Abstract
Proteolipid protein (PLP) and DM20, the most abundant myelin proteins, are coded by the human PLP1 and non-human Plp1 proteolipid protein gene. Mutations in the PLP1 gene cause Pelizaeus-Merzbacher Disease (PMD) with duplications of the native PLP1 gene accounting for 70% of PLP1 mutations. Humans with PLP1 duplications and mice with extra Plp1 copies have extensive neuronal degeneration. The mechanism that causes neuronal degeneration is unknown. We show that native PLP traffics to mitochondria when the gene is duplicated in mice and in humans. This report is the first demonstration of a specific cellular defect in brains of PMD patients; it validates rodent models as ideal models to study PMD. Insertion of nuclear-encoded mitochondrial proteins requires specific import pathways; we show that specific cysteine motifs, part of the Mia40/Erv1 mitochondrial import pathway, are present in PLP and are required for its insertion into mitochondria. Insertion of native PLP into mitochondria of transfected cells acidifies media, partially due to increased lactate; it also increases ATP in the media. The same abnormalities are found in the extracellular space of mouse brains with extra copies of Plp1. These physiological abnormalities are preventable by mutations in PLP cysteine motifs, a hallmark of the Mia40/Erv1 pathway. Increased extracellular ATP and acidosis lead to neuronal degeneration. Our findings may be the mechanism by which microglia are activated and pro-inflammatory molecules are up-regulated in Plp1 transgenic mice (Tatar et al., 2010). Manipulation of this metabolic pathway may restore normal metabolism and provide therapy for PMD patients.
Keywords: metabolism, neuroglia, dysmyelinating disorders, leukodystrophies, myelin
INTRODUCTION
Proteolipid protein (PLP) and its smaller isoform DM20 constitute about 50% of the total proteins in myelin (Eng et al., 1968; Norton and Poduslo, 1973). The stability of the myelin sheath is thought to be their major function (Braun, 1977; Duncan et al., 1987; Boison et al., 1995). Both PLP and DM20 are coded by the proteolipid protein PLP1 (human) and Plp1 (non-human) gene. PLP1 mutations cause Pelizaeus-Merzbacher Disease (PMD) and spastic paraplegia type II (SPG2) (Boespflug Tanguy et al., 1994; Ellis and Malcolm, 1994). In PMD, wtPLP1 duplications and missense mutations lead to shortened lifespan (Renier et al., 1981; Hodes et al., 1993; Ellis and Malcolm, 1994) including infant death in connatal PMD. Surprisingly, men with null mutations do not exhibit motor and sensory symptoms until their 20's and they survive into their 50's (Raskind et al., 1991; Garbern et al., 1997; Inoue et al., 2002). Similarly, PLP deficient mice lack behavioral signs in their first year and have a fairly normal life span (Boison and Stoffel 1994; Boison et al., 1995; Klugmann et al., 1997; Griffiths et al., 1998; Stecca et al., 2000; Yool et al., 2002). Thus, animals with a null mutation of the PLP1/Plp1 gene (and lack of PLP) have better outcomes compared to animals with extra copies or to missense mutations of the wtPLP1/Plp1 gene (and altered PLP levels). These findings indicate that duplications/missense mutations of the PLP1/Plp1 gene exert a downstream toxic gain of function.
Cellular abnormalities in PLP1/Plp1 mutations are not limited to oligodendrocytes (Olgs) but include astrocytes (Skoff, 1976), microglia (Tatar et al., 2010) and neurons (see Discussion). Factors that trigger astrocyte and microglia activation and the pathway that leads to neuronal degeneration are unknown. Co-culture of neurons with cells that over-express wtPlp1 lead to accelerated neuronal degeneration (Boucher et al., 2002). These findings demonstrate that over-expression of Plp1, in the absence of myelin, directly causes neuronal death. Moreover, wtPlp1 over-expressing cells cause a dramatic acidification of media (Boucher et al., 2002) and transgenic mice (Plp1tg) with extra copies of wtPlp1 have a dramatic acidification of extracellular fluid (ECF) (Skoff et al., 2004a). Clearly, cells that over-express wtPlp1 and oligodendrocytes (Olgs) in vivo are capable of altering their extracellular milieu, by acidification and/or secretion of solutes that are toxic to neurons.
Our lab recently showed that wtPLP, when over-expressed in COS7 cells and in the Plp1tg mice, co-localizes with mitochondria (Hüttemann et al., 2009). These data suggest causal links between the insertion of PLP into mitochondria, metabolic abnormalities, alterations in the extracellular milieu, and neuronal degeneration. Here, we identify specific motifs in the N-terminus of PLP that permit its insertion into mitochondria. This PLP mitochondrial import pathway involves CX3C and/or CX9C motifs in PLP that interact with the CX3C and/or CX9C motifs identified in the Mia40/Erv1 pathway (Mesecke et al., 2005, 2008; Bihlmaier et al., 2007; Longen et al., 2009). The essential component of this pathway is the interaction of an unfolded substrate with an oxidized form of Mia40. A disulfide intermediate is formed between the unfolded substrate and oxidized Mia40 generating a folded protein in a reduced state (Hell, 2008). We further show that mutations of these PLP cysteines prevents insertion of PLP into mitochondria, and prevents the acidification of tissue culture media and decreases ATP levels in the media. These findings are highly relevant to neuronal degeneration because increased ATP levels in media and in ECF activate cytokines via receptors on microglia and astrocytes that, in turn, lead to neuronal degeneration (Shigemoto-Mogami et al., 2001; Honda et al., 2001; Samuels, et al., 2010; Neary et al., 1996; Delarasse et al., 2009; Tu and Wang, 2009; Agteresch et al., 1999; Melani et al., 2005).
METHODS AND MATERIALS
Immunocytochemistry of Plp1tg mice and C57 mice brain sections
Breeding pairs of heterozygous and homozygous Plp1tg line 66 (provided by K-A Nave, Max Planck Institute of Experimental Medicine, Göttingen, Germany) mice are maintained in Wayne State University DLAR facilities, a federally approved AAALAC facility. To confirm the presence of the transgene, tails from newborns of either sex were clipped and processed for the presence or absence of the T7 promoter in the inserted transgene sequence (Readhead et al., 1994). Only mice that expressed the T7 promoter and showed tremors/seizures were used in this study. Plp1 copy number, determined by the delta delta CT method, averaged 4-5 when normalized to GAPDH. Plp1tg mice between 3 weeks and 7 months and C57BL/6J wild type mice between 2 weeks and 7 months were perfused with 4% paraformaldehyde. Their brains were removed and placed in 4% paraformaldehyde at 4°C for 72 h and then placed in 20% sucrose for 72 h. Fifty-micron sections were cut on a Vibratome (St. Louis, MO) and stored in 0.1M phosphate buffered saline (PBS) and processed for immunocytochemistry as follows. Sections were rinsed several times in 0.1M PBS, incubated in methanol at −20°C for 20 min. rinsed in PBS and simultaneously incubated with anti-PLP, a rabbit polyclonal antibody that specifically recognizes PLP and was generated for us by Affinity BioReagents (ABR, Rockford, IL) at 1:500 and mouse monoclonal anti-COX1 (Invitrogen, Grand Island, NY) at 1:100 in PBS overnight at room temperature (RT). The next day sections were rinsed in PBS, incubated in a goat anti-rabbit IgG conjugated to Texas Red at 1:100 and goat anti-mouse IgG conjugated to FITC at 1:100 for 4 h at RT, rinsed in PBS, stained with a 1:10,000 dilution of DAPI in methanol for 5mins at RT, rinsed in PBS and mounted in Aquamont. Sections were viewed and imaged using a Leica TCS SP5 confocal microscope. Sections from white and gray areas of cerebrum were systematically scanned in x-y and z-axis using a x63 oil objective. Images of Olgs were taken at a zoom setting of 4, exported as tiff files and imported into Adobe Photoshop for printing. Selected images were deconvolved with the DECONVOLUTION program, Huygens Essential, by Scientific Volume Imaging B.V.
Volocity (PerkinElmer), a program with co-localization features, was used to determine overlap of two or more fluorochromatic dyes in a single voxel of images obtained on the Leica TCS SP5 microscope. Volocity calculates measurement statistics known as the Pearson Correlation Coefficient (PCC) and the Mander's Coeffficients (Mx and My) based on the presence and intensity of two different fluorochromes in each voxel of an image. The Pearson Coefficient equals 1 for perfect co-localization, 0 for random co-localization and a negative 1 for perfect exclusion. The Manders coefficients equal 1 for perfect co-localization and 0 for no co-localization. The range of voxels for each fluorochrome were set to 0 and 255 (minimum and maximum values) according to statistical tests within the Volocity program (Costes et al. (2004) as this eliminates user bias. Rectangles were scribed around several mitochondria and surrounding cytoplasm that contained on average 6,000 voxels for analysis. A PCC was generated by Volocity for each analyzed rectangle. The significance of the PCCs generated was determined from the critical value of t for all rectangles measured. Average PCCs generated from multiple images with mitochondrial co-localization of (16-27 day old) Plp1tg mice had a significant positive PCC for the co-localization of PLP and COX1 (PCC>+0.6, P<0.001 (n=39). In a young 14DPN wt-mouse, the PCC for areas where PLP and COX1 were closely associated was smaller and not significant (PCC=+0.2, P>0.3 n=17) and for older (7 month) wt-mice (PCC=-0.072, P>0.3 n=6) showing little and no co-localization of PLP and COX1 respectively. Similarly, PCC values were calculated from 2 PMD duplication patients. Rectangles scribed around mitochondria of Olgs exhibiting yellow fluorescence had an average PCC value of 0.741 whereas rectangles scribed around either red or green fluorescence had an average PCC value of 0.281.
PMD Tisssue
Frozen cerebral autopsied tissue (kindly provided by Dr. J. Garbern while at Wayne State University) from 47, 50, and 54yr old PMD patients with a duplication of the PLP1 gene was used for this study (patients 1-3 respectively; Sima et al., 2009). Small blocks of tissue were dissected from corpus callosum and base of cortex, thawed in 4% paraformaldehyde in 0.1M PBS for 72 hrs and placed in PBS containing 20% sucrose for 72 hrs. Fifty-micron sections were cut with a Vibratome (St. Louis, MO) and sections immunostained for PLP and COX1 as described above. Imaging of the tissue was done on a Leica TCS SP5 Confocal Microscope. Images were analyzed for co-localization by measuring the Pearson's Correlation Coefficient using the Volocity as described above in areas of yellow staining and analyzed for non-co-localization in areas of red or green alone.
DNA constructs
Plasmid clone 68 of pDM100 (pDM100.68) contained a full-length cDNA for mouse Plp1 (kindly provided A. T. Campagnoni, University of California at Los Angeles, Los Angeles, CA). Full-length cDNA of mouse Plp1 was amplified by PCR and cloned into the pEGFP-N1 and pAcGFPC1 vectors (Clontech, Mountain View, CA) at the EcoRI/BamHI site to produce two different constructs. The PCR cycling conditions were one cycle at 94°C for 2 min, 29 cycles at 94°C for 15 sec, 58°C for 30 sec and 68°C for 1 min, and then one cycle at 68°C for 6 min. The constructs were PLP–EGFP and pAcGFP–PLP. The resulting plasmid constructs were propagated by standard procedures and purified using a Maxi-Prep Plasmid Kit (Qiagen, Valencia, CA). Restriction mapping and sequencing (performed at the Wayne State University Applied Genomics Technology Center) confirmed the correct sequence and orientation of the construct (Applied Biosystems, Carlsbad, CA) (Table 1).
Table 1.
PLP plasmids used for transfections and primers used to construct them.
| Construct | Primers used to make the constructs and check sequences | |
|---|---|---|
| Pln1 | PLP-EGFP | Sense 5′ATATATGAATTCGATGGGCTTGTTAGAGTGT3′ |
| pAcGFP-PLP | Antisense 5′ ATATATGGATCCCAGAACTTGGTGCCTCG 3′ | |
| C6SPlp1EGFP | Sense 5′ATGGGCTTGTTAGAGTCTTGTGCTAG3′ | |
| Antisense 5′CTAGCACAAGACTCTAACAAGCCCAT3′ | ||
| C7A Plp1HA | Sense 5′ATGGGCTTGTTAGAGTGTGCGGCTAG3′ | |
| Antisense 5′CTAGCCGCACACTCTAACAAGCCCAT3′ | ||
| C10SPlp1EGFP | Sense 5′ATGGGCTTGTTAGAGTGTTGTGTGCTAGATCTCTGGAT3′ | |
| Antisense 5′TACCAGAGATCTAGCACAACACTCTAACAAGCGCAT3′ | ||
| C24SPlp1HA | Sense 5′TTCCCTGGTGGCCACTGGATTGTGTTTCTTTGG3′ | |
| Antisense 5′CCAAACAAACACAATCCAGTGGCCACCGGGAA3′ | ||
| C34SPlp1HA | Sense 5′GGATGTGGACATGAAGCTCTCACTGGT3′ | |
| Antisense 5′ACCAGTGGAGCTTCATGTGCCACATCC3′ | ||
| C200SPlp1HA | Sense 5′ATAGGCAGTCTCTCCGCTGATGCCAGA3′ | |
| Antisense 5′TCTGGCATCAGCGAGAGACTGC | ||
| C219SPlp1HA | Sense 5′CCTGGCAAGGTTTCTGGCTCCACCTT3′ | |
| Antisense 5′AAGGTTGGGAGCCAGAAACCTTGCCAGG3′ | ||
DM20-AcGFP (Aequorea coerulescens) and Ds-Red (Discosoma sp.)-DM20 constructs were also generated, partially as controls for PLP but mainly for a separate study to exam interactions of PLP and DM20. Briefly, a DM20 cDNA was synthesized by Blue Heron (Bothell, Washington) and fused to the N-terminus of AcGFP (Clontech). DM20 cloning was verified by DNA sequencing. A DsRed-monomer-C1 (Clontech) was fused to the N-terminus of the DM20 cDNA.
Site-directed mutagenesis
PLP mutants C6SPlp1EGFP, C10SPlp1EGFP were created by site–directed mutagenesis, where cysteine is mutated to serine. Mutations were created by PCR site-directed mutagenesis with the High Fidelity DNA Polymerase enzyme Pfu (Quick change® site-directed mutagenesis, Stratagene, La Jolla, CA). Table 1 shows the constructs created and the sequences used to generate them. Highlighted bases indicate the point of mutation from a cysteine to a serine or an alanine. The cycling conditions were 95°C for 5 min, followed by 17 cycles of 95°C for 50 sec and 58°C for 50 sec followed by one cycle of 68°C for 6 min. The HA tag was introduced into the C-terminus of PLP and sub-cloned into a pCGN mammalian expression vector (Tanaka and Herr, 1990) to generate C7APlp1HA, C25SPlp1HA, C35SPlp1HA, C201SPlp1HA, C220SPlp1HA cDNAs which were kindly provided by Dr. Kamholz (Wayne State University). All constructs were sequenced at the Wayne State University Applied Genomics Technology Center to confirm correct sequence and orientation.
COS7, 158N Immortalized and Primary Oligodendrocyte Cell Cultures
COS7 cells were cultured in DMEM (GIBCO BRL, Grand Island, NY) containing 10% fetal bovine serum (FBS) (GIBCO BRL) supplemented with 100U/ml penicillin and 100μg/ml streptomycin (Life Technologies, USA). Immortalized 158N Olgs (Ghandour et al., 2002) were maintained in DMEM (GIBCO BRL) containing 10% newborn calf serum (GIBCO BRL) supplemented with 20ng/ml of triiodothyronine (T3), 100U/ml penicillin and 100μg/ml streptomycin at 37°C in 5% CO2. Primary Olg cultures were prepared as previously described (Swamydas et al., 2009) with the addition of 20ng/ml of T3. Enriched Olgs were harvested from primary glial cultures established from 2 DPN pups on 12mm coverslips coated with poly-L-lysine (PLL, 20mg/ml) and kept in culture for 4 days before being used for immunocytochemistry. Olg cultures were >85% pure.
Transfection of COS7, 293 and 158N Cells
Cells were transiently transfected 3-4 days after plating with Lipofectamine™2000 reagent containing 100ng of plasmid cDNAs (wtPlp1EGFP, wtPAcGFPPlp1, C6SPlp1EGFP, C7APlp1HA, C10SPlp1EGFP, C25SPlp1HA, C35Plp1HA, C201SPlp1HA and C220SPlp1HA) according to the manufacturer's instructions (Life Technologies). Transfection efficiency ranged between 65-70%.
Immunocytochemistry of Cultured Cells
COS7 or 158N cells were grown on 12 mm coverslips in 6 well plates until they were 70-75% confluent. At 36 hr post-transfection, cells were stained live with 90μM MitoTracker Red (Invitrogen) for 20min at 37°C. Cells were fixed in 4% paraformaldehyde for 10min at 4°C, rinsed in PBS, permeabilized with 0.3% Triton or chilled methanol for 15 min), rinsed in PBS and immunostained with a PLP specific antibody, diluted 1:100 or with an anti-HA chicken IgG antibody (Millipore Corp. Billerica, MA), diluted 1:200 in PBS for 1 h at RT. Coverslips were rinsed in PBS and incubated with goat anti-rabbit IgG antibody conjugated to Texas Red (Jackson ImmunoResearch, West Grove, PA), or with an anti-chicken IgG antibody conjugated to Alexa 488 (Life Technologies) diluted 1:100 in PBS for 1 hr at RT. For immunostaining with the antibody against the C-terminal end of PLP (1:250) and antibody to COX subunit IV (1:300) for mitochondria (Life Technologies), cells were processed for antigen retrieval (0.1M Tris, pH 8.0 for 10 min at 95°C, and then 30 min at RT), rinsed and incubated with primary antibody for 1 h, rinsed and then incubated in goat anti-rabbit IgG Texas Red (Jackson Labs) 1:300 and goat anti-mouse IgM Cy2, rinsed, DAPI stained and mounted on slides with Aquamount (Polysciences, Warrington, PA). All steps were carried out at RT. In some experiments, transfected cells were live-stained with the O10 antibody that recognizes the extracellular epitope of PLP. O10 antibody was added to coverslips on ice for 45 min in the dark, followed by fixation in 4% paraformaldehyde for 45 min, rinsed, incubated in anti-mouse IgM Texas Red for 45 min at RT, rinsed, DAPI stained and coverslips mounted. Cells were imaged on a Leica TCS S5P microscope and images were combined in Photoshop. 3D images were compiled from z-stack images consisting of 14 planes along the z-axis using Volocity 3D Image Analysis Software (Perkin Elmer). 3D images are displayed showing the XY-Z axis. Merged images were deconvolved using the Huygens Essential Deconvolution Software (Scientific Volume Imaging).
Four day Olg cultures were either fixed with 4% paraformaldehyde and stained with a PLP specific antibody or live-stained with the O10 antibody and then fixed. Cells were then stained with the COX1 antibody. Secondary antibodies used were a goat anti-rabbit IgG Texas Red (Jackson ImmunoResearch Laboratories, Inc) or goat anti-mouse IgM Texas Red and a donkey anti-mouse IgG Cy2 (Jackson ImmunoResearch Laboratories, Inc). More than 55% of Olgs showed PLP and COX1 co-localization.
pH, ATP and Lactate Measurements
COS7 cells were plated at a density of 10,000 cells on sterile 12mm diameter glass coverslips coated with PLL with 4 coverslips per well in a 6 well plate. When cells were 70% confluent, they were transfected with 100ng wtPlp1EGFP cDNA, C6SPlp1EGFP and non-transfected COS7 cells served as controls. Similar and simultaneous experiments were performed using C10SPlp1EGFP and C7APlp1HA cDNA. The media was changed 10-16 h post-transfection and media was allowed to remain on the cells for 5 days. After 5 days, the media from each well was separately collected. An aliquot of the collected media was equilibrated to RT, and the pH measured using a standard calibrated pH meter. The remaining media was stored at −80°C. After the media was collected, coverslips were live-stained for the O10 antibody, fixed and then DAPI stained and the percentage of transfected cells calculated. The cells were imaged with a Zeiss ApoTome Imaging System. In subsequent experiments, COS7 cells were cultured in 6 well plates until confluent, transfected with wtPlp1EGFP cDNA, C6SPlp1EGFP, and C10SPlp1EGFP cDNAs in triplicate and non-transfected cells served as controls. Media was changed the next day and plates were cultured for an additional 5 days without changing the media. On day 6, 200μl of culture media was collected in 1.5ml Eppendorf tubes and immediately frozen in dry ice; the remaining media was aspirated, cells were washed in 0.1M PBS and immediately frozen. Coverslips and media were later processed to measure cellular and extracellular ATP levels respectively using the ATP Bioluminescence Assay Kit HS II following the manufacturers instructions (Roche Applied Sciences, Indianapolis IN).
The ex vivo measurement of ATP in brain was done using a previously published procedure for measuring pH in brain (Skoff et al., 2004a). Briefly, Plp1tg mice at 3-11 wks and C57BL6/J and B6129/J wild type mice at 3-11 wks were sacrificed by cervical dislocation, brains removed and 2-2.5mm coronal sections were dissected using a razor blade, weighed, and cut into 16 cubes and placed in a vial on ice containing 1ml ice-cold artificial cerebral spinal fluid (aCSF) (124mM NaCl, 3mM KCl, 3mM CaCl2, 1.5mM MgCl2, 26mM NaHCO3, 1mM NaH2PO4, 10mM D-glucose, and 75μM HEPES) and a magnetic stir bar. Fluid and tissue were gently stirred on a magnetic stir plate for 5 min. Afterwards the contents of the vials were transferred to 1.5ml centrifuge tubes and centrifuged for 1 min. Brain tissue and aCSF was separated and immediately frozen in liquid nitrogen and ATP measured as above. ATP of each sample was measured in triplicate, averaged, and then averaged for each group of brains.
The ex vivo measurement of lactate was made from 2-2.5mm coronal brain slices cut into 16 cubes and immersed in 1ml of aCSF and stirred for 5 min at RT as described above. A 10μL sample of aCSF was added to 1ml of Lactate Reagent (Sigma, St. Louis, MO, USA) incubated for 10 min at RT in the dark and the absorbance at 540 nm was measured. Each sample was measured in duplicate and lactate concentration of aCSF determined by extrapolation from a standard curve made from measuring the absorbance of a lactate standard (Sigma, St. Louis, MO, USA) for each assay. Averages were plotted from 4 experiments using a total of 9 C57BL6/J and B6129 wild type mice at 4-11 wks and 13 Plp1tg mice at 4-11 wks.
The in vitro measurement of lactate was performed on the media and cell lysates of 293 cells transfected with PLP, DM20 and EGFP. Cells were seeded at 5000 cells per well in 12 well plates and the next day the media was removed and replaced with fresh media. One μl of media was added to 100μl lactate reagent as described above and absorbance measured at 540nm. This was repeated 2 and 3 days after plating. After 3 days, the media was removed and the cells were lysed, and lactate in the cells measured by adding 10 μl of the cell lysate to 1 ml lactate reagent and measuring absorbance at 540 nm. LDH-Cytotoxicity Assay Kit (Biovision, Milpitas, CA) was used to determine if the increase in lactate was due to massive cell death. Lactate dehydrogenase (LDH) in the media and cells was measured to determine an estimate for the percentage of live cells in culture. The measurements were performed according to the manufacturer's protocol. Briefly, supernatant was collected and the LDH measured using Sigma colorimetric standards. The cells attached to the plate were lysed and the LDH in the cell lysate was measured using the same standards. Total LDH was calculated as the sum of LDH in the media and LDH in the cell lysate. The ratio of LDH in the cell lysate to total LDH was used to represent the percentage of viable cells in each plate.
RESULTS
Co-localization of PLP with Mitochondria in Plp1tg and Normal Mouse Brain
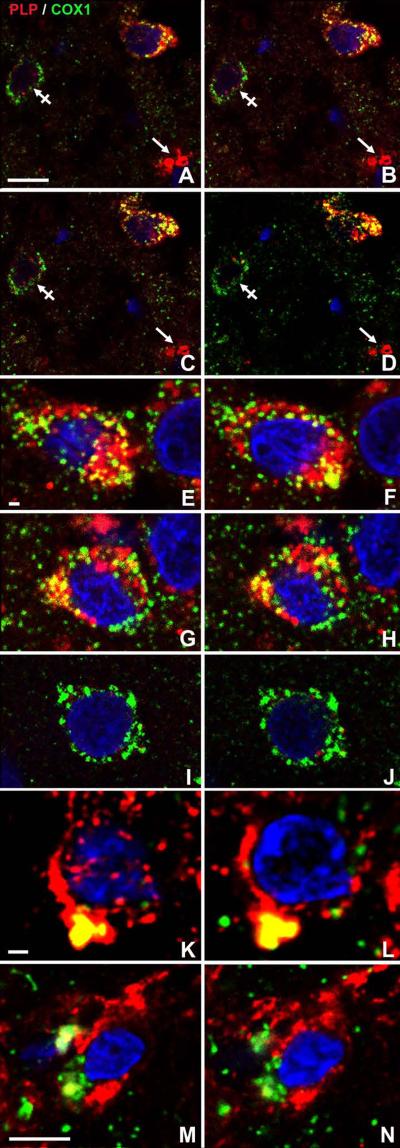
Since PLP co-localizes with mitochondria in COS7 cells transfected with wtPlp1 cDNAs (Hüttemann et al., 2009), we next investigated whether PLP co-localizes with mitochondria in Plp1tg mice with extra copies of the wtPlp1 gene. Mouse cerebra were stained with an antibody that recognizes PLP but not DM20 and an antibody to cytochrome c oxidase subunit 1 (COX1), a mitochondrial encoded protein. Z stack images from a 35 days postnatal (DPN) Plp1tg mouse shows two sets of Olgs (Fig. 1 A-C, D-E) in which PLP co-localizes with mitochondria (yellow). The low magnification picture (Fig. 1A) shows the vast majority of mitochondria are exclusively stained green for COX1; these intensely stained mitochondria are not Olgs because perikarya of Olgs are intensely stained for PLP (red). By scanning through Z stacks, it is important to document that co-localization of PLP and COX1 often changes within an individual mitochondrion. This is not unexpected because the imaged plane of section may ideally pass transversely through a mitochondrion such that the outer mitochondrial membrane, the inner mitochondrial space (IMM), inner membrane and then matrix are visualized. Alternatively, the imaged plane of section may obliquely pass through the mitochondrion such that only the outer membrane is grazed, etc. Frequently and as expected, the central core of a mitochondrion immunostains green (COX1), then the central core is surrounded by yellow fluorescence (COX1 and PLP), and the mitochondrial boundary surrounded by red fluorescence (PLP). We interpret this pattern to be COX1 at the interface between the mitochondrial matrix and the inner mitochondrial membrane, PLP and COX1 in the IMM space as it co-localizes with creatine kinase (Hüttemann et al., 2009) and PLP adjacent to mitochondria trafficking from ER/Golgi. Another set of imaged slices through several Olgs (Fig. 1 D-E) shows PLP co-localizes in most but not all Olg mitochondria; this is a common finding. This observation may be due to a combination of factors including the sensitivity of imaging, the immunocytochemical procedure, and the likelihood that every Olg mitochondrion does not contain PLP. PLP abundantly co-localizes with mitochondria in Olg perikarya but it is also present in the myelin sheaths (Figs. 1D-E).
Figure 1.
Confocal images from a 35DPN Plp1tg mouse cerebrum (A-E) and a 17 DPN wt-C57Bl6 mouse cerebrum (F) double immunostained with a PLP specific antibody and the mitochondrial encoded protein COX1. (A) Low magnification scan shows the vast majority of mitochondria are immunostained for COX1 (green) but not for PLP (red). (B-C) Higher magnification of this area in a different Z-axis shows two Olgs in which most mitochondria are double stained. (C) In the deconvolved image of B, co-localization of PLP in mitochondria of myelin sheaths (arrowhead) is common. The same PLP+ mitochondria (arrows) are shown in the different images. (D-E) Two deconvolved images in slightly different focal places show many but not all mitochondria in Olgs contain PLP (arrows). Note abundance of PLP throughout cytoplasm of these Olgs. (F) Co-localization of PLP with mitochondria also occurs to a very limited extent of normal mouse brains during myelination and in aged mice. In a 17DPN mouse cerebral cortex, 2 Olgs show PLP co-localizes with COX1+ mitochondria (arrows). The vast majority of DAPI stained cells (lower right) are not Olgs as shown by lack of PLP staining and presence of COX1+ mitochondria. Magnification bar=10μm (A-E) and 5μm (F).
Three Plp1tg mice have been examined with confocal microscopy for co-localization of PLP in mitochondria. In a 20 DPN moribund mouse, approximately 90% of Olgs contained mitochondria with PLP; in a 30 DPN mouse, illustrated here, approximately 50% of Olgs contained mitochondria with PLP; and in a 21DPN mouse, less than 10% of Olgs had mitochondria with PLP. These percentages are estimates based upon low magnification scans of cerebral corpus callosa, striata and cortices. In contrast to Plp1tg mice, where co-localization of PLP in Olg mitochondria is easily observed, co-localization is extremely rare in wild-type mice, even after extensive scans of cerebral white matter. The lack of co-localization in normal brain is not a function of poor immunostaining as red (PLP) and green (COX1) fluorescence is bright in normal tissue (Fig. 1F). An important caveat to the above statement is that PLP co-localizes to Olg mitochondria in normal brain but only in a narrow window of time and space. Pre-myelinating Olgs in young mice (Fig. 1F) and occasionally in dying Olgs of aged mice show co-localization. We are presently quantifying the frequency and location of this event (see Discussion).
To confirm that PLP and COX1 are co-localized within mitochondria, Volocity (Perkin Elmer), a program with co-localization features, was used to statistically determine whether PLP and COX1 are co-localized within individual pixels. Co-localization of PLP and COX1 in Plp1tg mice was highly significant but insignificant in wt-mice (see Materials and Methods for imaging, data details, and statistical methods utilized by the Volocity program).
Co-localization of PLP with Mitochondria in PLP1 PMD Patient Brains with Duplications
Because PLP in humans and rodents is identical at the amino acid level and the PLP1/Plp1 coding sequence itself is highly conserved (95%) in humans and rodents (Hudson and Nadon, 1992), it is reasonable to predict that cellular mechanisms, including insertion of PLP into mitochondria, are similar in both rodents and PMD patients with duplications. Similar pathology would seem especially likely in PMD cases and transgenic rodents with extra copies of wtPLP1/Plp1 gene. To confirm that PLP is also transported to mitochondria in PMD patients with duplications, we processed autopsy tissue for ICC from the cerebra of 3 PMD patients who had duplications (Woodward et al., 2005; Sima et al., 2009). A low magnification Z-axis stack of an Olg from a 54 year-old PMD patient (Fig. 2A-D) shows co-localization of PLP with COX1. Another cell in this stack lacks PLP but contains numerous mitochondria immunostained for COX1. Myelin sheaths, immunostained red for PLP, are also present, indicating specificity of PLP for myelin and Olgs. A higher magnification Z-axis stack (Fig. 2E-H) shows 2 different Olgs in which PLP co-localizes with mitochondria. As with mitochondria in Plp1tg mice, not all Olg mitochondria in this PMD patient showed co-localization with PLP. As expected, the vast majority of cells contain COX1 mitochondria but no PLP (Fig. 2I-J). The 47 and 50 year-old PMD duplication patients also showed co-localization of PLP with mitochondria (Fig. 2K-N) Volocity, used to determine extent of co-localization of PLP and COX1 in areas of yellow fluorescence, yielded similar Pearson's Correlation Coefficients (PCC) as that seen in the Plp1tg mice with extra copies of the wtPlp1 gene. The average PCC for the PMD samples was 0.741 for areas of yellow fluorescence compared to 0.6 in the over-expressers. In contrast, in areas of red or green fluorescence of the PMD samples, the PCC was 0.281 compared to 0.2 in the over-expressers. These images are the first report of co-localization of PLP in mitochondria of PMD patients. Toluidine Blue 1μm plastic sections of cerebral white matter from these patients show a dearth of myelin with extensive axonal degeneration (data not shown; see Sima et al. 2009 for light microscopic histology of these PMD patients with mutations).
Figure 2.
Confocal images of cortex adjacent to corpus callosum from 3 PMD patients with duplications (A-J) 54 years old; (K-L) 47 years old and (M-N) 50 years old (patients 1-3, Sima et al., 2009). Sections were immunostained with a PLP specific antibody and a mitochondrial encoded protein COX1. Nuclei are stained with DAPI. All 3 patients show PLP co-localizes with mitochondria (A-D) Low magnification Z-axis confocal stack of autopsied brain from a 54yr PMD patient with a PLP1 duplication. An Olg, identified by red PLP staining in cytoplasm, has many COX1+ mitochondria that contain PLP. Some mitochondria in this cell are not stained for PLP. A non-Olg cell (crossed arrow) lacks PLP but has many COX1+ mitochondria. Many mitochondria scattered throughout the neuropil stain only green for COX1+, indicating they are in cytoplasm of non-Olgs. Myelin sheaths (arrow) cut transversely, are only immunostained red. (E-J) High magnification of cerebral white matter cells from the same PMD patient illustrated above. (E-F, G-H) Serial, confocal images of two different Olgs have mitochondria in which PLP co-localizes (yellow). Not all COX1+ mitochondria in these two cells contain PLP as many are stained only green. The cytoplasm is packed with organelles that contain PLP, indicating this cell is an Olg. (I-J) Most cells have mitochondria that immunostain green for COX1 but show no red immunostaining for PLP. (K-L, M-N) Two Olgs from PMD patients with duplications, identified by abundant PLP in their cytoplasm, show PLP is present in some but not all mitochondria. Magnification bar=10μm (A-D) 1 μm (E-L) 5μm (M-N).
Co-localization of PLP in Mitochondria of Primary Cultured Olgs
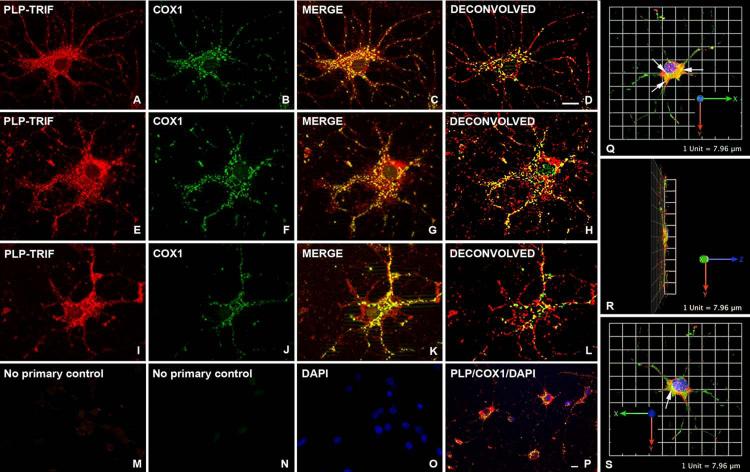
We next asked whether PLP co-localizes with mitochondria in cultured Olgs. Olgs derived from 2DPN B6CBA wild-type mice were cultured for up to 5 days in serum-free media supplemented with T3 following shake-off from mixed brain cultures. One-to-2 days after plating, PLP is not detectable or it exhibits low fluorescence throughout Olg cytoplasm and, consequently, little co-localization is observed (not shown). Five days after plating, the Olgs have matured and express abundant perinuclear PLP (Fig. 3). These Olgs are packed with mitochondria in which PLP co-localizes. The percentage of Olgs that showed co-localization of PLP with mitochondria at this time-point was >55%. Again, as with Olgs in vivo, PLP does not co-localize with all mitochondria because mitochondria, which are green only (COX1 stained), are visible (see especially Fig. 3K-L). The fluorescence from PLP and COX1 ICC is not artefactual because elimination of the primary PLP and COX1 antibodies but inclusion of secondary antibodies, shows negligible background staining (Fig. 3M-N). This control was processed together with the experiments that generated the pictures for Fig. 3. Co-localization of PLP in mitochondria of immortalized Olgs has previously been shown (Hüttemann et al., 2009; Baarine et al., 2009). 3D images (Fig. 3Q-S) produced using the Volocity 3D Image Analysis software demonstrates that PLP co-localizes with COX1 in mitochondria and does so mainly in the perikaryon between the nucleus and the apical portion of the cell.
Figure 3.
Co-localization of PLP in mitochondria of cultured primary Olgs 5 days after shake-off from primary glial cultures. (A-L) Olgs were double immunostained with a PLP specific polyclonal antibody and a COX1 monoclonal antibody. PLP is abundant throughout Olg cytoplasm whereas COX1 immunostaining is restricted to spherical and rod shaped structures. Merged images show abundant co-localization of PLP to mitochondria. (K) This Olg exhibits less co-localization than Olgs above, even though PLP and COX1 immunostaining are of similar intensities. (D, H, L) Deconvolved images of merged images show PLP and COX1 co-localization. (M-N) Elimination of primary PLP and COX1 antibodies and incubation with secondary antibodies shows virtually no staining, indicating red and green fluorescence are not due to secondary antibody staining. (O) DAPI staining of same field shows cells were present on the coverslip. (P) Low magnification of an Olg enriched culture shows the majority of PLP+ cells have mitochondria containing PLP. Cultures were grown in defined media including T3 that seems to facilitate Olg differentiation and co-localization. (Q-S) 3-D reconstructions obtained from the Volocity program show a cultured Olg spread out on the coverslip in the X-Y plane. Co-localization of PLP and COX1 in mitochondria (arrows) occurs mainly in the perikaryon in a plane between the nucleus and the apical portion of the cell. Magnification bar = 10μm
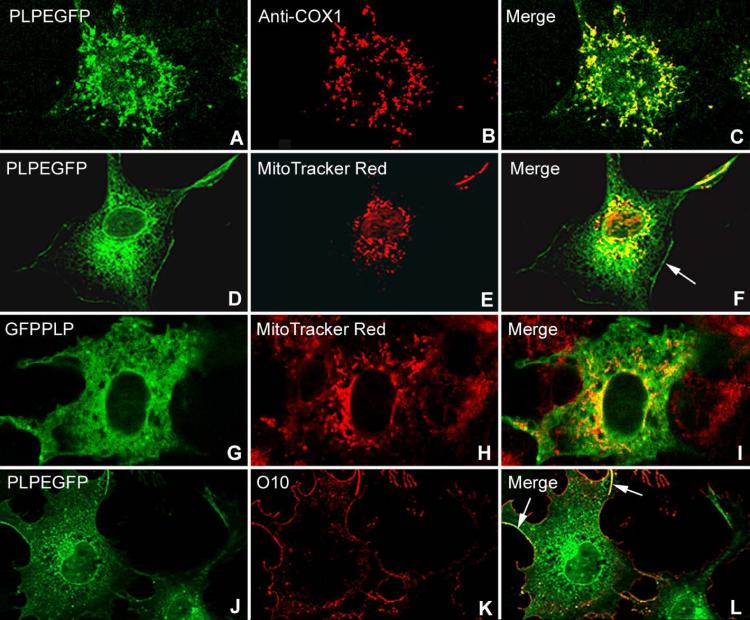
Co-localization of wt Plp1EGFP and GFPPlp1 cDNAs in COS7 Cells
To investigate the insertion of PLP into mitochondria, COS7 cells were transiently transfected with 2 different cDNA constructs; one encoding a wtPLP1 cDNA with EGFP at the COOH terminus and another with GFP tagged in front of the N-terminus (Fig. 4). Each construct was made as a control for the other; namely, to show that the tags did not interfere with the insertion of PLP into membranes. Mitochondria were immunostained with the COX1 antibody or with MitoTracker Red. Both constructs, when combined with either mitochondrial marker, show co-localization of wtPLP with mitochondria (Fig. 4A-I). wtPLP is also correctly inserted into the plasma membrane, confirmed by live staining with the O10 antibody that detects a conformationally specific epitope in the extracellular loop (Fig. 4 J-L) (Jung et al., 1996). Additional images that show insertion of wtPLP into mitochondria of COS7 cells are published in Hüttemann et al. (2009). Transfection of several other CNS and PNS myelin protein cDNAs, including myelin basic protein isoforms, do not co-localize with mitochondria (Hüttemann et al., 2009).
Figure 4.
(A-I). Co-localization of PLP to mitochondria in COS7 cells 48 h after transfection of different wtPlp1 cDNA constructs. Cells were stained with MitoTracker Red or with the COX1 antibody specific for mitochondria. Both Plp1EGFP (A-F) and GFPPlp1 (G-I) constructs show abundant co-localization with mitochondria. PLP green fluorescence is brightest around the nucleus and then diminishes in intensity as it is transported into processes. PLP is transported to the plasma membrane (F) (arrow) and correctly inserted into it as shown by staining live COS7 cells (J-L) with the O10 antibody (yellow-orange fluorescence) (arrows). Several non-transfected cells (G-I, margin area) have bright red mitochondria but lack green fluorescence. Differences in quality of images are due to different “confocal” microscopes, the Leica TCS SP5 providing sharper images than the Zeiss ApoTome.
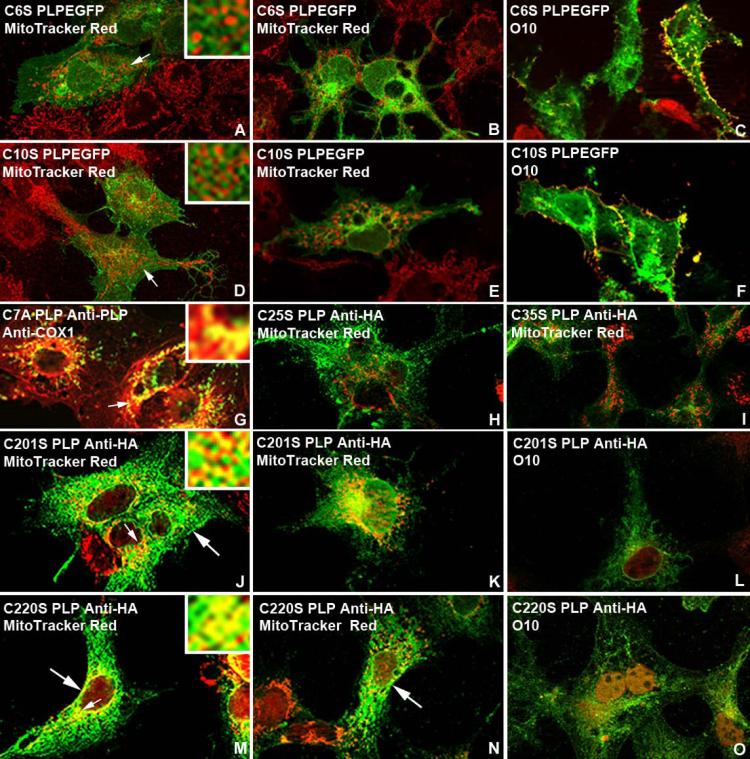
Cysteine Mutations of PLP Demonstrate Specific Mitochondrial and Plasma Membrane Targeting Motifs
Specificity of insertion of PLP into mitochondria is shown by site-directed mutagenesis of cysteines in the N-terminus, first trans-membrane domain, and extraceullular loop. PLP contains CX3C and CX9C twin disulfide cysteine motifs (C6, C10 and C25, C35) that are required for the Mia40/Erv1 redox-coupled mitochondrial protein import pathway (Messecke et al., 2005, 2008; Herrmann and Riemer, 2012). To determine whether specific cysteine residues in PLP are required for import into mitochondria using the Mia40/Erv1 pathway, cysteine residues were substituted to serine or alanine by site directed mutagenesis (Fig. 5). C6SPlp1EGFP, C7APlp1HA, C10SPlp1EGFP, C25SPlp1HA, C35SPlp1HA C201SPlp1HA, and C220SPlp1HA cDNAs were transfected into COS7 and 158N Olg (data not shown) cells. Thirty-six hours post-transfection, the coverslips were processed with MitoTracker Red or anti-COX1 and anti-PLP carboxy terminus or anti-HA antibodies as appropriate. PLP with the C6SPlp1EGFP, C10SPlp1EGFP, C25SPlp1HA, and C35SPlp1HA mutations does not co-localize to mitochondria (Fig. 6A-B, D-E, H-I). However, these constructs do not prevent PLP from its correct insertion into the plasma membrane as shown by live staining with the O10 antibody (Fig. 6C, F). The mitochondria in both transfected and non-transfected cells are brightly stained and the EGFP is brightly fluorescent, indicating the lack of co-localization is not a staining artefact. In striking contrast, PLP with the C7A mutation, which theoretically is not a component of the disulfide relay system, strongly co-localizes with mitochondria (Fig. 6G).
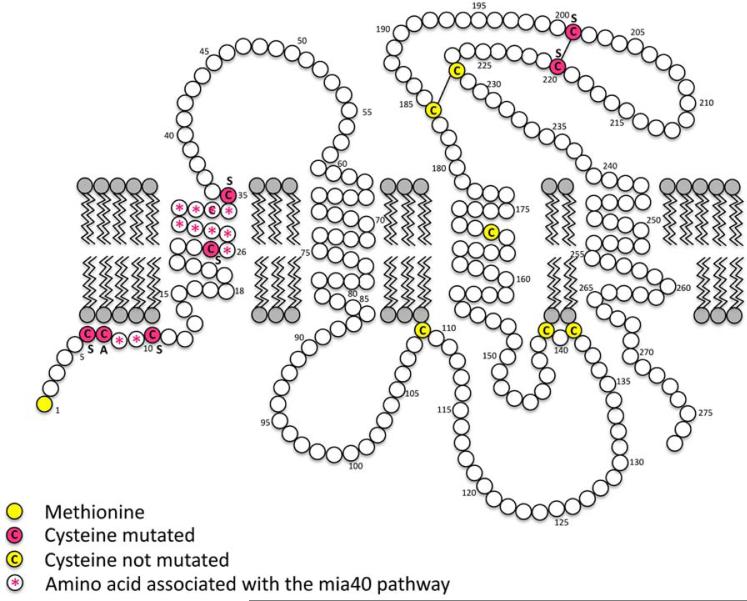
Figure 5.
Orientation of PLP in the plasma membrane. Site-directed mutagenesis of cysteines are in red; all other cysteines are in yellow; disulfide bridges are marked with a line. Diagram adapted from Hudson et al. (2004).
Figure 6.
Transient transfection of COS7 cells with C6SPlp1EGFP (A-C), C10SPlp1EGFP (D-F), C7APlp1HA (G), C25SPlp1HA (H), C35SPlp1HA (I), C201SPlp1HA (J-L) and C220SPlp1HA (M-O) cDNAs C6SPlp1EGFP, C10SPlp1EGFP, C25SPlp1HA, and C35SPlp1HA constructs do not co-localize with mitochondria stained with MitoTracker Red. Failure to co-localize with mitochondria does not prevent their proper cell surface expression in the plasma membrane as shown by live O10 immunostaining (C, F). The C7APlp1HA cDNA, C201SPlp1HA, and C220SPlp1HA constructs co-localize with mitochondria. No positive PLP plasma membrane staining is detected with O10 antibody (L, O) in the C201SPlp1HA and C220SPlp1HA constructs even though PLP is transported to the membrane (large arrows). Insets in A,D,G,J, M show at higher magnification areas of cytoplasm (small arrows) where PLP either does or does not co-localize with mitochondria.
PLP with cysteine mutations in the extracellular loop, C201SPlpHA and C220SPlpHA, co-localizes to mitochondria of COS7 cells (Fig. 6J-K, M-N). However, live O10 staining of the same COS7 cells demonstrates that PLP with these mutations does not co-localize to the plasma membrane (Fig. 6L, O). This staining pattern for C201SPlp1EGFP and C220SPlp1EGFP mutants is predicted because disruption of the disulphide bridges is required for proper folding and orientation of the protein into the plasma membrane (Dhaunchak and Nave, 2007). Transfection of C6SPlpEGFP and C10SPlpEGFP cDNAS into 158N Olg cells shows limited co-localization of EGFP-PLP with mitochondria but this is expected due to endogenous PLP co-localization with mitochondria (data not shown).
Transfection of wtDM20 that expresses either dsRed-Plp1 or Plp1-EAcGFP cDNAs into COS7 cells (see Materials and Methods) shows no or very limited co-localization with COX1 or MitoTracker Red stained mitochondria (data not shown). (The transport and distribution of DM20 is part of an other study). Failure to find co-localization of DM20 may be due to many factors that include its orientation into the plasma membrane (see Discussion).
pH Acidification: PLP Insertion into Mitochondria Regulates Extra-cellular pH in COS 7 Cultured Media
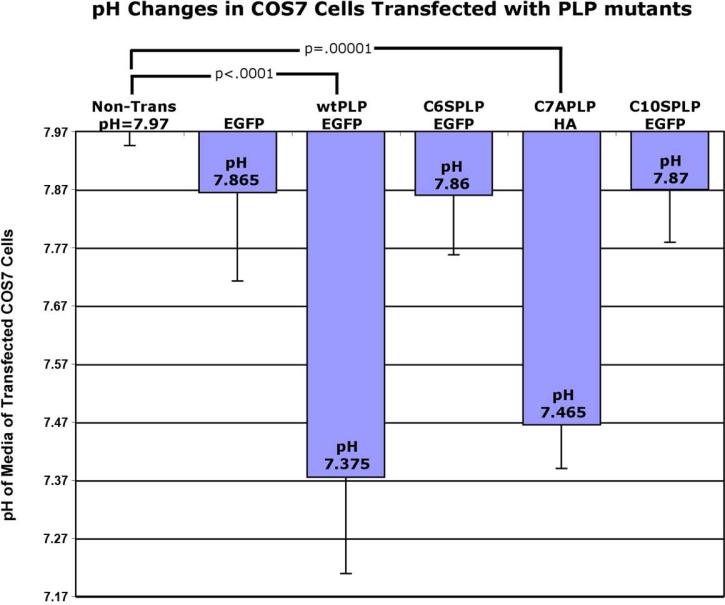
We previously showed that cells over-expressing wtPlp1 caused a dramatic acidic shift in their media (Boucher et al., 2002) and in artificial extracellular fluid (aECF) from Plp1tg brains (Skoff et al., 2004a). However, we did not know the mechanism that led to this acidic shift. Here, we ask whether the acidic shift is due to translocation of wtPLP into mitochondria. Nontransfected COS7 cells or COS7 cells transfected with EGFP have normal pH values of 7.97 and 7.87, respectively. Transient transfection of Plp1EGFP cDNA into COS7 cells leads to significant (p<0.0001, unpaired t test) media acidification with a pH of 7.4, a value similar to our previous findings with 293 cells transfected with wtPlp1cDNAs (Boucher et al., 2002). Transfection of a C7APlp1HA cDNA that permits insertion of PLP into mitochondria also reduces pH to 7.5 (P<0.00001, unpaired t test). In contrast, the C6S and C10S Plp1EGFP constructs had no significant effect on the pH of the media (pH 7.9), identical to controls (Fig. 7). It is important to note that the media in both our previous and present experiments was unchanged for 3-5 days, permitting the cells to alter their extracellular milieu. Plating of cells and transfection efficiency was controlled at the beginning of the experiment by starting with the same number of cells per coverslip, beginning transfections when coverslips were 70% confluent and allowing cells to grow 5 days after transfection. The percentage of transfected cells at the end of each experiment was 65% for all permutations. Clearly, insertion of wtPLP into mitochondria explains the acidification of extracellular media.
Figure 7.
pH of media of COS7 cells transfected with different PLP1 constructs. Non-transfected and EGFP transfected COS7 cells have normal pH values of 8.0 and 7.9, respectively. COS7 cells transfected with C6SPlp1EGFP and C10SPlp1EGFP, which prevent insertion of PLP into mitochondria, have pH values of 7.9, identical to non-transfected COS7 cells. In contrast, COS7 cells transfected with a wtPLP1EGFP and C7APlp1HA cDNAs, which permit insertion into mitochondria, have a pH of 7.4. The pH measurement for each cell group is based upon 2-4 different experiments with 4-8 wells measured for each group. Two-tailed T-test between nontransfected and transfected groups. Error bars are SD.
ATP Levels in vitro and in vivo
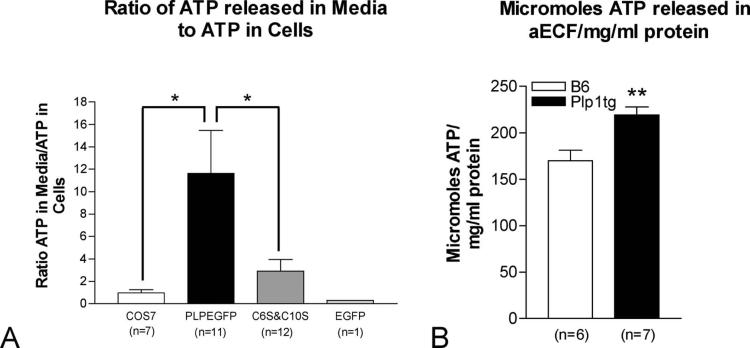
ATP levels in Plp1tg total brain homogenates are severely reduced by 50% compared to wt mice (Hüttemann et al., 2009). Similarly, other markers of oxidative phosphorylation are grossly abnormal in Plp1tg mice, indicative of whole brain metabolic deficiencies. Since increased extracellular ATP levels activate microglia by up-regulating cytokines, and microglia and cytokines are highly up-regulated in the Plp1tg mice (Tatar et al., 2010), we asked whether insertion of PLP into mitochondria caused altered intra- and extra-cellular ATP levels. Non-transfected COS7 cells, and COS7 cells transfected with EGFP, Plp1EGFP, C6SPlp1 and C10SPlp1 cDNAs were analyzed 4 days after transfection. First, all the media was aspirated from the wells to measure ATP levels in the media in μmol/L and, second, homogenates were prepared from the cells to measure cytosolic ATP in μmol/mg protein. The ratio of ATP in the media to cytosolic ATP significantly (p<0.05, unpaired t test) increased about 11-fold in the over-expressing Plp1EGFP cells compared to non-transfected cells, indicating more ATP is released into the media by these cells (Fig. 8A). The ATP ratio between the Plp1EGFP and the C6S and C10S constructs, which prevent insertion of PLP into mitochondria, was likewise significantly (P<0.05, unpaired t test) increased 3-4 fold. It is important to note that the amount of ATP in the media is extremely low compared to the amount in the cytosol.
Figure 8.
The ratio of ATP released into media by different COS7 cell groups to levels of ATP in homogenates (A) and the measurement of ATP in micromoles in aECF per amount of brain protein in brain homogenate in mg/ml (B). ATP in cell homogenates was measured in μm/mg protein and ATP in media was measured in μmol/l. (A) The fold change in the ratio between non-transfected COS7 and COS7 cells transfected with EGFP cDNA is about 1. In contrast, the ratio in cells transfected with the wtPlp1EGFP construct is roughly 11 fold greater than in nontransfected COS7 cells. This indicates more ATP is released into the media by wtPlp1EGFP expressing cells. The ratio of ATP released in media to ATP in cells for the COS7 cells transfected with C6SPlp1EGFP and C10SPlp1EGFP, which prevent insertion of PLP into mitochondria, is nearly 4-fold less than for the wtPlp1EGFP expressing cells. (B) Micromoles of ATP released into aECF corrected for protein concentration is significantly increased in Plp1tg compared to COS7 controls. Error bars are SEM (A) and SD (B). Significance p < .05 (A) and p=.0052 (B).
We also measured ATP released into aECF from brain slices of Plp1tg and B6129 wild type mice, similar to methods for measuring pH. ATP levels in Plp1tg mouse aECF, corrected for mg/ml of total brain protein, is increased approximately 30% compared to wild type (Fig. 8B). The in vitro experiment shows that the insertion of PLP into mitochondria contributes to higher levels of ATP in the media.
Lactate Levels in vivo and in vitro
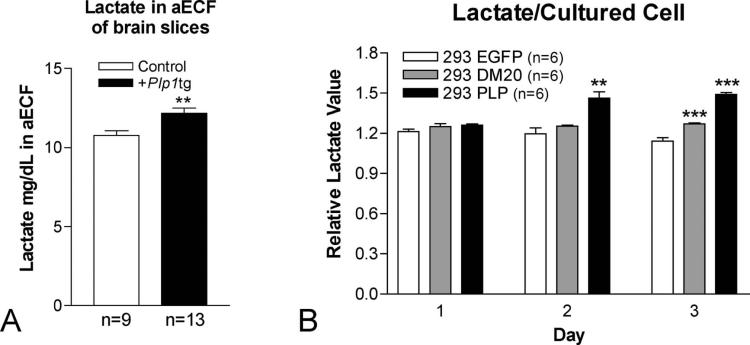
Since pH of aECF in vivo and media in vitro is significantly lowered due to over-expression of the wtPlp1 gene, we hypothesized that lactate is a possible source of increased protons. Lactate levels, measured in aECF prepared from brain slices are significantly (p<0.01, unpaired t test) increased approximately 13% in Plp1tg mice compared to C57 or B6129 wild type mice (Fig. 9A). While the percent increase is low, the concentration of lactate measured in aECF is not the concentration in ECF because it is significantly diluted in the aECF (see Fig.6 and Table 1 in Horn and Klein, 2010). Our measurement of lactate in aECF derived from brain slices, when converted to mM, is approximately 10-fold greater than that measured in mM in aECF of brain slices using a different procedure (Horn and Klein, 2010). However, our slices were 5x thicker and the buffer 10x more concentrated than in the Horn and Klein (2010) study, possibly accounting for the differences in concentration.
Figure 9.
Measurement of lactate from (A) brain slices incubated in aECF and lactate (B) in media measured in non-transfected 293 cells and 293 cells transfected with DM20 and with PLP. Lactate was measured in mg/dL when extrapolated from a standard curve generated using lactate standard. Measurements were averaged from 4 sets of experiments consisting of a total of 9 control and 13 Plp1tg mice. Two-tailed T-test between control and Plp1tg mice. p=.0052 Error bars are SEM (A). T-test between non-transfected and transfected groups. ** p<.01 ***p<.001. n=6 for all groups. Error bars are SEM (B).
In cultured 293 cells that express EGFP, DM20, or Plp1 constructs, lactate values in media are normal 1 day after plating for all 3 permutations. Lactate levels in 293EGFP expressing cells remain constant over the 3 days of culture whereas lactate levels in media of the Plp1 expressing cells at 2 and 3 days after plating were significantly increased (p<0.01 at 2 days; p<0.001 at 3 days, unpaired t test). Lactate levels in DM20 expressing cells 3 days after plating are also significantly increased compared to controls, suggesting DM20 expression modestly increases lactate efflux (Fig. 9B). The increase in lactate is not due to increased cell death as lactate dehydrogenase (LDH) in the lysate compared to total LDH is essentially the same at all 3 time points (Supplemental Data, Fig. 1). This experiment was stopped 4 days after the cells were plated because cell death sharply increases 5 days after plating when transfection efficiency is above 50%.
DISCUSSION
Both PMD patients and animal models exhibit a complex phenotype that shows abnormalities in macroglia, microglia, and neurons (eg., Skoff, 1976; Garbern et al., 1999; Anderson et al, 1998; Sima et al., 2009). Certainly, mutations of the PLP1/Plp1 gene, which is expressed in Olgs, are the primary cause but the cascade of events that leads to subsequent glial and neuronal abnormalities is poorly understood. Which of the multitude of abnormalities described in PMD and in the animal models causes premature death is unknown. Importantly, PLP1/Plp1 mutations fall into two broad categories, extra copies and missense mutations, and it is likely that the cascade of events leading to the animal's death are different within the two major types of mutations. In rodents with missense mutations, such as the jimpy mouse (jp) an unfolded protein response (UPR) and accumulation of misfolded PLP in the endoplasmic reticulum (ER) in Olgs has been well documented (Gow and Lazzarini, 1996; Beesley et al., 2001; Southwood et al., 2002; Sharma and Gow, 2007, Dhaunchak and Nave, 2007; Roboti et al., 2009). Whether an activated UPR and its constitutive components are the main drivers of the animals’ death in the Plp1 mutants is still unclear (Sharma and Gow, 2007). In contrast, rodents with extra copies of wtPlp1 exhibit more modest changes in markers of UPR and apoptosis (Cerghet et al., 2001). Physiologic parameters between these two mutations are also drastically different. Intracellular ATP and mitochondrial membrane potential are normal in jp mice but abnormal in Plp1tg mice (Hüttemann et al., 2009). These data strongly suggest that Olgs in the mice with extra copies of the wtPlp1 gene exhibit mainly a primary metabolic dysfunction whereas Olgs in the mice with missense mutations are mainly characterized by an up-regulated UPR.
It is essential to decipher the cause of degeneration of neurons/axons in PLP1 and Plp1 mutants because it is undoubtedly linked to dysfunction. Neuronal loss is more prevalent in patients with PLP1 duplications, particularly in their sub-cortical grey matter systems, than in patients with deletions and missense mutations (Sima et al., 2009). Axonal degeneration and transport deficiencies have been reported in rodents with all different Plp1 mutations including PLP1 deletions (Readhead et al., 1994, Anderson,et al., 1998, Klugmann et al., 1997; Boison and Stoffel 1994,1995,1997; Klugmann et al.,1997, Garbern et al, 1999). However, duplications of wtPLP1 are much more devastating than a PLP1 deletion (Garbern et al., 2002). Importantly, neuronal degeneration due to over-expression of wtPlp1 occurs independently of the presence of myelin. Co-culture of dorsal root ganglion (DRG) neurons with cells transfected with wtPlp1 cDNAs led to increased neuronal death (Boucher et al., 2002) when compared to DRG neurons co-cultured with different types of control cells. Moreover, cells transfected with wtPlp1 cDNAs but not DM20 cDNAs caused major acidification of the media (Boucher et al., 2002). This phenomenon is not a tissue culture artefact because brain slices from PLP1tg mice also dramatically acidified the artificial ECF in which they were incubated (Skoff et al., 2004a). In this study, we generated different constructs from those used in the previous study, and we were readily able to reproduce the acidification effect of over-expressed PLP in transfected cells. From this data, we hypothesized that protons and/or acidic molecules released into the media/extracellular space directly or indirectly contribute to neuronal death. In the present manuscript, we reproduce the pH acidification data and show that insertion of PLP into mitochondria is required for acidification and lactate release into media.
In our previous report (Hüttemann et al., 2009), we identified different potential mitochondrial targeting sequences in PLP and its likelihood for co-localization and enrichment in the inter-membrane space (IMM) based upon algorithms. Intriguingly, a recently described pathway for protein localization to the IMM known as the Mia40-Erv1 redox-coupled import pathway (Mesecke et al 2005; Bihlmaier et al., 2007; Terziyska et al., 2005; Longen et al., 2009; Herrmann and Riemer, 2012) utilizes unpaired twin CX3C or CX9C motifs that are present in the N-terminus of PLP. The 26kD PLP has both a CX3C and CX9C motif at C6, C10, C25, and C35. These cysteines may form transient disulfide bonds through intra-molecular disulfide bonds or protein cross-links that utilize the Mia40-Erv1 pathway permitting PLP's insertion into mitochondria. Using confocal, deconvolved imaging, we show that PLP co-localizes with Olg mitochondria in autopsied brain tissue from PMD patients with duplications (Sima et al., 2009), in Plp1tg mice and in normal mice, in cultured Olgs, and in COS7 cells transfected with wtPlp1 cDNAs. These findings are the first demonstration of a cellular defect common to both patients and rodent models, strongly suggesting that metabolic abnormalities in rodents are likely applicable to patients with duplications.
The insertion of native PLP into Olg mitochondria of animals with PLP1/Plp1 duplications begs the question of whether native PLP is inserted into Olg mitochondria of wild-type animals. It is well known that PLP-mRNAs and protein increase 10-20 fold in rodents during the first few weeks of postnatal development and are then reduced in adults (Gardinier et al., 1986; Manukian and Kirakosian, 1985). During postnatal development, the cell bodies of premyelinating Olgs in postnatal cerebral cortex are intensely stained for PLP/DM20, and approximately 20% of these Olgs are apoptotic (Trapp et al., 1997). Based upon that study and in line with our previous studies that showed increased levels of native and missense PLP correlated with increased cell death (Skoff et al., 2004a; Skoff et al., 2004b), we investigated whether PLP is inserted into Olg mitochondria of normal mice during postnatal development. In cerebral cortex adjacent to the corpus callosum, we found Olgs in which PLP is inserted into mitochondria (Fig. 1F). In contrast, in the striatum, where myelination is more advanced, we have not found co-localization. An exhaustive study is underway in normal mice to determine when and where in the brain PLP co-localizes with Olg mitochondria, its frequency, and whether these Olgs are dying.
To confirm that co-localization of PLP and the COX1 antibodies is not a staining or imaging artefact, we used the Perkin Elmer Volocity program to determine overlap of fluorochromatic dyes in a single voxel of an image of brain tissue (see Materials and Methods for details). Statistical analysis of this data shows the co-localization of PLP antibody with COX1 antibody in Olgs is statistically significant in the Plp1tg mice and in Olgs from PMD patients but not in Olgs of normal adult mice. The co-localization of the PLP and COX1 antibodies is not due to physical proximity of proteins (eg., PLP) in the endoplasmic reticulum and mitochondrial markers because endoplasmic reticulum and mitochondrial markers did not produce yellow fluorescence (Hüttemann et al., 2009). Additionally, attempts to co-localize other myelin proteins with mitochondria were unsuccessful (Hüttemann et al., 2009). Surprisingly, co-localization of DM20, a smaller isoform of PLP, with mitochondria is rare in COS7 cells transfected with a dsRedDM20 cDNA construct. The explanation for this phenomenon is not clear but may involve differences in multi-merization between PLP and DM20 (Ng and Deber, 2010; Daffu et al., 2012) in the transmembrane helices and accessibility of cysteines for disulphide bond formation. This finding is not surprising because DM20 is thought to function differently from PLP (Stecca et al., 2000).
Substitution of C6, C10, C25 or C35 to a serine prevents co-localization of PLP in mitochondria, indicating that any one of these 4 cysteines is required for insertion into mitochondria. However, it does not prevent the proper insertion of PLP into the plasma membrane of COS7 cells, demonstrated by live staining with the conformationally sensitive O10 antibody (Jung et al., 1996). In contrast, C201S and C220S substitutions do not prevent the insertion of PLP into mitochondria but prevent correct insertion into the plasma membrane, a finding previously reported (Dhaunchak and Nave, 2007). These findings demonstrate that the transport of PLP into the plasma membrane and mitochondria utilize two different import pathways.
To test whether the abnormalities associated with Plp1tg mice with extra copies are due to the insertion of PLP into mitochondria, we transfected (1) wtPlp1 cDNAs, (2) Plp1 cDNAs in which cysteines required for trafficking to mitochondria were mutated and (3) Plp1 cDNAs in which cysteines not required for insertion into mitochondria were mutated into COS7 cells. Transfected cells with the wtPlp1 construct or the Plp1C7A construct that permits its insertion into mitochondria have significantly reduced pH, whereas cells transfected with Plp1C6S and Plp1C10S, which prevent co-localization of PLP to mitochondria, have normal pH values. This data demonstrates that in vitro the acidification is due to insertion of PLP into mitochondria. We show that lactate is significantly increased in both extracellular media of 293 cells transfected with the wtPlp1 cDNA and in aCSF of Plp1tg mice. How lactate secretion into the extracellular space is increased in the Plp1tg mice and how it's regulated in vitro is unknown but may be due to the hypothesized role of PLP as a proton channel (see below) or changes in the lactate transporter monocarboxylate transporter 1 (MCT1). Alterations in transporters are intriguing because a mechanism by which Olgs modulate neuronal/axonal loss was recently published that shows MCT1 modulates axonal damage (Lee et al., 2012). This transporter is abundantly expressed in Olgs (Rinholm et al., 2011), and its absence and/or blockade by inhibitors leads to neuronal/axonal damage in experimental conditions. Other mechanisms for altering pH and ATP in aECF and in media have been described (Yamamura et al., 2008).
ATP, by itself, is not a major contributor of protons because pH was essentially stable when ATP was added in concentrations from 10μM to 100μM to aECF (data not shown). These levels are multifold greater than in normal ECF where they are in the nM range (Agteresch et al., 1999; Melani et al., 2005). We emphasize that the absolute amount of ATP released into the media (50μmoles/liter) is always low compared to the amount found within the cell: the ratio of intracellular ATP to media ATP was orders of magnitude different (about 600 to 1). Only slight increases of ATP in ECF or in media are sufficient to initiate microglial activation. Microglia move to areas of extracellular ATP/ADP that cause mitogenesis and cytokine release (Shigemoto-Mogami et al., 2001; Honda et al., 2001; Samuels, et al., 2010; Neary et al., 1996; Delarasse et al., 2009; Tu and Wang, 2009; Agteresch et al., 1999; Melani et al., 2005). Similarly, extracellular acidosis in many different types of tissues, including brain, initiates a host of molecular reactions that may culminate in tumor progression, pain, and irreversible brain damage (eg., Chesler, 2003; Dube et al, 2009; Wang et al., 2013).
The metabolic effects due to the insertion of PLP into mitochondria are quite different from the commonly accepted view that PLP is exclusively a myelin adhesive protein and that the pathology in the PLP mutants is due to demyelination and/or hypomyelination. Interestingly, the classical hypothesis is that PLP, based upon black membrane studies, functions as a membrane pore and/or a proton channel (Ting-Beall et al., 1979; Lin and Lees, 1982; Helynck et al., 1983; Kitagawa et al., 1993; Lees and Bizzozero, 1992). Our study supports the classical concept that PLP functions as a channel, permitting leakage of ATP and lactate into media and ECF when the gene is over-expressed. Alternatively, over-expression of the PLP gene may activate other types of channels that include mono-carboxylate transporters and acid sensing ion channels. Given the known effects of increased extracellular ATP and acidosis upon abnormal brain function, we hypothesize that the metabolic changes found in the Plp1tg mice and in the tissue culture studies contribute to their pathology. It is fair to state that the neuronal degeneration in the Plp1 mutants and in PMD patients is likely caused by many different cellular events besides myelin abnormalities. This statement is supported by the les rat that shows no signs of axonal degeneration up to 9 months even though it is chronically demyelinated (Smith et al., 2013). Accordingly, other non-myelin factors must be involved in maintaining axonal viability in myelin mutants and they need investigation.
ACKNOWLEDGEMENTS
We thank J. Kamholz and J. Sohi (Wayne State University School of Medicine) for providing PLP-HA tagged constructs; K-A Nave (Max Planck Institute of Experimental Medicine, Goettingen, Germany) for providing Plp1tg mice; and members of the Microscopy, Imaging and Cytometry Resources Core Facility (Wayne State University School of Medicine). This work was supported by NIH grant R01NS38236 and European Leukodystrophy Association Grant 2009-GATSK.
REFERENCES
- Agteresch HJ, Dagnelie PC, van den Berg JW, Wilson JH. Adenosine triphosphate: established and potential clinical applications. Drugs. 1999;58:211–232. doi: 10.2165/00003495-199958020-00002. [DOI] [PubMed] [Google Scholar]
- Anderson TJ, Schneider A, Barrie JA, Klugmann M, McCulloch MC, Kirkham D, Kyriakides E, Nave KA, Griffiths IR. Late-onset neurodegeneration in mice with increased dosage of the proteolipid protein gene. J Comp Neurol. 1998;394:506–519. doi: 10.1002/(sici)1096-9861(19980518)394:4<506::aid-cne8>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- Baarine M, Ragot K, Genin EC, El Hajj H, Trompier D, Andreoletti P, Ghandour MS, Menetrier F, Cherkaoui-Malki M, Savary S, Lizard G. Peroxisomal and mitochondrial status of two murine oligodendrocytic cell lines (158N, 158JP): potential models for the study of peroxisomal disorders associated with dysmyelination processes. J Neurochem. 2009;111:119–131. doi: 10.1111/j.1471-4159.2009.06311.x. [DOI] [PubMed] [Google Scholar]
- Beesley JS, Lavy L, Eraydin NB, Siman R, Grinspan JB. Caspase-3 activation in oligodendrocytes from the myelin-deficient rat. J Neurosci Res. 2001;64:371–379. doi: 10.1002/jnr.1088. [DOI] [PubMed] [Google Scholar]
- Bihlmaier K, Mesecke N, Terziyska N, Bien M, Hell K, Herrmann JM. The disulfide relay system of mitochondria is connected to the respiratory chain. J Cell Biol. 2007;79:389–395. doi: 10.1083/jcb.200707123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boespflug-Tanguy O, Mimault C, Melki J, Cavagna A, Giraud G, Pham Dinh D, Dastugue B, Dautigny A. Genetic homogeneity of Pelizaeus-Merzbacher disease: tight linkage to the proteolipoprotein locus in 16 affected families. PMD Clinical Group. Am J Hum Genet. 1994;55:461–467. [PMC free article] [PubMed] [Google Scholar]
- Boison D, Bussow H, D'Urso D, Muller HW, Stoffel W. Adhesive properties of proteolipid protein are responsible for the compaction of CNS myelin sheaths. J Neurosci. 1995;15:5502–5513. doi: 10.1523/JNEUROSCI.15-08-05502.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boison D, Stoffel W. Disruption of the compacted myelin sheath of axons of the central nervous system in proteolipid protein-deficient mice. Proc Natl Acad Sci (USA) 1994;91:11709–11713. doi: 10.1073/pnas.91.24.11709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher SE, Cypher MA, Carlock LR, Skoff RP. Proteolipid protein gene modulates viability and phenotype of neurons. J Neurosci. 2002;22:1772–1783. doi: 10.1523/JNEUROSCI.22-05-01772.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun PE. Molecular architecture of myelin. In: Morell Pierre., editor. Myelin. Plenum Press; New York, NY: 1977. pp. 91–115. [Google Scholar]
- Cerghet M, Bessert DA, Nave KA, Skoff RP. Differential expression of apoptotic markers in jimpy and in Plp overexpressors: evidence for different apoptotic pathways. J Neurocytol. 2001;30:841–855. doi: 10.1023/a:1019697506757. [DOI] [PubMed] [Google Scholar]
- Chesler M. Regulation and modulation of pH in the brain. Physiol Rev. 2003;83:1183–1221. doi: 10.1152/physrev.00010.2003. [DOI] [PubMed] [Google Scholar]
- Costes SV, Dailemans D, Cho EH, Dobbin Z, Pavlakis G, Lockett S. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophysical J. 2004;86:3339–4003. doi: 10.1529/biophysj.103.038422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daffu G, Sohi J, Kamholz J. Proteolipid protein dimerization at cysteine 108: Implications for protein structure. Neurosci Res. 2012;74:144–155. doi: 10.1016/j.neures.2012.07.009. [DOI] [PubMed] [Google Scholar]
- Delarasse C, Gonnord P, Galante M, Auger R, Daniel H, Motta I, Kanellopoulos JM. Neural progenitor cell death is induced by extracellular ATP via ligation of P2X7 receptor. J Neurochem. 2009;109:846–857. doi: 10.1111/j.1471-4159.2009.06008.x. [DOI] [PubMed] [Google Scholar]
- Dhaunchak AS, Nave KA. A common mechanism of PLP/DM20 misfolding causes cysteine-mediated endoplasmic reticulum retention in oligodendrocytes and Pelizaeus-Merzbacher disease. Proc Natl Acad Sci (USA) 2007;104:17813–17818. doi: 10.1073/pnas.0704975104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubé GR, Elagoz A, Mangat H. Acid sensing ion channels and acid nociception. Curr Pharm Des. 2009;15:1750–1766. doi: 10.2174/138161209788186263. [DOI] [PubMed] [Google Scholar]
- Duncan ID, Hammang JP, Trapp BD. Abnormal compact myelin in the myelin-deficient rat: absence of proteolipid protein correlates with a defect in the intraperiod line. Proc Natl Acad Sci (USA) 1987;84:6287–6291. doi: 10.1073/pnas.84.17.6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis D, Malcolm S. Proteolipid protein gene dosage effect in Pelizaeus-Merzbacher disease. Nat Genet. 1994;6:333–334. doi: 10.1038/ng0494-333. [DOI] [PubMed] [Google Scholar]
- Eng LF, Chao FC, Gerstl B, Pratt D, Tavaststjerna MG. The maturation of human white matter myelin. Fractionation of the myelin membrane proteins. Biochemistry. 1968;7:4455–4465. doi: 10.1021/bi00852a042. [DOI] [PubMed] [Google Scholar]
- Garbern JY, Cambi F, Tang XM, Sima AA, Vallat JM, Bosch EP, Lewis R, Shy M, Sohi J, Kraft G, Chen KL, Joshi I, Leonard DG, Johnson W, Raskind W, Dlouhy SR, Pratt V, Hodes ME, Bird T, Kamholz J. Proteolipid protein is necessary in peripheral as well as central myelin. Neuron. 1997;19:205–218. doi: 10.1016/s0896-6273(00)80360-8. [DOI] [PubMed] [Google Scholar]
- Garbern J, Cambi F, Shy M, Kamholz J. The molecular pathogenesis of Pelizaeus-Merzbacher disease. Arch Neurol. 1999;56:1210–1214. doi: 10.1001/archneur.56.10.1210. [DOI] [PubMed] [Google Scholar]
- Garbern JY, Yool DA, Moore GJ, Wilds IB, Faulk MW, Klugmann M, Nave K-A, Sistermans EA, van der Knaap MS, Bird TD, Shy ME, Kamholz JA, Griffiths IR. Patients lacking the major CNS myelin protein, proteolipid protein 1, develop length-dependent axonal degeneration in the absence of dymyelination and inflammation. Brain. 2002;125:551–561. doi: 10.1093/brain/awf043. [DOI] [PubMed] [Google Scholar]
- Gardinier MV, Macklin WB, Diniak AJ, Deininger PL. Characterization of myelin proteolipid mRNAs in normal and jimpy mice. Mol Cell Biol. 1986;6:3755–3762. doi: 10.1128/mcb.6.11.3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghandour MS, Feutz AC, Jalabi W, Taleb O, Bessert D, Cypher M, Carlock L, Skoff RP. Trafficking of PLP/DM20 and cAMP signaling in immortalized jimpy oligodendrocytes. Glia. 2002;40:300–311. doi: 10.1002/glia.10122. [DOI] [PubMed] [Google Scholar]
- Gow A, Lazzarini RA. A cellular mechanism governing the severity of Pelizaeus-Merzbacher disease. Nat Genet. 1996;13:422–428. doi: 10.1038/ng0896-422. [DOI] [PubMed] [Google Scholar]
- Griffiths I, Klugmann M, Anderson T, Yool D, Thomson C, Schwab MH, Schneider A, Zimmermann F, McCulloch M, Nadon N, Nave KA. Axonal swellings and degeneration in mice lacking the major proteolipid of myelin. Science. 1998;280:1610–1613. doi: 10.1126/science.280.5369.1610. [DOI] [PubMed] [Google Scholar]
- Hell K. The Erv1-Mia40 disulfide relay system in the intermembrane space of mitochondria. Biochimica et Biophysica Acta. 2008;1783:601–609. doi: 10.1016/j.bbamcr.2007.12.005. [DOI] [PubMed] [Google Scholar]
- Helynck G, Luu B, Nussbaum JL, Picken D, Skalidis G, Trifilieff E, Van Dorsselaer A, Seta P, Sandeaux R, Gavach C, Heitz F, Simon D, Spach G. Brain proteolipids. Isolation, purification and effect on ionic permeablilty of membranes. Eur J Biochem. 1983;133:689–695. doi: 10.1111/j.1432-1033.1983.tb07518.x. [DOI] [PubMed] [Google Scholar]
- Herrmann JM, Riemer J. Mitochondrial disulfide relay: redox-regulated protein import into the intermembrane space. J Biol Chem. 2012;228:4426–4433. doi: 10.1074/jbc.R111.270678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodes ME, Pratt VM, Dlouhy SR. Genetics of Pelizaeus-Merzbacher disease. Dev Neurosci. 1993;15:383–394. doi: 10.1159/000111361. [DOI] [PubMed] [Google Scholar]
- Honda S, Sasaki Y, Ohsawa K, Imai Y, Nakamura Y, Inoue K, Kohsaka S. Extracellular ATP or ADP induce chemotaxis of cultured microglia through Gi/o-coupled P2Y receptors. J Neurosci. 2001;21:1975–1982. doi: 10.1523/JNEUROSCI.21-06-01975.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn T, Klein J. Lactate levels in the brain are elevated upon exposure to volatile anesthetics: a microdialysis study. Neurochem Int. 2010;57:940–947. doi: 10.1016/j.neuint.2010.09.014. [DOI] [PubMed] [Google Scholar]
- Hudson L, Nadon N. Amino acid substitutions in proteolipid protein that cause dysmyelination. In: Martenson RE, editor. Myelin: Biology and Chemistry. CRC Press Inc.; Boca Raton, FL: 1992. pp. 677–702. [Google Scholar]
- Huttemann M, Zhang Z, Mullins C, Bessert D, Lee I, Nave KA, Appikatla S, Skoff RP. Different proteolipid protein mutants exhibit unique metabolic defects. ASN NEURO. 2009;1(3) doi: 10.1042/AN20090028. art:e00014.doi.10.1042/AN20090028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Osaka H, Thurston VC, Clarke JT, Yoneyama A, Rosenbarker L, Bird TD, Hodes ME, Shaffer LG, Lupski JR. Genomic rearrangements resulting in PLP1 deletion occur by nonhomologous end joining and cause different dysmyelinating phenotypes in males and females. Am J Hum Genet. 2002;71:838–853. doi: 10.1086/342728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung M, Sommer I, Schachner M, Nave KA. Monoclonal antibody O10 defines a conformationally sensitive cell-surface epitope of proteolipid protein (PLP): evidence that PLP misfolding underlies dysmyelination in mutant mice. J Neurosci. 1996;16:7920–7929. doi: 10.1523/JNEUROSCI.16-24-07920.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa K, Sinoway MP, Yang C, Gould RM, Colman DR. A proteolipid protein gene family: expression in sharks and rays and possible evolution from an ancestral gene encoding a poreforming polypeptide. Neuron. 1993;11:433–448. doi: 10.1016/0896-6273(93)90148-k. [DOI] [PubMed] [Google Scholar]
- Klugmann M, Schwab MH, Pühlhofer A, Schneider A, Zimmermann F, Griffiths IR, Nave KA. Assembly of CNS myelin in the absence of proteolipid protein. Neuron. 1997;18:59–70. doi: 10.1016/s0896-6273(01)80046-5. [DOI] [PubMed] [Google Scholar]
- Lee Y, Morrison BM, Li Y, Lengacher S, Farah MH, Hoffman PN, Liu Y, Tsingalia A, Jin L, Zhang PW, Pellerin L, Magistretti PJ, Rothstein JD. Oligodendroglia metabotically support axons and contribute to neurodegeneration. Nature. 2012;487:443–448. doi: 10.1038/nature11314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees MB, Bizzeozero OA. Structure and acylation of proteolipid protein. In: Martenson RE, editor. Myelin: Biology and Chemistry. CRC Press Inc.; Boca Raton, FL: 1992. pp. 237–235. [Google Scholar]
- Lin L-FH, Lees MB. Interactions of dicyclohexylcarbodiimide with myelin proteolipid. Proc Natl Acad Sci. 1982;79:941–945. doi: 10.1073/pnas.79.3.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longen S, Bien M, Bihlmaler K, Kloeppel C, Kauff F, Hammermeister M, Westermann B, Herrmann JM, Riemer J. Systematic analysis of the twin cx(9)c protein family. J Mol Biol. 2009;393:356–368. doi: 10.1016/j.jmb.2009.08.041. [DOI] [PubMed] [Google Scholar]
- Manukian KH, Kirakosian LG. Proteolipids in developing rat brain. Neurochem Res. 1985;10:1533–1545. [PubMed] [Google Scholar]
- Melani A, Turchi D, Vannucchi MG, Cipriani S, Gianfriddo M, Pedata F. ATP extracellular concentrations are increased in the rat striatum during in vivo ischemia. Neurochem Int. 2005;47:442–448. doi: 10.1016/j.neuint.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Mesecke N, Terziyska N, Kozany C, Baumann F, Neupert W, Hell K, Herrmann JM. A disulfide relay system in the intermembrane space of mitochondria that mediates protein import. Cell. 2005;121:1059–1069. doi: 10.1016/j.cell.2005.04.011. [DOI] [PubMed] [Google Scholar]
- Mesecke N, Bihlmaier K, Grumbt B, Longen S, Terziyska N, Hell K, Herrmann JM. The zinc-binding protein Hot13 promotes oxidation of the mitochondrial import receptor Mia40. EMBO Rep. 2008;9:1107–1113. doi: 10.1038/embor.2008.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neary JT, Zhu Q, Kang Y, Dash PK. Extracellular ATP induces formation of AP-1 complexes in astrocytes via P2 purinoceptors. Neuroreport. 1996;7:2893–2896. doi: 10.1097/00001756-199611250-00017. [DOI] [PubMed] [Google Scholar]
- Ng DP, Deber CM. Modulation of the oligomerization of myelin proteolipid protein by transmembrane helix interaction motifs. Biochemistry. 2010;49:6896–6902. doi: 10.1021/bi100739r. [DOI] [PubMed] [Google Scholar]
- Norton WT, Poduslo SE. Myelination in rat brain: changes in myelin composition during brain maturation. J Neurochem. 1973;21:759–773. doi: 10.1111/j.1471-4159.1973.tb07520.x. [DOI] [PubMed] [Google Scholar]
- Raskind WH, Williams CA, Hudson LD, Bird TD. Complete deletion of the proteolipid protein gene (PLP) in a family with X-linked Pelizaeus-Merzbacher disease. Am J Hum Genet. 1991;49:1355–1360. [PMC free article] [PubMed] [Google Scholar]
- Readhead C, Schneider A, Griffiths I, Nave KA. Premature arrest of myelin formation in transgenic mice with increased proteolipid protein gene dosage. Neuron. 1994;12:583–595. doi: 10.1016/0896-6273(94)90214-3. [DOI] [PubMed] [Google Scholar]
- Renier WO, Garbreë FJ, Hustinx TW, Jaspar HH, Geelen JA, Van Haelst UJ, Lommen EJ, Ter Haar BG. Connatal Pelizaeus-Merzbacher disease with congenital stridor in two maternal cousins. Acta Neuropathol. 1981;54:11–17. doi: 10.1007/BF00691328. [DOI] [PubMed] [Google Scholar]
- Rinholm JE, Hamilton NB, Kessaris N, Richardson WD, Bergersen LH, Attwell D. Regulation of oligodendrocyte development and myelination by glucose and lactate. J Neurosci. 2011;31:538–548. doi: 10.1523/JNEUROSCI.3516-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roboti P, Swanton E, High S. Differences in endoplasmic-reticulum quality control determine the cellular response to disease-associated mutants of proteolipid protein. J Cell Sci. 2009;122:3942–3953. doi: 10.1242/jcs.055160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels SE, Lipitz JB, Dahl G, Muller KJ. Neuroglial ATP release through innexin channels controls mircroglial cell movement to a nerve injury. J Gen Physiol. 2010;136:425–442. doi: 10.1085/jgp.201010476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma R, Gow A. Minimal role for caspase 12 in the unfolded protein response in oligodendrocytes in vivo. J Neurochem. 2007;101:889–897. doi: 10.1111/j.1471-4159.2007.04541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigemoto-Mogami Y, Koizumi S, Tsuda M, Ohsawa K, Kohsaka S, Inoue K. Mechanisms underlying extracellular ATP-evoked interleukin-6 release in mouse microglial cell line, MG-5. J Neurochem. 2001;78:1339–1349. doi: 10.1046/j.1471-4159.2001.00514.x. [DOI] [PubMed] [Google Scholar]
- Sima AA, Pierson CR, Woltjer RL, Hobson GM, Golden JA, Kupsky WJ, Schauer GM, Bird TD, Skoff RP, Garbern JY. Neuronal loss in Pelizaeus-Merzbacher disease differs in various mutations of the proteolipid protein 1. Acta Neuropathol. 2009;118:531–539. doi: 10.1007/s00401-009-0562-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoff RP. Myelin deficit in the Jimpy mouse may be due to cellular abnormalities in astroglia. Nature. 1976;264:560–562. doi: 10.1038/264560a0. [DOI] [PubMed] [Google Scholar]
- Skoff RP, Bessert DA, Cerghet M, Franklin MJ, Rout UK, Nave KA, Carlock L, Ghandour MS, Armant DR. The myelin proteolipid protein gene modulates apoptosis in neural and non-neural tissues. Cell Death Differ. 2004;11:1247–1257. doi: 10.1038/sj.cdd.4401498. [DOI] [PubMed] [Google Scholar]
- Skoff RP, Saluja I, Bessert D, Yang X. Analysis of proteolipid protein mutants show levels of proteolipid protein regulate oligodendrocyte number and cell death in vitro and in vivo. Neurochem Res. 2004;29:2095–2103. doi: 10.1007/s11064-004-6882-0. [DOI] [PubMed] [Google Scholar]
- Smith CM, Cooksey E, Duncan ID. Myelin loss does not lead to axonal degeneration in a long-live model of chronic demyelination. J Neurosci. 2013;27:2718–2727. doi: 10.1523/JNEUROSCI.4627-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southwood CM, Garbern J, Jiang W, Gow A. The unfolded protein response modulates disease severity in Pelizaeus-Merzbacher disease. Neuron. 2002;36:585–596. doi: 10.1016/s0896-6273(02)01045-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecca B, Southwood CM, Gragerov A, Kelley KA, Friedrich VL, Jr, Gow A. The evolution of lipophilin genes from invertebrates to tetrapods: DM-20 cannot replace proteolipid protein in CNS myelin. J Neurosci. 2000;20:4002–4010. doi: 10.1523/JNEUROSCI.20-11-04002.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swamydas M, Bessert D, Skoff R. Sexual dimorphism of oligodendrocytes is mediated by differential regulation of signaling pathways. J Neurosci Res. 2009;87:3306–3319. doi: 10.1002/jnr.21943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Herr W. Differential transcriptional activation by Oct-1 and Oct-2: interdependent activation domains induce Oct-2 phosphorylation. Cell. 1990;60:375–386. doi: 10.1016/0092-8674(90)90589-7. [DOI] [PubMed] [Google Scholar]
- Tatar CL, Appikatla S, Bessert DA, Paintlia AS, Singh I, Skoff RP. Increased Plp1 gene expression leads to massive microglial cell activation and inflammation throughout the brain. ASN NEURO. 2010;2(4) doi: 10.1042/AN20100016. art:e00043.doi:10.1042/AN2010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terziyska N, Lutz T, Kozany C, Mokranjac D, Mesecke N, Neupert W, Herrmann JM, Hell K. Mia40, a novel factor for protein import into the intermembrane space of mitochondria is able to bind metal ions. FEBS Lett. 2005;579:179–184. doi: 10.1016/j.febslet.2004.11.072. [DOI] [PubMed] [Google Scholar]
- Ting-Beall HP, Lees MB, Robertson JD. Interactions of Folch-Lees proteolipid apoprotein with planar lipid bilayers. J Membr Biol. 1979;51:33–46. doi: 10.1007/BF01869342. [DOI] [PubMed] [Google Scholar]
- Trapp BD, Nishiyama A, Cheng D, Macklin W. Differentiation and death of premyelinating oligodendrocytes in developing rodent brain. J Cell Biol. 1997;137:459–468. doi: 10.1083/jcb.137.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu J, Wang LP. Therapeutic potential of extracellular ATP and P2 receptors in nervous system diseases. Neurosci Bull. 2009;25:27–32. doi: 10.1007/s12264-009-1020-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Xu Y, Lian Z, Zhang J, Zhu T, Li M, Wei Yi, Dong B. Does closure of acid-sensing ion channels reduce ischemia/reperfusion injury in the rat brain? Neural Regeneration Res. 2013;8:1169–1179. doi: 10.3969/j.issn.1673-5374.2013.13.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward KJ, Cundall M, Sperle K, Sistermans EA, Ross M, Howell G, Gribble SM, Burford DC, Carter NP, Hobson DL, Garbern JY, Kamholz J, Heng H, Hodes ME, Malcolm S, Hobson GM. Heterogeneous duplications in patients with Pelizaeus-Merzbacher Disease suggest a mechanism of coupled homologous and nonhomologous recombination. Am J Hum Genet. 2005;77:966–987. doi: 10.1086/498048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamura H, Ugawa S, Ueda T, Nagao M, Shimada S. Epithelial Na+ channel delta subunit mediates acid-induced ATP release in the human skin. Biochem Biophys Res Comm. 2008;373:155–158. doi: 10.1016/j.bbrc.2008.06.008. [DOI] [PubMed] [Google Scholar]
- Yool D, Klugmann M, Barrie JA, McCulloch MC, Nave KA, Griffiths IR. Observations on the structure of myelin lacking the major proteolipid protein. Neuropathol Appl Neurobiol. 2002;28:75–78. doi: 10.1046/j.0305-1846.2001.00370.x. [DOI] [PubMed] [Google Scholar]