Abstract
Bacteriophages are the most abundant biological entities in the biosphere, and this dynamic and old population is, not surprisingly, highly diverse genetically. Relative to bacterial genomics, phage genomics has advanced slowly, and a higher-resolution picture of the phagosphere is only just emerging. This view reveals substantial diversity even among phages known to infect a common host strain, but the relationships are complex, with mosaic genomic architectures generated by illegitimate recombination over a long period of evolutionary history.
INTRODUCTION
Bacteriophages are the dark matter of the biological world (1); a vastness of ill-defined genetic variation whose impacts we observe on the microbial population but of which we have little understanding. The phage population is estimated to contain approximately 1031 particles and is highly dynamic, with the population turning over every few days. Moreover, this rolling boil of evolution has been churning away for perhaps two billion years or more, giving rise to fantastic genetic diversity (2, 3).
It is noteworthy that these estimations of phage population size and turnover emerged primarily from observations made with water samples that are simple to collect and quantify (4). Although terrestrial environments may contribute a relatively minor part of the total numbers of phage particles in the biosphere, prokaryotic diversity in soil samples is very high (5), which is anticipated to be reflected in the companion phage populations. Quantifying the phage populations in soil and terrestrial samples can be tricky, but phages are estimated to be present at levels approaching 109 particles per gram of soil (6).
Two key approaches to defining viral diversity are metagenomics of total concentrated phage samples collected from the environment and a genome-by-genome strategy of analyzing individually isolated phages. The two approaches are compatible but have distinct outcomes. Metagenomics generates a large amount of sequence data and provides good indicators of diversity. Analysis of individually isolated phages generates smaller data sets, but they are structured into whole genomes. Because phage genomes are architecturally mosaic, the availability of complete genomes contextualizes the complexities of the relationships among phages (7). Moreover, individual phages are available for genetic, biochemical, and microbiological analyses, a critical resource given the evident functional and regulatory novelty within the phage population. Metagenomics typically does not offer a confident linkage between viral sequences and potential bacterial hosts, although methods for enrichment with particular hosts have been described (8). For individually recovered phages, host ranges can be defined empirically.
Perhaps not surprisingly, phage diversity is sufficiently high that phages infecting phylogenetically distant hosts share little genetic information (9). The question then arises as to what is the diversity of viruses that infect a single host strain, in which they can be assumed to be in direct genetic contact with each other. Progress has been made in addressing this question using the host Mycobacterium smegmatis mc2155 and phages isolated from soil and other environmental sources, and integrated research and education programs have rapidly advanced the field (7).
The collection of individual mycobacteriophages has grown to about 850 (http://phagesdb.org), of which 627 have been comparatively analyzed (7). Some of these phages share extensive nucleotide sequence information, and this has been used to assort them into clusters (e.g., cluster A, B, C, etc.), some of which can be readily subdivided into subclusters (e.g., subcluster A1, A2, A3, etc.). The 627 genomes form 20 clusters (A to T) and eight singletons (phages with no close relatives), but with a highly skewed distribution, with 50% of the phages within two major clusters (A and B), although both span considerable diversity and many subclusters. Although assortment into clusters and subclusters is practical and recognizes the close relationships among some of the phages, the cluster and subcluster divisions clearly do not represent firm boundaries that prevent genetic exchange. The population thus generally appears as though it spans a continuum of genetic relationships, albeit with some phage types more prevalent than others, a different conclusion than was drawn from a metagenomic analysis of a single sample of Synechococcus phages (10). Interestingly, rarefaction analysis of mycobacteriophage gene contents suggests that this population is not closed and that sequencing new phage isolates will continue to reveal new genes (7). Enterobacteriophages can be similarly grouped into clusters according to nucleotide sequence similarity, suggesting that these relationships reflect a broader feature of phage populations (11).
What is the basis for all this diversity? And if the phages isolated on M. smegmatis are all in direct genetic contact with each other, why isn't mosaicism more prevalent at the nucleotide sequence level? We hypothesize that the phage population is migratory and moves across the microbial landscape relatively rapidly provided that two conditions are met (12): first, that there is a highly diverse bacterial population without large phylogenetic spaces between bacteria, and second, that the phages can mutate relatively rapidly to move from one host to another (12). At least for the soil/compost samples from which many of the mycobacteriophages have been isolated, both parameters are satisfied (12). Thus, phages that have closely related sequences have followed similar paths across the microbial landscape, accessing similar hosts and sampling similar parts of the microbial gene pool. Phages in different clusters, for example, would have taken very different paths using different hosts and thus are accessing different subsets of the gene pool. In this model, the reason why phages do not show more extensive nucleotide sequence mosaicism is that they have likely only “arrived” at the use of M. smegmatis as a host in relatively recent evolutionary time. The mosaic relationships are clearly evident from amino acid sequence comparisons, where more distant and older relationships are revealed (7).
This model supposes that, provided that richness of bacterial hosts is available, phages can skate across the microbial landscape faster than their genomes can acquire the sequence biases and characteristics of any given bacterial host. Thus, in addition to the diversity in sequence information, there is considerable variation in mycobacteriophage percent GC (GC%) content, ranging from 50% to 70%, compared to that in M. smegmatis, which is 67.3% (7). In this view, the GC% composition along with associated codon usage biases do not necessarily reflect that of a known host, but reflect the variety of hosts encountered in their evolutionary past. Mycobacteriophages such as Patience, with a GC% of 50.3%, thus likely evolved through environments where it encountered hosts with moderate GC% contents and only recently migrated to higher-GC% mycobacterial hosts (13). Given the disparity between the GC% contents of Patience and its mycobacterial hosts, it is not surprising that their codon usage profiles are markedly different too. The mycobacteria in general have limited repertoires that include only about 44 tRNAs, and 15 of the 16 5′-NNU codons are read by wobble base pairing; these codons are also used rarely. In contrast, there are nine codon sets in Patience where 5′-NNU is the most commonly used codon. Patience grows perfectly well and to high titer in M. smegmatis, so this has little impact on its growth and replication. Conceivably, Patience could easily manage this codon nonoptimization by acquisition of the cognate tRNA genes, but this is not the case, and Patience encodes only a single tRNAGln. Interestingly, some mycobacteriophages do have large sets of tRNA genes spanning almost the entire genetic code (1, 14), perhaps reflecting passage through hosts in which either these were needed for efficient expression or to counter antiphage tactics. Patience, however, has clearly not responded by tRNA acquisition. Identification and semiquantification of Patience gene products by liquid chromatography tandem mass spectrometry (LC-MS-MS) show a correlation between expression levels and codon usage bias, with more highly expressed genes having codon usage that is more similar to that of M. smegmatis (13). This is consistent with Patience being in the process of adapting to its higher-GC% mycobacterial hosts and aligns well with the general model for evolution of diversity described above. Patience does not infect any other hosts that have been tested, including moderate-GC% corynebacteria, and the genome space of actinobacteriophages is sufficiently sparse that it is little surprise that no other phages genomically related to Patience have been identified.
The general diversity of the phage population is reflected in the large number of novel genes that are unrelated to extant GenBank entries, and most genes are of unknown function. For the vast majority of the mycobacteriophages with siphoviral morphologies, the virion structure and assembly genes are syntenically conserved such that gene functions can be predicted even in the absence of sequence similarity (15). Mycobacteriophage genome lengths vary considerably (40 to 160 kbp), but the coding requirements for the virion structure and assembly genes are similar (20 to 25 kbp); thus, the size of the nonstructural genome segments is variable and appears to be a veritable playground for genetic exchange of small (average of 470 bp) genes of unknown function, presumably by a process of illegitimate recombination (9). Confident annotation of these genes can be tricky because of the small size of the genes, although there is good evidence that they are biologically relevant and not just pseudogenes, not only because of a plethora of examples of mosaicism (Fig. 1), but because many of the gene products were identified by LC-MS-MS of Patience-infected cells.
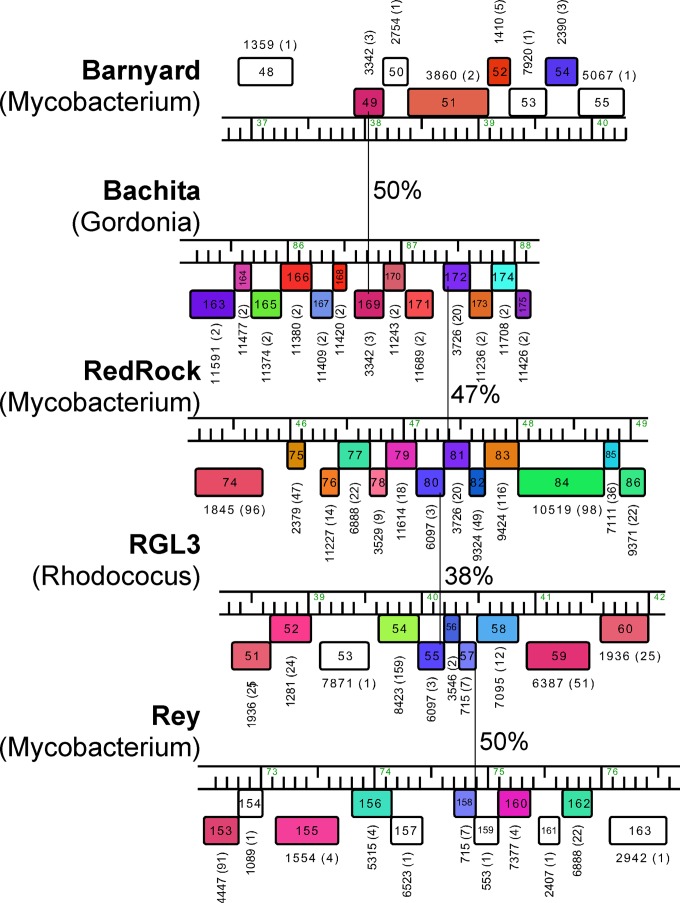
FIG 1.
Mosaic relationships among actinobacteriophages. Segments of five actinobacteriophage genomes are shown, with predicted genes represented as colored boxes (with gene numbers inside the boxes) transcribed either rightwards (shown above the genome) or leftwards (shown below the genome). Each of the 109,120 genes in a database (Actino_Draft, which contains 1,050 actinobacteriophage genomes) was assembled into phamilies (phams) of related genes in the program Phamerator; the Pham designation is shown above or below each gene, with the number of phamily members in parentheses. Vertical lines (with percent amino acid identity of their gene products) indicate genes that are related (members of the same Pham) but in different genomic contexts, illustrating the mosaic nature of phage genome architectures, which presumably occurred through a series of illegitimate recombination events. Thus, Barnyard gp49 (gene product 49) shares 50% amino acid identity with Bachita gp169, but the flanking genes are unrelated. Similar relationships between Bachita gp172 and Redrock gp81 (47% amino acid [aa] identity), between Redrock gp80 and RGL3 gp55 (38% aa identity), and between RGL3 gp57 and Rey gp158 (50% aa identity) are indicated. Interestingly, these relationships span phages isolated on hosts of different bacterial genera (as indicated in parentheses below the phage name). Note that the genes encoding these products and many of the surrounding genes are small (e.g., RGL3 57 is only 48 codons), and all are of unknown function with the exceptions of RGL3 gp53 and RGL3 gp59, which have predicted DinB-like and antirestriction functions, respectively.
What do all these phage gene products do? This is a much tougher question to address than the simpler discovery aspects of genome diversity. For the mycobacteriophages, a simple methodology for constructing defined non-polar-knockout mutants (16) showed that about two-thirds of the nonstructural genes are not required for lytic growth, and DNA replication roles could be assigned to at least two genes that were of unknown function (17). In view of the model for phage evolution described above, it is plausible that many of the genes were acquired in response to some particular need for growth in a host recently visited in their evolutionary past but not currently under selection for growth in M. smegmatis. In addition, because of the constant battle for survival between bacteria and their phages, there is likely to be a strong selection for phage acquisition of genes that confer protection from infection by other phages. Thus, phages encode restriction modification systems and clustered regularly interspaced short palindromic repeats (CRISPR) loci (18), and in the mycobacteriophages we see examples of immune mimicry (15). It is plausible that many other genes are involved in either countering host defense mechanisms, including anti-CRISPR activity (19), or protection from invasion (15).
Phages are likely to continue to be of considerable broad interest, not only because of their roles in global environmental cycling and bacterial pathogenesis, but because of their potential therapeutic use in a world in which antibiotic resistance is becoming ever more prevalent. Moreover, phage biodiversity can be readily tapped for biotechnological exploitation. It is noteworthy that two major biotechnological developments—restriction enzymes and CRISPRs—evolved as mechanisms for bacterial resistance to phage assault, and it is tempting to speculate that there are similar unidentified systems that may find similar broad biotechnological applications. The rich genomic information to be found in phages thus adds substantial fuel to this biotechnological fire.
What's ahead in phage genomics? Even with recent progress, we have barely scratched the proverbial surface of phage diversity, and the total phage sequence information is over 1,000-fold less than the total bacterial genome sequence information. The approximately 2,000 completely sequenced double-stranded DNA phage genomes have a median representation of two phages per host, and given the numbers of available bacterial host species (millions?) and the phage diversity on a single strain of one species, there is much darkness to be illuminated. Integrated research and education programs focusing on phage discovery and genomics can be expected to continue to assist in the isolation and comparative genomics of novel bacteriophages (20), and we anticipate a rich gold mine of new insights into bacteriophage diversity in the coming years.
ACKNOWLEDGMENTS
I thank David Asai, Deborah Jacobs-Sera, Welkin Pope, and Daniel Russell for comments on the manuscript.
REFERENCES
- 1.Pedulla ML, Ford ME, Houtz JM, Karthikeyan T, Wadsworth C, Lewis JA, Jacobs-Sera D, Falbo J, Gross J, Pannunzio NR, Brucker W, Kumar V, Kandasamy J, Keenan L, Bardarov S, Kriakov J, Lawrence JG, Jacobs WR, Hendrix RW, Hatfull GF. 2003. Origins of highly mosaic mycobacteriophage genomes. Cell 113:171–182. doi: 10.1016/S0092-8674(03)00233-2. [DOI] [PubMed] [Google Scholar]
- 2.Hendrix RW. 2002. Bacteriophages: evolution of the majority. Theor Popul Biol 61:471–480. doi: 10.1006/tpbi.2002.1590. [DOI] [PubMed] [Google Scholar]
- 3.Hatfull GF, Hendrix RW. 2011. Bacteriophages and their genomes. Curr Opin Virol 1:298–303. doi: 10.1016/j.coviro.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suttle CA. 2007. Marine viruses—major players in the global ecosystem. Nat Rev Microbiol 5:801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 5.Fierer N, Jackson RB. 2006. The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci U S A 103:626–631. doi: 10.1073/pnas.0507535103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williamson KE, Wommack KE, Radosevich M. 2003. Sampling natural viral communities from soil for culture-independent analyses. Appl Environ Microbiol 69:6628–6633. doi: 10.1128/AEM.69.11.6628-6633.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pope WH, Bowman CA, Russell DA, Jacobs-Sera D, Asai DJ, Cresawn SG, Jacobs WR, Hendrix RW, Lawrence JG, Hatfull GF, Science Education Alliance Phage Hunters Advancing Genomics and Evolutionary Science, Phage Hunters Integrating Research and Education, Mycobacterial Genetics Course. 2015. Whole genome comparison of a large collection of mycobacteriophages reveals a continuum of phage genetic diversity. Elife 4:e06416. doi: 10.7554/eLife.06416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng L, Gregory A, Yilmaz S, Poulos BT, Hugenholtz P, Sullivan MB. 2012. Contrasting life strategies of viruses that infect photo- and heterotrophic bacteria, as revealed by viral tagging. mBio 3(6):e00373-12. doi: 10.1128/mBio.00373-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hendrix RW, Smith MC, Burns RN, Ford ME, Hatfull GF. 1999. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc Natl Acad Sci U S A 96:2192–2197. doi: 10.1073/pnas.96.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng L, Ignacio-Espinoza JC, Gregory AC, Poulos BT, Weitz JS, Hugenholtz P, Sullivan MB. 2014. Viral tagging reveals discrete populations in Synechococcus viral genome sequence space. Nature 513:242–245. doi: 10.1038/nature13459. [DOI] [PubMed] [Google Scholar]
- 11.Grose JH, Casjens SR. 2014. Understanding the enormous diversity of bacteriophages: the tailed phages that infect the bacterial family Enterobacteriaceae. Virology 468-470:421–443. doi: 10.1016/j.virol.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacobs-Sera D, Marinelli LJ, Bowman C, Broussard GW, Guerrero Bustamante C, Boyle MM, Petrova ZO, Dedrick RM, Pope WH, Science Education Alliance Phage Hunters Advancing Genome and Evolutionary Science Sea-Phages Program, Modlin RL, Hendrix RW, Hatfull GF. 2012. On the nature of mycobacteriophage diversity and host preference. Virology 434:187–201 doi: 10.1016/j.virol.2012.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pope WH, Jacobs-Sera D, Russell DA, Rubin DH, Kajee A, Msibi ZN, Larsen MH, Jacobs WR Jr, Lawrence JG, Hendrix RW, Hatfull GF. 2014. Genomics and proteomics of mycobacteriophage patience, an accidental tourist in the mycobacterium neighborhood. mBio 5(6):e02145-14. doi: 10.1128/mBio.02145-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pope WH, Anders KR, Baird M, Bowman CA, Boyle MM, Broussard GW, Chow T, Clase KL, Cooper S, Cornely KA, Dejong RJ, Delesalle VA, Deng L, Dunbar D, Edgington NP, Ferreira CM, Weston Hafer K, Hartzog GA, Hatherill JR, Hughes LE, Ipapo K, Krukonis GP, Meier CG, Monti DL, Olm MR, Page ST, Peebles CL, Rinehart CA, Rubin MR, Russell DA, Sanders ER, Schoer M, Shaffer CD, Wherley J, Vazquez E, Yuan H, Zhang D, Cresawn SG, Jacobs-Sera D, Hendrix RW, Hatfull GF. 2014. Cluster M mycobacteriophages Bongo, PegLeg, and Rey with unusually large repertoires of tRNA isotypes. J Virol 88:2461–2480. doi: 10.1128/JVI.03363-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hatfull GF. 2012. The secret lives of mycobacteriophages. Adv Virus Res 82:179–288. doi: 10.1016/B978-0-12-394621-8.00015-7. [DOI] [PubMed] [Google Scholar]
- 16.Marinelli LJ, Piuri M, Swigonova Z, Balachandran A, Oldfield LM, van Kessel JC, Hatfull GF. 2008. BRED: a simple and powerful tool for constructing mutant and recombinant bacteriophage genomes. PLoS One 3:e3957. doi: 10.1371/journal.pone.0003957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dedrick RM, Marinelli LJ, Newton GL, Pogliano K, Pogliano J, Hatfull GF. 2013. Functional requirements for bacteriophage growth: gene essentiality and expression in mycobacteriophage Giles. Mol Microbiol 88:577–589. doi: 10.1111/mmi.12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seed KD, Lazinski DW, Calderwood SB, Camilli A. 2013. A bacteriophage encodes its own CRISPR/Cas adaptive response to evade host innate immunity. Nature 494:489–491. doi: 10.1038/nature11927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bondy-Denomy J, Pawluk A, Maxwell KL, Davidson AR. 2013. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature 493:429–432. doi: 10.1038/nature11723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jordan TC, Burnett SH, Carson S, Caruso SM, Clase K, DeJong RJ, Dennehy JJ, Denver DR, Dunbar D, Elgin SC, Findley AM, Gissendanner CR, Golebiewska UP, Guild N, Hartzog GA, Grillo WH, Hollowell GP, Hughes LE, Johnson A, King RA, Lewis LO, Li W, Rosenzweig F, Rubin MR, Saha MS, Sandoz J, Shaffer CD, Taylor B, Temple L, Vazquez E, Ware VC, Barker LP, Bradley KW, Jacobs-Sera D, Pope WH, Russell DA, Cresawn SG, Lopatto D, Bailey CP, Hatfull GF. 2014. A broadly implementable research course in phage discovery and genomics for first-year undergraduate students. mBio 5(1):e01051-13. doi: 10.1128/mBio.01051-13. [DOI] [PMC free article] [PubMed] [Google Scholar]