture. Statistical significance was determined for 9 replicate samples using multiple Student t tests and the Holm-Šidák method for correcting for multiple comparisons (*, P < 0.05). Sa, S. aureus; Pa, P. aeruginosa.
Upregulation of S. aureus 8325-4 fermentation pathway gene expression in coculture with P. aeruginosa PA14 on plastic was confirmed by qRT-PCR. Representative genes for formate (pflB, SAOUHSC_00187), lactate (ldh, SAOUHSC_00206), and ethanol (adh, SAOUHSC_00113) fermentation pathways were selected based on the RNA-seq results and quantified relative to the expression of a reference gene, rpoB (SAOUHSC_00524). S. aureus expression of pflB and ldh was upregulated >1,000-fold, and adh was upregulated >200-fold in coculture with P. aeruginosa PA14 compared to S. aureus 8325-4 monoculture in biofilm samples (Fig. 2B).
S. aureus shifts from aerobic respiration to lactic acid fermentation in the presence of P. aeruginosa secreted products.
The RNA-seq and qRT-PCR analyses strongly indicated that S. aureus upregulates fermentation pathways in the presence of P. aeruginosa. However, expression data alone do not definitively confirm a metabolic transition away from aerobic respiration. HPLC was performed to quantify the accumulation of metabolic products in culture supernatants and, thus, identify changes in S. aureus 8325-4 metabolism. Due to the complexity of the coculture system, simply analyzing monoculture versus coculture supernatants does not distinguish production and/or consumption of metabolic products by the two separate species. Therefore, a series of monoculture assays were performed using culture supernatants of P. aeruginosa PA14 or S. aureus 8325-4 in the mixing experiments outlined in Fig. 3A. By separating the two organisms and sequentially analyzing culture supernatants, we were able to identify production, accumulation, and consumption of metabolites by the individual bacterial species.
When S. aureus 8325-4 was grown as a monoculture without the addition of P. aeruginosa PA14 supernatant over 16 h, S. aureus 8325-4 produced over 6 mM acetate, indicative of aerobic respiration (59, 60) (Fig. 3B). However, S. aureus 8325-4 treated with P. aeruginosa PA14 culture supernatants showed greatly reduced production of acetate, accumulating to only 1.5 mM rather than 6 mM over 16 h. Instead, S. aureus 8325-4 cultures treated with P. aeruginosa PA14 supernatants accumulated lactate in a linear fashion over the 16-h culture (to about 7 mM), whereas S. aureus 8325-4 cultures not treated with P. aeruginosa PA14 supernatants accumulated less than 1 mM lactate over 16 h (Fig. 3C). Accumulation of lactate in the S. aureus 8325-4 culture treated with P. aeruginosa PA14 supernatants demonstrates a metabolic transition to fermentation in the presence of P. aeruginosa PA14 exoproducts.
Surprisingly, although formate and alcohol fermentation gene expression pathways were highly upregulated, formate and ethanol did not accumulate in S. aureus 8325-4 cultures treated with P. aeruginosa PA14 supernatants. After 16 h, formate and ethanol concentrations were detected at 0.5 mM and 0.6 mM, respectively (near the limit of detection for our instrument). These results are consistent with those seen in the S. aureus Col hemB mutant, previously reported by Kohler et al. (61).
In a coculture system, metabolites produced by one organism can serve as signals or nutrients for the other organism. Therefore, we were interested in whether P. aeruginosa utilizes the fermentation products secreted by S. aureus. When P. aeruginosa PA14 was grown in medium containing the supernatants from S. aureus 8325-4 cocultured with P. aeruginosa supernatants, the S. aureus-produced lactate was consumed by P. aeruginosa to below detectable levels (Fig. 3D). In fact, P. aeruginosa PA14 consumed the S. aureus 8325-4-produced lactate preferentially over the glucose in the medium (see Fig. S6 in the supplemental material), which was consistent with previous reports (62). P. aeruginosa can also consume acetate (63); however, it was not the preferred carbon source under the culture conditions used here; rather, additional acetate accumulated over time (Fig. 3E). The additional accumulation of acetate by P. aeruginosa monocultures was unexpected, given the lack of pyruvate in MEM. However, as shown previously, P. aeruginosa can produce pyruvic acid as an organic acid by-product when grown in glucose-based minimal medium, in the absence of pyruvate (64). Perhaps, this glucose-derived pyruvate serves as the substrate for acetate production by P. aeruginosa.
Together, these results suggest interesting metabolic dynamics wherein as a monoculture, S. aureus primarily respires aerobically and produces acetate; however, in the presence of P. aeruginosa exoproducts, S. aureus is shifted to a fermentative metabolism and primarily produces lactate, which P. aeruginosa then preferentially consumes.
HQNO and P. aeruginosa siderophores are required for S. aureus killing by P. aeruginosa.
While RNA expression and HPLC analyses yielded great insight into the S. aureus response to the presence of P. aeruginosa under the conditions of our coculture model, it did not inform the mechanism(s) by which P. aeruginosa causes altered viability of S. aureus. To investigate the mechanism behind P. aeruginosa-mediated killing of S. aureus in this coculture assay, a targeted candidate gene approach was used. Previously, P. aeruginosa-produced HQNO was reported to inhibit normal growth of S. aureus (44, 48, 51). Additionally, coculture of P. aeruginosa with S. aureus shifts the gene expression of P. aeruginosa toward that of a less iron-starved profile in a rat peritoneal coculture model (43). Therefore, we targeted the Pseudomonas quinolone signal (PQS) (Fig. 4A) pathway and Pseudomonas iron acquisition via siderophore production in our initial experiments.
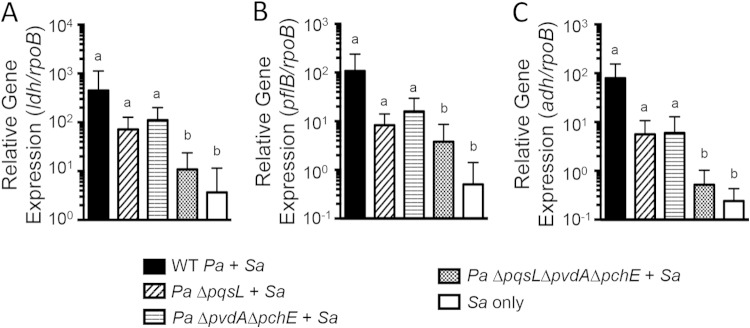
FIG 4.
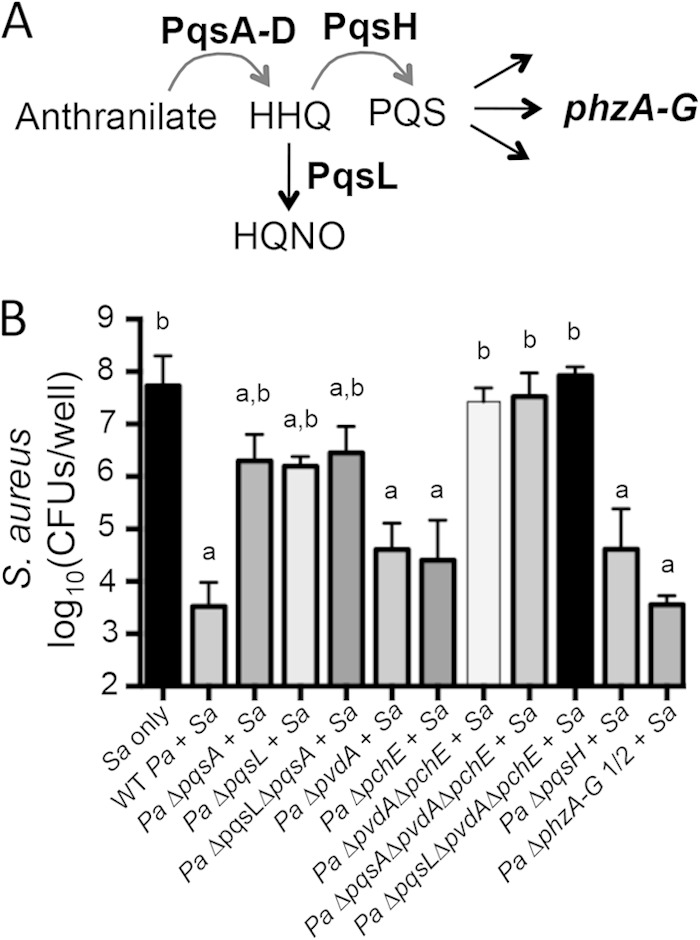
HQNO and siderophores produced by P. aeruginosa are required for late-stage killing of S. aureus in vitro. (A) Depiction of the PQS pathway in P. aeruginosa focusing on the genes/gene products analyzed in the coculture assays. (B) Coculture assays on a monolayer of epithelial cells were performed with S. aureus 8325-4 and P. aeruginosa PA14 WT or specified deletion mutant. S. aureus biofilm viability was measured at 16 h. Averages and errors (standard errors of the means) from three separate experiments, each run in triplicate, are shown. Differential survival of S. aureus in each coculture was compared to S. aureus monoculture using ANOVA and Dunnett's multiple-comparison posttest (a, P < 0.05). Differential survival of S. aureus monoculture or S. aureus in coculture with a P. aeruginosa mutant is compared to S. aureus coculture with WT P. aeruginosa also using ANOVA and Dunnett's multiple-comparison posttest (b, P < 0.05). Sa, S. aureus; Pa, P. aeruginosa.
P. aeruginosa PA14 strains with deletions in PQS pathway genes and/or genes required for siderophore biosynthesis were used in the airway epithelial cell and biofilm coculture assay. Similarly to wild-type (WT) P. aeruginosa PA14, survival and growth of mutant strains of P. aeruginosa PA14 with the targeted deletions were not altered by the presence of S. aureus 8325-4 (see Fig. S7 in the supplemental material). In contrast, S. aureus 8325-4 survival was differentially impacted by coculture with various mutant strains of P. aeruginosa PA14 (Fig. 4B). As seen previously, after 16 h in the CFBE coculture biofilm, WT P. aeruginosa PA14 kills S. aureus 8325-4, resulting in about a 10,000-fold reduction in S. aureus 8325-4 CFU/well compared to S. aureus 8325-4 monoculture (Fig. 4B). Coculture with single mutants incapable of synthesizing pyoverdine (ΔpvdA) or pyochelin (ΔpchE), the two major siderophores produced by P. aeruginosa PA14, trends toward decreased killing of S. aureus 8325-4 compared to coculture with WT P. aeruginosa PA14. However, coculture with a mutant P. aeruginosa strain that lacks synthesis of both siderophores, the ΔpvdA ΔpchE strain, markedly reduced P. aeruginosa-mediated killing of S. aureus 8325-4, allowing survival and growth of S. aureus to levels similar to those when this organism was grown in a monoculture. Interestingly, in a plastic-grown coculture biofilm, both siderophores are required for efficient P. aeruginosa-mediated killing of S. aureus 8325-4, as coculture with either single mutant, the ΔpvdA or ΔpchE strain, or the double mutant, the ΔpvdA ΔpchE strain, results in greatly reduced killing of S. aureus compared to WT P. aeruginosa PA14 coculture (see Fig. S8 in the supplemental material). Thus, to kill S. aureus 8325-4 most efficiently in both the CFBE- and plastic-grown coculture models, P. aeruginosa PA14 requires both major siderophores, pyoverdine and pyochelin.
Coculture on CFBE monolayers with the P. aeruginosa PA14 ΔpqsA mutant, which reduces all downstream products of the PQS biosynthetic pathway, similarly results in reduced S. aureus-killing (Fig. 4). Consistent with other previously reported coculture models of P. aeruginosa and S. aureus (51), HQNO plays an important role in S. aureus killing in our CFBE-coculture model; a P. aeruginosa PA14 ΔpqsL mutant, which does not produce HQNO (Fig. 4A), does not efficiently kill S. aureus 8325-4 (Fig. 4B). While S. aureus killing is significantly reduced in coculture with P. aeruginosa PA14 ΔpqsA or ΔpqsL mutants compared to WT P. aeruginosa PA14, S. aureus 8325-4 survival is not recovered to monoculture levels. Further, coculture with P. aeruginosa PA14 ΔpqsA ΔpqsL does not further reduce killing compared to either the P. aeruginosa PA14 ΔpqsA or the P. aeruginosa PA14 ΔpqsL single mutant. Similarly to the pattern observed in coculture with the siderophore mutants, the requirements for P. aeruginosa-mediated killing of S. aureus on plastic are more stringent than in the CFBE coculture model; that is, coculture with P. aeruginosa PA14 ΔpqsA, ΔpqsL, or ΔpqsA ΔpqsL mutants supports survival of S. aureus 8325-4, comparable to that of S. aureus 8325-4 monoculture (see Fig. S8 in the supplemental material). Interestingly, PQS plays a variable role in S. aureus 8325-4 killing in these coculture models. The P. aeruginosa PA14 ΔpqsH mutant, which does not produce PQS but instead accumulates its precursor 2-heptyl-4-quinolone (HHQ), trends toward a reduced ability to kill S. aureus 8325-4 in the CFBE coculture model, indicating a possible moderate impact of PQS or downstream signals in S. aureus killing (Fig. 4B). However, in our plastic-grown coculture model P. aeruginosa PA14 ΔpqsH trends toward more effective killing of S. aureus 8325-4 than that by WT P. aeruginosa PA14 (see Fig. S8 in the supplemental material). Together, our data are consistent with the model that it is the accumulation of HQNO that is primarily responsible for the role of the PQS pathway in P. aeruginosa-mediated killing of S. aureus.
Some redox-active phenazines produced by P. aeruginosa, like pyocyanin, inhibit respiration and have been reported to be antistaphylococcal (47, 65). In both our CFBE- and plastic-grown coculture models, P. aeruginosa PA14 ΔphzA-G1/2, a mutant lacking both phenazine biosynthetic clusters and thus unable to produce phenazines, including pyocyanin, still killed S. aureus 8325-4 to levels similar to that observed for the WT P. aeruginosa PA14 (Fig. 4B; see also Fig. S8 in the supplemental material). This result further indicates that HQNO is the primary effector in the PQS pathway required for S. aureus 8325-4-killing by P. aeruginosa PA14 in this coculture model. Finally, consistent with the requirement for both siderophores and HQNO for P. aeruginosa-mediated killing of S. aureus, a triple mutant of P. aeruginosa PA14, the ΔpqsA ΔpvdA ΔpchE or ΔpqsL ΔpvdA ΔpchE strain, results in comparable survival of S. aureus 8325-4 in coculture and monoculture on CFBE monolayers and plastic (Fig. 4; see also Fig. S8 in the supplemental material).
The concentration of iron in the MEM (0.055 μM) is similar to that detected in bronchoalveolar lavage fluid from healthy patients (<0.1 μM) (66). Patients with CF have increased iron concentrations in the lower respiratory tract. The concentration of lower airway total iron varies between patients but has been detected around 8 μM on average (67). Although total iron can be easily measured, the concentration of iron readily available for microbial use is less clear. Under coculture conditions supplemented with 8 μM ferric chloride (FeCl3), WT P. aeruginosa PA14 killed S. aureus 8325-4 more efficiently than in low-iron medium (see Fig. S9 in the supplemental material). Additionally, when supplemented with iron, P. aeruginosa PA14 ΔpvdA or ΔpchE single mutant strains did not experience a defect in S. aureus 8325-4 killing. P. aeruginosa PA14 did still require siderophores and HQNO for efficient killing of S. aureus 8325-4 under 8 μM iron conditions; however, loss of these components (as displayed through coculture with the ΔpvdA ΔpchE, ΔpqsL, or ΔpqsL ΔpvdA ΔpchE mutant of P. aeruginosa PA14) did not yield restoration of growth/survival of S. aureus 8325-4 to levels equivalent to those of monoculture. In the presence of additional iron, P. aeruginosa siderophores, in particular, played a reduced role in P. aeruginosa-mediated S. aureus killing compared to that under low-iron (unsupplemented) conditions. Further, the lack of complete recovery of S. aureus growth/survival in coculture with P. aeruginosa PA14 ΔpqsL ΔpvdA ΔpchE in 8 μM iron medium indicates a role for additional Pseudomonas aeruginosa factors or nutrient competition in mediating P. aeruginosa-S. aureus interbacterial interactions. Taken together, the production of HQNO and both siderophores is required for efficient killing of S. aureus by P. aeruginosa.
HQNO and P. aeruginosa siderophores induce mixed acid fermentation gene expression in S. aureus.
Based on the RNA expression results, HPLC metabolite quantification, and mutant P. aeruginosa coculture analyses, we hypothesized that molecules produced by P. aeruginosa lead to induction of fermentation pathway expression in S. aureus. To test this hypothesis, RNA expression of fermentation pathways was compared by qRT-PCR in S. aureus 8325-4 monoculture with that in S. aureus 8325-4–P. aeruginosa PA14 coculture biofilms on CFBE monolayers using key mutants identified from the late-stage killing phenotype (Fig. 5). Markers of gene expression of the three main fermentation pathways are the same as with those investigated in the plastic-grown coculture biofilms in Fig. 2: lactate fermentation (ldh, SAOUHSC_00206), formate fermentation (pflB, SAOUHSC_00187), and alcohol fermentation (adh, SAOUHSC_00113). Similarly to plastic-grown cocultures, S. aureus 8325-4 ldh expression was strongly induced when in coculture with WT P. aeruginosa PA14 on CFBE cells with >120-fold-increased expression compared to S. aureus 8325-4 monoculture (Fig. 5A). Interestingly, coculture with either the P. aeruginosa PA14 ΔpqsL strain, which does not produce HQNO, or the P. aeruginosa PA14 ΔpvdA ΔpchE double mutant, lacking two major siderophores, resulted in a trend toward decreased induction of ldh, compared to the coculture with WT P. aeruginosa PA14 (about 20-fold- and 30-fold-increased expression, respectively, compared to S. aureus 8325-4 monoculture). Coculture with the triple mutant P. aeruginosa PA14 ΔpqsL ΔpvdA ΔpchE, lacking HQNO and siderophore production, greatly decreased the induction of S. aureus 8325-4 ldh expression in coculture to a nonsignificant increase of about 3-fold compared to S. aureus 8325-4 monoculture. Thus, both HQNO and P. aeruginosa siderophores appear to contribute to the induction of the lactate fermentation pathway expression when S. aureus 8325-4 is in coculture with P. aeruginosa PA14 and do so in an additive manner. Similarly, induction of pflB and adh in S. aureus 8325-4 was dependent on the production of both HQNO and P. aeruginosa siderophores (Fig. 5B and C). This additive impact of P. aeruginosa-produced HQNO and siderophores on S. aureus fermentation gene induction in CFBE-grown coculture biofilms was also consistent in a second tested strain, S. aureus USA300 (see Fig. S10A to C in the supplemental material). Finally, similar to what was observed in CFBE-grown coculture biofilms, an additive role of HQNO and P. aeruginosa siderophores in the induction of S. aureus 8325-4 ldh, pflB, and adh expression was detected in coculture biofilms grown on plastic (see Fig. S10D to F in the supplemental material).
FIG 5.
P. aeruginosa-produced HQNO and siderophores are required to induce S. aureus fermentation gene expression. S. aureus 8325-4 biofilms were grown on CFBE monolayers. After 6 h, expression of marker genes from lactate (ldh) (A), formate (pflB) (B), and ethanol (adh) (C) fermentation pathways in S. aureus was measured using qRT-PCR and was normalized to the expression of an S. aureus control gene (rpoB). Relative fermentation gene expression in S. aureus was compared between S. aureus monoculture and coculture with WT or specified deletion mutants of P. aeruginosa PA14. Gene expression levels from 7 replicate experiments were compared using the Friedman rank test and Dunn's multiple-comparison posttest. Differential S. aureus gene expression when in coculture with P. aeruginosa was compared to that in S. aureus monoculture (a, P < 0.05). Differential S. aureus gene expression from S. aureus monoculture or S. aureus in coculture with a P. aeruginosa mutant was compared to that in S. aureus coculture with WT P. aeruginosa (b, P < 0.05). Error bars represent standard errors of the means. Sa, S. aureus; Pa, P. aeruginosa.
While HQNO and P. aeruginosa siderophores significantly contribute to the induction of fermentation gene expression, over replicate experiments, coculture of S. aureus 8325-4 with P. aeruginosa PA14 ΔpqsL ΔpvdA ΔpchE displayed a nonsignificant trend toward elevated expression of ldh, pflB, and adh compared to S. aureus 8325-4 monoculture. Thus, it is likely that other factors associated with coculture of S. aureus and P. aeruginosa contribute to S. aureus 8325-4 fermentation pathway induction. Namely, competition for oxygen likely makes the coculture environment less oxic as indicated by a >10-fold reduction in catalase expression by S. aureus 8325-4 when in coculture with P. aeruginosa PA14 and a >10-fold increase in expression of nrdG, a class III ribonucleotide reductase upregulated under anaerobic conditions (68) (see Table S5 in the supplemental material). Overall, P. aeruginosa production of HQNO and siderophores results in induction of lactate, formate, and alcohol fermentation pathway expression in S. aureus.
S. aureus small-colony variants are partially protected from P. aeruginosa killing.
Respiratory inhibitors like HQNO and pyocyanin were previously reported to select for S. aureus SCVs (50, 51). SCVs characteristically have a deficient electron transport chain and provide a survival mechanism that may help S. aureus persist in the presence of P. aeruginosa (50). We introduced S. aureus SCV strains with stable mutations in the electron transport chain into the coculture assay mixture on plastic to assess SCV susceptibility to P. aeruginosa-mediated killing of S. aureus in our biofilm model. Survival rates of S. aureus USA300 hemB and menD transposon mutants and an S. aureus Col hemB mutant, which generate SCVs, were compared to those of their respective WT parent strains in the presence of P. aeruginosa PA14 or in monoculture (Fig. 6). S. aureus USA300 hemB::Tn and menD::Tn mutants survived significantly better than WT S. aureus USA300 in the presence of P. aeruginosa PA14. However, protection from killing was partial as survival in coculture was 10- to 150-fold reduced compared to S. aureus monoculture. Interestingly, the S. aureus Col hemB mutant was completely protected from killing by P. aeruginosa PA14. Thus, growth as an SCV appears to provide a mechanism for S. aureus persistence in coculture; however, there is heterogeneity in the degree of protection from P. aeruginosa PA14 between S. aureus strains.
FIG 6.
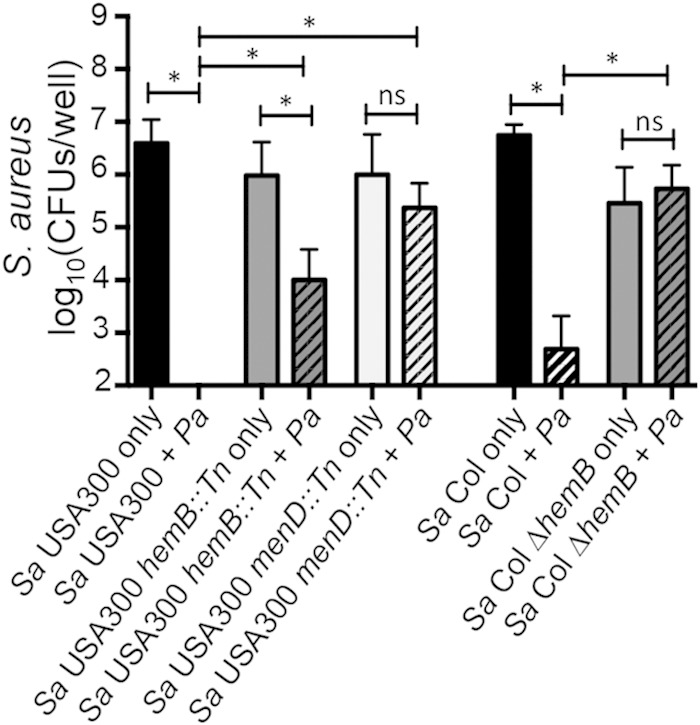
S. aureus SCV strains have increased resistance to killing by P. aeruginosa. Plastic-grown biofilm coculture assays were performed with P. aeruginosa PA14 and S. aureus USA300 and Col parent strains or genetic mutants with stable SCV phenotypes. S. aureus viability was measured at 16 h. Differential survival rates of S. aureus SCV strains were compared in coculture with P. aeruginosa versus monoculture and compared to parent S. aureus strain coculture viability using ANOVA and the Šidák multiple-comparison test for six replicate samples (*, P < 0.05; ns, not significant). Error bars represent standard errors of the means. Sa, S. aureus; Pa, P. aeruginosa.
DISCUSSION
It is well known in reports of the respiratory microbiota of patients with CF that Pseudomonas is a low-abundance, sometimes transient, organism during childhood. In pediatric patients, S. aureus is the primary pathogen, and it is not until late adolescence and adulthood that P. aeruginosa becomes a prevalent pathogen (20). Two main interaction phenotypes seen in these CF patients, the period of coexistence early in life and later-stage displacement of Staphylococcus by Pseudomonas, appear to be broadly modeled in our simplified, in vitro CFBE- and plastic-grown cocultures. While these in vitro coculture models clearly have limitations, they can be easily modified to address important questions pertaining to CF microbial interactions. Here, we showed the ability to study differential interactions as they occur over time in a single assay, including host cell colonization, biofilm formation, biofilm dispersion, and planktonic interactions. While not addressed here, we have also employed this coculture model to investigate the impacts of antibiotics and nutrient availability on interbacterial interactions, as well as the differential impact of multiple versus single bacterial species on host epithelial cell cytokine response (L. M. Filkins and G. A. O'Toole, unpublished data). Further, this simple model sets the foundation for building more complex in vitro polymicrobial communities with multiple bacterial species and additional host factors.
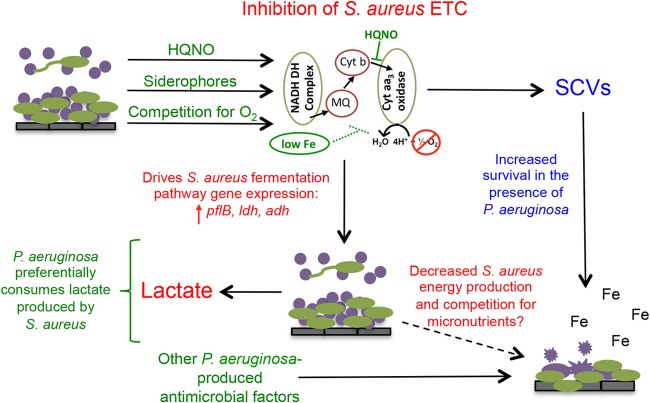
Using our CFBE-grown and plastic-grown coculture systems, we were able to further elucidate the multifactorial mechanism of S. aureus killing by P. aeruginosa, which we propose by the model presented in Fig. 7. We propose that P. aeruginosa secretes exoproducts, including siderophores and HQNO, and competes for oxygen, all of which negatively impact S. aureus growth and survival. Specifically, production of these exoproducts and oxygen utilization by P. aeruginosa inhibit the electron transport chain of S. aureus by sequestering iron that is a necessary heme cofactor of cytochromes, by inhibiting the oxidation of cytochrome b and reduction of cytochrome aa3 (65, 69), and by competing for the terminal electron acceptor. S. aureus responds to inhibition of the electron transport chain by increasing gene expression of fermentation pathways (formate, lactate, and ethanol). As a result, S. aureus metabolism is shifted away from acetate production and toward lactate fermentation. P. aeruginosa can preferentially consume S. aureus-produced lactate. We propose that over time the combination of a poisoned electron transport chain, and thus markedly slower growth (70, 71), competition for micronutrients, and additional antimicrobial products produced by P. aeruginosa leads to S. aureus cell death. As previously reported, P. aeruginosa-induced S. aureus cell lysis causes the release of iron, which can be further utilized by P. aeruginosa (43) and thus provide an additional competitive advantage to P. aeruginosa over S. aureus. The accompanying paper by Nguyen et al. (72) further explores the role of iron in the transition from S. aureus to P. aeruginosa predominance.
FIG 7.
P. aeruginosa-S. aureus interaction model. P. aeruginosa produces HQNO and siderophores and competes for oxygen which directly or indirectly inhibits the electron transport chain (ETC) of S. aureus. Inhibition of the electron transport chain induces fermentation gene expression in S. aureus and increases production of lactate. S. aureus-produced lactate is preferentially consumed by P. aeruginosa over medium-provided glucose. We propose that, over time, inhibition of the electron transport chain leads to decreased energy production in S. aureus, which in combination with competition for micronutrients and additional antimicrobial factors produced by P. aeruginosa leads to S. aureus killing. S. aureus lysis further benefits P. aeruginosa by providing an additional iron source (43). Selection of S. aureus SCVs in the presence of P. aeruginosa allows a subset of S. aureus cells to persist in the hostile coculture environment. NADH DH, NADH dehydrogenase; MQ, menaquinone; Cyt aa3, cytochrome aa3; Cyt b, cytochrome b.
The RNA expression analyses performed here strongly indicate a shift toward fermentative metabolism for S. aureus in the presence of P. aeruginosa. This observation was further confirmed by the accumulation of lactate and decrease in acetate accumulation in S. aureus culture supernatants in the presence of P. aeruginosa exoproducts. Although S. aureus was previously demonstrated to also produce ethanol and formate under anaerobic conditions (73), these additional metabolites were produced only to levels near the limit of detection under the culture conditions here, despite strong upregulation of genes from their respective fermentation pathway genes (adh and pflB). The gene expression and metabolite profile that we observed here when S. aureus was grown with P. aeruginosa are similar to those of an S. aureus Col hemB mutant, previously reported by Kohler et al. (61). In the Col hemB mutant, lactate was the primary fermentation metabolite detected, while no detectable formate accumulated and very low levels of ethanol were produced. Kohler et al. further determined that, although formate pyruvate lyase is expressed and translated at very high levels, the protein is inactive in the hemB mutant, explaining the lack of formate production. In our in vitro S. aureus-P. aeruginosa coculture model and sequential supernatant-treated monocultures, S. aureus metabolism mimics that seen in a stable, genetic mutant of the electron transport chain.
Although P. aeruginosa displaces S. aureus as the predominant organism and pathogen during adulthood, S. aureus remains a minor community member in some patients. Additionally, the prevalence of S. aureus SCVs correlates with worse disease state and increases with age (21, 74). The in vitro coculture interactions reported previously (50) and described here indicate that mutations in the electron transport chain (characteristic of S. aureus SCVs) partially protect S. aureus from P. aeruginosa killing (Fig. 7). Similarly, S. aureus SCVs may be selected for in respiratory communities where P. aeruginosa is abundant, contributing to their persistence in a hostile environment.
The precise mechanism of S. aureus cell death remains an open question. Based on the work here, it is evident that S. aureus cell death in this coculture model involves a shift to fermentation; however, S. aureus is equipped to grow under anaerobic conditions; thus, it is unlikely that the shift to lactate metabolism is the direct cause of death. Further, while P. aeruginosa exoproducts were sufficient to induce the shift in S. aureus metabolism, treating S. aureus monocultures with P. aeruginosa culture supernatants did not efficiently kill S. aureus and S. aureus actively metabolizes in the presence of P. aeruginosa supernatants (Fig. 3B and C). One possible indication from the requirement of viable P. aeruginosa for efficient killing of S. aureus is the role of oxygen consumption and competition in S. aureus killing. Some aerotolerant anaerobes can grow in oxic environments if cocultured with rapidly respiring aerobic organisms (75), such as P. aeruginosa. It is hypothesized that P. aeruginosa consumption of oxygen is rapid enough to maintain sufficiently low levels for the growth of such anaerobes. Thus, while competition for oxygen is evidently not sufficient to decrease S. aureus viability alone, in combination with HQNO and Pseudomonas aeruginosa siderophores, reduced oxygen availability may put S. aureus at a significant coculture disadvantage.
In addition to the proposed role of oxygen competition, there are several possible mechanisms by which the shift to lactate fermentation promotes S. aureus cell death in the presence of P. aeruginosa cells. One model is that reduced energy production by fermentation, compared to respiration, puts S. aureus at a competitive growth disadvantage in coculture. Alternatively, the switch of S. aureus to a fermentative lifestyle might increase S. aureus susceptibility to other antimicrobial factors produced by P. aeruginosa; however, the increased persistence of S. aureus SCVs, which also rely on fermentation for their primary energy generation, in coculture with P. aeruginosa argues against this simple model. Thus, the SCV phenotype may indicate a key role for S. aureus being properly adapted to a fermentative lifestyle before encountering P. aeruginosa in coculture in order for S. aureus to persist. Another possible model is that P. aeruginosa competes for/depletes a micronutrient required by S. aureus specifically under conditions of fermentative growth. Finally, S. aureus cell death may be impacted by pH balance, likely perturbed by increased lactate accumulation and accumulation of reactive oxygen species. At this time, it is not possible to distinguish among these (or alternative) models for P. aeruginosa-mediated killing of S. aureus. The difficulty of determining a precise mechanism of killing is not an unprecedented challenge. Analogous to our question of P. aeruginosa-mediated killing, the mechanism of antibiotic-mediated killing has been poorly understood for years and was recently brought back into focus for continued debate (76–79).
The respiratory microbial communities of CF patients are complex and dynamic throughout childhood and adolescence. Despite a strong focus on P. aeruginosa and S. aureus in CF lung infections, the driving factors that result in P. aeruginosa predominance during adulthood are not well understood. Here, we determined several possible mechanisms by which P. aeruginosa may displace S. aureus in respiratory communities. However, the immune response, host factors, and the other microbial community members undeniably also impact the transition from S. aureus to P. aeruginosa being the predominant pathogen in CF and the associated impact on patient outcome.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dianne Newman, Ambrose Cheung, and Deborah Hogan for generously providing bacterial strains. We also thank Thomas Hampton for advice on statistical analyses.
This work was supported by NIH grant R01 2 R37 AI83256-06 to G.A.O. and a Renal Function and Disease Training Grant fellowship (T32 DK007301) to L.M.F. The cell biology studies utilized the resources of the Host Pathogen Interaction Core, supported by the National Institute of General Medical Sciences of the NIH under award P20-GM103413 and the Cystic Fibrosis Research Development Program (STANTO07R0).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00059-15.
REFERENCES
- 1.Madan JC, Koestler DC, Stanton BA, Davidson L, Moulton LA, Housman ML, Moore JH, Guill MF, Morrison HG, Sogin ML, Hampton TH, Karagas MR, Palumbo PE, Foster JA, Hibberd PL, O'Toole GA. 2012. Serial analysis of the gut and respiratory microbiome in cystic fibrosis in infancy: interaction between intestinal and respiratory tracts and impact of nutritional exposures. mBio 3(4):e00251-12. doi: 10.1128/mBio.00251-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Delhaes L, Monchy S, Frealle E, Hubans C, Salleron J, Leroy S, Prevotat A, Wallet F, Wallaert B, Dei-Cas E, Sime-Ngando T, Chabe M, Viscogliosi E. 2012. The airway microbiota in cystic fibrosis: a complex fungal and bacterial community—implications for therapeutic management. PLoS One 7:e36313. doi: 10.1371/journal.pone.0036313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Filkins LM, Hampton TH, Gifford AH, Gross MJ, Hogan DA, Sogin ML, Morrison HG, Paster BJ, O'Toole GA. 2012. Prevalence of streptococci and increased polymicrobial diversity associated with cystic fibrosis patient stability. J Bacteriol 194:4709–4717. doi: 10.1128/JB.00566-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rogers GB, Hart CA, Mason JR, Hughes M, Walshaw MJ, Bruce KD. 2003. Bacterial diversity in cases of lung infection in cystic fibrosis patients: 16S ribosomal DNA (rDNA) length heterogeneity PCR and 16S rDNA terminal restriction fragment length polymorphism profiling. J Clin Microbiol 41:3548–3558. doi: 10.1128/JCM.41.8.3548-3558.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Willner D, Furlan M, Haynes M, Schmieder R, Angly FE, Silva J, Tammadoni S, Nosrat B, Conrad D, Rohwer F. 2009. Metagenomic analysis of respiratory tract DNA viral communities in cystic fibrosis and non-cystic fibrosis individuals. PLoS One 4:e7370. doi: 10.1371/journal.pone.0007370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zemanick ET, Harris JK, Wagner BD, Robertson CE, Sagel SD, Stevens MJ, Accurso FJ, Laguna TA. 2013. Inflammation and airway microbiota during cystic fibrosis pulmonary exacerbations. PLoS One 8:e62917. doi: 10.1371/journal.pone.0062917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao J, Schloss PD, Kalikin LM, Carmody LA, Foster BK, Petrosino JF, Cavalcoli JD, VanDevanter DR, Murray S, Li JZ, Young VB, LiPuma JJ. 2012. Decade-long bacterial community dynamics in cystic fibrosis airways. Proc Natl Acad Sci U S A 109:5809–5814. doi: 10.1073/pnas.1120577109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Willger SD, Grim SL, Dolben EL, Shipunova A, Hampton TH, Morrison HG, Filkins LM, O'Toole GA, Moulton LA, Ashare A, Sogin ML, Hogan DA. 2014. Characterization and quantification of the fungal microbiome in serial samples from individuals with cystic fibrosis. Microbiome 2:40. doi: 10.1186/2049-2618-2-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boucher RC. 2007. Airway surface dehydration in cystic fibrosis: pathogenesis and therapy. Annu Rev Med 58:157–170. doi: 10.1146/annurev.med.58.071905.105316. [DOI] [PubMed] [Google Scholar]
- 10.Matsui H, Grubb BR, Tarran R, Randell SH, Gatzy JT, Davis CW, Boucher RC. 1998. Evidence for periciliary liquid layer depletion, not abnormal ion composition, in the pathogenesis of cystic fibrosis airways disease. Cell 95:1005–1015. doi: 10.1016/S0092-8674(00)81724-9. [DOI] [PubMed] [Google Scholar]
- 11.Matsui H, Wagner VE, Hill DB, Schwab UE, Rogers TD, Button B, Taylor RM II, Superfine R, Rubinstein M, Iglewski BH, Boucher RC. 2006. A physical linkage between cystic fibrosis airway surface dehydration and Pseudomonas aeruginosa biofilms. Proc Natl Acad Sci U S A 103:18131–18136. doi: 10.1073/pnas.0606428103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cox MJ, Allgaier M, Taylor B, Baek MS, Huang YJ, Daly RA, Karaoz U, Andersen GL, Brown R, Fujimura KE, Wu B, Tran D, Koff J, Kleinhenz ME, Nielson D, Brodie EL, Lynch SV. 2010. Airway microbiota and pathogen abundance in age-stratified cystic fibrosis patients. PLoS One 5:e11044. doi: 10.1371/journal.pone.0011044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fodor AA, Klem ER, Gilpin DF, Elborn JS, Boucher RC, Tunney MM, Wolfgang MC. 2012. The adult cystic fibrosis airway microbiota is stable over time and infection type, and highly resilient to antibiotic treatment of exacerbations. PLoS One 7:e45001. doi: 10.1371/journal.pone.0045001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Price KE, Hampton TH, Gifford AH, Dolben EL, Hogan DA, Morrison HG, Sogin ML, O'Toole GA. 2013. Unique microbial communities persist in individual cystic fibrosis patients throughout a clinical exacerbation. Microbiome 1:27. doi: 10.1186/2049-2618-1-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stressmann FA, Rogers GB, van der Gast CJ, Marsh P, Vermeer LS, Carroll MP, Hoffman L, Daniels TW, Patel N, Forbes B, Bruce KD. 2012. Long-term cultivation-independent microbial diversity analysis demonstrates that bacterial communities infecting the adult cystic fibrosis lung show stability and resilience. Thorax 67:867–873. doi: 10.1136/thoraxjnl-2011-200932. [DOI] [PubMed] [Google Scholar]
- 16.van der Gast CJ, Walker AW, Stressmann FA, Rogers GB, Scott P, Daniels TW, Carroll MP, Parkhill J, Bruce KD. 2011. Partitioning core and satellite taxa from within cystic fibrosis lung bacterial communities. ISME J 5:780–791. doi: 10.1038/ismej.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mahenthiralingam E. 2014. Emerging cystic fibrosis pathogens and the microbiome. Paediatr Respir Rev doi: 10.1016/j.prrv.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 18.Hill UG, Floto RA, Haworth CS. 2012. Non-tuberculous mycobacteria in cystic fibrosis. J R Soc Med 105(Suppl 2):S14–S18. doi: 10.1258/jrsm.2012.12s003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sibley CD, Parkins MD, Rabin HR, Duan K, Norgaard JC, Surette MG. 2008. A polymicrobial perspective of pulmonary infections exposes an enigmatic pathogen in cystic fibrosis patients. Proc Natl Acad Sci U S A 105:15070–15075. doi: 10.1073/pnas.0804326105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cystic Fibrosis Foundation. 2011. Cystic Fibrosis Foundation patient registry 2010 annual data report. Cystic Fibrosis Foundation, Bethesda, MD. [Google Scholar]
- 21.Wolter DJ, Emerson JC, McNamara S, Buccat AM, Qin X, Cochrane E, Houston LS, Rogers GB, Marsh P, Prehar K, Pope CE, Blackledge M, Deziel E, Bruce KD, Ramsey BW, Gibson RL, Burns JL, Hoffman LR. 2013. Staphylococcus aureus small-colony variants are independently associated with worse lung disease in children with cystic fibrosis. Clin Infect Dis 57:384–391. doi: 10.1093/cid/cit270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong JK, Ranganathan SC, Hart E. 2013. Staphylococcus aureus in early cystic fibrosis lung disease. Pediatr Pulmonol 48:1151–1159. doi: 10.1002/ppul.22863. [DOI] [PubMed] [Google Scholar]
- 23.Hirschhausen N, Block D, Bianconi I, Bragonzi A, Birtel J, Lee JC, Dubbers A, Kuster P, Kahl J, Peters G, Kahl BC. 2013. Extended Staphylococcus aureus persistence in cystic fibrosis is associated with bacterial adaptation. Int J Med Microbiol 303:685–692. doi: 10.1016/j.ijmm.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 24.McNamara PJ, Proctor RA. 2000. Staphylococcus aureus small colony variants, electron transport and persistent infections. Int J Antimicrob Agents 14:117–122. doi: 10.1016/S0924-8579(99)00170-3. [DOI] [PubMed] [Google Scholar]
- 25.Com G, Carroll JL, Castro MM, Tang X, Jambhekar S, Berlinski A. 2014. Predictors and outcome of low initial forced expiratory volume in 1 second measurement in children with cystic fibrosis. J Pediatr 164:832–838. doi: 10.1016/j.jpeds.2013.11.064. [DOI] [PubMed] [Google Scholar]
- 26.Sagel SD, Gibson RL, Emerson J, McNamara S, Burns JL, Wagener JS, Ramsey BW. 2009. Impact of Pseudomonas and Staphylococcus infection on inflammation and clinical status in young children with cystic fibrosis. J Pediatr 154:183–188. doi: 10.1016/j.jpeds.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emerson J, Rosenfeld M, McNamara S, Ramsey B, Gibson RL. 2002. Pseudomonas aeruginosa and other predictors of mortality and morbidity in young children with cystic fibrosis. Pediatr Pulmonol 34:91–100. doi: 10.1002/ppul.10127. [DOI] [PubMed] [Google Scholar]
- 28.Zemanick ET, Emerson J, Thompson V, McNamara S, Morgan W, Gibson RL, Rosenfeld M. 2014. Clinical outcomes after initial Pseudomonas acquisition in cystic fibrosis. Pediatr Pulmonol doi: 10.1002/ppul.23036. [DOI] [PubMed] [Google Scholar]
- 29.Manos J, Hu H, Rose BR, Wainwright CE, Zablotska IB, Cheney J, Turnbull L, Whitchurch CB, Grimwood K, Harmer C, Anuj SN, Harbour C. 2013. Virulence factor expression patterns in Pseudomonas aeruginosa strains from infants with cystic fibrosis. Eur J Clin Microbiol Infect Dis 32:1583–1592. doi: 10.1007/s10096-013-1916-7. [DOI] [PubMed] [Google Scholar]
- 30.Deretic V, Govan JR, Konyecsni WM, Martin DW. 1990. Mucoid Pseudomonas aeruginosa in cystic fibrosis: mutations in the muc loci affect transcription of the algR and algD genes in response to environmental stimuli. Mol Microbiol 4:189–196. doi: 10.1111/j.1365-2958.1990.tb00586.x. [DOI] [PubMed] [Google Scholar]
- 31.Govan JR, Fyfe JA. 1978. Mucoid Pseudomonas aeruginosa and cystic fibrosis: resistance of the mucoid from to carbenicillin, flucloxacillin and tobramycin and the isolation of mucoid variants in vitro. J Antimicrob Chemother 4:233–240. doi: 10.1093/jac/4.3.233. [DOI] [PubMed] [Google Scholar]
- 32.Macia MD, Blanquer D, Togores B, Sauleda J, Perez JL, Oliver A. 2005. Hypermutation is a key factor in development of multiple-antimicrobial resistance in Pseudomonas aeruginosa strains causing chronic lung infections. Antimicrob Agents Chemother 49:3382–3386. doi: 10.1128/AAC.49.8.3382-3386.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oliver A, Mena A. 2010. Bacterial hypermutation in cystic fibrosis, not only for antibiotic resistance. Clin Microbiol Infect 16:798–808. doi: 10.1111/j.1469-0691.2010.03250.x. [DOI] [PubMed] [Google Scholar]
- 34.Harmer C, Alnassafi K, Hu H, Elkins M, Bye P, Rose B, Cordwell S, Triccas JA, Harbour C, Manos J. 2013. Modulation of gene expression by Pseudomonas aeruginosa during chronic infection in the adult cystic fibrosis lung. Microbiology 159:2354–2363. doi: 10.1099/mic.0.066985-0. [DOI] [PubMed] [Google Scholar]
- 35.Rajan S, Saiman L. 2002. Pulmonary infections in patients with cystic fibrosis. Semin Respir Infect 17:47–56. doi: 10.1053/srin.2002.31690. [DOI] [PubMed] [Google Scholar]
- 36.Haley CL, Colmer-Hamood JA, Hamood AN. 2012. Characterization of biofilm-like structures formed by Pseudomonas aeruginosa in a synthetic mucus medium. BMC Microbiol 12:181. doi: 10.1186/1471-2180-12-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kluge S, Hoffmann M, Benndorf D, Rapp E, Reichl U. 2012. Proteomic tracking and analysis of a bacterial mixed culture. Proteomics 12:1893–1901. doi: 10.1002/pmic.201100362. [DOI] [PubMed] [Google Scholar]
- 38.Ruger M, Ackermann M, Reichl U. 2014. Species-specific viability analysis of Pseudomonas aeruginosa, Burkholderia cepacia and Staphylococcus aureus in mixed culture by flow cytometry. BMC Microbiol 14:56. doi: 10.1186/1471-2180-14-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang L, Liu Y, Markussen T, Hoiby N, Tolker-Nielsen T, Molin S. 2011. Pattern differentiation in co-culture biofilms formed by Staphylococcus aureus and Pseudomonas aeruginosa. FEMS Immunol Med Microbiol 62:339–347. doi: 10.1111/j.1574-695X.2011.00820.x. [DOI] [PubMed] [Google Scholar]
- 40.Baldan R, Cigana C, Testa F, Bianconi I, De Simone M, Pellin D, Di Serio C, Bragonzi A, Cirillo DM. 2014. Adaptation of Pseudomonas aeruginosa in cystic fibrosis airways influences virulence of Staphylococcus aureus in vitro and murine models of co-infection. PLoS One 9:e89614. doi: 10.1371/journal.pone.0089614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fugere A, Lalonde Seguin D, Mitchell G, Deziel E, Dekimpe V, Cantin AM, Frost E, Malouin F. 2014. Interspecific small molecule interactions between clinical isolates of Pseudomonas aeruginosa and Staphylococcus aureus from adult cystic fibrosis patients. PLoS One 9:e86705. doi: 10.1371/journal.pone.0086705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park JH, Lee JH, Cho MH, Herzberg M, Lee J. 2012. Acceleration of protease effect on Staphylococcus aureus biofilm dispersal. FEMS Microbiol Lett 335:31–38. doi: 10.1111/j.1574-6968.2012.02635.x. [DOI] [PubMed] [Google Scholar]
- 43.Mashburn LM, Jett AM, Akins DR, Whiteley M. 2005. Staphylococcus aureus serves as an iron source for Pseudomonas aeruginosa during in vivo coculture. J Bacteriol 187:554–566. doi: 10.1128/JB.187.2.554-566.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mitchell G, Seguin DL, Asselin AE, Deziel E, Cantin AM, Frost EH, Michaud S, Malouin F. 2010. Staphylococcus aureus sigma B-dependent emergence of small-colony variants and biofilm production following exposure to Pseudomonas aeruginosa 4-hydroxy-2-heptylquinoline-N-oxide. BMC Microbiol 10:33. doi: 10.1186/1471-2180-10-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kessler E, Safrin M, Olson JC, Ohman DE. 1993. Secreted LasA of Pseudomonas aeruginosa is a staphylolytic protease. J Biol Chem 268:7503–7508. [PubMed] [Google Scholar]
- 46.Castric PA. 1975. Hydrogen cyanide, a secondary metabolite of Pseudomonas aeruginosa. Can J Microbiol 21:613–618. doi: 10.1139/m75-088. [DOI] [PubMed] [Google Scholar]
- 47.Hassan HM, Fridovich I. 1980. Mechanism of the antibiotic action pyocyanine. J Bacteriol 141:156–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Machan ZA, Taylor GW, Pitt TL, Cole PJ, Wilson R. 1992. 2-Heptyl-4-hydroxyquinoline N-oxide, an antistaphylococcal agent produced by Pseudomonas aeruginosa. J Antimicrob Chemother 30:615–623. doi: 10.1093/jac/30.5.615. [DOI] [PubMed] [Google Scholar]
- 49.Korgaonkar A, Trivedi U, Rumbaugh KP, Whiteley M. 2013. Community surveillance enhances Pseudomonas aeruginosa virulence during polymicrobial infection. Proc Natl Acad Sci U S A 110:1059–1064. doi: 10.1073/pnas.1214550110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Biswas L, Biswas R, Schlag M, Bertram R, Gotz F. 2009. Small-colony variant selection as a survival strategy for Staphylococcus aureus in the presence of Pseudomonas aeruginosa. Appl Environ Microbiol 75:6910–6912. doi: 10.1128/AEM.01211-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hoffman LR, Deziel E, D'Argenio DA, Lepine F, Emerson J, McNamara S, Gibson RL, Ramsey BW, Miller SI. 2006. Selection for Staphylococcus aureus small-colony variants due to growth in the presence of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 103:19890–19895. doi: 10.1073/pnas.0606756104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pernet E, Guillemot L, Burgel PR, Martin C, Lambeau G, Sermet-Gaudelus I, Sands D, Leduc D, Morand PC, Jeammet L, Chignard M, Wu Y, Touqui L. 2014. Pseudomonas aeruginosa eradicates Staphylococcus aureus by manipulating the host immunity. Nat Commun 5:5105. doi: 10.1038/ncomms6105. [DOI] [PubMed] [Google Scholar]
- 53.Anderson GG, Moreau-Marquis S, Stanton BA, O'Toole GA. 2008. In vitro analysis of tobramycin-treated Pseudomonas aeruginosa biofilms on cystic fibrosis-derived airway epithelial cells. Infect Immun 76:1423–1433. doi: 10.1128/IAI.01373-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moreau-Marquis S, Redelman CV, Stanton BA, Anderson GG. 2010. Co-culture models of Pseudomonas aeruginosa biofilms grown on live human airway cells. J Vis Exp 2010(44):2186. doi: 10.3791/2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sihto HM, Tasara T, Stephan R, Johler S. 2014. Validation of reference genes for normalization of qPCR mRNA expression levels in Staphylococcus aureus exposed to osmotic and lactic acid stress conditions encountered during food production and preservation. FEMS Microbiol Lett 356:134–140. doi: 10.1111/1574-6968.12491. [DOI] [PubMed] [Google Scholar]
- 57.Zhang YH, Lynd LR. 2005. Regulation of cellulase synthesis in batch and continuous cultures of Clostridium thermocellum. J Bacteriol 187:99–106. doi: 10.1128/JB.187.1.99-106.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hampton TH, Green DM, Cutting GR, Morrison HG, Sogin ML, Gifford AH, Stanton BA, O'Toole GA. 2014. The microbiome in pediatric cystic fibrosis patients: the role of shared environment suggests a window of intervention. Microbiome 2:14. doi: 10.1186/2049-2618-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gardner JF, Lascelles J. 1962. The requirement for acetate of a streptomycin-resistant strain of Staphylococcus aureus. J Gen Microbiol 29:157–164. doi: 10.1099/00221287-29-1-157. [DOI] [PubMed] [Google Scholar]
- 60.Strasters KC, Winkler KC. 1963. Carbohydrate metabolism of Staphylococcus aureus. J Gen Microbiol 33:213–229. doi: 10.1099/00221287-33-2-213. [DOI] [PubMed] [Google Scholar]
- 61.Kohler C, von Eiff C, Peters G, Proctor RA, Hecker M, Engelmann S. 2003. Physiological characterization of a heme-deficient mutant of Staphylococcus aureus by a proteomic approach. J Bacteriol 185:6928–6937. doi: 10.1128/JB.185.23.6928-6937.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hunt JC, Phibbs PV Jr. 1983. Regulation of alternate peripheral pathways of glucose catabolism during aerobic and anaerobic growth of Pseudomonas aeruginosa. J Bacteriol 154:793–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van der Kooij D, Oranje JP, Hijnen WA. 1982. Growth of Pseudomonas aeruginosa in tap water in relation to utilization of substrates at concentrations of a few micrograms per liter. Appl Environ Microbiol 44:1086–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Buch A, Archana G, Naresh Kumar G. 2008. Metabolic channeling of glucose towards gluconate in phosphate-solubilizing Pseudomonas aeruginosa P4 under phosphorus deficiency. Res Microbiol 159:635–642. doi: 10.1016/j.resmic.2008.09.012. [DOI] [PubMed] [Google Scholar]
- 65.Voggu L, Schlag S, Biswas R, Rosenstein R, Rausch C, Gotz F. 2006. Microevolution of cytochrome bd oxidase in Staphylococci and its implication in resistance to respiratory toxins released by Pseudomonas. J Bacteriol 188:8079–8086. doi: 10.1128/JB.00858-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moreau-Marquis S, Bomberger JM, Anderson GG, Swiatecka-Urban A, Ye S, O'Toole GA, Stanton BA. 2008. The DeltaF508-CFTR mutation results in increased biofilm formation by Pseudomonas aeruginosa by increasing iron availability. Am J Physiol Lung Cell Mol Physiol 295:L25–L37. doi: 10.1152/ajplung.00391.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stites SW, Plautz MW, Bailey K, O'Brien-Ladner AR, Wesselius LJ. 1999. Increased concentrations of iron and isoferritins in the lower respiratory tract of patients with stable cystic fibrosis. Am J Respir Crit Care Med 160:796–801. doi: 10.1164/ajrccm.160.3.9811018. [DOI] [PubMed] [Google Scholar]
- 68.Masalha M, Borovok I, Schreiber R, Aharonowitz Y, Cohen G. 2001. Analysis of transcription of the Staphylococcus aureus aerobic class Ib and anaerobic class III ribonucleotide reductase genes in response to oxygen. J Bacteriol 183:7260–7272. doi: 10.1128/JB.183.24.7260-7272.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kogut M, Lightbown JW. 1962. Selective inhibition by 2-heptyl-4-hydroxyquinoline N-oxide of certain oxidation-reduction reactions. Biochem J 84:368–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Belay N, Rasooly A. 2002. Staphylococcus aureus growth and enterotoxin A production in an anaerobic environment. J Food Prot 65:199–204. [DOI] [PubMed] [Google Scholar]
- 71.Ferreira MT, Manso AS, Gaspar P, Pinho MG, Neves AR. 2013. Effect of oxygen on glucose metabolism: utilization of lactate in Staphylococcus aureus as revealed by in vivo NMR studies. PLoS One 8:e58277. doi: 10.1371/journal.pone.0058277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nguyen AT, Jones JW, Ruge MA, Kane MA, Oglesby-Sherrouse AG. 2015. Iron depletion enhances production of antimicrobials by Pseudomonas aeruginosa. J Bacteriol 197:2265–2275. doi: 10.1128/JB.00072-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sun JL, Zhang SK, Chen JY, Han BZ. 2012. Metabolic profiling of Staphylococcus aureus cultivated under aerobic and anaerobic conditions with (1)H NMR-based nontargeted analysis. Can J Microbiol 58:709–718. doi: 10.1139/w2012-046. [DOI] [PubMed] [Google Scholar]
- 74.Besier S, Smaczny C, von Mallinckrodt C, Krahl A, Ackermann H, Brade V, Wichelhaus TA. 2007. Prevalence and clinical significance of Staphylococcus aureus small-colony variants in cystic fibrosis lung disease. J Clin Microbiol 45:168–172. doi: 10.1128/JCM.01510-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gerritse J, Schut F, Gottschal JC. 1992. Modelling of mixed chemostat cultures of an aerobic bacterium, Comamonas testosteroni, and an anaerobic bacterium, Veillonella alcalescens: comparison with experimental data. Appl Environ Microbiol 58:1466–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 77.Liu Y, Imlay JA. 2013. Cell death from antibiotics without the involvement of reactive oxygen species. Science 339:1210–1213. doi: 10.1126/science.1232751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Keren I, Wu Y, Inocencio J, Mulcahy LR, Lewis K. 2013. Killing by bactericidal antibiotics does not depend on reactive oxygen species. Science 339:1213–1216. doi: 10.1126/science.1232688. [DOI] [PubMed] [Google Scholar]
- 79.Ezraty B, Vergnes A, Banzhaf M, Duverger Y, Huguenot A, Brochado AR, Su SY, Espinosa L, Loiseau L, Py B, Typas A, Barras F. 2013. Fe-S cluster biosynthesis controls uptake of aminoglycosides in a ROS-less death pathway. Science 340:1583–1587. doi: 10.1126/science.1238328. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.