Abstract
Influenza A virus PA-X comprises an N-terminal PA endonuclease domain and a C-terminal PA-X-specific domain. PA-X reduces host and viral mRNA accumulation via its endonuclease function. Here, we found that the N-terminal 15 amino acids, particularly six basic amino acids, in the C-terminal PA-X-specific region are important for PA-X shutoff activity. These six basic amino acids enabled a PA deletion mutant to suppress protein expression at a level comparable to that of wild-type PA-X.
TEXT
PA-X was found to be expressed from the PA segment of influenza A viruses as an alternative product (1). It consists of an N-terminal endonuclease domain of PA (191 amino acids) and a C-terminal PA-X-specific domain (61 amino acids), which is encoded by an overlapping open reading frame generated by a ribosomal frameshift (1, 2). PA-X decreases host and viral mRNA accumulation via its shutoff activity, which is dependent on its endonuclease activity (1, 3). Although PA-X is not essential for viral replication in vitro, it modulates host responses in vivo (1, 4–7).
PA has lower shutoff activity than PA-X, although it has the N-terminal endonuclease domain (1, 3, 8–12), suggesting that the C-terminal PA-X-specific region is also important for the shutoff activity. To examine this possibility, we constructed two C-terminal deletion mutant forms of PA [A/WSN/33(H1N1)] (Fig. 1A). PA, PA-X, PA_191, or PA_252 was expressed in 293 cells together with Renilla luciferase by means of plasmid transfection with Trans IT-293 (Mirus). Renilla luciferase activities were measured with the Renilla-Glo luciferase assay system (Promega) at 24 h posttransfection (Fig. 1B). Renilla luciferase activities were suppressed by PA (0.6-log-fold), PA-X (2.0-log-fold), PA_191 (1.3-log-fold), and PA_252 (0.7-log-fold). To confirm these results, we analyzed the levels of expression of Renilla luciferase and the PA-X and PA variants by Western blotting with an anti-Renilla luciferase polyclonal antibody (Promega) (Fig. 1C) and an anti-PA monoclonal antibody, clone 55/2 (13) (Fig. 1D). Renilla luciferase was detected in the mock lane but not in the PA-X lane. In the PA, PA_191, and PA_252 lanes, the Renilla luciferase expression levels were lower than in the mock lane (43.6, 53.7, and 52.7%, respectively). PA-X was barely detected because of self-suppression, whereas PA, PA_191, and PA_252 were expressed to similar extents. These results indicate that the shutoff activity of PA_191 and PA_252 is lower than that of wild-type PA-X, suggesting that the C-terminal PA-X-specific region is important for PA-X shutoff activity.
FIG 1.
Comparisons of the shutoff activities of wild-type PA, wild-type PA-X, and C-terminal deletion mutant forms of PA. (A) Schematic representation of PA, PA-X, and C-terminal deletion mutant forms of PA. PA-X is composed of the N-terminal 191 amino acids of PA and a C-terminal PA-X-specific region of 61 amino acids. PA_191 and PA_252 contain the N-terminal 191 and 252 amino acids of PA, respectively. (B to D) Renilla luciferase activity (B), expression of Renilla luciferase (C), and expression of each viral protein (D) in cells transfected with pGL4.74[hRluc/TK] together with an empty plasmid or a plasmid encoding wild-type PA, wild-type PA-X, or deletion mutant forms of PA were evaluated by using a luciferase assay and Western blotting with an anti-Renilla luciferase and an anti-PA monoclonal antibody. The luminescence data in panel B are mean values ± standard deviations (n = 3). **, P < 0.01 (one-way analysis of variance, followed by Bonferroni correction). (C) The intensities of the Renilla luciferase and β-actin bands were quantified, and their ratios were calculated. The value of the mock lane was set to 100%. In panels C and D, β-actin served as a loading control. ORF, open reading frame; RLU, relative light units.
To pinpoint the amino acid residues in the C-terminal PA-X-specific region that are critical for PA-X activity, we prepared a series of C-terminal deletion mutant forms of PA-X (Fig. 2A) and examined their shutoff activities in the luciferase assay (Fig. 2B). Compared with wild-type PA-X (which exhibited 1.8-log-fold less than the mock sample), PA-X_X10, PA-X_X5, and PA_191 showed significantly lower shutoff activity (1.6-, 1.1-, and 1.1-log-fold less than the mock sample, respectively), whereas the PA-X_X15 shutoff activity was similar to that of wild-type PA-X. These results indicate that the N-terminal 15 amino acids in the C-terminal PA-X-specific region are important for PA-X shutoff activity.
FIG 2.

Importance of the N-terminal 15 amino acids in the PA-X-specific region for the shutoff activity of PA-X. (A) Schematic diagram of the C-terminal deletion mutant forms of PA-X. A series of C-terminal deletion mutant forms of PA-X was constructed to identify the region(s) important for the shutoff activity of PA-X. PA-X_X50, PA-X_X40, PA-X_X30, PA-X_X20, PA-X_X15, PA-X_X10, PA-X_X5, and PA_191 possessed 50, 40, 30, 20, 15, 10, 5, and no PA-X-specific amino acids, respectively. (B) Suppression of Renilla luciferase activity by each C-terminal deletion mutant form of PA-X in 293 cells. Renilla luciferase activity in cells cotransfected with wild-type PA-X, a series of C-terminal deletion mutant proteins, or PA_191 was measured with a luciferase assay. The luminescence in the each C-terminal deletion mutant sample was compared with that in the wild-type PA-X sample. Luminescence data are mean values ± standard deviations (n = 3). **, P < 0.01 (one-way analysis of variance, followed by Dunnett's test). RLU, relative light units.
Next, we compared the N-terminal portion of the PA-X-specific 15 amino acids with that of PA (Fig. 3A). We found a characteristic difference: six basic amino acids (lysine [K] and arginine [R]) in PA-X were acidic amino acids (glutamic acid [E]) in PA. To evaluate the significance of these six basic amino acids for PA-X shutoff activity, we constructed mutant PA-X proteins in which we replaced these six basic amino acids (K and R) with E (PA-X_6E), aspartic acid (D) (PA-X_6D), alanine (A) (PA-X_6A), R (PA-X_6R), K (PA-X_6K), or R and K (PA-X_KR) (Fig. 3B) and examined their shutoff activities (Fig. 3C). PA-X_6E, PA-X_6D, and PA-X_6A showed greater Renilla luciferase activities than wild-type PA-X, whereas the Renilla luciferase activities of PA-X_6R, PA-X_6K, and PA-X_KR were similar to that of wild-type PA-X. The trend of the levels of expression of wild-type and mutant PA-X (Fig. 3D) and the corresponding Renilla luciferase level (Fig. 3E) was similar, with the exception of PA-X_6A; although PA-X_6A suppressed Renilla luciferase and its own expression when analyzed by Western blotting (Fig. 3D and E), in the luciferase assay, the suppression of Renilla luciferase activity by PA-X_6A (Fig. 3C) was limited for an unknown reason. The six amino acid mutations with D or E affected the mobility of the mutant PA-X proteins (Fig. 3D). To identify the amino acid essential for PA-X shutoff activity, we introduced a single point mutation (K or R to E) at each of the six basic amino acids. The mutant proteins produced had similar shutoff activities, all of which were slightly lower than that of wild-type PA-X (data not shown). These data imply that the basic amino acids at these six positions contribute equally to the shutoff activity of PA-X.
FIG 3.
Identification of amino acids in the PA-X-specific region that are important for the shutoff activity of PA-X. (A) Comparison of the N-terminal PA-X-specific 15 amino acids of PA-X with the identical region of PA. All six basic amino acids (K or R) in PA-X were the acidic amino acid E in PA. The six amino acids are underlined. (B) Substitution of the six basic amino acids in the PA-X-specific region. The six basic amino acids of PA-X were replaced with acidic amino acids (PA-X_6E or PA-X_6D), a neutral amino acid (PA-X_6A), or a basic amino acid (PA-X_6R, PA-X_6K, or PA-X_KR). (C to E) Shutoff activities of mutant PA-X proteins. In 293 cells, Renilla luciferase was coexpressed with wild-type PA-X or each mutant PA-X protein. Renilla luciferase activity (C) and Renilla luciferase (E) and wild-type and mutant PA-X (D) expression levels were evaluated by means of a luciferase assay or Western blotting. (F) Comparison of the N-terminal PA-X-specific 15 amino acids of PA-X with the identical region of PA_252. Six acidic amino acids in PA_252 (underlined) were replaced with those of PA-X (PA_252_6KR). (G to I) Shutoff activity of PA_252_6KR. In 293 cells, Renilla luciferase was coexpressed without (mock) or with wild-type PA-X, PA_252, or PA_252_6KR. Renilla luciferase activity (G) and levels of expression of Renilla luciferase (H) and each viral protein (I) were evaluated by means of a luciferase assay or Western blotting. The luminescence data in panels C and G are mean values ± standard deviations (n = 3). **, P < 0.01 (t test, followed by Bonferroni correction). The intensities of the Renilla luciferase and β-actin bands were quantified and their ratios were calculated (E and H). The value of the mock lane was set to 100%. In panels D, E, H, and I, β-actin served as a loading control. RLU, relative light units.
To confirm the importance of these six basic amino acids, we replaced all six E residues in PA_252 with K or R (PA_252_6KR) (Fig. 3F) and assessed the shutoff activity (Fig. 3G). PA_252_6KR showed high shutoff activity at a level comparable to that of wild-type PA-X. When we assessed the levels of expression of Renilla luciferase and mutant and wild-type PA-X and related viral proteins (Fig. 3H and I), the results were similar to those obtained in the luciferase assay (Fig. 3G). These results demonstrate that the six basic amino acids at these positions are essential for the shutoff activity of PA-X.
We then asked whether these six basic amino acids in the PA-X-specific region were conserved among other influenza virus isolates deposited in the Influenza Research Database in December 2014. PA-X proteins of different viruses had substitutions between K and R at these six positions; however, basic amino acids (K or R) at these six positions were highly conserved among human (99.9% of 3,417 isolates), avian (99.3% of 4,047), swine (96.7% of 1,030), and equine (100% of 6) isolates. This information further supports the concept that these basic amino acids are required for the shutoff activity of PA-X.
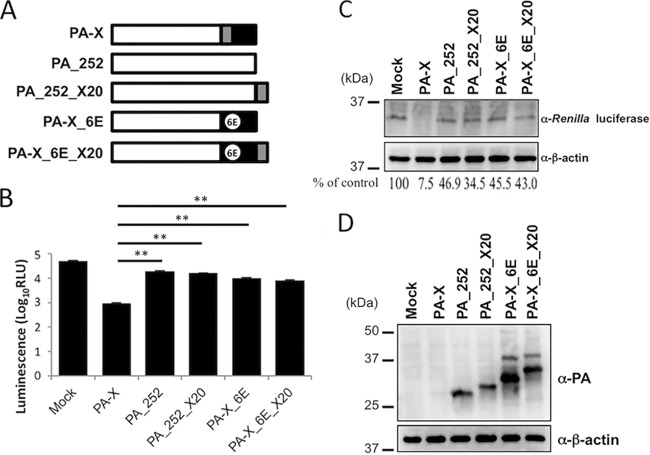
To examine whether the N-terminal 20 amino acids in the PA-X-specific region could increase the shutoff activity of PA_252 or PA-X_6E, we prepared PA_252_X20 and PA-X_6E_X20 (Fig. 4A) and evaluated their shutoff activities (Fig. 4B). PA_252, PA_252_X20, PA-X_6E, and PA-X_6E_X20 showed shutoff activity lower than that of wild-type PA-X. We confirmed that the levels of expression of Renilla luciferase and each viral protein were comparable by Western blotting (Fig. 4C and D). These results indicate that the N-terminal 20 amino acids of the PA-X-specific region contribute to the shutoff activity only when they are located in their original position.
FIG 4.
Effect of the N-terminal 20 amino acids in the PA-X-specific region on the shutoff activity of PA_252 and PA-X_6E. (A) Schematic diagram of PA_252_X20 and PA-X_6E_X20. These proteins contain the N-terminal 20 amino acids of the PA-X-specific region (gray) at the C terminus of PA_252 or PA-X_6E, respectively. (B to D) Shutoff activities of PA_252_X20 and PA-X_6E_X20. In 293 cells, Renilla luciferase was coexpressed without (mock) or with wild-type PA-X, PA_252, PA_252_X20, PA-X_6E, or PA-X_6E_X20. Renilla luciferase activity (B) and the expression of Renilla luciferase (C) and each viral protein (D) were evaluated by means of a luciferase assay or Western blotting. (C) The intensities of the Renilla luciferase and β-actin bands were quantified, and their ratios were calculated. The value of the mock lane was set to 100%. In panels C and D, β-actin served as a loading control. RLU, relative light units.
In summary, we found that the N-terminal 15 amino acids, especially six basic amino acids, in the C-terminal PA-X-specific region are required for the full shutoff activity of PA-X. Most influenza A viruses isolated from humans, equines, and avian species encode a PA-X protein that is identical in length to the PA-X protein of the WSN strain, which possesses 61 PA-X-specific amino acids at its C terminus (14). However, most human H1N1pdm and swine viruses possess a stop codon upstream of the stop codon present in other PA-X proteins, leading to a C-terminally truncated PA-X protein that possesses only 41 PA-X-specific amino acids (14). It has previously been reported that the C-terminally truncated PA-X protein has shutoff activity lower than that of full-length PA-X (4). However, our data demonstrate that PA-X proteins with the C-terminal 20 amino acids truncated would have adequate shutoff activity because the N-terminal 15 amino acids in the PA-X-specific region are sufficient for full shutoff activity.
ACKNOWLEDGMENTS
We thank Susan Watson for editing the manuscript.
This work was supported by the Japan Initiative for Global Research Network on Infectious Diseases from the Ministry of Education, Culture, Sports, Science, and Technology, Japan; by grants-in-aid from the Ministry of Health, Labor, and Welfare, Japan; by ERATO (Japan Science and Technology Agency); by Strategic Basic Research Programs of the Japan Science and Technology Agency; and by an NIH Functional Genomics award (Characterization of Novel Genes Encoded by RNA and DNA Viruses; U19 AI 107810).
REFERENCES
- 1.Jagger BW, Wise HM, Kash JC, Walters KA, Wills NM, Xiao YL, Dunfee RL, Schwartzman LM, Ozinsky A, Bell GL, Dalton RM, Lo A, Efstathiou S, Atkins JF, Firth AE, Taubenberger JK, Digard P. 2012. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science 337:199–204. doi: 10.1126/science.1222213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Firth AE, Jagger BW, Wise HM, Nelson CC, Parsawar K, Wills NM, Napthine S, Taubenberger JK, Digard P, Atkins JF. 2012. Ribosomal frameshifting used in influenza A virus expression occurs within the sequence UCC_UUU_CGU and is in the +1 direction. Open Biol 2:120109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Desmet EA, Bussey KA, Stone R, Takimoto T. 2013. Identification of the N-terminal domain of the influenza virus PA responsible for the suppression of host protein synthesis. J Virol 87:3108–3118. doi: 10.1128/JVI.02826-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao H, Sun H, Hu J, Qi L, Wang J, Xiong X, Wang Y, He Q, Lin Y, Kong W, Seng LG, Pu J, Chang KC, Liu X, Liu J, Sun Y. 15 April 2015. The 20 amino acids at the C-terminus of PA-X are associated with increased influenza A virus replication and pathogenicity. J Gen Virol doi: 10.1099/vir.0.000143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao H, Sun Y, Hu J, Qi L, Wang J, Xiong X, Wang Y, He Q, Lin Y, Kong W, Seng LG, Sun H, Pu J, Chang KC, Liu X, Liu J. 2015. The contribution of PA-X to the virulence of pandemic 2009 H1N1 and highly pathogenic H5N1 avian influenza viruses. Sci Rep 5:8262. doi: 10.1038/srep08262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hayashi T, MacDonald LA, Takimoto T. 2015. Influenza A virus protein PA-X contributes to viral growth and suppression of the host antiviral and immune responses. J Virol 89:6442–6452. doi: 10.1128/JVI.00319-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu J, Mo Y, Wang X, Gu M, Hu Z, Zhong L, Wu Q, Hao X, Hu S, Liu W, Liu H, Liu X, Liu X. 2015. PA-X decreases the pathogenicity of highly pathogenic H5N1 influenza A virus in avian species by inhibiting virus replication and host response. J Virol 89:4126–4142. doi: 10.1128/JVI.02132-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khaperskyy DA, Emara MM, Johnston BP, Anderson P, Hatchette TF, McCormick C. 2014. Influenza A virus host shutoff disables antiviral stress-induced translation arrest. PLoS Pathog 10:e1004217. doi: 10.1371/journal.ppat.1004217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuan PW, Bartlam M, Lou ZY, Chen SD, Zhou J, He XJ, Lv ZY, Ge RW, Li XM, Deng T, Fodor E, Rao ZH, Liu YF. 2009. Crystal structure of an avian influenza polymerase PA(N) reveals an endonuclease active site. Nature 458:909-U912. doi: 10.1038/nature07720. [DOI] [PubMed] [Google Scholar]
- 10.Dias A, Bouvier D, Crepin T, McCarthy AA, Hart DJ, Baudin F, Cusack S, Ruigrok RWH. 2009. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature 458:914–918. doi: 10.1038/nature07745. [DOI] [PubMed] [Google Scholar]
- 11.Sanz-Ezquerro JJ, Zurcher T, delaLuna S, Ortin J, Nieto A. 1996. The amino-terminal one-third of the influenza virus PA protein is responsible for the induction of proteolysis. J Virol 70:1905–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanz-Ezquerro JJ, Delaluna S, Ortin J, Nieto A. 1995. Individual expression of influenza virus PA protein induces degradation of coexpressed proteins. J Virol 69:2420–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hatta M, Asano Y, Masunaga K, Ito T, Okazaki K, Toyoda T, Kawaoka Y, Ishihama A, Kida H. 2000. Epitope mapping of the influenza A virus RNA polymerase PA using monoclonal antibodies. Arch Virol 145:895–903. doi: 10.1007/s007050050682. [DOI] [PubMed] [Google Scholar]
- 14.Shi M, Jagger BW, Wise HM, Digard P, Holmes EC, Taubenberger JK. 2012. Evolutionary conservation of the PA-X open reading frame in segment 3 of influenza A virus. J Virol 86:12411–12413. doi: 10.1128/JVI.01677-12. [DOI] [PMC free article] [PubMed] [Google Scholar]