ABSTRACT
Certain members of the Arenaviridae family are category A agents capable of causing severe hemorrhagic fevers in humans. Specific antiviral treatments do not exist, and the only commonly used drug, ribavirin, has limited efficacy and can cause severe side effects. The discovery and development of new antivirals are inhibited by the biohazardous nature of the viruses, making them a relatively poorly understood group of human pathogens. We therefore adapted a reverse-genetics minigenome (MG) rescue system based on Junin virus, the causative agent of Argentine hemorrhagic fever, for high-throughput screening (HTS). The MG rescue system recapitulates all stages of the virus life cycle and enables screening of small-molecule libraries under biosafety containment level 2 (BSL2) conditions. The HTS resulted in the identification of four candidate compounds with potent activity against a broad panel of arenaviruses, three of which were completely novel. The target for all 4 compounds was the stage of viral entry, which positions the compounds as potentially important leads for future development.
IMPORTANCE The arenavirus family includes several members that are highly pathogenic, causing acute viral hemorrhagic fevers with high mortality rates. No specific effective treatments exist, and although a vaccine is available for Junin virus, the causative agent of Argentine hemorrhagic fever, it is licensed for use only in areas where Argentine hemorrhagic fever is endemic. For these reasons, it is important to identify specific compounds that could be developed as antivirals against these deadly viruses.
INTRODUCTION
The Arenaviridae are enveloped, negative-strand RNA viruses (1). The mammalian arenaviruses are separated serologically and geographically into Old World (OW) and New World (NW) complexes, with the NW complex being further subdivided into clades A, B, A/B (recombinant), and C. The OW viruses Lassa virus (LASV) and Lujo virus and several members of the NW clade B group are capable of causing severe hemorrhagic fevers in humans, with fatality rates ranging from 15 to 80% (1–6). Accordingly, these viruses are considered category A agents, posing the highest threat to public health and biosecurity.
Junin virus (JUNV) is a NW clade B virus that is endemic in Argentina, where it causes Argentine hemorrhagic fever (AHF). An attenuated vaccine strain of JUNV, called Candid#1 (Cd1), has proven to be 95% effective but is licensed for use only in Argentina (7). There are currently no FDA-approved treatments for AHF. Although the nonspecific antiviral drug ribavirin has been used off-label to treat these infections, it has demonstrated limited efficacy and severe side effects, such as anemia and teratogenesis (8, 9). New, more specific drugs are therefore needed to combat JUNV and other pathogenic members of this family.
Studies of the most hazardous arenaviruses require laboratory facilities at biosafety containment level 4 (BSL4), which imposes many limitations for basic and translational studies (10). To address this issue, surrogate genetic systems have been created, which recapitulate certain aspects of the viral life cycle but can be used under more standard BSL2 laboratory conditions. These systems include glycoprotein (GP)-pseudotyped retroviral vectors (11, 12) and minigenome (MG) replicons (13, 14). The arenaviruses have two genomic strands, S and L, and use an ambisense coding strategy to express four viral proteins: GP precursor (GPC), nucleoprotein (NP), RNA-dependent RNA polymerase (L), and matrix protein (Z) (15). GPC is further processed to its subunit proteins GP1 and GP2 and a stable signal peptide (SSP), which form the mature GP spike on the surface of the virion (1). GP-pseudotyped retroviral vectors express the mature GP complex on the surface of the vector particles and thereby mimic the arenavirus entry process. In contrast, MG replicon reverse-genetics systems use exogenous elements such as T7 RNA polymerase to generate an RNA genome that mimics the natural viral genome. When the viral L and NP proteins are provided in trans, the MG is transcribed and replicated, allowing the expression of any reporter genes carried by the MG. Coexpression of all 4 viral proteins with the MG RNA species will also produce virus-like particles that are capable of budding from the cell and infecting a new cell.
MG systems have provided powerful tools to investigate the biology and pathogenesis of many hazardous viruses (14, 16–18). They are also highly amenable to drug discovery applications using high-throughput screening (HTS) (19, 20). In the present study, we used an MG system based on elements of JUNV and Cd1 to interrogate all stages of the arenavirus life cycle (13). This system was used to screen libraries of synthetic and repurposed small molecules, ultimately resulting in the discovery of four compounds that were highly effective against both JUNV and other members of the arenavirus family. It was determined that all four compounds targeted the entry step of the arenavirus life cycle.
MATERIALS AND METHODS
Cell lines and viruses.
HEK293T/17 (293T), BHK-21, NIH 3T3, K562, and Vero E6 cells were obtained from the American Type Culture Collection (ATCC) (Manassas, VA); 293A cells were obtained from Qbiogene/MP Biomedicals (Irvine, CA). 293T, BHK-21, NIH 3T3, Vero E6, and 293A cells were maintained in Dulbecco's modified Eagle's medium (DMEM) (Cellgro, Manassas, VA) supplemented with 10% fetal bovine serum (FBS) (Denville, Metuchen, NJ), and K562 cells were maintained in RPMI 1640 (Cellgro) supplemented with 10% FBS and 1% penicillin-streptomycin (JR Scientific, Woodland, CA).
Virus stocks of lymphocytic choriomeningitis virus Armstrong clone 4 (LCMV-4) (21) and Candid#1 were kindly provided by M. Buchmeier (University of California, Irvine) and amplified on BHK-21 (22) and Vero E6 cells, respectively.
Minigenome system.
The previously described JUNV/Cd1 MG system (13) contains a reporter genome derived from the S RNA strand of Cd1, where the GP and NP genes have been replaced with enhanced green fluorescent protein (eGFP) and Gaussia luciferase (GLuc), respectively. T7 polymerase promoter and terminator elements and a hepatitis delta virus (HDV) ribozyme flank the MG cassette, allowing the production of an analog of the native Cd1 S strand genome. To generate arenavirus particles, the MG plasmid was cotransfected into 293T cells by calcium phosphate transfection, together with expression plasmids for the Cd1 proteins NP, L, and Z; JUNV GPC; and T7 RNA polymerase, as described previously (13), at a ratio of 1.25:1:1.5:0.1:1.04:1.25, respectively. For 96-well-scale experiments, 293T cells were first transfected in 10-cm dishes by using 20 μg total plasmid DNA; the growth medium was replaced after 16 h; and, following a further 8-h incubation, the cells were then harvested and plated into 96-well plates at a density of 2 × 104 cells per well. Antiarenavirus inhibitors, including ribavirin (Sigma-Aldrich, St. Louis, MO), 2-hydroxymyristic acid (OHM) (Santa Cruz Biotechnology, Dallas, TX), or an antibody against human transferrin receptor 1 (hTfR1) (BD Biosciences, San Jose, CA), or a dimethyl sulfoxide (DMSO) control was added at the plating stage. GLuc reporter gene expression was evaluated 96 h after transfection by measuring luciferase activity in culture supernatants using the Biolux Gaussia luciferase assay kit (New England BioLabs, Ipswich, MA), according to the manufacturer's protocol, with samples being read on a Mithras LB 940 multimode microplate reader (Berthold Technologies, Bad Wildbad, Germany). The Z′ factor was calculated by using the following formula: Z′ = 1 − [(3σc+ + 3σc−)/|μc+ − μc−|], where σ is the standard deviation, μ is the mean, c+ is the positive control, and c− is the negative control (23).
Compound libraries and HTS.
The chemical libraries to be screened were curated by the Molecular Screening Shared Resource (MSSR) of UCLA (http://www.mssr.ucla.edu/lib.html), with components selected to be lead-like, and filtered against predicted problematic properties. Among the 87,321 compounds screened were novel sets as well as FDA-approved and clinically tested compounds. All stages of the HTS were performed at the MSSR.
The MG system was adapted for a 384-well format, suitable for HTS. A total of 30 μg of DNA was prepared for transfection by using the calcium phosphate method and added to 12.6 × 106 293T cells in suspension, which were then plated into 14.5-cm dishes. Sixteen hours later, the cells were washed with phosphate-buffered saline (PBS), and the medium was replaced. After a further 8 h, cells were harvested, and 3,500 cells in 30 μl of medium were added to previously prepared compound-containing white 384-well plates (Thermo Scientific, Glen Burnie, MD). The plates contained 20 μl of growth medium and 0.5 μl of library compounds per well, with transfer of the compounds from stock compound plates being performed by using a Biomek FX pin tool in a Sagian Core System I (Beckman Coulter, Brea, CA). Plates were then transferred to a Cytomat 6001 incubator (Thermo Scientific) by using an Orca robotic arm (Beckman Coulter). The final concentration of inhibitors was 10 μM in 1% DMSO. The positive control was ribavirin at a concentration of 100 μM, and the negative control was 1% DMSO.
The plates were incubated at 37°C in 5% CO2 for 72 h and then assayed for luminescence by the addition of 10 μl per well of a prepared Biolux Gaussia Luciferase Flex assay substrate (New England BioLabs), followed by incubation for 3 min 30 s. Luminescence from each well was measured with a 0.1-s integration time on a Victor 3V plate reader (PerkinElmer, Waltham, MA). Each compound was screened in duplicate for the primary screen and in triplicate for subsequent assays.
Secondary screening was performed by making customized stock compound plates based on the hits from the primary screen. These compounds were screened in triplicate by using the same methodology as the one used for the primary screen. As a counterscreen, an off-target assay was used to recapitulate the nonviral elements of the MG assay by directing the expression of GLuc mRNA by T7 polymerase. The T7-GLuc plasmid contained an internal ribosomal entry site (IRES) sequence and a GLuc cassette downstream of a T7 promoter in plasmid pBluescript SK− (Agilent Technologies, Santa Clara, CA). T7-GLuc was transfected into cells (30 μg) by using the calcium phosphate method, with subsequent processing steps as described above for the primary screen. The cutoff for the secondary assay was ≥75% inhibition in the MG assay, with no inhibition in the off-target assay, for all three replicates.
Compounds passing the secondary screen were further interrogated by determining the 50% inhibitory concentration (IC50) and 50% cytotoxic concentration (CC50) doses for each compound by making stock plates of serially diluted (1:2) inhibitors using Precision 2000 equipment (BioTek, Winooski, VT). The doses ranged from 100 μM to 0.19 nM. By using the same procedure as the one used for the primary screen, compound doses were used to test inhibition and cell viability in the MG system with compounds, each in triplicate. Cell viability was determined by using the Cell TiterGlo assay (Promega, Madison, WI), according to the manufacturer's protocol, with luminescence being detected as described above.
All data were analyzed in Excel (Microsoft, Redmond, WA), by determining the percent inhibition of each compound, compared to DMSO treatment, as well as by evaluating the z score (number of standard deviations from the mean). Cutoff values for the z score were <−3. IC50 and CC50 values were determined by nonlinear regression curve-fitting analysis using a four-parameter variable slope in Prism 5 (GraphPad, La Jolla, CA). Values for the selectivity index were determined by dividing the CC50 by the IC50.
Compound handling and resuspension.
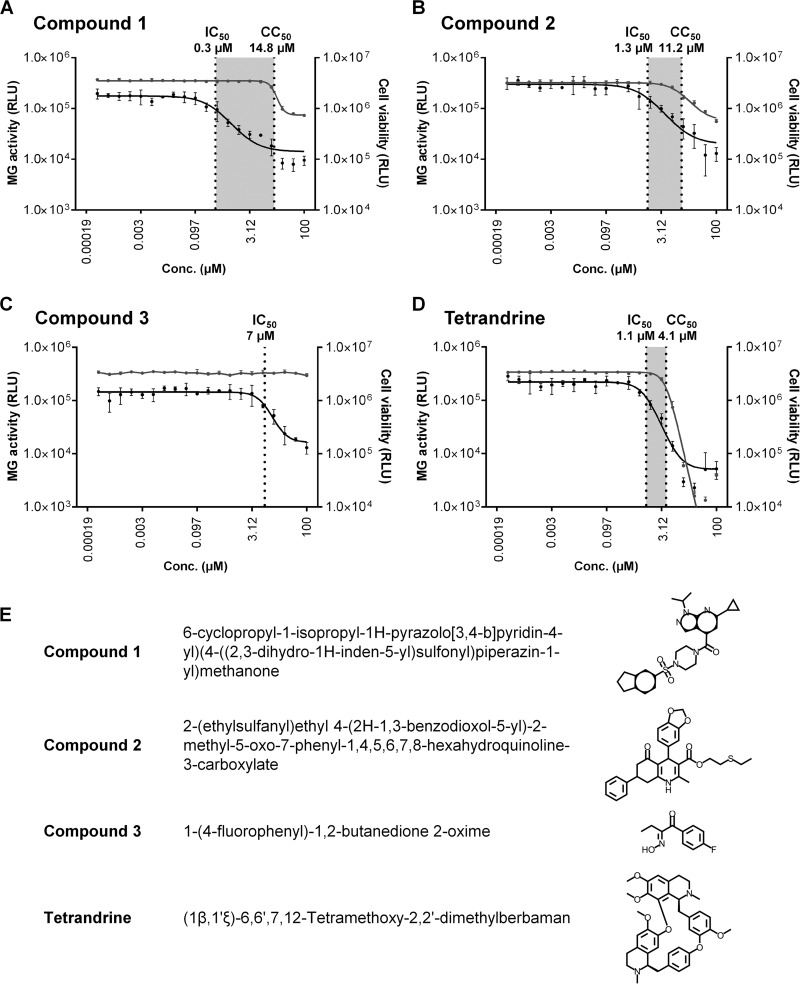
Ribavirin, OHM, and trans-Ned 19 (Ned19) (Santa Cruz Biotechnology) were resuspended in DMSO (Sigma-Aldrich) to make a 100 mM stock and stored at −20°C or 4°C. Compounds purchased for postscreening tests were compound 1 {(6-cyclopropyl-1-isopropyl-1H-pyrazolo[3,4-b]pyridin-4-yl)(4-((2,3-dihydro-1H-inden-5-yl)sulfonyl)piperazin-1-yl)methanone) (Z52089667)}, purchased from Enamine (Monmouth Junction, NJ); compound 2 [2-(ethylsulfanyl)ethyl 4-(2H-1,3-benzodioxol-5-yl)-2-methyl-5-oxo-7-phenyl-1,4,5,6,7,8-hexahydroquinoline-3-carboxylate (STK363677)], purchased from Vitas-M Laboratory (Apeldoorn, Netherlands); compound 3 [1-(4-fluorophenyl)-1,2-butanedione 2-oxime) (5348017)], purchased from ChemBridge Corporation (San Diego, CA); and tetrandrine, purchased from Santa Cruz Biotechnology. These four compounds were resuspended in DMSO to make a 10 mM stock and stored at 4°C.
Compound IUPAC names were determined by accessing the Chemicalize website (ChemAxon, Budapest, Hungary [http://www.chemicalize.org/]). Structure images were produced in ChemBioDraw Ultra (PerkinElmer). Structural similarity between compounds was evaluated by using the Tanimoto coefficient, which compares the 512-bit chemical fingerprints of two structures. This comparison was done by using the Collaborative Drug Discovery website (Collaborative Drug Discovery, Burlingame, CA).
Virus infectivity focus-forming assay.
Vero E6 cells were seeded into poly-l- or poly-d-lysine coated, black, clear-bottom, 96-well plates (BD Biosciences) at 7 × 103 cells per well, 24 h before virus infection. Cells were incubated for 1 h with compound-containing medium, with 0.2% DMSO (final volume). Compound-containing medium was removed, and 50 to 100 μl of the LCMV (∼1.1 × 103 to 2.2 × 103 focus-forming units [FFU] per well) or 100 μl of the Candid#1 (∼1 × 103 FFU per well) virus stock was added to cells for 1.5 h. Virus was removed, compound-containing medium was added to cells, and cells were further incubated for 48 h until they were assayed. In the case of tetrandrine, the compound was present throughout the experiment. Infected cells were fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100 (Sigma-Aldrich), and then blocked in 1% bovine serum albumin (BSA). Mouse immune ascites fluid against LCMV (ATCC VR-1271AF) or JUNV (ATCC VR-1270AF) was used at a 1:500 dilution in 1% BSA and added to cells, followed by a donkey anti-mouse secondary antibody conjugated with Alexa Fluor 488 (Life Technologies, Carlsbad, CA) at a dilution of 1:400. Cells were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (Life Technologies). Plates were read by using a BD Pathway high-content imager (Becton Dickinson, Franklin Lakes, NJ) to detect Alexa Fluor 488 and DAPI stains. Nuclei and virus foci were counted using the Fiji Software 3D objects counter (http://fiji.sc/Fiji) (24). Titers in focus-forming units per ml were calculated by extrapolating the number of foci detected during imaging to the whole well surface area and multiplying by the virus stock dilution factor. Statistical analysis was performed by using one-way analysis of variance (ANOVA) followed by Dunnett's multiple-comparison test in Prism 5 (GraphPad).
Pseudotyped retroviral and lentiviral vector production and transduction.
Pseudotyped retroviral and lentiviral vectors displaying viral fusion proteins were produced by calcium phosphate transfection of 293T cells, as described previously (11, 12). Retroviral vectors contained the vector genome pMND-eGFP (25), which transfers the eGFP gene to target cells. For lentiviral vectors, the genome was FUGW (a gift from D. Baltimore, California Institute of Technology), which also transfers the eGFP gene (26). Vector stocks were used as unconcentrated culture supernatants.
Expression plasmids for the GPCs of the following arenaviruses were previously described: JUNV (XJ; Parodi strain), Cd1, Machupo virus (MACV) (Carvallo strain), Tacaribe virus (TCRV) (TRVL 11598 strain), LCMV (Armstrong 53b and 13 variants), LASV (Josiah strain), Guanarito virus (GTOV) (INH-95551 strain), Amapari virus (AMAV) (BeAn 70563 strain), and Whitewater Arroyo virus (WWAV) (strain AV 9310135) (11–13, 27–29). JUNV GPC derivatives were the F427I mutant and chimeric proteins C1X2 and X1C2 (13). Expression plasmids for the GPCs from Big Brushy Tank virus (BBTV), Skinner Tank virus (SKTV), Pichinde virus (PICV) (PICV-18, a guinea pig-adapted strain), and Oliveros virus (OLVV) were used (30–32). Latino virus (LATV) GPC (GenBank accession no. YP_001936021) was synthesized by GenScript (Piscataway, NJ), and Flexal virus (FLEV) (accession no. AAN_09937.1) GPC was synthesized by Bio Basic Inc. (Amherst, NY) and then cloned into pCAGGS. Mutant JUNV GPCs (K33H, N37A, T418N, L420T, A435I, and F438I) were made by splice overlap PCR using wild-type JUNV GPC as a template. Previously described expression plasmids for other viral fusion proteins were pHIT123 and pHIT456 for ecotropic and amphotropic murine leukemia virus (MLV) Env proteins, respectively (33); pHCMV-G for vesicular stomatitis virus G (VSV-G) (34); pEboGP for full-length Ebola virus (Zaire strain) GP-8A (35); pCMVHA for influenza virus hemagglutinin (HA) (subtype H7) (36); and Hendra and Nipah virus F and G, which were kindly provided by B. Lee (Icahn School of Medicine at Mount Sinai) (37, 38). HA-pseudotyped vectors were prepared as stated above, with the addition of 0.025 U/ml Vibrio cholerae neuraminidase (Sigma-Aldrich) to cell supernatants at the time of medium change. All pseudotyped vectors were made as lentiviral vector stocks, except for WWAV, BBTV, SKTV, LATV, LCMV53b, Ebola virus, amphotropic and ecotropic MLVs, influenza virus, and Hendra virus, which were retroviral vectors. Vector titers were determined by incubation of serially diluted vector stocks with target cells for 48 to 72 h. For compound studies, 0.2% DMSO or specific compounds were incubated on target cells 1 to 5 h prior to transduction, and the compound and vectors were then placed onto cells and left for 48 to 72 h until analysis. Cellular GFP expression was determined by using a Canto II (BD Biosciences) or Guava easyCyte 6-2L (EMD Millipore, Billerica, MA) flow cytometer. The percentage of GFP-positive cells was determined by using FlowJo analysis software (TreeStar Software, Ashland, OR). The titer of vector stocks was expressed as transducing units (TU) per milliliter. Statistical analysis was performed by using one-way ANOVA followed by Dunnett's multiple-comparison test in Prism 5 (GraphPad).
Cell-cell fusion assay.
293T cells were transfected with GPC expression plasmids by using the calcium phosphate method. At 24 h posttransfection, cells were seeded into 6-well plates and allowed to recover for 16 h. Cells were then treated with compound- or DMSO-containing medium for 1 h and pulsed for 20 min with 25 mM 2-(N-morpholino)ethanesulfonic acid (MES) buffer (Sigma-Aldrich) containing compounds or 0.2% DMSO, at pH 4.00 or 7.00. After the pH pulse, cells were washed, and compound- or DMSO-containing medium was added for 7 h. Cells were fixed in 1% methylene blue (Sigma-Aldrich) in methanol for 15 min, washed, and air dried. Wells were visually scored for cell-cell fusion, using a scale of no visible fusion (−) to maximal fusion (++++). Images of stained cells were acquired on an IX71 microscope (Olympus, Center Valley, PA), using a 20× objective with a Digital Sight DS-Fi1 camera (Nikon, Melville, NY).
MACV GP1 immunoadhesin binding assay.
MACV GP1 immunoadhesin (IM) supernatants were prepared as described previously (27). A total of 5 × 105 K562 cells were incubated in a 0.5% BSA blocking solution with compounds or 0.2% DMSO for 20 min on ice and then incubated with 30 μl of 100×-concentrated IM with compounds or DMSO for 30 min on ice. The cells were then stained with a donkey anti-rabbit antibody conjugated with Alexa Fluor 488 (Life Technologies) and fixed with 4% paraformaldehyde in PBS. Binding was determined by detecting Alexa Fluor 488 on cells by flow cytometry, as described above.
Compound-virus interaction studies.
JUNV GP-pseudotyped lentiviral vectors were produced and concentrated 100× by ultracentrifugation through a 20% sucrose cushion for 2 h at 25,000 rpm on an SW28 or SW41 rotor (Beckman Coulter). A total of 1 × 107 TU of pseudotyped vectors was incubated with compounds at 2 μM (compounds 1 and 2) or 3 μM (compound 3), 1 h prior to transduction. The vector-compound mix was then diluted with growth medium to final compound concentrations of 0.05 μM (compounds 1 and 2) and 0.07 μM (compound 3) and added to 4 × 105 to 6 × 105 293A cells. Alternatively, 4 × 105 to 6 × 105 293A cells were incubated for 1 h prior to transduction with compounds at concentrations of 0.05 μM (compounds 1 and 2) and 0.07 μM (compound 3), which represent noninhibitory doses, or 2 μM (compounds 1 and 2) and 3 μM (compound 3), which represent inhibitory doses. The cells were maintained in compound-containing medium at these doses while being transduced with 1 × 107 TU of JUNV GP-pseudotyped vectors. Cells were analyzed for eGFP expression by flow cytometry at 48 to 72 h postransduction, as described above.
RESULTS
A high-throughput screen interrogating all stages of the JUNV/Cd1 life cycle identifies four inhibitors.
We previously described a JUNV/Cd1-based reverse-genetics system comprising a MG reporter construct expressing eGFP and Gaussia luciferase (GLuc) (Fig. 1A) (13). Following transcription by T7 RNA polymerase and processing by a ribozyme, the MG so generated can be recognized by viral proteins provided in trans. This results in replication and transcription of the MG by the NP and L proteins and packaging into virus-like particles formed by the action of the Z protein. If GPC is also present, the particles can infect new cells, including those transfected by NP and L, leading to amplification of the MG reporter gene signals. In this way, the MG rescue system recapitulates all stages of the JUNV/Cd1 life cycle, including RNA replication and transcription, viral particle assembly and release, and GP-mediated cell entry (Fig. 1B). Since GLuc is secreted into the culture supernatants, it also provides a quantitative readout of the efficiency of MG rescue that is amenable to HTS.
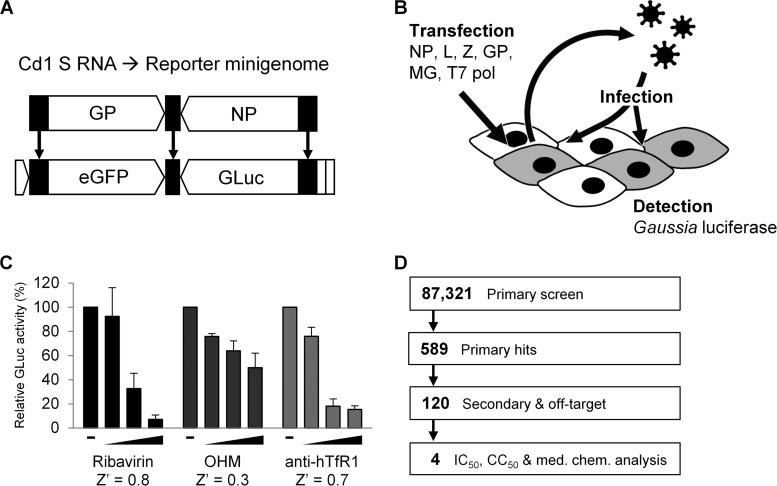
FIG 1.
HTS based on the JUNV/Cd1 MG rescue system. (A) Derivation of the reporter MG from the Cd1 genomic S strand. Black segments represent viral noncoding regions, and flanking open segments represent T7 polymerase promoter/terminator and HDV ribozyme sequences. GP and NP genes were replaced by eGFP and GLuc reporter genes, respectively. (B) Schematic of the MG rescue system. 293T cells are transfected with the indicated plasmids, resulting in virus-like particles being released from the initial producer cells. The virions can infect neighboring cells, including other producer cells, where replication and transcription of the reporter genes results in reporter protein expression. GLuc is secreted and can be detected in the culture supernatant. (C) Response of the MG rescue assay to arenavirus inhibitors, including ribavirin (to a maximum dose of 100 μM), OHM (to a maximum dose of 50 μM), and anti-human transferrin receptor 1 antibody (to a maximum dose of 50 nM). The Z′ factor value of the assay for each inhibitor at the maximum dose used is indicated. (D) Schematic of HTS. Primary hits were subjected to secondary and off-target screens, and 117 of the 120 secondary hits were evaluated in more detail for potency, toxicity, and medicinal chemistry properties, resulting in 4 candidates.
We confirmed the expected properties of the MG system by demonstrating dose-dependent sensitivity to the following arenavirus inhibitors: ribavirin, which blocks viral RNA synthesis (39–41); 2-hydroxymyristic acid (OHM), which inhibits arenavirus budding (42, 43); and an antibody against human transferrin receptor 1, to prevent interaction with the primary JUNV receptor (44) (Fig. 1C). The Z′ factor value (23), a measure of the robustness of the assay, varied between 0.3 and 0.8 for these compounds at a 96-well scale.
For HTS, we scaled the assay to 384-well plates. In pilot MG rescue experiments in a 384-well-plate format using 100 μM ribavirin, the Z′ factor value of the assay was reduced to 0.2, with greater variability well to well and a decreased signal output per well compared to that observed at the 96-well scale. To compensate for the lower score and increase the reliability of the results, HTS was performed in duplicate. The HTS that we performed is summarized in Fig. 1D. A well-balanced library of 87,321 compounds was curated by combining novel drug-like and FDA-approved small-molecule libraries. Compounds inhibiting the MG signal by at least 50% in both replicates, and having a z score (number of standard deviations below the mean for the samples in a plate) at or below −3 in at least one replicate, were considered hits. Using these criteria, 589 compounds, 0.67% of the compounds tested, were classified as hits.
All 589 compounds identified in the primary screen were then subjected to more rigorous secondary screening to confirm activity and rule out compounds that nonspecifically inhibited GLuc expression. This was achieved by counterscreening against a T7-GLuc expression plasmid. A successful compound in these secondary screens was defined as a compound that decreased the GLuc activity at least 75% in all three replicates of the MG assay but that did not inhibit the control T7-GLuc plasmid. These secondary screens resulted in a further reduction of hits to 120 total compounds.
Of the 120 compounds deemed successful from the secondary screen, 117 were available for purchase and further tested to determine both the 50% inhibitory concentration (IC50) and 50% cytotoxic concentration (CC50). The ratio of the IC50 to the CC50 was used to determine a selectivity index (SI) score. Analysis of the SI (>5) and predicted medicinal chemistry properties narrowed the list of compounds to 4 drug-like and selective candidates for further investigation (Fig. 2). These candidates comprised three novel compounds (designated compounds 1 to 3) with SIs of >5 and tetrandrine (Fig. 2E), a bisbenzylisoquinoline alkaloid derived from the root of Stephania tetrandra, a Chinese herb (45), that was recently reported to have activity against Ebola virus (46). Although tetrandrine had an SI of only 3.5, it was selected because it has previously been used in humans, and its mechanism of action is understood (45). Dose-response plots showing the IC50 and CC50 values for each compound are presented in Fig. 2.
FIG 2.
(A to D) IC50 and CC50 plots. Dose-response curves for each compound show MG rescue activity (GLuc activity in supernatants) (plotted on the left y axis with black circles and lines) and cell viability (luminescence in cell lysates) (plotted on the right y axis with gray circles and lines). IC50 and CC50 values are indicated by vertical dotted lines, with the shaded area between these values representing the selectivity index values, which are 49.3, 8.6, and 3.5, for compound 1 (A), compound 2 (B), and tetrandrine (D), respectively. For compound 3 (C), no toxicity occurred, even at the maximum assay dose of 100 μM, so the CC50 and SI were not calculated. Each graph point is the mean of data from three replicates ± standard deviations, joined by a fitted curve. RLU, relative light units. (E) Compounds 1 to 3 and tetrandrine. IUPAC names and structures are shown.
Effect of novel compounds 1 to 3 on Cd1 and LCMV infectivity.
We next evaluated the three novel compounds identified from the MG screen for their ability to inhibit infection by representative arenaviruses. For this, we selected the BSL2 viruses LCMV-4 (21), which is an OW virus, and Cd1, which is an attenuated vaccine strain of JUNV (7). We and others have previously noted amino acid differences between JUNV and Cd1, including some that change the properties of GP and may contribute to its lack of virulence (13, 47).
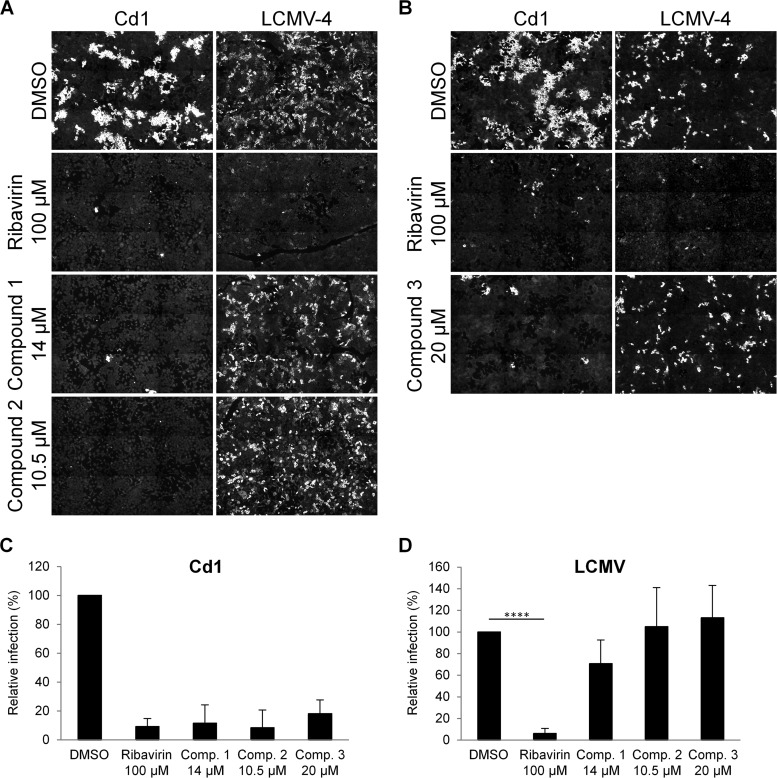
Vero E6 cells were pretreated with each compound, using concentrations selected as being the highest doses tolerated without toxicity in 293T cells (i.e., 14 μM compound 1, 10.5 μM compound 2, and 20 μM compound 3) (Fig. 2), and then infected with Cd1 or LCMV-4. As a control, we also included ribavirin (100 μM). Following incubation, the virus was removed and replaced with fresh compound-containing medium. Using these conditions, we confirmed a lack of toxicity for Vero E6 cells and further found that all three compounds inhibited Cd1 infectivity, decreasing the observed foci of infection to 11.5%, 8.3%, and 18.1% (all P < 0.0001), respectively, compared to the DMSO-only control infection (Fig. 3A to C). In contrast, none of the compounds significantly inhibited LCMV-4 infectivity (Fig. 3A, B, and D), even when the compounds were maintained throughout the experiment (data not shown). These observations indicated that the three compounds were likely not paninhibitors of the family but displayed specificity for the virus used to construct the MG system.
FIG 3.
Effects of compounds 1 to 3 on arenavirus replication. (A and B) Focus-forming assays. Vero E6 monolayers were infected with Cd1 or LCMV-4, with the indicated compounds or the DMSO control being present before and after virus incubation. Cells were analyzed 48 h later by using specific antibodies and immunofluorescence to detect foci of infection. Images are representative of data from 2 to 5 experiments. (C and D) Quantitation of virus infections (FFU per milliliter) of cells treated with DMSO or compounds compared to the DMSO-treated control ± standard deviations from 2 to 5 independent experiments. Mean titers in DMSO-treated control cells were 1 × 104 FFU/ml (Cd1) and 2.3 × 104 FFU/ml (LCMV-4). All data from panel C were statistically significant compared to the DMSO control (P < 0.0001), and in panel D, statistical significance compared to the DMSO control is indicated (****, P < 0.0001).
Effects of compounds 1 to 3 on arenavirus entry pathway.
The OW and NW arenaviruses differ in the entry pathways that they use to enter host cells, recognizing distinct cell surface receptors (29, 30, 44, 48–55) and following different intracellular pathways to reach the late endosomes, where virus-cell fusion occurs (reviewed in reference 56). For example, NW clade B viruses use TfR1 as a primary receptor (30, 44, 48), while both OW and NW clade C viruses use α-dystroglycan (α-DG) (53, 55). The lack of an effect of compounds 1 to 3 on LCMV compared to Cd1 therefore led us to hypothesize that they could be acting on the specific Cd1 entry pathway. To test this, we examined the effects of the compounds on GP-pseudotyped lentiviral and retroviral vectors, which provide a means to assay the entry pathway of viral fusion proteins. We found that all three compounds potently inhibited transduction by JUNV GP-pseudotyped vectors, indicating that this was indeed the stage of the life cycle at which the compounds were acting (Fig. 4A). As controls to rule out any effects on other aspects of vector biology, none of the three compounds inhibited entry by VSV-G-pseudotyped retroviral or lentiviral vectors (data not shown).
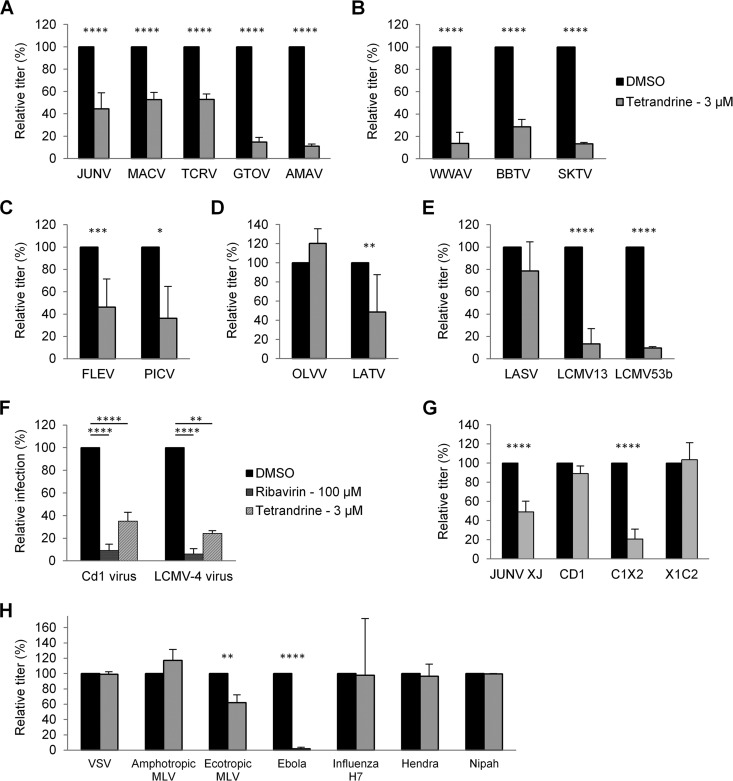
FIG 4.
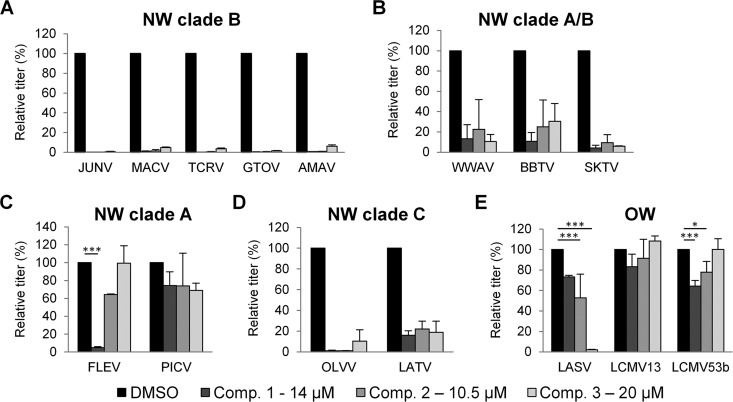
Effects of compounds 1 to 3 on the entry pathway of different arenaviruses. 293A cells were transduced with retroviral or lentiviral vectors pseudotyped with GPs from the indicated viruses. Transductions were performed in the presence of DMSO alone or the indicated concentrations of compound 1, 2, or 3. Relative titers are shown as percentages of the control titer (DMSO treated). (A) NW clade B GP vectors. (B) NW clade A/B GP vectors. (C) NW clade A GP vectors. (D) NW clade C GP vectors. (E) OW GP vectors. Mean titers (in TU per milliliter) on DMSO-treated cells were as follows: 2.0 × 105 for JUNV, 3.4 × 104 for MACV, 9.3 × 104 for TCRV, 4.6 × 104 for GTOV, 4.6 × 104 for AMAV, 8.1 × 103 for WWAV, 8.2 × 103 for BBTV, 8.8 × 103 for SKTV, 6.7 × 103 for FLEV, 2.8 × 105 for PICV, 1.3 × 105 for OLVV, 6.3 × 103 for LATV, 9.0 × 105 for LASV, 2.8 × 105 for LCMV13, and 3.3 × 105 for LCMV53b. All data represent the means ± standard deviations from 3 to 7 independent experiments. All data from panels A, B, and D were statistically significant compared to the DMSO control (P < 0.001), and in panels C and E, statistical significance compared to the DMSO control is indicated (*, P < 0.05; ***, P < 0.001).
Using a broader panel of GP-pseudotyped vectors, we next tested the compounds for activity against GPs from other NW viruses. These viruses included other members of clade B; the closely related clade A/B recombinant viruses, which have a clade B-like GP (57); and the more distantly related viruses from clades A and C (57) (Fig. 4A to D). Within clade B, we found that all three compounds retained broad and potent activity against each of the GPs tested, including those from human-pathogenic strains (MACV and GTOV) as well as nonpathogenic members (TCRV and AMAV) (28). Similarly, GPs from clade A/B recombinant viruses, which also use species-specific versions of TfR1 as the primary receptor (29, 30, 44, 48–52), were inhibited by all three compounds, although the levels of inhibition were not as severe as those observed for clade B GPs (Fig. 4B). This pattern was also true for the two clade C GPs that we tested, despite the fact that clade C viruses use α-DG as a receptor (53, 54) (Fig. 4D). In contrast, when we tested the compounds against clade A GPs, we found a different pattern: FLEV was highly sensitive to compound 1, partially sensitive to compound 2, and completely insensitive to compound 3, while PICV exhibited low but similar degrees of sensitivity to all three compounds (Fig. 4C). The cell surface receptor used by clade A viruses is currently unknown.
Finally, we tested the compounds against OW GPs, including two different strains of LCMV and the significant human pathogen LASV. Like the clade C viruses, OW viruses use α-DG as a receptor (55). However, in contrast to the clade C results, we found that the GPs from LCMV13 and LCMV53b were insensitive to all three compounds, while the LASV GP-pseudotyped vectors were strongly inhibited (97.8%; P < 0.0001) by compound 3 alone (Fig. 4E). The lack of an effect on the LCMV GP vectors mirrored the observations from the viral infections with LCMV-4 (Fig. 3).
In summary, we observed varied responses to the compounds from different arenaviruses, ranging from sensitivity to all three compounds by the clade B, clade A/B, and clade C GPs tested to relative insensitivity to all three compounds for the GPs from certain clade A and OW viruses (PICV and LCMV). Since these patterns do not track with the cell surface receptor used, or the subsequent endosomal trafficking pathway followed (56), they instead suggest that the target for the compounds could be a common conserved feature in the arenavirus GP that is more or less available in different virus strains. Furthermore, the activity of compound 3 against LASV GP indicates that the MG assay, although based on NW clade B components, is capable of identifying broadly acting inhibitors.
Compounds 1 to 3 are specific for the arenaviruses.
We next addressed whether the entry pathways of other enveloped RNA virus families were inhibited by any of the three compounds. We generated a panel of pseudotyped retroviral or lentiviral vectors, carrying fusion proteins from amphotropic and ecotropic MLVs; from the nonsegmented negative-sense RNA viruses VSV, Ebola virus, Hendra virus, and Nipah virus; and from influenza virus (subtype H7). Apart from compound 1, which had a modest effect against Ebola virus GP entry, inhibiting it by 62% (P < 0.0001), we found that the compounds were inactive against the panel of viral fusion proteins (Fig. 5). These results indicate that the inhibitory effects of the three compounds are specific to the arenaviruses.
FIG 5.
Effects of compounds 1 to 3 on the entry pathway of other viruses. 293A or NIH 3T3 cells (for ecotropic MLV GP vectors) were transduced with retroviral or lentiviral vectors pseudotyped with the indicated viral attachment/fusion proteins. Transductions were performed in the presence of DMSO alone or the indicated concentrations of compound 1, 2, or 3. Relative titers are shown as percentages of the control titer (DMSO treated). Mean titers (in TU per milliliter) on DMSO-treated cells were as follows: 3.0 × 106 for VSV, 4.8 × 104 for amphotropic MLV, 8.2 × 104 for ecotropic MLV, 9.0 × 104 for Ebola virus, 2.5 × 103 for influenza virus H7, 5.2 × 104 for Hendra virus, and 4.2 × 105 for Nipah virus. All data represent the means ± standard deviations from 3 to 7 independent experiments. Statistical significance compared to the DMSO control is indicated (****, P < 0.0001).
Compounds 1 to 3 prevent arenavirus GP-mediated cell-cell fusion.
In common with other pH-dependent viral fusion proteins, arenavirus entry involves several steps that can be investigated independently. These steps include binding to a cellular receptor(s), internalization into an intracellular vesicle, a conformational change in GP driven by low pH in a late endosome, and eventual fusion between the viral envelope and a cellular membrane. In order to investigate the involvement of endocytosis as the target of the compounds, we investigated the effects of the compounds in a GP-mediated cell-cell fusion assay. Here, treatment of the cells with a low-pH pulse provided the necessary trigger to promote membrane (cell-cell) fusion so that the assay could evaluate the stages of GP-receptor interactions and downstream conformational changes leading to fusion, without requiring internal trafficking steps.
293T cells were transfected with the GPC genes from candidate viruses that had been proven to be sensitive to the compounds in the GP vector assays (JUNV, MACV, TCRV, OLVV, and LASV) and subjected to a low-pH pulse to promote fusion. We found that compounds 1 and 2 completely inhibited syncytium formation by all of the GPs tested (Fig. 6 and Table 1). In contrast, compound 3 was less potent against the clade B GPs while retaining full apparent activity against clade C OLVV. This seemingly greater activity of compound 3 against OLVV GP may reflect the overall smaller amount of cell-cell fusion induced by OLVV in this system. In agreement with the LASV GP vector results, we found that compounds 1 and 2 had no effect on LASV GP-mediated fusion, while compound 3 was completely inhibitory. Taken together, these results suggest that the compounds are not acting to prevent internalization, trafficking through intracellular vesicular pathways, or low-pH acidification but instead are acting to prevent either the initial GP-receptor interaction or the subsequent conformational changes in GP that lead to membrane fusion.
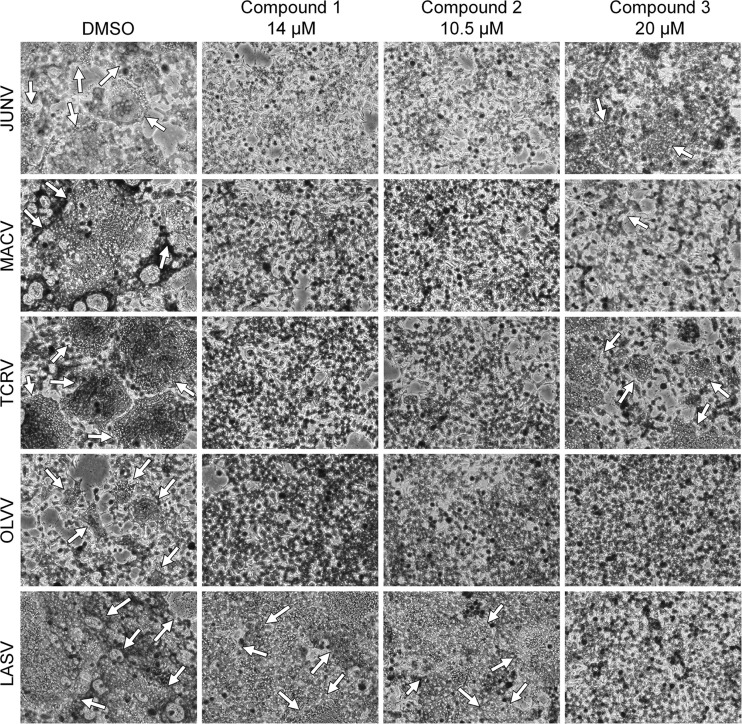
FIG 6.
Effects of compounds 1 to 3 on cell-cell fusion. Shown is the extent of syncytium formation (arrows) triggered by low-pH-pulse treatment of 293T cell monolayers expressing the indicated viral GPs. DMSO or compounds were added 1 h before the pH pulse and maintained throughout the experiment. Images are representative of images from 2 to 3 independent experiments.
TABLE 1.
Visual scoring of cell-cell fusiona
| GP | Visual score |
|||
|---|---|---|---|---|
| DMSO | Compound 1 (14 μM) | Compound 2 (10.5 μM) | Compound 3 (20 μM) | |
| JUNV | +++ | − | − | + |
| MACV | ++++ | − | − | +/− |
| TCRV | +++ | − | − | ++ |
| OLVV | ++ | − | − | − |
| LASV | ++++ | ++++ | ++++ | − |
−, no syncytia detected; +, minimal amount detected; ++++, maximum amount detected.
Compounds 1 to 3 do not block MACV GP1 interaction with human TfR1.
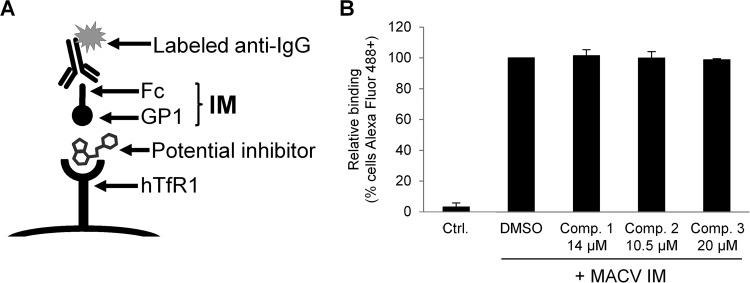
We next addressed whether the compounds could be blocking GP-receptor interactions (Fig. 7A). Using a previously described MACV GP immunoadhesin (IM) reagent (27), we performed cell binding assays in the absence and presence of the compounds and found no effect on receptor binding (Fig. 7B), indicating that for MACV GP at least, none of the compounds interfered with this initial stage of the virus entry process. This agrees with our previous observations that viruses using both TfR1 (clade B) and α-DG (clade C) as cell surface receptors could be inhibited by all three compounds, suggesting that the initial interaction with the receptor was likely not the step being impacted.
FIG 7.
Effects of compounds 1 to 3 on MACV IM binding to cells. (A) Schematic representation of the IM binding assay. (B) A total of 5 × 105 K562 cells were incubated with MACV IM supernatants, or a control supernatant (Ctrl.) from cells transfected with an Fc-only construct, in the presence of DMSO or compounds, as indicated. IM binding was determined by flow cytometry for Alexa Fluor 488 and is shown relative to that of DMSO-treated cells (100%). The mean percentage of Alexa Fluor 488-positive cells on DMSO-treated IM cells was 93.7%. All data represent the means ± standard deviations from 2 to 3 independent experiments.
Compounds 1 to 3 do not irreversibly bind to GP-pseudotyped vectors.
The sum of our data so far suggested that the compounds could be binding to a relatively conserved region of the arenavirus GP and acting to block the low-pH-induced transition that leads to fusion. We therefore asked whether the compounds could bind to GPs in the native, neutral-pH conformation that is present on the pseudotyped vectors or whether they were instead binding to a structure that was exposed only following triggering by low pH. As a precedent, arenavirus entry inhibitors that bind to native neutral-pH forms of GP have previously been described (58, 59).
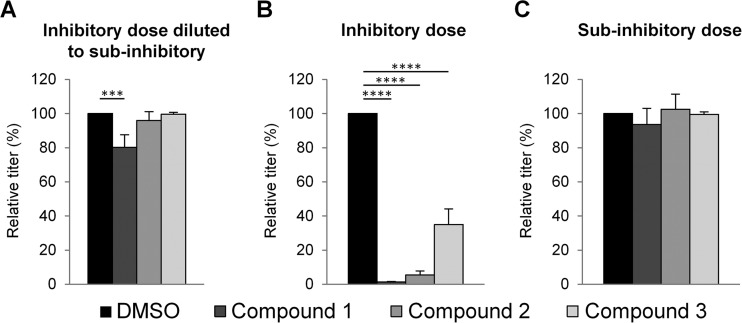
To determine if the compounds were capable of irreversibly binding to GP, we first incubated stocks of JUNV GP-pseudotyped vectors with the three different compounds and then added this mixture to cells plus medium containing no compounds. We used concentrations of the compounds that would be inhibitory if maintained throughout the experiment (Fig. 8B) but would be diluted to noninhibitory levels (Fig. 2) by the 1:41 dilution that occurred when the vector stock-compound mixture was added to the cells (Fig. 8A). As controls, the compounds were maintained at the designated inhibitory (Fig. 8B) or noninhibitory (Fig. 8C) concentrations throughout the experiment. We found that inhibition of transduction occurred mainly when the compounds were present at inhibitory concentrations throughout the experiment, suggesting that none of the compounds were able to bind irreversibly to the native form of GP displayed on the vectors. Although we observed statistically significant differences between the control (DMSO) and dilution (inhibitory to subinhibitory) treatments for compound 1 (P < 0.001) (Fig. 8A), the difference in percent inhibitions when the compound was present throughout the experiment (98.7%) and when it was diluted out (19.8%) was also significantly different (P < 0.0001), indicating dissimilar potencies of this compound with these two different treatments. Finally, we note that this assay cannot distinguish between effects due to reversible binding to GP and an alternative scenario where the targets of the compounds in GP are exposed only following entry into an endosome and low-pH-induced conformational changes.
FIG 8.
Compound-virus interactions. (A) JUNV GP-pseudotyped lentiviral vectors were incubated with DMSO or compounds at an inhibitory concentration of 2 μM (compounds 1 and 2) or 3 μM (compound 3) for 1 h and then added to 293A cells and medium so that dilution resulted in a subinhibitory concentration of 0.05 μM (compounds 1 and 2) or 0.075 μM (compound 3). (B and C) 293A cells were incubated with DMSO or compounds 1 h before the addition of JUNV GP-pseudotyped lentiviral vectors, at the inhibitory concentration (B) or subinhibitory dose (C). All cells were assayed for GFP at 48 to 72 h. Graphs show the resulting inhibition of JUNV GP entry after treatments. Relative titers are shown as percentages of the control titer (DMSO treated). The mean titer on DMSO-treated cells was 1.6 × 107 TU/ml. All data are the means ± standard deviations of results from 2 to 3 independent experiments. Statistical significance compared to the DMSO control is indicated (***, P < 0.001; ****, P < 0.0001).
JUNV GP2 transmembrane and SSP mutations confer resistance to compounds 1 to 3.
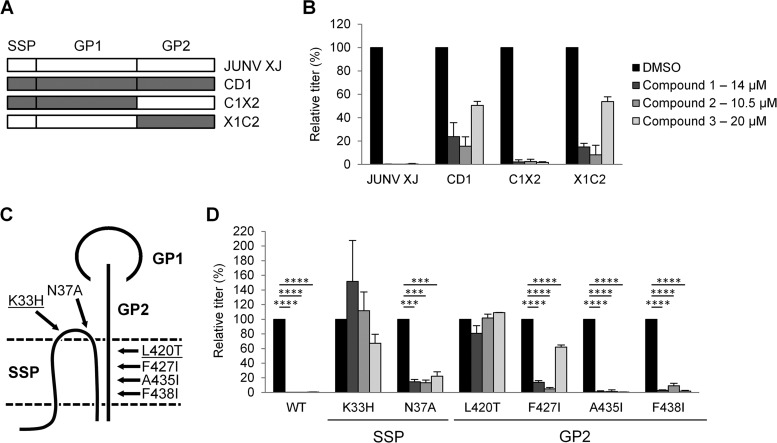
The GPs from JUNV and Cd1 vary by only 6 amino acids (60), and both proteins recognize the human TfR1 receptor (13). However, we noted that all three compounds were less effective against Cd1 GP-pseudotyped vectors than against JUNV GP vectors (Fig. 9B). To map the determinants responsible for these differences, we tested reciprocal GP1/GP2 chimeras formed between JUNV and Cd1 (Fig. 9A) (13). This analysis revealed that sensitivity to all three compounds mapped to the GP2 subunit (Fig. 9B).
FIG 9.
Mapping of compound sensitivity in JUNV GP. (A) Schematic representation of GP chimeras between JUNV (strain XJ) and Cd1. (B) 293A cells incubated with JUNV XJ, Cd1, or chimera (C1X2 and X1C2) GP-pseudotyped lentiviral vectors, with DMSO or the indicated compounds being present. Relative titers are shown as percentages of the control titer (DMSO treated). Mean titers (in TU per milliliter) in DMSO-treated cells were as follows: 1.9 × 105 for JUNV XJ, 5.3 × 103 for Cd1, 2.4 × 104 C1X2, and 1.9 × 104 for X1C2. (C) Schematic representation of JUNV GP and the relative positions of the point mutations tested. The point mutations K33H and L420T (underlined) conferred the greatest degree of resistance to all three compounds. The drawing is not to scale and illustrates the interactions described previously by York et al. (61, 62). (D) 293A cells were incubated with JUNV GP-pseudotyped lentiviral vectors, either the wild type (WT) or the indicated mutants, in the presence of DMSO or the indicated compounds. Relative titers are shown as percentages of the control titer (DMSO treated). Mean titers (in TU per milliliter) on DMSO-treated cells were as follows: 1.9 × 105 for the wild type, 1.0 × 104 for the K33H mutant, 3.0 × 105 for the N37A mutant, 3.1 × 105 for the L420T mutant, 8.6 × 104 for the F427I mutant, 6.6 × 104 for the A435I mutant, and 2.9 × 105 for the F438I mutant. All data are the means ± standard deviations of results from 2 to 5 independent experiments. All data from panel B was statistically significant compared to the DMSO control (P < 0.001), and in panel D, statistical significance compared to the DMSO control is indicated (***, P < 0.001; ****, P < 0.0001).
The mature GP complex on the surface of arenavirus particles contains noncovalently linked GP1, GP2, and SSP subunits (Fig. 9C), further organized as trimers (1). Residues in both an ectodomain loop of SSP and the transmembrane (TM) region of GP2 have been shown to be important for maintaining the stability of the GP complex (61–63). Upon encountering low pH, this interaction is disrupted, allowing GP1 dissociation and triggering conformational changes in GP2 that eventually lead to virus-cell fusion (61–63). Furthermore, there is evidence that these interactions between GP2 and SSP have created a highly druggable target, and several independently identified arenavirus inhibitors are hypothesized to target this interaction (58, 59, 61, 64, 65). Specifically, resistance to two of the compounds, designated ST-294 and ST-193, could be conferred by mutating specific residues in SSP and GP2 (59, 61, 64). We therefore asked whether compounds 1 to 3 could also be binding to this same target site by evaluating their sensitivity to the same mutations.
We generated a panel of SSP and GP2 TM mutants of JUNV GP (Fig. 9C). These mutants were previously reported to result in no adverse effects on GP cell surface expression and retain the ability to promote cell-cell fusion (13, 61). We independently confirmed that all mutants retained cell-cell fusion capabilities following a low-pH pulse (data not shown). The mutant GPs were incorporated into lentiviral vectors and tested for their ability to transduce 293A cells in the absence and presence of each compound (Fig. 9D). We found that the A435I and F438I substitutions in GP2 had no effect on compound action, and an N37A substitution in the ectodomain of SSP led to only a minor reduction in compound potencies. In contrast, significant reductions in sensitivity to all three compounds occurred with the K33H substitution in SSP and the L420T substitution in GP2. Residue K33 is thought to be particularly critical for pH sensing, thereby linking the action of these compounds to a conserved component of the arenavirus fusion machinery (62).
We also observed an intermediate phenotype for the F427I substitution in GP2, which abrogated 50% of the activity of compound 3 while having only minimal effects on compounds 1 and 2. We previously hypothesized that F427I is an important mutation involved in the attenuated phenotype of Cd1, since it promotes the premature transitioning of GP to a fusogenic conformation at neutral pH (13). By mutating this residue alone, we obtained a compound insensitivity profile that more closely matched that of Cd1 (Fig. 9B), indicating the importance of this residue for the decreased potency of the compounds against Cd1 compared to JUNV GP. Taken together, these results provide strong evidence that compounds 1 to 3 inhibit arenavirus entry by targeting a crucial interaction between SSP and GP2 that regulates the transition to a fusogenic conformation for the whole GP complex.
Tetrandrine, a calcium channel inhibitor, has broad activity against arenaviruses.
The HTS also identified tetrandrine, a naturally occurring compound used to treat multiple ailments, including hypertension, and possessing calcium channel-blocking and anti-inflammatory properties (45, 66). In the arenaviruses, inhibition of voltage-gated calcium channels by the drugs nifedipine, verapamil, and gabapentin has also been shown to inhibit entry, indicating that calcium channels play a role in the entry pathway of arenaviruses (67). One mechanism of action of tetrandrine has been described as blocking two-pore channels (TPCs) in endosomal vesicles (46). TPCs are the target of NAADP (nicotinic acid adenine dinucleotide phosphate) (68), which promotes the release of calcium from lysosomes and endosomes into the cytoplasm (reviewed in reference 69). Interestingly, tetrandrine has also been reported to be a potent inhibitor of Ebola virus entry, with the mechanism being through inhibition of TPCs (46).
Using pseudotyped vectors, we tested tetrandrine against a panel of arenavirus GPs. We found that the drug was active against at least 1 member of each subgroup but exhibited a range of responses, even with closely related viruses (Fig. 10A to E). For example, in clade B, tetrandrine blocked the subset B1 (57) viruses JUNV, MACV, and TCRV by 50% but caused closer to 90% inhibition of the B2 viruses AMAV and GTOV (Fig. 10A). Similarly, in the OW viruses, tetrandrine was potent against LCMV but not LASV (Fig. 10E), while in the clade C viruses, it had activity against OLVV but not LATV (Fig. 10D). When the drug was tested in focus-forming infection assays, we observed that it was active against both LCMV-4 and Cd1, although the effects were not as potent as those that we observed with ribavirin (Fig. 10F).
FIG 10.
Effects of tetrandrine. (A to E, G, and H) 293A cells (or NIH 3T3 cells for ecotropic MLV) were incubated with pseudotyped lentiviral or retroviral vectors expressing fusion proteins from arenavirus NW clade B (A); NW recombinant clade A/B (B); NW clade A (C); NW clade C (D); OW arenaviruses (E); JUNV, Cd1, or chimeras (G); and fusion proteins from other virus families (H). Cells were treated with DMSO or tetrandrine, and relative titers are shown as percentages of the control titer (DMSO treated). Mean titers (in TU per milliliter) on DMSO-treated cells were as follows: 2.7 × 105 for JUNV, 5.0 × 104 for MACV, 1.1 × 105 for TCRV, 5.5 × 104 for GTOV, 5.6 × 104 for AMAV, 8.1 × 103 for WWAV, 8.2 × 103 for BBTV, 8.8 × 103 for SKTV, 7.5 × 103 for FLEV, 3.4 × 105 for PICV, 3.9 × 104 for OLVV, 7.5 × 103 for LATV, 2.1 × 106 for LASV, 4.0 × 105 for LCMV13, 3.3 × 105 for LCMV53b, 8.4 × 104 for JUNV XJ, 7.9 × 103 for Cd1, 1.7 × 104 for C1X2, 8.2 × 103 for X1C2, 1 × 106 for VSV, 3.7 × 104 for amphotropic MLV, 8.2 × 104 for ecotropic MLV, 4.7 × 104 for Ebola virus, 1.9 × 103 for influenza virus, 2.8 × 104 for Hendra virus, and 3.4 × 105 for Nipah virus. All data are the means ± standard deviations of results from 2 to 4 independent experiments. (F) Quantitation of Cd1 and LCMV-4 infection (number of foci) of Vero E6 cells treated with DMSO alone, ribavirin, or tetrandrine. Mean titers on DMSO-treated cells were 1 × 104 FFU/ml and 2.3 × 104 FFU/ml for Cd1 and LCMV, respectively. All data are the means ± standard deviations of results from 2 to 7 independent experiments. Statistical significance compared to the DMSO control is indicated (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
Although Cd1 GP is very closely related to JUNV GP, and the drug partially inhibited Cd1 infection in the focus-forming assay, we detected no activity of tetrandrine against vectors pseudotyped with Cd1 GP (Fig. 10G). This discrepancy between the sensitivities of the virus and GP vectors likely reflects the fact that the virus assay involves multiple rounds of a spreading infection, which could amplify even a small effect that might not be apparent in the single-round vector assays. Analysis of JUNV/Cd1 GP chimeras mapped the differential sensitivity to tetrandrine to the GP2 subunit (Fig. 10G). Finally, we tested tetrandrine for activity against other viral fusion proteins and found no effect, except for the previously noted potent effect against Ebola virus reported by Sakurai et al. (46) (Fig. 10H).
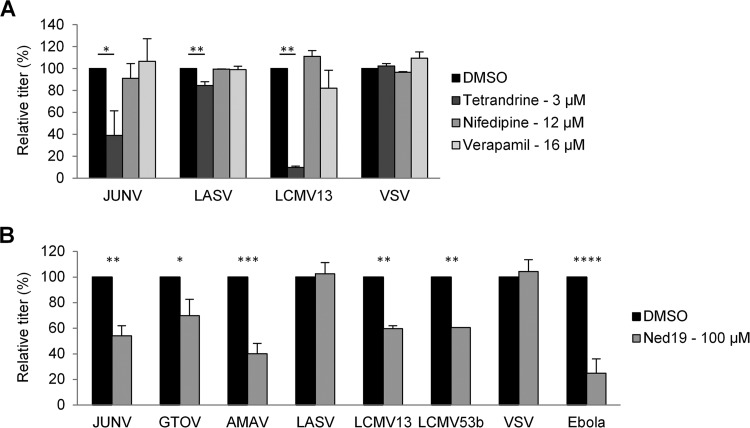
In addition to TPCs, tetrandrine inhibits other calcium channels such as voltage-dependent L and T types as well as calcium-dependent potassium channels (reviewed in reference 70). To evaluate whether its effects on arenavirus entry were related to TPC inhibition, or another class of channels, we tested the effects of the L-type calcium channel inhibitors verapamil and nifedipine (Fig. 11A) and also Ned19 (Fig. 11B), which is a specific inhibitor of the TCP-stimulating molecule NAADP (68, 71). Nifedipine and verapamil were previously reported to inhibit arenavirus entry, resulting in 60% inhibition of LCMV GP vectors and ∼80 to 90% inhibition of both JUNV GP vectors and Cd1 (67). However, different from the data in that report, our assays did not find either drug to have an effect on JUNV or LCMV GP vectors (Fig. 11A). Nonetheless, when we examined the ability of Ned19 to inhibit an expanded panel of tetrandrine-sensitive fusion proteins, we observed a similar pattern of inhibition for both drugs albeit with a lower overall degree of inhibition by Ned19 (Fig. 11B). In the same way, Ebola virus GP has been reported to be less sensitive to Ned19 than to tetrandrine (46).
FIG 11.
Effects of calcium channel pathway inhibitors on virus entry. (A) 293A cells were incubated with pseudotyped lentiviral vectors expressing the tetrandrine-sensitive fusion proteins from JUNV and LCMV13, together with the insensitive fusion proteins from LASV and VSV, in the presence of DMSO or the indicated drugs. Relative titers are shown as percentages of the control titer (DMSO treated). Mean titers (in TU per milliliter) in DMSO-treated cells were as follows: 1.3 × 105 for JUNV, 3.1 × 106 for LASV, 1.8 × 105 for LCMV13, and 2.3 × 106 for VSV. (B) 293A cells were incubated with pseudotyped lentiviral or retroviral vectors expressing the indicated viral fusion proteins, in the presence of DMSO or Ned19. Relative titers are shown as percentages of the control titer (DMSO treated). Mean titers (in TU per milliliter) in DMSO-treated cells were as follows: 2.9 × 105 for JUNV, 4.7 × 104 for GTOV, 3.5 × 104 for AMAV, 1.5 × 107 for LASV, 2.9 × 105 for LCMV13, 2.1 × 105 for LCMV53b, 6.4 × 106 for VSV, and 2.5 × 104 for Ebola virus. All data are the means ± standard deviations of results from 2 independent experiments. Statistical significance compared to the DMSO control is indicated (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
If tetrandrine was inhibiting arenavirus entry through an effect on TPCs, a prediction of the model is that the drug would impact a late stage of virus entry and thus would not block cell-cell fusion. In agreement, we found that tetrandrine did not inhibit cell-cell fusion when JUNV, GTOV, or LCMV GPs were expressed on the surface of 293T cells and subjected to a low-pH pulse (data not shown).
Together, these observations suggest that, as has recently been described for Ebola virus GP (46), TPCs play a role in the entry of certain arenavirus. Since sensitivity to tetrandrine and Ned19 was not determined by the relatedness of the arenavirus GPs, or the cell receptor used, this suggests that the use of this pathway in the trafficking of internalized arenavirus particles has likely been acquired and lost several times during arenavirus evolution.
DISCUSSION
We used a JUNV/Cd1-based MG rescue system to interrogate a library of 87,321 compounds, from which we identified 4 potent hit compounds with favorable predicted medicinal chemistry profiles. Despite the fact that all aspects of the arenavirus life cycle were represented in the MG rescue system, the 4 compounds identified acted at the entry stage. One of the compounds that we identified was tetrandrine, a calcium channel blocker that inhibits TPCs and has activity against Ebola virus entry (46); the remaining three compounds were novel.
The novel compounds (compounds 1 to 3) proved to be broadly acting against the NW arenaviruses, with the most activity against clade B, of which JUNV is a member. Compound 1 had the broadest host range within the NW viruses, as it was the only compound to inhibit at least one member of every clade. In contrast, compound 3 was the only compound to inhibit the more distantly related OW viruses, having activity against the important human pathogen LASV. Compound sensitivity did not correlate with the cellular receptor used, since NW clade C viruses were sensitive to all three compounds, while LCMV, which also uses α-DG as a receptor, was resistant to all three compounds. Additionally, comparison of the GP sequences of resistant and sensitive GPs did not identify any obvious candidates for points of compound interactions.
More detailed assays interrogating the binding and low-pH-triggered fusion events of entry narrowed the stage of action for compounds 1 to 3 to fusion. This, together with the results from specific mutational analyses, strongly suggested that the compounds were similar to previously identified fusion inhibitors, such as ST-294 and ST-193, among others (58, 59, 61, 64, 65). These compounds have been hypothesized to target an interaction between SSP and GP2 that controls the conformational changes that occur under low-pH conditions, when internalized virions reach late endosomes (63, 72, 73).
Although functionally similar, compounds 1 to 3 were structurally quite distinct (Fig. 2E) and also different from the other previously described fusion inhibitors. Therefore, if a common molecular interface between SSP and GP2 is being targeted by all of these compounds, it is likely that the compounds make different specific contacts. Of note, when 4 mutations in JUNV GP that confer >50% resistance to ST-294 were tested, only two of the mutations resulted in complete resistance to compounds 1 to 3, and only one additional mutation led to >50% resistance to compound 3. This included a substitution at residue K33, which is highly conserved across the entire arenavirus family and therefore likely to be a functionally important residue. These observations also suggest that compounds that target this interaction in the GP complex could be developed with improved profiles.
The HTS also identified tetrandrine, an inhibitor of various types of calcium channels (70), as being active against JUNV GP, and we further found that it had activity against a broad panel of arenaviruses. However, similar to our observations with compounds 1 to 3, we found that the absolute levels of sensitivity to tetrandrine varied among the family members, with LASV in particular being resistant. Tetrandrine was recently identified as an inhibitor of Ebola virus entry, in agreement with previous reports that this virus has a requirement for calcium channels during entry (46, 74). Using pseudotyped vectors, we also confirmed that tetrandrine inhibited Ebola virus GP, but we were unable to identify any other sensitive viral fusion proteins from the panel that we tested. This suggests that tetrandrine might be relatively specific for these two groups of important human pathogens that cause hemorrhagic fevers.
The action of tetrandrine against Ebola virus GP was further reported to involve the TPC subtype of calcium channels, as evidenced by sensitivity to Ned19, an inhibitor of the TPC-stimulating molecule NAADP (46). We further found that the tetrandrine-sensitive arenavirus GPs were also sensitive to Ned19, suggesting that the drug's activity against the arenaviruses may also involve TPCs. Interestingly, Sakurai et al. (46) did not identify tetrandrine as an inhibitor of the arenaviruses, but this was because they tested only LASV GP, which we also found to be only weakly sensitive to the drug. Our results therefore extend their observations and suggest that the TPC pathway may be a common pathway used during entry by human hemorrhagic fever viruses and that this pathway and class of compounds could provide a viable path to developing broadly acting anti-hemorrhagic fever drugs.
ACKNOWLEDGMENTS
We thank Michael Buchmeier and Benhur Lee, who generously provided reagents. We also thank Nick Llewellyn for helpful discussions and Su Chiang and Kyungae Lee for medicinal chemistry assessments.
This work was supported by PHS grant U54 AI065359 (NIAD, NIH) to the Pacific Southwest Regional Center of Excellence for Biodefense and Emerging Infectious Diseases and grant U54 AI057159 (NIAD, NIH) to the New England Regional Center of Excellence for Biodefense and Emerging Infectious Diseases.
REFERENCES
- 1.Buchmeier MJ, Peters CJ, De la Torre JC. 2007. Arenaviridae: the viruses and their replication, p 1791–1828. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Childs J, Peters C. 1993. Ecology and epidemiology of arenaviruses and their hosts, p 331–384. In Salvato M. (ed), The Arenaviridae. Springer, New York, NY. [Google Scholar]
- 3.Cajimat MN, Milazzo ML, Bradley RD, Fulhorst CF. 2012. Ocozocoautla de Espinosa virus and hemorrhagic fever, Mexico. Emerg Infect Dis 18:401–405. doi: 10.3201/eid1803.111602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maiztegui JI. 1975. Clinical and epidemiological patterns of Argentine haemorrhagic fever. Bull World Health Organ 52:567–575. [PMC free article] [PubMed] [Google Scholar]
- 5.Asogun DA, Adomeh DI, Ehimuan J, Odia I, Hass M, Gabriel M, Olschläger S, Becker-Ziaja B, Folarin O, Phelan E, Ehiane PE, Ifeh VE, Uyigue EA, Oladapo YT, Muoebonam EB, Osunde O, Dongo A, Okokhere PO, Okogbenin SA, Momoh M, Alikah SO, Akhuemokhan OC, Imomeh P, Odike MA, Gire S, Andersen K, Sabeti PC, Happi CT, Akpede GO, Günther S. 2012. Molecular diagnostics for Lassa fever at Irrua specialist teaching hospital, Nigeria: lessons learnt from two years of laboratory operation. PLoS Negl Trop Dis 6:e1839. doi: 10.1371/journal.pntd.0001839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Briese T, Paweska JT, McMullan LK, Hutchison SK, Street C, Palacios G, Khristova ML, Weyer J, Swanepoel R, Egholm M, Nichol ST, Lipkin WI. 2009. Genetic detection and characterization of Lujo virus, a new hemorrhagic fever-associated arenavirus from southern Africa. PLoS Pathog 5:e1000455. doi: 10.1371/journal.ppat.1000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Enria DA, Ambrosio AM, Briggiler AM, Feuillade MR, Crivelli E, Study Group on Argentine Hemorrhagic Fever Vaccine. 2010. Candid#1 vaccine against Argentine hemorrhagic fever produced in Argentina. Immunogenicity and safety. Medicina (B Aires) 70:215–222. (In Spanish.) [PubMed] [Google Scholar]
- 8.Enria DA, Maiztegui JI. 1994. Antiviral treatment of Argentine hemorrhagic fever. Antiviral Res 23:23–31. doi: 10.1016/0166-3542(94)90030-2. [DOI] [PubMed] [Google Scholar]
- 9.Enria DA, Briggiler AM, Sánchez Z. 2008. Treatment of Argentine hemorrhagic fever. Antiviral Res 78:132–139. doi: 10.1016/j.antiviral.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson DE, Chosewood LC (ed). 2009. Biosafety in microbiological and biomedical laboratories, 5th ed Centers for Disease Control and Prevention, Atlanta, GA. http://www.cdc.gov/biosafety/publications/bmbl5/BMBL.pdf: Accessed 4 February 2015. [Google Scholar]
- 11.Christodoulopoulos I, Cannon PM. 2001. Sequences in the cytoplasmic tail of the gibbon ape leukemia virus envelope protein that prevent its incorporation into lentivirus vectors. J Virol 75:4129–4138. doi: 10.1128/JVI.75.9.4129-4138.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reignier T, Oldenburg J, Noble B, Lamb E, Romanowski V, Buchmeier MJ, Cannon PM. 2006. Receptor use by pathogenic arenaviruses. Virology 353:111–120. doi: 10.1016/j.virol.2006.05.018. [DOI] [PubMed] [Google Scholar]
- 13.Droniou-Bonzom ME, Reignier T, Oldenburg JE, Cox AU, Exline CM, Rathbun JY, Cannon PM. 2011. Substitutions in the glycoprotein (GP) of the Candid#1 vaccine strain of Junin virus increase dependence on human transferrin receptor 1 for entry and destabilize the metastable conformation of GP. J Virol 85:13457–13462. doi: 10.1128/JVI.05616-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Albariño CG, Bergeron E, Erickson BR, Khristova ML, Rollin PE, Nichol ST. 2009. Efficient reverse genetics generation of infectious Junin viruses differing in glycoprotein processing. J Virol 83:5606–5614. doi: 10.1128/JVI.00276-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee KJ, Novella IS, Teng MN, Oldstone MBA, de la Torre JC. 2000. NP and L proteins of lymphocytic choriomeningitis virus (LCMV) are sufficient for efficient transcription and replication of LCMV genomic RNA analogs. J Virol 74:3470–3477. doi: 10.1128/JVI.74.8.3470-3477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freiberg A, Dolores LK, Enterlein S, Flick R. 2008. Establishment and characterization of plasmid-driven minigenome rescue systems for Nipah virus: RNA polymerase I- and T7-catalyzed generation of functional paramyxoviral RNA. Virology 370:33–44. doi: 10.1016/j.virol.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Groseth A, Feldmann H, Theriault S, Mehmetoglu G, Flick R. 2005. RNA polymerase I-driven minigenome system for Ebola viruses. J Virol 79:4425–4433. doi: 10.1128/JVI.79.7.4425-4433.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hass M, Gölnitz U, Müller S, Becker-Ziaja B, Günther S. 2004. Replicon system for Lassa virus. J Virol 78:13793–13803. doi: 10.1128/JVI.78.24.13793-13803.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jasenosky LD, Neumann G, Kawaoka Y. 2010. Minigenome-based reporter system suitable for high-throughput screening of compounds able to inhibit ebolavirus replication and/or transcription. Antimicrob Agents Chemother 54:3007–3010. doi: 10.1128/AAC.00138-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uebelhoer LS, Albariño CG, McMullan LK, Chakrabarti AK, Vincent JP, Nichol ST, Towner JS. 2014. High-throughput, luciferase-based reverse genetics systems for identifying inhibitors of Marburg and Ebola viruses. Antiviral Res 106:86–94. doi: 10.1016/j.antiviral.2014.03.018. [DOI] [PubMed] [Google Scholar]
- 21.Wright KE, Salvato MS, Buchmeier MJ. 1989. Neutralizing epitopes of lymphocytic choriomeningitis virus are conformational and require both glycosylation and disulfide bonds for expression. Virology 171:417–426. doi: 10.1016/0042-6822(89)90610-7. [DOI] [PubMed] [Google Scholar]
- 22.Welsh RM, Seedhom MO. 2008. Lymphocytic choriomeningitis virus (LCMV): propagation, quantitation, and storage. Curr Protoc Microbiol Chapter 15:Unit 15A.1. doi: 10.1002/9780471729259.mc15a01s8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang JH, Chung TD, Oldenburg KR. 1999. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen 4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 24.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng Y, Ryazantsev S, Ohmi K, Zhao H-Z, Rozengurt N, Kohn DB, Neufeld EF. 2004. Retrovirally transduced bone marrow has a therapeutic effect on brain in the mouse model of mucopolysaccharidosis IIIB. Mol Genet Metab 82:286–295. doi: 10.1016/j.ymgme.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 26.Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. 2002. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science 295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- 27.Oldenburg J, Reignier T, Flanagan ML, Hamilton GA, Cannon PM. 2007. Differences in tropism and pH dependence for glycoproteins from the clade B1 arenaviruses: implications for receptor usage and pathogenicity. Virology 364:132–139. doi: 10.1016/j.virol.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flanagan ML, Oldenburg J, Reignier T, Holt N, Hamilton GA, Martin VK, Cannon PM. 2008. New World clade B arenaviruses can use transferrin receptor 1 (TfR1)-dependent and -independent entry pathways, and glycoproteins from human pathogenic strains are associated with the use of TfR1. J Virol 82:938–948. doi: 10.1128/JVI.01397-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reignier T, Oldenburg J, Flanagan ML, Hamilton GA, Martin VK, Cannon PM. 2008. Receptor use by the Whitewater Arroyo virus glycoprotein. Virology 371:439–446. doi: 10.1016/j.virol.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zong M, Fofana I, Choe H. 2014. Human and host species transferrin receptor 1 use by North American arenaviruses. J Virol 88:9418–9428. doi: 10.1128/JVI.01112-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar N, Wang J, Lan S, Danzy S, McLay Schelde L, Seladi-Schulman J, Ly H, Liang Y. 2012. Characterization of virulence-associated determinants in the envelope glycoprotein of Pichinde virus. Virology 433:97–103. doi: 10.1016/j.virol.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jemielity S, Wang JJ, Chan YK, Ahmed AA, Li W, Monahan S, Bu X, Farzan M, Freeman GJ, Umetsu DT, Dekruyff RH, Choe H. 2013. TIM-family proteins promote infection of multiple enveloped viruses through virion-associated phosphatidylserine. PLoS Pathog 9:e1003232. doi: 10.1371/journal.ppat.1003232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soneoka Y, Cannon PM, Ramsdale EE, Griffiths JC, Romano G, Kingsman SM, Kingsman AJ. 1995. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res 23:628–633. doi: 10.1093/nar/23.4.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yee JK, Friedmann T, Burns JC. 1994. Generation of high-titer pseudotyped retroviral vectors with very broad host range. Methods Cell Biol 43(Part A):99–112. [DOI] [PubMed] [Google Scholar]
- 35.Lopez LA, Yang SJ, Hauser H, Exline CM, Haworth KG, Oldenburg J, Cannon PM. 2010. Ebola virus glycoprotein counteracts BST-2/tetherin restriction in a sequence-independent manner that does not require tetherin surface removal. J Virol 84:7243–7255. doi: 10.1128/JVI.02636-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin AH, Cannon PM. 2002. Use of pseudotyped retroviral vectors to analyze the receptor-binding pocket of hemagglutinin from a pathogenic avian influenza A virus (H7 subtype). Virus Res 83:43–56. doi: 10.1016/S0168-1702(01)00407-5. [DOI] [PubMed] [Google Scholar]
- 37.Levroney EL, Aguilar HC, Fulcher JA, Kohatsu L, Pace KE, Pang M, Gurney KB, Baum LG, Lee B. 2005. Novel innate immune functions for galectin-1: galectin-1 inhibits cell fusion by Nipah virus envelope glycoproteins and augments dendritic cell secretion of proinflammatory cytokines. J Immunol 175:413–420. doi: 10.4049/jimmunol.175.1.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Negrete OA, Chu D, Aguilar HC, Lee B. 2007. Single amino acid changes in the Nipah and Hendra virus attachment glycoproteins distinguish ephrinB2 from ephrinB3 usage. J Virol 81:10804–10814. doi: 10.1128/JVI.00999-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moreno H, Gallego I, Sevilla N, de la Torre JC, Domingo E, Martín V. 2011. Ribavirin can be mutagenic for arenaviruses. J Virol 85:7246–7255. doi: 10.1128/JVI.00614-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ölschläger S, Neyts J, Günther S. 2011. Depletion of GTP pool is not the predominant mechanism by which ribavirin exerts its antiviral effect on Lassa virus. Antiviral Res 91:89–93. doi: 10.1016/j.antiviral.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 41.Sepúlveda CS, García CC, Fascio ML, D'Accorso NB, Docampo Palacios ML, Pellón RF, Damonte EB. 2012. Inhibition of Junin virus RNA synthesis by an antiviral acridone derivative. Antiviral Res 93:16–22. doi: 10.1016/j.antiviral.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 42.Perez M, Greenwald DL, de La Torre JC. 2004. Myristoylation of the RING finger Z protein is essential for arenavirus budding. J Virol 78:11443–11448. doi: 10.1128/JVI.78.20.11443-11448.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Urata S, Yasuda J, de la Torre JC. 2009. The z protein of the new world arenavirus Tacaribe virus has bona fide budding activity that does not depend on known late domain motifs. J Virol 83:12651–12655. doi: 10.1128/JVI.01012-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Radoshitzky SR, Abraham J, Spiropoulou CF, Kuhn JH, Nguyen D, Li W, Nagel J, Schmidt PJ, Nunberg JH, Andrews NC, Farzan M, Choe H. 2007. Transferrin receptor 1 is a cellular receptor for New World haemorrhagic fever arenaviruses. Nature 446:92–96. doi: 10.1038/nature05539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu QY, Li B, Gang JM, Karpinski E, Pang PK. 1995. Tetrandrine, a Ca++ antagonist: effects and mechanisms of action in vascular smooth muscle cells. J Pharmacol Exp Ther 273:32–39. [PubMed] [Google Scholar]
- 46.Sakurai Y, Kolokoltsov AA, Chen CC, Tidwell MW, Bauta WE, Klugbauer N, Grimm C, Wahl-Schott C, Biel M, Davey RA. 2015. Ebola virus. Two-pore channels control Ebola virus host cell entry and are drug targets for disease treatment. Science 347:995–998. doi: 10.1126/science.1258758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Albariño CG, Bird BH, Chakrabarti AK, Dodd KA, Flint M, Bergeron E, White DM, Nichol ST. 2011. The major determinant of attenuation in mice of the Candid1 vaccine for Argentine hemorrhagic fever is located in the G2 glycoprotein transmembrane domain. J Virol 85:10404–10408. doi: 10.1128/JVI.00856-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abraham J, Kwong JA, Albariño CG, Lu JG, Radoshitzky SR, Salazar-Bravo J, Farzan M, Spiropoulou CF, Choe H. 2009. Host-species transferrin receptor 1 orthologs are cellular receptors for nonpathogenic New World clade B arenaviruses. PLoS Pathog 5:e1000358. doi: 10.1371/journal.ppat.1000358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Caì Y, Yú S, Mazur S, Dong L, Janosko K, Zhang T, Müller MA, Hensley LE, Bavari S, Jahrling PB, Radoshitzky SR, Kuhn JH. 2013. Nonhuman transferrin receptor 1 is an efficient cell entry receptor for Ocozocoautla de Espinosa virus. J Virol 87:13930–13935. doi: 10.1128/JVI.02701-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Choe H, Jemielity S, Abraham J, Radoshitzky SR, Farzan M. 2011. Transferrin receptor 1 in the zoonosis and pathogenesis of New World hemorrhagic fever arenaviruses. Curr Opin Microbiol 14:476–482. doi: 10.1016/j.mib.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Radoshitzky SR, Kuhn JH, Spiropoulou CF, Albariño CG, Nguyen DP, Salazar-Bravo J, Dorfman T, Lee AS, Wang E, Ross SR, Choe H, Farzan M. 2008. Receptor determinants of zoonotic transmission of New World hemorrhagic fever arenaviruses. Proc Natl Acad Sci U S A 105:2664–2669. doi: 10.1073/pnas.0709254105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Radoshitzky SR, Longobardi LE, Kuhn JH, Retterer C, Dong L, Clester JC, Kota K, Carra J, Bavari S. 2011. Machupo virus glycoprotein determinants for human transferrin receptor 1 binding and cell entry. PLoS One 6:e21398. doi: 10.1371/journal.pone.0021398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spiropoulou CF, Kunz S, Rollin PE, Campbell KP, Oldstone MB. 2002. New World arenavirus clade C, but not clade A and B viruses, utilizes alpha-dystroglycan as its major receptor. J Virol 76:5140–5146. doi: 10.1128/JVI.76.10.5140-5146.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rojek JM, Spiropoulou CF, Campbell KP, Kunz S. 2007. Old World and clade C New World arenaviruses mimic the molecular mechanism of receptor recognition used by alpha-dystroglycan's host-derived ligands. J Virol 81:5685–5695. doi: 10.1128/JVI.02574-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cao W, Henry MD, Borrow P, Yamada H, Elder JH, Ravkov EV, Nichol ST, Compans RW, Campbell KP, Oldstone MB. 1998. Identification of alpha-dystroglycan as a receptor for lymphocytic choriomeningitis virus and Lassa fever virus. Science 282:2079–2081. doi: 10.1126/science.282.5396.2079. [DOI] [PubMed] [Google Scholar]
- 56.Kunz S. 2009. Receptor binding and cell entry of Old World arenaviruses reveal novel aspects of virus-host interaction. Virology 387:245–249. doi: 10.1016/j.virol.2009.02.042. [DOI] [PubMed] [Google Scholar]
- 57.Charrel RN, Feldmann H, Fulhorst CF, Khelifa R, de Chesse R, de Lamballerie X. 2002. Phylogeny of New World arenaviruses based on the complete coding sequences of the small genomic segment identified an evolutionary lineage produced by intrasegmental recombination. Biochem Biophys Res Commun 296:1118–1124. doi: 10.1016/S0006-291X(02)02053-3. [DOI] [PubMed] [Google Scholar]
- 58.Thomas CJ, Casquilho-Gray HE, York J, DeCamp DL, Dai D, Petrilli EB, Boger DL, Slayden RA, Amberg SM, Sprang SR, Nunberg JH. 2011. A specific interaction of small molecule entry inhibitors with the envelope glycoprotein complex of the Junín hemorrhagic fever arenavirus. J Biol Chem 286:6192–6200. doi: 10.1074/jbc.M110.196428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bolken TC, Laquerre S, Zhang Y, Bailey TR, Pevear DC, Kickner SS, Sperzel LE, Jones KF, Warren TK, Lund SA, Kirkwood-Watts DL, King DS, Shurtleff AC, Guttieri MC, Deng Y, Bleam M, Hruby DE. 2006. Identification and characterization of potent small molecule inhibitor of hemorrhagic fever New World arenaviruses. Antiviral Res 69:86–97. doi: 10.1016/j.antiviral.2005.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goñi SE, Iserte JA, Ambrosio AM, Romanowski V, Ghiringhelli PD, Lozano ME. 2006. Genomic features of attenuated Junín virus vaccine strain candidate. Virus Genes 32:37–41. doi: 10.1007/s11262-005-5843-2. [DOI] [PubMed] [Google Scholar]
- 61.York J, Dai D, Amberg SM, Nunberg JH. 2008. pH-induced activation of arenavirus membrane fusion is antagonized by small-molecule inhibitors. J Virol 82:10932–10939. doi: 10.1128/JVI.01140-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.York J, Nunberg JH. 2006. Role of the stable signal peptide of Junín arenavirus envelope glycoprotein in pH-dependent membrane fusion. J Virol 80:7775–7780. doi: 10.1128/JVI.00642-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.York J, Nunberg JH. 2009. Intersubunit interactions modulate pH-induced activation of membrane fusion by the Junin virus envelope glycoprotein GPC. J Virol 83:4121–4126. doi: 10.1128/JVI.02410-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Larson RA, Dai D, Hosack VT, Tan Y, Bolken TC, Hruby DE, Amberg SM. 2008. Identification of a broad-spectrum arenavirus entry inhibitor. J Virol 82:10768–10775. doi: 10.1128/JVI.00941-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee AM, Rojek JM, Spiropoulou CF, Gundersen AT, Jin W, Shaginian A, York J, Nunberg JH, Boger DL, Oldstone MB, Kunz S. 2008. Unique small molecule entry inhibitors of hemorrhagic fever arenaviruses. J Biol Chem 283:18734–18742. doi: 10.1074/jbc.M802089200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li S-Y, Ling L-H, Teh BS, Seow WK, Thong YH. 1989. Anti-inflammatory and immunosuppressive properties of the bis-benzylisoquinolines: in vitro comparisons of tetrandrine and berbamine. Int J Immunopharmacol 11:395–401. [DOI] [PubMed] [Google Scholar]
- 67.Lavanya M, Cuevas CD, Thomas M, Cherry S, Ross SR. 2013. siRNA screen for genes that affect Junín virus entry uncovers voltage-gated calcium channels as a therapeutic target. Sci Transl Med 5:204ra131. doi: 10.1126/scitranslmed.3006827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brailoiu E, Rahman T, Churamani D, Prole DL, Brailoiu GC, Hooper R, Taylor CW, Patel S. 2010. An NAADP-gated two-pore channel targeted to the plasma membrane uncouples triggering from amplifying Ca2+ signals. J Biol Chem 285:38511–38516. doi: 10.1074/jbc.M110.162073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Galione A. 2011. NAADP receptors. Cold Spring Harb Perspect Biol 3:a004036. doi: 10.1101/cshperspect.a004036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang G, Lemos J. 1995. Tetrandrine: a new ligand to block voltage-dependent Ca2+ and Ca2+-activated K+ channels. Life Sci 56:295–306. doi: 10.1016/0024-3205(94)00952-X. [DOI] [PubMed] [Google Scholar]
- 71.Naylor E, Arredouani A, Vasudevan SR, Lewis AM, Parkesh R, Mizote A, Rosen D, Thomas JM, Izumi M, Ganesan A, Galione A, Churchill GC. 2009. Identification of a chemical probe for NAADP by virtual screening. Nat Chem Biol 5:220–226. doi: 10.1038/nchembio.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Borrow P, Oldstone MB. 1994. Mechanism of lymphocytic choriomeningitis virus entry into cells. Virology 198:1–9. doi: 10.1006/viro.1994.1001. [DOI] [PubMed] [Google Scholar]
- 73.Castilla V, Mersich SE, Candurra NA, Damonte EB. 1994. The entry of Junin virus into Vero cells. Arch Virol 136:363–374. doi: 10.1007/BF01321064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kolokoltsov AA, Saeed MF, Freiberg AN, Holbrook MR, Davey RA. 2009. Identification of novel cellular targets for therapeutic intervention against Ebola virus infection by siRNA screening. Drug Dev Res 70:255–265. doi: 10.1002/ddr.20303. [DOI] [PMC free article] [PubMed] [Google Scholar]