Abstract
The human genome contains millions of DNA regulatory elements and a large number of gene clusters, most of which have not been tested experimentally. The clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated nuclease 9 (Cas9) programed with a synthetic single-guide RNA (sgRNA) emerges as a method for genome editing in virtually any organisms. Here we report that targeted DNA fragment inversions and duplications could easily be achieved in human and mouse genomes by CRISPR with two sgRNAs. Specifically, we found that, in cultured human cells and mice, efficient precise inversions of DNA fragments ranging in size from a few tens of bp to hundreds of kb could be generated. In addition, DNA fragment duplications and deletions could also be generated by CRISPR through trans-allelic recombination between the Cas9-induced double-strand breaks (DSBs) on two homologous chromosomes (chromatids). Moreover, junctions of combinatorial inversions and duplications of the protocadherin (Pcdh) gene clusters induced by Cas9 with four sgRNAs could be detected. In mice, we obtained founders with alleles of precise inversions, duplications, and deletions of DNA fragments of variable sizes by CRISPR. Interestingly, we found that very efficient inversions were mediated by microhomology-mediated end joining (MMEJ) through short inverted repeats. We showed for the first time that DNA fragment inversions could be transmitted through germlines in mice. Finally, we applied this CRISPR method to a regulatory element of the Pcdhα cluster and found a new role in the regulation of members of the Pcdhγ cluster. This simple and efficient method should be useful in manipulating mammalian genomes to study millions of regulatory DNA elements as well as vast numbers of gene clusters.
Keywords: DNA regulatory element inversion, duplication, deletion, CRISPR/Cas9, enhancer, genome manipulation, gene cluster
Introduction
With the completion of the human Encyclopedia of DNA Elements (ENCODE) project, an arduous task is to elucidate the function of the vast number of regulatory DNA elements identified in the human genome (The ENCODE Project Consortium, 2012; Stamatoyannopoulos, 2012). There are millions of regulatory DNA elements in the human genome, such as enhancers (activators), silencers (repressors), promoters, insulators, and locus control regions. These DNA elements play an important role in tissue- and cell-specific gene regulation; however, vast majorities of them have not been characterized experimentally (Banerji et al., 1983; Zhang et al., 2004; The ENCODE Project Consortium, 2012; Neph et al., 2012; Shen et al., 2012; Thurman et al., 2012; de Laat and Duboule, 2013). In addition, DNA is a double-stranded molecule with polarity (Watson and Crick, 1953; The ENCODE Project Consortium, 2012). The chromatin environments of regulatory DNA elements are usually asymmetrical (The ENCODE Project Consortium, 2012; Kundaje et al., 2012). The role of polarity or orientation in the function of regulatory DNA elements, such as enhancers and insulators, has long been controversial (Blackwood and Kadonaga, 1998; Tanimoto et al., 1999; Wei and Brennan, 2000; West et al., 2002). Moreover, structural variations such as inversions and duplications are common in human genomes (Sharp et al., 2006; Stankiewicz and Lupski, 2010), and DNA rearrangements frequently occur in cancers (Stephens et al., 2011; Baca et al., 2013). Finally, mammalian genomes contain a large number of gene clusters whose members often have redundant functions. For example, there are >50 highly-similar Pcdh genes organized into three closely-linked clusters, named Pcdh α, β, and γ gene clusters (Wu and Maniatis, 1999; Wu et al., 2001; Wu, 2005). The Pcdh α and γ clusters are organized into a tandem array of more than a dozen variable exons and a single set of downstream constant exons (Figure 1A), similar to that of the UDP glucuronosyltransferase, immunoglobulin, and T-cell receptor gene clusters (Wu and Maniatis, 1999; Zhang et al., 2004; Wu, 2005). Thus, an efficient method to manipulate DNA fragments in mice is central for investigating gene expression and modeling human diseases.
Figure 1.
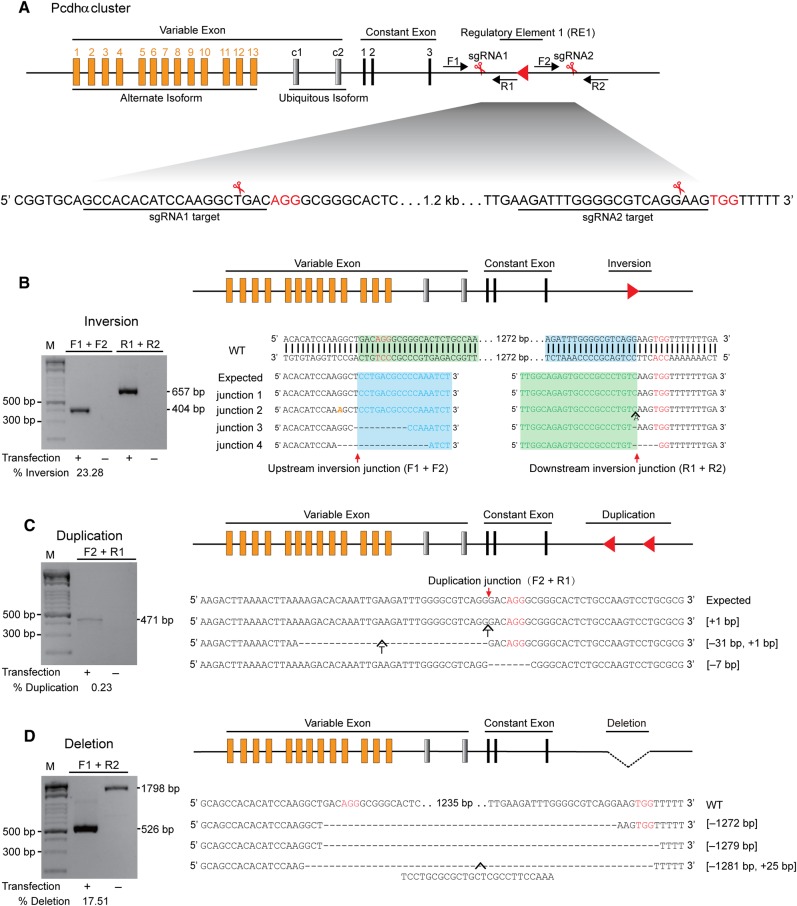
Inversion, duplication, and deletion of a DNA fragment in the Pcdhα regulatory region by CRISPR with a pair of sgRNAs. (A) Diagram of the Pcdhα gene cluster with a DNA fragment targeted by CRISPR with two sgRNAs. The human Pcdhα gene cluster is organized into a variable region of 15 exons (13 alternate isoforms and 2 ubiquitous isoforms) followed by a constant region of three exons. The locus regulatory region is located downstream. The sgRNA-targeting sequences are underlined. The protospacer adjacent motif (PAM) sequences of NGG are highlighted in red. The positions of the forward and reverse PCR primers are indicated by arrows. (B) Inversion of the DNA fragment is indicated by a red arrowhead. Shown are the amplified upstream and downstream junctions as well as their sequences. Deleted bases are indicated by dashes. Mutated bases are shown in yellow italics. Inserted bases are also shown. (C) Duplication of the DNA fragment. The sequences at the duplication junctions are shown. (D) Deletion of the DNA fragment. PCR products at the deletion junctions are sequenced. The 25-bp inserted T-nucleotides are also shown. The efficiency of inversion, duplication, and deletion is shown under each panel.
Forward genetics by screening for mutants of interests with specific phenotypes has shed insights into many biological processes (Müller, 1927), especially in the mouse model (Castle and Little, 1909). Reverse genetics with targeted mutations in chosen sequences has become possible since the development of DNA sequencing technology (Capecchi, 2005; Carroll, 2014). In particular, targeted gene modifications by homologous recombination in mice have produced thousands of knockout mouse lines (Smithies et al., 1985; Thomas and Capecchi, 1986; Capecchi, 2005). In addition, inversion and duplication methods are developed in mice and zebrafish for targeted structural modifications (Zheng et al., 2000; Wu et al., 2007; Gupta et al., 2013; Xiao et al., 2013; Kraft et al., 2015). In particular, tandem duplications could be generated from trans-allelic recombination involving inter-chromosomal ligation of two concurrent DSBs, each within one of the two homologous chromosomes or chromatids (Wu et al., 2007; Lee et al., 2012). Finally, targeted mutations engineered by sequence-specific nucleases such as zinc finger nucleases (ZFN) and transcription activator-like effector nucleases (TALEN) turn out to be useful for genome editing (Carroll, 2014). However, these methods are time-consuming and of low efficiency (Yu and Bradley, 2001; Capecchi, 2005; Carroll, 2014).
Recently, the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated nuclease (Cas) system emerged to be very efficient in inducing insertion/deletion mutations (indels) or targeted alterations in mammalian genomes (Mali et al., 2013b; Harrison et al., 2014; Hsu et al., 2014). CRISPR repeats were initially identified as mysterious repetitive extragenic palindromic sequences in bacteria 30 years ago (Stern et al., 1984; Ishino et al., 1987). These sequences include direct partial palindromic repeats that are interspaced with constant-sized, non-repetitive, spacers (Haft et al., 2005). CRISPR clusters exist in many prokaryotes and often have Cas genes located in the vicinity (Jansen et al., 2002; Haft et al., 2005). They provide an adaptive immune system for bacteria and archaea to defend themselves against invading viruses, phages, and plasmids (Barrangou et al., 2007) through directly targeting the matching protospacer of the invader (Marraffini and Sontheimer, 2008; Garneau et al., 2010). The protospacer adjacent motif (PAM) in the target DNA of the invader is essential for the host CRISPR system to distinguish self/non-self and to license the RNA-guided nuclease (RGN) to cleave the target DNA sequences (Mojica et al., 2009; Marraffini and Sontheimer, 2010; Anders et al., 2014; Sternberg et al., 2014). The CRISPR/Cas system can be classified into three types (Makarova et al., 2011). In the type II system, Cas9 endonuclease (also known as Cas5 or Csn1) guided by a complex of two non-coding RNAs, the CRISPR RNA (crRNA) and the trans-activating crRNA (tracrRNA), in a bilobed structure (Anders et al., 2014; Jinek et al., 2014; Nishimasu et al., 2014; Sternberg et al., 2014), directly cleaves the PAM-containing cognate target DNA at 3-bp upstream to generate double-strand breaks (DSBs) with blunt ends (Mojica et al., 2009; Garneau et al., 2010; Deltcheva et al., 2011). The complex of the two non-coding RNAs can be programed into a single-guide RNA (sgRNA) (Jinek et al., 2012). This sgRNA can guide the Cas9 enzyme to cleave specific sites in mammalian genomes based on principles of Watson-Crick base pairing, generating a blunt-end DSB that is thought to be repaired by the NHEJ DNA repair pathway (Cho et al., 2013; Cong et al., 2013; Jinek et al., 2013; Mali et al., 2013c). These transformative findings pave the way for unprecedented applications in life science studies (Gilbert et al., 2013; Li et al., 2013; Malina et al., 2013; Qi et al., 2013; Ran et al., 2013; Wang et al., 2013, 2014a, b; Wei et al., 2013; Yang et al., 2013; Cai and Yang, 2014; Doudna and Charpentier, 2014; González et al., 2014; Harrison et al., 2014; Kiani et al., 2014; Niu et al., 2014; Shalem et al., 2014; Shen et al., 2014; Zhou et al., 2014; Wan et al., 2015; Wu et al., 2015).
Here we developed an efficient simple method for inverting, duplicating, and deleting mammalian DNA fragments of small regulatory elements and large gene clusters by CRISPR/Cas9 with a pair of sgRNAs. Our data suggested that this method could be applied continuously for precise inversions, duplications, and deletions of DNA fragments of any size ranging from a few tens of bp up to a million of bp, and should be very useful for manipulating DNA fragments to control gene expression and to model copy number variations in human diseases.
Results
Targeted DNA fragment inversions and duplications by CRISPR with a pair of sgRNAs
We first designed a pair of sgRNAs for two sites flanking a 1272-bp DNA fragment within a 3-kb regulatory region of the Pcdhα gene cluster (Figure 1A and Supplementary Tables S1 and S2) (Ribich et al., 2006; Guo et al., 2012). We tested whether efficient inversion of this DNA regulatory element (RE1) could be generated with two DSBs induced by the Cas9 nuclease guided by these two sgRNAs (Figure 1A). Genomic DNA was isolated from the human HEC-1-B cells transfected with plasmids encoding Cas9 and a pair of sgRNAs. Given that DNA inversions have two junctions, we designed two pairs of specific PCR primers near the cleavage sites of Cas9 to identify upstream and downstream inversion junctions (Figure 1B). We found exact inversion of the regulatory DNA element between the two DSBs induced by Cas9 at 3 bp upstream of the PAM sites (Figure 1B). In addition, we also detected indels at both inversion junctions (Figure 1B). Thus, inversions induced by Cas9 with a pair of sgRNAs were probably generated by the Ku70-Ku80 and DNA ligase IV-dependent repair mechanism of non-homologous end joining (NHEJ) (Chapman et al., 2012; Jasin and Rothstein, 2013; Carroll, 2014).
We next tested whether duplications could also be generated by CRISPR with two sgRNAs and found tandem duplications of DNA fragments between DSBs induced by Cas9 at 3 bp upstream of PAM (Figure 1C). Indels were also detected near the duplication junctions, suggesting that these tandem duplications are generated by NHEJ between two DSBs each located in a homologous chromosome (chromatid) (Figure 1C). Finally, we found DNA fragment deletions generated by a pair of sgRNAs (Figure 1D), consistent with recent studies (Canver et al., 2014; Byrne et al., 2015; Ho et al., 2015). Interestingly, we observed a string of templated nucleotides (T nucleotides) in one of the deletion alleles (McVey and Lee, 2008). Taken together, inversions, duplications, and deletions of a DNA fragment were successfully generated by CRISPR with a pair of sgRNAs.
Inversions of DNA fragments of defined length by CRISPR in mice
To see whether inversions could be generated in mice by CRISPR, we co-injected a pair of sgRNAs into one-cell embryos, for two sites flanking a DNA fragment of 1241 bp, together with the Cas9 mRNA (Figure 2A). We prepared total genomic DNA from a batch of 68 blastocysts cultured in vitro. We successfully amplified a DNA fragment with the size equal to the expected upstream inversion junction. Cloning and sequencing confirmed the upstream junction of the inversion event (Figure 2A and Supplementary Figure S1A). Similarly, we amplified and confirmed the sequences of the downstream inversion junction (Figure 2A and Supplementary Figure S1A). In addition, we detected DNA fragment deletions in blastocysts with a pair of sgRNAs (Supplementary Figure S1A). Thus, CRISPR-mediated inversions with a pair of sgRNAs occur in mouse blastocysts.
Figure 2.
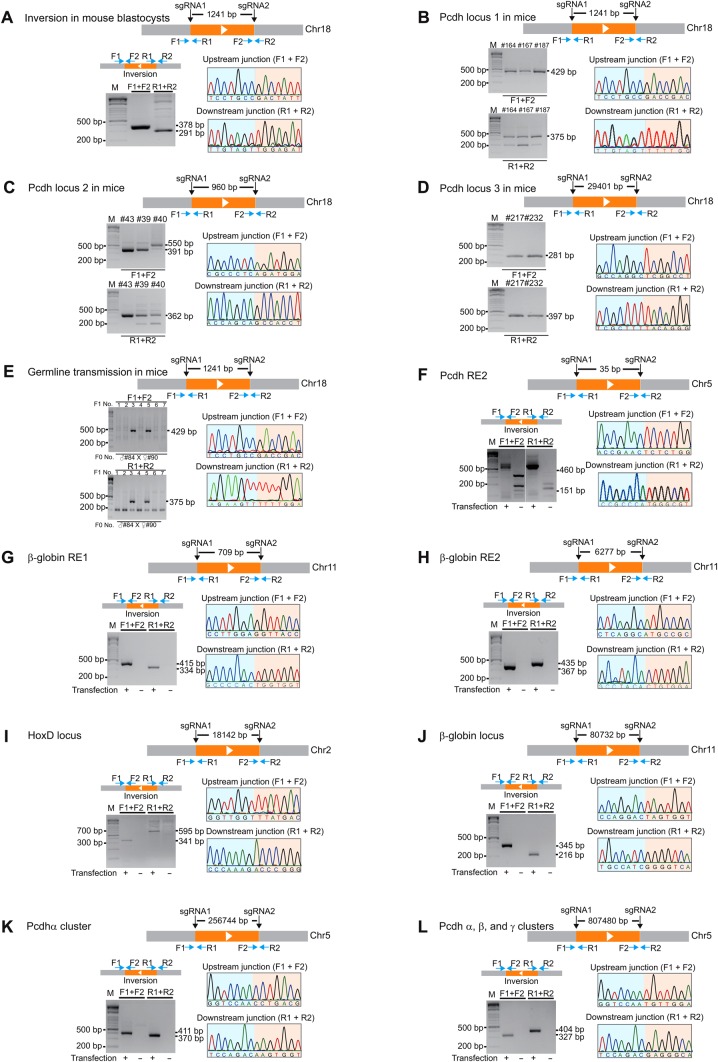
Targeted inversions of DNA fragments of different sizes in mice and in human cells. (A) Diagram of CRISPR with a pair of sgRNAs for two sites flanking a 1241-bp DNA fragment in the Pcdh locus. Shown are inversion junctions amplified by PCR from mouse blastocysts with specific primer pairs. An example of sequence chromatograms of the upstream and downstream junctions is shown. (B) Inversion in F0 founder mice genotyped by tail clipping. (C) Diagram of inversion of a 960-bp DNA fragment in the Pcdh locus. Shown are F0 inversion mice generated by CRISPR with a pair of sgRNAs. The upstream and downstream junctions of inversions were confirmed by PCR and Sanger sequencing. (D) Diagram of inversion of a 29401-bp DNA fragment. F0 inversion mice were genotyped by PCR with specific primer pairs and confirmed by Sanger sequencing. (E) Germline transmission in mice of the DNA fragment inversion induced by CRISPR with a pair of sgRNAs. Shown is the genotyping of DNA fragment inversion in F1 mice from the crossing of two founder mice. Chromatograms of the upstream and downstream junctions by Sanger sequencing confirmed the germline transmission of inversions in mice. (F) Inversion of a short Pcdh regulatory element (RE2) in human cells. Each inversion junction in human cells is confirmed by Sanger sequencing. Inversion of 709-bp (G) and 6277-bp (H) DNA fragments at the β-globin locus. (I) Inversion of an 18142-bp DNA fragment at the HoxD locus. (J) Inversion of an 80732-bp DNA fragment at the β-globin locus. (K) Inversion of a 256744-bp DNA fragment spanning the Pcdhα gene cluster. (L) Inversion of an 807480-bp DNA fragment spanning the Pcdh α, β, and γ gene clusters.
To see whether inversion could occur in F0 founder mice, we screened 120 founders and found inversions in 6 mice (5% founders) (Figure 2B and Supplementary Figure S1B). In addition, we found DNA fragment deletion in 26 F0 founder mice (21.7% founders) (Supplementary Figure S1C), consistent with recent studies (Fujii et al., 2013; Wang et al., 2013). Finally, we detected indels at the two targeted sites in 13 out of the 15 mice examined (Supplementary Figure S2), consistent with previous studies (Wang et al., 2013).
To further demonstrate the applicability of inversion by CRISPR with two sgRNAs in other sites of the mouse genome, we co-injected Cas9 mRNA with another pair of sgRNAs separated by 960 bp (Figure 2C). Strikingly, we found eight inversions in eight F0 founder mice (100% founders) (Figure 2C and Supplementary Figure S3A). Out of these eight F0 founder mice, we found six mice also with alleles of DNA fragment deletion (75% founders) (Supplementary Figure S3B). Finally, we tested a pair of sgRNAs separated by about 30 kb and obtained 2 mice with inversions and 5 mice with deletions out of 26 mice examined (7.7% inversion and 19.2% deletion in founders) (Figure 2D and Supplementary Figure S4). Together, we demonstrated that founder mice with inversions and deletions of DNA fragments of different sizes can be efficiently generated by CRISPR with a pair of sgRNAs.
Efficient germline transmission of DNA fragment inversion in mice
The high efficiency of founder mice with inversions and deletions encouraged us to investigate whether these CRISPR alleles could be germline transmitted. We crossed founder mice of inversion with deletion for the Pcdh locus 1 (Figure 2B and Supplementary Figure S5A). In a litter of seven F1 mice, we found germline transmissions of two mice with DNA fragment inversions (Figure 2E) and of one mouse with DNA fragment deletions (Supplementary Figure S5B). Sequencing confirmed the germline transmission for inversion in two F1 mice (28.6%) and for deletion in one F1 mouse (14.3%) (Supplementary Figure S5C). In another two litters of F1 mice from F0 founders with deletions of a different DNA fragment, we detected the germline transmission of five different deletion alleles (Supplementary Figure S5D and E). Thus, we demonstrated for the first time that DNA fragment inversions and deletions by CRISPR with a pair of sgRNAs can be transmitted through germline in mice.
Efficient inversions of a repertoire of human DNA fragments with defined length
To investigate whether inversions of DNA fragments have size limitations, we designed seven pairs of sgRNAs to invert DNA fragments on three different human chromosomes ranging in size from as small as 35 bp to as large as 807480 bp in the HEK293T cell line (Figure 2F–L and Supplementary Table S2). These include regulatory DNA elements of 35 bp in the Pcdh locus (RE2: regulatory element 2) (Figure 2F and Supplementary Figure S6A) and of 709 bp (Figure 2G and Supplementary Figure S6B) and 6277 bp (Figure 2H and Supplementary Figure S6C) in the β-globin locus, as well as large gene clusters of HoxD (18142 bp), β-globin (80732 bp), and Pcdh (256744 and 807480 bp) (Figure 2I–L and Supplementary Figure S6D–G). Sequencing of both upstream and downstream junctions for each inverted DNA fragment demonstrated that all of the inversion junctions contain precise ligations except those of the Pcdh RE2 (Figure 2F and Supplementary Figure S6A) and the upstream junction of the β-globin locus (Figure 2J and Supplementary Figure S6E). The imprecise junctions of the seven inversions were also detected at these loci, including micro-deletions, micro-insertions, and mutations (Supplementary Figure S6A–G), suggesting a mechanism of NHEJ. Together, we concluded that CRISPR-induced precise inversions could be achieved for DNA fragments in size ranging from about three dozens of bp to a million of bp and that precise inversions could occur in distinct loci on different chromosomes in human cells.
Segmental duplications by CRISPR with a pair of sgRNAs in mice and human cells
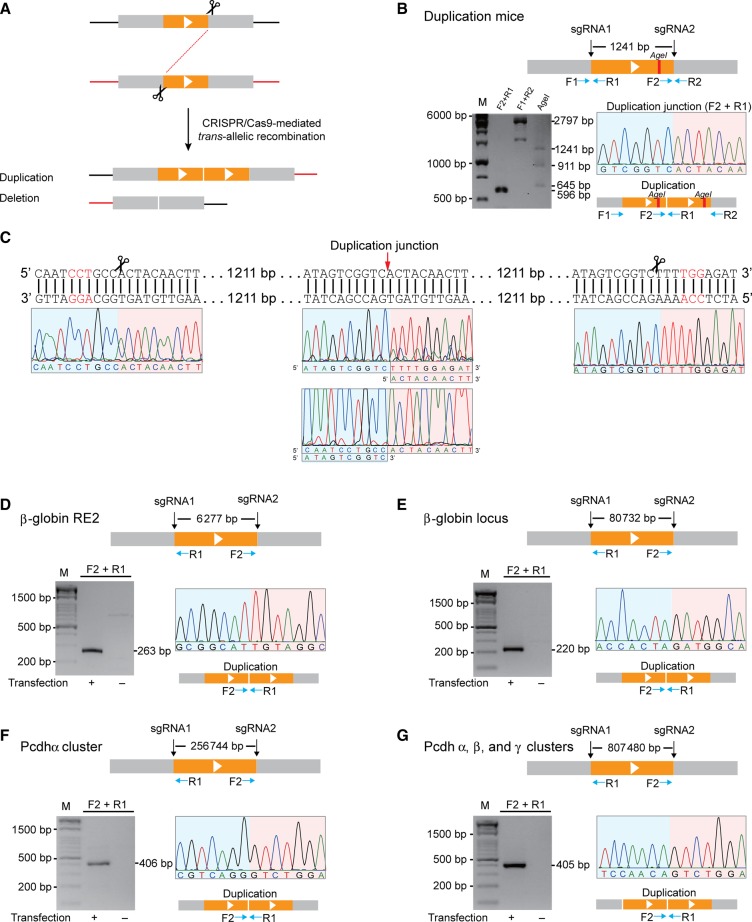
There are two scenarios for generating DNA fragment deletions from two concurrent DSBs: intra-chromosomal recombination between two DSBs on a single chromosome and trans-allelic recombination between two DSBs each on one of the two homologous chromosomes (chromatids) (Figure 3A). In the second scenario, a tandem (segmental) duplication will simultaneously be generated by trans-allelic recombination between the two DSBs (Figure 3A) (Wu et al., 2007; Lee et al., 2012). To see whether segmental duplication could be generated by CRISPR with a pair of sgRNA, we first designed a pair of PCR primers to amplify the duplication junction and screened 26 F0 founder mice with DNA fragment deletions. We found one founder mouse with the expected duplication junction and confirmed the junction by Sanger sequencing (Figure 3B and Supplementary Figure S7A). We then designed a pair of flanking PCR primers to amplifying the entire duplicated region. Restriction enzyme digestion of the duplicated region generates three fragments with expected size (Figure 3B). Finally, Sanger sequencing of the entire duplication region confirmed the segmental duplication (Figure 3C). In particular, Sanger sequencing with either forward or reverse primers, upstream of the sgRNA2 target site or downstream of the sgRNA1 target site, respectively, generated the expected chromatogram degeneracy (Figure 3C). Together, these data definitively demonstrated for the first time that segmental duplication occurs in mice by CRISPR with a pair of sgRNAs.
Figure 3.
Segmental duplications by CRISPR through trans-allelic recombination in mice and human cells. (A) Diagram of CRISPR-mediated duplication by trans-allelic recombination between two DSBs in homologous chromosomes (chromatids). (B) The duplication junction (F2+R1 primer pair) as well as the entire length of segmental duplicated region (F1+R2 primer pair) were amplified from F0 mice. The segmental duplication was first confirmed by the AgeI digestion, resulting in three fragments. (C) The entire fragment of segmental duplication was confirmed by Sanger sequencing. (D) Duplication of a 6277-bp DNA fragment at the β-globin locus in human cells. Duplication junctions were identified by PCR with a pair of specific primers. An example of sequence chromatograms at the duplication junction is shown. (E) Duplication of an 80732-bp DNA fragment at the β-globin locus. (F) Duplication of a 256744-bp DNA fragment spanning the Pcdhα gene cluster. (G) Duplication of an 807480-bp DNA fragment spanning the Pcdh α, β, and γ gene clusters.
To investigate whether segmental duplications could also be generated in human HEK293T cells by CRISPR, we designed PCR primer pairs to detect segmental duplications in the seven loci examined above and identified duplication junctions in four loci (Figure 3D–G). By cloning and sequencing, we confirmed precise junctions of segmental duplications (Figure 3D–G). The imprecise junctions of indels were also detected at these four loci (Supplementary Figure S7B–E). We also detected DNA fragment deletions (Supplementary Figures S8 and S9). Interestingly, in addition to indels, we observed precise deletions in all of the seven loci tested (Supplementary Figures S8 and S9). These data suggest that precise segmental duplications and deletions could be generated by CRISPR with a pair of sgRNAs.
Frequencies of inversions and duplications by CRISPR
We estimated the frequencies of inversion and duplication events by a quantification method to correct differences in PCR efficiencies of primer pairs, similar to that of the chromosome conformation capture method (Hagège et al., 2007; Guo et al., 2012). First, templates for each PCR fragment were amplified and quantified. After mixing equal molar of all templates, a standard curve was generated by quantitative PCR (qPCR) for each pair of primers with a series of dilutions. Six days after transfecting HEK293T cells with plasmids encoding Cas9 and a pair of sgRNAs, the amount of the inverted or duplicated DNA fragments was estimated by fitting on the standard curves. The efficiencies of inversions, duplications, and deletions at these loci ranged from 0.71% to 23.28%, 0.17% to 5.97%, and 0.47% to 34.49%, respectively (Table 1). These efficiencies are much higher than those of gene targeting by homologous recombination or by ZFN and TALEN (Wu et al., 2007; Lee et al., 2012). Potential off-target sites of sgRNAs were predicted by searching the human genome sequences (Supplementary Table S3) (Mali et al., 2013c; Wang et al., 2013; Lin et al., 2014). We tested some candidates by PCR with specific primer pairs (Supplementary Table S1). After sequencing, we did not find any modification in these potential off-target sites (Supplementary Table S4).
Table 1.
The efficiency of inversions, duplications, and deletions by CRISPR with a pair of sgRNAs.
| DNA fragment | β-globin RE1 | Pcdh RE1 | β-globin RE2 | HoxD locus | β-globin locus | Pcdhα cluster | Pcdh α, β, and γ clusters |
|---|---|---|---|---|---|---|---|
| (size in bp) | (709) | (1272) | (6277) | (18142) | (80732) | (256744) | (807480) |
| Inversion (%) | 21.12 ± 4.99 | 23.28 ± 2.42 | 23.13 ± 1.13 | 7.28 ± 1.60 | 5.96 ± 0.28 | 5.48 ± 0.37 | 0.71 ± 0.12 |
| Duplication (%) | ND | 0.23 ± 0.12 | 5.30 ± 1.19 | ND | 5.97 ± 0.33 | 0.61 ± 0.02 | 0.17 ± 0.03 |
| Deletion (%) | 28.33 ± 6.19 | 17.51 ± 1.04 | 34.49 ± 3.57 | 9.15 ± 0.11 | 13.39 ± 0.80 | 8.46 ± 0.24 | 0.47 ± 0.08 |
ND, not detectable. Efficiency = mean ± SD.
Combinatorial inversions and duplications by CRISPR with four sgRNAs
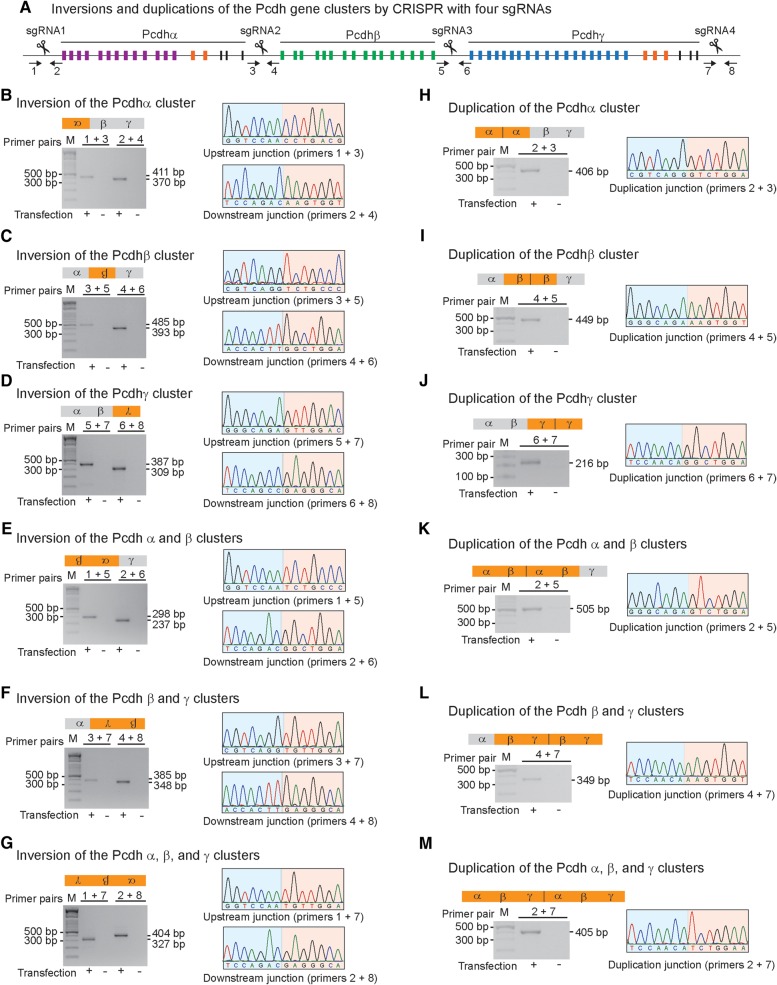
To investigate whether combinatorial inversions and duplications could be generated by CRISPR with multiple sgRNAs, we used four sgRNAs targeting the Pcdh clusters in HEK293T cells. These four sgRNAs were designed to target specific sites at the boundaries of the Pcdh α, β, and γ gene clusters (Figure 4A). PCR with specific primer pairs was used to detect six combinatorial inversions of DNA fragments induced by CRISPR (Figure 4B–G). Upstream and downstream junctions for each inversion event were analyzed by PCR and sequencing. We detected inversion junctions of individual α (Figure 4B), β (Figure 4C), or γ (Figure 4D) gene cluster as well as of combinations of these clusters, such as the Pcdh α/β (Figure 4E), β/γ (Figure 4F), or the entire Pcdh α/β/γ gene clusters (Figure 4G). We confirmed the combinatorial inversion junctions by Sanger sequencing (Supplementary Figure S10). Therefore, combinatorial inversions of large DNA fragments of the Pcdh gene clusters can be generated by CRISPR with four sgRNAs.
Figure 4.
Combinatorial genomic inversions and duplications by CRISPR with four sgRNAs. (A) Diagram of CRISPR with four sgRNAs targeted at the Pcdh α, β, and γ gene clusters. Shown are the inversions of the Pcdh α (B), β (C), γ (D), α/β (E), β/γ (F), and α/β/γ (G) gene clusters, as well as segmental duplications of the Pcdh α (H), β (I), γ (J), α/β (K), β/γ (L), and α/β/γ (M). The amplified upstream and downstream inversion junctions or segmental duplication junctions were sequenced. An example of Sanger sequencing chromatograms for each inversion or duplication is shown.
Previous studies revealed that copy number variations by segmental duplications of the Pcdh gene clusters are frequent in human populations and may associate with neurodevelopmental diseases or high-order brain functions (Noonan et al., 2003; Schmutz et al., 2004; Ukkola-Vuoti et al., 2013). Interestingly, we also detected segmental duplications generated by trans-allelic recombination between two concurrent DSBs induced by Cas9 with combinations of all four sgRNAs (Figure 4H–M). Specifically, segmental duplication junctions of individual Pcdh α, β, or γ gene cluster were amplified by PCR and confirmed by Sanger sequencing (Supplementary Figure S11). In addition, segmental duplication junctions of the α/β or β/γ gene clusters were also detected as well as potential duplication of all three Pcdh α/β/γ gene clusters. Finally, we detected indels at the duplication junctions (Supplementary Figure S11), suggesting that the segmental duplications are generated by NHEJ.
In addition to inversions and duplications, six combinatorial deletions by CRISPR were also identified and confirmed by Sanger sequencing. Both precise and imprecise deletion junctions were found at the cleavage site induced by Cas9 (Supplementary Figures S12 and S13). In summary, we found combinatorial inversions, duplications, and deletions generated by CRISPR with four sgRNAs. Their efficiencies range from 0.550% to 8.574%, 0.228% to 6.347%, and 0.318% to 20.989%, respectively (Supplementary Table S5). These data suggest that this efficient CRISPR method may be useful to model human diseases due to segmental DNA fragment variations.
Single-cell screening for DNA fragment inversion, duplication, and deletion by CRISPR
We screened single-cell CRISPR clones for inversions, duplications, and deletions of the β-globin RE1, Pcdh enhancer, β-globin RE2, and HoxD cluster, and found that they have high efficiency in cultured human cells (Supplementary Table S6). Specifically, for the 709-bp DNA fragment of the β-globin RE1, from a total of 78 single-cell clones isolated from limiting dilutions of cells transfected with Cas9 and a pair of sgRNAs, we obtained 38 clones with at least one inversion allele and 46 clones with at least one deletion alleles. Thus, the inversion and deletion efficiencies per cell are 48.72% and 58.97%, respectively. However, we did not find any clone with duplications. For the 1272-bp DNA fragment of the Pcdh enhancer, the inversion and deletion frequencies per cell were 12.50% and 37.50%, respectively. For the 6277-bp DNA fragment of the β-globin RE2, the inversion and deletion frequencies were 50.00% and 72.58%, respectively. In this case, we detected one clone with duplication. Finally, for the 18142-bp DNA fragment from the HoxD locus, the inversion and deletion frequencies per cell were 0.89% and 3.57%, respectively. These frequencies are much higher than those obtained by homologous recombination or by ZFN and TALEN (Wu et al., 2007; Lee et al., 2012). This suggests that one should be able to easily obtain inversion and deletion clones by CRISPR with a pair of sgRNAs.
A new role of the Pcdhα regulatory element in the regulation of the Pcdhγ cluster
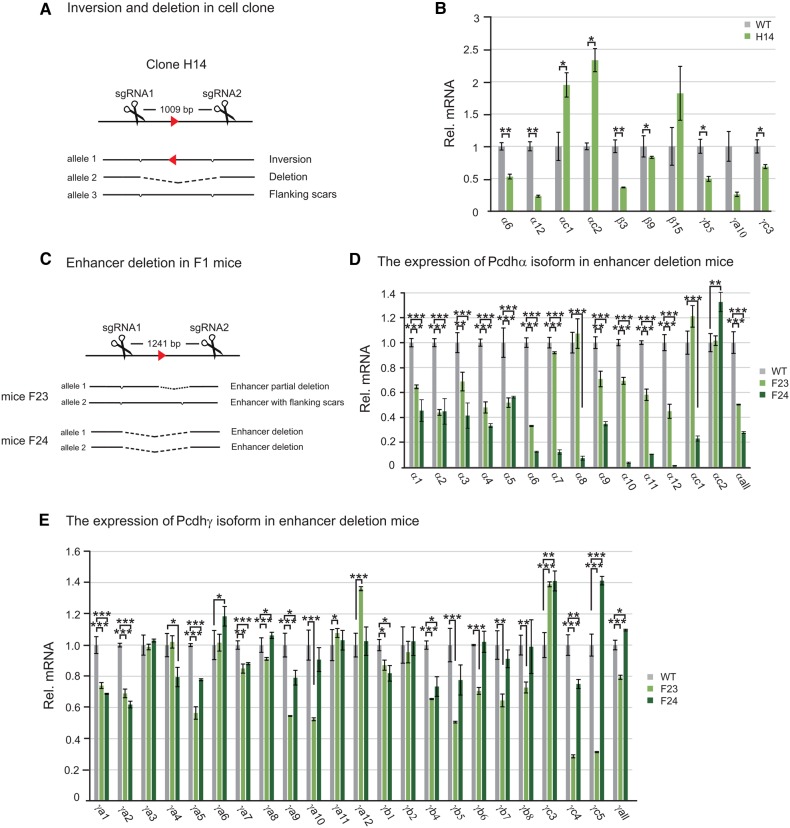
To demonstrate the usefulness of the CRISPR-mediated inversions and deletions in manipulating regulatory DNA elements, we transfected Hec-1-B cells (which is triploid for the Pcdh locus) with a pair of sgRNAs for two sites flanking a DNA fragment within a known Pcdhα enhancer (Ribich et al., 2006) and screened for single-cell CRISPR clones with deletion and inversion alleles. After screening 51 single-cell clones, we obtained a CRISPR cell line with one inversion and one deletion alleles (Figure 5A and Supplementary Figure S14A). Because Hec-1-B cells express two ‘alternate isoforms’ (α6 and α12 variable genes) of the Pcdhα gene cluster (Tasic et al., 2002; Guo et al., 2012), we measured expression levels of these two Pcdhα genes in the CRISPR cell lines. Quantitative real-time RT–PCR experiments revealed a significant decrease of their expression levels compared with the wild-type (WT) control (Figure 5B). Surprisingly, the expression of the two ubiquitous isoforms of the Pcdhα cluster is significantly increased (Figure 5B), probably because the targeted element contains a NRSF/REST suppressor binding site (Kehayova et al., 2011). Interestingly, this element also regulates expression members of the Pcdh β and γ clusters (Figure 5B).
Figure 5.
The application of inversion and deletion by CRISPR to an enhancer of the Pcdhα gene cluster reveals a new role in the regulation of the Pcdhγ cluster. (A) Single-cell Hec-1-B clone with enhancer deletion and inversion alleles obtained by CRISPR with a pair of sgRNAs. (B) Significant decreases of the Pcdh α6, α12, β3, β9, γb5, and γc3 gene expression in enhancer-deleted and inverted CRISPR cell line. (C) Enhancer deletion in F1 mice. Expression profiles of Pcdh α (D) and γ (E) clusters were measured by real-time RT–PCR using mouse brain tissues. Statistical analysis was performed by Student's t-test from three independent experiments.
To further investigate mechanisms of Pcdh gene regulation, we generated mice with deletions of the NRSF/REST binding site (473 bp deletion) as well as the large deletion of 1241 bp region (Figure 5C and Supplementary Figure S14B). Expression analysis revealed similar patterns of expression changes to previous large deletions of the 3 kb enhancer region (Figure 5D) (Kehayova et al., 2011). Surprisingly, we found significant expression changes of members of the Pcdhγ gene cluster, suggesting a new role of this element in the regulation of the Pcdhγ gene cluster (Figure 5E). Together, these experiments demonstrated the usefulness of DNA fragment inversion and deletion by CRISPR in human cells and mice.
Discussion
Since the discovery that one bacterial Cas9 enzyme and a single synthetic RNA molecule are the only two required heterologous components for achieving targeted genome editing in human cells (Cong et al., 2013; Jinek et al., 2013; Mali et al., 2013c), CRISPR/Cas9 technology has rapidly been utilized in broad engineering and biology, such as genetics, synthetic biology, neurosciences, and tumor biology (Mali et al., 2013b; Doudna and Charpentier, 2014; Harrison et al., 2014; Hsu et al., 2014; Platt et al., 2014). Here we found that DNA fragment inversions ranging from 35 to 807480 bp can easily be achieved in human cells and mice by CRISPR. In addition, segmental duplications can also be achieved by trans-allelic recombination between two DSBs induced by Cas9 with a pair of sgRNAs. Moreover, by using four sgRNAs targeting to the Pcdh gene clusters, we showed that combinatorial inversions and duplications of the Pcdh α, β, and γ gene clusters can be easily achieved in human cells. Finally, we demonstrated for the first time that mice with germline transmission of DNA fragment inversions can be obtained by injecting embryos with Cas9 mRNA and a pair of sgRNAs. These data demonstrated the usefulness of CRISPR technology in manipulating mammalian genomes to study gene regulatory elements as well as gene clusters.
Gene targeting technology in mice through homologous recombination has revolutionized mammalian biology (Capecchi, 2005). Gene editing utilizing NHEJ with artificial DSBs induced by ZFN, TALEN, or CRISPR has advanced rapidly (Carroll, 2014; Doudna and Charpentier, 2014; Harrison et al., 2014). Although ZFN and TALEN are powerful and effective, their usage is limited because of intrinsic difficulty of protein design and validation. In addition, laborious construction of a different fusion nuclease is required for each targeting site (Carroll, 2014; Doudna and Charpentier, 2014; Hsu et al., 2014). However, owning to its simplicity, reliability, and easiness to design, CRISPR has emerged as the method of choice for targeted genome engineering in various species (Mali et al., 2013b; Harrison et al., 2014; Hsu et al., 2014).
Our method of generating DNA fragment inversions and duplications in cultured cells and mice by CRISPR/Cas9 is simple and efficient. The efficiency of inversions and duplications by CRISPR/Cas9 is much higher than those obtained by methods through recombinases and nucleases (Ramirez-Solis et al., 1995; Wu et al., 2007; Lee et al., 2012; Gupta et al., 2013), paving the way for efficient DNA fragment inversion and duplication in mice in vivo. Owning to the high efficiency of generating blunt end by Cas9, we showed that precise inversion and duplication of DNA fragments with defined length ranging in size from tens of bp to hundreds of kb could be easily achieved. When designing experiments for DNA fragment editing in mammalian genomes, one should try to target both sgRNAs to G/C-rich sites in DNaseI hypersensitive regions. On one hand, editing of small DNA fragments could be applied to millions of regulatory DNA elements such as enhancers and insulators in the human genome, which are usually several hundred bp long and occupied asymmetrically by a variety of regulatory protein complexes (The ENCODE Project Consortium, 2012; Kundaje et al., 2012; Stamatoyannopoulos, 2012). On the other hand, editing of large DNA fragments of several hundred kb could be used to study complex gene clusters or to model DNA segmental duplications, which are common in mammalian genomes (Pan and Zhang, 2008). For example, genome editing of inversion of a large DNA fragment by Cre recombinase has shed light on the regulatory landscape of the HoxD cluster (Spitz et al., 2005). Finally, potential unintended modification from off-target sites could be alleviated by engineering Cas9 enzymes or using truncated sgRNAs, as well as using orthogonal Cas9 nucleases with different specificities (Esvelt et al., 2013; Mali et al., 2013a; Ran et al., 2013; Fu et al., 2014; Harrison et al., 2014; Nishimasu et al., 2014; Shen et al., 2014).
Mammals have evolved many mechanisms to repair DSBs, among which NHEJ and HR are the major pathways to maintain genome stability (Chapman et al., 2012; Jasin and Rothstein, 2013). DSBs repaired by HR are essential for meiotic recombination during meiosis, whereas DSBs repaired by NHEJ are essential for DNA rearrangement during normal mammalian immune development (Chapman et al., 2012). The blunt-ended DSBs generated by Cas9 are usually repaired by NHEJ (Carroll, 2014). Since we detected both precise inversions and inversions with small indels, they are most likely resulted from NHEJ. An ‘alternative end joining’ repair pathway, known as microhomology-mediated end joining (MMEJ), involves short repeats of 4–25 bp near the DSBs and causes genomic deletions, inversions, and other DNA rearrangements (McVey and Lee, 2008). For the human Pcdh RE2, we observed that the inversion is from the downstream microhomology repeat of ‘CTGG’ to the upstream double-strand breakpoint. For the mouse Pcdh locus 1, we observed that the downstream inversion junctions in two mice have deletions near a 6-bp inverted repeat of ‘CTAGAA’ (Supplementary Figure S1B). Finally, for the mouse Pcdh locus 2, we observed that the upstream inversion junctions in five mice have deletions near a 7-bp inverted repeat of ‘GGGTGGT’ (Supplementary Figure S3A). In all these cases, DSBs induced by Cas9 near the inverted microhomologous repeats stimulate the inversions of DNA fragments. Thus, these inversions are likely resulted from MMEJ.
Cas9 guided by multiple sgRNAs causes simultaneous multiplexed genome editing in cultured cells and mice (Cong et al., 2013; Wang et al., 2013). Furthermore, large chromosomal rearrangements can also be obtained in human cells through CRISPR (Choi and Meyerson, 2014; Maddalo et al., 2014; Torres et al., 2014). Given the complex nature of detected combinatorial inversions and duplications of the Pcdh gene clusters with four sgRNAs, attention should be paid to potential combinations of complex genome rearrangements by CRISPR with multiple sgRNAs. Finally, our observation of combinatorial inversions, duplications, and deletions by CRISPR with four sgRNAs is consistent with the idea that all of the ends of DSBs induced by Cas9 with multiple sgRNAs are brought into a single giga-Dalton repair center and ligated stochastically (Jasin and Rothstein, 2013).
In summary, precise inversions, duplications, and deletions of DNA fragments of variable sizes could be easily generated in mammals with Cas9 guided by two sgRNAs. In addition, multiplex targeted genomic inversions, duplications, and deletions of very large gene clusters could easily be obtained. This CRISPR method should be useful in mammalian genome engineering for studying millions of regulatory DNA elements as well as large gene clusters whose members often have redundant functions.
Materials and methods
Construction of targeting sgRNA vectors
A pair of complementary oligos (Supplementary Table S1) for each CRISPR targeting sgRNA was annealed with 5′ overhangs of ‘ACCG’ and ‘AAAC’. The annealed DNA is inserted into a BsaI-linearized pGL3 vector with the U6 promoter (Shen et al., 2014). SgRNA sequences were designed to target specific genomic sites (Supplementary Table S2).
Cell culture, transfection, and PCR analysis
HEK293T cells were maintained in Dulbecco's modified Eagle's medium (HyClone) supplemented with 10% (v/v) FBS (Gibco) and 1% penicillin–streptomycin (Gibco). HEC-1-B cells were maintained in Eagle's Minimum Essential Medium (Gibco) supplemented with 10% (v/v) FBS (Gibco), 1% penicillin–streptomycin (Gibco), 1 mM sodium pyruvate (Sigma) and 2 mM GlutaMAX (Gibco). Cells were cultured at 37°C in a 5% (v/v) CO2 incubator. Cells were transfected with the Cas9 (2 μg) (Chang et al., 2013) and sgRNA plasmids (2.5 μg) by Lipofectamine 2000 (Life Technology) 1 day after plating 9 × 105 cells per well in a 6-well plate. On Day 2 post-transfection, puromycin (2 μg/ml) was added and cells were cultured for additional 4 days. After harvesting cells, 80 μl alkaline lysis buffer (25 mM NaOH and 0.2 mM disodium EDTA, pH12.0) was added and heated at 98°C for 40 min. Equal volume of neutralizing buffer (40 mM Tris–HCl, pH5.0) was then added. One microliter was then used as template genomic DNA for PCR in a total volume of 20 μl with a specific pair of primers (Supplementary Table S1) to screen for DNA fragment inversions, duplications, and deletions. The PCR conditions are: predenaturing at 94°C for 4 min; followed by 35 cycles of 94°C denaturing for 30 sec, 60°C annealing for 30 sec, and 72°C extension for 50 sec; followed by a final extension at 72°C for 3 min.
In vitro transcription of Cas9 mRNA and sgRNAs
The Cas9 vector (Chang et al., 2013) with T7 promoter was first linearized with XbaI for use as a template for in vitro transcription with T7 polymerase. Cas9 mRNA was in vitro transcribed using mMESSAGE mMACHINE T7 Ultra Kit according to the manufacturer's manual (Life Technologies). SgRNAs templates with T7 promoter were obtained by PCR amplification with primers (Supplementary Table S1). The MEGAshortscript Kit (Life Technologies) was used to transcribe sgRNAs from the PCR product template. Cas9 mRNA and sgRNAs were purified with the MEGAclear Kit (Life Technologies) and eluted in TE buffer (0.2 mM EDTA) for microinjections.
One-cell embryos injection
All mice were housed at 23°C on a 12/12 h light-dark cycle (7:00 am–19:00 pm) in the SPF facilities. All experiments were carried out in accordance with Institutional Animal Care and Use Committee of Shanghai Jiao Tong University. C57BL/6 female mice and ICR mouse strains were used as embryo donors and foster mothers, respectively. Superovulated female C57BL/6 mice (26–30 days old) were mated to the C57BL/6 stud males. After 20 h, mouse embryos were obtained from the oviducts of superovulated female C57BL/6 mice. Cas9 mRNAs (100 ng/μl) and sgRNAs (50 ng/μl) were injected into the cytoplasm of one-cell embryos. Next, the injected embryos were in vitro cultured in the Ksom medium (Millipore) for half an hour at 37°C in a 5% CO2 incubator. The survivors of the injected embryos were implanted into the oviducts of pseudo-pregnant ICR mice. For blastocysts experiments, the survivors of the injected embryos were in vitro cultured in the Ksom medium for 4 days at 37°C in a 5% CO2 incubator. The blastocyst embryos of normal development were selected for experiments.
Mouse genotyping for detecting DNA fragment inversions, duplications, and deletions
The genotyping method was described previously (Wu et al., 2007). PCR was used to identify inversions, duplications, and deletions in mice with appropriate primer pairs (Supplementary Table S1).
Prediction and analysis of potential off-target sites
Potential off-target sites of sgRNAs were predicted (Mali et al., 2013c). For a 23-nt sequence of sgRNA target and PAM, the first seven nucleotides of sgRNA target sequences allow mismatches and the first nucleotide of PAM (NGG) also allows mismatches. However, thirteen nucleotides of seed sequences of sgRNAs must fully match. Potential off-target sites were searched within the human genome (hg19) and mouse genome (mm9) by a customized Python program. Potential off-target sites were amplified by PCR with specific primer pairs (Supplementary Table S1) and the amplified DNAs were sequenced.
RT–PCR and real-time PCR
Total RNA was prepared from cultured HEC-1-B cells by using TRIzol Reagent (Ambion). The reverse-transcription reactions were carried out by using the Reverse Transcription Systems (Promega) with 1 μg total RNA. The appropriate primers for Real-Time PCR were listed in Supplementary Table S1. Statistical analysis was performed by student t-test from three independent experiments.
Quantitative PCR analysis of the efficiency for DNA fragment deletions, inversions, and duplications
A q-PCR based method was used to analyze the efficiency of inversions, duplications, and deletions of DNA fragments. Owing to the limitation of q-PCR product length and indels produced by NHEJ-mediated repair of DSBs, the primers flanking each pair of sgRNA target sites were designed at least 50 bp away from the cleavage site of Cas9 and used to amplify an expected PCR product of less than 600 bp. Genomic DNA of transfected cells were used to amplify the PCR products of wild-type (WT), deletion (Del), inversion (Inv), and duplication (Dup) for reference templates. These PCR product templates were first quantified and diluted to equal amount of molars in TE buffer. A series of dilutions of these template DNAs were used to construct a standard curve. The appropriate primers for q-PCR analysis were listed in Supplementary Table S1. Q-PCR reactions were performed with the Faststart Universal SYBR Green Master system (Roche) under conditions of 10 min at 95°C for activation, 40 cycles of 15 sec at 95°C and 90 sec at 60°C. The quantities of DNA fragment inversions, duplications, and deletions were obtained by plotting against the standard curve. The efficiencies were calculated using the following formulas:
Single-cell clone screening
HEK293T cells were cultured and transfected with plasmids encoding Cas9 and two sgRNAs as described above. On Day 2 post-transfection, puromycin (2 μg/ml) was added and continued culturing for additional 4 days. Then cells were cultured in fresh medium for 8 days to grow to confluency before harvesting. To obtain single-cell CRISPR clone, the harvested cells were seeded and cultured in 96-well plates at a density of one cell per well. Positive wells with one single-cell clone were identified on Day 6 and continued culturing for additional 8 days. Single-cell clones were then screened by PCR analyses for inversion, duplication, and deletion events.
Supplementary material
Supplementary material is available at Journal of Molecular Cell Biology online.
Funding
This study was supported by grants to Q.W. from the National Natural Science Foundation of China (31171015 and 31470820) and the Science and Technology Commission of Shanghai Municipality (13XD1402000 and 14JC1403600). Q.W. is a Shanghai Subject Chief Scientist.
Conflict of interest: none declared.
Supplementary Material
Acknowledgements
We thank H. Huang and L. Peng from our lab for reading the manuscript, X. Huang (Nanjing University) and J. Xi (Peking University) for Cas9 and pGL3 plasmids, and members of our lab for discussion.
References
- Anders C., Niewoehner O., Duerst A., et al. (2014). Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 513, 569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baca S.C., Prandi D., Lawrence M.S., et al. (2013). Punctuated evolution of prostate cancer genomes. Cell 153, 666–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerji J., Olson L., Schaffner W. (1983). A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell 33, 729–740. [DOI] [PubMed] [Google Scholar]
- Barrangou R., Fremaux C., Deveau H., et al. (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712. [DOI] [PubMed] [Google Scholar]
- Blackwood E.M., Kadonaga J.T. (1998). Going the distance: a current view of enhancer action. Science 281, 60–63. [DOI] [PubMed] [Google Scholar]
- Byrne S.M., Ortiz L., Mali P., et al. (2015). Multi-kilobase homozygous targeted gene replacement in human induced pluripotent stem cells. Nucleic Acids Res. 43, e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai M., Yang Y. (2014). Targeted genome editing tools for disease modeling and gene therapy. Curr. Gene Ther. 14, 2–9. [DOI] [PubMed] [Google Scholar]
- Canver M.C., Bauer D.E., Dass A., et al. (2014). Characterization of genomic deletion efficiency mediated by clustered regularly interspaced palindromic repeats (CRISPR)/Cas9 nuclease system in mammalian cells. J. Biol. Chem. 289, 21312–21324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capecchi M.R. (2005). Gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century. Nat. Rev. Genet. 6, 507–512. [DOI] [PubMed] [Google Scholar]
- Carroll D. (2014). Genome engineering with targetable nucleases. Annu. Rev. Biochem. 83, 409–439. [DOI] [PubMed] [Google Scholar]
- Castle W.E., Little C.C. (1909). The peculiar inheritance of pink eyes among colored mice. Science 30, 313–314. [DOI] [PubMed] [Google Scholar]
- Chang N., Sun C., Gao L., et al. (2013). Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 23, 465–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman J.R., Taylor M.R., Boulton S.J. (2012). Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell 47, 497–510. [DOI] [PubMed] [Google Scholar]
- Cho S.W., Kim S., Kim J.M., et al. (2013). Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 31, 230–232. [DOI] [PubMed] [Google Scholar]
- Choi P.S., Meyerson M. (2014). Targeted genomic rearrangements using CRISPR/Cas technology. Nat. Commun. 5, 3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F.A., Cox D., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laat W., Duboule D. (2013). Topology of mammalian developmental enhancers and their regulatory landscapes. Nature 502, 499–506. [DOI] [PubMed] [Google Scholar]
- Deltcheva E., Chylinski K., Sharma C.M., et al. (2011). CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471, 602–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna J.A., Charpentier E. (2014). Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esvelt K.M., Mali P., Braff J.L., et al. (2013). Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat. Methods 10, 1116–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y.F., Sander J.D., Reyon D., et al. (2014). Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 32, 279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii W., Kawasaki K., Sugiura K., et al. (2013). Efficient generation of large-scale genome-modified mice using gRNA and CAS9 endonuclease. Nucleic Acids Res. 41, e187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau J.E., Dupuis M.E., Villion M., et al. (2010). The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468, 67–71. [DOI] [PubMed] [Google Scholar]
- Gilbert L.A., Larson M.H., Morsut L., et al. (2013). CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González F., Zhu Z., Shi Z.D., et al. (2014). An iCRISPR platform for rapid, multiplexable, and inducible genome editing in human pluripotent stem cells. Cell Stem Cell 15, 215–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Monahan K., Wu H., et al. (2012). CTCF/cohesin-mediated DNA looping is required for protocadherin α promoter choice. Proc. Natl Acad. Sci. USA 109, 21081–21086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A., Hall V.L., Kok F.O., et al. (2013). Targeted chromosomal deletions and inversions in zebrafish. Genome Res. 23, 1008–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft D.H., Selengut J., Mongodin E.F., et al. (2005). A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput. Biol. 1, e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagège H., Klous P., Braem C., et al. (2007). Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nat. Protoc. 2, 1722–1733. [DOI] [PubMed] [Google Scholar]
- Harrison M.M., Jenkins B.V., O'Connor-Giles K.M., et al. (2014). A CRISPR view of development. Genes Dev. 28, 1859–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho T.T., Zhou N., Huang J., et al. (2015). Targeting non-coding RNAs with the CRISPR/Cas9 system in human cell lines. Nucleic Acids Res. 43, e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P.D., Lander E.S., Zhang F. (2014). Development and applications of CRISPR-Cas9 for genome engineering. Cell 157, 1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishino Y., Shinagawa H., Makino K., et al. (1987). Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 169, 5429–5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R., van Embden J.D., Gaastra W., et al. (2002). Identification of a novel family of sequence repeats among prokaryotes. OMICS 6, 23–33. [DOI] [PubMed] [Google Scholar]
- Jasin M., Rothstein R. (2013). Repair of strand breaks by homologous recombination. Cold Spring Harb. Perspect. Biol. 5, a012740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., et al. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., East A., Cheng A., et al. (2013). RNA-programmed genome editing in human cells. Elife 2, e00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Jiang F., Taylor D.W., et al. (2014). Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science 343, 1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehayova P., Monahan K., Chen W., et al. (2011). Regulatory elements required for the activation and repression of the protocadherin-α gene cluster. Proc. Natl Acad. Sci. USA 108, 17195–17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiani S., Beal J., Ebrahimkhani M.R., et al. (2014). CRISPR transcriptional repression devices and layered circuits in mammalian cells. Nat. Methods 11, 723–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft K., Geuer S., Will A.J., et al. (2015). Deletions, inversions, duplications: engineering of structural variants using CRISPR/Cas in mice. Cell Rep. 10, 833–839. [DOI] [PubMed] [Google Scholar]
- Kundaje A., Kyriazopoulou-Panagiotopoulou S., Libbrecht M., et al. (2012). Ubiquitous heterogeneity and asymmetry of the chromatin environment at regulatory elements. Genome Res. 22, 1735–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.J., Kweon J., Kim E., et al. (2012). Targeted chromosomal duplications and inversions in the human genome using zinc finger nucleases. Genome Res. 22, 539–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Qiu Z., Shao Y., et al. (2013). Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat. Biotechnol. 31, 681–683. [DOI] [PubMed] [Google Scholar]
- Lin Y., Cradick T.J., Brown M.T., et al. (2014). CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res. 42, 7473–7485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddalo D., Manchado E., Concepcion C.P., et al. (2014). In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature 516, 423–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova K.S., Haft D.H., Barrangou R., et al. (2011). Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 9, 467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Aach J., Stranges P.B., et al. (2013a). CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 31, 833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Esvelt K.M., Church G.M. (2013b). Cas9 as a versatile tool for engineering biology. Nat. Methods 10, 957–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K.M., et al. (2013c). RNA-guided human genome engineering via Cas9. Science 339, 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malina A., Mills J.R., Cencic R., et al. (2013). Repurposing CRISPR/Cas9 for in situ functional assays. Genes Dev. 27, 2602–2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini L.A., Sontheimer E.J. (2008). CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322, 1843–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini L.A., Sontheimer E.J. (2010). Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature 463, 568–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey M., Lee S.E. (2008). MMEJ repair of double-strand breaks (director's cut): deleted sequences and alternative endings. Trends Genet. 24, 529–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica F.J., Díez-Villaseñor C., García-Martínez J., et al. (2009). Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155, 733–740. [DOI] [PubMed] [Google Scholar]
- Müller H.J. (1927). Artificial transmutation of the gene. Science 66, 84–87. [DOI] [PubMed] [Google Scholar]
- Neph S., Vierstra J., Stergachis A.B., et al. (2012). An expansive human regulatory lexicon encoded in transcription factor footprints. Nature 489, 83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H., Ran F.A., Hsu P.D., et al. (2014). Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 156, 935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y., Shen B., Cui Y., et al. (2014). Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell 156, 836–843. [DOI] [PubMed] [Google Scholar]
- Noonan J.P., Li J., Nguyen L., et al. (2003). Extensive linkage disequilibrium, a common 16.7-kilobase deletion, and evidence of balancing selection in the human protocadherin α cluster. Am. J. Hum. Genet. 72, 621–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, D., and Zhang, L. (2008). Tandemly arrayed genes in vertebrate genomes. Comp. Funct. Genomics, doi:10.1155/2008/545269. [DOI] [PMC free article] [PubMed]
- Platt R.J., Chen S., Zhou Y., et al. (2014). CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159, 440–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L.S., Larson M.H., Gilbert L.A., et al. (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Solis R., Liu P., Bradley A. (1995). Chromosome engineering in mice. Nature 378, 720–724. [DOI] [PubMed] [Google Scholar]
- Ran F.A., Hsu P.D., Lin C.Y., et al. (2013). Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154, 1380–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribich S., Tasic B., Maniatis T. (2006). Identification of long-range regulatory elements in the protocadherin-α gene cluster. Proc. Natl Acad. Sci. USA 103, 19719–19724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J., Martin J., Terry A., et al. (2004). The DNA sequence and comparative analysis of human chromosome 5. Nature 431, 268–274. [DOI] [PubMed] [Google Scholar]
- Shalem O., Sanjana N.E., Hartenian E., et al. (2014). Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp A.J., Cheng Z., Eichler E.E. (2006). Structural variation of the human genome. Annu. Rev. Genomics Hum. Genet. 7, 407–442. [DOI] [PubMed] [Google Scholar]
- Shen Y., Yue F., McCleary D.F., et al. (2012). A map of the cis-regulatory sequences in the mouse genome. Nature 488, 116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B., Zhang W., Zhang J., et al. (2014). Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat. Methods 11, 399–402. [DOI] [PubMed] [Google Scholar]
- Smithies O., Gregg R.G., Boggs S.S., et al. (1985). Insertion of DNA sequences into the human chromosomal β-globin locus by homologous recombination. Nature 317, 230–234. [DOI] [PubMed] [Google Scholar]
- Spitz F., Herkenne C., Morris M.A., et al. (2005). Inversion-induced disruption of the Hoxd cluster leads to the partition of regulatory landscapes. Nat. Genet. 37, 889–893. [DOI] [PubMed] [Google Scholar]
- Stamatoyannopoulos J.A. (2012). What does our genome encode? Genome Res. 22, 1602–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz P., Lupski J.R. (2010). Structural variation in the human genome and its role in disease. Annu. Rev. Med. 61, 437–455. [DOI] [PubMed] [Google Scholar]
- Stephens P.J., Greenman C.D., Fu B., et al. (2011). Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 144, 27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern M.J., Ames G.F., Smith N.H., et al. (1984). Repetitive extragenic palindromic sequences: a major component of the bacterial genome. Cell 37, 1015–1026. [DOI] [PubMed] [Google Scholar]
- Sternberg S.H., Redding S., Jinek M., et al. (2014). DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507, 62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimoto K., Liu Q., Bungert J., et al. (1999). Effects of altered gene order or orientation of the locus control region on human β-globin gene expression in mice. Nature 398, 344–348. [DOI] [PubMed] [Google Scholar]
- Tasic B., Nabholz C.E., Baldwin K.K., et al. (2002). Promoter choice determines splice site selection in protocadherin α and γ pre-mRNA splicing. Mol. Cell 10, 21–33. [DOI] [PubMed] [Google Scholar]
- Thomas K.R., Capecchi M.R. (1986). Introduction of homologous DNA sequences into mammalian cells induces mutations in the cognate gene. Nature 324, 34–38. [DOI] [PubMed] [Google Scholar]
- Thurman R.E., Rynes E., Humbert R., et al. (2012). The accessible chromatin landscape of the human genome. Nature 489, 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres R., Martin M.C., Garcia A., et al. (2014). Engineering human tumour-associated chromosomal translocations with the RNA-guided CRISPR-Cas9 system. Nat. Commun. 5, 3964. [DOI] [PubMed] [Google Scholar]
- Ukkola-Vuoti L., Kanduri C., Oikkonen J., et al. (2013). Genome-wide copy number variation analysis in extended families and unrelated individuals characterized for musical aptitude and creativity in music. PLoS One 8, e56356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan H., Feng C., Teng F., et al. (2015). One-step generation of p53 gene biallelic mutant Cynomolgus monkey via the CRISPR/Cas system. Cell Res. 25, 258–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Yang H., Shivalila C.S., et al. (2013). One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Sengel C., Emerson M.M., et al. (2014a). A gene regulatory network controls the binary fate decision of rod and bipolar cells in the vertebrate retina. Dev. Cell 30, 513–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Wei J.J., Sabatini D.M., et al. (2014b). Genetic screens in human cells using the CRISPR-Cas9 system. Science 343, 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson J.D., Crick F.H. (1953). Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature 171, 737–738. [DOI] [PubMed] [Google Scholar]
- Wei W., Brennan M.D. (2000). Polarity of transcriptional enhancement revealed by an insulator element. Proc. Natl Acad. Sci. USA 97, 14518–14523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C., Liu J., Yu Z., et al. (2013). TALEN or Cas9—rapid, efficient and specific choices for genome modifications. J. Genet. Genomics 40, 281–289. [DOI] [PubMed] [Google Scholar]
- West A.G., Gaszner M., Felsenfeld G. (2002). Insulators: many functions, many mechanisms. Genes Dev. 16, 271–288. [DOI] [PubMed] [Google Scholar]
- Wu Q. (2005). Comparative genomics and diversifying selection of the clustered vertebrate protocadherin genes. Genetics 169, 2179–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q., Maniatis T. (1999). A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell 97, 779–790. [DOI] [PubMed] [Google Scholar]
- Wu Q., Zhang T., Cheng J.F., et al. (2001). Comparative DNA sequence analysis of mouse and human protocadherin gene clusters. Genome Res. 11, 389–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Ying G., Wu Q., et al. (2007). Toward simpler and faster genome-wide mutagenesis in mice. Nat. Genet. 39, 922–930. [DOI] [PubMed] [Google Scholar]
- Wu Y., Zhou H., Fan X., et al. (2015). Correction of a genetic disease by CRISPR-Cas9-mediated gene editing in mouse spermatogonial stem cells. Cell Res. 25, 67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao A., Wang Z., Hu Y., et al. (2013). Chromosomal deletions and inversions mediated by TALENs and CRISPR/Cas in zebrafish. Nucleic Acids Res. 41, e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Wang H., Shivalila C.S., et al. (2013). One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154, 1370–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Bradley A. (2001). Engineering chromosomal rearrangements in mice. Nat. Rev. Genet. 2, 780–790. [DOI] [PubMed] [Google Scholar]
- Zhang T., Haws P., Wu Q. (2004). Multiple variable first exons: a mechanism for cell- and tissue-specific gene regulation. Genome Res. 14, 79–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B., Sage M., Sheppeard E.A., et al. (2000). Engineering mouse chromosomes with Cre-loxP: range, efficiency, and somatic applications. Mol. Cell. Biol. 20, 648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Zhu S., Cai C., et al. (2014). High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature 509, 487–491. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.