ABSTRACT
Alphaviruses are known to possess a unique viral mRNA capping mechanism involving the viral nonstructural protein nsP1. This enzyme harbors methyltransferase (MTase) and nsP1 guanylylation (GT) activities catalyzing the transfer of the methyl group from S-adenosylmethionine (AdoMet) to the N7 position of a GTP molecule followed by the formation of an m7GMP-nsP1 adduct. Subsequent transfer of m7GMP onto the 5′ end of the viral mRNA has not been demonstrated in vitro yet. Here we report the biochemical characterization of Venezuelan equine encephalitis virus (VEEV) nsP1. We have developed enzymatic assays uncoupling the different reactions steps catalyzed by nsP1. The MTase and GT reaction activities were followed using a nonhydrolyzable GTP (GIDP) substrate and an original Western blot assay using anti-m3G/m7G-cap monoclonal antibody, respectively. The GT reaction is stimulated by S-adenosyl-l-homocysteine (Ado-Hcy), the product of the preceding MTase reaction, and metallic ions. The covalent linking between nsP1 and m7GMP involves a phosphamide bond between the nucleotide and a histidine residue. Final guanylyltransfer onto RNA was observed for the first time with an alphavirus nsP1 using a 5′-diphosphate RNA oligonucleotide whose sequence corresponds to the 5′ end of the viral genome. Alanine scanning mutagenesis of residues H37, H45, D63, E118, Y285, D354, R365, N369, and N375 revealed their respective roles in MT and GT reactions. Finally, the inhibitory effects of sinefungin, aurintricarboxylic acid (ATA), and ribavirin triphosphate on MTase and capping reactions were investigated, providing possible avenues for antiviral research.
IMPORTANCE Emergence or reemergence of alphaviruses represents a serious health concern, and the elucidation of their replication mechanisms is a prerequisite for the development of specific inhibitors targeting viral enzymes. In particular, alphaviruses are able, through an original reaction sequence, to add to their mRNA a cap required for their protection against cellular nucleases and initiation of viral proteins translation. In this study, the capping of a 5′ diphosphate synthetic RNA mimicking the 5′ end of an alphavirus mRNA was observed in vitro for the first time. The different steps for this capping are performed by the nonstructural protein 1 (nsP1). Reference compounds known to target the viral capping inhibited nsP1 enzymatic functions, highlighting the value of this enzyme in antiviral development.
INTRODUCTION
Emergence or reemergence of alphaviruses represents a serious health concern, as exemplified by the worldwide epidemics of Chikungunya virus during the last 10 years (1). The Alphavirus genus comprises 31 species that together with the genus Rubivirus forms the Togaviridae family. Alphaviruses have been classified on the basis of their geographical distribution. Alphaviruses circulating in the Old World (OW) most commonly cause febrile illness and painful arthralgias or polyarthralgias, particularly in the small joints (2). In contrast, New World (NW) alphaviruses target the central nervous system, except Mayaro virus, which is considered an arthritogenic agent like OW alphaviruses (3). Among NW alphaviruses, Venezuelan equine encephalitis virus (VEEV) is an important pathogen present in the Americas from Texas to Argentina (4, 5). In 1995, a major outbreak in Venezuela and Colombia resulted in roughly 100,000 human cases, with more than 300 fatal encephalitis cases (6). Other epidemics have since been reported, indicating that VEEV still represents a serious public health problem (7). Moreover, it is noteworthy that VEEV infection symptoms resemble those of dengue fever, leading to an underestimation of the number of cases related to this virus in the regions where both dengue virus and VEEV circulate (8). In humans, while the overall mortality rate is low (<1%), neurological disease, including disorientation, ataxia, mental depression, and convulsions, can be detected in up to 14% of infected individuals, especially children (9). Neurological sequelae in humans are also common (10). An attenuated vaccine strain exists, consisting of either a living or inactivated form of the virus. Although it showed efficacy in protecting equines by decreasing viral amplification, it did not contain VEEV outbreaks. Moreover, the vaccine showed important adverse effects in humans and failed to be completely protective (11).
Antiviral drugs against VEEV are lacking, and antiviral compounds in development are still at the first stages of evaluation (12–14). In this context, a better understanding of how viral mRNA is transcribed and capped is essential for the development of screening tools for agents against VEEV and other alphaviruses.
To date, the mRNA capping mechanism in alphaviruses has been studied only for OW viruses, such as Sindbis virus (SINV) or Semliki Forest virus (SFV). Alphavirus mRNAs bear a cap-0 structure [m7G(5′)ppp(5′)N] at the 5′ end. This structure is essential for mRNA translation and prevents viral mRNA from degradation by cellular 5′ exonucleases. Unlike other (+) single-strand RNA viruses such as flaviviruses, the first RNA nucleotide is not methylated at its 2′O position. Despite this absence of 2′O methylation, it is likely that viral RNA are barely detected by innate immune sensors since a short viral RNA structure prevents their recognition as non-self RNA (15).
The alphavirus RNA capping mechanism is not fully unraveled. It supposedly requires an unconventional sequence of four reactions (capping mechanisms reviewed in reference 16): (i) methylation (via methyltransferase [MTase]) of GTP into m7GTP (GTP + S-adenosylmethionine [AdoMet] → m7GTP + S-adenosyl-l-homocysteine [AdoHcy]), (ii) guanylylation (GT) of nonstructural protein 1 (nsP1) by m7GMP, releasing pyrophosphate (m7GTP + nsP1→ m7GMP-nsP1 + PPi), (iii) 5′-triphosphate RNA dephosphorylation (RTPase activity) into 5′-diphosphate RNA (pppRNA + RNA triphosphatase → ppRNA + Pi), and (iv) transfer of m7GMP onto 5′-diphosphate RNA to produce the final capped RNA product m7G(5′)ppp(5′)N (ppRNA + m7GMP-nsP1 → m7G(5′)ppp(5′)RNA).
The alphavirus MTase activity of nsP1 was first identified in a SINV nsP1 mutant replicating in insect cells depleted of methionine, the precursor of AdoMet (17). The MTase activity of the SINV and SFV nsP1 was next confirmed by in vitro enzymatic assays (18, 19). Critical residues for the MTase activity were then identified by comparative sequence analysis, which demonstrated that this enzyme is conserved in the alphavirus-like supergroup (20). In contrast to most viral MTases, the substrate of nsP1 is unlikely to be the cap structure present at the 5′ end of RNA but rather a GTP molecule which becomes methylated at its N7 position (m7GTP). The m7GTP is then used for the guanylylation of nsP1 (m7GMP-nsP1), with concomitant release of inorganic pyrophosphate (21). Chemical stability, site-directed mutagenesis, and sequence conservation studies suggest that the amino acid residue bound to m7GMP is histidine 38 of SFV nsP1 (22, 23). In addition, SFV nsP1 activity was also shown to be dependent on membrane association (24). This interaction is mediated by an amphipathic helix located in the middle of the nsP1 sequence (25). The helix contains hydrophobic amino acids that can interact with acyl groups of the membrane and a cluster of positively charged residues that contact the phospholipid polar heads (26). Several palmitoylated cysteines of nsP1 were also proposed to contribute to membrane binding and cellular localization of the replication complex (27, 28). However, SINV nsP1 and SFV nsP1 expressed as recombinant proteins in Escherichia coli remain catalytically active for methylation of GTP and covalent binding to nsP1, suggesting that this posttranslational modification, which cannot occur in bacteria, has probably a limited effect on both MTase and guanylylation activities of nsp1 (18, 29). In addition, nsP1 is probably not the only viral nsP involved in the capping mechanism. The N-terminal region of nsP2, which was predicted to carry a helicase domain (30, 31), displays both nucleotide and RNA triphosphatase activities and is therefore a good candidate for the catalysis of the third step of the capping (32, 33). The last step of the capping reaction, which is the guanylyltransfer of m7GMP linked to nspP1 onto RNA, has not yet been experimentally demonstrated for the alphavirus family. This last step (step iv), however, has been demonstrated in vitro using the capping enzyme of bamboo mosaic virus (BaMV), a member of the alphavirus-like group (34). Thus, despite extensive studies on OW alphavirus nsP1, the capping mechanism is ill defined, a statement especially valid for NW alphaviruses such as VEEV.
Here we report the first demonstration of a functional RNA capping reaction displayed by NW alphavirus nsP1. In this study, we use a purified recombinant protein to dissect the MTase activity and m7GMP covalent binding properties of VEEV nsP1, as well as the guanylyltransfer of m7GMP on a 5′-diphosphate RNA. Using an original immunoassay to uncouple MTase activity from GTP binding reactions, we deciphered by means of site-directed mutagenesis the respective roles of conserved amino acids in the RNA capping pathway. Finally, we used nsp1-associated MTase, GT, and RNA capping activities to assess the effect of known RNA capping inhibitors.
MATERIALS AND METHODS
Chemicals.
General chemicals such as GTP, m7GTP, m7GDP, S-adenosyl-l-homocysteine (AdoHcy), aurintricarboxylic acid (ATA), sinefungin, and guanylyl-imido-diphosphate (GIDP) were purchased from Sigma, and ribavirin triphosphate was from Jena Biosciences. m7GpppG, m7GpppA, and S-adenosylmethionine (AdoMet) were from New England Biolabs, whereas S-adenosyl-[methyl-3H]methionine, [α-32P]GTP, [α-32P]ATP, and [γ-32P]GTP are products of PerkinElmer and [β-32P]GTP was from Bioactif.
Expression and purification of VEEV nsP1 wt and mutants.
The codon-optimized DNA encoding nsP1 of VEEV (strain P676, amino acid 1 to 535) was synthesized by GenScript. The DNA sequence was cloned into the expression vector pET28b (Novagen) to enable the fusion of a 6×His tag at the C terminus of nsP1. Site-directed mutagenesis was performed using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). All constructs were confirmed by DNA sequencing. The proteins (wild type [wt] and mutants) were then produced in E. coli Rosetta pLysS (DE3) (Novagen) cells. Expression was induced for 2.5 h at 25°C in Terrific Broth medium with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) after the optical density at 600 nm (OD600) of the cultures reached 0.6. Cells were harvested by centrifugation at 5,000 × g for 10 min. The cell pellets were then resuspended in lysis buffer (20 mM Tris [pH 7.5], 100 mM NaCl, 5% glycerol) supplemented with 5 mM β-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride, 10 mM benzamidine, 10 μg/ml of DNase I, and 0.25 mg/ml of lysozyme prior to sonication. After centrifugation for 30 min at 16,000 × g, the soluble fraction was purified by immobilized-metal affinity chromatography (IMAC) on a 5-ml His TrapFF crude column (GE Healthcare). The proteins were eluted in a buffer containing 50 mM Tris (pH 7.5), 300 mM NaCl, and 250 mM imidazole and then desalted and concentrated using an Ultrafree-30 centrifugal filter unit (Millipore) in a buffer containing 20 mM Tris (pH 7.5), 100 mM NaCl, and 10% glycerol. Proteins were further purified onto a HiTrap heparin column (GE Healthcare) equilibrated with 20 mM Tris (pH 7.5), 100 mM NaCl, and 10% glycerol. The elution was performed using a salt gradient from 100 to 500 mM NaCl. Elution occurred with approximately 300 mM NaCl.
Formation of the covalent nsP1-m7GMP intermediate (MTase and GT).
The standard reactions were carried out at 30°C for different incubation times ranging from 0 to 180 min in 20-μl reaction solutions, each containing 5 μM enzyme, 5 μCi of [α-32P]GTP (83 nM), 50 mM Tris (pH 7.0), 2 mM dithiothreitol (DTT), 10 mM KCl, 2 mM MgCl2, and 100 μM AdoMet, unless otherwise stated, following previously described protocols (22). The reaction was stopped by adding Laemmli sample buffer, followed by 2 min of heating at 95°C. The reaction mixture was resolved by 10% SDS-PAGE, and radiolabeled proteins were visualized by autoradiography (Fujifilm FLA-3000). The number of photostimulated luminescence events (PSL-BG) per square millimeter was determined using Image Gauge 4.0. Based on the time course experiment results, 1 h of incubation was chosen to analyze AdoMet concentration, pH, and divalent ion dependence.
Methyltransferase assay.
The methylation reaction mixture containing 50 mM Tris (pH 7.0), 2 mM DTT, 10 mM KCl, 2 mM MgCl, 2 mM GTP or GIDP, 0.55 μCi of S-adenosyl-[methyl-3H]methionine (0.33 μM), 10 μM AdoMet, and 5 μM enzyme was incubated at 30°C for different periods ranging from 0 to 120 min. The reaction was stopped by the addition of an equal volume of 0.2% SDS and 20 mM EDTA, and then the samples were loaded on DEAE-cellulose filters (PerkinElmer). Unincorporated label was removed by washing the filters twice with 20 mM ammonium formate, once with H2O, and once with absolute ethanol. The filters were dried, and the radioactivity was measured by scintillation counting with SCINT BETAPLATE solution in a Wallac MicroBeta Trilux 1450 counter (PerkinElmer) (18).
Immunoblotting of nsP1 guanylylation reaction.
Typical reactions were performed for 1 h at 30°C in buffer 50 mM Tris (pH 7.0), 10 mM NaCl, 2 mM MgCl2, 2 mM DTT, 10 μM m7GTP, 100 μM AdoHcy, and 5 μM nsP1. Then, reactions were stopped after 1 h of incubation with Laemmli sample buffer, followed by 2 min of heating at 95°C, and then the products were subjected to 10% SDS-PAGE. Proteins were then transferred to a polyvinylidene difluoride (PVDF) membrane (Amersham Hybond-P). The latter was blocked overnight at 4°C in 5% (wt/vol) skim milk solution in TBS-Tween (50 mM Tris [pH 7.6[, 150 mM NaCl, 0.1% [vol/vol] Tween) and subsequently incubated for 1 h at room temperature (RT) with a 1:1,000 dilution of reconstituted anti-m3G/m7G-cap monoclonal antibody (Synaptic Systems, Göttingen, Germany). After 3 washes of 10 min each in the TBS-Tween solution, the membrane was incubated with a secondary peroxidase-conjugated rabbit anti-mouse antibody (Sigma; number A9044) at a dilution of 1:2,000 in TBS at RT for 1 h and then rinsed sequentially twice for 10 min with TBS-Tween and once for 10 min with TBS. The immunoreactive proteins were revealed by the ECL detection system (Pierce ECL Western blotting substrate; Thermo Scientific) and detected using a Kodak Image Station 4000MM Pro (Carestream Health, Inc.). The signal was quantified using ImageJ software as previously described (35). For inhibition assays, half-maximal inhibitory concentrations (IC50s) were determined using a logistic equation with Sigmaplot software.
Chemical stability of the nsP1-m7GMP complex.
The guanylylation was carried out at 30°C for 2 h in a 20-μl solution that contained 5 μM enzyme, 25 μCi of [α-32P]GTP (415 nM), 50 mM Tris (pH 7.0), 2 mM DTT, 10 mM KCl, 2 mM MgCl2, and 100 μM AdoMet. The reaction was stopped by the addition of 1 μl of 0.5 M EDTA and 1 μl of 10% SDS. Two-microliter aliquots of the reaction mixture were incubated with 20 μl of various reagents (0.1 M Tris-HCl buffer [pH 7.5], 0.1 M HCl, 0.1 M NaOH, and 0.1 or 1 M hydroxylamine [pH 7.5]) at 37°C for 45 min. After the incubation, the acidic and alkaline solutions were immediately neutralized by the addition of 2 μl of 1 M NaOH and HCl, respectively. The samples were chilled on ice, mixed with Laemmli sample buffer, and subsequently analyzed by 10% SDS-PAGE followed by autoradiography.
Synthesis of RNA substrates.
RNA sequence AUGGGCGGCGCAAGA was chemically synthesized on a solid support using an ABI 394 synthesizer (36). After RNA assembly with 2′-O-pivaloyloxymethyl phosphoramidite monomers (Chemgenes, USA) (37), the 5′-hydroxyl group was phosphorylated and the resulting H-phosphonate derivative was oxidized and activated into a phosphoroimidazolidate derivative to react with either phosphoric acid (for ppRNA synthesis) or pyrophosphate (for pppRNA) (38). To prepare monophosphate RNA (pRNA), the 5′-H-phosphonate RNA was treated with a mixture of N,O-bis-trimethylacetamide (0.4 ml), CH3CN (0.8 ml), and triethylamine (0.1 ml) at 35°C for 15 min and then oxidized with a tert-butyl hydroperoxide solution (5 M to 6 M in decane, 0.4 ml; 35°C, 15 min). After deprotection and release from solid support with an aqueous ammonia solution (28%, 3 h, RT), phosphorylated RNAs were purified by ion exchange (IEX)–high-performance liquid chromatography (HPLC) (purity > 95%) and characterized by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) spectrometry.
Cy-5-labeled RNA binding assays.
The 5′ end of the VEEV RNA genome, corresponding to the sequence 5′-AUGGGCGGCGCAAGA-3′, was labeled with cyanine 5-cytidine-5′-phosphate-3′-(6-aminohexyl)phosphate (Jena Biosciences; NU1706) by a ligation reaction. RNA and pCpCy5 were incubated in the presence of ATP for 16 h at 16°C in the dark with 80 U of T4 ligase 1 (Biolabs). T4 RNA ligase was removed with StrataClean resin (Agilent Technologies), and Cy5-labeled RNA was separated from unincorporated Cy5 dye by centrifugation through a MicroSpin G-25 column (GE Healthcare). Fluorescence polarization (FP) measurements were performed in a microplate reader (PHERAstar FS; BMG Labtech) with an optical module equipped with polarizers and using excitation and emission wavelengths of 590 and 675 nm, respectively.
Equilibrium binding data were subjected to a nonlinear, least-squares analysis using the single-independent-site equation ΔP = ΔPmax(x/KD,app + x), where ΔPmax represents the calculated maximal fluorescence polarization change, x represents the free RNA concentration, and KD,app represents the apparent equilibrium dissociation constant for RNA binding.
Transfer of m7GMP from the covalent m7GMP-nsP1 to RNAs (RNA capping).
The covalent nsP1-m7GMP intermediate was prepared at 30°C for 2 h in a 20-μl solution that contained 2 μM enzyme, 50 μCi of [α-32P]GTP (830 nM), 50 mM Tris (pH 7.0), 2 mM DTT, 10 mM KCl, 2 mM MgCl2, and 100 μM AdoMet. Then 2 μl of the 32P-labeled covalent intermediate was mixed with 18 μl of solution that contained 50 mM Tris (pH 7.0), 2 mM DTT, 2 mM MgCl2, and 2 μM RNA molecule, and the mixture was incubated at 30°C for 1 h. When the reaction was over, half of the reaction mixture was subjected to SDS-PAGE (10%) analysis, and the other half was subjected to 7 M urea-PAGE (20%). To examine the cap structures formed at the 5′ end of RNA molecules, the radiolabeled RNA product was precipitated with ethanol in the presence of glycogen (Sigma; number G1767). Then the precipitated RNA was dissolved in a 30-μl solution containing 50 mM sodium acetate (pH 5.3), 1 mM ZnCl2, and 1 U of nuclease P1. Twenty microliters of the reaction mixture was incubated at 37°C for 60 min in the presence of 1 U of nuclease P1 (Sigma; number N8630). The remaining 10 μl of reaction mixture treated with nuclease P1 was subsequently incubated at 37°C for 60 min in the presence of 1 μl of 10× dephosphorylation buffer and 1 μl of calf intestinal alkaline phosphatase (CIAP) (Life Technologies; number 18009). The resulting products were loaded on a 7 M urea-PAGE gel (20%) to assess the presence of m7GpppAUGGGCGGCGCAAGA before treatment and released m7GpppA. The vaccinia virus (VV) capping system (vac-CE) (New England Biolabs; number M2080S) was used as a positive control for RNA capping and cap detection.
RESULTS
Production and purification of VEEV nsP1.
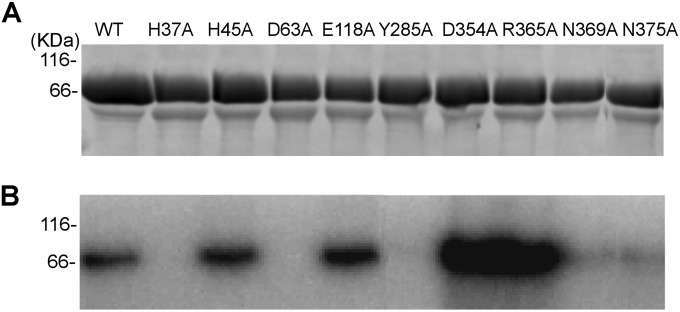
To explore the mechanism of the VEEV mRNA capping reaction, the wild type (wt) and nine mutants of VEEV nsP1 were produced and purified with a 6×His tag fused to their C termini. All the residues selected for alanine substitution are strictly conserved within the alphavirus genus except E118, which is replaced by another acidic amino acid (D) in most versions of nsP1 (Fig. 1). Among the mutants, H37 is supposedly involved in m7GMP binding, while D63 would participate in AdoMet binding (22). Residues E118 and N375 were reported to be important for minus-strand RNA synthesis during virus replication of other alphaviruses (39–41). No functional information is available for H45, Y285, D354, R365, and N369 so far, but they are located in highly conserved sequence stretches within alphavirus nsP1. After two steps of affinity purification (IMAC and heparin), the purified proteins were visualized by SDS-PAGE after Coomassie blue staining. The main protein band corresponding to nsP1 (95%) was detected at 65 kDa, and a minor contaminant was detected around 60 kDa (5%) (Fig. 2A). The identities of both proteins were determined by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry after trypsin digestion. They were full-length nsP1 and the E. coli heat shock protein Hsp60 (data not shown). Attempts to get rid of Hsp60 by including an additional purification step were unsuccessful.
FIG 1.
Amino acid sequence comparison of selected alphavirus nsP1 proteins. nsP1 sequences were aligned by T-coffee (70). Secondary-structure predictions for VEEV nsP1 were obtained by PSIPRED (71). Secondary structural elements with a reliability are marked with squiggles (α helices) or arrows (β-strands). If reliability is low, helices appear as small squiggles, β-strands appear as dotted lines, and labels are not written. Amino acids mutated in the course of this study are marked with asterisks. SINV, Sindbis virus; SFV, Semliki Forest virus; ONNV, O'nyong-nyong virus; CHIKV, Chikungunya virus; SPDV, salmon pancreas disease virus.
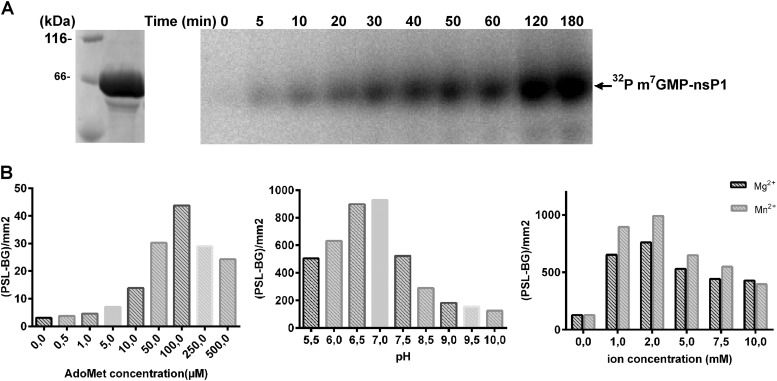
FIG 2.
Preliminary characterization of VEEV nsP1. (A) Purified wt nsP1 loaded on a 10% Coomassie blue-stained SDS-PAGE gel (left) and time course analysis of the formation of the covalent m7GMP-nsP1 intermediate (right). The activity assays were carried out at 30°C by incubating nsP1 with [α-32P]GTP and AdoMet for various times as described in Materials and Methods. The reaction products were separated by SDS-PAGE followed by autoradiography analysis. (B) Effects of AdoMet, pH, and divalent ions on the formation of the covalent m7GMP-nsP1 intermediate. The numbers of photostimulated luminescence events (PSL-BG) per square millimeter were determined by Image Gauge software.
Formation of a covalent m7GMP-nsP1 complex.
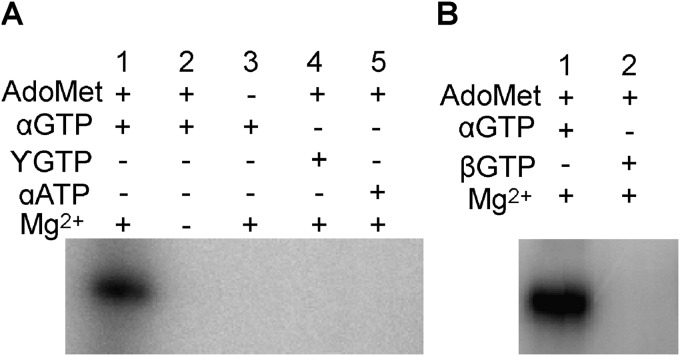
The putative formation of a covalent m7GMP-nsP1 intermediate was first investigated using SDS-PAGE after incubation of VEEV nsP1 with radiolabeled [α-32P]GTP in the presence of the methyl donor AdoMet. The time course analysis results presented in Fig. 2A indicates a progressive accumulation of a radiolabeled complex migrating at the apparent molecular size of nsP1 (∼65 kDa). Divalent cations, Mn2+ or Mg2+, as well as AdoMet are essential for the formation of the covalent complex (Fig. 2B). The optimal pH was around 7, and the standard reaction buffer contained 50 mM Tris (pH 7.0), 100 μM AdoMet, and 2 mM magnesium. The formation of radiolabeled complex was also analyzed in the presence of [β-32P]GTP, [γ-32P]GTP, or [α-32P]ATP in order to investigate the specificity of the reaction. Figure 3 indicates that labeled covalent complex is specific for [α-32P]GTP. The absence of labeling in the presence of [β-32P]GTP or [γ-32P]GTP indicates that although covalent bonding may occur, both β and γ phosphates are released during the formation of a GMP- or presumably m7GMP-nsP1 adduct. As presented below (see Fig. 6), an antibody recognizing the m3G/m7G-cap but not G-cap structures (42) is able to detect the guanylylated nsP1 in Western blotting, suggesting that the complex is m7GMP-nsP1 as described in the case of SINV and SFV nsP1 (21). Together, these data suggest that VEEV nsP1 covalently binds m7GMP in an AdoMet-dependent reaction.
FIG 3.
Substrate and cofactor specificity for m7GMP-nsP1 formation. Shown are the results of analysis of the covalent m7GMP-nsP1 intermediate formed with [α-32P]GTP, [β-32P]GTP, [γ-32P]GTP, or [α-32P]ATP. The activity assays were carried out at 30°C by incubating the nsP1 with [α-32P]GTP, [β-32P]GTP, [γ-32P]GTP, or [α-32P]ATP in the presence of AdoMet and/or Mg2+. The reaction products were separated by SDS-PAGE, followed by autoradiography analysis.
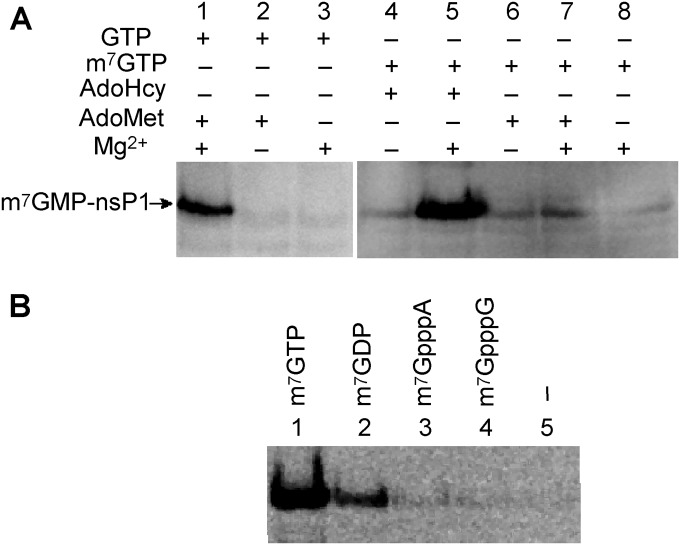
FIG 6.
Substrate and cofactor specificity of the GT reaction. (A) The guanylylation mixture containing m7GTP or GTP, AdoHcy or AdoMet, with or without 2 mM MgCl2, and nsP1 was incubated at 30°C for 2 h. (B) Guanylylation of nsP1 with AdoHcy, MgCl2, using different possible substrates (m7GTP, m7GDP, m7GpppA, m7GpppG, or no substrate), was analyzed by Western blotting using anti-m3G/m7G-Cap antibody as described in Materials and Methods.
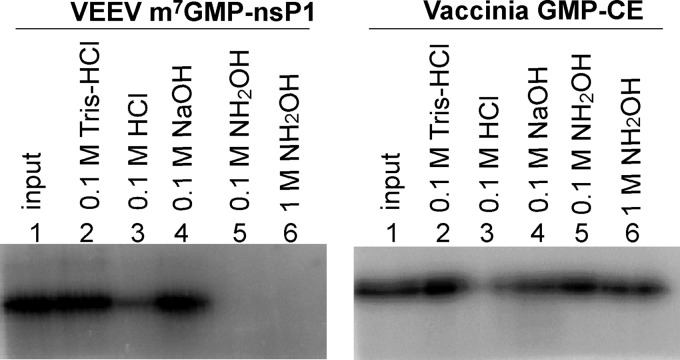
To further investigate the nature of the covalent link between m7GMP and nsP1, the stability of the m7GMP-nsP1 intermediate was compared to that of vaccinia virus (VV) capping enzyme (CE) linking GMP by phosphoamide bond with the ε-amino group of a lysine [CE-(lysyl-N)-GMP]. Guanylylated intermediates were formed by incubation of nsP1 or VV CE with [32P]GTP in the presence or absence of AdoMet. The complexes were next incubated in various buffers in order to follow their stability under different reaction conditions. Figure 4 indicates that the m7GMP-nsP1 intermediate is stable at neutral pH (lane 2) or at alkaline pH (lane 4) but sensitive to low pH like for VV CE (Fig. 4) (43). These results suggest that a phosphoamide bond links GMP to a histidine or a lysine residue (44). The nature of the linkage was further examined by treatment of the m7GMP-nsP1 intermediate with hydroxylamine (0.1 M and 1 M) at pH 7.5. As shown in Fig. 4, the m7GMP-nsP1 intermediate was labile when incubated with neutral hydroxylamine, whereas the VV CE-GMP complex containing a phosphoamide bond with a lysine was resistant under these experimental conditions. Hydroxylamine at neutral pH was reported to be able to hydrolyze phosphoamide bonds involving histidine residues in several proteins (43–46), whereas efficient hydrolysis of the phosphoamide bond with lysine, like in the VV CE-GMP complex, requires a high concentration (4 M) of hydroxylamine under a mildly acidic condition (pH 4.8) (43, 47, 48). We therefore conclude that the m7GMP is linked to a histidine residue through a phosphoramide bond.
FIG 4.
Characterization of the phosphoamide bond between VEEV nsP1 and m7GMP by chemical treatments. The m7GMP-nsP1 or vaccinia virus GMP-CE intermediates formed as described in Materials and Methods were incubated separately with 0.1 M Tris-HCl buffer (pH 7.5), 0.1 M HCl, 0.1 M NaOH, or 0.1 M or 1 M hydroxylamine (pH 7.5) at 37°C for 45 min, then separated by 10% SDS-PAGE, and subjected to autoradiography analysis.
Specific requirements of the MTase and the formation of m7GMP-nsP1 complex.
Uncoupling the MTase from the GT would allow the specific characterization of these two enzymatic steps leading to the formation of m7GMP-nsP1 adduct. To make it possible, we set up a filter-based assay in order to specifically follow the methylation step using a nonhydrolyzable GTP molecule (GIDP). Since the formation of enzyme-NMP intermediates using a β-γ nonhydrolyzable bond has already been reported (49), we first demonstrated that GIDP is unable to make a covalent link with nsP1 (data not shown). Additionally, an immunoassay based on the detection of m7G by specific antibodies was developed to further depict the guanylylation of nsP1.
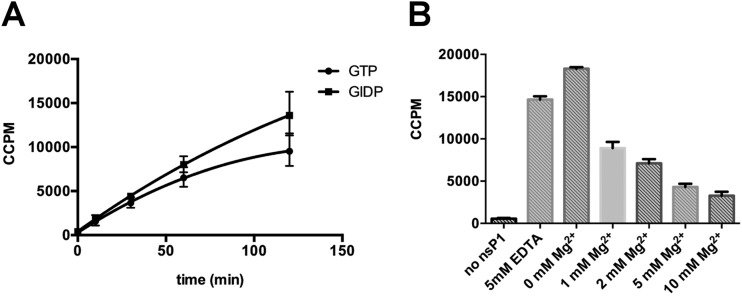
The AdoMet-dependent MTase activity of nsP1 was first characterized by measuring the transfer of a 3H-labeled methyl group from [3H]AdoMet to the methyl acceptor GIDP. After the MTase reaction, the unbound [3H]AdoMet was separated from the acceptor molecule by DEAE filter assay and the radioactivity associated with the substrate bound to the filter was counted. The time course experiment results presented in Fig. 5A indicate that the reaction efficacy is slightly higher for GIDP than for GTP. One can also conclude that the methylation can be followed using GIDP instead of GTP in order to decouple MTase and GT reaction, with no risk that the resulting product can guanylylate nsP1 and consequently affect MTase activity. The GIDP-based assay was used to investigate the Mg2+ dependency of the methylation reaction. Interestingly, Fig. 5B indicates that the MTase activity of nsP1 is Mg2+ independent, as 5 mM EDTA does not block the reaction. We also noticed that the addition of increasing concentrations of Mg2+ slightly decreases the MTase activity in a dose-dependent manner.
FIG 5.
nsP1 MTase activity. (A) Time course experiment. The methylation reaction mixture containing GTP or guanylylimidodiphosphate (GIDP), S-adenosyl-[methyl-3H]methionine, AdoMet, and nsP1 was incubated at 30°C. Reaction samples were collected at different times, and the radioactivity (corrected counts per minute [CCPM]) associated with GTP or GIDP was measured by filter binding assay as described in Materials and Methods. The data presented are means from three independent experiments, with standard deviations indicated. (B) Effect of magnesium on nsP1 MTase activity. The methylation reaction was performed at 30°C for 2 h in the presence of GIDP, S-adenosyl-[methyl-3H]methionine, AdoMet, nsP1, and 5 mM EDTA or increasing concentrations of magnesium. Reaction samples were collected and the radioactivity associated with of GIDP was measured by filter binding assay as described in Materials and Methods. The data presented are means from three independent experiments, with standard deviations indicated.
We next explored the Mg2+ dependency of the formation of the m7GMP-nsP1 complex (GT) starting from m7GTP. Since [α-32P]m7GTP was not commercially available, we developed an immunoassay to follow the formation of m7GMP-nsP1 complex. Briefly, VEEV nsP1 was incubated with GTP plus AdoMet or m7GTP, and the reaction products were then separated on a 10% SDS-PAGE gel before immunoblotting. The m7GMP-nsP1 complex was detected by using an antibody recognizing specifically the m3G/m7G cap structures, although m7GMP-nsP1 intermediate does not constitute an RNA cap per se. Figure 6A shows that with GTP, AdoMet, and Mg2+ (lane 1), a complex migrating at the size of nsP1 can be detected by the antibodies. However, in the absence of Mg2+ or AdoMet, only a slight fraction of m7GMP-nsP1 was detected. Figure 6A also indicates that the bonding of m7GMP onto nsP1 can be efficiently obtained using directly m7GTP, but only in the presence of Mg2+ ions and AdoHcy (lane 5), the by-product of the MTase reaction. AdoMet does not play the activating role when incubated with nsP1 and m7GTP. The requirement of AdoHcy and not AdoMet for the GT reaction suggests that the MTase reaction producing AdoHcy has to occur to allow the downstream GT reaction. Additionally, we also show that m7GTP is the preferred substrate for nsP1 guanylylation (Fig. 6B) and that cap analogs mimicking VEEV mRNA (m7GpppA and m7GpppG) cannot form a covalent binding with nsP1 as expected, suggesting that the viral RNA capping is probably not a reversible mechanism.
Mutagenesis analysis of the MTase and GT activities.
To determine the role of conserved amino acid residues (Fig. 1) in both nsP1 activities (MTase and GT), we first examined the formation of the covalent intermediate by incubating the nsP1 mutants with [α-32P]GTP in the presence of AdoMet. The effects of the mutations, presented in Table 1 and Fig. 7, could be grouped into four categories: (i) mutations H37A and D63A completely abolish the formation of m7Gp-nsp1 complex; (ii) Y285A, N369A, and N375A substitutions lead to a decrease in the formation of the covalent complex; (iii) D354A and R365A mutations increase the complex formation; and (iv) H45A and E118A substitutions barely affect the nsP1 activity. To further understand the role of the mutations in each enzymatic activity, the effects of the nsP1 mutations on the MTase and GT activities were characterized separately.
TABLE 1.
Relative activities of various nsP1 mutants compared to wild-type nsP1
| nsP1 | m7GMP-nsP1 formationa (%) | Methylation (%) | Guanylylation (%) |
|---|---|---|---|
| Wild type | 100 | 100 | 100 |
| Mutants | |||
| H37A | 1 ± 1 | 154 ± 15 | 0 |
| H45A | 90 ± 3 | 50 ± 1 | 128 ± 18 |
| D63A | 0 | 21 ± 3 | 2 ± 2 |
| E118A | 113 ± 18 | 74 ± 14 | 87 ± 18 |
| Y285A | 16 ± 4 | 33 ± 12 | 20 ± 14 |
| D354A | 371 ± 12 | 132 ± 12 | 187 ± 14 |
| R365A | 379 ± 51 | 125 ± 9 | 233 ± 24 |
| N369A | 51 ± 12 | 52 ± 10 | 61 ± 21 |
| N375A | 47 ± 12 | 63 ± 14 | 53 ± 14 |
“m7GMP-nsP1 formation” refers to the detection of guanylylated nsP1 in the presence of [α-32P]GTP and AdoMet. Values are means ± standard deviations.
FIG 7.
Effect of mutations on coupled MTase and GT. (A) Purified nsP1 mutants loaded on a 10% Coomassie blue-stained SDS-PAGE gel. (B) Formation of the m7GMP-enzyme intermediate after incubation of the nsP1 mutants with [α-32P]GTP in the presence of AdoMet as described in Materials and Methods.
The methyltransferase activities of the VEEV nsP1 mutants were determined as described above, by following the transfer of 3H-methyl group from [3H]AdoMet to GIDP (Table 1). D63A and Y285A mutants show the weakest MTase activity, the H45A, N369A, and N375A mutants showed more than 50% of the MTase activity of the wild type, and the H37A, D354A, and R365A mutants were about 30% more efficient than the wild type for the methyltransfer on GIDP.
The nsP1 mutant proteins were also analyzed for the ability to form the covalent intermediate m7GMP-nsP1 in the presence of m7GTP and AdoHcy. The immunoassay results presented in Table 1 indicate that the H37A, D63A, and Y285A mutants block the formation of the m7GMP-nsP1 complex, whereas the N369A and N375A mutants resulted in only moderately reduced activities and mutations at D354 and R365 increased the activity 2- to ∼3-fold. Together, these results indicate that H37 plays a crucial role in the formation of the phosphoramide link with m7GMP, with no significant effect on the MTase activity of nsP1. The mutagenesis experiments also demonstrate that D63A and Y285A limit both MTase and GT, suggesting a strong interplay between the two enzymatic activities. The latter observation is also supported by the stimulation of m7GMP binding by AdoHcy, a product of the MTase reaction (Fig. 6A).
RNA capping: the guanylyltransferase activity.
The RNA-capping mechanism in alphaviruses should involve the recruitment of an RNA to nsP1 in order to allow the m7GMP transfer onto its 5′-diphosphate end, generating a capped RNA. We first compared the RNA binding properties of nsP1 with a 15-nucleotide RNA mimicking the 5′ end of the VEEV genome using the fluorescence polarization (FP) technique. The 5′ tri-, di-, and monophosphate, capped, and hydroxyl RNAs were synthesized and labeled with Cy5 at the 3′ end. The different RNAs were then incubated with increasing concentration of nsP1, and the change in fluorescence polarization resulting from RNA-protein interaction was followed at 675 nm (Fig. 8). Higher polarization values were obtained with 5′-phosphorylated RNAs, yielding very similar KDs (2.6, 3.5, and 2.2 μM for tri-, di-, and monophosphorylated RNAs, respectively). In contrast, the presence of a cap structure or a hydroxyl group at the 5′ end of RNA strongly reduced the interaction and the plateau of fluorescence was not reached under our experimental conditions, highlighting the requirement of the phosphorylated 5′ end for RNA recruitment.
FIG 8.
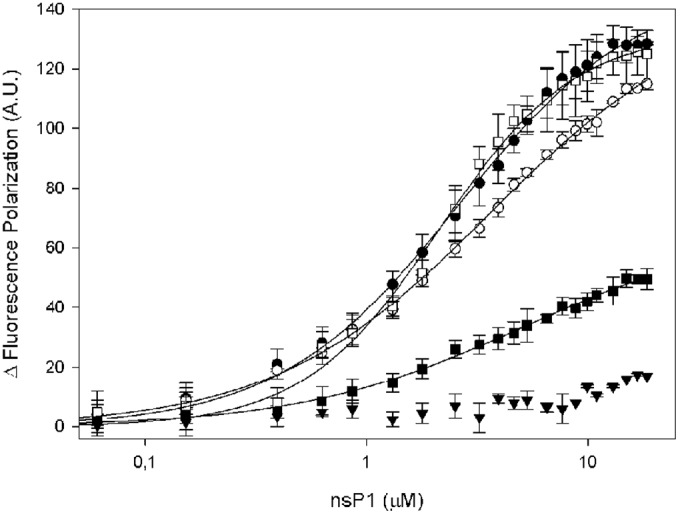
Binding of RNA to VEEV nsP1 assessed by fluorescence polarization. 3′ Cy5-labeled 15-mer RNAs with different 5′ ends, i.e., 5′pppRNA (●), 5′ppRNA (○), 5′pRNA (□), 5′RNA (■), and 5′GpppRNA (▼), were incubated in increasing concentrations of VEEV nsP1.
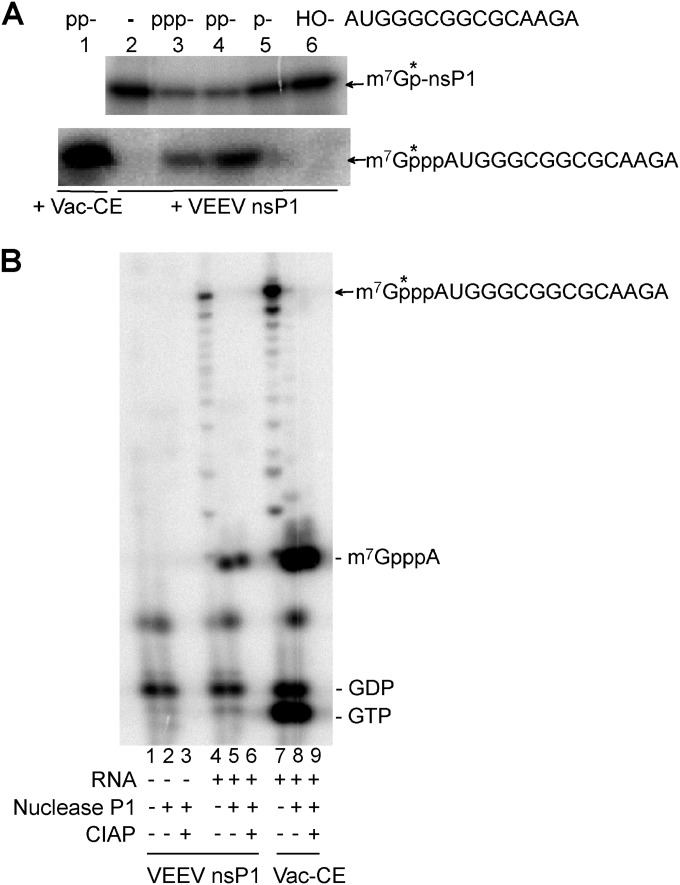
To demonstrate the guanylyltransfer onto the RNA, the different RNAs were then incubated with the preformed [α-32P]m7GMP-nsP1 complex (Fig. 9A). The incubation resulted in a decrease of the [α-32P]m7GMP-nsP1 complex when di- and triphosphate RNAs were added and a concomitant radiolabeling of RNA on a urea-PAGE gel was observed, but the radiolabeling transfer was more efficient for the 5′-diphosphate RNA than for triphosphate and monophosphate RNAs. No transfer was observed using the 5′ hydroxyl RNA, as expected. The four nsP1 mutants which are able to form the m7GMP-nsP1complex (H45A, E118A, D345A, and R365A mutants) were also tested for the ability to transfer m7Gp on the 5′ end of RNA. They all displayed cap binding activity (data not shown). As for the MTase and GT activities, D345A and R365A mutations resulted in an efficiency higher than that of the wild type. Regarding H45A and E118A, no major difference in GTase activity from the wild type nsP1 protein was observed (data not shown).
FIG 9.
Transfer of m7GMP from the covalent nsP1-m7GMP intermediate to RNA and characterization of the cap structure. (A) m7GMP transfer from nsP1 to the RNA. 5′-Diphosphate RNA was incubated with the VV CE in the presence of AdoMet and [α-32P]GTP (lane 1), no substrate (lane 2), 5′-triphosphate RNA (lane 3), 5′-diphosphate RNA (lane 4), 5′-monophosphate RNA (lane 5), and 5′-hydroxyl RNA (lane 6) and with the 32P-labeled covalent nsP1-m7GMP intermediate at 30°C. Then half of the reaction mixture was subjected to SDS-PAGE (10%) analysis (top) to check the decrease of the 32P-labeled covalent nsP1-m7GMP intermediate, and the other half was subjected to 7 M urea-PAGE (20%) (bottom) to show the produced m7Gp*ppAUGGGCGGCGCAAGA RNA. (B) Characterization of the cap structure. No RNA (lanes 1, 2, and 3) and 5′-diphosphate AUGGGCGGCGCAAGA RNA were incubated with the 32P-labeled covalent nsP1-m7GMP intermediate (lanes 4, 5, and 6) at 30°C for 2 h. 5′-Diphosphate AUGGGCGGCGCAAGA RNA was also incubated with vaccinia virus (Vac) CE in the presence of AdoMet and [α-32P]GTP at 37°C for 60 min (lanes 7, 8, and 9). The reaction products were recovered by ethanol precipitation and then treated with nuclease P1 (lanes 2, 5, and 8) or with nuclease P1 then CIAP (lanes 3, 6, and 9). The resulting products were loaded on a 7 M urea-PAGE (20%) to assess the presence of m7GpppAUGGGCGGCGCAAGA and released m7GpppA. Lines along the edge of the urea-PAGE gel indicate the migration positions of markers GTP, GDP, and m7GpppA.
In order to further characterize the cap structure at the 5′ end, the product of the RNA capping reaction by VEEV nsP1 was digested with P1 nuclease, and the cap structures, resistant to nuclease P1 digestion, were analyzed (Fig. 9B). An additional treatment with CIAP was performed to confirm the presence of a cap structure. The resulting products were compared to a cap-0 structure obtained using the vaccinia virus capping system followed by nuclease P1 and CIAP treatments. A bona fide m7GpppA cap structure could be observed after incubations with P1 nuclease and CIAP. These results demonstrate for the first time that a cap can be formed on a 5′-diphosphate VEEV RNA end by an alphavirus nsP1. In addition to the cap, another nuclease P1 product could be observed, corresponding to labeled GMP or m7GMP. This product, also present in the vaccinia virus capping system, could correspond to released m7GMP from remaining VEEV nsP1 and VV CE, since the nuclease P1 reaction was performed at low pH (5.3), a condition under which the phosphoramide bonds were shown to be unstable (Fig. 4).
Effects of inhibitors on VEEV nsP1.
Since RNA capping is essential for the translation initiation and the control of innate immunity, nsP1 is a key enzyme in alphavirus replication and thus an attractive target for the development of antiviral compounds limiting nsP1 functions related to RNA capping. Three reference compounds were assessed for their potencies to inhibit the MTase reaction and/or the GT. We selected sinefungin, an AdoMet analog which was previously shown to be a potent inhibitor of viral cap MTases (50–52); aurintricarboxylic acid (ATA), a known inhibitor of flavivirus mRNA capping (53); and the triphosphate form of ribavirin, known for its broad spectral antiviral effect against alphaviruses and for which resistance substitutions were observed in the nsP1 sequence (54).
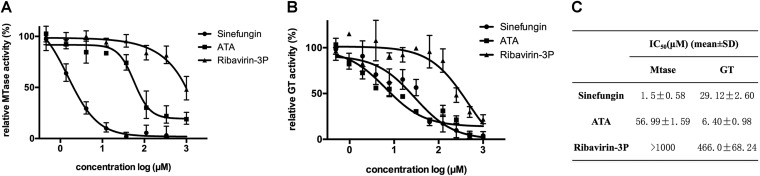
The inhibition of the MTase activity was first addressed. For this purpose, VEEV nsP1 was incubated with [3H]AdoMet and GIDP with increasing concentrations of sinefungin, ATA, and ribavirin triphosphate. Figure 10A shows that the MTase activity was efficiently inhibited by sinefungin, with an IC50 at 1.5 μM, and to a lower extent by ATA (IC50 = 57 μM). In contrast, ribavirin 5′-triphosphate poorly inhibited the MTase activity, with an IC50 above 1 mM.
FIG 10.
Inhibition of VEEV nsP1 methylation and guanylylation activities. (A) Inhibition of VEEV nsP1 methylation was tested by incubating nsP1 with guanylylimidodiphosphate (GIDP), S-adenosyl-[methyl-3H]methionine, and AdoMet in the presence of increasing concentrations of sinefungin, ATA, or ribavirin triphosphate (3P) at 30°C for 1 h. (B) Inhibition of VEEV nsP1 guanylylation was tested by incubating nsP1 with m7GTP, AdoHcy, and MgCl2 in the presence of increasing concentrations of sinefungin, ATA, or ribavirin 3P at 30°C for 1 h. Reaction samples were collected and analyzed by experimental procedures given in Materials and Methods. The data presented are means from three independent experiments, with standard deviations indicted. (C) IC50s of the three compounds are illustrated.
The effects of these molecules on nsP1 guanylylation were then tested. nsP1 was incubated together with m7GTP and AdoHcy in the presence of increasing concentrations of sinefungin, ATA, and ribavirin triphosphate, and the formation of the covalent complex was followed by Western blotting using the antibody anti-m3G/m7G-cap. As for the MTase reaction, sinefungin and ATA inhibited nsP1 GT with IC50s of 29 and 6.4 μM, respectively, whereas ribavirin 5′-triphosphate was a poor inhibitor, with an IC50 close to 500 μM (Fig. 10B). If both sinefungin and ATA are active against both nsP1 functions, their highest potencies are against the MTase activity and the nsP1 GT, respectively.
DISCUSSION
Cytoplasmic RNA viruses have evolved unconventional mRNA capping mechanisms to efficiently protect their RNA from nuclease degradation, initiate protein translation, and escape innate immunity (16). Since the 1990s, many studies have reported the functional characterization of OW alphavirus nsP1, identifying two main functions in the viral mRNA capping, namely, the methylation of GTP due to the MTase activity and the nsP1 guanylylation (GT). However, the last putative capping step, i.e., the transfer of the m7GMP from nsP1 on the 5′-diphosphate RNA, had never been demonstrated in vitro. In order to get insight into alphavirus RNA capping, we selected VEEV, the model of NW alphavirus for which nsP1 enzymatic activities had not been explored yet. In this work, we demonstrate that purified VEEV nsP1 carries both Mtase and GT activities and performed the guanylyltransfer on a ppRNA. We show that the first step of the VEEV capping process consists of the AdoMet-dependent methylation of the GTP molecule by nsP1 (MTase activity). m7GTP subsequently forms a covalent link with nsP1 (m7GMP-nsP1), releasing PPi (GT reaction). In the presence of ppRNA mimicking the 5′ end of the VEEV genome, the m7Gp can be next transferred to the 5′ end (RNA capping), generating capped RNA.
The uncoupling of the MTase from the GT shows that the MTase activity does not require divalent ions, whereas GT is Mg2+ or Mn2+ dependent. This feature is conserved among alphavirus nsP1 enzymes (17–19) and alphavirus-like capping enzymes (34). The AdoMet-AdoHcy balance on the nsP1 functions was also shown to have a regulatory effect on the MTase and GT activities. First, we show that AdoHcy, the by-product of the methyltransfer reaction, poorly inhibits nsP1 MTase activity (data not shown). Similar weak inhibition effect was already reported for other viral MTases, for which AdoHcy has an IC50 in the range of 10 to 100 μM (50, 55, 56). In contrast, we also demonstrated that as for SFV nsP1 and bamboo mosaic virus (BaMV) capping enzyme (22, 57), the GT activity is strongly stimulated by AdoHcy. Indeed, when the MTase reaction was bypassed by using m7GTP as the substrate, the nsP1 guanylylation could not occur, except when the reaction buffer was supplemented with AdoHcy. AdoHcy thus plays a crucial role in the interplay between the nsP1 functions, probably not by competition with AdoMet for the MTase inhibition but rather by inducing conformational changes favoring the GT reaction. During the preparation of the manuscript, Gullberg and coworkers showed that AdoMet and consequently AdoHcy are not necessary to produce the m7GMP-nsP1 product (58). However, the study was performed in the presence of GTP-ATTO 680, a GTP analog whose modification in position 8 could possibly affect the chemical properties of GTP and favor the guanylylation reaction.
In an effort to further characterize the covalent link formed during GT reaction, we used [α-32P]GTP, [β-32P]GTP, and [γ-32P]GTP as substrates. The covalent binding assay demonstrated that β and γ phosphates are hydrolyzed during formation of the m7Gp-nsP1 adduct. Using cap analogs, GTP, m7GDP, or m7GTP, we also confirmed that the preferred substrate for the guanylylation is GTP (in the presence of AdoMet) or m7GTP. Strikingly, m7GDP is also able to covalently bind to nsP1, also to a lesser extent than m7GTP, but since nsP1 is poorly active as a methyltransferase on GDP (59), one can wonder if m7GDP is a biologically relevant substrate in the cell. Western blotting demonstrated that the resulting product of GT reaction is m7GMP linked with nsP1. The nature of the linkage was also assessed by hydrolysis in different buffers and by mutagenesis analysis. Together, the results demonstrate that the m7GMP should be linked to histidine 37 through a phosphoramide bond, an observation in agreement with previous studies performed with OW alphavirus (21). This result confirms again the unique feature of mRNA capping within the alphavirus-like supergroup (20), accomplishing the methyltransfer reaction before cap formation and thus opening a narrow window for specific antiviral targeting.
The functional study reported herein provided also the first experimental evidence that in vitro the m7GMP moiety can be ultimately transferred onto the viral ppRNA to form the capped RNA in alphavirus (Fig. 9). Fluorescence polarization experiments with labeled 5′ ends of VEEV mRNA showed that nsP1 is able to bind specifically 5′-phosphorylated RNA. When these RNA are added to pre-prepared m7GMP-nsP1, m7GMP is released from nsP1 and transferred to an RNA, with the greatest efficacy for 5′-diphosphate RNA. However, a poor transfer on a triphosphate RNA could also be observed, suggesting that either an unconventional cap (m7GppppRNA) is formed in vitro, as observed for in vesicular stomatitis virus (VSV) mRNA capping (60), or the triphosphate group of the RNA is not stable under the reaction conditions. In the presence of the diphosphate RNA, the presence of a cap-0 structure was confirmed after RNA digestion by P1 nuclease and CIAP treatments.
Point mutation experiments are powerful tools to assess the contributions of specific residues in different enzymatic activities carried out by nsP1, especially in the absence of structural data. The mutants generated in our case revealed that the MTase and the nsP1 GT reactions can be to a certain extent uncoupled in terms of residues involved in each activity. Hence, the H37A substitution inhibits the guanylylation reaction probably by abolishing the phosphamide bond formation with m7GMP, which could involve this specific amino acid. In contrast, the H37A mutant displayed enhanced MTase activity. A similar effect was observed in SFV, where the corresponding mutant conserved the ability to bind to the AdoMet cofactor and perform the methyltransfer on GTP without forming the complex intermediate (22). These results, together with the conservation of this histidine within alphaviruses and alphavirus-like viruses, support strongly that the H37 located in the GTP binding site forms the phosphoramide bond. The substitution of H45, which might be also a candidate for guanylylation, exhibits an opposite effect, i.e., a decreased MTase activity with a slight increase in the GT reaction, and cannot be the m7GMP acceptor. The D63A mutation yielded a reduction of the MTase and abolished the GT activity. This mutation is located in the consensus site UU[D/E]Uooxo (ILDIGSAP sequence in VEEV nsP1), where U represents hydrophobic acids, “o” small residues (often a G), and x any amino acid (22, 61). The mutation of the aspartic acid might prevent the recruitment of AdoMet and AdoHcy, known to be stabilized in MTase by a bridge network depending on D63 (62), and thus the activation of the MTase and GT, respectively. So far, the role of the C-terminal part of nsP1 in the capping reactions has been poorly investigated. Y285A, N369A, and N375A substitutions decrease but do not abolish both MTase and nsP1 GT activities, whereas D354A and R365A stimulate the two functions. In SINV, the last four mutations are located in a region found to be involved in minus-strand RNA synthesis (63), and this role would result from an interaction between nsP1 and nsP4 (39, 41, 64). Since the mutations affect the MTase and GT in the same direction—positively or negatively—and they are located in a putative interacting domain far from the catalytic site, these results suggest that the substitutions either destabilize further the recombinant nsP1 in the absence of nsP4 (N369A and N375A) or stabilize nsP1 by modifying the charge at its surface (D354A and R365A). The two stabilizing substitutions, D354A and R365A, also improve the guanylyltransfer on RNA, reinforcing the role of these mutations in protein stabilization (data not shown).
Because of its unconventional mechanism, alphavirus mRNA capping is an attractive target for the design of specific inhibitors. However, nsP1 has been poorly addressed to date as an antiviral target, and except several guanosine or cap analogs (59), no small molecules have been tested against nsP1. We took advantage of the protocols we developed for the functional characterization of VEEV nsP1 to assess the inhibitory effects of three molecules. Sinefungin, an AdoMet/AdoHcy analog, was tested as a competitive inhibitor. Sinefungin strongly inhibits nsP1 MTase and also limits the formation of the m7GMP-nsP1 complex. It is likely that sinefungin can affect nsP1 functions at two levels: it inhibits the methyltransfer by competing with the methyl donor AdoMet and prevents from the activation of AdoHcy in the GT reaction. This result confirms that the two enzymatic functions are closely intertwined. Although ATA results in the same effects on the MTase and GT, its role is more elusive since it can inhibit a large range of proteins, including hepatitis C virus (HCV) NS3 or influenza virus neuraminidase (65, 66). Concerning ribavirin and its phosphorylated derivatives, it is commonly accepted that it is a broad-spectrum antiviral molecule that probably inhibits the cellular enzyme IMP dehydrogenase (IMPDH) through 5′-monophosphate ribavirin, thus depleting the cellular GTP pool required for virus replication. However, for Lassa virus, the antiviral effect of ribavirin is not predominantly a result of IMPDH inhibition (67). Among alphaviruses, viruses resistant to ribavirin possess mutations at the N terminus of nsP1, in contrast to the sensitive ones (54). Since we could not find any potent inhibition of the MTase, GT, or RNA capping by ribavirin 5′-triphosphate (data not shown for the last step), we can exclude that its antiviral effect is directly due to capping inhibition and deduce that it is more likely an indirect effect on the cellular GTP concentration (68).
In conclusion, this study showed for the first time an enzymatic characterization of the peculiar RNA capping processed by alphavirus nsP1. The MTase and GT activities already reported for nsP1 for several OW alphaviruses were confirmed and further characterized for the NW alphavirus model. Moreover, cap formation was observed in the presence of m7GMP-nsP1 and 5′-diphosphate RNA, confirming the proposed capping model (21). The RNA structure, length, and sequence specificity driving the cap formation still remain to be elucidated (69), and this can now be addressed by testing a larger set of RNAs. The functional enzymatic assays developed for uncoupling MTase, GT, and guanylyltransfer on the RNA for the functional characterization will also benefit research to help understand the mechanism of action of compounds targeting nsP1.
ACKNOWLEDGMENTS
This work was supported by European Union Seventh Framework Programme FP7/2007-2013 (Project SILVER grant 260644 and Project EUVIRNA grant 264286). J.G. was the recipient of a postdoctoral fellowship from MEC (BFS-2010-1217) and an Intra-European Marie Curie fellowship (FP7-PEOPLE-2011-IEF, grant 299753).
REFERENCES
- 1.Charrel RN, Leparc-Goffart I, Gallian P, de Lamballerie X. 2014. Globalization of Chikungunya: 10 years to invade the world. Clin Microbiol Infect 20:662–663. doi: 10.1111/1469-0691.12694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gould EA, Coutard B, Malet H, Morin B, Jamal S, Weaver S, Gorbalenya A, Moureau G, Baronti C, Delogu I, Forrester N, Khasnatinov M, Gritsun T, de Lamballerie X, Canard B. 2010. Understanding the alphaviruses: recent research on important emerging pathogens and progress towards their control. Antiviral Res 87:111–124. doi: 10.1016/j.antiviral.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suhrbier A, Jaffar-Bandjee MC, Gasque P. 2012. Arthritogenic alphaviruses—an overview. Nat Rev Rheumatol 8:420–429. doi: 10.1038/nrrheum.2012.64. [DOI] [PubMed] [Google Scholar]
- 4.Adams AP, Navarro-Lopez R, Ramirez-Aguilar FJ, Lopez-Gonzalez I, Leal G, Flores-Mayorga JM, Travassos da Rosa AP, Saxton-Shaw KD, Singh AJ, Borland EM, Powers AM, Tesh RB, Weaver SC, Estrada-Franco JG. 2012. Venezuelan equine encephalitis virus activity in the Gulf Coast region of Mexico, 2003–2010. PLoS Negl Trop Dis 6:e1875. doi: 10.1371/journal.pntd.0001875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pisano MB, Oria G, Beskow G, Aguilar J, Konigheim B, Cacace ML, Aguirre L, Stein M, Contigiani MS. 2013. Venezuelan equine encephalitis viruses (VEEV) in Argentina: serological evidence of human infection. PLoS Negl Trop Dis 7:e2551. doi: 10.1371/journal.pntd.0002551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rivas F, Diaz LA, Cardenas VM, Daza E, Bruzon L, Alcala A, De la Hoz O, Caceres FM, Aristizabal G, Martinez JW, Revelo D, De la Hoz F, Boshell J, Camacho T, Calderon L, Olano VA, Villarreal LI, Roselli D, Alvarez G, Ludwig G, Tsai T. 1997. Epidemic Venezuelan equine encephalitis in La Guajira, Colombia, 1995. J Infect Dis 175:828–832. doi: 10.1086/513978. [DOI] [PubMed] [Google Scholar]
- 7.Weaver SC, Salas R, Rico-Hesse R, Ludwig GV, Oberste MS, Boshell J, Tesh RB. 1996. Re-emergence of epidemic Venezuelan equine encephalomyelitis in South America. VEE Study Group. Lancet 348:436–440. [DOI] [PubMed] [Google Scholar]
- 8.Aguilar PV, Estrada-Franco JG, Navarro-Lopez R, Ferro C, Haddow AD, Weaver SC. 2011. Endemic Venezuelan equine encephalitis in the Americas: hidden under the dengue umbrella. Future Virol 6:721–740. doi: 10.2217/fvl.11.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson KM, Martin DH. 1974. Venezuelan equine encephalitis. Adv Vet Sci Comp Med 18:79–116. [PubMed] [Google Scholar]
- 10.León CA. 1975. Sequelae of Venezuelan equine encephalitis in humans: a four year follow-up. Int J Epidemiol 4:131–140. doi: 10.1093/ije/4.2.131. [DOI] [PubMed] [Google Scholar]
- 11.Taylor KG, Paessler S. 2013. Pathogenesis of Venezuelan equine encephalitis. Vet Microbiol 167:145–150. doi: 10.1016/j.vetmic.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 12.Chung D, Schroeder CE, Sotsky J, Yao T, Roy S, Smith RA, Tower NA, Noah JW, McKellip S, Sosa M, Rasmussen L, White EL, Aube J, Golden JE. 2013. ML336: development of quinazolinone-based inhibitors against Venezuelan equine encephalitis virus (VEEV). Probe reports from the NIH Molecular Libraries Program. National Center for Biotechnology Information, Bethesda, MD: http://www.ncbi.nlm.nih.gov/books/NBK179829/?report=reader#!po=1.85185. [PubMed] [Google Scholar]
- 13.Chung DH, Jonsson CB, Tower NA, Chu YK, Sahin E, Golden JE, Noah JW, Schroeder CE, Sotsky JB, Sosa MI, Cramer DE, McKellip SN, Rasmussen L, White EL, Schmaljohn CS, Julander JG, Smith JM, Filone CM, Connor JH, Sakurai Y, Davey RA. 2014. Discovery of a novel compound with anti-Venezuelan equine encephalitis virus activity that targets the nonstructural protein 2. PLoS Pathog 10:e1004213. doi: 10.1371/journal.ppat.1004213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Julander JG, Bowen RA, Rao JR, Day C, Shafer K, Smee DF, Morrey JD, Chu CK. 2008. Treatment of Venezuelan equine encephalitis virus infection with (−)-carbodine. Antiviral Res 80:309–315. doi: 10.1016/j.antiviral.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hyde JL, Gardner CL, Kimura T, White JP, Liu G, Trobaugh DW, Huang C, Tonelli M, Paessler S, Takeda K, Klimstra WB, Amarasinghe GK, Diamond MS. 2014. A viral RNA structural element alters host recognition of nonself RNA. Science 343:783–787. doi: 10.1126/science.1248465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Decroly E, Ferron F, Lescar J, Canard B. 2012. Conventional and unconventional mechanisms for capping viral mRNA. Nat Rev Microbiol 10:51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mi S, Durbin R, Huang HV, Rice CM, Stollar V. 1989. Association of the Sindbis virus RNA methyltransferase activity with the nonstructural protein nsP1. Virology 170:385–391. doi: 10.1016/0042-6822(89)90429-7. [DOI] [PubMed] [Google Scholar]
- 18.Laakkonen P, Hyvonen M, Peranen J, Kaariainen L. 1994. Expression of Semliki Forest virus nsP1-specific methyltransferase in insect cells and in Escherichia coli. J Virol 68:7418–7425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mi S, Stollar V. 1991. Expression of Sindbis virus nsP1 and methyltransferase activity in Escherichia coli. Virology 184:423–427. doi: 10.1016/0042-6822(91)90862-6. [DOI] [PubMed] [Google Scholar]
- 20.Rozanov MN, Koonin EV, Gorbalenya AE. 1992. Conservation of the putative methyltransferase domain: a hallmark of the ‘Sindbis-like’ supergroup of positive-strand RNA viruses. J Gen Virol 73(Part 8):2129–2134. [DOI] [PubMed] [Google Scholar]
- 21.Ahola T, Kaariainen L. 1995. Reaction in alphavirus mRNA capping: formation of a covalent complex of nonstructural protein nsP1 with 7-methyl-GMP. Proc Natl Acad Sci U S A 92:507–511. doi: 10.1073/pnas.92.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahola T, Laakkonen P, Vihinen H, Kaariainen L. 1997. Critical residues of Semliki Forest virus RNA capping enzyme involved in methyltransferase and guanylyltransferase-like activities. J Virol 71:392–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin HY, Yu CY, Hsu YH, Meng M. 2012. Functional analysis of the conserved histidine residue of Bamboo mosaic virus capping enzyme in the activity for the formation of the covalent enzyme-m7GMP intermediate. FEBS Lett 586:2326–2331. doi: 10.1016/j.febslet.2012.05.024. [DOI] [PubMed] [Google Scholar]
- 24.Ahola T, Lampio A, Auvinen P, Kaariainen L. 1999. Semliki Forest virus mRNA capping enzyme requires association with anionic membrane phospholipids for activity. EMBO J 18:3164–3172. doi: 10.1093/emboj/18.11.3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spuul P, Salonen A, Merits A, Jokitalo E, Kaariainen L, Ahola T. 2007. Role of the amphipathic peptide of Semliki Forest virus replicase protein nsP1 in membrane association and virus replication. J Virol 81:872–883. doi: 10.1128/JVI.01785-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lampio A, Kilpelainen I, Pesonen S, Karhi K, Auvinen P, Somerharju P, Kaariainen L. 2000. Membrane binding mechanism of an RNA virus-capping enzyme. J Biol Chem 275:37853–37859. doi: 10.1074/jbc.M004865200. [DOI] [PubMed] [Google Scholar]
- 27.Ahola T, Kujala P, Tuittila M, Blom T, Laakkonen P, Hinkkanen A, Auvinen P. 2000. Effects of palmitoylation of replicase protein nsP1 on alphavirus infection. J Virol 74:6725–6733. doi: 10.1128/JVI.74.15.6725-6733.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laakkonen P, Ahola T, Kaariainen L. 1996. The effects of palmitoylation on membrane association of Semliki Forest virus RNA capping enzyme. J Biol Chem 271:28567–28571. doi: 10.1074/jbc.271.45.28567. [DOI] [PubMed] [Google Scholar]
- 29.Tomar S, Narwal M, Harms E, Smith JL, Kuhn RJ. 2011. Heterologous production, purification and characterization of enzymatically active Sindbis virus nonstructural protein nsP1. Protein Expr Purif 79:277–284. doi: 10.1016/j.pep.2011.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gorbalenya AE, Koonin EV, Donchenko AP, Blinov VM. 1988. A novel superfamily of nucleoside triphosphate-binding motif containing proteins which are probably involved in duplex unwinding in DNA and RNA replication and recombination. FEBS Lett 235:16–24. doi: 10.1016/0014-5793(88)81226-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hodgman TC. 1988. A new superfamily of replicative proteins. Nature 333:22–23. [DOI] [PubMed] [Google Scholar]
- 32.Karpe YA, Aher PP, Lole KS. 2011. NTPase and 5′-RNA triphosphatase activities of Chikungunya virus nsP2 protein. PLoS One 6:e22336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vasiljeva L, Merits A, Auvinen P, Kaariainen L. 2000. Identification of a novel function of the alphavirus capping apparatus. RNA 5′-triphosphatase activity of Nsp2. J Biol Chem 275:17281–17287. [DOI] [PubMed] [Google Scholar]
- 34.Huang YL, Hsu YH, Han YT, Meng M. 2005. mRNA guanylation catalyzed by the S-adenosylmethionine-dependent guanylyltransferase of bamboo mosaic virus. J Biol Chem 280:13153–13162. doi: 10.1074/jbc.M412619200. [DOI] [PubMed] [Google Scholar]
- 35.Lantez V, Dalle K, Charrel R, Baronti C, Canard B, Coutard B. 2011. Comparative production analysis of three phlebovirus nucleoproteins under denaturing or non-denaturing conditions for crystallographic studies. PLoS Negl Trop Dis 5:e936. doi: 10.1371/journal.pntd.0000936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lavergne T, Bertrand JR, Vasseur JJ, Debart F. 2008. A base-labile group for 2′-OH protection of ribonucleosides: a major challenge for RNA synthesis. Chemistry 14:9135–9138. doi: 10.1002/chem.200801392. [DOI] [PubMed] [Google Scholar]
- 37.Lavergne T, Janin M, Dupouy C, Vasseur JJ, Debart F. 2010. Chemical synthesis of RNA with base-labile 2′-O-(pivaloyloxymethyl)-protected ribonucleoside phosphoramidites. Curr Protoc Nucleic Acid Chem Chapter 3:Unit 3.19. doi: 10.1002/0471142700.nc0319s43. [DOI] [PubMed] [Google Scholar]
- 38.Zlatev I, Lavergne T, Debart F, Vasseur JJ, Manoharan M, Morvan F. 2010. Efficient solid-phase chemical synthesis of 5′-triphosphates of DNA, RNA, and their analogues. Org Lett 12:2190–2193. doi: 10.1021/ol1004214. [DOI] [PubMed] [Google Scholar]
- 39.Fata CL, Sawicki SG, Sawicki DL. 2002. Modification of Asn374 of nsP1 suppresses a Sindbis virus nsP4 minus-strand polymerase mutant. J Virol 76:8641–8649. doi: 10.1128/JVI.76.17.8641-8649.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lulla V, Sawicki DL, Sawicki SG, Lulla A, Merits A, Ahola T. 2008. Molecular defects caused by temperature-sensitive mutations in Semliki Forest virus nsP1. J Virol 82:9236–9244. doi: 10.1128/JVI.00711-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rupp JC, Jundt N, Hardy RW. 2011. Requirement for the amino-terminal domain of Sindbis virus nsP4 during virus infection. J Virol 85:3449–3460. doi: 10.1128/JVI.02058-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bochnig P, Reuter R, Bringmann P, Luhrmann R. 1987. A monoclonal antibody against 2,2,7-trimethylguanosine that reacts with intact, class U, small nuclear ribonucleoproteins as well as with 7-methylguanosine-capped RNAs. Eur J Biochem 168:461–467. doi: 10.1111/j.1432-1033.1987.tb13439.x. [DOI] [PubMed] [Google Scholar]
- 43.Shuman S, Hurwitz J. 1981. Mechanism of mRNA capping by vaccinia virus guanylyltransferase: characterization of an enzyme-guanylate intermediate. Proc Natl Acad Sci U S A 78:187–191. doi: 10.1073/pnas.78.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duclos B, Marcandier S, Cozzone AJ. 1991. Chemical properties and separation of phosphoamino acids by thin-layer chromatography and/or electrophoresis. Methods Enzymol 201:10–21. [DOI] [PubMed] [Google Scholar]
- 45.Gottlin EB, Rudolph AE, Zhao Y, Matthews HR, Dixon JE. 1998. Catalytic mechanism of the phospholipase D superfamily proceeds via a covalent phosphohistidine intermediate. Proc Natl Acad Sci U S A 95:9202–9207. doi: 10.1073/pnas.95.16.9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ogino T, Banerjee AK. 2007. Unconventional mechanism of mRNA capping by the RNA-dependent RNA polymerase of vesicular stomatitis virus. Mol Cell 25:85–97. doi: 10.1016/j.molcel.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 47.Toyama R, Mizumoto K, Nakahara Y, Tatsuno T, Kaziro Y. 1983. Mechanism of the mRNA guanylyltransferase reaction: isolation of N epsilon-phospholysine and GMP (5′ leads to N epsilon) lysine from the guanylyl-enzyme intermediate. EMBO J 2:2195–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Venkatesan S, Moss B. 1982. Eukaryotic mRNA capping enzyme-guanylate covalent intermediate. Proc Natl Acad Sci U S A 79:340–344. doi: 10.1073/pnas.79.2.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raae AJ, Kleppe K. 1978. Effect of ATP analogues on T4 polynucleotide ligase. Biochem Biophys Res Commun 81:24–27. doi: 10.1016/0006-291X(78)91625-X. [DOI] [PubMed] [Google Scholar]
- 50.Barral K, Sallamand C, Petzold C, Coutard B, Collet A, Thillier Y, Zimmermann J, Vasseur JJ, Canard B, Rohayem J, Debart F, Decroly E. 2013. Development of specific dengue virus 2′-O- and N7-methyltransferase assays for antiviral drug screening. Antiviral Res 99:292–300. doi: 10.1016/j.antiviral.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 51.Decroly E, Debarnot C, Ferron F, Bouvet M, Coutard B, Imbert I, Gluais L, Papageorgiou N, Sharff A, Bricogne G, Ortiz-Lombardia M, Lescar J, Canard B. 2011. Crystal structure and functional analysis of the SARS-coronavirus RNA cap 2′-O-methyltransferase nsp10/nsp16 complex. PLoS Pathog 7:e1002059. doi: 10.1371/journal.ppat.1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pugh CS, Borchardt RT, Stone HO. 1977. Inhibition of Newcastle disease virion messenger RNA (guanine-7-)-methyltransferase by analogues of S-adenosylhomocysteine. Biochemistry 16:3928–3932. doi: 10.1021/bi00636a032. [DOI] [PubMed] [Google Scholar]
- 53.Milani M, Mastrangelo E, Bollati M, Selisko B, Decroly E, Bouvet M, Canard B, Bolognesi M. 2009. Flaviviral methyltransferase/RNA interaction: structural basis for enzyme inhibition. Antiviral Res 83:28–34. doi: 10.1016/j.antiviral.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scheidel LM, Stollar V. 1991. Mutations that confer resistance to mycophenolic acid and ribavirin on Sindbis virus map to the nonstructural protein nsP1. Virology 181:490–499. doi: 10.1016/0042-6822(91)90881-B. [DOI] [PubMed] [Google Scholar]
- 55.Decroly E, Imbert I, Coutard B, Bouvet M, Selisko B, Alvarez K, Gorbalenya AE, Snijder EJ, Canard B. 2008. Coronavirus nonstructural protein 16 is a cap-0 binding enzyme possessing (nucleoside-2′O)-methyltransferase activity. J Virol 82:8071–8084. doi: 10.1128/JVI.00407-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pugh CS, Borchardt RT, Stone HO. 1978. Sinefungin, a potent inhibitor of virion mRNA(guanine-7-)-methyltransferase, mRNA(nucleoside-2′-)-methyltransferase, and viral multiplication. J Biol Chem 253:4075–4077. [PubMed] [Google Scholar]
- 57.Huang YL, Han YT, Chang YT, Hsu YH, Meng M. 2004. Critical residues for GTP methylation and formation of the covalent m7GMP-enzyme intermediate in the capping enzyme domain of bamboo mosaic virus. J Virol 78:1271–1280. doi: 10.1128/JVI.78.3.1271-1280.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gullberg RC, Jordan Steel J, Moon SL, Soltani E, Geiss BJ. 2015. Oxidative stress influences positive strand RNA virus genome synthesis and capping. Virology 475:219–229. doi: 10.1016/j.virol.2014.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lampio A, Ahola T, Darzynkiewicz E, Stepinski J, Jankowska-Anyszka M, Kaariainen L. 1999. Guanosine nucleotide analogs as inhibitors of alphavirus mRNA capping enzyme. Antiviral Res 42:35–46. doi: 10.1016/S0166-3542(99)00011-X. [DOI] [PubMed] [Google Scholar]
- 60.Ogino T, Banerjee AK. 2008. Formation of guanosine(5′)tetraphospho(5′)adenosine cap structure by an unconventional mRNA capping enzyme of vesicular stomatitis virus. J Virol 82:7729–7734. doi: 10.1128/JVI.00326-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ingrosso D, Fowler AV, Bleibaum J, Clarke S. 1989. Sequence of the d-aspartyl/l-isoaspartyl protein methyltransferase from human erythrocytes. Common sequence motifs for protein, DNA, RNA, and small molecule S-adenosylmethionine-dependent methyltransferases. J Biol Chem 264:20131–20139. [PubMed] [Google Scholar]
- 62.Rutherford K, Daggett V. 2009. A hotspot of inactivation: the A22S and V108M polymorphisms individually destabilize the active site structure of catechol O-methyltransferase. Biochemistry 48:6450–6460. doi: 10.1021/bi900174v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang YF, Sawicki SG, Sawicki DL. 1991. Sindbis virus nsP1 functions in negative-strand RNA synthesis. J Virol 65:985–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shirako Y, Strauss EG, Strauss JH. 2000. Suppressor mutations that allow Sindbis virus RNA polymerase to function with nonaromatic amino acids at the N-terminus: evidence for interaction between nsP1 and nsP4 in minus-strand RNA synthesis. Virology 276:148–160. doi: 10.1006/viro.2000.0544. [DOI] [PubMed] [Google Scholar]
- 65.Hashem AM, Flaman AS, Farnsworth A, Brown EG, Van Domselaar G, He R, Li X. 2009. Aurintricarboxylic acid is a potent inhibitor of influenza A and B virus neuraminidases. PLoS One 4:e8350. doi: 10.1371/journal.pone.0008350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shadrick WR, Mukherjee S, Hanson AM, Sweeney NL, Frick DN. 2013. Aurintricarboxylic acid modulates the affinity of hepatitis C virus NS3 helicase for both nucleic acid and ATP. Biochemistry 52:6151–6159. doi: 10.1021/bi4006495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ölschläger S, Neyts J, Günther S. 2011. Depletion of GTP pool is not the predominant mechanism by which ribavirin exerts its antiviral effect on Lassa virus. Antiviral Res 91:89–93. [DOI] [PubMed] [Google Scholar]
- 68.Liao HJ, Stollar V. 1993. Reversal of the antiviral activity of ribavirin against Sindbis virus in Ae. albopictus mosquito cells. Antiviral Res 22:285–294. doi: 10.1016/0166-3542(93)90038-K. [DOI] [PubMed] [Google Scholar]
- 69.Hyde JL, Chen R, Trobaugh DW, Diamond MS, Weaver SC, Klimstra WB, Wilusz J. 25 January 2015. The 5′ and 3′ ends of alphavirus RNAs—non-coding is not non-functional. Virus Res doi: 10.1016/j.virusres.2015.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Notredame C, Higgins DG, Heringa J. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol 302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 71.McGuffin LJ, Bryson K, Jones DT. 2000. The PSIPRED protein structure prediction server. Bioinformatics 16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]