ABSTRACT
To identify host factors relevant for severe acute respiratory syndrome-coronavirus (SARS-CoV) replication, we performed a small interfering RNA (siRNA) library screen targeting the human kinome. Protein kinases are key regulators of many cellular functions, and the systematic knockdown of their expression should provide a broad perspective on factors and pathways promoting or antagonizing coronavirus replication. In addition to 40 proteins that promote SARS-CoV replication, our study identified 90 factors exhibiting an antiviral effect. Pathway analysis grouped subsets of these factors in specific cellular processes, including the innate immune response and the metabolism of complex lipids, which appear to play a role in SARS-CoV infection. Several factors were selected for in-depth validation in follow-up experiments. In cells depleted for the β2 subunit of the coatomer protein complex (COPB2), the strongest proviral hit, we observed reduced SARS-CoV protein expression and a >2-log reduction in virus yield. Knockdown of the COPB2-related proteins COPB1 and Golgi-specific brefeldin A-resistant guanine nucleotide exchange factor 1 (GBF1) also suggested that COPI-coated vesicles and/or the early secretory pathway are important for SARS-CoV replication. Depletion of the antiviral double-stranded RNA-activated protein kinase (PKR) enhanced virus replication in the primary screen, and validation experiments confirmed increased SARS-CoV protein expression and virus production upon PKR depletion. In addition, cyclin-dependent kinase 6 (CDK6) was identified as a novel antiviral host factor in SARS-CoV replication. The inventory of pro- and antiviral host factors and pathways described here substantiates and expands our understanding of SARS-CoV replication and may contribute to the identification of novel targets for antiviral therapy.
IMPORTANCE Replication of all viruses, including SARS-CoV, depends on and is influenced by cellular pathways. Although substantial progress has been made in dissecting the coronavirus replicative cycle, our understanding of the host factors that stimulate (proviral factors) or restrict (antiviral factors) infection remains far from complete. To study the role of host proteins in SARS-CoV infection, we set out to systematically identify kinase-regulated processes that influence virus replication. Protein kinases are key regulators in signal transduction, controlling a wide variety of cellular processes, and many of them are targets of approved drugs and other compounds. Our screen identified a variety of hits and will form the basis for more detailed follow-up studies that should contribute to a better understanding of SARS-CoV replication and coronavirus-host interactions in general. The identified factors could be interesting targets for the development of host-directed antiviral therapy to treat infections with SARS-CoV or other pathogenic coronaviruses.
INTRODUCTION
Positive-stranded RNA (+RNA) viruses interact with the infected host cell at many levels during their replicative cycle, and thus far numerous host cell proteins that influence virus infection have been identified (1, 2). These include, for example, host factors recruited by the virus during the various stages of its replicative cycle and those involved in the host's defense against virus infection. Such proteins may constitute interesting targets for the development of novel antiviral strategies, as drug resistance is less likely to develop when cellular rather than viral functions are targeted. Antiviral drug resistance is a serious problem, particularly when combating RNA viruses, due to their high mutation rate and potential for rapid adaptation.
Systems biology approaches have been instrumental in advancing our knowledge of the proteins and cellular pathways that influence +RNA virus infection. For example, systematic functional genomics screens using small interfering RNA (siRNA) libraries have identified numerous host proteins with a role in the replication of important human pathogens, like West Nile virus (3), Dengue virus (4, 5), human immunodeficiency virus 1 (6), hepatitis C virus (7–12), and influenza virus (8, 13, 14). For coronaviruses, a number of relevant host proteins have been described already (15–17 and reviewed in references 2 and 18), but the use of siRNA screens to systematically identify such factors has not been reported thus far.
Coronaviruses, and some other members of the order Nidovirales (19), have the largest RNA genomes known to date (25 to 34 kb) (20), and the complexity of their molecular biology clearly distinguishes them from other +RNA virus groups. Although infection with most established human coronaviruses is associated with relatively mild respiratory symptoms (21, 22), the 2003 outbreak of severe acute respiratory syndrome (SARS) highlighted the potential of zoonotic coronaviruses to cause lethal disease in humans. The emergence of SARS-coronavirus (SARS-CoV), which likely originated from bats, initiated an outbreak that affected about 8,000 humans, with a mortality rate of approximately 10% (23). Strikingly, a similar outbreak of coronavirus-induced severe respiratory disease has been developing in a number of Arab countries since April 2012, with ∼420 of the >1,100 confirmed cases having a fatal outcome by April 2015 (http://www.who.int/). The causative agent, Middle East respiratory syndrome-coronavirus (MERS-CoV), was identified as a previously unknown member of the betacoronavirus subgroup 2c (24, 25). These recent developments stress the importance of developing antiviral approaches to combat coronavirus infections and highlight the relevance of the systematic dissection of coronavirus-host interactions.
SARS-CoV RNA synthesis, like that of many +RNA viruses (26), takes place at virus-induced membrane structures (27, 28), which in this case comprise a reticulovesicular network (RVN) of modified endoplasmic reticulum (28; reviewed in reference 29). The viral replication and transcription complexes (RTCs) are associated with this RVN, which is thought to create a suitable microenvironment for RNA synthesis and possibly also provides protection against cellular antiviral activities. The biogenesis of the RVN and the functional details of the RTC, in particular the role of cellular factors and pathways, are far from understood.
Previous studies addressed coronavirus-induced immune responses, as well as a number of specific interactions between coronaviruses and the antiviral immune response (reviewed in reference 2). Several immune evasion mechanisms were attributed to protein functions that are either conserved across CoVs or specific for certain CoV lineages. Proteins such as nonstructural protein 1 (nsp1) (30), the nsp3 papain-like proteinase (31), the nsp16 2′-O-methyltransferase (32), the nucleocapsid (N) protein (33), and the products of SARS-CoV open reading frame 3b (ORF3b) and ORF6 (34–37) have been reported to interfere with interferon (IFN) induction and/or signaling. In addition, the SARS-CoV E protein has been shown to manipulate the cellular stress response in cell culture, including the unfolded protein response and apoptosis (38).
To gain more insight into the role of host factors in the SARS-CoV replicative cycle, we set out to systematically identify kinase-regulated cellular processes that influence virus replication. Protein kinases are key regulators in signal transduction and control a wide variety of cellular processes. Thus, assessing their relevance for virus replication can provide a broad perspective on factors and pathways relevant for SARS-CoV replication, as illustrated by previous studies identifying cellular kinases as host factors influencing various stages of the replicative cycle of other +RNA viruses (5, 10, 11, 39, 40).
In this study, we have screened an siRNA library that targets the cellular kinome (779 genes) and identified 40 proviral and 90 antiviral factors whose depletion significantly reduced or enhanced SARS-CoV replication, respectively. Pathway analysis grouped several subsets of hits in specific cellular pathways, suggesting that these play an important role in the SARS-CoV-infected cell. Two strong hits from the siRNA screen, the proviral β2 subunit of the coatomer complex (COPB2) and the antiviral double-stranded RNA-activated protein kinase (PKR), were selected for independent validation and follow-up analysis, which confirmed their importance for SARS-CoV replication. In addition, several other hits from the primary screen were evaluated, and the relevance of the antiviral factor CDK6 and the proviral factor PRKCι could be confirmed. Our data offer a glimpse into the complex interplay between SARS-CoV and its host cell and provide a basis for in-depth studies that will enhance our understanding of coronavirus replication and coronavirus-host interactions.
MATERIALS AND METHODS
Cell culture, compound, viruses, and virus titration.
293/ACE2 (41) and Vero E6 cells were cultured as described previously (42). Although 293/ACE2 cells have been described as a human 293 cell-derived cell line (41), our recent work established that these cells actually must have originated from a nonhuman primate species that is closely related to the rhesus monkeys Macaca mulatta and Papio anubis (43). Cells were infected with SARS-CoV strain Frankfurt-1 (44) or green fluorescent protein (GFP)-expressing recombinant SARS-CoV (Urbani strain) (45) as described previously (42). Sodium aurothiomalate (ATM; no. 157201; Sigma) was dissolved in phosphate-buffered saline (PBS) and stored as 100 mM stock at −20°C. Virus titrations were performed essentially as described before (46). All work with infectious wild-type (wt) SARS-CoV and SARS-CoV-GFP was performed inside biosafety cabinets in a biosafety level 3 facility at Leiden University Medical Center.
siRNA library and transfection reagents.
The ON-TARGETplus SMARTpool protein kinases siRNA library that targets the mRNAs of 779 genes, comprising the complete human kinome and some additional targets, was obtained from Dharmacon. Each individual siRNA SMARTpool consisted of four siRNAs targeting the same gene. A nontargeting (scrambled) siRNA (no. D-001810-10; Dharmacon) served as a negative control, and a glyceraldehyde-3-phosphate dehydrogenase (GAPDH)-targeting siRNA (no. D-001830-10; Dharmacon) was used to routinely monitor transfection and knockdown efficiency. Stock solutions (2 μM) of siRNA SMARTpools were prepared by dissolving 0.5 nmol of an siRNA SMARTpool in 250 μl of 1× siRNA buffer (Dharmacon) according to the manufacturer's instructions. Using a 96-well pipettor (Rainin Liquidator 96), the contents of the siRNA library master plates were aliquoted into volumes appropriate for individual screening experiments. The resulting sets of 10 deep-well 96-well library plates (Greiner Bio-One) were stored at −80°C until further use.
siRNA library screening and validation.
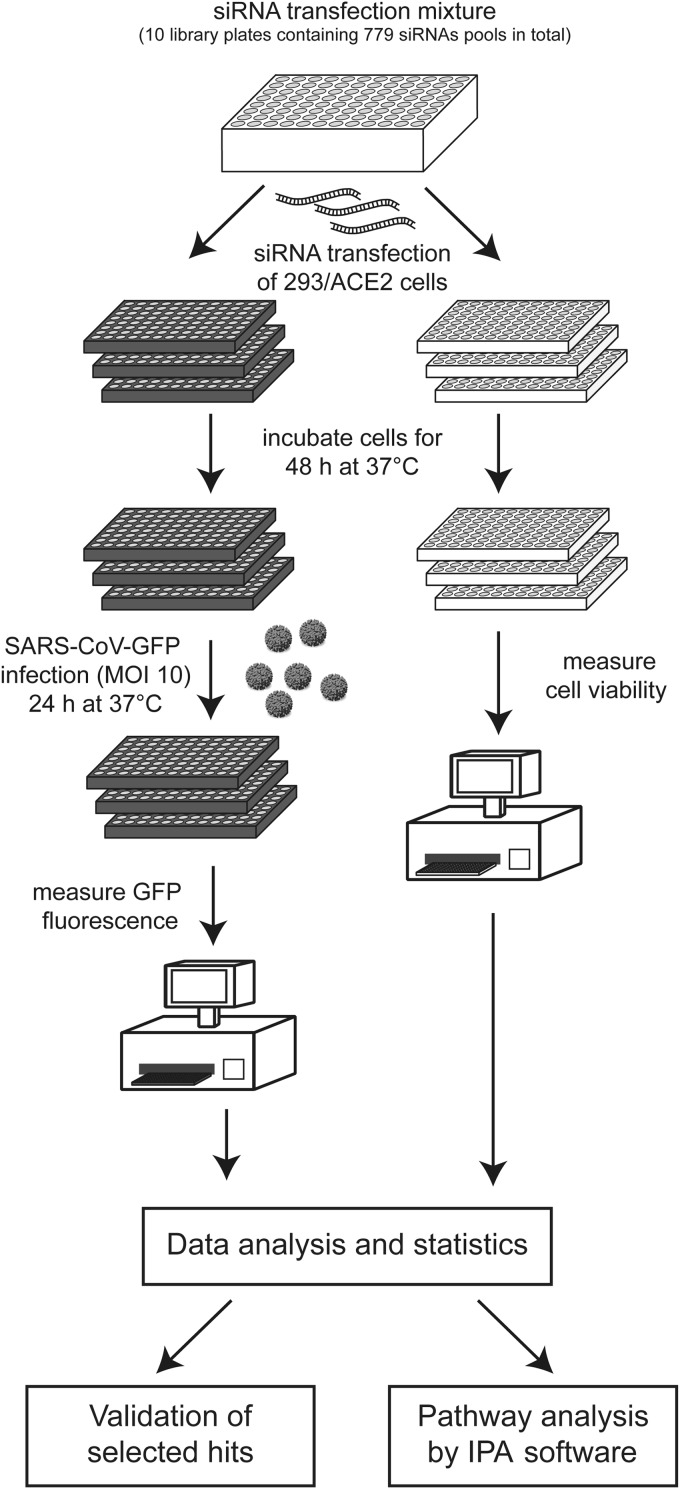
In each siRNA screen, 293/ACE2 cells in 96-well plates containing ∼104 cells per well were transfected with a 100-μl mixture containing 100 nM siRNA, 0.2 μg DharmaFECT1 (Dharmacon), Opti-MEM (Invitrogen), and antibiotic-free cell culture medium, supplemented with 8% fetal calf serum (FCS) and 2.5 mM l-glutamine, according to Dharmacon's instructions. Transfection mixes were prepared in the 10 deep-well 96-well plates that together contained the complete library of 779 siRNA SMARTpools (described above). Using the contents of these library plates, we transfected 293/ACE2 cells in black (3 wells per target) and transparent (3 wells per target) 96-well plates. For a schematic representation of the experimental setup, see Fig. 2. Transfection of individual siRNAs (ON-TARGETplus siRNAs; Dharmacon) targeting CDK6 (no. LU-003240-00), MAP2K1 (no. LU-003571-00), MAP2K3, (no. LU-003509-00), PKR (no. LU-003527-00), or siRNA SMARTpools targeting COPB1 (no. L-017940-01) and GBF1 (no. L-019783-00) was performed as described previously (42). Twenty-four hours posttransfection (p.t.), the medium was replaced, and cells were incubated for another 24 h at 37°C. At 48 h p.t., cells were infected with SARS-CoV-GFP at an MOI of 10, and 24 h later they were fixed with 3% paraformaldehyde (PFA) in PBS. GFP expression was quantified by measuring fluorescence in a 96-well plate reader (Berthold Mithras LB 940) using excitation and emission wavelengths of 485 and 535 nm, respectively. The fluorescence in wells containing mock-infected cells was used to correct for background signal.
FIG 2.
Design of siRNA library screening procedure. See the text for details.
GAPDH and cell viability assays.
At 48 h p.t., GAPDH enzyme activity in lysates of siRNA-transfected cells was measured using the KDalert GAPDH assay kit (Ambion) according to the manufacturer's instructions. Possible cytotoxic effects of siRNA transfection were analyzed (in triplicate) at 48 h p.t., using the CellTiter 96 AQueous non-radioactive cell proliferation assay (Promega). After 90 min, the reaction was terminated by the addition of 25 μl of 10% SDS, and absorbance at 490 nm (A490) was measured using a 96-well plate reader (Berthold).
Data analysis.
Raw data from GFP fluorescence and cell viability measurements were analyzed per individual screen with the Bioconductor/R package CellHTS2 (47) with minor modifications (see Results for details). Average GFP expression (n = 3) and cell viability were calculated and normalized to the signals of scrambled siRNA-transfected (control) cells. A two-sided one-sample Student's t test was used on the log2-transformed normalized values to determine the significance (P < 0.05) of the changes in GFP expression caused by siRNA transfection. The siRNA transfection was considered noncytotoxic when the normalized cell viability assay readings (A490) were above 0.85 (P < 0.05). Significance was determined using a one-sided one-sample Student's t test on the log2-transformed normalized values using μ ≤ 0.85 as the null hypothesis.
Gene silencing using lentivirus-expressed shRNAs.
Vectors for expression of short hairpin RNAs (shRNAs) targeting human COPB2 (no. TRCN-065114; accession no. NM_004766) or expression of a nontargeting (scrambled) control shRNA (no. SHC-002) were picked from the Mission TRC-1 library of shRNA-expressing lentiviruses (Sigma), and lentivirus stocks were prepared according to the manufacturer's instructions. Lentivirus particle titers were determined using a p24 enzyme-linked immunosorbent assay (ELISA) (Zeptometrix) according to the manufacturer's instructions. Wells (4 cm2) containing 8 × 104 293/ACE2 cells were transduced with shRNA-expressing lentiviruses at an MOI of 3 in culture medium containing 8 μg/ml Polybrene, and after 24 h fresh medium was given. At 72 h p.t., cells were infected with wt SARS-CoV or SARS-CoV-GFP (MOI, 0.01), and depletion of COPB2 was validated by Western blotting.
Protein analysis and antibodies.
Total cell lysates were prepared in 4× Laemmli sample buffer (100 mM Tris-HCl, pH 6.8, 40% glycerol, 8% sodium dodecyl sulfate [SDS], 40 mM DTT, 0.04 mg/ml bromophenol blue), after which samples were heated at 95°C for 15 min. Following SDS-PAGE, proteins were transferred to Hybond-LFP membranes (GE Healthcare) by semidry blotting, and membranes were blocked with 1% casein in PBS containing 0.1% Tween 20 (PBST). The following antisera against cellular proteins were used: rabbit anti-PKR (no. 610764; BD Biosciences), goat anti-COPB2 (sc-13332; Santa-Cruz), rabbit anti-CDK6 (sc-177; Santa Cruz), rabbit anti-MAP2K1 (710446; Life Technologies), rabbit anti-MAP2K3 (sc-961; Santa Cruz), and mouse monoclonal antibodies against β-actin (A5316; Sigma) and the transferrin receptor (TfR; no. 13-6890; Invitrogen). Rabbit antisera against SARS-CoV nsp8 and N protein (28, 48) were used to analyze viral protein expression. After overnight incubation with the primary antibody, membranes were probed with biotinylated secondary antibodies (rabbit anti-goat, swine anti-rabbit, or goat anti-mouse) for 1 h at RT, after which a tertiary mouse anti-biotin-Cy3 antibody was used to visualize protein bands using a Typhoon 9410 scanner (GE Healthcare).
Canonical pathway analysis.
The Ingenuity Pathway Analysis (IPA) package was used to assign hits to canonical cellular pathways. The significance of the association between the data set and the respective pathways was determined in two ways: (i) the number of molecules from the data set that mapped to a specific pathway divided by the total number of molecules in that canonical pathway (the higher the percentage of hits identified in a specific pathway, the higher the likelihood it plays a role in the viral replicative cycle); (ii) Fisher's exact test was used to determine the probability that the association between the genes in the data set and the canonical pathway is explained by chance alone.
RESULTS
Development of an siRNA screening protocol for host factors involved in nidovirus replication.
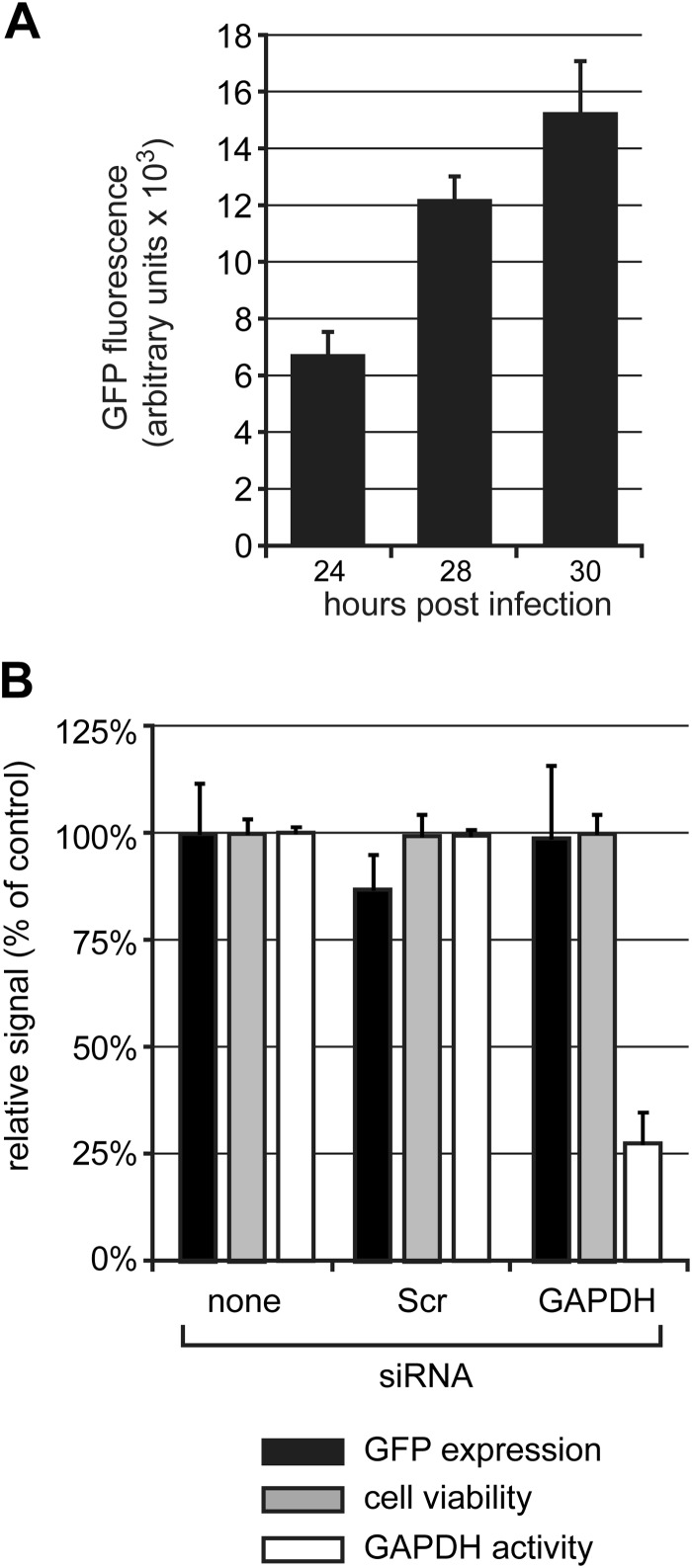
A robust protocol was developed to assess the effect of systematic knockdown of individual host kinases on the replicative cycle of SARS-CoV (this study) and the distantly related arterivirus equine arteritis virus (EAV; K. F. Wannee, A. H. de Wilde, C. Beugeling, P. ten Dijke, E. J. Snijder, M. J. van Hemert, and M. Kikkert, unpublished data), which was applied to the screening of a commercial human kinome-directed siRNA library (779 targets). We performed our siRNA screens in 293/ACE2 cells (41), which express the SARS-CoV receptor angiotensin-converting enzyme 2 and, in contrast to other cell lines tested, were found to be permissive to a combination of siRNA transfection and infection with either SARS-CoV or EAV. This property facilitated direct comparative studies between these two distantly related nidoviruses. Unfortunately, after completion of the siRNA screens, it was discovered that these cells are not of human origin but most likely have originated from an Old World monkey closely related to Papio anubis and Macaca mulatta (43). Nevertheless, because the sequence identity between the human genome and that of several Old World monkeys is 94% (49) and because pools of four siRNAs were used for each target, we believe that the consequences of the misidentification of this cell line are limited, although the chance of false-negative hits may have been somewhat increased (49). Infection of 293/ACE2 cells with SARS-CoV-GFP at an MOI of 10 yielded a robust and readily detectable GFP signal at 24 h p.i. (Fig. 1A). The GFP signal was stronger at 28 and 30 h p.i., indicating that it had not yet reached a plateau at 24 h p.i. (Fig. 1A). Therefore, we chose the latter time point to fix cells and measure GFP fluorescence, as it also should allow the identification of antiviral factors whose knockdown would increase reporter gene expression. The 293/ACE2 cells could be efficiently transfected with siRNAs, as illustrated by a consistent ∼75% reduction of GAPDH activity at 48 h p.t. using an siRNA SMARTpool targeting the GAPDH mRNA (Fig. 1B, white bars). No change in cell viability was detected by 48 h p.t. following transfection with either a scrambled siRNA or the GAPDH-specific siRNA (Fig. 1B, gray bars). When these cells subsequently were infected with SARS-CoV-GFP (MOI, 10), no significant differences in GFP expression were observed at 24 h p.i. compared to control cells that had not been transfected with siRNAs. This demonstrated that the siRNA transfection procedure per se did not adversely affect SARS-CoV-GFP replication (Fig. 1B, black bars).
FIG 1.
Viability and susceptibility to SARS-CoV infection of siRNA-transfected 293/ACE2 cells. (A) 293/ACE2 cells were infected with SARS-CoV-GFP (MOI, 10), and at 24, 28, and 30 h p.i., cells were fixed and GFP fluorescence was measured. (B) 293/ACE2 cells were transfected with siRNAs targeting GAPDH mRNA or a scrambled control siRNA (Scr). At 48 h p.t., cells were infected with SARS-CoV-GFP (MOI, 10), and 24 h later cells were fixed and GFP expression was measured (black bars). Cell viability (dark gray bars) was analyzed at 48 h after siRNA transfection, and knockdown of GADPH expression was monitored by measuring enzymatic activity (light gray bars). All values were normalized to those obtained with nontransfected control cells (100%).
Kinome-wide siRNA screens for host factors involved in SARS-CoV replication.
A human kinome-directed siRNA screen was performed to identify host cell kinases that affect SARS-CoV-GFP replication according to the experimental set-up outlined in Fig. 2. For each independent siRNA screening experiment, we used a set of ten 96-well library plates, each containing approximately 80 specific siRNA SMARTpools and several controls. Transfection mixes (final concentration of 100 nM siRNA) were prepared in these library plates and their contents was used to transfect (per library plate) 293/ACE2 cells in three black and three transparent 96-well plates. Forty-eight hours after siRNA transfection, the black plates were infected with the SARS-CoV-GFP reporter virus (MOI, 10), and at 24 h p.i. GFP expression was measured by fluorometry. At the moment of infection, the transparent plates were used to monitor (potential) cytotoxic effects of siRNA transfection using a colorimetric cell viability assay. The complete siRNA screen, i.e., the viability controls (in triplicate for each siRNA SMARTpool) and the quantitation of SARS-CoV-driven GFP expression (in triplicate), was repeated in three independent experiments. The data, obtained from a 96-well plate reader, was processed with the Bioconductor/R package CellHTS2 as described previously (47). Experimental controls were assigned, and the NPI method (normalized percent of inhibition) was used to normalize GFP fluorescence values to those of scrambled siRNA-transfected cells and to correct for plate-to-plate variation. Subsequently, the GFP data were transformed to a multiplicative scale (the value obtained using scrambled siRNA-transfected cells was set to 1). The results for each replicate library screen next were summarized and used for further data analysis, including the assignment of gene identifiers to each well. Finally, the data from the three independent library screens were combined and summarized.
Host cell kinases were considered to have a proviral effect when their siRNA-mediated knockdown reduced the GFP signal (negative score values), and kinases were considered antiviral when the GFP signal increased upon their knockdown (positive score values). Graphical representations of the hit distribution per plate were visually inspected in order to minimize the chance of false-positive or false-negative hits due to major (technical) artifacts (data not shown).
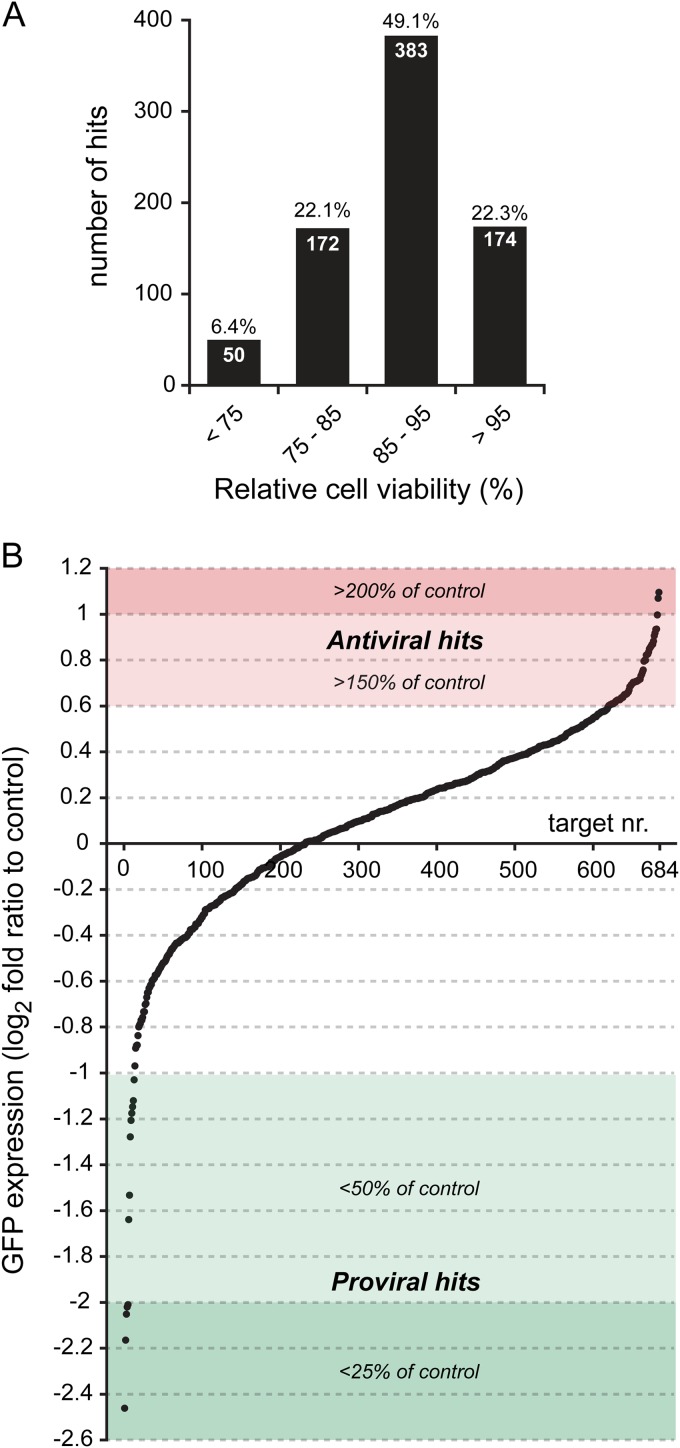
Using scrambled siRNA-transfected control cells as a reference, the knockdown of most cellular kinases was found to be noncytotoxic within the time frame of this experiment (Fig. 3A; also see Data Set S1 in the supplemental material). Transfection of siRNAs was considered to be cytotoxic when the viability of cells transfected with a target-specific siRNA pool was <85% of the viability of control cells transfected with scrambled siRNAs (Fig. 3A). Using this criterion, 222 out of 779 (28.5%) transfections with the specific siRNA pools appeared to be toxic to the cells. A minor fraction (50 targets; 6.4%) appeared to be highly detrimental (normalized viability value below 75%). To prevent false-positive proviral hits due to a general negative effect on cell viability or cell division, we excluded all targets whose knockdown was associated with viability measurements below 85%. Such data filtering was not applied for antiviral hits (i.e., hits whose knockdown enhanced GFP expression), since siRNA-induced cytotoxicity is expected to inhibit virus replication and should not give rise to false-positive antiviral hits.
FIG 3.
Results of the siRNA screens for host factors influencing SARS-CoV replication. (A) Viability assays were done at 48 h p.t., and data were normalized to the measurements for control cells transfected with scrambled siRNA (100%). Data were binned into 4 viability categories, as indicated below the x axis, and the number in each bar is the absolute number of siRNA targets within that category. The fraction of the total number of targets (779) in each category is indicated above each bar. For each siRNA pool in the library, the viability data are the averages from nine measurements, resulting from three independent library screens. (B) The plot shows the distribution of the log2-transformed values of GFP reporter gene expression by SARS-CoV-GFP in siRNA-transfected cells, normalized to the GFP signal of infected control cells that were transfected with scrambled siRNA. Targets were ranked based on the magnitude of the effect of their knockdown on SARS-CoV replication. Targets were considered to have a robust antiviral effect when their knockdown increased reporter gene expression to at least 150% (red area above the x axis). Proviral hits, whose knockdown induced an at least 2-fold reduction in GFP expression, are depicted in the green area below the x axis. Proviral targets whose knockdown reduced cell viability to below 85% were excluded (see main text), leaving a total of 684 targets included in the final analysis. The plot represents the averages from three library screens (each done in triplicate).
Proviral and antiviral proteins and pathways in SARS-CoV-GFP infection.
After exclusion of cytotoxic siRNA SMARTpools that decreased GFP expression (described above), the remaining 684 targets were ranked on the basis of the GFP signal in host factor-depleted SARS-CoV-GFP-infected cells compared to control cells (Fig. 3B). Targets were qualified as antiviral or proviral hits if GFP expression differed significantly from that in infected control cells transfected with the scrambled siRNA pool (P < 0.05). Knockdown of the majority of the targets (552 proteins) did not significantly alter GFP reporter gene expression (P > 0.05). However, as is not uncommon in this type of screening experiment and considering the issue with the origin of the cell line used (described above), we cannot formally exclude that our results were influenced to some extent by insufficient knockdown of certain target genes by the siRNA pools in the library.
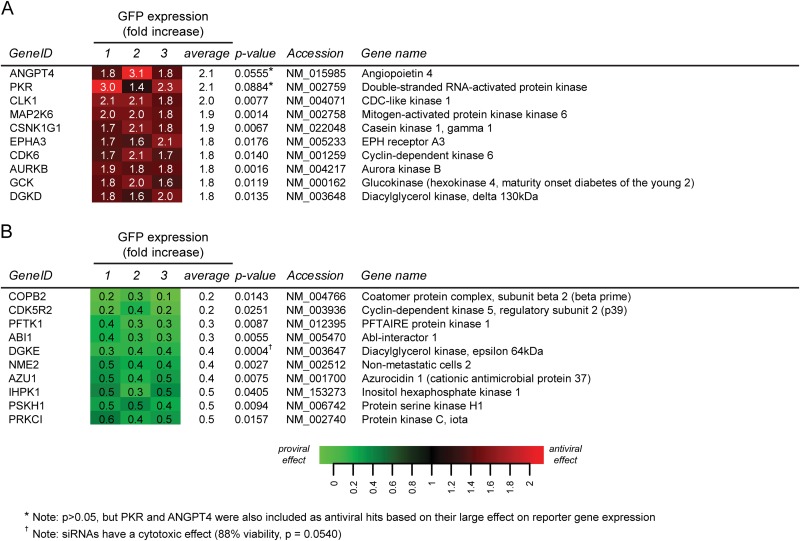
Using the criteria outlined above, a total of 90 cellular proteins (19.4% of all targets) were identified as antiviral factors, since their depletion significantly increased GFP expression, although for most of them it did so less than 2-fold. The 10 best antiviral hits are depicted in Fig. 4A (the complete data set is provided in Data Set S1 in the supplemental material). Forty proviral factors were identified, and the knockdown of nine of those reduced GFP expression by more than 2-fold (Fig. 4B) (for the complete data set, see Data Set S1). Although according to the criteria formulated above (P < 0.05), ANGPT4 (214%; P = 0.0555) and PKR (210%; P = 0.0884) formally did not qualify as antiviral hits, we have included these proteins in view of the exceptionally strong stimulation of GFP expression triggered by their knockdown (Fig. 4A). Furthermore, since its knockdown resulted in an almost 3-fold decrease of the GFP signal (35%; P = 0.0004), DGKE was included as a proviral hit, despite the fact that the viability assay did not rigorously exclude cytotoxic effects for this siRNA pool (viability, 88%; P = 0.0540).
FIG 4.
Heat maps of pro- and antiviral hits identified in this study. (A) List of the 10 most prominent antiviral (A) and proviral (B) hits. For each target, the P value, accession number, and gene name are shown. Each data point represents the result of a single library screen and is the average from 3 replicates that were done in each screen. The full hit lists can be found in Data Set S1 in the supplemental material.
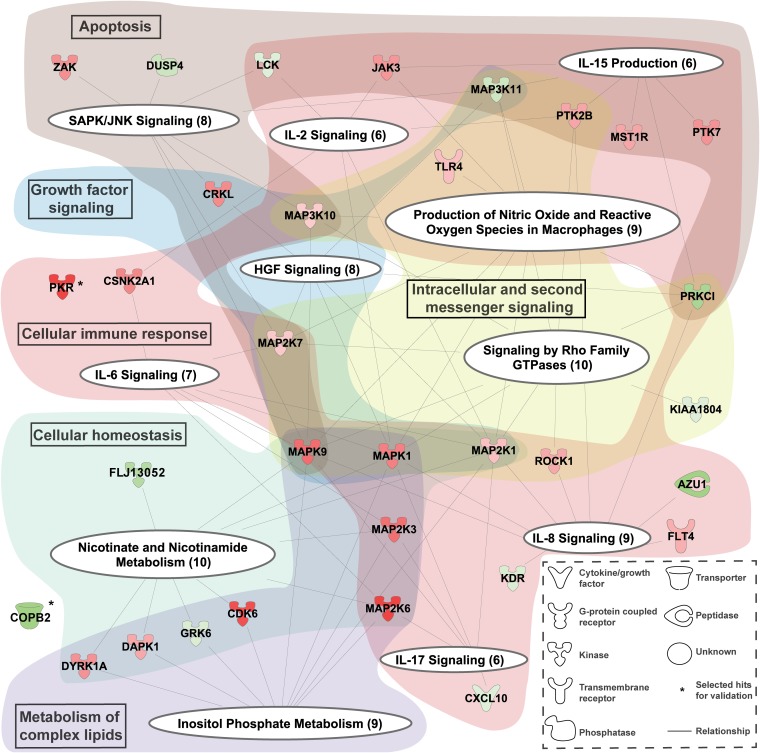
The pro- and antiviral hits identified in the siRNA screen were mapped to cellular pathways using the IPA software package. Figure 5 shows the canonical pathways and more general functional categories (highlighted in color) in which the proviral (green) and antiviral (red) hits were strongly represented (P < 0.05). These pathways included apoptosis, cellular immune response, growth factor signaling, cellular homeostasis, metabolism of complex lipids, and intracellular and second messenger signaling.
FIG 5.
Cellular pathways influencing SARS-CoV-GFP replication. Graphical representation of the canonical pathways (white ellipses) identified in the siRNA library screen for cellular factors affecting SARS-CoV replication. All proviral (green) and antiviral (red) hits (for a complete list, see Data Set S1 in the supplemental material) for which depletion significantly altered SARS-CoV-GFP replication were used to identify cellular pathways by IPA in which the hits were clearly overrepresented, and only those pathways and the hits represented in them are shown here. The hits are represented by nodes with lines linking them to one or more canonical pathways. The color intensity of the nodes indicates the strength of the pro- or antiviral effect (log2 ratio of GFP expression normalized to infected control cells), e.g., factors with a stronger antiviral effect have a more intense red color. The identified canonical pathways were clustered into more general categories that are indicated by text boxes in the colored background shading.
Evaluation of antiviral hits.
An unexpectedly large number of antiviral hits was identified in the primary siRNA screen, although for most of them knockdown resulted in a less than 2-fold increase in SARS-CoV-driven GFP expression. To assess the overall quality of our siRNA screen and the reliability of the identification of antiviral hits in particular, we selected a set of strong and weak antiviral hits for further evaluation, namely, PKR, ANGPT4, CLK1 (>2-fold increase in GFP signal), CDK6 (1.8-fold increase), MAP2K3 (1.6-fold increase), and MAP2K1 (1.2-fold increase). Per target, a deconvoluted set of four individual siRNAs was used for additional knockdown experiments, after which SARS-CoV-GFP replication was quantified, and then the knockdown efficiency at the protein level for each target was evaluated by Western blotting (Fig. 6). We checked whether there was strong similarity between the target sequence of our (human) siRNAs and the corresponding Macaca mulatta sequence, considering the origin of the 293/ACE2 cells (described above). This was the case for all siRNAs except for siRNA CDK6 number 4, which was excluded from further analysis. Using commercially available antisera against the human proteins, we were unable to reliably detect endogenous expression of ANGPT4 and CLK1. Therefore, it remains uncertain whether ANGPT4 and CLK1 are true antiviral or false-positive hits, as we could not determine knockdown levels and correlate these to effects on virus replication (data not shown).
FIG 6.
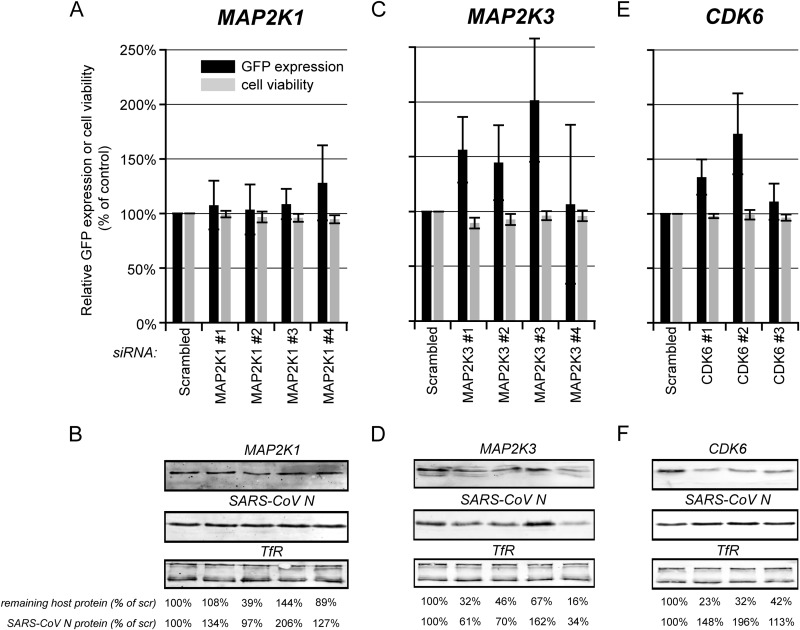
Evaluation of the antiviral hits CDK6, MAP2K1, and MAP2K3. 293/ACE2 cells were transfected with four individual siRNAs targeting, MAP2K1 (A and B), or MAP2K3 (C and D) or three CDK6-specific siRNAs (E and F). A nontargeting scrambled siRNA was used as a control. At 48 h p.t. cells were infected with SARS-CoV-GFP at an MOI of 10 (A, C, and E) and fixed 24 h later, and GFP fluorescence (black bars) was quantified and normalized to the value measured in infected, scrambled siRNA-transfected cells (100%). The effect of siRNA transfection on cell viability was analyzed in parallel (gray bars), and values were normalized to those of scrambled siRNA-transfected control cells (100%). Each experiment was repeated at least three times (averages ± standard deviations [SD]). In parallel, siRNA-transfected cells were infected with wt SARS-CoV (MOI, 5), and at 8 h p.i., SARS-CoV N expression was monitored by Western blotting (B, D, and F). TfR was used as a loading control. Knockdown levels of the host proteins were analyzed by Western blotting (B, D, and F). The amount of SARS-CoV N protein and remaining quantity of host protein compared to that of scrambled siRNA-transfected cells (100%) is shown below each lane. All experiments were repeated at least twice.
The mitogen-activated protein kinases (MAPKs) were relatively highly represented among the antiviral hits (Fig. 5; also see Data Set S1 in the supplemental material); therefore, we included MAP2K1 and MAP2K3 in our secondary evaluation. MAP2K1 was a weak antiviral hit in the primary screen, as its depletion led to an ∼1.2-fold increased GFP expression. In validation experiments with individual siRNAs, we also observed a small but nonsignificant increase in SARS-CoV-driven GFP expression (Fig. 6A). Western blot analysis of siRNA-transfected cells that were infected with wt SARS-CoV revealed poor knockdown efficiencies, and a clear correlation between the level of MAP2K1 and SARS-CoV N protein expression could not be established (Fig. 6B). The siRNA that gave the best knockdown of MAP2K1 (number 2) had no effect on GFP expression, suggesting that this weak antiviral hit was a false positive in the primary screen. Knockdown of MAP2K3 resulted in a >1.6-fold increased SARS-CoV-driven GFP expression in the primary screen (see Data Set S1). In cells transfected with a deconvoluted set of individual siRNAs targeting the MAP2K3 mRNA, we observed a significant increase in GFP expression for 3 out of 4 siRNAs (Fig. 6C). However, in siRNA-transfected cells that were infected with wt SARS-CoV, only the introduction of siRNA number 3 led to clearly enhanced SARS-CoV N protein expression (Fig. 6D). The other siRNAs actually reduced expression of N protein, with an apparent correlation between the remaining percentage of MAP2K3 and N protein levels. Based on these results, MAP2K3 could not be confirmed as an antiviral hit.
We were able to detect expression of cyclin-dependent kinase 6 (CDK6; 1.8-fold increase in GFP expression in the primary screen) and found that CDK6 siRNA numbers 1 and 2 reduced protein levels by at least two-thirds (Fig. 6F). Transfection with the same siRNAs significantly enhanced SARS-CoV-GFP replication (Fig. 6E), and in cells infected with wt SARS-CoV, this led to an ∼1.5- to 2-fold increase in N protein levels (Fig. 6F). These results suggest CDK6 is a bona fide antiviral hit, as its depletion leads to a moderate but significant and reproducible increase in SARS-CoV replication.
Taken together, the results of our validation experiments suggest that the antiviral hits identified in the primary screen should be considered with caution, as several of them may have been false positives, especially those that had a moderate (but significant) effect in the primary screen.
Validation of PKR as an antiviral factor in SARS-CoV replication.
PKR was one of the strongest of the 90 antiviral hits that were identified in the primary siRNA library screen. In two independent follow-up experiments with reordered PKR-specific siRNA SMARTpools, a more than 2-fold increase in GFP expression by SARS-CoV-GFP was observed (data not shown), suggesting that PKR is a bona fide antiviral hit. PKR is a serine/threonine protein kinase that is activated by double-stranded RNA (dsRNA), a hallmark of RNA virus infection, and the activated form of PKR blocks translation initiation through eIF-2α phosphorylation (reviewed in reference 50).
To further validate the antiviral role of PKR in SARS-CoV replication, a deconvoluted set of four single PKR-directed siRNAs was used, and transfection of 293/ACE2 cells with three of these siRNAs (numbers 2, 3, and 4) significantly increased SARS-CoV-driven GFP expression (Fig. 7A, black bars). Cell viability was slightly reduced after transfection with these PKR-directed siRNAs, in particular using siRNA number 2, which caused a 14% reduction in cell viability (Fig. 7A, gray bars). Nevertheless, despite the fact that this siRNA adversely affected cell viability, an increase rather than a decrease of SARS-CoV-driven GFP expression was observed.
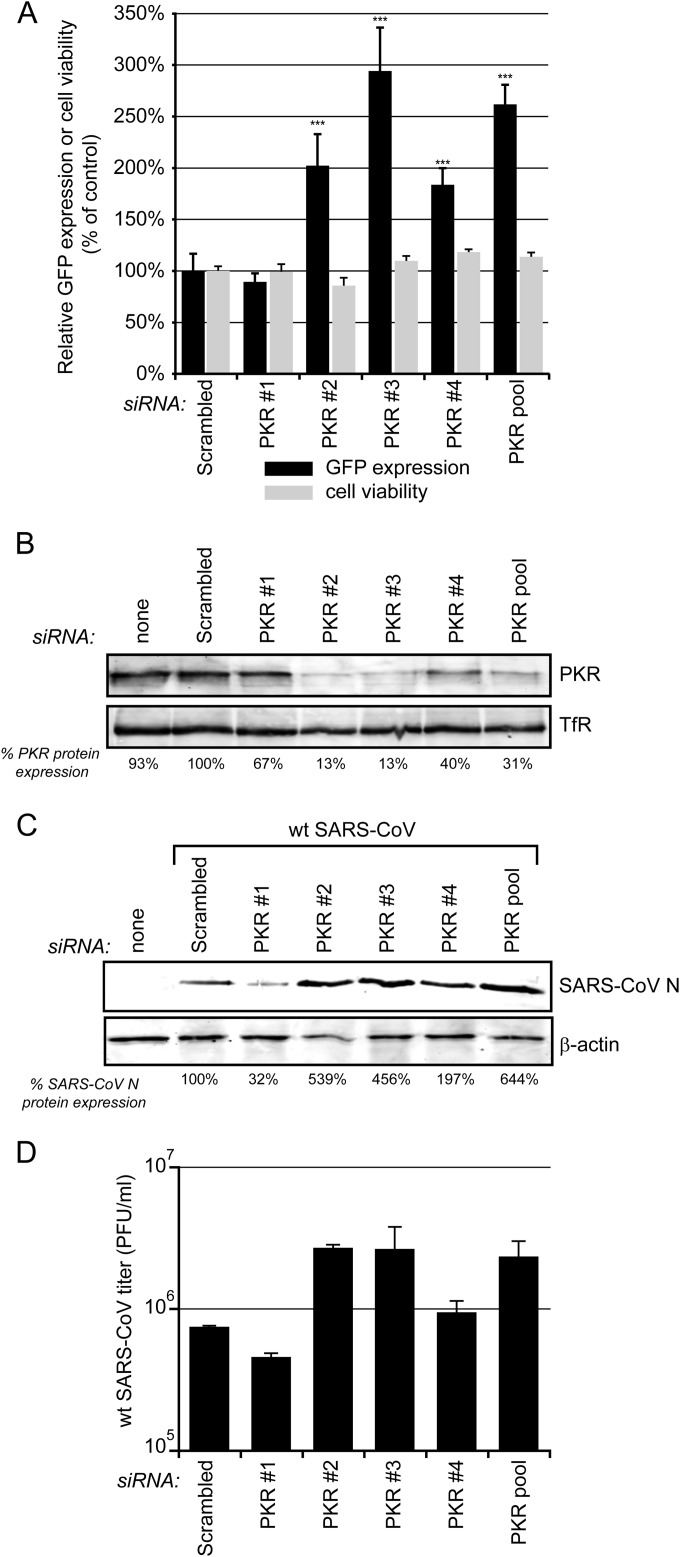
FIG 7.
Validation of PKR as an antiviral factor in SARS-CoV replication. 293/ACE2 cells were transfected with four individual siRNAs targeting PKR or a scrambled control siRNA. (A) At 48 h p.t. cells were infected with SARS-CoV-GFP (MOI, 10) and fixed 24 later, and GFP fluorescence (black bars) was quantified and normalized to the value measured in infected, scrambled siRNA-transfected cells (100%). The effect of siRNA transfection on cell viability was analyzed in parallel (gray bars), and values were normalized to those of scrambled siRNA-transfected control cells (100%). Averages ± SD are given (***, P < 0.001). (B) Knockdown of PKR expression at 48 h p.t. was monitored by Western blotting, and the percentage of PKR remaining compared to that of scrambled siRNA-transfected cells is shown below each lane. TfR was used as a loading control. (C) Cells transfected with PKR-specific siRNAs and control cells were infected with SARS-CoV (MOI, 0.01), and 24 h later these cells were lysed to assess SARS-CoV N levels by Western blotting (shown below the panels as a percentage of the control), using β-actin as the loading control. (D) Virus titers in the 24-h p.i. culture supernatants of wt SARS-CoV-infected cells (MOI of 0.01) transfected with PKR-specific or scrambled siRNA. All experiments were repeated at least twice.
Transfection with PKR-specific siRNAs reduced PKR levels in 293/ACE2 cells up to 87% compared to control cells, depending on the siRNA used (Fig. 7B). To verify that PKR knockdown increased wt SARS-CoV replication, siRNA-transfected 293/ACE2 cells were infected with wt SARS-CoV and viral protein expression was analyzed by Western blotting. In line with the effect of PKR siRNA number 2 on 293/ACE2 cell viability (Fig. 7A), cells transfected with this siRNA contained reduced levels of β-actin, which was used as loading control (Fig. 7C, lower). Transfection with two of the four individual PKR-directed siRNAs (2 and 3) clearly increased the expression of SARS-CoV N protein (Fig. 7C, upper) and also led to an ∼1-log increase in infectious progeny titers (Fig. 7D). Taken together, the increases in GFP signal, N expression, and infectious progeny titer correlate well with the magnitude of PKR knockdown, which confirms a strong antiviral role for PKR in SARS-CoV-infected cells.
Confirmation of a proviral role for protein kinase C iota in SARS-CoV replication.
For the evaluation of the proviral hits of the primary screen (Fig. 4B), four hits were selected that caused either a 5-fold reduction (COPB2 and CDK5R2) or a more moderate 2-fold reduction in SARS-CoV-driven GFP expression (IHPK1 and PRKCι). We were unable to detect endogenous CDK5R2 and IHPK1 by Western blotting; therefore, we could not validate the proviral role of these two host factors (data not shown). The proviral role of PRKCι and COPB2 could be validated as discussed in the sections below.
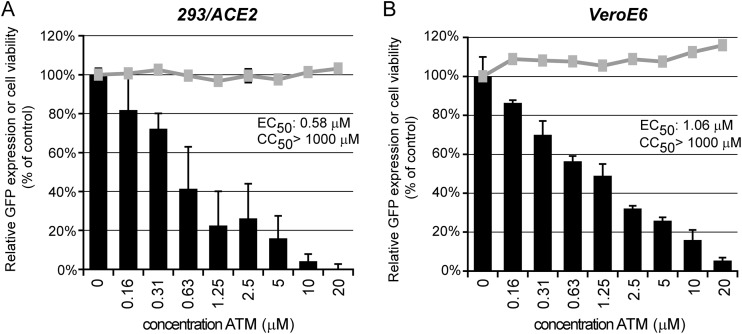
The proviral effect of protein kinase C iota (PRKCι) was validated using the chemical inhibitor sodium aurothiomalate (ATM), which blocks the interaction between PRKCι and other PB1 domain-containing proteins (51, 52). VeroE6 and 293/ACE2 cells infected with SARS-CoV-GFP were treated with 0.13 to 20 μM ATM, starting 2 h prior to infection. In both 293/ACE2 (Fig. 8A) and in VeroE6 cells (Fig. 8B), SARS-CoV-mediated GFP expression was efficiently inhibited by ATM in a dose-dependent manner, with 50% effective concentrations (EC50s) of 0.58 and 1.06 μM, respectively. No cytotoxicity was observed at the ATM concentrations used (Fig. 8).
FIG 8.
Validation of PRKCι as proviral host factor. 293/ACE2 (A) or VeroE6 (B) cells in 96-well plates were infected with SARS-CoV-GFP (MOI, 10). Treatment with 0.13 to 20 μM sodium aurothiomalate (ATM) was started 2 h prior to infection, and cells were fixed at 18 h p.i. (VeroE6) or 24 h p.i. (293/ACE2). GFP reporter gene expression (black bars) was measured and normalized to the signal in untreated control cells (100%). The gray lines show the effect of ATM on the viability of mock-infected cells normalized to the viability of solvent-treated control cells. Graphs show the results (averages and SD) from a representative experiment performed in quadruplicate. Both experiments were repeated at least twice.
COPB2 and proteins of the early secretory pathway are important for SARS-CoV replication.
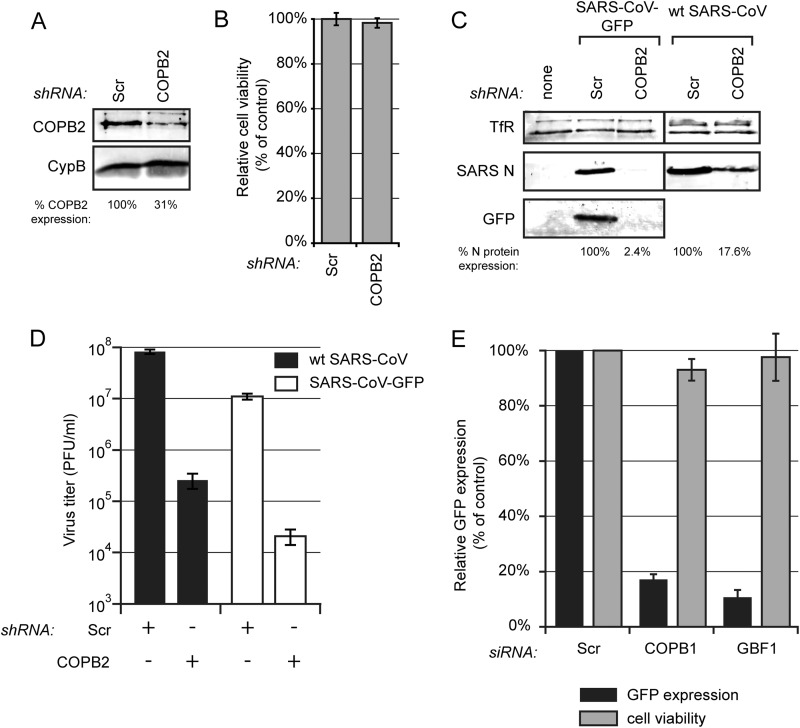
COPB2 (or β′-COP) was identified as the strongest proviral hit in our screen, as its knockdown resulted in an 82% decrease of GFP expression (Fig. 4B). The coatomer protein complex, of which COPB2 is a subunit, contains a total of seven protein subunits (α-, β-, β′-, γ-, δ-, ε-, and ζ-COP) and drives the formation of COPI-coated vesicles, which function in retrograde transport in the early secretory pathway (53). To validate its role as a proviral host factor in SARS-CoV replication, COPB2 was depleted by transducing 293/ACE2 cells with lentiviruses expressing COPB2 mRNA-specific shRNAs. This reduced COPB2 levels by ∼70% compared to levels of control cells transduced with a lentivirus expressing a scrambled shRNA (Fig. 9A), a reduction that did not affect cell viability (Fig. 9B). Subsequent infection of COPB2-depleted cells with SARS-CoV-GFP resulted in a strong decrease of N protein and GFP expression (Fig. 9C, left). To exclude that the observed effect was an artifact caused by the use of the GFP reporter virus, we also analyzed viral protein expression and virus yield in COPB2-depleted cells infected with wt SARS-CoV. As for SARS-CoV-GFP, a clear reduction in N protein expression then also was observed in COPB2-depleted cells compared to the level in cells transduced with a lentivirus expressing a scrambled shRNA (Fig. 9C, right). Titration of culture supernatants from SARS-CoV-GFP-infected cells and wt SARS-CoV-infected cells revealed a 2- to 3-log reduction for both viruses upon COPB2 depletion (Fig. 9D).
FIG 9.
Proteins of the early secretory pathway are important for SARS-CoV replication. (A) 293/ACE2 cells were transduced with lentiviruses expressing a COPB2 mRNA-specific or a scrambled shRNA. Knockdown of COPB2 expression at 48 h p.t. was monitored by Western blotting with a COPB2-specific antiserum, and cyclophilin B (CypB) was used as a loading control. (B) Viability of COPB2-depleted 293/ACE2 cells was analyzed at 48 h after transduction (percentage of control cells transduced with lentiviruses expressing a scrambled shRNA). (C and D) COPB2-depleted and control cells were infected with either SARS-CoV-GFP or wt SARS-CoV (MOI of 0.01). (C) SARS-CoV protein expression at 32 h p.i. (SARS-CoV-GFP) or 24 h p.i. (wt SARS-CoV) was analyzed by Western blotting with N-specific and GFP-specific antisera, using the TfR protein as a loading control. SARS-CoV N protein expression was quantified and normalized to that in scrambled siRNA-transfected cells (100%) as indicated under each lane. (D) SARS-CoV-GFP (white bars) and wt SARS-CoV (black bars) progeny titers in the culture supernatants of control or COPB2-depleted cells at 32 h p.i. (SARS-CoV-GFP) or 24 h p.i. (wt SARS-CoV). (E) Normalized GFP expression by SARS-CoV-GFP in 293/ACE2 cells transfected with siRNA SMARTpools targeting COPB1, GBF1, or a scrambled control siRNA. Cells were infected 48 h p.t. at an MOI of 10, and 24 h later GFP fluorescence was quantified and normalized to that in infected cells transfected with a scrambled siRNA. GFP fluorescence data are the averages from three independent experiments.
To further substantiate the importance of COPI-coated vesicles for SARS-CoV replication, another component of the coatomer protein complex, subunit β1 (COPB1), was depleted by transfection of 293/ACE2 cells with a COPB1 mRNA-specific siRNA SMARTpool. Depletion of COPB1 resulted in an 83% reduction of SARS-CoV-driven GFP expression (Fig. 9E). The formation of COPI-coated vesicles is mediated through activation of ADP ribosylation factor 1 (Arf1) by Golgi-specific brefeldin A-resistant guanine nucleotide exchange factor 1 (GBF1) (54). Therefore, we also analyzed the importance of GBF1. GFP reporter gene expression by SARS-CoV-GFP was reduced by 89% in 293/ACE2 cells that had been depleted for GBF1 (Fig. 9E). GBF1 and COPB1 depletion had no significant effect on cell viability (Fig. 9E). Taken together, these data suggest that COPB2 and COPI-coated vesicles play an essential role in SARS-CoV replication.
DISCUSSION
In the past decade, functional genomics studies have systematically identified host factors that can influence the replication of diverse +RNA viruses (3, 4, 8–10, 39, 55). Here, we describe a human kinome-wide siRNA screen that aimed to identify factors influencing the entry and replication of SARS-CoV. To our knowledge, this is the first report on a systematic functional genomics study of this kind for any coronavirus. As kinases are key regulators of many cellular processes, the pro- and antiviral factors identified in this study should pinpoint cellular pathways that are important for SARS-CoV replication.
After we had completed our screen, we unfortunately discovered that the 293/ACE2 cells used were not of human origin but must have derived from a nonhuman primate, probably an Old World monkey closely related to Macaca mulatta. This may have increased the number of false-negative hits due to mismatches between siRNAs designed to target human genes and the sequence of the homologous monkey mRNAs. However, the human and Macaca mulatta genomes are 94% identical, and even a nucleotide mismatch in an siRNA would not automatically render it inactive, as it might still silence gene expression by blocking translation of the mRNA (56). Therefore, we concluded that, despite this postscreening complication, we should still be able to identify host factors that are relevant for coronavirus infection. Moreover, it is important to stress that the use of a nonhuman primate cell line should not have increased the number of false-positive hits. The cell line was highly susceptible to both SARS-CoV and EAV infection and could be efficiently transfected with siRNAs. This allowed us to perform siRNA screens for host factors involved in the replication of these two distantly related nidoviruses and to directly compare hits (Wannee et al., unpublished).
A recombinant SARS-CoV-GFP reporter virus, in which ORF7a and ORF7b were replaced by the GFP gene, was used in our screen in order to conveniently quantify the effect of gene knockdown on virus replication. The SARS-CoV ORF7a protein is known to interact with the structural envelope (E), membrane (M), and spike (S) proteins (57), and some studies suggest that it is involved in specific virus-host interactions (35, 58, 59). However, the replication efficiency in cell culture of SARS-CoV mutants lacking both ORF7a and ORF7b is unchanged (60, 61), and there were none of the differences in replication kinetics, morbidity, and mortality observed in a hamster infection model (61). Although the deletion of the two accessory protein genes likely has affected the results of our primary screen only marginally, a wt SARS-CoV isolate was used in several of the validation experiments to rule out artifacts caused by the lack of expression of ORF7a and ORF7b. In all cases tested, we did not find major differences between wild-type virus and the deletion mutant.
For SARS-CoV, screening of the kinome-directed library of 779 siRNA SMARTpools resulted in the identification of 90 antiviral and 40 proviral proteins. Canonical cellular processes and pathways in which these factors were represented strongly include inositol phosphate metabolism, signaling by Rho family GTPases, and stress-activated protein kinase/Jun N-terminal protein kinase (SAPK/JNK) signaling (Fig. 5). Many of the hits also could be mapped to the interleukin-2 (IL-2), IL-6, IL-8, and IL-17 signaling pathways, which have been implicated previously in controlling coronavirus infection and coronavirus-induced inflammation (reviewed in reference 2). For example, the SARS-CoV spike (S) protein was shown to induce the expression of the proinflammatory cytokine IL-8 (62), and IL-6 and IL-8 levels were elevated in the serum of SARS-CoV-infected patients (62, 63). Furthermore, mouse hepatitis virus (MHV) and infectious bronchitis virus (IBV) infections were reported to upregulate the synthesis of these same cytokines (64, 65). Although our siRNA library screen did not target interleukins directly, the identification of (kinase-regulated) interleukin signaling pathways is in line with these earlier studies and emphasizes their importance in SARS-CoV infection.
A list of host proteins involved in SARS-CoV infection is no more than a good starting point for follow-up studies into the role of individual host protein or pathways in the virus replication cycle. Previous studies on other viruses, e.g., HIV-1, showed a very limited overlap between hits from independent siRNA screens performed in different laboratories (66), highlighting the importance of validation and follow-up studies. To judge the overall quality of our siRNA screen, six hits with various impacts on SARS-CoV-driven GFP expression were chosen for validation. Four of these, PKR, CDK6, COPB2, and PRKCι (Fig. 6 to 9), could be confirmed. The weak antiviral hits MAP2K1 and MAP2K3 could not be confirmed, as in follow-up experiments knockdown could not be achieved or did not convincingly affect SARS-CoV replication, respectively. Interestingly, the diacylglycerol kinase was highly represented as hits in the primary screen (6 of 8 targets; see Data Set S1 in the supplemental material). Although not included in follow-up experiments presented here, a related study that aimed to identify host factors with a general effect in nidovirus infection (Fig. 4 and Wannee et al., unpublished) confirmed that one of these hits, diacylglycerol kinase epsilon (DGKE), plays a role in the SARS-CoV replication cycle.
MAP2K3, a kinase that acts in the p38 MAPK module, was a moderate hit in our primary screen. This MAP kinase signaling pathway is involved in multiple processes, like regulation of inflammatory responses, cell proliferation, and cell cycle progression (reviewed in reference 67). This pathway has been implicated in the replication of other coronaviruses, but its exact role still is not fully understood. Activation of p38 MAPK promotes MHV replication (65), and chemical inhibition of the p38 MAPK pathway restricts HCoV-229E replication (68). Overexpression of the SARS-CoV ORF3a (69) and ORF7a (35) proteins activates the p38 MAPK signaling pathway, but the role of this pathway in SARS-CoV-infected cells remains unclear. Our screen identified several proteins from MAPK signaling cascades, but our validation studies suggested that MAP2K3 was a false-positive hit. This is supported by the fact that the inhibitor SB203580 had no effect on SARS-CoV replication in cell culture (data not shown). This compound was previously shown to block HCoV-229E infection in cell culture (68) and to increase the survival of SARS-CoV-infected mice through reducing the SARS-CoV-induced inflammatory response (70).
CDK6, a kinase involved in cell cycle progression from G1 to S phase (71), was confirmed as an antiviral hit. Depletion of CDK6 results in G1 phase cell cycle arrest. In addition, CDK6 also is involved in NF-κB signaling and coregulation of inflammatory genes by binding and activation of the p65 subunit of NF-κB (72, 73). Consequently, besides the effect on the cell cycle, depletion of CDK6 also might reduce the inflammatory response against virus infection. A recent study by DeDiego et al. highlighted the relevance of NF-κB-mediated inflammation in SARS-CoV-infected mice (74). Several laboratories studied the (antiviral) role of CDK6, and cell cycle progression in general, in coronavirus replication. For MHV, it has been shown that upon high-MOI inoculation, the virus induces cell cycle arrest in the G0/G1 phase to promote its replication. In addition, CDK6 is downregulated in MHV-infected 17Cl1 cells (75). Similar observations have been made for SARS-CoV, with the N protein limiting cell cycle progression by reducing CDK4 and CDK6 kinase activity (76). Overexpression of the SARS-CoV ORF7a protein induced cell cycle arrest in the G0/G1 phase; however, this was not associated with inhibition of CDK4 and CDK6 activity (59). In our study, the antiviral role of CDK6 was confirmed, and the observed antiviral effect is in line with previous studies.
As pointed out above, our screen yielded a relatively high proportion of antiviral hits, although their effect on SARS-CoV replication, while being statistically significant, generally was limited. Based on our assessment of some of these moderate hits, at least some of them must have been false positives. Knockdown of PKR had the strongest effect (∼2-fold increase in GFP expression) on SARS-CoV replication, and this hit could be confirmed independently, as three out of four individual PKR-directed siRNAs induced a clear increase in SARS-CoV protein expression and virus yield (Fig. 7C and D). PKR is one of four mammalian kinases that can phosphorylate eIF-2α in response to stress signals (the others being the PKR-like endoplasmic reticulum kinase [PERK], GCN2, and HRI). Many virus families have evolved gene products and strategies to counteract or evade the antiviral action of PKR, highlighting the importance of this kinase in the antiviral defense. Previously, it was found that PKR inhibits the replication of the coronavirus IBV, as overexpression of a dominant-negative kinase-defective PKR mutant enhanced IBV replication by almost 2-fold. Furthermore, IBV appeared to (weakly) antagonize the antiviral activity of PKR through two independent mechanisms, including a partial block of PKR activation (77). Interestingly, MHV-A59 infection in L2 or 17Cl1 cells did not induce PKR activation, and the sensitivity of MHV to IFN treatment appeared to be PKR independent (33, 78, 79). Transmissible gastroenteritis coronavirus (TGEV) protein 7 was shown to counteract PKR activation by binding protein phosphatase 1 to dephosphorylate eIF-2α (80), which, in support of our findings, also suggests that PKR is involved in controlling coronavirus replication. Krahling et al. showed that PKR was activated in SARS-CoV-infected 293/ACE2 cells but concluded that PKR knockdown did not significantly affect virus replication, despite the fact that an ∼1-log increase in SARS-CoV titer was observed in their experiments (81). This is in contrast to our PKR knockdown experiments, which clearly pointed to an antiviral role for PKR (Fig. 8). In our hands, depletion of PKR significantly increased SARS-CoV-driven GFP expression (Fig. 8A) and also enhanced N protein expression (Fig. 8C) and the production of infectious progeny (Fig. 8D). This discrepancy cannot be due to host cell differences, as the same 293/ACE2 cells were used in both studies (81); thus, they might be attributed to differences in the experimental setup, choice of controls, or normalization and interpretation of the data.
In line with the findings for PKR, reducing the expression of PERK (or EIF2AK3), one of the other kinases known to phosphorylate eIF-2α, resulted in an increase of SARS-CoV-GFP reporter gene expression to 57% (P < 0.01) (see Data Set S1 in the supplemental material). The unfolded protein response, i.e., the detection of misfolded proteins within the ER lumen, activates PERK, which in turn phosphorylates eIF2α and ultimately triggers apoptosis. The relatively strong antiviral effect of PERK observed in this study is in line with previous studies suggesting that the phosphorylation of eIF-2α in SARS-CoV-infected cells is mediated by PERK activation (81). Our findings support the hypothesis that upon SARS-CoV infection, the unfolded protein response is activated as an antiviral strategy. In fact, countering SARS-CoV infection may involve multiple cellular responses that induce apoptosis, including the activation of PKR and PERK, which also could explain the identification of several other hits involved in apoptosis, like those from the SAPK/JNK pathway.
Among the proviral hits, PRKCι had a relatively moderate effect in the primary screen (Fig. 4B), but its proviral role was validated using the inhibitor ATM (Fig. 8). Members of the protein kinase C family are serine/threonine protein kinases and are involved in several signaling pathways that regulate, e.g., cell proliferation, cell cycle progression, and cell survival (reviewed in reference 82). Interestingly, PRKCι can be activated by phosphoinositol lipids involved in microtubule dynamics within the early secretory pathway (83). PRKCι contains an N-terminal PB1 domain that ensures the signaling specificity (reviewed in reference 84), and ATM affects the interaction of PRKCι with other PB1 domain-containing proteins, like Par6, MEK5, and p62 (51, 52, 84). Therefore, blocking the PRKCι PB1 domain could decrease MEK5 (85) and NF-κB signaling (via p62) and affect cell polarity (via Par6) (reviewed in reference 84). In addition, PRKCι plays an essential role in COPI vesicle formation, since the GTPase Rab2 binds PRKCι and ultimately promotes recruitment of β-COP to pre-Golgi membrane structures for the formation of early secretory vesicles (83, 86, 87). As discussed below, COPB2 and the early secretory pathway play a crucial role in SARS-CoV replication, and in this manner PRKCι may affect SARS-CoV replication as well. Tisdale et al. have shown that PRKCι kinase activity is essential for the generation of retrograde transport vesicles (87). However, the role of the PRKCι PB1 domain, the main target of the drug ATM, in COPI vesicle formation was not directly investigated in this study. Nonetheless, this hypothesis is substantiated by the ATM concentration that blocked SARS-CoV replication. Our EC50s are similar to the reported 50% inhibitory concentration (IC50; ∼1 μM) for inhibition of the PRKCι PB-mediated interactions (51), while the IC50 for inhibition of the kinase activity was ∼100-fold higher (88). The exact mechanism by which PRKCι influences the SARS-CoV replicative cycle remains an interesting topic for future research, and it could even be an interesting target for the development of host-directed antiviral therapy for pathogenic coronaviruses.
Coronavirus replication is associated with a cytoplasmic reticulovesicular network of modified ER, including double-membrane vesicles and convoluted membranes (28). Despite the in-depth characterization of their ultrastructure, the biogenesis of these membrane structures and the cellular factors involved have remained largely uncharacterized. For example, the membrane source of these coronavirus-induced replication structures still is controversial, with advanced EM analyses showing the RVN to be derived from and continuous with the ER (28, 44, 89) and other studies implicating the autophagy pathway (90) or EDEMosomes (91) as the primary membrane donor. Our earlier work already suggested that the integrity of the early secretory pathway is important for efficient SARS-CoV replication, as brefeldin A treatment of SARS-CoV-infected cells significantly reduced replication as well as the accumulation of virus-induced membrane structures (89). In line with these findings, COPI-coated vesicles were implicated in the biogenesis of MHV replication structures (92, 93), and SARS-CoV nsp3 was shown to interact with three COPI subunits (94). In none of these previous SARS-CoV and MHV studies could a complete block of virus replication be achieved, either by reducing COPI vesicle formation by depletion of one of the coatomer subunits or by treatment with brefeldin A. These results may in part be explained by incomplete knockdown or the presence of residual COPI vesicles (complete knockdown probably is not possible due to its detrimental effect on intracellular trafficking and cell viability). Although our present study clearly demonstrates the importance of COPI vesicles in SARS-CoV replication (Fig. 9), their role in the formation or function of the SARS-CoV-induced RVN remains elusive. The importance of COPI-coated vesicles is further supported by their essential role in the replication of many other RNA viruses, such as poliovirus (95, 96) and other picornaviruses (40, 97–100).
In conclusion, our kinome-wide siRNA screen has identified several cellular proteins and pathways that influence SARS-CoV replication and possibly coronavirus infections in general. Thus, our data provide a starting point for further validation and in-depth mechanistic studies which should enhance our understanding of the complex interplay between coronaviruses and their host.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ali Tas, Corrine Beugeling, Dennis Ninaber, Emmely Treffers, and Maarten van Dinther for helpful discussions and excellent technical assistance and Shinji Makino for providing 293/ACE2 cells. We are grateful to Martijn Rabelink and Rob Hoeben (Department of Molecular Cell Biology) for sharing their lentivirus expertise and for providing plasmids and reagents for generating shRNA-expressing lentiviruses.
This research was supported by TOP grant 700.57.301 from the Council for Chemical Sciences of the Netherlands Organization for Scientific Research (NWO-CW).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01029-15.
REFERENCES
- 1.Nagy PD, Pogany J. 2012. The dependence of viral RNA replication on co-opted host factors. Nat Rev Microbiol 10:137–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhong Y, Tan YW, Liu DX. 2012. Recent progress in studies of arterivirus- and coronavirus-host interactions. Viruses 4:980–1010. doi: 10.3390/v4060980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krishnan MN, Ng A, Sukumaran B, Gilfoy FD, Uchil PD, Sultana H, Brass AL, Adametz R, Tsui M, Qian F, Montgomery RR, Lev S, Mason PW, Koski RA, Elledge SJ, Xavier RJ, Agaisse H, Fikrig E. 2008. RNA interference screen for human genes associated with West Nile virus infection. Nature 455:242–245. doi: 10.1038/nature07207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sessions OM, Barrows NJ, Souza-Neto JA, Robinson TJ, Hershey CL, Rodgers MA, Ramirez JL, Dimopoulos G, Yang PL, Pearson JL, Garcia-Blanco MA. 2009. Discovery of insect and human dengue virus host factors. Nature 458:1047–1050. doi: 10.1038/nature07967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kwon YJ, Heo J, Wong HE, Cruz DJ, Velumani S, da Silva CT, Mosimann AL, Duarte Dos Santos CN, Freitas-Junior LH, Fink K. 2014. Kinome siRNA screen identifies novel cell-type specific dengue host target genes. Antiviral Res 110:20–30. doi: 10.1016/j.antiviral.2014.07.006. [DOI] [PubMed] [Google Scholar]
- 6.Zhou H, Xu M, Huang Q, Gates AT, Zhang XD, Castle JC, Stec E, Ferrer M, Strulovici B, Hazuda DJ, Espeseth AS. 2008. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe 4:495–504. doi: 10.1016/j.chom.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 7.Ng TI, Mo H, Pilot-Matias T, He Y, Koev G, Krishnan P, Mondal R, Pithawalla R, He W, Dekhtyar T, Packer J, Schurdak M, Molla A. 2007. Identification of host genes involved in hepatitis C virus replication by small interfering RNA technology. Hepatology 45:1413–1421. doi: 10.1002/hep.21608. [DOI] [PubMed] [Google Scholar]
- 8.Tai AW, Benita Y, Peng LF, Kim SS, Sakamoto N, Xavier RJ, Chung RT. 2009. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe 5:298–307. doi: 10.1016/j.chom.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Q, Brass AL, Ng A, Hu Z, Xavier RJ, Liang TJ, Elledge SJ. 2009. A genome-wide genetic screen for host factors required for hepatitis C virus propagation. Proc Natl Acad Sci U S A 106:16410–16415. doi: 10.1073/pnas.0907439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reiss S, Rebhan I, Backes P, Romero-Brey I, Erfle H, Matula P, Kaderali L, Poenisch M, Blankenburg H, Hiet MS, Longerich T, Diehl S, Ramirez F, Balla T, Rohr K, Kaul A, Buhler S, Pepperkok R, Lengauer T, Albrecht M, Eils R, Schirmacher P, Lohmann V, Bartenschlager R. 2011. Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe 9:32–45. doi: 10.1016/j.chom.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Supekova L, Supek F, Lee J, Chen S, Gray N, Pezacki JP, Schlapbach A, Schultz PG. 2008. Identification of human kinases involved in hepatitis C virus replication by small interference RNA library screening. J Biol Chem 283:29–36. doi: 10.1074/jbc.M703988200. [DOI] [PubMed] [Google Scholar]
- 12.Randall G, Panis M, Cooper JD, Tellinghuisen TL, Sukhodolets KE, Pfeffer S, Landthaler M, Landgraf P, Kan S, Lindenbach BD, Chien M, Weir DB, Russo JJ, Ju J, Brownstein MJ, Sheridan R, Sander C, Zavolan M, Tuschl T, Rice CM. 2007. Cellular cofactors affecting hepatitis C virus infection and replication. Proc Natl Acad Sci U S A 104:12884–12889. doi: 10.1073/pnas.0704894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hao L, Sakurai A, Watanabe T, Sorensen E, Nidom CA, Newton MA, Ahlquist P, Kawaoka Y. 2008. Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature 454:890–893. doi: 10.1038/nature07151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karlas A, Machuy N, Shin Y, Pleissner KP, Artarini A, Heuer D, Becker D, Khalil H, Ogilvie LA, Hess S, Maurer AP, Muller E, Wolff T, Rudel T, Meyer TF. 2010. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature 463:818–822. doi: 10.1038/nature08760. [DOI] [PubMed] [Google Scholar]
- 15.Vogels MW, van Balkom BW, Kaloyanova DV, Batenburg JJ, Heck AJ, Helms JB, Rottier PJ, de Haan CA. 2011. Identification of host factors involved in coronavirus replication by quantitative proteomics analysis. Proteomics 11:64–80. doi: 10.1002/pmic.201000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitchell HD, Eisfeld AJ, Sims AC, McDermott JE, Matzke MM, Webb-Robertson BJ, Tilton SC, Tchitchek N, Josset L, Li C, Ellis AL, Chang JH, Heegel RA, Luna ML, Schepmoes AA, Shukla AK, Metz TO, Neumann G, Benecke AG, Smith RD, Baric RS, Kawaoka Y, Katze MG, Waters KM. 2013. A network integration approach to predict conserved regulators related to pathogenicity of influenza and SARS-CoV respiratory viruses. PLoS One 8:e69374. doi: 10.1371/journal.pone.0069374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burkard C, Verheije MH, Wicht O, van Kasteren SI, van Kuppeveld FJ, Haagmans BL, Pelkmans L, Rottier PJ, Bosch BJ, de Haan CA. 2014. Coronavirus cell entry occurs through the endo-/lysosomal pathway in a proteolysis-dependent manner. PLoS Pathog 10:e1004502. doi: 10.1371/journal.ppat.1004502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Haan CA, Rottier PJ. 2006. Hosting the severe acute respiratory syndrome coronavirus: specific cell factors required for infection. Cell Microbiol 8:1211–1218. doi: 10.1111/j.1462-5822.2006.00744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Groot RJ, Cowley JA, Enjuanes L, Faaberg KS, Perlman S, Rottier PJ, Snijder EJ, Ziebuhr J, Gorbalenya AE. 2012. Order of Nidovirales, p 785–795. In King A, Adams M, Carstens E, Lefkowitz EJ (ed), Virus taxonomy, the 9th report of the international committee on taxonomy of viruses. Academic Press, New York, NY. [Google Scholar]
- 20.Gorbalenya AE, Enjuanes L, Ziebuhr J, Snijder EJ. 2006. Nidovirales: evolving the largest RNA virus genome. Virus Res 117:17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van der Hoek L. 2007. Human coronaviruses: what do they cause? Antiviral Ther 12:651–658. [PubMed] [Google Scholar]
- 22.Pyrc K, Berkhout B, van der Hoek L. 2007. The novel human coronaviruses NL63 and HKU1. J Virol 81:3051–3057. doi: 10.1128/JVI.01466-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perlman S, Netland J. 2009. Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol 7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Boheemen S, de Graaf M, Lauber C, Bestebroer TM, Raj VS, Zaki AM, Osterhaus AD, Haagmans BL, Gorbalenya AE, Snijder EJ, Fouchier RA. 2012. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. mBio 3:e00473–00412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. 2012. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 26.Miller S, Krijnse-Locker J. 2008. Modification of intracellular membrane structures for virus replication. Nat Rev Microbiol 6:363–374. doi: 10.1038/nrmicro1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Hemert MJ, van den Worm SH, Knoops K, Mommaas AM, Gorbalenya AE, Snijder EJ. 2008. SARS-coronavirus replication/transcription complexes are membrane-protected and need a host factor for activity in vitro. PLoS Pathog 4:e1000054. doi: 10.1371/journal.ppat.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knoops K, Kikkert M, van den Worm SH, Zevenhoven-Dobbe JC, van der Meer Y, Koster AJ, Mommaas AM, Snijder EJ. 2008. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol 6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neuman BW, Angelini MM, Buchmeier MJ. 2014. Does form meet function in the coronavirus replicative organelle? Trends Microbiol 22:642–647. doi: 10.1016/j.tim.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Narayanan K, Huang C, Makino S. 2008. SARS coronavirus accessory proteins. Virus Res 133:113–121. doi: 10.1016/j.virusres.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ratia K, Saikatendu KS, Santarsiero BD, Barretto N, Baker SC, Stevens RC, Mesecar AD. 2006. Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proc Natl Acad Sci U S A 103:5717–5722. doi: 10.1073/pnas.0510851103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zust R, Cervantes-Barragan L, Habjan M, Maier R, Neuman BW, Ziebuhr J, Szretter KJ, Baker SC, Barchet W, Diamond MS, Siddell SG, Ludewig B, Thiel V. 2011. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol 12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ye Y, Hauns K, Langland JO, Jacobs BL, Hogue BG. 2007. Mouse hepatitis coronavirus A59 nucleocapsid protein is a type I interferon antagonist. J Virol 81:2554–2563. doi: 10.1128/JVI.01634-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frieman M, Yount B, Heise M, Kopecky-Bromberg SA, Palese P, Baric RS. 2007. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J Virol 81:9812–9824. doi: 10.1128/JVI.01012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kopecky-Bromberg SA, Martinez-Sobrido L, Palese P. 2006. 7a protein of severe acute respiratory syndrome coronavirus inhibits cellular protein synthesis and activates p38 mitogen-activated protein kinase. J Virol 80:785–793. doi: 10.1128/JVI.80.2.785-793.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hussain S, Perlman S, Gallagher TM. 2008. Severe acute respiratory syndrome coronavirus protein 6 accelerates murine hepatitis virus infections by more than one mechanism. J Virol 82:7212–7222. doi: 10.1128/JVI.02406-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou P, Li H, Wang H, Wang LF, Shi Z. 2012. Bat severe acute respiratory syndrome-like coronavirus ORF3b homologues display different interferon antagonist activities. J Gen Virol 93:275–281. doi: 10.1099/vir.0.033589-0. [DOI] [PubMed] [Google Scholar]
- 38.DeDiego ML, Nieto-Torres JL, Jimenez-Guardeno JM, Regla-Nava JA, Alvarez E, Oliveros JC, Zhao J, Fett C, Perlman S, Enjuanes L. 2011. Severe acute respiratory syndrome coronavirus envelope protein regulates cell stress response and apoptosis. PLoS Pathog 7:e1002315. doi: 10.1371/journal.ppat.1002315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coyne CB, Bozym R, Morosky SA, Hanna SL, Mukherjee A, Tudor M, Kim KS, Cherry S. 2011. Comparative RNAi screening reveals host factors involved in enterovirus infection of polarized endothelial monolayers. Cell Host Microbe 9:70–82. doi: 10.1016/j.chom.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hsu NY, Ilnytska O, Belov G, Santiana M, Chen YH, Takvorian PM, Pau C, van der Schaar H, Kaushik-Basu N, Balla T, Cameron CE, Ehrenfeld E, van Kuppeveld FJ, Altan-Bonnet N. 2010. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell 141:799–811. doi: 10.1016/j.cell.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kamitani W, Narayanan K, Huang C, Lokugamage K, Ikegami T, Ito N, Kubo H, Makino S. 2006. Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. Proc Natl Acad Sci U S A 103:12885–12890. doi: 10.1073/pnas.0603144103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Wilde AH, Li Y, van der Meer Y, Vuagniaux G, Lysek R, Fang Y, Snijder EJ, van Hemert MJ. 2013. Cyclophilin inhibitors block arterivirus replication by interfering with viral RNA synthesis. J Virol 87:1454–1464. doi: 10.1128/JVI.02078-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Treffers EE, Tas A, Scholte FE, Van MN, Heemskerk MT, de Ru AH, Snijder EJ, van Hemert MJ, van Veelen PA. 12 March 2015. Temporal SILAC-based quantitative proteomics identifies host factors involved in chikungunya virus replication. Proteomics doi: 10.1002/pmic.201400581. [DOI] [PubMed] [Google Scholar]
- 44.Snijder EJ, van der Meer Y, Zevenhoven-Dobbe J, Onderwater JJ, van der Meulen J, Koerten HK, Mommaas AM. 2006. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J Virol 80:5927–5940. doi: 10.1128/JVI.02501-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sims AC, Burkett SE, Yount B, Pickles RJ. 2008. SARS-CoV replication and pathogenesis in an in vitro model of the human conducting airway epithelium. Virus Res 133:33–44. doi: 10.1016/j.virusres.2007.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van den Worm SH, Eriksson KK, Zevenhoven JC, Weber F, Zust R, Kuri T, Dijkman R, Chang G, Siddell SG, Snijder EJ, Thiel V, Davidson AD. 2012. Reverse genetics of SARS-related coronavirus using vaccinia virus-based recombination. PLoS One 7:e32857. doi: 10.1371/journal.pone.0032857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boutros M, Bras LP, Huber W. 2006. Analysis of cell-based RNAi screens. Genome Biol 7:R66. doi: 10.1186/gb-2006-7-7-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Hemert MJ, de Wilde AH, Gorbalenya AE, Snijder EJ. 2008. The in vitro RNA synthesizing activity of the isolated arterivirus replication/transcription complex is dependent on a host factor. J Biol Chem 283:16525–16536. doi: 10.1074/jbc.M708136200. [DOI] [PubMed] [Google Scholar]
- 49.Rhesus Macaque Genome Sequencing and Analysis Consortium, Gibbs RA, Rogers J, Katze MG, Bumgarner R, Weinstock GM, Mardis ER, Remington KA, Strausberg RL, Venter JC, Wilson RK, Batzer MA, Bustamante CD, Eichler EE, Hahn MW, Hardison RC, Makova KD, Miller W, Milosavljevic A, Palermo RE, Siepel A, Sikela JM, Attaway T, Bell S, Bernard KE, Buhay CJ, Chandrabose MN, Dao M, Davis C, Delehaunty KD, Ding Y, Dinh HH, Dugan-Rocha S, Fulton LA, Gabisi RA, Garner TT, Godfrey J, Hawes AC, Hernandez J, Hines S, Holder M, Hume J, Jhangiani SN, Joshi V, Khan ZM, Kirkness EF, Cree A, Fowler RG, Lee S, Lewis LR, Li Z, Liu YS, Moore SM, Muzny D, Nazareth LV, Ngo DN, Okwuonu GO, Pai G, Parker D, Paul HA, Pfannkoch C, Pohl CS, Rogers YH, Ruiz SJ, Sabo A, Santibanez J, Schneider BW, Smith SM, Sodergren E, Svatek AF, Utterback TR, Vattathil S, Warren W, White CS, Chinwalla AT, Feng Y, Halpern AL, Hillier LW, Huang X, Minx P, Nelson JO, Pepin KH, Qin X, Sutton GG, Venter E, Walenz BP, Wallis JW, Worley KC, Yang SP, Jones SM, Marra MA, Rocchi M, Schein JE, Baertsch R, Clarke L, Csuros M, Glasscock J, Harris RA, Havlak P, Jackson AR, Jiang H, Liu Y, Messina DN, Shen Y, Song HX, Wylie T, Zhang L, Birney E, Han K, Konkel MK, Lee J, Smit AF, Ullmer B, Wang H, Xing J, Burhans R, Cheng Z, Karro JE, Ma J, Raney B, She X, Cox MJ, Demuth JP, Dumas LJ, Han SG, Hopkins J, Karimpour-Fard A, Kim YH, Pollack JR, Vinar T, Addo-Quaye C, Degenhardt J, Denby A, Hubisz MJ, Indap A, Kosiol C, Lahn BT, Lawson HA, Marklein A, Nielsen R, Vallender EJ, Clark AG, Ferguson B, Hernandez RD, Hirani K, Kehrer-Sawatzki H, Kolb J, Patil S, Pu LL, Ren Y, Smith DG, Wheeler DA, Schenck I, Ball EV, Chen R, Cooper DN, Giardine B, Hsu F, Kent WJ, Lesk A, Nelson DL, O'Brien WE, Prufer K, Stenson PD, Wallace JC, Ke H, Liu XM, Wang P, Xiang AP, Yang F, Barber GP, Haussler D, Karolchik D, Kern AD, Kuhn RM, Smith KE, Zwieg AS. 2007. Evolutionary and biomedical insights from the rhesus macaque genome. Science 316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- 50.Dauber B, Wolff T. 2009. Activation of the antiviral kinase PKR and viral countermeasures. Viruses 1:523–544. doi: 10.3390/v1030523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stallings-Mann M, Jamieson L, Regala RP, Weems C, Murray NR, Fields AP. 2006. A novel small-molecule inhibitor of protein kinase Ciota blocks transformed growth of non-small-cell lung cancer cells. Cancer Res 66:1767–1774. doi: 10.1158/0008-5472.CAN-05-3405. [DOI] [PubMed] [Google Scholar]
- 52.Erdogan E, Lamark T, Stallings-Mann M, Lee J, Pellecchia M, Thompson EA, Johansen T, Fields AP. 2006. Aurothiomalate inhibits transformed growth by targeting the PB1 domain of protein kinase Ciota. J Biol Chem 281:28450–28459. doi: 10.1074/jbc.M606054200. [DOI] [PubMed] [Google Scholar]
- 53.Beck R, Rawet M, Wieland FT, Cassel D. 2009. The COPI system: molecular mechanisms and function. FEBS Lett 583:2701–2709. doi: 10.1016/j.febslet.2009.07.032. [DOI] [PubMed] [Google Scholar]
- 54.Niu TK, Pfeifer AC, Lippincott-Schwartz J, Jackson CL. 2005. Dynamics of GBF1, a brefeldin A-sensitive Arf1 exchange factor at the Golgi. Mol Biol Cell 16:1213–1222. doi: 10.1091/mbc.E04-07-0599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cherry S, Doukas T, Armknecht S, Whelan S, Wang H, Sarnow P, Perrimon N. 2005. Genome-wide RNAi screen reveals a specific sensitivity of IRES-containing RNA viruses to host translation inhibition. Genes Dev 19:445–452. doi: 10.1101/gad.1267905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. 2006. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev 20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 57.Tan YJ, Teng E, Shen S, Tan TH, Goh PY, Fielding BC, Ooi EE, Tan HC, Lim SG, Hong W. 2004. A novel severe acute respiratory syndrome coronavirus protein, U274, is transported to the cell surface and undergoes endocytosis. J Virol 78:6723–6734. doi: 10.1128/JVI.78.13.6723-6734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tan YJ, Fielding BC, Goh PY, Shen S, Tan TH, Lim SG, Hong W. 2004. Overexpression of 7a, a protein specifically encoded by the severe acute respiratory syndrome coronavirus, induces apoptosis via a caspase-dependent pathway. J Virol 78:14043–14047. doi: 10.1128/JVI.78.24.14043-14047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yuan X, Wu J, Shan Y, Yao Z, Dong B, Chen B, Zhao Z, Wang S, Chen J, Cong Y. 2006. SARS coronavirus 7a protein blocks cell cycle progression at G0/G1 phase via the cyclin D3/pRb pathway. Virology 346:74–85. doi: 10.1016/j.virol.2005.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sims AC, Baric RS, Yount B, Burkett SE, Collins PL, Pickles RJ. 2005. Severe acute respiratory syndrome coronavirus infection of human ciliated airway epithelia: role of ciliated cells in viral spread in the conducting airways of the lungs. J Virol 79:15511–15524. doi: 10.1128/JVI.79.24.15511-15524.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schaecher SR, Stabenow J, Oberle C, Schriewer J, Buller RM, Sagartz JE, Pekosz A. 2008. An immunosuppressed Syrian golden hamster model for SARS-CoV infection. Virology 380:312–321. doi: 10.1016/j.virol.2008.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chang YJ, Liu CY, Chiang BL, Chao YC, Chen CC. 2004. Induction of IL-8 release in lung cells via activator protein-1 by recombinant baculovirus displaying severe acute respiratory syndrome-coronavirus spike proteins: identification of two functional regions. J Immunol 173:7602–7614. doi: 10.4049/jimmunol.173.12.7602. [DOI] [PubMed] [Google Scholar]
- 63.Zhang X, Wu K, Wang D, Yue X, Song D, Zhu Y, Wu J. 2007. Nucleocapsid protein of SARS-CoV activates interleukin-6 expression through cellular transcription factor NF-kappaB. Virology 365:324–335. doi: 10.1016/j.virol.2007.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liao Y, Wang X, Huang M, Tam JP, Liu DX. 2011. Regulation of the p38 mitogen-activated protein kinase and dual-specificity phosphatase 1 feedback loop modulates the induction of interleukin 6 and 8 in cells infected with coronavirus infectious bronchitis virus. Virology 420:106–116. doi: 10.1016/j.virol.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Banerjee S, Narayanan K, Mizutani T, Makino S. 2002. Murine coronavirus replication-induced p38 mitogen-activated protein kinase activation promotes interleukin-6 production and virus replication in cultured cells. J Virol 76:5937–5948. doi: 10.1128/JVI.76.12.5937-5948.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bushman FD, Malani N, Fernandes J, D'Orso I, Cagney G, Diamond TL, Zhou H, Hazuda DJ, Espeseth AS, Konig R, Bandyopadhyay S, Ideker T, Goff SP, Krogan NJ, Frankel AD, Young JA, Chanda SK. 2009. Host cell factors in HIV replication: meta-analysis of genome-wide studies. PLoS Pathog 5:e1000437. doi: 10.1371/journal.ppat.1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cargnello M, Roux PP. 2011. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol Mol Biol Rev 75:50–83. doi: 10.1128/MMBR.00031-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kono M, Tatsumi K, Imai AM, Saito K, Kuriyama T, Shirasawa H. 2008. Inhibition of human coronavirus 229E infection in human epithelial lung cells (L132) by chloroquine: involvement of p38 MAPK and ERK. Antiviral Res 77:150–152. doi: 10.1016/j.antiviral.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Padhan K, Minakshi R, Towheed MA, Jameel S. 2008. Severe acute respiratory syndrome coronavirus 3a protein activates the mitochondrial death pathway through p38 MAP kinase activation. J Gen Virol 89:1960–1969. doi: 10.1099/vir.0.83665-0. [DOI] [PubMed] [Google Scholar]
- 70.Jimenez-Guardeno JM, Nieto-Torres JL, DeDiego ML, Regla-Nava JA, Fernandez-Delgado R, Castano-Rodriguez C, Enjuanes L. 2014. The PDZ-binding motif of severe acute respiratory syndrome coronavirus envelope protein is a determinant of viral pathogenesis. PLoS Pathog 10:e1004320. doi: 10.1371/journal.ppat.1004320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Donjerkovic D, Scott DW. 2000. Regulation of the G1 phase of the mammalian cell cycle. Cell Res 10:1–16. doi: 10.1038/sj.cr.7290031. [DOI] [PubMed] [Google Scholar]
- 72.Handschick K, Beuerlein K, Jurida L, Bartkuhn M, Muller H, Soelch J, Weber A, Dittrich-Breiholz O, Schneider H, Scharfe M, Jarek M, Stellzig J, Schmitz ML, Kracht M. 2014. Cyclin-dependent kinase 6 is a chromatin-bound cofactor for NF-kappaB-dependent gene expression. Mol Cell 53:193–208. doi: 10.1016/j.molcel.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 73.Buss H, Handschick K, Jurrmann N, Pekkonen P, Beuerlein K, Muller H, Wait R, Saklatvala J, Ojala PM, Schmitz ML, Naumann M, Kracht M. 2012. Cyclin-dependent kinase 6 phosphorylates NF-kappaB P65 at serine 536 and contributes to the regulation of inflammatory gene expression. PLoS One 7:e51847. doi: 10.1371/journal.pone.0051847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.DeDiego ML, Nieto-Torres JL, Regla-Nava JA, Jimenez-Guardeno JM, Fernandez-Delgado R, Fett C, Castano-Rodriguez C, Perlman S, Enjuanes L. 2014. Inhibition of NF-kappaB-mediated inflammation in severe acute respiratory syndrome coronavirus-infected mice increases survival. J Virol 88:913–924. doi: 10.1128/JVI.02576-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen CJ, Makino S. 2004. Murine coronavirus replication induces cell cycle arrest in G0/G1 phase. J Virol 78:5658–5669. doi: 10.1128/JVI.78.11.5658-5669.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Surjit M, Liu B, Chow VT, Lal SK. 2006. The nucleocapsid protein of severe acute respiratory syndrome-coronavirus inhibits the activity of cyclin-cyclin-dependent kinase complex and blocks S phase progression in mammalian cells. J Biol Chem 281:10669–10681. doi: 10.1074/jbc.M509233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang X, Liao Y, Yap PL, Png KJ, Tam JP, Liu DX. 2009. Inhibition of protein kinase R activation and upregulation of GADD34 expression play a synergistic role in facilitating coronavirus replication by maintaining de novo protein synthesis in virus-infected cells. J Virol 83:12462–12472. doi: 10.1128/JVI.01546-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Koetzner CA, Kuo L, Goebel SJ, Dean AB, Parker MM, Masters PS. 2010. Accessory protein 5a is a major antagonist of the antiviral action of interferon against murine coronavirus. J Virol 84:8262–8274. doi: 10.1128/JVI.00385-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zorzitto J, Galligan CL, Ueng JJ, Fish EN. 2006. Characterization of the antiviral effects of interferon-alpha against a SARS-like coronavirus infection in vitro. Cell Res 16:220–229. doi: 10.1038/sj.cr.7310030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cruz JL, Sola I, Becares M, Alberca B, Plana J, Enjuanes L, Zuniga S. 2011. Coronavirus gene 7 counteracts host defenses and modulates virus virulence. PLoS Pathog 7:e1002090. doi: 10.1371/journal.ppat.1002090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Krahling V, Stein DA, Spiegel M, Weber F, Muhlberger E. 2009. Severe acute respiratory syndrome coronavirus triggers apoptosis via protein kinase R but is resistant to its antiviral activity. J Virol 83:2298–2309. doi: 10.1128/JVI.01245-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Reyland ME. 2009. Protein kinase C isoforms: multi-functional regulators of cell life and death. Front Biosci 14:2386–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tisdale EJ. 2002. Glyceraldehyde-3-phosphate dehydrogenase is phosphorylated by protein kinase Ciota/lambda and plays a role in microtubule dynamics in the early secretory pathway. J Biol Chem 277:3334–3341. doi: 10.1074/jbc.M109744200. [DOI] [PubMed] [Google Scholar]
- 84.Moscat J, Diaz-Meco MT, Albert A, Campuzano S. 2006. Cell signaling and function organized by PB1 domain interactions. Mol Cell 23:631–640. doi: 10.1016/j.molcel.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 85.Diaz-Meco MT, Moscat J. 2001. MEK5, a new target of the atypical protein kinase C isoforms in mitogenic signaling. Mol Cell Biol 21:1218–1227. doi: 10.1128/MCB.21.4.1218-1227.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tisdale EJ, Artalejo CR. 2006. Src-dependent aprotein kinase C iota/lambda (aPKCiota/lambda) tyrosine phosphorylation is required for aPKCiota/lambda association with Rab2 and glyceraldehyde-3-phosphate dehydrogenase on pre-Golgi intermediates. J Biol Chem 281:8436–8442. doi: 10.1074/jbc.M513031200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tisdale EJ. 2000. Rab2 requires PKC iota/lambda to recruit beta-COP for vesicle formation. Traffic 1:702–712. doi: 10.1034/j.1600-0854.2000.010903.x. [DOI] [PubMed] [Google Scholar]
- 88.Pillai P, Desai S, Patel R, Sajan M, Farese R, Ostrov D, Acevedo-Duncan M. 2011. A novel PKC-iota inhibitor abrogates cell proliferation and induces apoptosis in neuroblastoma. Int J Biochem Cell Biol 43:784–794. doi: 10.1016/j.biocel.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 89.Knoops K, Swett-Tapia C, van den Worm SH, te Velthuis AJ, Koster AJ, Mommaas AM, Snijder EJ, Kikkert M. 2010. Integrity of the early secretory pathway promotes, but is not required for, severe acute respiratory syndrome coronavirus RNA synthesis and virus-induced remodeling of endoplasmic reticulum membranes. J Virol 84:833–846. doi: 10.1128/JVI.01826-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Prentice E, Jerome WG, Yoshimori T, Mizushima N, Denison MR. 2004. Coronavirus replication complex formation utilizes components of cellular autophagy. J Biol Chem 279:10136–10141. doi: 10.1074/jbc.M306124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Reggiori F, Monastyrska I, Verheije MH, Cali T, Ulasli M, Bianchi S, Bernasconi R, de Haan CA, Molinari M. 2010. Coronaviruses hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe 7:500–508. doi: 10.1016/j.chom.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Oostra M, te Lintelo EG, Deijs M, Verheije MH, Rottier PJ, de Haan CA. 2007. Localization and membrane topology of coronavirus nonstructural protein 4: involvement of the early secretory pathway in replication. J Virol 81:12323–12336. doi: 10.1128/JVI.01506-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Verheije MH, Raaben M, Mari M, Te Lintelo EG, Reggiori F, van Kuppeveld FJ, Rottier PJ, de Haan CA. 2008. Mouse hepatitis coronavirus RNA replication depends on GBF1-mediated ARF1 activation. PLoS Pathog 4:e1000088. doi: 10.1371/journal.ppat.1000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Neuman BW, Joseph JS, Saikatendu KS, Serrano P, Chatterjee A, Johnson MA, Liao L, Klaus JP, Yates JR III, Wuthrich K, Stevens RC, Buchmeier MJ, Kuhn P. 2008. Proteomics analysis unravels the functional repertoire of coronavirus nonstructural protein 3. J Virol 82:5279–5294. doi: 10.1128/JVI.02631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Belov GA, Altan-Bonnet N, Kovtunovych G, Jackson CL, Lippincott-Schwartz J, Ehrenfeld E. 2007. Hijacking components of the cellular secretory pathway for replication of poliovirus RNA. J Virol 81:558–567. doi: 10.1128/JVI.01820-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Belov GA, Fogg MH, Ehrenfeld E. 2005. Poliovirus proteins induce membrane association of GTPase ADP-ribosylation factor. J Virol 79:7207–7216. doi: 10.1128/JVI.79.11.7207-7216.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gazina EV, Mackenzie JM, Gorrell RJ, Anderson DA. 2002. Differential requirements for COPI coats in formation of replication complexes among three genera of Picornaviridae. J Virol 76:11113–11122. doi: 10.1128/JVI.76.21.11113-11122.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang J, Wu Z, Jin Q. 2012. COPI is required for enterovirus 71 replication. PLoS One 7:e38035. doi: 10.1371/journal.pone.0038035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.van der Linden L, van der Schaar HM, Lanke KH, Neyts J, van Kuppeveld FJ. 2010. Differential effects of the putative GBF1 inhibitors Golgicide A and AG1478 on enterovirus replication. J Virol 84:7535–7542. doi: 10.1128/JVI.02684-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cherry S, Kunte A, Wang H, Coyne C, Rawson RB, Perrimon N. 2006. COPI activity coupled with fatty acid biosynthesis is required for viral replication. PLoS Pathog 2:e102. doi: 10.1371/journal.ppat.0020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.