Abstract
Recently, novel arenaviruses were found in snakes with boid inclusion body disease (BIBD); these form the new genus Reptarenavirus within the family Arenaviridae. We used next-generation sequencing and de novo sequence assembly to investigate reptarenavirus isolates from our previous study. Four of the six isolates and all of the samples from snakes with BIBD contained at least two reptarenavirus species. The viruses sequenced comprise four novel reptarenavirus species and a representative of a new arenavirus genus.
TEXT
Until very recently, arenaviruses were known as a group of mainly rodent-borne zoonotic viruses (1). The negative-sense RNA genome of arenaviruses is divided into two segments, which are designated small (S, approximately 3.5 kb) and large (L, approximately 7 to 7.5 kb); both use an ambisense coding strategy (1). The S segment encodes the glycoprotein precursor (GPC) and the nucleoprotein (NP), whereas the RNA-dependent RNA polymerase (RdRp) and the Z protein (ZP) are encoded in the L segment (1). The discovery of arenaviruses in snakes with boid inclusion body disease (BIBD) by three independent groups (2–4) has expanded the family Arenaviridae by a new group of viruses. In fact, the BIBD-associated arenaviruses (BIBDAVs) have been suggested to form a new arenavirus genus called Reptarenavirus (5). At the same time, the arenavirus study group of the International Committee on Taxonomy of Viruses (ICTV) has suggested that the genus Arenavirus, harboring the “classical” Old and New World arenaviruses, be renamed Mammarenavirus (6). According to a recent proposal, the genus Reptarenavirus would contain three species: alethinophid reptarenaviruses 1 (member virus: Golden Gate virus), 2 (CAS virus, CASV), and 3 (University of Helsinki virus 1 [UHV-1], boa AV NL B3 virus). While in vitro evidence suggests a causal relationship between arenavirus infection and BIBD (4), the in vivo proof is still missing. Also, the reservoir host(s) of the reptarenaviruses has not yet been confirmed; however, our recent study suggests that these viruses preferentially grow in organisms with body temperatures close to 30°C (7).
To study the diversity of reptarenaviruses, we applied next-generation sequencing (NGS) to characterize certain isolates described in our previous report (4). On the basis of phylogeny, we selected isolates originating from six Boa constrictor snakes and used a continuous B. constrictor kidney cell line, I/1Ki (4), for their propagation. While the virus isolate of snake 1 has already been almost fully sequenced (GenBank accession numbers KF297880.1 and KF297881.1), only partial L segment sequences were available for isolates from snakes 5 (KF564801.1), 8 (KF564796.1), 9 (KF564800.1), 11, and 41 (KF564797.1). We infected clean I/1Ki cultures with tissue homogenates and collected the viruses produced during the first passage by pelleting through a sucrose cushion as previously described (4). Viral RNAs were extracted from the pelleted viruses with the QIAmp Viral RNA minikit (Qiagen) according to the manufacturer's instructions, without carrier RNA. The isolated RNA was further purified and concentrated with SPRISelect beads (Agencourt). Indexed Illumina sequencing libraries were prepared with the NEBNext Ultra RNA Library Prep kit (New England BioLabs). Pooled libraries were sequenced on the Illumina MiSeq with 161-bp paired-end reads. Reads were demultiplexed, adapter sequences were removed, and sample FASTQ files were produced with the MiSeq reporter. De novo contiguous sequence (contig) assembly was performed with MIRA version 4.0.2 (http://mira-assembler.sourceforge.net/) and CSC (IT Center for Science Ltd., Finland) Taito supercluster. Initial contigs were used to remove the cellular RNA background with the mirabait tool, after which de novo assemblies were run with a subset of reads extracted with Chipster v.3.1.0 (8).
For most virus preparations, contigs of full-length or almost full-length S and L segments of arenaviruses were obtained (Table 1). The contig coverages, determined with Bowtie2 (9) in Unipro UGENE 1.14.2 (10), are presented in Table 1. The virus isolate from snake 1 served as a positive control for this study, since it represents the UHV-1 species that we have characterized in detail (4). Bowtie2 alignment of the NGS data in Unipro UGENE 1.14.2 to the UHV-1 reference L segment (KF297881.1) revealed that the sequence did not exist in the sample. Instead, two nearly complete L segment sequences were recovered from the purified UHV preparation by de novo sequence assembly. The database UHV sequence KF297881.1 was assembled from NGS data and by Sanger sequencing of a PCR-cloned fragment from the pGEM-T vector. At the time, we did not expect coinfection with multiple arenaviruses and accidentally generated an “in silico” recombinant of the two viruses in the preparation. The sequence with accession no. KF297881.1 combines the first ∼2,200 nucleotides (nt) from the 5′ end of the first L segment and ∼5,000 nt from the 3′ end of the second L segment found in the preparation. In the region where the mismatch occurred, 24/26 nt are identical in both sequences. The fact that an S segment nearly identical (3367/3393 nt, >99% nucleotide sequence identity) to the UHV-1 reference S segment (KF297881.1) was recovered from the NGS data provides further evidence of the reliability of the de novo assembly approach chosen. Interestingly, three of the other virus preparations studied contained more than one L and S segment, indicating that several snakes were co- or superinfected with two or more reptarenaviruses. For most (3/4) samples with several reptarenaviruses, two L segment sequences were constructed; however, one of the isolates (from snake 8) contained six different L segments. Curiously, at maximum, two full-length (or nearly full-length) S segments were constructed per virus preparation, which may reflect stronger selection pressure or an unknown method-induced bias.
TABLE 1.
Lengths, coverages, and sequence identities of the de novo constructed BIBDAV S and L segmentsa
| Origin and genome (segment) | Length (nt) | Coverage (min-max) | No. of reads aligned | % PASC identity to other contigs (BLAST vs global alignment) | % PASC identity to database sequences (BLAST vs global alignment) | L segment prevalence in prepn (%) | GenBank accession no. |
|---|---|---|---|---|---|---|---|
| Snake 1 | |||||||
| UHV-1 (S) | 3,393 | 7,071–35,430 | 523,677 | ABV-2 (74.5 vs 66.4) | UHV-1 (99.0 vs 99.0) | KR870011 | |
| ABV-1 (S) | 2,725 | 746–30,609 | 381,847 | UHV-3 (71.8) vs UHV-2 (72.6) | GGV (71.7 vs 64.7) | KR870010 | |
| UHV-1 (L) | 6,834 | 40–20,629 | 647,002 | UHV-4 (96.0 vs 95.4) | Boa Av NL B3 (78.7 vs 79.6) | 2.6 | KR870020 |
| ABV-1 (L) | 6,892 | 688–17,159 | 413,735 | ABV-2 (85.1 vs 85.0) | GGV (82.2 vs 82.4) | 97.4 | KR870021 |
| Snake 5 | |||||||
| UGV-1 (S) | 3,433 | 1,055–79,695 | 1096,065 | UGV-3 (96.7 vs 95.7) | GGV (74.9 vs 75.6) | KR870012 | |
| UGV-1 (L) | 6,787 | 1,310–32,480 | 698,732 | UGV-3 (81.5 vs 81.3) | GGV (68.1 vs 70.0) | KR870022 | |
| Snake 8 | |||||||
| UGV-3 (S) | 3,455 | 68–41,472 | 612,439 | UGV-2 (99.0 vs 98.9) | GGV (73.0 vs 75.8) | KR870013 | |
| UGV-4 (S) | 2,774 | 167–41,725 | 510,392 | UGV-2 (86.9 vs 77.9) | GGV (66.5 vs 60.8) | KR870014 | |
| UGV-3 (L) | 6,830 | 91–3,228 | 85,261 | UGV-2 (99.2 vs 54.0) | GGV (67.8) vs Cupixi (54.0) | 14.9 | KR870023 |
| SVaV (L) | 6,506 | 289–2,547 | 78,412 | UGV-1 (69.7 vs 69.6) | GGV (65.3 vs 66.2) | 0.3 | KR870024 |
| ABV-3 (L) | 6,641 | 73–2,048 | 50,159 | ABV-1 (79.4 vs 78.3) | GGV (79.1 vs 78.1) | 59.4 | KR870025 |
| TSMV (L) | 6,928 | 11–2,439 | 73,643 | ABV-2 (70.8 vs 72.7) | GGV (70.5 vs 72.3) | 20.2 | KR870026 |
| UHV-4 (L) | 6,882 | 127–3,210 | 69,701 | UHV-3 (98.3 vs 98.3) | Boa Av NL B3 (79.5 vs 81.5) | 0.6 | KR870027 |
| HKV (L) | 6,906 | 3–1,655 | 37,354 | ABV-1 (70.3 vs 72.8) | GGV (69.5 vs 73.0) | 4.6 | KR870028 |
| Snake 9 | |||||||
| UGV-2 (S) | 3,465 | 194–31,376 | 468,578 | UGV-3 (99.0 vs 98.9) | GGV (73.5 vs 76.0) | KR870015 | |
| UGV-2 (L) | 6,903 | 123–14,415 | 401,021 | UGV-3 (99.2) vs Sn5 (81.3) | GGV (68.7 vs 71.6) | KR870029 | |
| Snake 11 | |||||||
| UHV-2 (S) | 3,383 | 37–4,780 | 62,970 | UHV-3 (93.8 vs 93.0) | Boa Av NL B3 (78.5 vs 80.0) | KR870016 | |
| HISV (S) | 3,376 | 11–27,092 | 350,355 | UHV-1 (15.2) vs ABV-1 (52.1) | Lassa (16.5 vs 54.0) | KR870017 | |
| UHV-2 (L) | 6,894 | 132–2,077 | 63,741 | UHV-4 (79.7 vs 80.0) | Boa Av NL B3 (70.4 vs 72.8) | 93.4 | KR870030 |
| HISV (L) | 5,913 | 357–4,493 | 115,633 | TSMV (15.3) vs ABV-3-3 (50.0) | AMPV (18.8) vs BCV (48.5) | 6.6 | KR870031 |
| Snake 41 | |||||||
| ABV-2 (S) | 3,413 | 28–3,460 | 42,023 | UHV-1 (74.6) vs Sn5 (76.1) | GGV (79.2 vs 79.1) | KR870018 | |
| UHV-3 (S) | 3,339 | 10–1,055 | 13,563 | UHV-2 (93.8 vs 93.0) | Boa Av NL B3 (80.4 vs 83.0) | KR870019 | |
| UHV-3 (L) | 6,925 | 3–4,572 | 36,844 | UHV-4 (98.3 vs 98.3) | Boa Av NL B3 (80.4 vs 81.9) | 99.7 | KR870032 |
| ABV-2 (L) | 6,938 | 111–2,966 | 49,692 | ABV-1 (85.0 vs 85.0) | GGV (82.3 vs 82.8) | 0.3 | KR870033 |
The viruses (abbreviations) are CAS virus (CASV), University of Helsinki viruses 1 to 4 (UHV-1 to -4), Boa Av NL B3, Aurora borealis viruses 1 to 3 (ABV-1 to -3), Golden Gate virus (GGV), Tavallinen suomalainen mies virus (TSMV), Hans Kompis virus (HKV), Suri Vanera virus (SVaV), University of Giessen viruses 1 to 4 (UGV-1 to -4), Haartman Institute snake virus (HISV), Amapari virus (AMPV), and Bear Canyon virus (BCV).
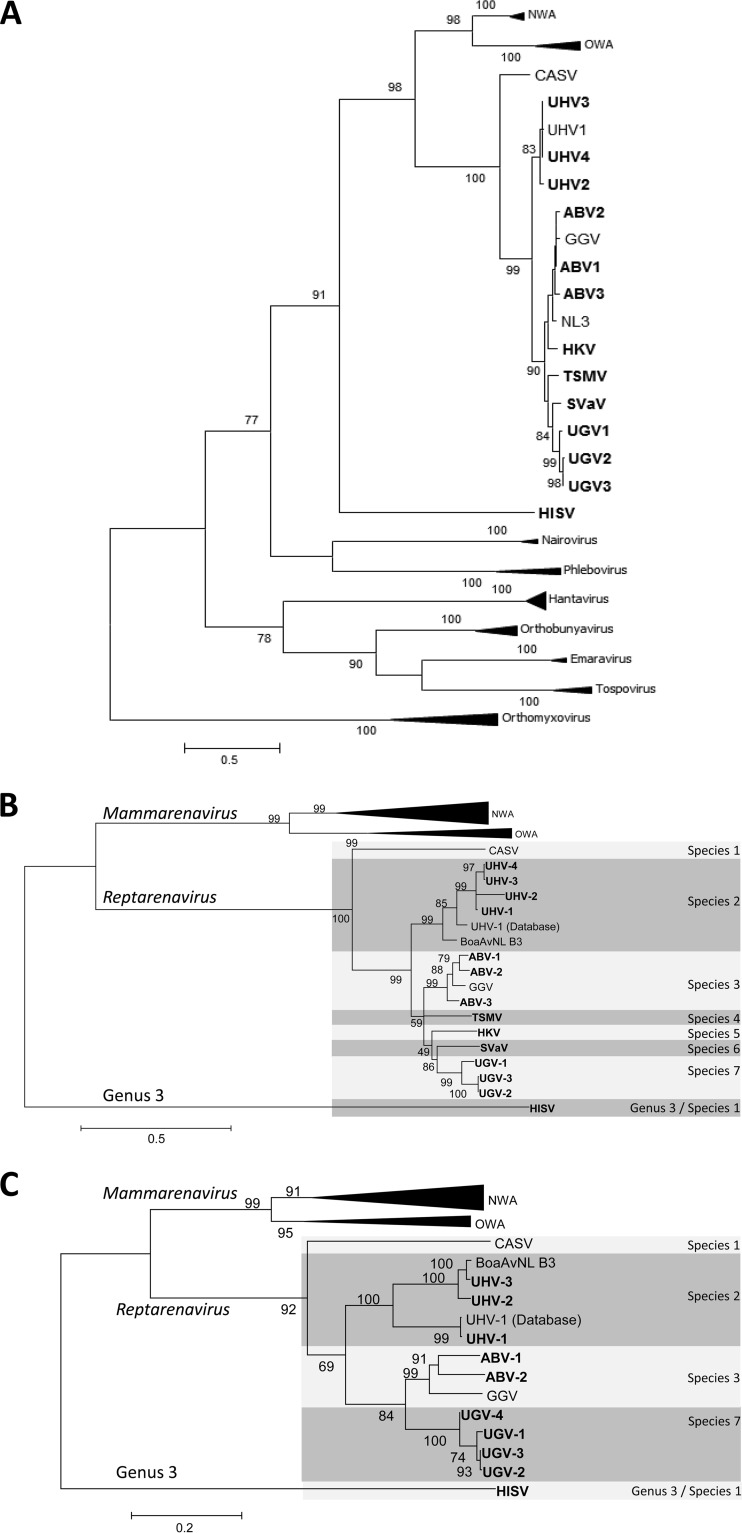
We then used pairwise sequence comparison (PASC [11], http://www.ncbi.nlm.nih.gov/sutils/pasc/viridty.cgi) to compare the sequences obtained to each other and to the arenavirus S and L segments in the databases (Table 1). According to PASC analysis and ICTV criteria, several of the viruses that we sequenced showed <76% nucleotide sequence identity to known arenaviruses and thus represent putative new alethinophid reptarenavirus species, as depicted by phylogeny (Fig. 1). For alethinophid reptarenavirus 4 (University of Giessen virus 1 [UGV-1] and UGV-2) we recovered both segments, since isolates from snakes 5 and 9 contained only a single virus each. However, for the putative representatives of alethinophid reptarenaviruses 5 to 7 (Tavallinen suomalainen mies virus [TSMV], Hans Kompis virus [HKV], and Suri Vanera virus [SVaV]), we obtained only the sequence of the L segment. Remarkably, according to the PASC analysis, one of our isolates, Haartman Institute snake virus (HISV), would be sufficiently distant from the known arenaviruses to represent a novel arenavirus genus. The S segment of HISV encodes GPC and NP by ambisense coding strategy, but despite our attempts, we were unable to obtain the full-length L segment (the ZP gene and part of the RdRp were missing). To roughly estimate the relative quantities of the viruses in each preparation, we selected primers targeting the L and S segments specific for each virus identified (the sequences of the primers used are available upon request). We performed quantitative reverse transcription (RT)-PCR with purified virus preparations and the respective primers and confirmed the presence of viruses and primer specificity by Sanger sequencing. The prevalence of L segments in the purified virus preparation is presented in Table 1.
FIG 1.
Phylogenetic relationships of the newly sequenced BIBDAVs. (A) Phylogenetic tree of aligned core polymerase domains of the RdRps of segmented negative-strand viruses. The tree was reconstructed by the maximum-likelihood method in MEGA 6.06 with 1,000 bootstrap replicates. The amino acid substitution model WAG was used as suggested by MEGA as the best-fitting model. Bootstrap support values of >70 are shown at the nodes. (B, C) Maximum-likelihood trees built on the basis of RdRp amino acid sequences (B) and nucleoprotein-encoding nucleotide sequences (C). The phylogeny was reconstructed with the MEGA 5.05 software with 1,000 bootstrap replicates. The sequence data set (including representatives of Old and New World arenaviruses (OWA and NWA, respectively) and the previously reported reptarenaviruses) was compiled from the Virus Pathogen Resource database. The nucleotide sequence alignment was guided by amino acid translations with Translator X (15) and the MAFFT algorithm. The viruses (abbreviations) are CAS virus (CASV), University of Helsinki viruses 1 to 4 (UHV-1 to -4), Boa Av NL B3, Aurora borealis viruses 1 to 3 (ABV-1 to -3), Golden Gate virus (GGV), Tavallinen suomalainen mies virus (TSMV), Hans Kompis virus (HKV), Suri Vanera virus (SVaV), University of Giessen viruses 1 to 4 (UGV-1 to -4), and Haartman Institute snake virus (HISV). The newly sequenced viruses are in bold.
We further tested the original tissues (stored frozen at −80°C) from the diseased constrictor snakes that had served to generate the isolates (liver tissue from snakes 5, 8, 9, and 11) by using the L segment primers for RT-PCR and confirmed the presence of these viruses in the original tissues by Sanger sequencing the products. To our surprise, the livers of snakes 5 and 9, unlike the respective isolates, were also found to have multiple reptarenaviruses (UGV-2, HKV, and ABV-3 in snake 5 and HKV and UGV-3 in snake 9) and snake 11 was found to be positive for all except ABV-2 and HISV. All of the snakes studied were from Europe. PASC analysis of the sequences revealed that the virus preparations of coinfected snakes always contained at least two different reptarenavirus species, and thus, co- or superinfection might be essential for the development of BIBD. In vitro studies with mammarenaviruses demonstrate that persistent arenavirus infection restricts the replication of a serologically similar but not more distant arenavirus (12). Some strains of lymphocytic choriomeningitis virus, the prototype arenavirus, induce acquired immunosuppression in their natural host (13), as also suggested for BIBD (14). We hypothesize that reptarenaviruses may establish a chronic infection in snakes. Co- or superinfection of a chronically infected snake with another reptarenavirus species might then result in amplified replication and eventually in BIBD due to immunosuppression induced by the chronically infecting virus.
The data presented here demonstrate that the recently established genus Reptarenavirus is likely to expand in the near future, since we identified four novel representatives of this genus from only six B. constrictors. Furthermore, our results suggest that co- or superinfection with reptarenaviruses is common and might be relevant for the pathogenesis of BIBD. Isolation of the individual viruses is required to confirm this hypothesis and to study the potential reassortment of reptarenaviruses.
Nucleotide sequence accession numbers.
The GenBank accession numbers of the assembled S and L segments are provided in Table 1.
ACKNOWLEDGMENTS
This work was supported by grants from the Academy of Finland (251836 [Arboviruses in Northern Europe] to O.V.) and the Finnish Foundation of Veterinary Research (to U.H.).
REFERENCES
- 1.Moraz ML, Kunz S. 2011. Pathogenesis of arenavirus hemorrhagic fevers. Expert Rev Anti Infect Ther 9:49–59. doi: 10.1586/eri.10.142. [DOI] [PubMed] [Google Scholar]
- 2.Stenglein MD, Sanders C, Kistler AL, Ruby JG, Franco JY, Reavill DR, Dunker F, Derisi JL. 2012. Identification, characterization, and in vitro culture of highly divergent arenaviruses from boa constrictors and annulated tree boas: candidate etiological agents for snake inclusion body disease. mBio 3:e00180–12. doi: 10.1128/mBio.00180-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bodewes R, Kik MJ, Raj VS, Schapendonk CM, Haagmans BL, Smits SL, Osterhaus AD. 2013. Detection of novel divergent arenaviruses in boid snakes with inclusion body disease in The Netherlands. J Gen Virol 94:1206–1210. doi: 10.1099/vir.0.051995-0. [DOI] [PubMed] [Google Scholar]
- 4.Hetzel U, Sironen T, Laurinmaki P, Liljeroos L, Patjas A, Henttonen H, Vaheri A, Artelt A, Kipar A, Butcher SJ, Vapalahti O, Hepojoki J. 2013. Isolation, identification, and characterization of novel arenaviruses, the etiological agents of boid inclusion body disease. J Virol 87:10918–10935. doi: 10.1128/JVI.01123-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anonymous. 2014. EC-approved taxonomic proposal 2014.011a-dV.A.v2.reptarenavirus.pdf awaiting ICTV ratification. http://talk.ictvonline.org/files/proposals/taxonomy_proposals_vertebrate1/m/vert04/5108.aspx Accessed 16 February 2015. [Google Scholar]
- 6.Anonymous. 2014. EC-approved taxonomic proposal 2014.012aV.A.v3.Arenavirus_ren.pdf awaiting ICTV ratification. http://talk.ictvonline.org/files/proposals/taxonomy_proposals_vertebrate1/m/vert04/5109.aspx Accessed 16 February 2015. [Google Scholar]
- 7.Hepojoki J, Kipar A, Korzyukov Y, Bell-Sakyi L, Vapalahti O, Hetzel U. 2015. Replication of boid inclusion body disease-associated arenaviruses is temperature sensitive in both boid and mammalian cells. J Virol 89:1119–1128. doi: 10.1128/JVI.03119-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kallio MA, Tuimala JT, Hupponen T, Klemela P, Gentile M, Scheinin I, Koski M, Kaki J, Korpelainen EI. 2011. Chipster: user-friendly analysis software for microarray and other high-throughput data. BMC Genomics 12:507. doi: 10.1186/1471-2164-12-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okonechnikov K, Golosova O, Fursov M, UGENE Team. 2012. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28:1166–1167. doi: 10.1093/bioinformatics/bts091. [DOI] [PubMed] [Google Scholar]
- 11.Bao Y, Chetvernin V, Tatusova T. 2014. Improvements to pairwise sequence comparison (PASC): a genome-based web tool for virus classification. Arch Virol 159:3293–3304. doi: 10.1007/s00705-014-2197-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Damonte EB, Mersich SE, Coto CE. 1983. Response of cells persistently infected with arenaviruses to superinfection with homotypic and heterotypic viruses. Virology 129:474–478. [DOI] [PubMed] [Google Scholar]
- 13.Botten J, Sidney J, Mothe BR, Peters B, Sette A, Kotturi MF. 2010. Coverage of related pathogenic species by multivalent and cross-protective vaccine design: arenaviruses as a model system. Microbiol Mol Biol Rev 74:157–170. doi: 10.1128/MMBR.00045-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schumacher J, Jacobson ER, Homer BL, Gaskin JM. 1994. Inclusion body disease in boid snakes. J Zoo Wildl Med 25:511–524. [Google Scholar]
- 15.Abascal F, Zardoya R, Telford MJ. 2010. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res 38:W7-13. doi: 10.1093/nar/gkq291. [DOI] [PMC free article] [PubMed] [Google Scholar]