ABSTRACT
Cystic fibrosis (CF) is a heritable disease characterized by chronic, polymicrobial lung infections. While Staphylococcus aureus is the dominant lung pathogen in young CF patients, Pseudomonas aeruginosa becomes predominant by adulthood. P. aeruginosa produces a variety of antimicrobials that likely contribute to this shift in microbial populations. In particular, secretion of 2-alkyl-4(1H)-quinolones (AQs) contributes to lysis of S. aureus in coculture, providing an iron source to P. aeruginosa both in vitro and in vivo. We previously showed that production of one such AQ, the Pseudomonas quinolone signal (PQS), is enhanced by iron depletion and that this induction is dependent upon the iron-responsive PrrF small RNAs (sRNAs). Here, we demonstrate that antimicrobial activity against S. aureus during coculture is also enhanced by iron depletion, and we provide evidence that multiple AQs contribute to this activity. Strikingly, a P. aeruginosa ΔprrF mutant, which produces very little PQS in monoculture, was capable of mediating iron-regulated growth suppression of S. aureus. We show that the presence of S. aureus suppresses the ΔprrF1,2 mutant's defect in iron-regulated PQS production, indicating that a PrrF-independent iron regulatory pathway mediates AQ production in coculture. We further demonstrate that iron-regulated antimicrobial production is conserved in multiple P. aeruginosa strains, including clinical isolates from CF patients. These results demonstrate that iron plays a central role in modulating interactions of P. aeruginosa with S. aureus. Moreover, our studies suggest that established iron regulatory pathways of these pathogens are significantly altered during polymicrobial infections.
IMPORTANCE Chronic polymicrobial infections involving Pseudomonas aeruginosa and Staphylococcus aureus are a significant cause of morbidity and mortality, as the interplay between these two organisms exacerbates infection. This is in part due to enhanced production of antimicrobial metabolites by P. aeruginosa when these two species are cocultured. Using both established and newly developed coculture techniques, this report demonstrates that iron depletion increases P. aeruginosa's ability to suppress growth of S. aureus. These findings present a novel role for iron in modulating microbial interaction and provide the basis for understanding how essential nutrients drive polymicrobial infections.
INTRODUCTION
Pseudomonas aeruginosa is a Gram-negative opportunistic pathogen that causes a variety of life-threatening infections. Many of these infections are polymicrobial, including those in diabetic foot ulcers (1, 2) and chronic lung infections in cystic fibrosis (CF) patients (3, 4). Several studies have noted that polymicrobial infections with P. aeruginosa are exacerbated compared to monoinfection (2, 5–10). Thus, defining the mechanisms that mediate these microbial interactions is paramount to understanding the pathogenesis of polymicrobial infections.
P. aeruginosa requires iron for growth and virulence (11–14). However, iron is a limiting nutrient in the environment and in the host (15, 16). To acquire this valuable nutrient, P. aeruginosa encodes several iron uptake systems. Among these are the production and secretion of siderophores, such as pyoverdine and pyochelin (17), which chelate ferric iron at high affinities and deliver it to the cell. Additionally, P. aeruginosa encodes a ferrous iron uptake system (Feo) to allow for import of soluble ferrous iron in oxygen-depleted environments (18). Finally, P. aeruginosa encodes two heme uptake systems, Has and Phu, which transport and degrade heme, releasing iron for use inside the cell (19).
Iron is also toxic at high concentrations; thus, P. aeruginosa regulates the expression of iron uptake systems in response to intracellular iron concentrations. In the presence of iron, genes for iron acquisition are repressed via the ferric uptake regulator (Fur) protein, which is essential in P. aeruginosa (20–22). The Fur protein also represses the transcription of the PrrF small RNAs (sRNAs), which regulate the expression of many iron-containing and iron storage proteins (23) and are required for virulence in an acute murine lung infection (24). Included in the PrrF regulon are genes encoding anthranilate (ANT) degradation enzymes (AntA and CatBCA), which allow catabolism of this metabolite by the tricarboxylic acid (TCA) cycle (25). Anthranilate also serves as a precursor for the Pseudomonas quinolone signal (PQS), a quorum-sensing molecule and virulence trait (26, 27). Thus, regulation by the PrrF sRNAs promotes production of PQS (25, 28).
PQS production is initiated by PqsA, a coenzyme ligase that converts anthranilate to anthraniloyl coenzyme A (anthraniloyl-CoA) (29). PqsA was previously reported to be required for lysis of Staphylococcus aureus, resulting in increased iron availability to P. aeruginosa (30). The specific mechanism for this activity remains unclear and is likely to be multifactorial, as anthraniloyl-CoA can serve as the precursor for at least 55 unique alkylquinolone (AQ) molecules (29, 31). One of these, 2-heptyl-4-hydroxyquinoline N-oxide (HQNO), is an N-oxide ubiquinone that inhibits growth of Gram-positive bacteria, including S. aureus (32–36). Additionally, PQS stimulates the production of redox active phenazines, which may contribute to iron acquisition due to (i) their ability to lyse bacterial and mammalian cells (37), thus liberating intracellular iron stores, and (ii) redox cycling, which reduces insoluble ferric iron to its more bioavailable, ferrous form (38). PQS has also been shown to possess iron-chelating activity, which may provide a competitive advantage in complex microbial environments (39, 40). However, PQS is not a siderophore, in that it cannot deliver iron to the cytosol of P. aeruginosa (39, 40). Instead, previous studies have hypothesized that PQS serves as an iron trap at the surface of the cell (39). The precursor to PQS, 4-hydroxy-2-heptylquinoline (HHQ), shares many of the same signaling activities as PQS, including the induction of phenazine production (36, 39). However, HHQ does not possess the 3-hydroxyl group of PQS and is therefore incapable of chelating ferric iron (40).
While several of the studies cited above provide a link between iron homeostasis and antimicrobial activity in P. aeruginosa, no studies have yet investigated the impact of iron availability on polymicrobial interactions involving P. aeruginosa. In this study, we demonstrate that iron depletion enhances antimicrobial activity of P. aeruginosa when grown in coculture with S. aureus. While this phenomenon is dependent upon PqsA, we found that this activity is independent of the PrrF sRNAs, indicating the presence of a distinct iron regulatory pathway that controls PQS production. We further show that this activity is conserved in multiple strains of P. aeruginosa, including two cystic fibrosis isolates. These studies demonstrate that iron availability impacts polymicrobial interactions of P. aeruginosa with S. aureus, presenting additional implications for the role of AQs in iron acquisition and pathogenesis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains used in this work are listed in Table 1. The pqs mutants were generated in our laboratory's PAO1 strain by allelic exchange (41) using the previously described ΔpqsE (42) and ΔpqsL (43) deletion constructs. Escherichia coli strains were routinely grown in L broth or on L agar plates, and P. aeruginosa strains were maintained in brain heart infusion (BHI) broth or on BHI agar plates. For iron-depleted medium, Chelex-treated, dialyzed Trypticase soy broth (DTSB) was prepared as previously described (44). Ferric chloride (FeCl3) was added to DTSB at a final concentration of 100 μM for iron-replete conditions.
TABLE 1.
Strains used in this study
| Strain | Description | Reference or source |
|---|---|---|
| PAO1 | Wild-type P. aeruginosa strain used for mutational analysis in this and previous studies | 55 |
| ΔpqsA mutant | Deletion of pqsA gene generated in PAO1 | 28 |
| ΔpqsD mutant | Deletion of pqsD gene generated in PAO1 | E. Pesci |
| ΔpqsE mutant | Deletion of pqsE gene generated in PAO1 | 42 |
| ΔpqsL mutant | Deletion of pqsL gene generated in PAO1 | 43 |
| PA14 | Burn wound isolate from 1995 at Massachusetts General Hospital, Boston, MA | 56 |
| ΔpqsH mutant | Deletion of pqsH gene generated in PA14 | D. Hogan |
| PAK | Wild-type P. aeruginosa strain | M. Vasil |
| JSRI-1 | CF P. aeruginosa lung isolate from 8-yr-old patient | 28 |
| JSRI-2 | CF P. aeruginosa lung isolate from 17-yr-old patient | 28 |
| MRSA-M2 | Methicillin-resistant isolate of S. aureus isolated from an osteomyelitis patient in Galveston, TX | 57 |
| UA159 | Streptococcus mutans serotype c strain | 49 |
| 1128 | Haemophilus influenzae strain originally isolated from a child with otitis media | 48 |
Coculture assays.
Antimicrobial activity against S. aureus was assayed as previously described (30) with some modifications. The indicated P. aeruginosa strains were grown in DTSB, supplemented or not supplemented with 100 μM FeCl3, for 18 h at 37°C. S. aureus strain MRSA-M2, grown overnight on BHI and diluted to an A600 of 0.1, was spread well with a cotton swab onto a BHI or DTSB plate and allowed to dry. Five microliters of the indicated P. aeruginosa DTSB culture was spotted onto the S. aureus lawn, and plates were allowed to dry at room temperature. Plates were incubated overnight at 37°C and visualized the next day for S. aureus growth.
To quantify antimicrobial activity against S. aureus, a liquid coculture system using transwell cell culture inserts (Corning Costar, NY, USA) was developed. Strains were grown overnight in DTSB for 18 h at 37°C. S. aureus cultures were diluted to an optical density at 600 nm (OD600) of 0.05 in DTSB supplemented with or without 100 μM FeCl3, and 600 μl of the resulting cell suspension was inoculated into the bottom of the transwell plate. The transwell insert with a 0.4-μm membrane was then placed onto the plate, and 100 μl of P. aeruginosa cultures, diluted to an OD600 of 0.05, was inoculated on top of the membrane. The transwell plates were incubated at 37°C for 18 h under static growth conditions, and S. aureus cell density (OD630) was measured spectroscopically in a BioTek Synergy HT plate reader.
Detection of PQS.
Bacteria were grown in DTSB for 18 h at 37°C, with and without 100 μM FeCl3 supplementation as indicated. Each culture was harvested and extracted with acidified ethyl acetate as described by Collier et al. (45). One-half of the resulting organic extract was transferred to a clean tube and evaporated to dryness. Samples were resuspended in 1:1 acidified ethyl acetate-acetonitrile and analyzed by thin-layer chromatography (TLC) (46).
UPLC-MS/MS.
Culture supernatants prepared as described above were suspended in water-acetonitrile (1:1) with 0.1% formic acid and analyzed as described below. Ultraperformance liquid chromatography (UPLC) was performed on a Waters Acquity UPLC system (Milford, MA). The separation was achieved using a C18 CSH (1.7 μm; 2.1- by 100-mm) column (Waters, Milford, MA). Mobile phase A was water with 0.1% formic acid, and mobile phase B was acetonitrile with 0.1% formic acid. The gradient was kept as 30% B for 0.5 min, ramped to 95% B over 9.5 min, held for 3 min, ramped to 30% B in 0.75 min, and equilibrated for 2.25 min for a total run time of 16 min. The flow rate was 0.3 ml/min. The column was maintained at 40°C, and the autosampler was kept at 5°C. A 5-μl injection was used for all samples. The tandem mass spectrometry (MS/MS) experiments were performed on a Waters Synapt G2-S quadrupole time of flight (QTOF) mass spectrometer (Milford, MA). The instrument was operated in positive ion mode electrospray. The capillary voltage was 2.5 kV, and the sampling cone voltage was 30 V. Nitrogen at a flow of 650 liters/h was used as the desolvation gas with a constant desolvation temperature of 400°C. The source temperature was set at 125°C. Data were acquired over the m/z range of 100 to 1,200. The mass spectrometer was operated in MSE mode with alternating low- and high-collision energies. The first scan was set at low-collision energy (4 eV) and used to collect precursor ion spectra. The second scan was set at high-collision energy and ramped from 25 to 40 eV, which was used for generation of product ion spectra. Argon gas was used for collision-induced dissociation (CID). Leucine enkephalin was used as the lock-mass to ensure high-mass-accuracy data acquisition. Data were acquired and analyzed with Waters MassLynx v4.1 software.
RT-PCR.
Real-time PCR (RT-PCR) (quantitative PCR [qPCR]) analysis of prrF, pqsA, antA, and antR gene expression in broth cultures was carried out using primers and probes as previously described (24, 25, 28), Expression of pqsH was detected using the following primers and probe: PqsH.for (TCG AGT TCA TCA GGA AGC AAT C), PqsH.rev (CGA GGG TAT TCC TCA GCC AGA), and PqsH.probe (CTT GGT CAG TGG GAA TCG CCC TCC). Samples were analyzed using the Applied Biosystems StepOne Plus real-time PCR system (Life Technologies). Relative amounts of cDNA were determined by the threshold cycle (ΔΔCT) method or by use of a standard curve generated from cDNA acquired from serial dilutions of PAO1 RNA samples. Expression was normalized to oprF cDNA detected in each sample.
RESULTS
Iron regulates pqsA-mediated growth suppression of S. aureus.
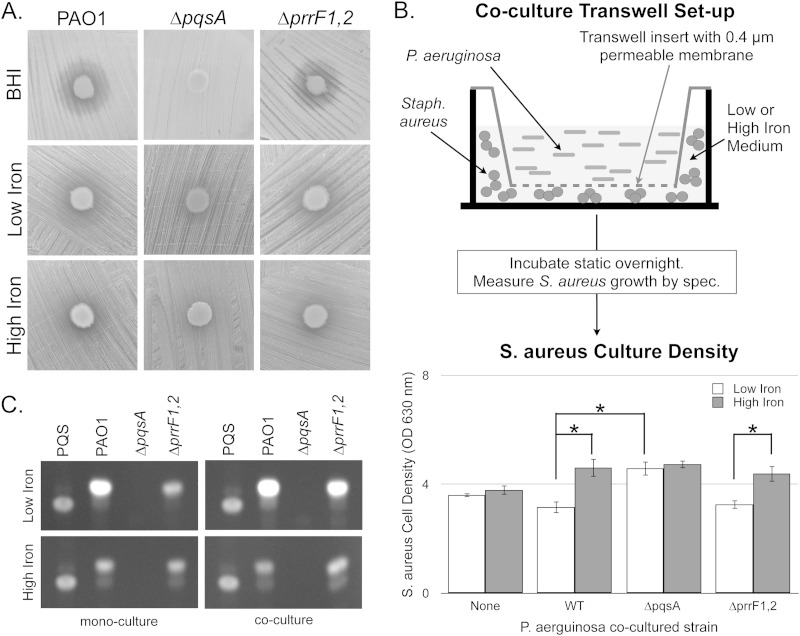
We previously showed that production of certain AQs is regulated by iron in a PrrF-dependent manner (28). Since AQ production is required for antimicrobial activity against S. aureus, we tested whether iron depletion would also affect the ability of P. aeruginosa to suppress growth of S. aureus. S. aureus was exposed to overnight low-iron cultures of either wild type (PAO1) or an isogenic ΔpqsA mutant of P. aeruginosa on agar plates containing rich, complex medium (BHI). As was previously shown for P. aeruginosa strain PA14 (30), PAO1, but not the isogenic ΔpqsA mutant, induced a halo of S. aureus growth inhibition after overnight incubation (Fig. 1A). This effect was also observed when we repeated the experiment with low-iron (DTSB) agar plates (Fig. 1A). However, supplementation of this medium with 100 μM FeCl3 significantly reduced the size of the growth inhibition halo induced by PAO1 (Fig. 1A), suggesting that P. aeruginosa antimicrobial activity against S. aureus is linked to iron availability.
FIG 1.
PQS-dependent growth stasis of S. aureus is enhanced by iron depletion. (A) The indicated P. aeruginosa strains were spotted onto confluent lawns of S. aureus on BHI, DTSB minus iron (“low iron”), or DTSB plus 100 μM FeCl3 (“high iron”) as described in Materials and Methods. Plates were incubated at 37°C overnight and visualized the next day. (B) S. aureus cell density was measured spectroscopically as OD630 after coculture with the indicated P. aeruginosa strains in transwell cell culture plates as described in Materials and Methods. Error bars indicate standard deviations from three biological replicates. Asterisks indicate a P value of less than 0.05 as determined by a two-tailed Student t test. (C) Overnight cultures of the indicated P. aeruginosa strains were grown in mono- or coculture with S. aureus in DTSB medium, with or without supplementation with 100 μM FeCl3. Cells were harvested, and supernatants were analyzed by TLC as described in Materials and Methods.
We next developed a transwell growth system to quantify the antimicrobial effects of P. aeruginosa coculture on S. aureus when the strains were separated by a 0.4-μm membrane. S. aureus was inoculated into low- or high-iron DTSB below the transwell membrane, and the indicated P. aeruginosa strain was inoculated on the top side of the membrane (diagram in Fig. 1B). The transwell cocultures were grown without shaking overnight, and S. aureus culture densities were determined spectroscopically. Notably, monocultures of S. aureus grew just as well in low-iron as in high-iron medium under these conditions (Fig. 1B). In contrast, iron depletion had a significant impact on S. aureus growth when cocultured with PAO1 (Fig. 1B). Moreover, this effect was eliminated in the ΔpqsA mutant (Fig. 1B), demonstrating that growth suppression of S. aureus in this system is due to AQ production. Addition of P. aeruginosa culture supernatants to S. aureus low- and high-iron cultures was not sufficient to produce this effect (see Fig. S1 in the supplemental material), indicating that coculture of these two strains is required for AQ-mediated growth suppression. Together, these data demonstrate that AQ-mediated antimicrobial activity against S. aureus is enhanced by iron depletion.
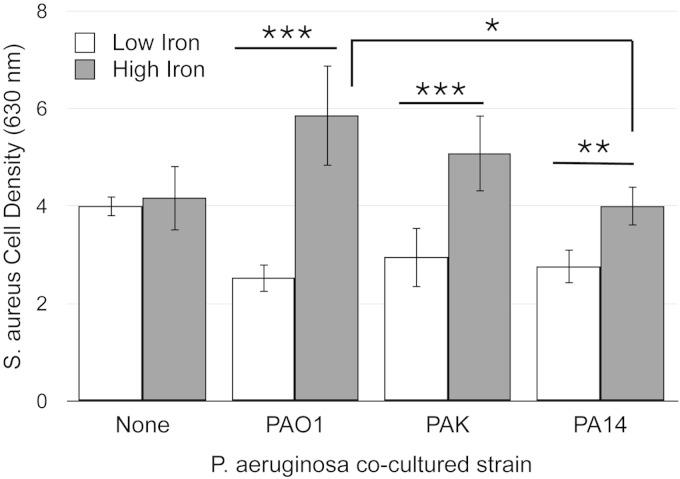
We next determined if iron-regulated antimicrobial activity was conserved among different P. aeruginosa strains using our transwell coculture assay. The wild-type P. aeruginosa strains PAO1, PAK, and PA14 all show iron-regulated growth suppression of S. aureus (Fig. 2). Variable degrees of iron-regulated growth suppression between PAO1 and PA14 were noted during these assays. However, culture densities of S. aureus in low iron were not significantly changed in comparison among the P. aeruginosa strains that were included in the coculture. Thus, our data indicate that iron-regulated antimicrobial activity against S. aureus is conserved among commonly used laboratory strains of P. aeruginosa.
FIG 2.
Iron-regulated antimicrobial activity is conserved in P. aeruginosa laboratory strains. S. aureus cell density was measured spectroscopically as OD630 after coculture with the indicated P. aeruginosa strains in transwell cell culture plates as described in Materials and Methods. Error bars indicate standard deviations from six biological replicates. Asterisks indicate the following P values as determined by a two-tailed Student t test: *, P < 0.05; **, P < 0.005; ***, P < 0.0005.
PqsA contributes to antimicrobial activity against both Gram-negative and Gram-positive bacterial species.
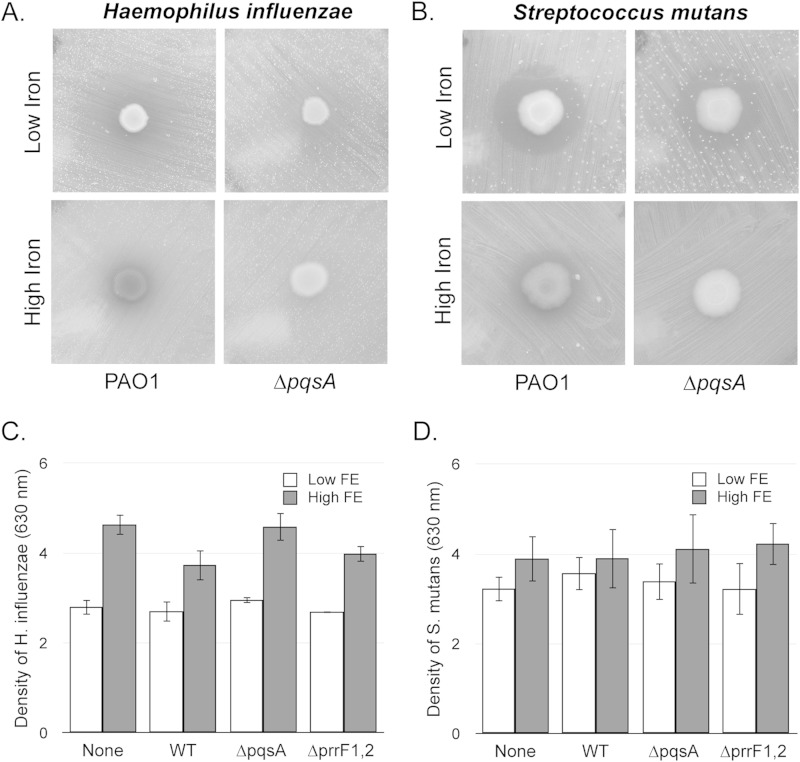
To determine if AQ-mediated antimicrobial activity was uniquely active against S. aureus, we analyzed the ability of PAO1 and the ΔpqsA mutant to suppress growth of two other bacterial pathogens that are often found in the sputum from cystic fibrosis (CF) patients (47): Haemophilus influenzae, a Gram-negative bacterium and normal flora of the upper respiratory tract (48), and Streptococcus mutans, a Gram-positive dental pathogen (49). PAO1 inhibited growth of each of these species on iron-depleted medium, and this inhibition was substantially diminished in iron-replete medium (Fig. 3A and B), suggesting that iron-regulated antimicrobial activity is not restricted to S. aureus or even to just Gram-positive bacteria.
FIG 3.
AQs mediate antimicrobial activity against Gram-negative and Gram-positive bacterial pathogens. (A and B) The indicated P. aeruginosa strains were spotted onto confluent lawns of H. influenzae (A) or S. mutans (B) on DTSB minus iron (“low iron”) or DTSB plus 100 μM FeCl3 (“high iron”) as described in Materials and Methods. Plates were incubated at 37°C overnight and visualized the next day. (C and D) H. influenzae (C) or S. mutans (D) cell density was measured spectroscopically as OD630 after coculture with the indicated P. aeruginosa strains in transwell cell culture plates as described in Materials and Methods. Error bars indicate standard deviations from three biological replicates.
As might be expected, the extent to which P. aeruginosa was capable of inhibiting growth of each of these species was variable—the zone of growth inhibition for S. mutans was well defined, whereas a gradient of clearance was observed for H. influenzae (Fig. 3A and B). Neither of these species was inhibited by the PAO1 ΔpqsA mutant when plated on high-iron medium, and the growth suppression halos of both species were smaller when exposed to the ΔpqsA mutant on low-iron medium (Fig. 3A and B). Thus, it appears that iron-regulated antimicrobial activity of these species is also dependent upon AQ production.
Unfortunately, attempts at quantifying this phenotype in our transwell coculture system were unsuccessful. H. influenzae demonstrated significantly reduced growth in low- versus high-iron medium when grown in monoculture (Fig. 3C), making it impossible to assign a specific role for P. aeruginosa in suppressing growth of this species in coculture. In contrast, no significant differences in S. mutans growth were observed in low- versus high-iron medium either in monoculture or in coculture by this assay (Fig. 3D). We reasoned that the lack of iron-regulated growth suppression of S. mutans could be due to reduced PQS production during static growth in the transwell plates, as PQS production is known to be dependent upon oxygen (50). However, iron-regulated AQ production not only was retained under these conditions but was significantly enhanced during static versus shaking growth (see Fig. S2 in the supplemental material). Thus, while PqsA appears to mediate antimicrobial activity against each of these organisms, we are not able at this time to make any further conclusions about the impact of iron on these interspecies relationships.
The ΔprrF1,2 mutant retains the ability to mediate iron-regulated growth suppression of S. aureus.
The PrrF sRNAs were previously shown to be required for optimal PQS production by P. aeruginosa grown under low-iron conditions (25). Since PQS is one of many AQs that promote growth suppression of S. aureus, we hypothesized that the ΔprrF1,2 mutant would also be defective for suppressing S. aureus growth. Surprisingly, growth suppression of S. aureus by the ΔprrF1,2 mutant was indistinguishable from that by PAO1, both on BHI and on low-iron DTSB (Fig. 1A). Moreover, antimicrobial activity of the ΔprrF1,2 mutant was reduced by supplementing DTSB medium with 100 μM FeCl3, again in a manner similar to that of PAO1. This finding was further corroborated by our transwell coculture assay (Fig. 1B). These data suggest that an alternate, PrrF-independent iron regulatory pathway mediates iron regulation of AQ production in the presence of S. aureus.
Based on previously published data showing that coculture with S. aureus can induce AQ production by P. aeruginosa (6), we reasoned that AQ production might similarly be restored to the ΔprrF1,2 mutant when grown in the presence of S. aureus. To test this idea, we analyzed PQS production in culture supernatants of PAO1 and the ΔprrF1,2 mutant grown in high- or low-iron media, either alone or in coculture with S. aureus. As previously shown by thin-layer chromatography (TLC) (28), production of PQS, as well as a distinct, fluorescent metabolite, was substantially reduced in the ΔprrF1,2 mutant compared to the wild type when grown in low-iron monocultures (Fig. 1C). Like PQS, production of this metabolite was dependent upon pqsA, indicating that it is likely to be an AQ. In contrast to what was observed in monoculture, production of this metabolite by the ΔprrF1,2 mutant was substantially increased by coculture with S. aureus (Fig. 1C). Moreover, iron-regulated production of this metabolite was restored to the ΔprrF1,2 mutant when cocultured with S. aureus (Fig. 1C). Thus, our data demonstrate that the AQ-deficiency phenotype of the ΔprrF1,2 mutant is overcome by growth with S. aureus. Moreover, these results suggest the presence of a PrrF-independent iron regulatory pathway that controls AQ production in P. aeruginosa.
Expression of anthranilate degradation genes is not altered by coculture with S. aureus.
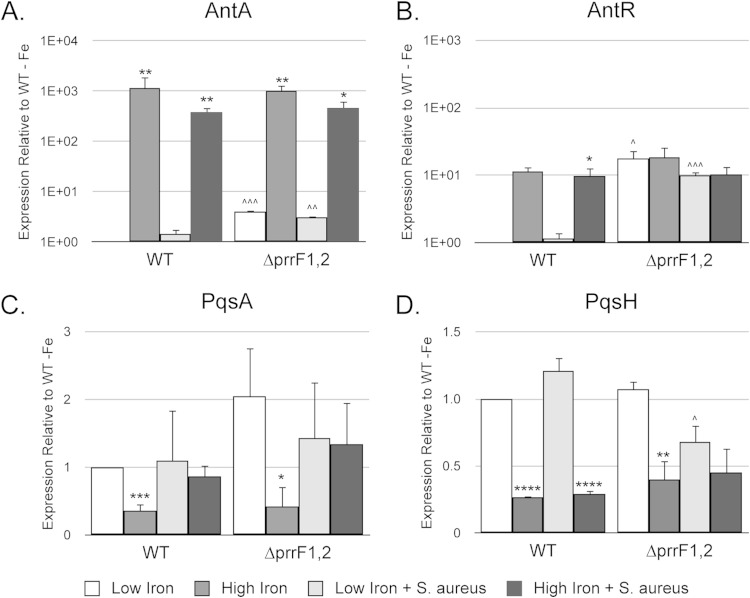
While the PrrF sRNAs positively affect AQ production through modulation of antA expression in monoculture (25), our results above show that PrrF is not required for production of these metabolites in coculture (Fig. 1C). We therefore hypothesized that coculture with S. aureus enhanced iron-regulated expression of antA in the ΔprrF1,2 mutant, allowing for iron-regulated AQ production. In agreement with previous work (25), real-time PCR (RT-PCR) analysis demonstrated that iron induces expression of antA in both a PrrF-dependent and a PrrF-independent manner (Fig. 4A). However, iron induction of antA was not enhanced by coculture with S. aureus, suggesting that modulation of this specific regulatory pathway is not the source of restored PQS production in the ΔprrF1,2 mutant. Iron was also previously shown to induce expression of antR, encoding a positive LysR-type activator of the antABC operon (25). In contrast to antA, iron induction of antR was almost completely dependent upon an intact prrF locus (25). Our current work corroborates this finding, as we observed significant iron induction of antR in the wild type but no induction of antR in the ΔprrF1,2 mutant (Fig. 4B). Notably, coculture of the ΔprrF1,2 mutant with S. aureus did not restore iron-regulated expression of antR (Fig. 4B). Thus, our data indicate that increased AQ production by the ΔprrF1,2 mutant upon coculture with S. aureus is not due to changes in antA or antR expression.
FIG 4.
Iron regulation of anthranilate degradation and pqs biosynthesis genes is not affected by coculture with S. aureus. RNA was isolated from the indicated strains grown for 18 h at 37°C in DTSB with and without iron supplementation, either in monoculture or cocultured with S. aureus. The resulting RNA was analyzed for expression of antA (A), antR (B), pqsA (C), and pqsH (D) by RT-PCR as described in Materials and Methods. Error bars indicate standard deviations from three biological replicates. Asterisks indicate the following P values for comparing expression under low- and high-iron conditions as determined by Student's t test: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Carets indicate the following P values for comparing expression in the wild type and ΔprrF1,2 mutant as determined by Student's t test: ∧, P < 0.05; ∧∧, P < 0.01; ∧∧∧, P < 0.001.
To determine if increased AQ production under iron-depleted conditions was due to increased expression of genes for AQ synthesis, we next analyzed pqsA and pqsH expression. As previously observed (28), pqsA mRNA levels were significantly increased by iron depletion in both the wild type and the ΔprrF1,2 mutant (Fig. 4C). Iron-regulated expression of pqsH was also observed in both strains (Fig. 4D). Unexpectedly, coculture with S. aureus eliminated iron-regulated expression of pqsA and pqsH, both in the wild type and in the ΔprrF1,2 mutant (Fig. 4C and D). While the rationale for these regulatory data remains unclear at this time, they demonstrate the capacity of S. aureus to modulate iron regulatory pathways in P. aeruginosa. Overall, our data indicate that additional iron regulatory pathways control AQ production in P. aeruginosa and present the potential for S. aureus to enhance regulation via these pathways during coculture.
Iron-regulated antimicrobial activity is retained in CF isolates.
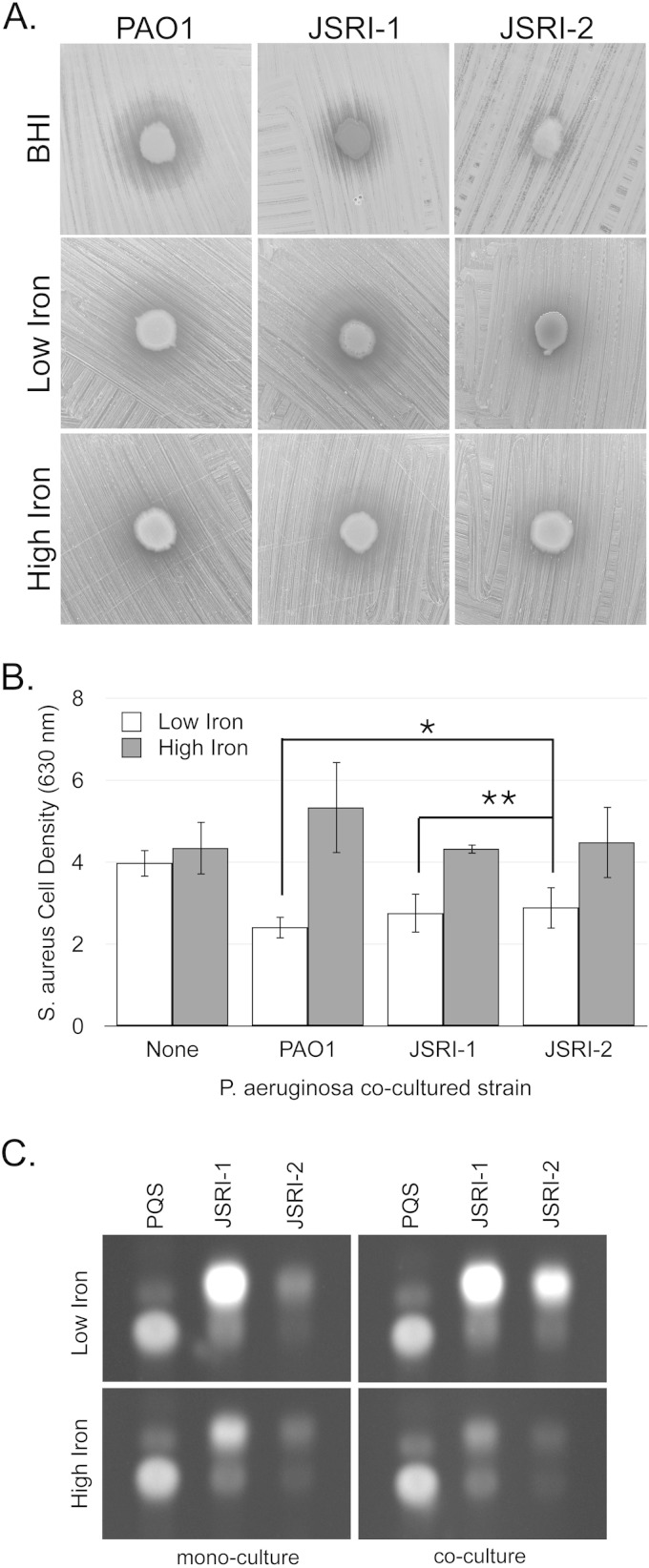
We previously analyzed a series of clonal, longitudinally collected CF isolates and demonstrated that their iron acquisition and regulatory pathways were substantially altered during CF lung infection (28). We additionally showed that one of the later CF isolates, termed JSRI-2, produced substantially less AQ than its “parent” isolate, termed JSRI-1. Moreover, AQ production by the JSRI-2 strain was not responsive to iron (28). We therefore determined if these strains retained the ability to suppress S. aureus growth. Our data show that, as expected, JSRI-2 was less efficient at suppressing S. aureus growth than either JSRI-1 or the PAO1 laboratory strain, regardless of growth medium (Fig. 5A). However, iron-regulated growth stasis of S. aureus was still observed in the JSRI-2 isolate (Fig. 5A), indicating that AQ production could be stimulated by growth in iron-depleted medium. When we quantified this activity using our transwell coculture assay, we observed significant growth suppression of S. aureus in low iron by both the JSRI-1 and JSRI-2 strains (Fig. 5B). Moreover, this assay showed a significantly reduced ability of JSRI-2 to suppress S. aureus growth compared to JSRI-1 or PAO1 (Fig. 5B). Thus, this later-stage CF isolate, while reduced for this activity, retained the ability to mediate iron-regulated antimicrobial activity against S. aureus.
FIG 5.
Iron-regulated antimicrobial activity is conserved in a CF lung infection isolate. The indicated P. aeruginosa strains were spotted onto confluent lawns of S. aureus on BHI, DTSB minus iron (“low iron”), or DTSB plus 100 μM FeCl3 (“high iron”) as described in Materials and Methods. Plates were incubated at 37°C overnight and visualized the next day. (B) S. aureus cell density was measured spectroscopically as OD630 after coculture with the indicated P. aeruginosa strains in transwell cell culture plates as described in Materials and Methods. Error bars indicate standard deviations from six biological replicates. Asterisks indicate P values as determined by a two-tailed Student t test: *, P < 0.05; **, P < 0.005. (C) Overnight cultures of the indicated P. aeruginosa strains were grown in mono- or coculture with S. aureus in DTSB medium, with or without supplementation with 100 μM FeCl3. Cells were harvested, and supernatants were analyzed by TLC as described in Materials and Methods.
Since AQ production by the JSRI-2 isolate was previously reported to be diminished and unresponsive to iron, we determined if this strain's defect in AQ production could be overcome by S. aureus coculture, as was observed in the ΔprrF1,2 mutant (Fig. 1C). TLC analysis of culture supernatants from these strains demonstrated that coculture with S. aureus greatly enhanced production of both PQS and the distinct pqsA-dependent metabolite by JSRI-2 (Fig. 5C). These data demonstrate that, despite our earlier findings that AQ production was substantially reduced in this isolate when grown in monoculture (28), its capacity for iron-regulated antimicrobial activity was conserved.
PAO1 produces an iron-regulated C9-PQS metabolite.
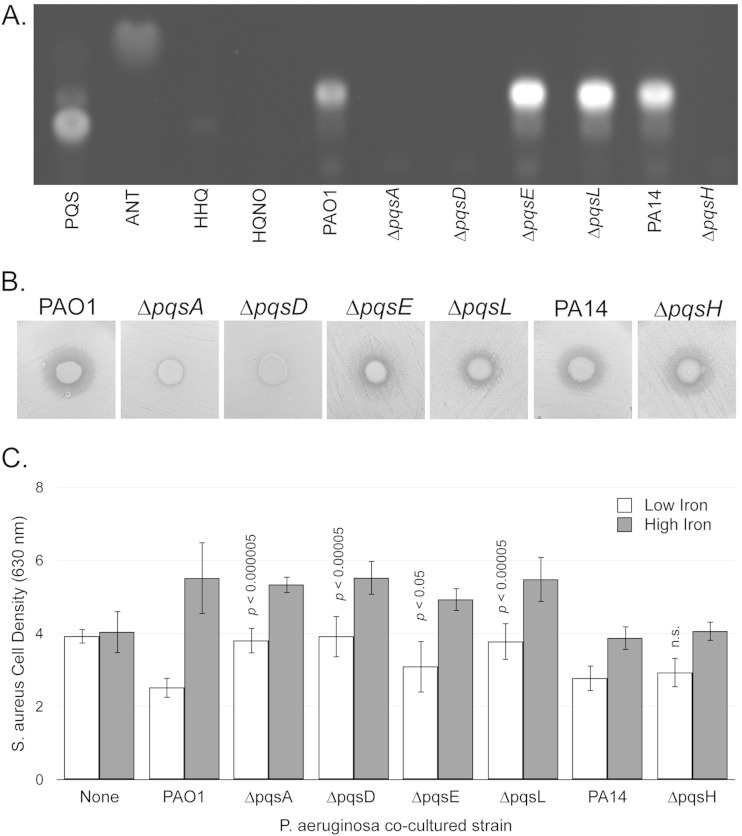
Previously and in our current report, we have observed an iron-regulated metabolite whose production is dependent upon pqsA but which migrates faster than PQS (28). TLC analysis of several AQ standards, which were verified by high-resolution tandem mass spectrometry (see Fig. S3 in the supplemental material), demonstrated that this metabolite was not HHQ, HQNO, or anthranilate (ANT) (Fig. 6A). Since the levels of this metabolite correlated with antimicrobial activity in the above-described studies, we analyzed the contribution of known PQS biosynthetic genes toward its production. As expected, TLC analysis of culture supernatants demonstrated that this metabolite was not produced by P. aeruginosa mutants deleted for either pqsA or pqsD (Fig. 6A), each of which catalyzes the first and second steps in HHQ and PQS biosynthesis, respectively (29, 31). Also as expected, deletion of pqsE, which is required for quorum signaling by HHQ and PQS, but not their synthesis, had no impact on the production of this metabolite (Fig. 6A). Moreover, pqsL, which allows for production of the Gram-positive antimicrobial HQNO, was not required for the production of this metabolite (Fig. 6A). Combined, these data suggest that this metabolite is a potential analog of either HHQ or PQS, each of which requires PqsA and PqsD for its synthesis.
FIG 6.
Multiple PQS genes contribute to iron-regulated growth suppression of S. aureus. (A) Strains were grown at 37°C for 18 h in DTSB. Cells were harvested, and supernatants were analyzed by TLC (A), S. aureus growth suppression on agar plates (B), or transwell growth suppression assay (C). Purchased standards PQS, anthranilate (ANT), HHQ, and HQNO were resuspended in 1:1 ethyl acetate-acetonitrile as described in Materials and Methods. Error bars indicate standard deviations from six biological replicates. The P values as determined by a two-tailed Student t test indicate significantly increased S. aureus culture density under low-iron conditions compared to PAO1.
We next analyzed a deletion mutant for pqsH generated in the PA14 P. aeruginosa laboratory strain background, a generous gift from the laboratory of Deborah Hogan. PqsH catalyzes the final step in PQS production (36, 51), adding a 3-hydroxyl to the quinolone moiety. Our analysis of the ΔpqsH mutant demonstrates that this gene is absolutely required for production of this metabolite (Fig. 6A), indicating that it is in fact an analog of PQS. We therefore hypothesized that this metabolite is a PQS analog with an alkyl chain of different length, such as 3-hydroxy-2-nonyl-4(1H)-quinolone (C9-PQS), which was previously reported to be produced by P. aeruginosa (36).
To further investigate the production of C9-PQS under our culture conditions, we analyzed PAO1 supernatants by ultraperformance liquid chromatography (UPLC) coupled with high-resolution tandem mass spectrometry (MS/MS). This analysis identified an ion with a mass-to-charge (m/z) value of 288.2 corresponding to C9-PQS (see Fig. S4A in the supplemental material). Analysis of the tandem mass spectrum of this precursor ion revealed the formation of product ions at m/z values 175.1 and 188.1, which were also observed in the PQS standard (see Fig. S3C), confirming the presence of C9-PQS in our samples (see Fig. S4B). Comparative analysis further showed that C9-PQS production was regulated by iron at levels similar to those of the unknown AQ detected by TLC (Table 2). Two other related AQ structures, one with an identical m/z of 288.2 and another with an m/z value of 286.2, were also detected via unique chromatographic retention times and diagnostic tandem mass spectra (see Fig. S4). These precursor ions fragmented into distinct product ions that were characteristic of the HQNO core quinolone structure (see Fig. S3B) and were assigned as C9-QNO (m/z 288.2) and monounsaturated C9:1-QNO (m/z 286.2) (see Fig. S4C and E). The unique chromatographic retention times, accurate mass measurements, and diagnostic fragmentation data obtained from the UPLC-MS/MS platform provided structural data that could distinguish C9-PQS from closely related AQ structures (see Fig. S4). Combined with our genetic analysis above, these data indicated that the iron-regulated fluorescent metabolite observed by TLC in this and previous reports was C9-PQS.
TABLE 2.
Quantification of PQS and C9-PQS
| Metabolite | Growth condition | Relative levelc as determined by TLC or LC-MS/MS in: |
|||||
|---|---|---|---|---|---|---|---|
| Monoculture |
Coculture |
||||||
| PAO1 | ΔpqsA mutant | ΔprrF1,2 mutant | PAO1 | ΔpqsA mutant | ΔprrF1,2 mutant | ||
| PQSa | Low iron | 2,551 ± 321** | 932 ± 104 | 2,405 ± 491* | 3,075 ± 428** | 743 ± 79 | 2,411 ± 503* |
| High iron | 935 ± 13 | 620 ± 59 | 912 ± 159 | 1,167 ± 57 | 595 ± 66 | 1,171 ± 82 | |
| C9-PQSa | Low iron | 11,481 ± 1,631** | 937 ± 107 | 5,722 ± 761** | 14,204 ± 1,342*** | 720 ± 71 | 9,626 ± 1,082**∧ |
| High iron | 1,827 ± 116 | 616 ± 64 | 1,896 ± 429 | 2,298 ± 380 | 610 ± 68 | 2,662 ± 749 | |
| C9-PQSb | Low iron | 1,251 ± 139*** | 0.5 ± 1 | 83 ± 48 | ND | ND | ND |
| High iron | 315 ± 119 | 0 ± 0 | 115 ± 67 | ND | ND | ND | |
Values for the indicated metabolite were determined by densitometry of thin-layer chromatography. C9-PQS refers to the unknown metabolite observed in Fig. 1C. Standard deviations are from 3 biological replicates.
Values for the indicated metabolite were determined by UPLC-MS/MS (described in Materials and Methods). Standard deviations are from 5 biological replicates.
Asterisks indicate the following P values in comparing high- and low-iron conditions: *, P < 0.05; **, P < 0.005; ***, P < 0.0005. The caret indicates significance between monoculture and coculture with S. aureus (P < 0.05). ND, not done.
Multiple pqs genes contribute to iron-regulated antimicrobial activity against S. aureus.
Since multiple AQs are produced by P. aeruginosa with a suite of different activities, we next determined antimicrobial activity by each of the pqs mutants against S. aureus. While antimicrobial activity was not observed for the ΔpqsA mutant on agar plates (Fig. 6B), we did observe some level of iron-regulated growth suppression by this mutant using our transwell assay in this study set (Fig. 6C). However, the ability of the ΔpqsA mutant to suppress S. aureus grow was significantly reduced compared to PAO1 (Fig. 6C) (P < 0.000005). The ΔpqsD, ΔpqsE, and ΔpqsL mutants also showed significant defects in suppressing S. aureus growth compared to the wild type (Fig. 6B and C), indicating that this phenotype is dependent upon the activity of multiple AQs. Surprisingly, the ΔpqsH mutant, which did not produce any PQS or C9-PQS (Fig. 6A), demonstrated iron-regulated growth suppression of S. aureus at levels similar to those of its PA14 parent strain (Fig. 6B and C), indicating that PQS is an indirect correlate of antistaphylococcal activity. Thus, our data demonstrate that iron-regulated antimicrobial activity of P. aeruginosa is multifactorial and yet is independent of the PQS metabolite produced by this pathway.
DISCUSSION
Iron is an essential nutrient and is limiting in most environments, especially in the context of infections. Therefore, it is not surprising that P. aeruginosa has adapted several mechanisms to acquire this nutrient in the presence of competing microorganisms, as well as from the human host. AQs play many important functions in P. aeruginosa, including signaling to induce expression of virulence determinants, as well as growth-suppressing activities (52). Here, we show that iron-regulated production of AQs contributes to P. aeruginosa's ability to suppress growth of another microbial pathogen, S. aureus. It was previously reported that S. aureus could serve as an iron source for P. aeruginosa, purportedly due to lysis and the subsequent releases of iron stores during coculture (30). Our finding that antimicrobial activity against S. aureus is enhanced by iron depletion further supports the idea that AQs contribute to iron acquisition, particularly in polymicrobial environments. In the context of CF lung infection, this iron competition strategy may be critical for P. aeruginosa's ability to supplant other microflora, allowing for the observed shift in microbial population.
Perhaps more strikingly, our data demonstrate that coculture with S. aureus alters the iron-regulatory pathways that control AQ production in P. aeruginosa. Specifically, coculture of the ΔprrF1,2 mutant with S. aureus restores iron-regulated AQ production, demonstrating that a PrrF-independent pathway mediates iron-regulated antimicrobial production. Our initial analysis of iron-regulated expression of both antA and antR demonstrates that S. aureus does not have a significant impact on these pathways (Fig. 4). Further confounding this analysis was the finding that coculture with S. aureus reduces iron-regulated expression of both pqsA and pqsH (Fig. 4). Thus, it is likely that coculture with S. aureus directs the expression of other genes involved in either AQ biosynthesis or catabolism. Alternatively, this phenomenon may be due to iron-dependent changes that occur posttranscriptionally. Regardless, our current understanding of iron regulation in P. aeruginosa is largely based on studies with pure cultures. Combined with the findings of earlier studies (6, 30), this work demonstrates the need to reevaluate well-established paradigms in iron regulation to determine how polymicrobial environments alter these pathways.
Our study has also provided evidence that the iron-regulated AQ observed in previous (28) and current studies is a C9 analog of PQS. Like PQS, this species is eliminated by deletion of pqsA, pqsD, or pqsH and yet is still present in the ΔpqsE and ΔpqsL mutants (Fig. 6A). Moreover, UPLC-MS/MS analysis identified C9-PQS in PAO1 supernatants and demonstrated that this species is induced by iron depletion (Table 2). However, it is not yet clear why this species is so much more predominant than the C7-PQS that has been observed in previous analyses (25, 27). This particular iron-regulated AQ has been detected in supernatants of every strain that we have analyzed to date, indicating that its presence is the result of our experimental conditions, rather than strain differences. The potential for this distinct AQ to have a biological impact is especially intriguing, and we are currently working toward further defining the regulation of this and other AQs by UPLC-MS/MS.
The iron-regulated C9-PQS analog observed in these studies correlated very strongly with antimicrobial activity against S. aureus (Fig. 1 and 5). Thus, it was surprising to find that the ΔpqsH mutant, which does not make this metabolite, is not defective for S. aureus growth suppression (Fig. 6). Instead, it appears that this activity is likely due to a combination of the signaling (ΔpqsE) and direct growth stasis (ΔpqsL) capabilities of these diverse AQ molecules. With this in mind, determining how iron affects the production of each of these AQ molecules is likely to shed further light on the mechanism by which P. aeruginosa regulates AQ-dependent antimicrobial activity.
The implications of this study for CF disease and the interplay between residents of the CF lung are important to consider. JSRI-2, which represents a later-stage CF isolate, was previously shown to have diminished AQ production in monoculture (28). This finding was notable in consideration of a previous report in which PQS was consistently detected in sputum from several late-stage CF patients (45). Here, we show that coculture with S. aureus can restore AQ production to strains previously thought to be unable to produce this metabolite (Fig. 5). Reduction in AQ production by the JSRI-2 isolate grown in monoculture may suggest that this activity becomes more dependent upon the presence of S. aureus in later stages of disease. In light of these data, reported loss of other virulence traits by late-stage CF isolates should be reconsidered, as polymicrobial cultures could have similar effects on the expression of additional virulence determinants.
Increased production of AQs by the JSRI-2 isolate in the presence of S. aureus has compelling implications for the mechanism underlying exacerbations in CF lung infections. Previous work has noted that coinfection with both of these pathogens correlated strongly with rapidly decreased lung function (8). Our studies suggest that iron availability in the CF lung is also likely to be a determinant for these exacerbations. As such, it will be interesting to consider how disruption of iron acquisition affects this activity in future studies. Our initial analysis of siderophore biosynthesis mutants indicates that eliminating this mode of iron uptake does not enhance antimicrobial activity (A. T. Nguyen and A. G. Oglesby-Sherrouse, unpublished data). Perhaps, there are differences in limiting environmental iron, such as through the activity of iron chelators, versus limiting P. aeruginosa's ability to acquire this element in iron-limiting environments.
A notable finding from our study is that coculture is required for iron-regulated antimicrobial activity (see Fig. S1 in the supplemental material), indicating that a complex interplay between P. aeruginosa and S. aureus allows for this activity. Several reports provide additional evidence for an exchange between these two bacterial species. Korgaonkar et al. demonstrated that PQS production by P. aeruginosa was enhanced by coculture with S. aureus (6). This report additionally found that agtR, a response regulator involved in metabolite sensing and uptake, was required for this activity, suggesting metabolite sensing as a possible means for bacterial cross talk. An additional report indicated that S. aureus secretes one or more proteins to manipulate the growth characteristics of P. aeruginosa (53). It is therefore likely that iron-regulated AQ production is not the sole driver of interactions between these two bacterial species. The accompanying paper by Filkins et al. (54) further explores the potential for metabolite sensing to drive these interactions. Future studies into how S. aureus alters the phenotypes of different P. aeruginosa strains are likely to increase our understanding of how these two microbial pathogens establish equilibrium and cause disease during polymicrobial infections.
Supplementary Material
ACKNOWLEDGMENTS
We thank Eb Pesci for providing isogenic pqs mutants and deletion constructs, Deborah Hogan for providing PA14 and ΔpqsH strains for our studies, and Mark Shirtliff for sharing the M2-MRSA, S. mutans, and H. influenzae strains. We also thank Geoffrey Heinzl and Luke Brewer for thoughtful discussion during preparation of the manuscript.
This work was supported by NIH-NIAID K22 grant AI089776 (to A.G.O.-S.), NIH-NIAID contract HHSN272201000046C (to M.A.K.), the University of Maryland School of Pharmacy Mass Spectrometry Center (SOP1841-IQB2014; to M.A.K.), and startup funds from the University of Maryland School of Pharmacy (to A.G.O.-S. and M.A.K.).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00072-15.
REFERENCES
- 1.Abdulrazak A, Bitar ZI, Al-Shamali AA, Mobasher LA. 2005. Bacteriological study of diabetic foot infections. J Diabetes Complications 19:138–141. doi: 10.1016/j.jdiacomp.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 2.Peters BM, Jabra-Rizk MA, O'May GA, Costerton JW, Shirtliff ME. 2012. Polymicrobial interactions: impact on pathogenesis and human disease. Clin Microbiol Rev 25:193–213. doi: 10.1128/CMR.00013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cystic Fibrosis Foundation. 2012. Cystic Fibrosis Foundation patient registry annual data report 2011. Cystic Fibrosis Foundation, Bethesda, MD. [Google Scholar]
- 4.Lyczak JB, Cannon CL, Pier GB. 2002. Lung infections associated with cystic fibrosis. Clin Microbiol Rev 15:194–222. doi: 10.1128/CMR.15.2.194-222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stacy A, Everett J, Jorth P, Trivedi U, Rumbaugh KP, Whiteley M. 2014. Bacterial fight-and-flight responses enhance virulence in a polymicrobial infection. Proc Natl Acad Sci U S A 111:7819–7824. doi: 10.1073/pnas.1400586111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Korgaonkar A, Trivedi U, Rumbaugh KP, Whiteley M. 2013. Community surveillance enhances Pseudomonas aeruginosa virulence during polymicrobial infection. Proc Natl Acad Sci U S A 110:1059–1064. doi: 10.1073/pnas.1214550110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pastar I, Nusbaum AG, Gil J, Patel SB, Chen J, Valdes J, Stojadinovic O, Plano LR, Tomic-Canic M, Davis SC. 2013. Interactions of methicillin resistant Staphylococcus aureus USA300 and Pseudomonas aeruginosa in polymicrobial wound infection. PLoS One 8:e56846. doi: 10.1371/journal.pone.0056846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosenbluth DB, Wilson K, Ferkol T, Schuster DP. 2004. Lung function decline in cystic fibrosis patients and timing for lung transplantation referral. Chest 126:412–419. doi: 10.1378/chest.126.2.412. [DOI] [PubMed] [Google Scholar]
- 9.DeLeon S, Clinton A, Fowler H, Everett J, Horswill AR, Rumbaugh KP. 2014. Synergistic interactions of Pseudomonas aeruginosa and Staphylococcus aureus in an in vitro wound model. Infect Immun 82:4718–4728. doi: 10.1128/IAI.02198-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hendricks KJ, Burd TA, Anglen JO, Simpson AW, Christensen GD, Gainor BJ. 2001. Synergy between Staphylococcus aureus and Pseudomonas aeruginosa in a rat model of complex orthopaedic wounds. J Bone Joint Surg Am 83-A:855–861. [DOI] [PubMed] [Google Scholar]
- 11.Meyer JM, Neely A, Stintzi A, Georges C, Holder IA. 1996. Pyoverdine is essential for virulence of Pseudomonas aeruginosa. Infect Immun 64:518–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takase H, Nitanai H, Hoshino K, Otani T. 2000. Impact of siderophore production on Pseudomonas aeruginosa infections in immunosuppressed mice. Infect Immun 68:1834–1839. doi: 10.1128/IAI.68.4.1834-1839.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nadal Jimenez P, Koch G, Papaioannou E, Wahjudi M, Krzeslak J, Coenye T, Cool RH, Quax WJ. 2010. Role of PvdQ in Pseudomonas aeruginosa virulence under iron-limiting conditions. Microbiology 156:49–59. doi: 10.1099/mic.0.030973-0. [DOI] [PubMed] [Google Scholar]
- 14.Xiong YQ, Vasil ML, Johnson Z, Ochsner UA, Bayer AS. 2000. The oxygen- and iron-dependent sigma factor pvdS of Pseudomonas aeruginosa is an important virulence factor in experimental infective endocarditis. J Infect Dis 181:1020–1026. doi: 10.1086/315338. [DOI] [PubMed] [Google Scholar]
- 15.Otto BR, Verweij-van Vught AM, MacLaren DM. 1992. Transferrins and heme-compounds as iron sources for pathogenic bacteria. Crit Rev Microbiol 18:217–233. doi: 10.3109/10408419209114559. [DOI] [PubMed] [Google Scholar]
- 16.Nairz M, Schroll A, Sonnweber T, Weiss G. 2010. The struggle for iron—a metal at the host-pathogen interface. Cell Microbiol 12:1691–1702. doi: 10.1111/j.1462-5822.2010.01529.x. [DOI] [PubMed] [Google Scholar]
- 17.Budzikiewicz H. 2001. Siderophores of the human pathogenic fluorescent pseudomonads. Curr Top Med Chem 1:1–6. doi: 10.2174/1568026013395560. [DOI] [PubMed] [Google Scholar]
- 18.Hunter RC, Asfour F, Dingemans J, Osuna BL, Samad T, Malfroot A, Cornelis P, Newman DK. 2013. Ferrous iron is a significant component of bioavailable iron in cystic fibrosis airways. mBio 4(4):e00557-13. doi: 10.1128/mBio.00557-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ochsner UA, Johnson Z, Vasil ML. 2000. Genetics and regulation of two distinct haem-uptake systems, phu and has, in Pseudomonas aeruginosa. Microbiology 146:185–198. [DOI] [PubMed] [Google Scholar]
- 20.Hassett DJ, Sokol PA, Howell ML, Ma JF, Schweizer HT, Ochsner U, Vasil ML. 1996. Ferric uptake regulator (Fur) mutants of Pseudomonas aeruginosa demonstrate defective siderophore-mediated iron uptake, altered aerobic growth, and decreased superoxide dismutase and catalase activities. J Bacteriol 178:3996–4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ochsner UA, Vasil AI, Johnson Z, Vasil ML. 1999. Pseudomonas aeruginosa fur overlaps with a gene encoding a novel outer membrane lipoprotein, OmlA. J Bacteriol 181:1099–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong SM, Mekalanos JJ. 2000. Genetic footprinting with mariner-based transposition in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 97:10191–10196. doi: 10.1073/pnas.97.18.10191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilderman PJ, Sowa NA, FitzGerald DJ, FitzGerald PC, Gottesman S, Ochsner UA, Vasil ML. 2004. Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc Natl Acad Sci U S A 101:9792–9797. doi: 10.1073/pnas.0403423101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reinhart AA, Powell DA, Nguyen AT, O'Neill M, Djapgne L, Wilks A, Ernst RK, Oglesby-Sherrouse AG. 2015. The prrF-encoded small regulatory RNAs are required for iron homeostasis and virulence of Pseudomonas aeruginosa. Infect Immun 83:863–875. doi: 10.1128/IAI.02707-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oglesby AG, Farrow JM III, Lee JH, Tomaras AP, Greenberg EP, Pesci EC, Vasil ML. 2008. The influence of iron on Pseudomonas aeruginosa physiology: a regulatory link between iron and quorum sensing. J Biol Chem 283:15558–15567. doi: 10.1074/jbc.M707840200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deziel E, Gopalan S, Tampakaki AP, Lepine F, Padfield KE, Saucier M, Xiao G, Rahme LG. 2005. The contribution of MvfR to Pseudomonas aeruginosa pathogenesis and quorum sensing circuitry regulation: multiple quorum sensing-regulated genes are modulated without affecting lasRI, rhlRI or the production of N-acyl-l-homoserine lactones Mol Microbiol 55:998–1014. doi: 10.1111/j.1365-2958.2004.04448.x. [DOI] [PubMed] [Google Scholar]
- 27.Calfee MW, Coleman JP, Pesci EC. 2001. Interference with Pseudomonas quinolone signal synthesis inhibits virulence factor expression by Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 98:11633–11637. doi: 10.1073/pnas.201328498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen AT, O'Neill MJ, Watts AM, Robson CL, Lamont IL, Wilks A, Oglesby-Sherrouse AG. 2014. Adaptation of iron homeostasis pathways by a Pseudomonas aeruginosa pyoverdine mutant in the cystic fibrosis lung. J Bacteriol 196:2265–2276. doi: 10.1128/JB.01491-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coleman JP, Hudson LL, McKnight SL, Farrow JM III, Calfee MW, Lindsey CA, Pesci EC. 2008. Pseudomonas aeruginosa PqsA is an anthranilate-coenzyme A ligase. J Bacteriol 190:1247–1255. doi: 10.1128/JB.01140-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mashburn LM, Jett AM, Akins DR, Whiteley M. 2005. Staphylococcus aureus serves as an iron source for Pseudomonas aeruginosa during in vivo coculture. J Bacteriol 187:554–566. doi: 10.1128/JB.187.2.554-566.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dulcey CE, Dekimpe V, Fauvelle DA, Milot S, Groleau MC, Doucet N, Rahme LG, Lepine F, Deziel E. 2013. The end of an old hypothesis: the pseudomonas signaling molecules 4-hydroxy-2-alkylquinolines derive from fatty acids, not 3-ketofatty acids. Chem Biol 20:1481–1491. doi: 10.1016/j.chembiol.2013.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Ark G, Berden JA. 1977. Binding of HQNO to beef-heart sub-mitochondrial particles. Biochim Biophys Acta 459:119–127. doi: 10.1016/0005-2728(77)90014-7. [DOI] [PubMed] [Google Scholar]
- 33.Smirnova IA, Hagerhall C, Konstantinov AA, Hederstedt L. 1995. HOQNO interaction with cytochrome b in succinate:menaquinone oxidoreductase from Bacillus subtilis. FEBS Lett 359:23–26. doi: 10.1016/0014-5793(94)01442-4. [DOI] [PubMed] [Google Scholar]
- 34.Rothery RA, Weiner JH. 1996. Interaction of an engineered [3Fe-4S] cluster with a menaquinol binding site of Escherichia coli DMSO reductase. Biochemistry 35:3247–3257. doi: 10.1021/bi951584y. [DOI] [PubMed] [Google Scholar]
- 35.Machan ZA, Taylor GW, Pitt TL, Cole PJ, Wilson R. 1992. 2-Heptyl-4-hydroxyquinoline N-oxide, an antistaphylococcal agent produced by Pseudomonas aeruginosa. J Antimicrob Chemother 30:615–623. doi: 10.1093/jac/30.5.615. [DOI] [PubMed] [Google Scholar]
- 36.Deziel E, Lepine F, Milot S, He J, Mindrinos MN, Tompkins RG, Rahme LG. 2004. Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc Natl Acad Sci U S A 101:1339–1344. doi: 10.1073/pnas.0307694100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lau GW, Hassett DJ, Ran H, Kong F. 2004. The role of pyocyanin in Pseudomonas aeruginosa infection. Trends Mol Med 10:599–606. doi: 10.1016/j.molmed.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 38.Wang Y, Wilks JC, Danhorn T, Ramos I, Croal L, Newman DK. 2011. Phenazine-1-carboxylic acid promotes bacterial biofilm development via ferrous iron acquisition. J Bacteriol 193:3606–3617. doi: 10.1128/JB.00396-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Diggle SP, Matthijs S, Wright VJ, Fletcher MP, Chhabra SR, Lamont IL, Kong X, Hider RC, Cornelis P, Camara M, Williams P. 2007. The Pseudomonas aeruginosa 4-quinolone signal molecules HHQ and PQS play multifunctional roles in quorum sensing and iron entrapment. Chem Biol 14:87–96. doi: 10.1016/j.chembiol.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 40.Bredenbruch F, Geffers R, Nimtz M, Buer J, Haussler S. 2006. The Pseudomonas aeruginosa quinolone signal (PQS) has an iron-chelating activity. Environ Microbiol 8:1318–1329. doi: 10.1111/j.1462-2920.2006.01025.x. [DOI] [PubMed] [Google Scholar]
- 41.Schweizer HP. 1992. Allelic exchange in Pseudomonas aeruginosa using novel ColE1-type vectors and a family of cassettes containing a portable oriT and the counter-selectable Bacillus subtilis sacB marker. Mol Microbiol 6:1195–1204. doi: 10.1111/j.1365-2958.1992.tb01558.x. [DOI] [PubMed] [Google Scholar]
- 42.Farrow JM III, Sund ZM, Ellison ML, Wade DS, Coleman JP, Pesci EC. 2008. PqsE functions independently of PqsR-Pseudomonas quinolone signal and enhances the rhl quorum-sensing system. J Bacteriol 190:7043–7051. doi: 10.1128/JB.00753-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.D'Argenio DA, Calfee MW, Rainey PB, Pesci EC. 2002. Autolysis and autoaggregation in Pseudomonas aeruginosa colony morphology mutants. J Bacteriol 184:6481–6489. doi: 10.1128/JB.184.23.6481-6489.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oglesby-Sherrouse AG, Djapgne L, Nguyen AT, Vasil AI, Vasil ML. 2014. The complex interplay of iron, biofilm formation, and mucoidy affecting antimicrobial resistance of Pseudomonas aeruginosa. Pathog Dis 70:307–320. doi: 10.1111/2049-632X.12132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Collier DN, Anderson L, McKnight SL, Noah TL, Knowles M, Boucher R, Schwab U, Gilligan P, Pesci EC. 2002. A bacterial cell to cell signal in the lungs of cystic fibrosis patients. FEMS Microbiol Lett 215:41–46. doi: 10.1111/j.1574-6968.2002.tb11367.x. [DOI] [PubMed] [Google Scholar]
- 46.Pesci EC, Milbank JB, Pearson JP, McKnight S, Kende AS, Greenberg EP, Iglewski BH. 1999. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 96:11229–11234. doi: 10.1073/pnas.96.20.11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harrison F. 2007. Microbial ecology of the cystic fibrosis lung. Microbiology 153:917–923. doi: 10.1099/mic.0.2006/004077-0. [DOI] [PubMed] [Google Scholar]
- 48.Ehrlich GD, Veeh R, Wang X, Costerton JW, Hayes JD, Hu FZ, Daigle BJ, Ehrlich MD, Post JC. 2002. Mucosal biofilm formation on middle-ear mucosa in the chinchilla model of otitis media. JAMA 287:1710–1715. doi: 10.1001/jama.287.13.1710. [DOI] [PubMed] [Google Scholar]
- 49.Ajdic D, McShan WM, McLaughlin RE, Savic G, Chang J, Carson MB, Primeaux C, Tian R, Kenton S, Jia H, Lin S, Qian Y, Li S, Zhu H, Najar F, Lai H, White J, Roe BA, Ferretti JJ. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc Natl Acad Sci U S A 99:14434–14439. doi: 10.1073/pnas.172501299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schertzer JW, Brown SA, Whiteley M. 2010. Oxygen levels rapidly modulate Pseudomonas aeruginosa social behaviours via substrate limitation of PqsH. Mol Microbiol 77:1527–1538. doi: 10.1111/j.1365-2958.2010.07303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gallagher LA, McKnight SL, Kuznetsova MS, Pesci EC, Manoil C. 2002. Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J Bacteriol 184:6472–6480. doi: 10.1128/JB.184.23.6472-6480.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heeb S, Fletcher MP, Chhabra SR, Diggle SP, Williams P, Camara M. 2011. Quinolones: from antibiotics to autoinducers. FEMS Microbiol Rev 35:247–274. doi: 10.1111/j.1574-6976.2010.00247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Michelsen CF, Christensen AM, Bojer MS, Hoiby N, Ingmer H, Jelsbak L. 2014. Staphylococcus aureus alters growth activity, autolysis, and antibiotic tolerance in a human host-adapted Pseudomonas aeruginosa lineage. J Bacteriol 196:3903–3911. doi: 10.1128/JB.02006-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Filkins LM, Graber JA, Olson DG, Dolben EL, Lynd LR, Bhuju S, O'Toole GA. 2015. Coculture of Staphylococcus aureus with Pseudomonas aeruginosa drives S. aureus towards fermentative metabolism and reduced viability in a cystic fibrosis model. J Bacteriol 197:2252–2264. doi: 10.1128/JB.00059-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Holloway BW. 1955. Genetic recombination in Pseudomonas aeruginosa. J Gen Microbiol 13:572–581. doi: 10.1099/00221287-13-3-572. [DOI] [PubMed] [Google Scholar]
- 56.Rahme LG, Stevens EJ, Wolfort SF, Shao J, Tompkins RG, Ausubel FM. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- 57.Harro JM, Daugherty S, Bruno VM, Jabra-Rizk MA, Rasko DA, Shirtliff ME. 2013. Draft genome sequence of the methicillin-resistant Staphylococcus aureus isolate MRSA-M2. Genome Announc 1(1):e00037-12. doi: 10.1128/genomeA.00037-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.