Abstract
Phage display is a powerful method for target discovery and selection of ligands for cancer treatment and diagnosis. Our goal was to select tumor-binding antibodies in cancer patients. Eligibility criteria included absence of preexisting anti-phage-antibodies and a Stage IV cancer status. All patients were intravenously administered 1 × 1011 TUs/kg of an scFv library 1 to 4 h before surgical resection of their tumors. No significant adverse events related to the phage library infusion were observed. Phage were successfully recovered from all tumors. Individual clones from each patient were assessed for binding to the tumor from which clones were recovered. Multiple tumor-binding phage-antibodies were identified. Soluble scFv antibodies were produced from the phage clones showing higher tumor binding. The tumor-homing phage-antibodies and derived soluble scFvs were found to bind varying numbers (0–5) of 8 tested normal human tissues (breast, cervix, colon, kidney, liver, spleen, skin, and uterus). The clones that showed high tumor-specificity were found to bind corresponding tumors from other patients also. Clone enrichment was observed based on tumor binding and DNA sequence data. Clone sequences of multiple variable regions showed significant matches to certain cancer-related antibodies. One of the clones (07-2,355) that was found to share a 12-amino-acid-long motif with a reported IL-17A antibody was further studied for competitive binding for possible antigen target identification. We conclude that these outcomes support the safety and utility of phage display library panning in cancer patients for ligand selection and target discovery for cancer treatment and diagnosis.
Electronic supplementary material
The online version of this article (doi:10.1007/s00262-013-1443-5) contains supplementary material, which is available to authorized users.
Keywords: Targeted therapy, Cancer-specific ligands, Antibodies, Target identification, Phage-displayed library, Human phage infusion safety
Introduction
The field of targeted therapy is providing an entirely new generation of highly active anticancer drugs including monoclonal antibodies [1–4]. A method to rapidly develop different sets of therapeutic antibodies would greatly contribute to the field of targeted anticancer therapy. Phage display is a powerful method to generate antibodies to known or unknown tumor targets. In this technology, the bacteriophage express the antibody library on their outer surface, and the antibodies of desired characteristics are selected by the process of panning on the targets [5]. Known targets typically are purified and used in the purified form for selection of phage-displayed ligands. More complex assemblage of biological material is typically used to screen for ligands to unknown targets. For example, whole tumor cells are typically screened in serial panning events for ligands. Identification of the target of a tumor-selective ligand can provide important information related to molecular features that differ between tumor and normal cells. Use of a complex tissue for screening can yield both a ligand to and identification of important targets.
The choice of the target tissue depends on the panning strategy used to identify target-selective ligands. For example, whole tumor cell-panning is typically accompanied by subtraction of the input phage with non-tumor cells [6, 7]. A number of investigators have used in vitro phage display approaches with innovative variations and identified tumor-specific ligands by panning established tumor cell lines [8–11]. We have focused on using clinical material for panning strategies [12, 13]. Patient-derived material has the advantage of clinical relevance, but has the disadvantage of limited and variable supply [14]. Also, other than blood elements, there are limited options to use a normal tissue counterpart from the same patient for subtraction of input phage.
Another approach utilizes an in vivo selection process in which animal models of cancer are injected with a phage display library and the tumor-homing phage are recovered and assessed for their binding to the tumors. Several research groups have identified tumor-specific ligands following phage library infusion in animals using this strategy [15–23]. It would be a great translational success if the same strategy could be successfully applied to human cancer patients. Tumors in a patient represent the most complex state of a tumor and the most clinically relevant. All the cellular components are present, and the tumor is in a dynamic state of interaction with the blood supply and the immune system. Based on the data from our preclinical study [24], we chose to develop a protocol for selection of phage-displayed ligands in cancer patients. In addition to the advantages associated with the presence of all the tumor elements, some level of subtraction to normal tissue elements should occur as the library circulates through the body. With all of the targets present and blood flowing through the tumor, this approach should provide the maximum opportunity for identifying unique tumor targets.
We previously reported the very first study related to a phage library infusion in human cancer patients and established toxicity profile of different doses and formats of phage-displayed libraries [25]. In this second study with phage display panning in 6 patients with Stage IV cancer, we have evaluated the binding of the tumor-homing phage-antibodies and derived soluble scFv antibodies to patients’ tumors and to a panel of normal human tissues in order to determine the cancer-specificity of the selected clones.
Materials and methods
Human subjects
This Phase 1 clinical study was performed in accordance with the US Federal Drug Administration (approval # FDA BB-IND 9145; Protocol Amendment: CHRMS 05–170, V0210, Protocol 815) and the University of Vermont Institutional Review Board Committee on Human Research in the Medical Sciences. Eligibility criteria included the following: Stage IV malignancy of any histology; >18 years of age; non-pregnant; a life expectancy of ≥4 months; Karnofsky status ≥70; Hgb ≥10 gm/dL Hct ≥30 %; ANC 1,500/μL; platelets ≥75,000/μL, creatinine<institutional upper limit of normal; hepatic function parameters less than 2 × upper limit of normal (serum albumin 3–5.5 g/dL, alkaline phosphatase 38–126 U/L, alanine aminotransferase 15–75 U/L, aspartate aminotransferase 8–50 U/L, direct bilirubin 0.0–0.3 mg/dL, total bilirubin 0.2–1.3 mg/dL, total protein 6–8.5 g/dL); cardiac NYHA Grade II or less; no evidence of impaired lung function on physical examination; no clinical symptoms suggestive of brain metastases unless ruled out by imaging studies; no psychiatric conditions that would prevent informed consent regarding this study; and no elevation of anti-bacteriophage antibodies. The schematic outline of the study has been summarized in Supplementary Table S1.
Six cancer patients were recruited for this study. The tumor types included malignant melanoma, granulocytic lymphoma, colorectal cancer, and chondrosarcoma (Table 1). Patients’ ages ranged from 43 to 72 years.
Table 1.
Summary of the phage-antibody treatment procedures conducted in the patients who participated in this study
| Patient ID | Tumor histology | Dose of phage scFv library, TUs/kg (Body weight, kg) | Location(s) of tumor resection | Size of excised tumor | Side effects during infusion | Side effects post-infusion days 2–60 |
|---|---|---|---|---|---|---|
| 180-12 | Granulocytic lymphoma | 1 × 1011 (91.08) | Left chest wall, high axillary area-left pectoralis muscle | 2 × 2 × 2 cm | None | Please see footnotea |
| 180-13 | Colorectal metastasis to liver | 1 × 1011 (95.79) | Pedunculated hanging tumor-right posterior lateral liver, segment VI | 6 × 5 × 4 cm | None | None |
| 180-14 | Chondrosarcoma metastasis to pelvis | 1 × 1011 (85.05) | Pelvic mass | 21 × 11.5 × 10.5 cm | None | None |
| 180-15 | Melanoma metastasis to lymph node | 1 × 1011 (83.00) | Left axillary mass | 16 × 12 × 12 cm | None | Please see footnoteb |
| 180-16 | Melanoma metastasis to liver | 1 × 1011 (53.10) | Liver resection partial segmentectomy of segment VI | 1.6 cm diameter × 1.9 cm length | None | None |
| 180-17 | Metastatic melanoma | 1 × 1011 (91.7) | Subcutaneous mass excised from right posterior thigh | 1.5 cm diameter × 1.5 cm length | None | None |
aOne-month follow-up: low values for blood HGB, HCT, ANC, platelets, albumin, and total protein; high levels of alkaline phosphatase. Two-month follow-up: ANC returned to normal levels, other side effects remained, attributed to progressive metastatic disease, and/or response to chemotherapy
bNo complications were reported at 1-week follow-up visit. The patient came to ER about 1.5 months after the infusion with nausea, vomiting, and neck pain. An obstruction was treated with a stent in the superior vena cava. At 2-month follow-up, it was noted that continued side effects were related to the growth of the malignant melanoma
Serum phage-antibody assay
Serum phage antibodies were determined using an ELISA-based method we previously developed for animal and human panning [24, 25]. Sera for positive controls were available from the patients that had previously been infused with a phage library. Briefly, a 96-well Maxisorp plate (NUNC, Rochester, NY) was coated with 1 × 107 TUs of filamentous phage library overnight at 4 °C. The unbound phage were removed with Tris-buffered saline (TBS) containing 0.1 % Tween, and the wells were blocked with 1 % casein in TBS. Phosphate buffered saline, pH 7.2 (PBS), human IgG (negative control), the sera from positive controls, and the test serum samples were added in triplicate to the appropriate wells (100 μL/well). These samples were incubated on the plate for 2 h with a slow shaking at room temperature. The plate was washed again to remove any unbound material. Goat anti-human IgG-HRP conjugate (1:10,000) was added to the wells and incubated for 2 h at room temperature. The plate was washed again to remove the unbound HRP conjugate. ABTS (2,2′-azinobis [3-ethylbenzothiazoline-6-sulfonic acid]-diammonium salt) was used for detection of the conjugate.
Phage library
The human single chain scFv Tomlinson I library [26], cloned in ampicillin-resistant phagemid vector pIT2 and transformed into TG1 Escherichia coli cells, was obtained from MRC, HGMP Resource Centre (Hinxton, Cambridge, UK). The scFv fragments were comprised of single polypeptides in which the VH and VL domains were attached together with a flexible glycine-serine linker. The scFv library amplification and titration were done as described earlier [26, 27], using KM13 helper phage [28] and TG1 E. coli bacteria.
Preparation of phage library for infusion
Endotoxins were removed from the library preparation by performing 1 % (v/v) Triton X-114 extractions until the preparation had endotoxin levels under the FDA limit of 5EU/kg as determined by Limulus Amebocyte Lysate gel clot assay (Endosafe, Charles River Labs, Wilmington, MA). The phage were concentrated with polyethylene glycol, and the resulting pellet was resuspended in PBS containing protease inhibitor aprotonin (Trasylol; Bayer Pharmaceuticals Corporation, West Haven, CT). The phage suspension was shaken for 10 min at 200 rpm on ice to completely solubilize the phage particles. The sample was centrifuged, and the supernatant containing the phage was passed through a pyrogen-free 0.22 μm polyethersulfone membrane filter. Sterility of the phage preparation that was injected into the human patients was tested according to the US FDA Code of Federal Regulations (21 CFR 610.12), using fluid thioglycolate and soybean casein media. All the infused phage preparations demonstrated no evidence of microbial growth after 2 weeks of the sterility testing.
Patient monitoring and intravenous phage administration
Patients had an intravenous line placed and baseline vital signs (blood pressure, pulse, temperature, and respiratory rate) determined before the infusion of phage. There were no concurrent cytotoxic drugs or radiation therapy during the week preceding phage infusion. The phage library at a dose of 1.0 × 1011 TU/kg was diluted in 100 mL saline and infused over 15 min. Patients were closely monitored for early signs of anaphylaxis. Personnel skilled in handling allergic reactions were present during infusions, and resuscitative equipment and drugs were immediately available. Tumors were surgically removed between 1 and 4 h after the phage infusion. A portion of the tumors was cut and taken by a surgical pathologist for evaluation, and the rest was transported immediately for processing and phage recovery. The vital signs, including blood pressure, pulse, respiratory rate, pulse oximetry, and level of consciousness were monitored closely throughout the infusion, biopsy, and for 2 h after the procedure. Patients were re-evaluated at 1 and 2 months following the procedure (Supplementary Table S1).
Tumor processing and phage recovery
A small portion of the resected tumor was snap frozen and stored at −80 °C, and the rest was used for the phage recovery. The tumor amounts available from patients 180-12, 180-16, and 180-17 were too small for an optimal phage recovery and binding analyses. After removing the adherent blood, the tumor tissue was cut into several small pieces and rinsed 5 times by repeated suspension and centrifugation steps. The rinsed tumor pellet was homogenized to release cell-internalized phage. The bacterial host infection, plating, titration, and amplification were done by the methods described earlier [24, 25].
Binding assessment
Tumor binding of the individual clones was evaluated using frozen sections of the same patient’s tumors from which the clones were recovered. Histological sections were mounted on 12-mm round coverslips and placed in 24-well plates (Thermo Fisher Scientific, Rochester, NY) as described earlier [13].
Phage clone
Selected individual phage clones were amplified and normalized by ELISA [29] and/or by OD269 measurements [30]. 1 × 1012 TU of individual phage clones in 200 μL binding buffer (PBS containing 0.2 % casein and 20 % goat serum) were incubated with the tissue sections at room temperature with a slow shaking for 2 h. After the incubation, slides were rinsed 10 times in PBST (PBS containing 0.1 % Tween-20) and once in PBS. Mouse anti-M13 antibody (GE Healthcare, Piscataway, NJ) and goat anti-mouse IgG-Alexa Fluor 488 (Molecular Probes, Invitrogen, CA) were used to visualize the phage using Nikon Eclipse Ti fluorescence microscope (MVI, Avon, MA). Negative phage controls included naïve library, KM13 helper phage, and a previously identified phage clone with no demonstrable binding. For patient cases in which large numbers of phage clones were recovered, initial binding analysis was done with 4 phage clones per tissue section followed by subsequent re-analysis of individual clones from the positive groups. Our control experiments indicated that 1 positive clone mixed with 9 negative clones in a well could be successfully screened in this manner. Phage clones with the strongest tumor-binding signal were chosen for further analysis with their soluble scFv preparations and characterization.
Soluble scFv antibody
Soluble scFvs were produced by infection in E. coli HB2151 [31]. E. coli HB2151culture (OD600nm = 0.4) was incubated with an aliquot of the selected phage clone at 37 °C for 1 h, then plated onto TYE agar containing 100 μg/mL ampicillin and 1 % (v/v) glucose for overnight growth at 30 °C. Individual colonies were amplified, and the overnight culture was diluted 1:100 into 200 mL 2× YT containing 100 μg/mL ampicillin and 0.1 % glucose. The bacterial culture was shaken at 37 °C until the culture’s optical density reached 0.9 at 600 nm. The expression of scFvs was then induced by the addition of 25 mL 2× TY containing 100 μg/mL ampicillin and 9 mM isopropyl-β-d-thio-galactoside (IPTG). The incubation was continued for 20 h at 30 °C. The bacterial culture was centrifuged at 3,000 rpm for 15 min, and the supernatants were used for the scFv purification and ELISA. Centrifuge columns packed with protein-l resin (Pierce, Rockford, IL) were used for scFv purification. scFv proteins were eluted with 0.1 M glycine at pH 3 and immediately neutralized by 1 M Tris buffer, pH 9. The eluted fractions were analyzed by SDS-PAGE. The fractions found to contain purified scFv were pooled, concentrated to~100-fold using Ultra-4 centrifugal filter (Amicon, Millipore, UK), and stored at −20 °C in 20 % glycerol.
A chemiluminescent ELISA was used to determine the amount of scFv in each preparation [ 29 ]. Briefly, the microtiter plates coated with protein-A (0.5 μg/well, Southern Biotech, Birmingham, AL) were blocked with casein and incubated with the scFv samples in PBST for 1 h at room temperature. scFvs were detected using a HisProbe-HRP in presence of SuperSignal ELISA chemiluminescent substrate (Thermo Scientific, Rockford, IL).
For tumor-binding studies, selected soluble scFvs from individual phage clones were normalized. A total of 200 μL of scFv solutions (0.2–1 μM) in PBS were incubated with the tissue sections in 24-well plates for 1 h with shaking at room temperature. Slides were rinsed in PBS after the incubation, and the bound scFvs were labeled with mouse anti-6xHis antibody (Sigma, St. Louis, MO). Alexa Fluor 568 or 488 conjugates of goat anti-mouse IgG (Molecular Probes, Invitrogen, CA) were used to visualize the scFvs. Alternatively, the mouse anti-6xHis antibody conjugated to Alexa Fluor 488 (Millipore, Temecula, CA) was used. Imaging was performed using the same methods as with phage clones. The scFv from a phage clone that was previously shown to have no demonstrable binding was used as a negative control.
Tumor-binding phage clones and soluble scFvs with the best binding intensity were subsequently assessed for binding to a panel of normal human tissues including breast, cervix, colon, kidney, liver, spleen, skin, and uterus that were obtained from the Vermont Cancer Center Tissue Procurement Facility at the University of Vermont.
The binding of selected tumor-specific clones was confirmed by lysate ELISA assay. The tumor and normal tissue lysates were prepared by 3 alternate cycles of freezing/thawing and ultrasonication. The binding of phage-scFvs to the immobilized tissue lysates in microtiter plates was monitored by using anti-M13 antibody-HRP conjugate in presence of TMB soluble (Millipore Corporation, Billerica, MA) as a substrate [12, 14]. Some of the tumor-specific phage-antibodies from patients 180-13 and 180-15 were also assessed for their binding to the archived melanoma (metastasis to axillary lymph node) and colorectal tumors, respectively, from other patients using immunofluorescence microscopy as described above for tumor and normal tissues.
DNA sequence analysis and bioinformatics
When a limited number of phage clones were recovered from a patient tumor, all the clones were sequenced (180-17). Otherwise, only the clones with demonstrated binding to the target tumor tissue were sequenced. PCR-amplified library inserts from the pIII region of the phage genome were used for sequencing with the pIII sequencing primer, 5′-CCC TCA TAG TTA GCG TAA CG-3′, designed by Dr. George Smith and the LMB3 primer from the Tomlinson library manual. The sequencing reactions were carried out using BigDye Ver.1 Dye Terminator kit (PE Biosystems) by the Vermont Cancer Center DNA Analysis Facility at the University of Vermont.
We extracted the DNA sequences of the variable regions of the scFvs and translated them into amino acid sequences for analysis. The proper alignment of the CDRH and CDRL regions was verified by CLUSTALW software (EMBL-EBI, Hinxton, UK) applied to the whole set of the resulting sequences [32]. CLUSTALW was also applied to search for the similarities between the obtained scFv sequences. The analysis consisted of alignments of CDRH2, CDRH2+CDRH3, and CDRL2+CDRL3 domains separately. The sequences of select scFvs were further analyzed by homology searches with BLASTp software [33] (NCBI, Bethesda, MD) to find similarities with the reported antibodies. The selection criteria for inclusion in this search were the following: (a) tumor binding on histological assay; (b) repeated clones in output phage from patient panning; and (c) motifs in the scFv groups.
One of the clones (07-2,355) that was found to share a very significant motif with a reported IL-17A antibody was further studied for its binding to the tumor in the presence of a mouse polyclonal anti-human IL-17A antibody (eBioscience Inc., San Diego, CA) or the negative control mouse irrelevant antibodies (anti-poly-His tag and anti-V5 tag, Sigma Chemical Co., St. Louis, MO). Clone 07–2,355 binding was localized with a rabbit anti-M13 phage-antibody followed by anti-rabbit secondary antibody conjugated to Alexa Fluor 488. The IL-17A expression of melanoma tumor from which this clone was recovered was compared to that of normal human skin by immunostaining with anti-human IL-17A antibody and anti-mouse secondary antibody-Alexa Fluor 568 conjugate.
Results
Adverse events
No immediate or delayed phage-related adverse events were observed during infusion or during the 8-week observation period. Specifically, no allergy-related reactions were observed. Patient 180-12 at 1-month follow-up showed low values for blood HGB, HCT, ANC, platelets, albumin and total protein, and high levels of alkaline phosphatase. At this patient’s 2-month follow-up, the absolute neutrophil count had returned to normal levels but other side effects remained, attributed to progressive metastatic disease and reaction to chemotherapy. Patient 180-15 reported no complications at the 1-week follow-up visit. At 1.5 months after the infusion, the patient developed nausea, vomiting, and neck pain. An obstruction from tumor growth was treated with a stent in the superior vena cava. At 2-month follow-up, it was noted that continued side effects were related to the growth of the malignant melanoma (Table 1).
Sterility tests
All sterility tests for the reagents used in connection with the phage infusion to patients demonstrated no growth of bacteria.
Anti-phage antibodies
Figure 1 shows the levels of anti-filamentous phage IgG in the archived positive control sera and in the test samples of enrolled patients, as determined by ELISA. The serum anti-filamentous phage IgG did not show elevation in their levels in any of the enrolled patients prior to the phage infusion.
Fig. 1.
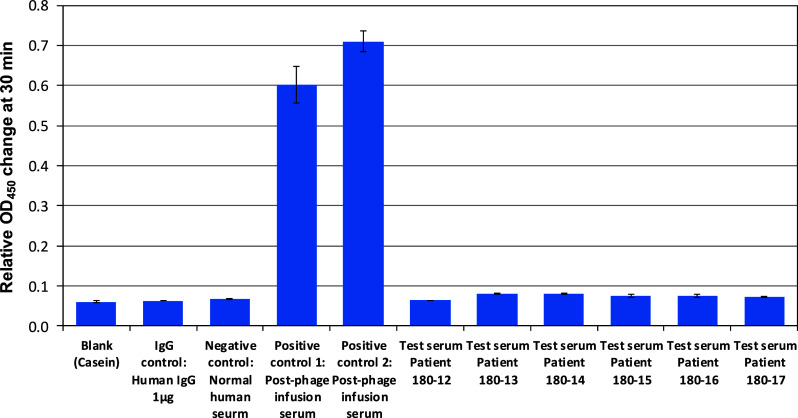
Anti-filamentous phage serum IgG levels in the patients before infusion of phage-displayed scFv library. The values represent mean ± SE (n = 3). The first 3 bars represent different types of negative controls, followed by archived positive controls from 2 post-fusion patients. The last 6 bars represent the prephage fusion test serums from patients presented in this report
Tumor phage recovery and tumor-binding assessment of phage-scFv clones and soluble scFvs
Tumor-homing phage were recovered from all the patients infused with phage-displayed antibody library. The phage recovery ranged from 2.9 × 10−4 to 3.1 × 10−3 percent of the injected phage dose (Table 2). The number of phage clones evaluated for tumor binding per patient ranged from 24 to 469. The lowest numbers of recovered phage were related to the small size of the surgical specimen. The binding studies of scFv antibodies to tumors and normal tissues in phage-displayed and soluble forms produced hundreds of images. We have summarized the image data in several Tables and presented only representative images for a better understanding of the results. Table 2 shows that the percentage of randomly selected phage clones per patient with detectable tumor binding ranged from 0 to 28 %. Our further studies were focused on patients 180-13, 180-14, and 180-15 because the quantity of these patients’ tumor tissues was sufficient for assays. A total of 78 selected scFvs from these 3 patients were expressed in sufficient quantity for tumor-binding assessment. More than half (58 %) of the selected scFvs had detectable tumor binding.
Table 2.
The tumor phage recovery, and number of phage clones and the soluble scFvs, derived from the higher tumor-binding phage clones, evaluated for tumor binding and the number that demonstrated tumor binding
| Patient ID | Tumor phage recovery (% of dose) | Phage clones | Soluble scFv | ||
|---|---|---|---|---|---|
| Number evaluated | Number binding to tumor (%) | Number evaluated | Number binding to tumor (%) | ||
| 180-12 | 1.2 × 10−3 | 40 | 11 (28) | 0 | 0 |
| 180-13 | 3.1 × 10−3 | 469 | 99 (21) | 58 | 32 (55) |
| 180-14 | 4.1 × 10−4 | 241 | 50 (21) | 9 | 4 (44) |
| 180-15 | 2.9 × 10−4 | 288 | 40 (14) | 11 | 9 (82) |
| 180-16 | 1.7 × 10−3 | 176 | 4 (2) | 0 | 0 |
| 180-17 | 7.6 × 10−4 | 24 | 0 | 0 | 0 |
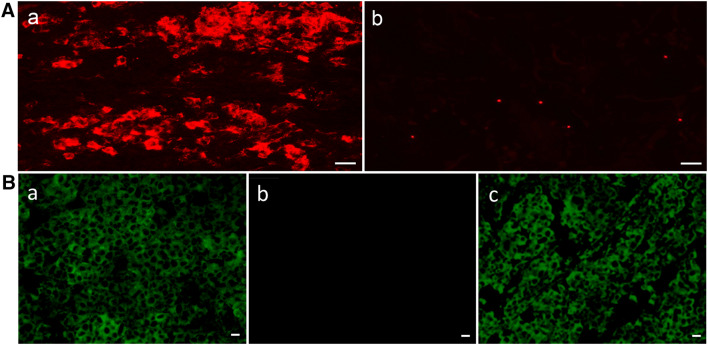
Representative images of scFv binding to cancer and normal tissues are shown in Fig. 2. These scFvs were derived from clones recovered from panning a patient with colon cancer metastatic to the liver. Clone 08-2,350 binds to the colon cancer and not the corresponding normal colon. Clone 08-2,035 binds to the colon cancer, but also binds to normal colon. Clone 07-0,093 binds to neither colon cancer nor normal colon tissue.
Fig. 2.
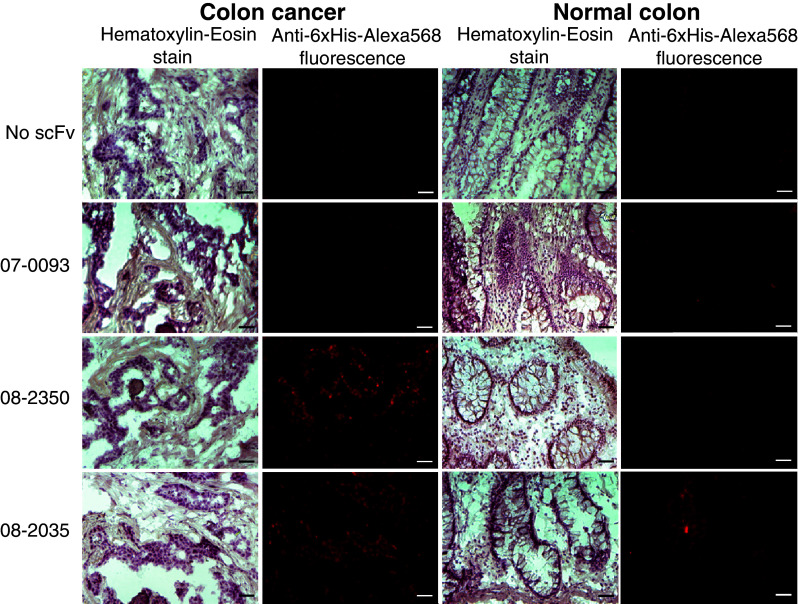
Representative images of 3 scFvs derived from phage clones recovered from panning a patient with colon cancer hepatic metastases. Columns 1 (Hematoxylin and Eosin stain) and 2 (anti-6xHis-Alexa Fluor 568 immunofluorescence) are colon cancer. Columns 3 and 4 are normal colon. Row 1 is negative control with no scFv (PBS only). Row 2 shows non-binding scFv clone 07-0,093. Row 3 shows scFv clone 08-2,350 binding to tumor only. Row 4 shows scFv clone 08-2,035 binding to colon cancer and also corresponding normal colon tissue. Scale bar length = 100 μm
The phage clones and soluble scFvs that demonstrated high tumor binding were further studied for their binding to a panel of 8 normal human tissues. A total of 2 of 49 tumor-binding phage clones evaluated for normal tissue binding did not bind to any of the tested normal tissues and 11 of 49 bound to only 1 normal human tissue (Table 3). The extent of normal tissue binding was variable, and the majority of clones bound to an intermediate number of normal tissues. None of the clones bound to more than 5 of the 8 normal tissues assessed. A similar variable pattern of binding to normal tissue was observed with the set of tumor-binding scFvs. These results show tumor specificity of several clones selected from in vivo panning.
Table 3.
The profile of normal tissue binding for selected tumor-binding phage and scFv clones grouped by patient case
| Binding agent | Patient ID | Total number of clones | Number of different normal tissues to which the set of tumor-binding clones bound | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | >5 | |||
| Phage clones | 180-13 | 15 | 0 | 0 | 4 | 6 | 4 | 1 | 0 |
| 180-14 | 12 | 0 | 3 | 2 | 6 | 1 | 0 | 0 | |
| 180-15 | 22 | 2 | 8 | 4 | 5 | 2 | 1 | 0 | |
| ScFvs | 180-13 | 15 | 1 | 2 | 4 | 4 | 2 | 2 | 0 |
| 180-14 | 4 | 0 | 2 | 2 | 0 | 0 | 0 | 0 | |
| 180-15 | 7 | 2 | 0 | 0 | 2 | 3 | 0 | 0 | |
Clones with the best binding intensity were evaluated for binding to 8 normal human tissues (breast, cervix, colon, kidney, liver, spleen, skin, and uterus). Each numbered column shows the number of tumor-binding clones for that case that bind to the indicated number of normal tissues
Binding to normal tissues by tumor-binding clones varied according to the type of normal tissue. Figure 3A shows the percentage of tumor-binding phage clones and soluble scFvs from individual patients that bound to each of the normal tissues tested. The liver had the highest frequency of binding by phage clones in all the patients. However, the scFvs from patients 180-13, 180-14, and 180-15 showed the highest binding frequencies in spleen, liver, and colon, respectively. Figure 3B presents the arithmetic mean of the data from the 3 individual patients along with standard deviations. The data demonstrate a trend almost similar to that observed in individual patients. Furthermore, for most normal tissues, phage clones bound at a higher frequency than soluble scFvs.
Fig. 3.
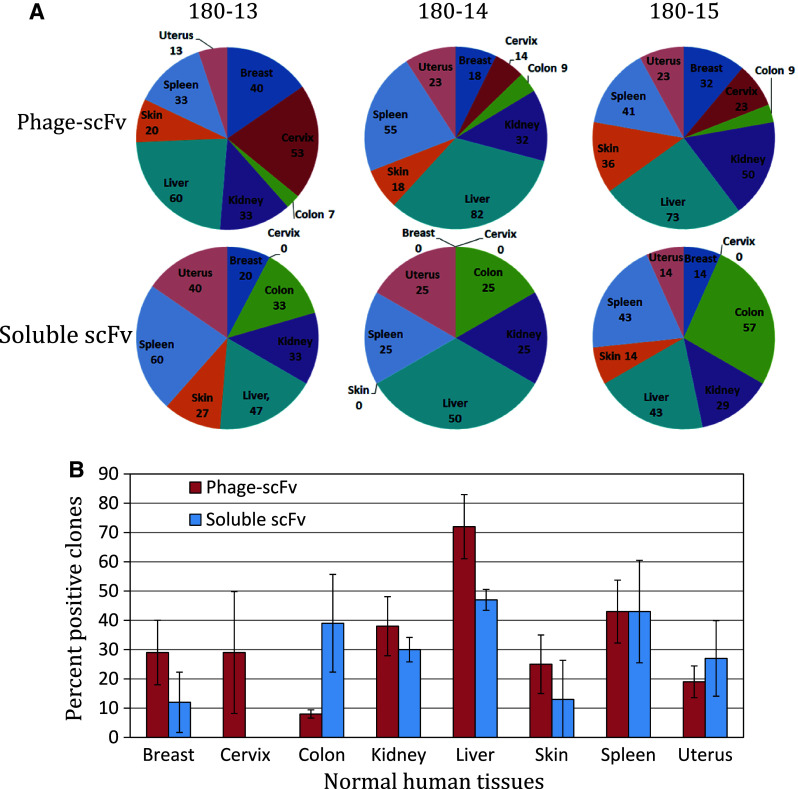
Profile of normal tissue binding of tumor-binding phage-scFv clones and tumor-binding soluble scFvs. A The pie charts of individual patients show clone binding to different normal human tissues. Each human organ in the pie charts shows percent of the analyzed phage clones (180-13 = 15; 180-14 = 12; 180-14 = 22) or soluble scFvs (180-13 = 15; 180-14 = 4; 180-14 = 7) from a patient binds to that tissue. B Bar chart shows the arithmetic mean ± SD (n = 3) of the data from the above 3 patients. The red and blue bars represent, respectively, the percentage of phage-scFv clones and soluble scFvs that bind to the indicated normal human tissue
The binding results of immunofluorescence microscopy for some of the tumor-specific clones were further confirmed by using tissue lysate ELISA. The ELISA results of melanoma-specific clones 07-2,355 and 09-8,126 and colorectal tumor-specific clone 08-2,350, which did not bind to any of the tested normal human tissues, are presented in Fig. 4A and B, respectively. We included the clone 08-2,350 from patient 180-13 in the list of 3 highly tumor-specific clones because the soluble scFv from this clone did not bind to any of the tested normal human tissues (Table 3). All three phage-scFv clones exhibited significantly higher binding to the lysates of patients’ own tumors, as compared to the corresponding control of normal human skin or colon lysate. In addition, the lysate ELISA on the 8 phage-scFvs from patient 180-15 that bound to one normal human organ tissue, and the 3 clones from patient 180-13 that bound to a maximum of two normal human organ tissues (Table 3), also demonstrated higher binding to the tumors from which they were recovered than to normal human skin or colon (Supplementary Fig. S1).
Fig. 4.
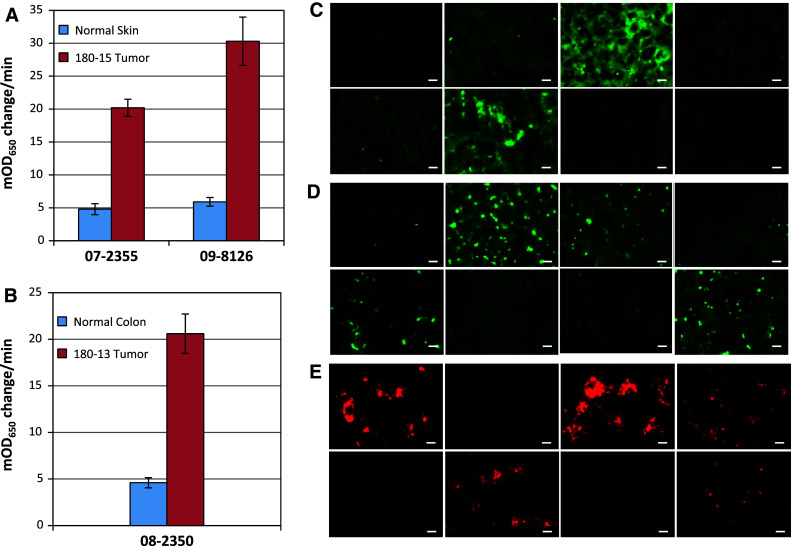
Binding of the selected tumor-specific phage-antibodies to the patient’s own tumor and to the archived tumors from other patients. Lysate ELISA was employed to confirm the binding of A melanoma-specific clones 07-2,355 and 09-8,126 to the melanoma tumor from which these clones were recovered and to normal human skin; B colorectal tumor-specific clone 08-2,350 for binding to the colorectal tumor from which this clone was recovered and to normal human colon. The bars represent the arithmetic mean ± SE (n = 3) of the experiment done in triplicate. For all the clones, the tumor lysate bindings were significantly (p < 0.001; Student t test) higher in comparison with corresponding control normal tissues. Immunofluorescence microscopy was used to examine the binding of melanoma-specific clones C 07-2,355 and D 09-8,126 to archived melanoma tumors from 8 patients. The binding was detected with anti-M13 phage-antibody followed by Alexa Fluor 488 conjugated secondary antibody. E The colorectal tumor-specific clone 08-2,350 was screened on archived colorectal tumors from 8 patients using anti-M13 phage-antibody followed by Alexa Fluor 568 conjugated secondary antibody. Scale bar length = 10 μm
The clones 07-2,355, 09-8,126, and 08-2,350 were further analyzed for binding to the archived melanoma and colorectal tumors from other patients. Immunofluorescence microscopy demonstrated the binding of melanoma-specific clone 07-2,355 (Fig. 4C) to the melanoma tumors from 2 of the 8 patients. The other melanoma-specific clone 09-8,126 (Fig. 4D) bound to the melanoma tumors from 4 of the 8 patients. However, the colorectal tumor-specific clone 08-2,350 was found to bind to the colorectal tumors from 5 of the 8 patients (Fig. 4E).
Sequence homology analysis of the variable regions of the phage-scFv clones
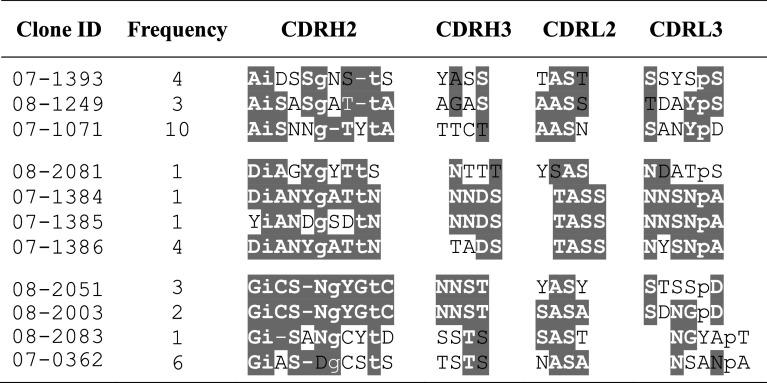
A total of 367 clones were sequenced. We identified motif sharing among amino acid sequences of tumor-homing clones and identified those motifs in human proteins. As an example, Table 4 shows amino acid sequences of the variable regions, their frequency, and possible motifs of phage clones obtained from surgically harvested colon cancer hepatic metastasis from patient 180-13. An enrichment of tumor-binding clones is indicated by the multiple occurrences of certain clones. The sequence homology analyses were done independent of normal tissue-binding characteristics of the clones.
Table 4.
Motif analysis of variable domains of scFv fusions in the phage clones recovered from colon cancer hepatic metastasis of patient 180-13
Amino acid sequences of CDRH2 and CDRH3 domains of heavy chains and CDRL2 and CDRL3 domains of light chains in the scFv fusions of selected phage clones are presented. The residues sharing consensus motifs are highlighted; white letters = exactly the same amino acids, black letters = conservative substitutions. Lower case letters in the CDRH2 sequences represent amino acids that are built-in by the library design
Search results identified several CDRH2 sequences of phage-scFv clones with significant matches to cancer-related antibodies [34–38]. Supplementary Table S2 shows examples of matches of tumor-binding scFv to several interesting antibodies. The variable region sequence of clone 07-2,355 that shared a 12-amino-acid-long motif (SAISGSGGSTYY) with a reported IL-17A antibody was studied further. The melanoma tumor from which this clone was recovered was found to show a higher staining with anti-human IL-17A antibody compared to that of normal human skin (Fig. 5A). Competitive study showed a complete blockage of clone 07-2,355 binding to the tumor by the polyclonal IL-17A antibody; however, the same concentration of an irrelevant antibody (poly-His tag or V5 tag) did not affect clone 07-2,355 binding (Fig. 5B).
Fig. 5.
Tumor IL 17A expression and competition of clone 07-2,355 binding with an anti-IL 17A antibody. A Immunofluorescence detection of higher IL-17A expression in the (a) metastatic melanoma tumor from which clone 07-2,355 was recovered; (b) no significant IL-17A staining is seen in normal human skin. IL-17A was detected with anti-human IL-17A antibody followed by Alexa Fluor 568 conjugated secondary antibody. Scale bar length = 10 μm. B Competition of clone 07-2,355 with the mouse anti-human polyclonal IL-17A antibody for binding to the patient’s metastatic melanoma tumor from which this clone was recovered. Tumor binding was studied in presence of (a) clone 07-2,355 only; (b) clone 07-2,355 + anti-IL-17A antibody; (c) clone 07–2,355 + a mouse irrelevant (anti-poly His) antibody. Clone 07-2,355 binding was localized with anti-M13 phage-antibody followed by Alexa Fluor 488 conjugated secondary antibody. Scale bar length = 10 μm
Discussion
In vivo phage display selection methods in animal models of cancers have successfully generated antibodies against tumor-associated antigens. The in vivo selection strategy involves selection against more complex targets with a greater number of possible binding sites that can result in recovery of important ligands and identification of unique disease-related targets. Although a purified molecule may have many possible binding sites and is complex from that perspective, here we use the term “complex” to refer to much larger assemblages of biological material including cells, tissues, organs, and organisms. On complex targets, potential binding sites include a wide array of different and often unknown molecules. Our goal is to use phage display to identify important cancer-related targets. Performing the selection in vivo has the advantage of the tumor being in the most native and intact state. Here, the tumor will be unchanged by artificial manipulations such as disaggregation, fixation, or serial growth in vitro. In vivo, all the cellular elements will be present such as blood vessels, matrix molecules, supporting stroma, and immune cells. In a tumor, all the non-malignant components may offer potentially useful targets for therapy.
It has been very challenging and time-consuming to achieve stepwise transfer of an in vivo selection strategy in human cancer patients. We previously reported a preclinical study [24], and based on that, we got US Federal Drug Administration approval (# FDA BB-IND 9145) for an initial toxicity study in human cancer patients. This clinical toxicity study observed no phage-related adverse events during infusion or during the 8-week follow-up period [25]. Specifically, no allergy-related reactions were observed. This indicates that in patients with no preexisting anti-phage antibodies, the toxicity profile is sufficiently safe to proceed for in vivo phage panning.
In the present investigation, the tumor phage recoveries were comparable to our earlier study [25]; however, they were much lower in comparison with the preclinical studies where animals were injected with a 50- to 500-time higher dose of a phage library on body weight basis [15–20, 23]. The tumor phage recoveries in our study were also lower than the recoveries reported in the 6 biopsied normal tissues from a brain-dead human following 1 × 1014 TU phage infusion for 15 min [39]. The lower phage recovery in our study may be resulted from using a comparatively lower phage dose and also from aggressive tumor washings in an effort to remove blood phage contamination. The results of the present study demonstrated tumor phage enrichment as determined by clone-binding studies and sequence analysis of tumor-homing phage. Clone binding was assessed using a tumor sample from the same patient that was infused with phage. In some cases, only a small amount of tumor tissue was available, which decreased the number of clones recovered and restricted subsequent clone analyses. In order to use the tumor samples efficiently, we sectioned the tumor into small pieces prior to rapid freezing. This process instantly preserved the histological architecture and molecular integrity, and maximized the number of histologic specimens that could be prepared for binding assays. Immunohistochemical staining procedures were facilitated by mounting tumor sections on round 12-mm cover slips. The cover slips were placed in 24-well plates for all subsequent incubation and rinsing steps [13]. Although this process was not high throughput, it did allow comparative binding results and sufficient throughput for up to several hundreds of clones per case. This technique of clone screening was successfully validated by confirming the binding results of the selected tumor-binding phage clones by lysate ELISA.
Between cases, there was variation in the number of clones that showed positive tumor binding. In all cases, the majority of the clones did not show immunohistological evidence of tumor binding. This indicates that these clones either derived from the blood contamination of the tumor samples or that their tumor binding was below the detection threshold of the immunohistochemical assay. Efforts are needed to minimize blood contamination of the tumors before the homogenization step. It may be achieved by introducing more stringent washes or by adopting other strategies, including density gradient separation [40]. Nevertheless, the possibility of a non-specific tumor-homing by these phage clones cannot be ruled out. Despite the relatively low percentage yield of tumor binders, several high tumor-binding clones were identified for further evaluation.
All the tumor-binding clones showed some degree of specificity; for example, 2 of 49 tumor-binding clones did not bind to any normal tissues and 11 of them showed binding to only one normal human tissue in the panel. This shows successful isolation of tumor-specific clones. There was a higher frequency of binding to liver in comparison with other normal tissues. Increased binding to organs of the reticuloendothelial system indicates that some of the binding was likely due to the phage particle and not to the displayed antibody [41, 42]. We could not find any data in the literature regarding M13 phage coat protein affinity to different human organ tissues. However, our observations with negative controls of helper phage and unselected random phage libraries have consistently shown higher binding of phage particles to human liver in comparison with other organs (unpublished data). The majority of the soluble scFvs prepared from tumor-binding clones had the same binding profile as when displayed by the phage particle. A significantly lower liver binding by soluble scFvs in comparison with phage-displayed scFvs further supports our conclusion that the phage particle itself may have some contribution in the observed higher phage-scFvs binding to normal liver. The clones that showed high tumor-specificity in cross-binding studies on normal human tissues were also found to bind corresponding tumors from other patients. These results indicate that the clones selected from this technique may possibly find their use in certain other cancer patients also, in addition to the patient in whom the in vivo selection was performed.
Sequencing is a strategy that has been used to identify phage clone enrichment during systemic panning of organisms [43]. The recovery of multiple copies of a number of clones and multiple motif-sharing events of variable regions of clones in our present study support our clone-binding data demonstrating tumor enrichment. Sequences of multiple variable regions in the present study showed significant matches to previously reported cancer-related antibodies [34–38]. These are interesting findings that suggest commonality of certain tumor antigens. It is true that sequence homology analysis can identify the right antigen or epitope sometimes; however, caution should be exercised while interpreting these findings, and such data should be regarded only as leads for further validation. One of the clones (07-2,355) that was found to share a 12-amino-acid-long motif with a reported IL-17A antibody was further studied. We found a higher expression of IL-17A in the melanoma tumor from which this clone was recovered. Elevated expressions of IL-17A have been reported in a variety of cancers [44, 45]. A complete blockage of clone 07-2,355 binding to the tumor by a polyclonal IL-17A antibody indicated IL-17A as a possible antigen target of this clone. Further work is needed to validate this finding employing other techniques, such as flow cytometry and co-immunoprecipitation, and to expand the work of target identification for the other tumor-binding clones using multiple proteomic approaches suitable for a very small amount of tumor tissues.
These results demonstrate a successful selection of several tumor-binding clones from the in vivo panning in cancer patients. There are several options available to further improve human cancer patient panning protocols by adopting the subtraction strategies used in vitro and animal studies; for example, negatively selecting normal tissue binding clones with blood cells from the same patient prior to in vivo panning [6, 46]. In addition, a library can be positively selected on the patient’s tumor in vitro, and the resulting library, enriched with tumor binders, can be infused to the same patient for in vivo selection [15, 47]. This in vitro selection step can be easily performed on the tumor biopsy sample obtained prior to the phage infusion [14]. Multiple options for alternative phage libraries are also available. For example, there are larger naïve libraries now available than those used in this study [26, 48, 49]. Customized libraries derived from patients with the same disease or even derived from the same patient are also alternative strategies for increasing the yield of tumor-binding antibodies [50].
In conclusion, intravenous administration of phage-antibody libraries to cancer patients resulted in enrichment of clones recovered from the surgically excised cancer tissue. Several tumor-binding clones were identified, and some of them demonstrated high tumor-specificity, but others showed some degree of binding to varying numbers of normal tissues. The clones that showed high tumor-specificity were found to bind corresponding tumors from other patients also. The selected tumor-binding clones from human panning also provide an opportunity to identify possible tumor targets. We conclude that these outcomes support the safety and utility of phage display library panning in human cancer patients for ligand selection and target discovery for cancer treatment and diagnosis.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
The authors thank Ms Susan Fuller and Ms Patricia Lutton for technical assistance, Ms Shelley Bissonnette for secretarial help, and Ms Sarah Howe for editing the manuscript. The automated DNA sequencing was performed in the VT Cancer Center DNA Analysis Facility and was supported by Vermont Cancer Center, Lake Champlain Cancer Research Organization, and the UVM College of Medicine. This study was financially supported by National Cancer Institute, USA (NCI award # PHS R21CA097679) and the SD Ireland Cancer Research Foundation.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Dancey JE, Chen HX. Strategies for optimizing combinations of molecularly targeted anticancer agents. Nat Rev Drug Discov. 2006;5:649–659. doi: 10.1038/nrd2089. [DOI] [PubMed] [Google Scholar]
- 2.Hurwitz H, Fehrenbacher L, Novotny W, Cartwright T, Hainsworth J, Heim W, Berlin J, Baron A, Griffing S, Holmgren E, Ferrara N, Fyfe G, Rogers B, Ross R, Kabbinavar F. Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. N Engl J Med. 2004;350:2335–2342. doi: 10.1056/NEJMoa032691. [DOI] [PubMed] [Google Scholar]
- 3.Mass RD, Press MF, Anderson S, Cobleigh MA, Vogel CL, Dybdal N, Leiberman G, Slamon DJ. Evaluation of clinical outcomes according to HER2 detection by fluorescence in situ hybridization in women with metastatic breast cancer treated with trastuzumab. Clin Breast Cancer. 2005;6:240–246. doi: 10.3816/CBC.2005.n.026. [DOI] [PubMed] [Google Scholar]
- 4.Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, Fleming T, Eiermann W, Wolter J, Pegram M, Baselga J, Norton L. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 5.Geyer CR, McCafferty J, Dubel S, Bradbury AR, Sidhu SS. Recombinant antibodies and in vitro selection technologies. Methods Mol Biol. 2012;901:11–32. doi: 10.1007/978-1-61779-931-0_2. [DOI] [PubMed] [Google Scholar]
- 6.Siva A, Kirkland R, Lin B, Maruyama T, McWhirter J, Yantiri-Wernimont F, Bowdish K, Xin H. Selection of anti-cancer antibodies from combinatorial libraries by whole-cell panning and stringent subtraction with human blood cells. J Immunol Methods. 2008;330:109–119. doi: 10.1016/j.jim.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 7.Ridgway JB, Ng E, Kern JA, Lee J, Brush J, Goddard A, Carter P. Identification of a human anti-CD55 single-chain Fv by subtractive panning of a phage library using tumor and nontumor cell lines. Cancer Res. 1999;59:2718–2723. [PubMed] [Google Scholar]
- 8.Noronha EJ, Wang X, Desai SA, Kageshita T, Ferrone S. Limited diversity of human scFv fragments isolated by panning a synthetic phage-display scFv library with cultured human melanoma cells. J Immunol. 1998;161:2968–2976. [PubMed] [Google Scholar]
- 9.Desai SA, Wang X, Noronha EJ, Kageshita T, Ferrone S. Characterization of human anti-high molecular weight-melanoma-associated antigen single-chain Fv fragments isolated from a phage display antibody library. Cancer Res. 1998;58:2417–2425. [PubMed] [Google Scholar]
- 10.Poul MA, Becerril B, Nielsen UB, Morisson P, Marks JD. Selection of tumor-specific internalizing human antibodies from phage libraries. J Mol Biol. 2000;301:1149–1161. doi: 10.1006/jmbi.2000.4026. [DOI] [PubMed] [Google Scholar]
- 11.Shukla GS, Krag DN. Cancer cell-specific internalizing ligands from phage displayed beta-lactamase-peptide fusion libraries. Protein Eng Des Sel. 2010;23:431–440. doi: 10.1093/protein/gzq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shukla G, Krag D. Selection of tumor-targeting agents on freshly excised human breast tumors using a phage display library. Oncol Rep. 2005;13:757–764. [PubMed] [Google Scholar]
- 13.Sun Y, Shukla GS, Weaver D, Pero SC, Krag DN. Phage-display selection on tumor histological specimens with laser capture microdissection. J Immunol Methods. 2009;347:46–53. doi: 10.1016/j.jim.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shukla G, Krag D. Phage display selection for cell-specific ligands: development of a screening procedure suitable for small tumor specimens. J Drug Target. 2005;13:7–18. doi: 10.1080/10611860400020464. [DOI] [PubMed] [Google Scholar]
- 15.Veleva AN, Nepal DB, Frederick CB, Schwab J, Lockyer P, Yuan H, Lalush DS, Patterson C. Efficient in vivo selection of a novel tumor-associated peptide from a phage display library. Molecules. 2011;16:900–914. doi: 10.3390/molecules16010900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newton-Northup JR, Figueroa SD, Deutscher SL. Streamlined in vivo selection and screening of human prostate carcinoma avid phage particles for development of peptide based in vivo tumor imaging agents. Comb Chem High Throughput Screen. 2011;14:9–21. doi: 10.2174/1386207311107010009. [DOI] [PubMed] [Google Scholar]
- 17.Li ZJ, Wu WK, Ng SS, Yu L, Li HT, Wong CC, Wu YC, Zhang L, Ren SX, Sun XG, Chan KM, Cho CH. A novel peptide specifically targeting the vasculature of orthotopic colorectal cancer for imaging detection and drug delivery. J Control Release. 2010;148:292–302. doi: 10.1016/j.jconrel.2010.09.015. [DOI] [PubMed] [Google Scholar]
- 18.Roodink I, Franssen M, Zuidscherwoude M, Verrijp K, van der Donk T, Raats J, Leenders WP. Isolation of targeting nanobodies against co-opted tumor vasculature. Lab Invest. 2010;90:61–67. doi: 10.1038/labinvest.2009.107. [DOI] [PubMed] [Google Scholar]
- 19.Zhao X, Hu J, Huang R, Yang L. Identification of one vasculature specific phage-displayed peptide in human colon cancer. J Exp Clin Cancer Res. 2007;26:509–514. [PubMed] [Google Scholar]
- 20.Newton JR, Kelly KA, Mahmood U, Weissleder R, Deutscher SL. In vivo selection of phage for the optical imaging of PC-3 human prostate carcinoma in mice. Neoplasia. 2006;8:772–780. doi: 10.1593/neo.06331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joyce JA, Laakkonen P, Bernasconi M, Bergers G, Ruoslahti E, Hanahan D. Stage-specific vascular markers revealed by phage display in a mouse model of pancreatic islet tumorigenesis. Cancer Cell. 2003;4:393–403. doi: 10.1016/S1535-6108(03)00271-X. [DOI] [PubMed] [Google Scholar]
- 22.Laakkonen P, Porkka K, Hoffman JA, Ruoslahti E. A tumor-homing peptide with a targeting specificity related to lymphatic vessels. Nat Med. 2002;8:751–755. doi: 10.1038/nm720. [DOI] [PubMed] [Google Scholar]
- 23.Schluesener HJ, Xianglin T. Selection of recombinant phages binding to pathological endothelial and tumor cells of rat glioblastoma by in vivo display. J Neurol Sci. 2004;224:77–82. doi: 10.1016/j.jns.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 24.Krag DN, Fuller S, Oligino L, Pero S, Weaver D, Soden A, Hebert C, Mills S, Liu C, Peterson D. Phage-displayed random peptide libraries in mice: toxicity after serial panning. Cancer Chemother Pharmacol. 2002;50:325–332. doi: 10.1007/s00280-002-0489-4. [DOI] [PubMed] [Google Scholar]
- 25.Krag DN, Shukla GS, Shen G, Pero S, Ashikaga T, Fuller S, Weaver D, Burdette-Radoux S, Thomas C. Selection of tumor-binding ligands in cancer patients with phage display libraries. Cancer Res. 2006;66:7724–7733. doi: 10.1158/0008-5472.CAN-05-4441. [DOI] [PubMed] [Google Scholar]
- 26.de Wildt R, Mundy C, Gorick B, Tomlinson I. Antibody arrays for high-throughput screening of antibody-antigen interactions. Nat Biotechnol. 2000;18:989–994. doi: 10.1038/79494. [DOI] [PubMed] [Google Scholar]
- 27.Griffiths A, Williams S, Hartley O, Tomlinson I, Waterhouse P, Crosby W, Kontermann R, Jones P, Low N, Allison T. Isolation of high affinity human antibodies directly from large synthetic repertoires. EMBO J. 1994;13:3245–3260. doi: 10.1002/j.1460-2075.1994.tb06626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kristensen P, Winter G. Proteolytic selection for protein folding using filamentous bacteriophages. Fold Des. 1998;3:321–328. doi: 10.1016/S1359-0278(98)00044-3. [DOI] [PubMed] [Google Scholar]
- 29.Shukla GS, Krag DN. A sensitive and rapid chemiluminescence ELISA for filamentous bacteriophages. J Immunoassay Immunochem. 2005;26:89–95. doi: 10.1081/IAS-200051990. [DOI] [PubMed] [Google Scholar]
- 30.Clackson TLB. Introduction to phage biology and phage display. London: Oxford University Press; 2004. [Google Scholar]
- 31.Golchin M, Aitken R. Isolation by phage display of recombinant antibodies able to block adherence of Escherichia coli mediated by the K99 colonisation factor. Vet Immunol Immunopathol. 2008;121:321–331. doi: 10.1016/j.vetimm.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 32.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altschul SF, Gish W. Local alignment statistics. Methods Enzymol. 1996;266:460–480. doi: 10.1016/S0076-6879(96)66029-7. [DOI] [PubMed] [Google Scholar]
- 34.Ford JW, McVicar DW. TREM and TREM-like receptors in inflammation and disease. Curr Opin Immunol. 2009;21:38–46. doi: 10.1016/j.coi.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Diez S, Navarro G, de Tros Ilarduya C. In vivo targeted gene delivery by cationic nanoparticles for treatment of hepatocellular carcinoma. J Gene Med. 2009;11:38–45. doi: 10.1002/jgm.1273. [DOI] [PubMed] [Google Scholar]
- 36.Birkle S, Zeng G, Gao L, Yu RK, Aubry J. Role of tumor-associated gangliosides in cancer progression. Biochimie. 2003;85:455–463. doi: 10.1016/S0300-9084(03)00006-3. [DOI] [PubMed] [Google Scholar]
- 37.Gerhardt S, Abbott WM, Hargreaves D, Pauptit RA, Davies RA, Needham MR, Langham C, Barker W, Aziz A, Snow MJ, Dawson S, Welsh F, Wilkinson T, Vaugan T, Beste G, Bishop S, Popovic B, Rees G, Sleeman M, Tuske SJ, Coales SJ, Hamuro Y, Russell C. Structure of IL-17A in complex with a potent, fully human neutralizing antibody. J Mol Biol. 2009;394:905–921. doi: 10.1016/j.jmb.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 38.von Budingen HC, Menge T, Hauser SL, Genain CP. Restrictive and diversifying elements of the anti-myelin/oligodendrocyte glycoprotein antibody response in primate experimental allergic encephalomyelitis. Immunogenetics. 2006;58:122–128. doi: 10.1007/s00251-006-0100-y. [DOI] [PubMed] [Google Scholar]
- 39.Arap W, Kolonin MG, Trepel M, Lahdenranta J, Cardo-Vila M, Giordano RJ, Mintz PJ, Ardelt PU, Yao VJ, Vidal CI, Chen L, Flamm A, Valtanen H, Weavind LM, Hicks ME, Pollock RE, Botz GH, Bucana CD, Koivunen E, Cahill D, Troncoso P, Baggerly KA, Pentz RD, Do KA, Logothetis CJ, Pasqualini R. Steps toward mapping the human vasculature by phage display. Nat Med. 2002;8:121–127. doi: 10.1038/nm0202-121. [DOI] [PubMed] [Google Scholar]
- 40.Williams BR, Sharon J. Polyclonal anti-colorectal cancer Fab phage display library selected in one round using density gradient centrifugation to separate antigen-bound and free phage. Immunol Lett. 2002;81:141–148. doi: 10.1016/S0165-2478(02)00004-4. [DOI] [PubMed] [Google Scholar]
- 41.Pasqualini R, Koivunen E, Ruoslahti E. Alpha v integrins as receptors for tumor targeting by circulating ligands. Nat Biotechnol. 1997;15:542–546. doi: 10.1038/nbt0697-542. [DOI] [PubMed] [Google Scholar]
- 42.Yip YL, Hawkins NJ, Smith G, Ward RL. Biodistribution of filamentous phage-Fab in nude mice. J Immunol Methods. 1999;225:171–178. doi: 10.1016/S0022-1759(99)00044-7. [DOI] [PubMed] [Google Scholar]
- 43.Staquicini FI, Cardo-Vila M, Kolonin MG, Trepel M, Edwards JK, Nunes DN, Sergeeva A, Efstathiou E, Sun J, Almeida NF, Tu SM, Botz GH, Wallace MJ, O’Connell DJ, Krajewski S, Gershenwald JE, Molldrem JJ, Flamm AL, Koivunen E, Pentz RD, Dias-Neto E, Setubal JC, Cahill DJ, Troncoso P, Do KA, Logothetis CJ, Sidman RL, Pasqualini R, Arap W vascular ligand-receptor mapping by direct combinatorial selection in cancer patients. Proc Natl Acad Sci USA 108:18637–18642 [DOI] [PMC free article] [PubMed]
- 44.Wang L, Yi T, Kortylewski M, Pardoll DM, Zeng D, Yu H. IL-17 can promote tumor growth through an IL-6-Stat3 signaling pathway. J Exp Med. 2009;206:1457–1464. doi: 10.1084/jem.20090207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quan Y, Zhou B, Wang Y, Duan R, Wang K, Gao Q, Shi S, Song Y, Zhang L, Xi M. Association between IL17 Polymorphisms and Risk of Cervical Cancer in Chinese Women. Clin Dev Immunol. 2012;2012:258293. doi: 10.1155/2012/258293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huie MA, Cheung MC, Muench MO, Becerril B, Kan YW, Marks JD. Antibodies to human fetal erythroid cells from a nonimmune phage antibody library. Proc Natl Acad Sci USA. 2001;98:2682–2687. doi: 10.1073/pnas.051631798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zahid M, Phillips BE, Albers SM, Giannoukakis N, Watkins SC, Robbins PD. Identification of a cardiac specific protein transduction domain by in vivo biopanning using a M13 phage peptide display library in mice. PLoS ONE. 2010;5:e12252. doi: 10.1371/journal.pone.0012252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.De Haard H, van Neer N, Reurs A, Hufton S, Roovers R, Henderikx P, De Bruïne A, Arends J, Hoogenboom H. A large non-immunized human Fab fragment phage library that permits rapid isolation and kinetic analysis of high affinity antibodies. J Biol Chem. 1999;274:18218–18230. doi: 10.1074/jbc.274.26.18218. [DOI] [PubMed] [Google Scholar]
- 49.Goletz S, Christensen P, Kristensen P, Blohm D, Tomlinson I, Winter G, Karsten U. Selection of large diversities of antiidiotypic antibody fragments by phage display. J Mol Biol. 2002;315:1087–1097. doi: 10.1006/jmbi.2001.5314. [DOI] [PubMed] [Google Scholar]
- 50.Thie H, Toleikis L, Li J, von Wasielewski R, Bastert G, Schirrmann T, Esteves IT, Behrens CK, Fournes B, Fournier N, de Romeuf C, Hust M, Dubel S. Rise and fall of an anti-MUC1 specific antibody. PLoS ONE. 2011;6:e15921. doi: 10.1371/journal.pone.0015921. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.