ABSTRACT
Anaerobic ammonium-oxidizing (anammox) bacteria oxidize ammonium with nitrite as the terminal electron acceptor to form dinitrogen gas in the absence of oxygen. Anammox bacteria have a compartmentalized cell plan with a central membrane-bound “prokaryotic organelle” called the anammoxosome. The anammoxosome occupies most of the cell volume, has a curved membrane, and contains conspicuous tubule-like structures of unknown identity and function. It was suggested previously that the catalytic reactions of the anammox pathway occur in the anammoxosome, and that proton motive force was established across its membrane. Here, we used antibodies raised against five key enzymes of the anammox catabolism to determine their cellular location. The antibodies were raised against purified native hydroxylamine oxidoreductase-like protein kustc0458 with its redox partner kustc0457, hydrazine dehydrogenase (HDH; kustc0694), hydroxylamine oxidase (HOX; kustc1061), nitrite oxidoreductase (NXR; kustd1700/03/04), and hydrazine synthase (HZS; kuste2859-61) of the anammox bacterium Kuenenia stuttgartiensis. We determined that all five protein complexes were exclusively located inside the anammoxosome matrix. Four of the protein complexes did not appear to form higher-order protein organizations. However, the present data indicated for the first time that NXR is part of the tubule-like structures, which may stretch the whole length of the anammoxosome. These findings support the anammoxosome as the locus of catabolic reactions of the anammox pathway.
IMPORTANCE Anammox bacteria are environmentally relevant microorganisms that contribute significantly to the release of fixed nitrogen in nature. Furthermore, the anammox process is applied for nitrogen removal from wastewater as an environment-friendly and cost-effective technology. These microorganisms feature a unique cellular organelle, the anammoxosome, which was proposed to contain the energy metabolism of the cell and tubule-like structures with hitherto unknown function. Here, we purified five native enzymes catalyzing key reactions in the anammox metabolism and raised antibodies against these in order to localize them within the cell. We showed that all enzymes were located within the anammoxosome, and nitrite oxidoreductase was located exclusively at the tubule-like structures, providing the first insights into the function of these subcellular structures.
INTRODUCTION
Anaerobic ammonium-oxidizing (anammox) bacteria oxidize ammonium to dinitrogen gas in the absence of oxygen, with nitrite as the electron acceptor and nitric oxide and hydrazine as intermediates (1). They do so through a unique set of subsequent reactions (equations 1 to 3) (Fig. 1):
| (1) |
| (2) |
| (3) |
where e stands for electron and E0′ stands for the midpoint potential of the reactions at pH 7.
FIG 1.
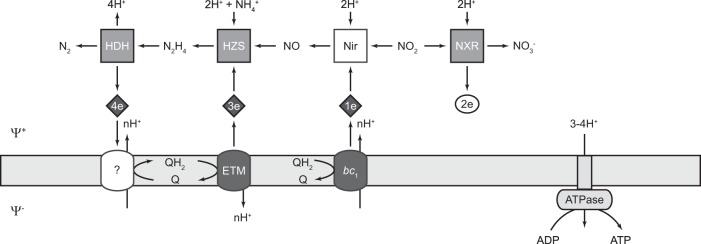
Schematic overview of anammox catabolism. Dinitrogen gas (N2) is formed from ammonium (NH4+) and nitrite (NO2−), with hydrazine (N2H4) and nitric oxide (NO) as intermediates. Diamonds represent c-type hemes involved in electron transfer between protein complexes. The number indicates the number of electrons transferred. Membrane-bound complexes are suggested to contribute to the proton motive force that is used by ATP synthase to generate ATP. Gray boxes indicate the soluble protein complexes whose intracellular locations were investigated in this study. HDH, hydrazine dehydrogenase (kustc0694); HZS, hydrazine synthase (kuste2859-60); NXR, nitrite oxidoreductase (kustd1700/03/04). NO-producing hydroxylamine oxidase (HOX; kustc1061) and the hydroxylamine oxidoreductase-like protein kustc0458 with its redox partner kustc0457, of unknown function, are not included in the scheme. The anammoxosome membrane contains the quinone (Q) pool and is energized. ψ+, positive side (anammoxosome); ψ−, negative side (cytoplasm). Other abbreviations: bc1, putative bc1 complex; ETM, electron transfer module; ATPase, F1F0-type ATP synthase; Nir, nitrite reductase. Adapted from reference 30.
In the first step, nitrite is reduced to nitric oxide (NO), which is catalyzed by a nitrite reductase (equation 1). The genome of Kuenenia stuttgartiensis, the anammox bacterium used in this study, encodes a cd1 nitrite:nitric oxide oxidoreductase (NirS, kuste4136), which was postulated to catalyze this reaction (2). NO and ammonium then are combined into hydrazine (N2H4) by the anammox-specific hydrazine synthase (HZS; kuste2859-61) (equation 2). This reaction is followed by the oxidation of hydrazine to N2 (equation 3), which is catalyzed by hydrazine dehydrogenase (HDH; kustc0694), an enzyme that is related to hydroxylamine oxidoreductase (HAO) of aerobic ammonium-oxidizing bacteria (1). The electrons released from this reaction (equation 3) ultimately are used to drive the first two reductive steps (equations 1 and 2).
HZS and HDH previously have been purified, and hydrazine synthesis and oxidation activities were shown in vitro (3). Apart from HDH, K. stuttgartiensis encodes nine additional HAO paralogs (2, 4). One of these, hydroxylamine oxidase (HOX; kustc1061), was purified and characterized in detail (5). This enzyme detoxified hydroxylamine to NO, the substrate of HZS (equation 4). Furthermore, another HAO-like protein encoded by kustc0458 has been purified from K. stuttgartiensis together with its redox partner, kustc0457, a c-type diheme-containing protein, but their function remains unknown (6).
| (4) |
| (5) |
Anammox bacteria are autotrophic microorganisms, and their growth is associated with the formation of nitrate. The oxidation of nitrite to nitrate (equation 5) was postulated to supply the cells with reducing equivalents for CO2 reduction (7, 8). The nxr gene cluster (kustd1699-1713) encodes a nitrite oxidoreductase (NXR; kustd1700/03/04) and additional electron transfer proteins that might aid in electron transport (2, 4).
Apart from nitrite reductase, all currently available anammox genomes encode close homologs of the proteins described above (2, 9–14), suggesting that they have the same energy metabolism as K. stuttgartiensis.
It was recently shown that the anammox process occurs inside a specific cellular compartment, called the anammoxosome (15). This prokaryotic organelle comprises approximately 60% of the cell volume (16) (Fig. 2). It contains iron-enriched particles and the majority of heme proteins (16–18). The anammoxosome also contains tubule-like structures, which at times stretch the whole length of the compartment. The function of these structures currently is unknown. The anammoxosome matrix is surrounded by a curved membrane which harbors an ATP synthase (19).
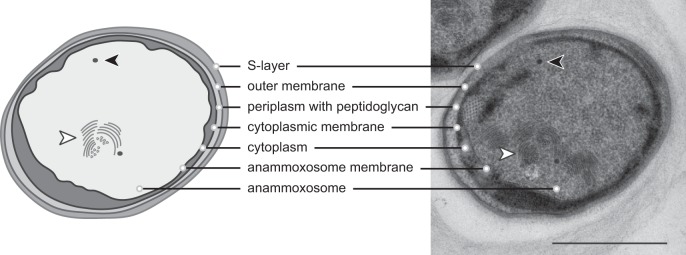
FIG 2.
Schematic overview (left) and transmission electron micrograph (right) of a Kuenenia stuttgartiensis cell. The cell plan is divided into three compartments, separated by three bilayer membranes. From outside to inside: surface-layer protein (35), outer membrane, peptidoglycan-containing periplasm (36), cytoplasmic membrane, cytoplasm, anammoxosome membrane, anammoxosome that contains tubule-like structures (white arrowheads), and electron-dense, iron-rich particles (black arrowheads). Note that not all tubule-like structures visible in the micrograph were visualized in the schematic overview. Scale bar, 500 nm.
In the present study, five key enzymes of the anammox bacterium K. stuttgartiensis were localized in the cell. To this end, antibodies against five key enzymes of anammox catabolism were generated in order to determine where these proteins were located. Labeling studies with protein A adsorbed to 10-nm gold particles showed that all five enzymes were located exclusively inside the anammoxosome. While four of the protein complexes were located in the anammoxosome matrix, NXR was specifically located at the tubule-like structures, providing the first insights into the role of these characteristic features.
MATERIALS AND METHODS
General procedures.
All chemicals used were purchased from Sigma-Aldrich, USA, unless stated otherwise. High-performance liquid chromatography (HPLC)-grade chemicals were purchased from Baker, USA. For desalting and concentration of samples, 100-kDa-molecular-mass-cutoff spin filters were used (Vivaspin 20 or 500; Sartorius Stedim Biotech, Germany). All fast-performance liquid chromatography (FPLC) steps were performed on an ÄKTA purifier system (GE Healthcare, Sweden). Depending on the column material and diameter, the FPLC columns were run with flow rates of 5 or 2 ml/min. The elution of proteins was monitored on-line by the absorbance at 280 nm, and samples were collected in 2-ml fractions. All steps apart from the FPLC runs were performed at 4°C.
Preparation of CE.
In brief, cell extract (CE) was obtained from an ∼95% pure enrichment culture of K. stuttgartiensis (3). Cells were harvested by centrifugation (5,000 × g for 15 min at 4°C). Cell pellets were resuspended in 20 mM potassium phosphate (KPi) buffer, pH 7.0, and French pressed in three passages at 138 MPa. Cell debris was removed by centrifugation (5,000 × g for 15 min at 4°C), and the supernatant was subjected to an ultracentrifugation step (180,000 × g for 1 h at 4°C). The resulting membrane pellet was dissolved by incubation with detergent for 1 h at 4°C (2% [wt/vol] β-laurylmaltoside for NXR and 1% [wt/vol] sodium deoxycholate for all other enzymes). After a second ultracentrifugation step, the two clear dark red supernatants were combined, representing the CE.
Protein purification.
All enzymes were purified from CE using liquid chromatography. Purification of HZS, HDH, and HOX was performed as described previously (1, 5). The HAO-like protein kustc0458, together with its partner kustc0457 and NXR, were purified as follows. CE was loaded onto a 60-ml Q Sepharose XL column (GE Healthcare) equilibrated with 20 mM Tris-HCl buffer, pH 8.0. NXR eluted isocratically with 20 mM Tris-HCl, pH 8.0, containing 200 mM NaCl. kustc0457/58 eluted at 400 mM NaCl in 20 mM Tris-HCl, pH 8.0. The collected fractions were subsequently loaded onto a 30-ml ceramic hydroxyapatite column (Bio-Rad, USA). In the case of kustc0457/58, the column was equilibrated with 20 mM KPi, pH 7.0, and protein elution was performed in a linear-gradient (20 to 500 mM) KPi buffer, pH 7.0. kustc0457/58 eluted with 44 mM KPi, pH 7.0. At this stage, kustc0457/58 was purified to homogeneity, as shown by SDS-PAGE. For NXR, the hydroxyapatite column was equilibrated with 75 mM KPi, pH 8.0, and proteins were eluted with a linear gradient of 75 to 500 mM KPi, pH 8.0. NXR eluted at ∼90 mM KPi, pH 8.0. The fraction containing NXR was loaded onto a 30-ml SP Sepharose fast-flow column (GE Healthcare, USA) previously equilibrated with 30 mM KPi, pH 6.0. The runoff was loaded onto a Sephacryl S-400 HR gel filtration column (GE Healthcare, USA) equilibrated in 50 mM KPi buffer, pH 7.3, containing 150 mM NaCl. NXR was purified to homogeneity after this step, as shown by SDS-PAGE. Identification of all subunits was performed by matrix-assisted laser desorption ionization–time-of-flight mass spectrometry (MALDI-TOF MS) analysis (see below).
Antibody generation.
Antisera containing the antibodies against the five protein complexes of K. stuttgartiensis were generated against liquid protein samples. For each enzyme, two rabbits were immunized in a 3-month immunization protocol (Eurogentec SA, Belgium). Anti-HOX, anti-HZS, and anti-NXR subsequently were affinity purified from both rabbit antisera. The total IgG fractions (crude sera) of one rabbit each were used as anti-HDH and anti-kustc0457/58. The obtained antisera were used as the primary antibody in immunoblot analysis and immunogold localization (see below).
Immunoblot analysis.
Blots were made from 10% SDS-PAGE gels containing 30 μg of CE. After separation, the proteins were blotted onto a Protran nitrocellulose transfer membrane (Whatman, Germany) using a semidry transfer cell blotting system (Bio-Rad, Netherlands). Membrane and cellulose chromatography papers (3MM Chr; Whatman, Germany) were soaked in transfer buffer (25 mM Tris, 192 mM glycine, and 20% methanol), after which the proteins were blotted onto the membrane for 45 min at 100 mA. After drying, the blots were washed in ultrapure water for 30 min and then soaked in blocking buffer (1% bovine serum albumin [BSA] in Tris-buffered saline, pH 7.4 [TBS]) for an hour. Here, the blots were incubated for 1 h with either preimmune serum in the case of kustc0457/58 and HDH (negative control), the total IgG fraction of rabbits (anti-kustc0457/58 and anti-HDH), or affinity-purified antibodies (HOX, NXR and HZS) diluted 2,000-fold in serum diluent (the same as the blocking buffer). As an additional negative control, no primary antibody was added. After washing the blots three times in TBS–0.05% Tween 20 for 10 min, blots were incubated with a monoclonal mouse anti-rabbit IgG–alkaline phosphatase conjugate diluted 150,000-fold in blocking buffer. After 1 h of incubation, the blots were washed twice in TBS–0.05% Tween 20, followed by two washing steps in TBS only for 10 min each. For immune detection, the blots were incubated in the 5-bromo-4-chloro-3-indolylphosphate–nitroblue tetrazolium (BCIP-NBT) liquid substrate system (Sigma, The Netherlands) until purple bands appeared (usually between 2 and 10 min). After washing in demineralized water, the blots were air dried before imaging and storage at 4°C.
Sample preparation for immunogold localization on Lowicryl sections.
Biofilm aggregates of K. stuttgartiensis were harvested from a continuous culture and cryofixed by high-pressure freezing (Leica EMHPF; Leica Microsystems, Austria). Samples were freeze substituted in a freeze substitution unit (Leica Microsystems, Austria) in anhydrous acetone (Seccosolv; Merck) containing 0.1% uranyl acetate (Merck) at −90°C for 46 h, at −60°C for 8 h, and at −40°C for 8 h in subsequent steps. Infiltration was performed at −40°C by gradually increasing concentrations of Lowicryl HM20 (EMS, USA) in acetone (1:3, 1:1, 3:1, and finally 3× pure HM20 resin). The duration of each step was 1 h. Polymerization under UV light was carried out at −40°C for 48 h with subsequent curing with UV light at room temperature for approximately 10 h. Ultrathin sections (approximately 60 nm) were incubated on 0.1% glycine–PHEM [60 mM piperazine-N,N′-bis(2-ethanesulfonic acid), 25 mM HEPES, 10 mM EGTA, 2 mM MgCl2, pH 6.9] before blocking with 1% BSA–PHEM for 15 min. Incubation with primary antibodies diluted 50 to 300 times in 1% BSA–PHEM was performed for 1 h. As negative controls, sections were incubated with preimmune sera (for anti-kustc0457/58 and anti-HDH) and without primary antibody. After washing with 0.1% BSA–PHEM, incubation with protein A gold (10 nm; PAG-10; CMC UMC Utrecht) in 0.1% BSA–PHEM was performed for 1 h. Sections were washed with 0.1% BSA–PHEM and with PHEM. Antibody interactions were fixed by 1% glutaraldehyde–PHEM for 10 min. Sections subsequently were washed with PHEM buffer and ultrapure water. Poststaining was performed in the dark with 4% aqueous uranyl acetate for 20 min and Reynolds lead citrate for 2 min and investigated with a JEOL (Japan) Jem1010 transmission electron microscope operating at 60 kV.
Sample preparation for immunogold localization on rehydrated cryosections.
K. stuttgartiensis cells were cryofixed by high-pressure freezing and freeze substituted in acetone containing 0.5% glutaraldehyde, 0.1% uranyl acetate, and 1% H2O as described previously (19). Uranyl acetate was removed by washing four times for 30 min in the AFS (Automated Freeze Substitution) at −30°C and once for 60 min on ice with acetone containing 0.5% glutaraldehyde and 1% H2O. Samples were rehydrated in a graded acetone series on ice: 95%, 90%, 80%, and 70% acetone in water containing 0.5% glutaraldehyde, 50% and 30% acetone in PHEM buffer containing 0.5% glutaraldehyde, and finally 0.5% glutaraldehyde in PHEM buffer. Samples were rinsed in PHEM buffer and embedded in 12% gelatin in PHEM buffer. The gelatin-embedded cells were cut into small cubes (1 to 2 mm3) under the stereo microscope, infiltrated overnight at 4°C with 2.3 M sucrose in PHEM buffer, and frozen in liquid nitrogen (20). Samples were cryosectioned using a cryoultramicrotome UC7/FC7 (Leica Microsystems, Vienna, Austria). Cryosections (65 nm) were picked up with a drop of 1% methylcellulose and 1.15 M sucrose in PHEM buffer and transferred to Formvar carbon-coated copper hexagonal 100-mesh grids for immunogold localization.
After the immunogold localization procedure, cryosections were poststained with 2% uranyl acetate in 0.15 M oxalic acid, pH 7.4, at room temperature and embedded in 1.8% methyl cellulose containing 0.4% aqueous uranyl acetate on ice, after which they were air dried.
Bioinformatics tools.
An in-house protein database was created by translating all open reading frames in the genome (PRJNA16685) of K. stuttgartiensis (2). SignalP 4.0 (21) and TMHMM 2.0 (22) software packages were used to evaluate signal peptides and putative transmembrane helices. For identification of tat leader sequences, tatP software was used (23).
Protein determination, polyacrylamide gel electrophoresis, and protein identification.
Protein quantification was performed with the Bio-Rad protein assay, based on the Bradford method (24), with bovine serum albumin as the standard. To check for purity, 30 μg of protein was separated on a 10% SDS-PAGE gel, as described previously (25), and was stained with a ready-to-use colloidal Coomassie blue stain (Severn Biotech Ltd., USA). The PageRuler Plus prestained protein ladder (Thermo Scientific, USA) was used as a molecular weight marker. Subunit identification of all enzymes was performed by MALDI-TOF MS as described previously (1, 26). In brief, gel plugs were picked from visible protein bands and subjected to tryptic digestion and MALDI-TOF MS analysis on a Bruker III mass spectrometer (Bruker Daltonik, Germany). Peptides were analyzed using the Mascot Peptide Fingerprint search software (Matrix Science, United Kingdom) against the in-house protein database. Search settings included carbamidomethylation (27) as a fixed and methionine oxidation as a variable peptide modification. One missed cleavage site and a mass difference of ±0.2 Da were tolerated.
RESULTS
Protein purification and subunit identification.
All five protein complexes were purified to homogeneity, as was shown by SDS-PAGE analysis (Fig. 3A to E). Tryptic digestion followed by MALDI-TOF MS analysis unambiguously identified all subunits of the purified protein complexes. Protein sequence analyses indicated that all HAO-like proteins used in this study and the three HZS subunits contained a transmembrane helix representing a signal peptide with predicted cleavage sites after the leader sequence at the N terminus of the proteins. While the C subunit of NXR (kustd1704) contained a signal peptide of the sec translocation pathway, the B subunit lacked such a signal peptide. kustd1700 (NxrA), the molybdopterin- and iron sulfur cluster-containing catalytic subunit, clearly had a conserved twin-arginine (RR) motif, suggesting translocation of this protein via the tat translocation system, presumably taking NxrB as a passenger protein (28).
FIG 3.
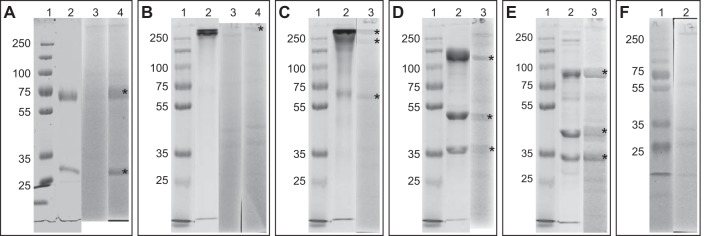
Ten percent SDS-PAGE gels of the five purified enzymes and immunoblot analyses of the five purified proteins. Lane 2 contained approximately 30 μg of purified protein. For immunoblot analyses (lanes 3 and 4), 30 μg cell extract separated by 10% SDS-PAGE was blotted onto a cellulose-nitrate membrane, and blots were incubated with the respective antibodies. Lane 1 in each panel is a 6-μl PageRuler plus prestained protein ladder (Thermo Scientific, USA). (A) Lane 2, purified kustc0457/58; lane 3, incubation with the preimmune serum of anti-kustc0457/58; lane 4, incubation with anti-kustc0457/58. (B) Lane 2, purified HDH (kustc0694); lane 3, incubation with the preimmune serum of anti-HDH; lane 4, incubation with anti-HDH. (C) Lane 2, purified HOX (kustc1061); lane 3, incubation with affinity-purified anti-HOX. (D) Lane 2, purified NXR (kustd1700/03/04); lane 3, incubation with affinity-purified anti-NXR. (E) Lane 2, purified HZS (kuste2859-61); lane 3, incubation with affinity-purified anti-HZS. (F) Lane 2, incubation with only secondary antibody. Asterisks indicate expected target subunits. It should be noted that the dominant heme-containing proteins of K. stuttgartiensis were clearly visible as yellowish (light gray in this figure) bands on the blots; therefore, they were distinguishable from a positive immunoblot reaction.
Antibody generation and immunoblotting.
Purified protein complexes were used to generate antibodies in rabbits. Incubation of blotted cell extract with the five obtained antisera resulted in specific bands of all subunits of the antigens (Fig. 3). For kustc0457/58 (anti-kustc0457/58) and HDH (anti-HDH), the total IgG fraction (crude sera) of one rabbit each was used for immunoblotting and subsequent intracellular localization of the complexes. Controls for these two protein complexes included preimmune sera of the immunized rabbits, which showed no reaction with Kuenenia proteins (Fig. 3A and B). For HOX (anti-HOX), NXR (anti-NXR), and HZS (anti-HZS), the generated antibodies were affinity purified from the antisera. Incubation of blots without primary antibody showed no reaction with Kuenenia proteins present in the cell extract (Fig. 3F). It should be noted that dominant heme-containing proteins of K. stuttgartiensis already were visible as yellowish bands on the blots and were clearly distinguishable from the positive (purple) immunoblot reaction.
Incubation of the blots with antiserum against kustc0457/58 resulted in two specific bands, at approximately 60 and 27 kDa, which was in agreement with the expected theoretical masses of the subunits (Fig. 3A). Although the HDH monomer has a molecular mass of approximately 65 kDa, only the covalently bound trimer was detected at >200 kDa (Fig. 3B), albeit with a weak signal (6). In the case of HOX, the covalently bound trimer (>200 kDa), and, to a lesser extent, also the dimeric (∼130 kDa) and monomeric forms (∼55 kDa) of the protein complex were detected after incubation with the antiserum (Fig. 3C) (5). Three specific bands were visible when incubating blotted crude extract with the antibody against NXR, representing the subunits NxrA (∼130 kDa), NxrB (∼50 kDa), and NxrC (∼35 kDa) (Fig. 3D). Similarly, all three subunits, HzsA (∼90 kDa), HzsC (∼42 kDa), and HzsB (∼39 kDa), were detected specifically when incubating the blots with anti-HZS (Fig. 3E).
Intracellular localization of the five key enzymes.
All five metabolic enzymes were unambiguously localized within the cell using immunogold localization on high-pressure-frozen, freeze-substituted, Lowicryl HM20-embedded, and ultrathin sectioned cells of K. stuttgartiensis. This was confirmed on cryosections of high-pressure-frozen, freeze-substituted, and rehydrated K. stuttgartiensis cells, which were embedded in methylcellulose-containing aqueous uranyl acetate after immunolabeling. With this method, membranes appear in negative contrast, providing clear discrimination between lipid-based and proteinaceous structures.
As negative controls, the sections were incubated with only protein A coupled to 10-nm gold particles (PAG-10), omitting the primary antibodies and including the preimmune sera (for anti-kustc0457/58 and anti-HDH). Negative controls showed only very few gold labels, derived from aspecific binding of either the preimmune sera (Fig. 4C; also see Fig. 6C) or PAG-10 to the fixed cells or the embedding medium (data not shown). The electron-dense, iron-containing anammoxosome particles seen in the immunogold localization images (fuzzy, 16 to 25 nm [16]) could be distinguished from the gold labels (sharp, 10 nm) based on their size and appearance.
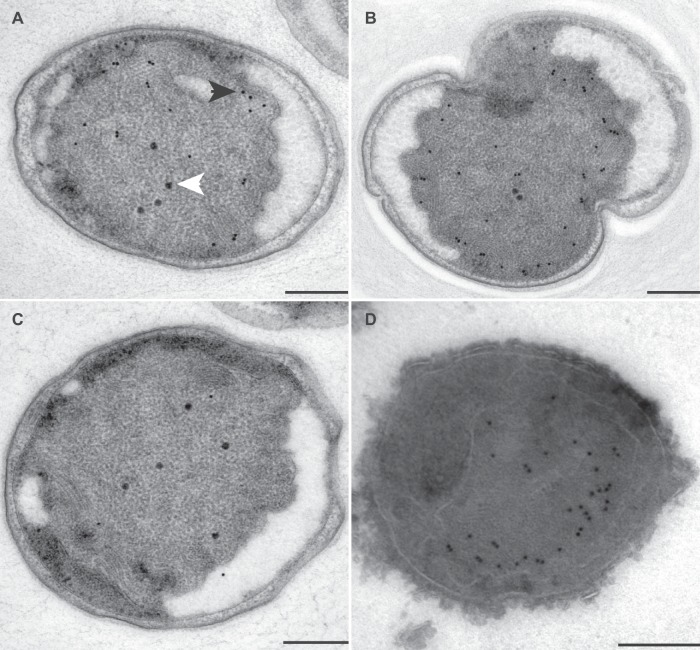
FIG 4.
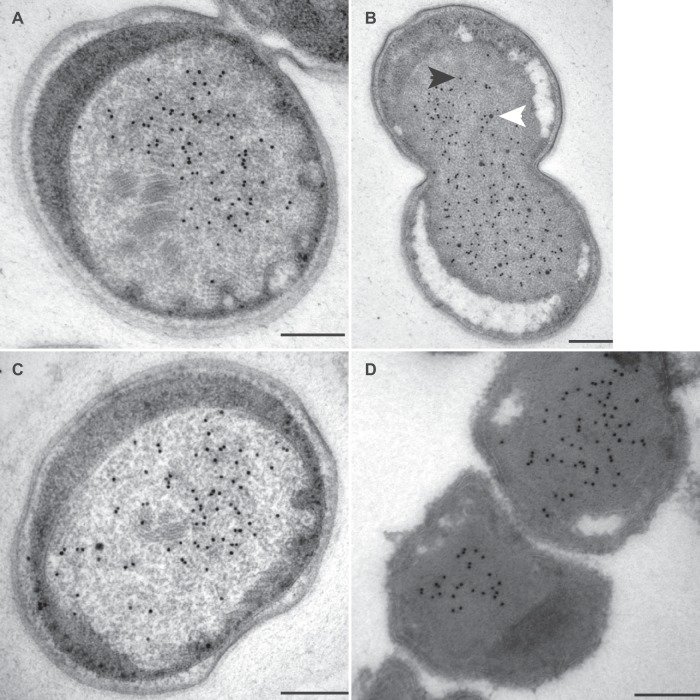
Transmission electron micrographs of ultrathin sections of high-pressure-frozen, freeze-substituted, Lowicryl-embedded (A to C) or rehydrated and cryosectioned (D) K. stuttgartiensis cells incubated with antibodies against kustc0457/58. Thin sections were incubated with 1:150-diluted anti-kustc0457/58 (A and B), 1:50-diluted preimmune serum (C), or 1:100-diluted anti-kustc0457/58 (D). Gold particles were visible only inside the anammoxosome, preferentially located toward the inner side of the anammoxosome membrane. Incubation with preimmune serum led to the detection of a few gold particles throughout the cell. Electron-dense, iron-containing particles (16 to 25 nm; white arrowhead) were distinguished from gold labels (10 nm; black arrowhead) by size and appearance. Scale bars, 200 nm.
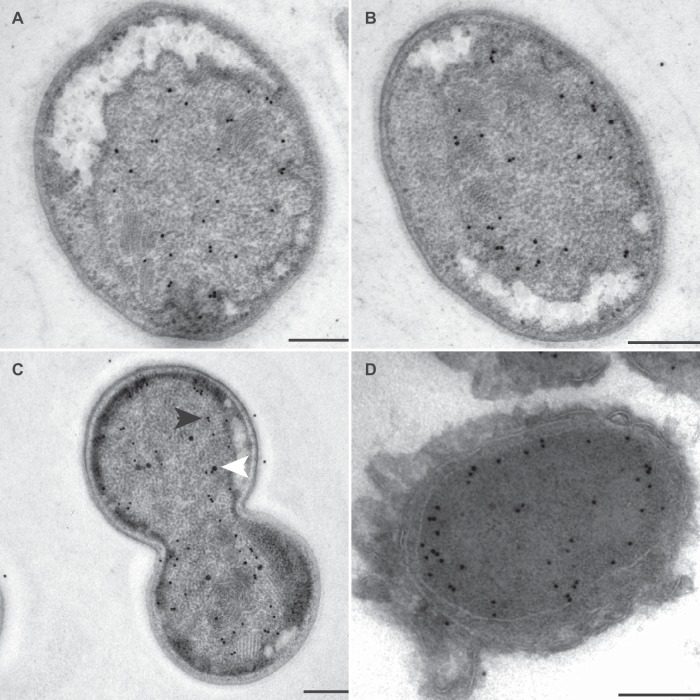
FIG 6.
Transmission electron micrographs of ultrathin sections of high-pressure-frozen, freeze-substituted, Lowicryl-embedded (A to C) or rehydrated and cryosectioned (D) K. stuttgartiensis cells incubated with antibodies against HDH. Thin sections were incubated with 1:100-diluted anti-HDH (A and B), 1:50-diluted preimmune serum (C), or 1:100-diluted anti-HDH (D). Gold particles were visible only inside the anammoxosome. Electron-dense, iron-containing particles (16 to 25 nm; white arrowhead) were distinguished from gold labels (10 nm; black arrowhead) by size and appearance. Scale bars, 200 nm.
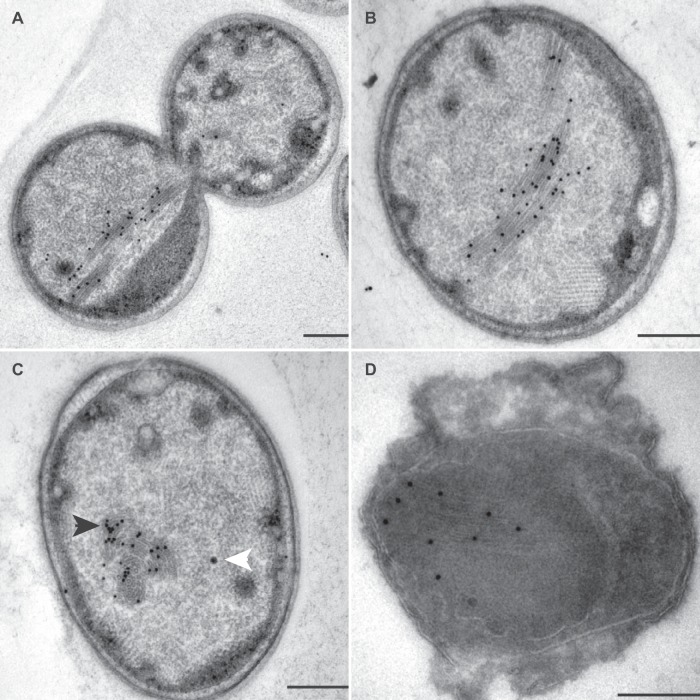
Incubation with anti-kustc0457/58 and anti-HOX localized these protein complexes to inside the anammoxosome, preferentially near the inner side of the membrane (Fig. 4 and 5). HDH (Fig. 6) and HZS (Fig. 7) were distributed across the anammoxosome. In contrast, anti-NXR was very specifically observed at the tubule-like structures in the anammoxosome (Fig. 8) in both transverse (Fig. 8C) and longitudinal (Fig. 8A, B, and D) sectioned tubules. Although all cells most probably contained the tubule-like structures, these were visible only if the cell was sectioned through that plane. If the structures were not in the plane of the section, only a little labeling with anti-NXR was observed (data not shown). The tubules were most visible in the lowicryl-embedded sections.
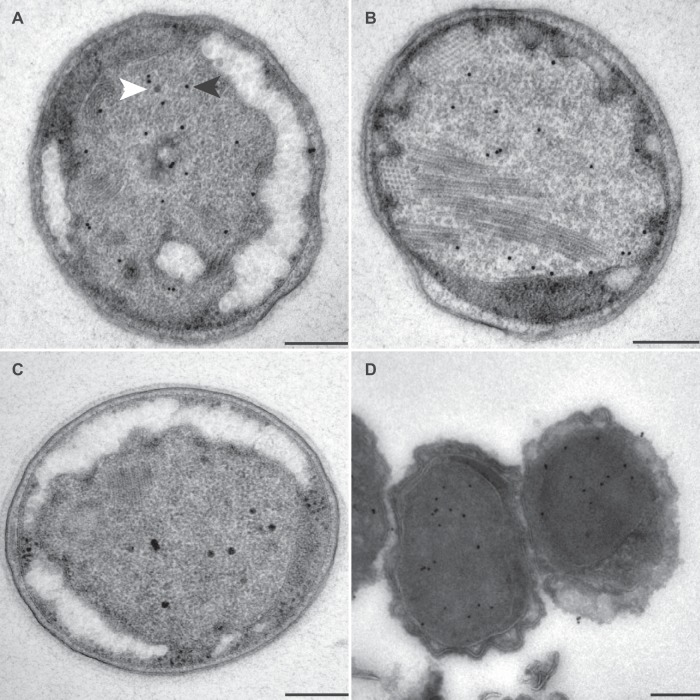
FIG 5.
Transmission electron micrographs of ultrathin sections of high-pressure-frozen, freeze-substituted, Lowicryl-embedded (A to C) or rehydrated and cryosectioned (D) K. stuttgartiensis cells incubated with affinity-purified antibodies against HOX. Thin sections were incubated with 1:300-diluted anti-HOX (A and B) or 1:100-diluted anti-HOX (C and D). Gold particles were visible only inside the anammoxosome. HOX appeared to be preferentially localized in the proximity of the anammoxosome membrane. Electron-dense, iron-containing particles (16 to 25 nm; white arrowhead) were distinguished from gold labels (10 nm; black arrowhead) by size and appearance. Scale bars, 200 nm.
FIG 7.
Transmission electron micrographs of ultrathin sections of high-pressure-frozen, freeze-substituted, Lowicryl-embedded (A to C) or rehydrated and cryosectioned (D) K. stuttgartiensis cells incubated with affinity-purified antibodies against HZS. Thin sections were incubated with 1:200-diluted anti-HZS (A to C) or 1:300-diluted anti-HZS (D). Gold particles were visible only inside the anammoxosome. Electron-dense, iron-containing particles (16 to 25 nm; white arrowhead) were distinguished from gold labels (10 nm; black arrowhead) by size and appearance. Scale bars, 200 nm.
FIG 8.
Transmission electron micrographs of ultrathin sections of high-pressure-frozen, freeze-substituted, Lowicryl-embedded (A to C) or rehydrated and cryosectioned (D) K. stuttgartiensis cells incubated with affinity-purified antibodies against NXR. Thin sections were incubated with 1:100-diluted anti-NXR (A), 1:200-diluted anti-NXR (B and C), and 1:100-diluted anti-NXR (D). In all cells, the labeling was specifically directed against the tubule-like structures present in this compartment (insert); longitudinal (A, B, and D) and transverse (C) sections of these structures are shown. Electron-dense, iron-containing particles (16 to 25 nm; white arrowhead) were distinguished from gold labels (10 nm; black arrowhead) by size and appearance. Scale bars, 200 nm.
DISCUSSION
Since its first observation in electron microscopy studies, the anammoxosome has been suggested to be the location of energy metabolism of the anammox cell (17, 29). Recent purification studies of intact anammoxosomes in combination with activity assays showed that the purified anammoxosome indeed could catalyze the anammox reaction (equations 1 to 3) (15). In the present study, we showed the intracellular localization of the hydroxylamine oxidoreductase (HAO)-like protein kustc0457/58, hydrazine dehydrogenase (HDH), the NO-producing HAO-like protein HOX, nitrite oxidoreductase (NXR), and hydrazine synthase (HZS) within an anammox cell. All of these proteins are encoded in all presently available anammox genomes and play key roles in anammox catabolism (1, 2, 9–14). The fact that all studied protein complexes were exclusively immunolocalized to the anammoxosome in the present study supported the hypothesis that this cell compartment was a true prokaryotic organelle with a dedicated function.
Antibodies used in this study were generated against the purified native protein complexes of the anammox bacterium K. stuttgartiensis and reacted specifically against Kuenenia crude extract on immunoblots and cells in immunogold localization. The localization studies confirmed all protein complexes to be soluble, as opposed to membrane bound, as labeling was observed exclusively inside the anammoxosome. This was in line with the identification of signal peptides either of the sec or of the tat translocation system, suggesting transportation of the expressed polypeptides from the ribosome-containing cytoplasm across the (anammoxosome) membrane.
The detection of substantial amounts of kustc0457/58 within the vicinity of the anammoxosome membrane indicated an important role for this protein complex in anammox metabolism. Unfortunately, the physiological function of this protein remains elusive, but sequence analysis of kustc0458 suggested a reductive function of this HAO-like protein (30). Interestingly, in the present study we could show that the antibodies against NXR were exclusively directed toward the tubule-like structures within the anammoxosome. These structures previously were suggested to support cell division or structural integrity of the cell (29, 31) or represent highly abundant, densely packed metabolic enzymes (16). The present data suggested that these tubule-like structures contained NXR, most likely facilitating high local concentrations of this protein complex. Such high local concentrations of NXR are also found in aerobic nitrite-oxidizing bacteria, where the protein complex formed higher-ordered two-dimensional structures (32–34). However, NXR was anchored to a membrane in these cells. Apparently, instead of using membrane stacks to increase local NXR concentrations, as observed in aerobic nitrite-oxidizing bacteria, anammox bacteria evolved to produce higher-order protein structures in a dedicated catabolic organelle to serve the same purpose. Further studies will focus on determining how these tubule-like structures form and whether NXR is the only protein involved in their formation.
ACKNOWLEDGMENTS
We thank Elly van Donselaar and Cveta Tomova for assistance with sample preparation for TEM. We thank Winnie Toonders for initial experiments.
N.M.D.A., B.K., and L.V.N. are supported by the Netherlands Organization for Scientific Research (ALW grant 818.02.015 and VENI grants 863.11.003 and 863.09.009, respectively); R.J.M. is supported by the 2012 Netherlands Organization for Scientific Research SPINOZA Award, awarded to M.S.M.J.; S.N., C.F., and M.S.M.J. are supported by the European Research Council (ERC232937).
REFERENCES
- 1.Kartal B, Maalcke WJ, de Almeida NM, Cirpus I, Gloerich J, Geerts W, Op den Camp HJM, Harhangi HR, Janssen-Megens EM, Francoijs K-J, Stunnenberg HG, Keltjens JT, Jetten MSM, Strous M. 2011. Molecular mechanism of anaerobic ammonium oxidation. Nature 479:127–130. doi: 10.1038/nature10453. [DOI] [PubMed] [Google Scholar]
- 2.Strous M, Pelletier E, Mangenot S, Rattei T, Lehner A, Taylor MW, Horn M, Daims H, Bartol-Mavel D, Wincker P, Barbe V, Fonknechten N, Vallenet D, Segurens B, Schenowitz-Truong C, Médigue C, Collingro A, Snel B, Dutilh BE, Op den Camp HJM, van der Drift C, Cirpus I, van de Pas-Schoonen KT, Harhangi HR, van Niftrik L, Schmid M, Keltjens J, van de Vossenberg J, Kartal B, Meier H, Frishman D, Huynen MA, Mewes H-W, Weissenbach J, Jetten MSM, Wagner M, Le Paslier D. 2006. Deciphering the evolution and metabolism of an anammox bacterium from a community genome. Nature 440:790–794. doi: 10.1038/nature04647. [DOI] [PubMed] [Google Scholar]
- 3.Kartal B, Geerts W, Jetten MSM. 2011. Cultivation, detection, and ecophysiology of anaerobic ammonium-oxidizing bacteria. Methods Enzymol 486:89–108. doi: 10.1016/B978-0-12-381294-0.00004-3. [DOI] [PubMed] [Google Scholar]
- 4.de Almeida NM, Maalcke WJ, Keltjens JT, Jetten MSM, Kartal B. 2011. Proteins and protein complexes involved in the biochemical reactions of anaerobic ammonium-oxidizing bacteria. Biochem Soc Trans 39:303–308. doi: 10.1042/BST0390303. [DOI] [PubMed] [Google Scholar]
- 5.Maalcke WJ, Dietl A, Marritt SJ, Butt JN, Jetten MSM, Keltjens JT, Barends TRM, Kartal B. 2014. Structural basis of biological NO generation by octaheme oxidoreductases. J Biol Chem 289:1228–1242. doi: 10.1074/jbc.M113.525147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maalcke WJ. 2012. Ph.D. thesis. Multiheme protein complexes of anaerobic ammonium-oxidizing bacteria. Radboud University Nijmegen, Nijmegen, the Netherlands. [Google Scholar]
- 7.Van de Graaf AA, de Bruijn P, Robertson LA, Jetten MS, Kuenen JG. 1996. Autotrophic growth of anaerobic ammonium-oxidizing micro-organisms in a fluidized bed reactor. Microbiology 142:2187–2196. doi: 10.1099/13500872-142-8-2187. [DOI] [Google Scholar]
- 8.Schouten S, Strous M, Kuypers MMM, Rijpstra WIC, Baas M, Schubert CJ, Jetten MSM, Sinninghe Damsté JS. 2004. Stable carbon isotopic fractionations associated with inorganic carbon fixation by anaerobic ammonium-oxidizing bacteria. Appl Environ Microbiol 70:3785–3788. doi: 10.1128/AEM.70.6.3785-3788.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gori F, Tringe SG, Kartal B, Marchiori E, Machiori E, Jetten MSM. 2011. The metagenomic basis of anammox metabolism in Candidatus “Brocadia fulgida.” Biochem Soc Trans 39:1799–1804. doi: 10.1042/BST20110707. [DOI] [PubMed] [Google Scholar]
- 10.Hira D, Toh H, Migita CT, Okubo H, Nishiyama T, Hattori M, Furukawa K, Fujii T. 2012. Anammox organism KSU-1 expresses a NirK-type copper-containing nitrite reductase instead of a NirS-type with cytochrome cd1. FEBS Lett 586:1658–1663. doi: 10.1016/j.febslet.2012.04.041. [DOI] [PubMed] [Google Scholar]
- 11.van de Vossenberg J, Woebken D, Maalcke WJ, Wessels HJCT, Dutilh BE, Kartal B, Janssen-Megens EM, Roeselers G, Yan J, Speth D, Gloerich J, Geerts W, van der Biezen E, Pluk W, Francoijs K-J, Russ L, Lam P, Malfatti SA, Tringe SG, Haaijer SCM, Op den Camp HJM, Stunnenberg HG, Amann R, Kuypers MMM, Jetten MSM. 2013. The metagenome of the marine anammox bacterium “Candidatus Scalindua profunda” illustrates the versatility of this globally important nitrogen cycle bacterium. Environ Microbiol 15:1275–1289. doi: 10.1111/j.1462-2920.2012.02774.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ali M, Oshiki M, Awata T, Isobe K, Kimura Z, Yoshikawa H, Hira D, Kindaichi T, Satoh H, Fujii T, Okabe S. 2014. Physiological characterization of anaerobic ammonium oxidizing bacterium “Candidatus Jettenia caeni.” Environ Microbiol doi: 10.1111/1462-2920.12674. [DOI] [PubMed] [Google Scholar]
- 13.Speth DR, Russ L, Kartal B, Op den Camp HJM, Dutilh BE, Jetten MSM. 2015. Draft genome sequence of anammox bacterium “Candidatus Scalindua brodae,” obtained using differential coverage binning of sequencing data from two reactor enrichments. Genome Announc 3:e01415-14. doi: 10.1128/genomeA.01415-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oshiki M, Shinyako-Hata K, Satoh H, Okabe S. 2015. Draft genome sequence of an anaerobic ammonium-oxidizing bacterium, “Candidatus Brocadia sinica.” Genome Announc 3:e00267-15. doi: 10.1128/genomeA.00267-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neumann S, Wessels HJ, Rijpstra WIC, Sinninghe Damsté JS, Kartal B, Jetten MS, Niftrik L. 2014. Isolation and characterization of a prokaryotic cell organelle from the anammox bacterium Kuenenia stuttgartiensis. Mol Microbiol 94:794–802. doi: 10.1111/mmi.12816. [DOI] [PubMed] [Google Scholar]
- 16.van Niftrik L, Geerts WJC, van Donselaar EG, Humbel BM, Yakushevska A, Verkleij AJ, Jetten MSM, Strous M. 2008. Combined structural and chemical analysis of the anammoxosome: a membrane-bounded intracytoplasmic compartment in anammox bacteria. J Struct Biol 161:401–410. doi: 10.1016/j.jsb.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 17.Lindsay M, Webb R, Strous M, Jetten M, Butler M, Forde R, Fuerst J. 2001. Cell compartmentalisation in planctomycetes: novel types of structural organisation for the bacterial cell. Arch Microbiol 175:413–429. doi: 10.1007/s002030100280. [DOI] [PubMed] [Google Scholar]
- 18.van Niftrik L, Geerts WJC, van Donselaar EG, Humbel BM, Webb RI, Fuerst JA, Verkleij AJ, Jetten MSM, Strous M. 2008. Linking ultrastructure and function in four genera of anaerobic ammonium-oxidizing bacteria: cell plan, glycogen storage, and localization of cytochrome c proteins. J Bacteriol 190:708–717. doi: 10.1128/JB.01449-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Niftrik L, van Helden M, Kirchen S, van Donselaar EG, Harhangi HR, Webb RI, Fuerst JA, Op den Camp HJM, Jetten MSM, Strous M. 2010. Intracellular localization of membrane-bound ATPases in the compartmentalized anammox bacterium “Candidatus Kuenenia stuttgartiensis.” Mol Microbiol 77:701–715. doi: 10.1111/j.1365-2958.2010.07242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Donselaar E, Posthuma G, Zeuschner D, Humbel BM, Slot JW. 2007. Immunogold labeling of cryosections from high-pressure frozen cells. Traffic 8:471–485. doi: 10.1111/j.1600-0854.2007.00552.x. [DOI] [PubMed] [Google Scholar]
- 21.Petersen TN, Brunak S, Heijne von G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 22.Krogh A, Larsson B, Heijne von G, Sonnhammer ELL. 2001. Predicting transmembrane protein topology with a Hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 23.Bendtsen JD, Nielsen H, Widdick D, Palmer T, Brunak S. 2005. Prediction of twin-arginine signal peptides. BMC Bioinformatics 6:167. doi: 10.1186/1471-2105-6-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 25.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 26.Farhoud MH, Wessels HJCT, Steenbakkers PJM, Mattijssen S, Wevers RA, van Engelen BG, Jetten MSM, Smeitink JA, van den Heuvel LP, Keltjens JT. 2005. Protein complexes in the archaeon Methanothermobacter thermautotrophicus analyzed by blue native/SDS-PAGE and mass spectrometry. Mol Cell Proteomics 4:1653–1663. doi: 10.1074/mcp.M500171-MCP200. [DOI] [PubMed] [Google Scholar]
- 27.Nielsen ML, Vermeulen M, Bonaldi T, Cox J, Moroder L, Mann M. 2008. Iodoacetamide-induced artifact mimics ubiquitination in mass spectrometry. Nat Methods 5:459–460. doi: 10.1038/nmeth0608-459. [DOI] [PubMed] [Google Scholar]
- 28.Rothery RA, Workun GJ, Weiner JH. 2008. The prokaryotic complex iron-sulfur molybdoenzyme family. Biochim Biophys Acta 1778:1897–1929. doi: 10.1016/j.bbamem.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 29.van Niftrik LA, Fuerst JA, Damsté JSS, Kuenen JG, Jetten MSM, Strous M. 2004. The anammoxosome: an intracytoplasmic compartment in anammox bacteria. FEMS Microbiol Lett 233:7–13. doi: 10.1016/j.femsle.2004.01.044. [DOI] [PubMed] [Google Scholar]
- 30.Kartal B, de Almeida NM, Maalcke WJ, Op den Camp HJM, Jetten MSM, Keltjens JT. 2013. How to make a living from anaerobic ammonium oxidation. FEMS Microbiol Rev 37:428–461. doi: 10.1111/1574-6976.12014. [DOI] [PubMed] [Google Scholar]
- 31.Fuerst JA. 2005. Intracellular compartmentation in planctomycetes. Annu Rev Microbiol 59:299–328. doi: 10.1146/annurev.micro.59.030804.121258. [DOI] [PubMed] [Google Scholar]
- 32.Spieck E, Aamand J, Bartosch S, Bock E. 1996. Immunocytochemical detection and location of the membrane-bound nitrite oxidoreductase in cells of Nitrobacter and Nitrospira. FEMS Microbiol Lett 139:71–76. doi: 10.1111/j.1574-6968.1996.tb08181.x. [DOI] [Google Scholar]
- 33.Spieck E, Muller S, Engel A, Mandelkow E, Patel H, Bock E. 1996. Two-dimensional structure of membrane-bound nitrite oxidoreductase from Nitrobacter hamburgensis. J Struct Biol 117:117–123. doi: 10.1006/jsbi.1996.0076. [DOI] [Google Scholar]
- 34.Spieck E, Ehrich S, Aamand J, Bock E. 1998. Isolation and immunocytochemical location of the nitrite-oxidizing system in Nitrospira moscoviensis. Arch Microbiol 169:225–230. doi: 10.1007/s002030050565. [DOI] [PubMed] [Google Scholar]
- 35.van Teeseling MCF, de Almeida NM, Klingl A, Speth DR, Op den Camp HJM, Rachel R, Jetten MSM, van Niftrik L. 2014. A new addition to the cell plan of anammox bacteria: “Candidatus Kuenenia stuttgartiensis” has a protein surface layer as the outermost layer of the cell. J Bacteriol 196:80–89. doi: 10.1128/JB.00988-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Teeseling MCF, Mesman RJ, Kuru E, Espaillat A, Cava F, Brun YV, VanNieuwenhze MS, Kartal MS, van Niftrik L. 2015. Anammox Planctomycetes have a peptidoglycan cell wall. Nat Commun 6:6878. doi: 10.1038/ncomms7878. [DOI] [PMC free article] [PubMed] [Google Scholar]