Abstract
Comparative genomics continues illuminating amniote genome evolution, but for many lineages our understanding remains incomplete. Here, we refine the assembly (CPI 3.0.3 NCBI AHGY00000000.2) and develop a cytogenetic map of the painted turtle (Chrysemys picta—CPI) genome, the first in turtles and in vertebrates with temperature-dependent sex determination. A comparison of turtle genomes with those of chicken, selected nonavian reptiles, and human revealed shared and novel genomic features, such as numerous chromosomal rearrangements. The largest conserved syntenic blocks between birds and turtles exist in four macrochromosomes, whereas rearrangements were evident in these and other chromosomes, disproving that turtles and birds retain fully conserved macrochromosomes for greater than 300 Myr. C-banding revealed large heterochromatic blocks in the centromeric region of only few chromosomes. The nucleolar-organizing region (NOR) mapped to a single CPI microchromosome, whereas in some turtles and lizards the NOR maps to nonhomologous sex-chromosomes, thus revealing independent translocations of the NOR in various reptilian lineages. There was no evidence for recent chromosomal fusions as interstitial telomeric-DNA was absent. Some repeat elements (CR1-like, Gypsy) were enriched in the centromeres of five chromosomes, whereas others were widespread in the CPI genome. Bacterial artificial chromosome (BAC) clones were hybridized to 18 of the 25 CPI chromosomes and anchored to a G-banded ideogram. Several CPI sex-determining genes mapped to five chromosomes, and homology was detected between yet other CPI autosomes and the globally nonhomologous sex chromosomes of chicken, other turtles, and squamates, underscoring the independent evolution of vertebrate sex-determining mechanisms.
Keywords: physical molecular cytogenetic BAC clone mapping, chromosomal rearrangements, genome and chromosome evolution, translocations and inversions, nonmodel vertebrates, turtles, chicken, human, temperature-dependent and genotypic sex determination
Introduction
Novel and recalcitrant questions in biology are elucidated at an increasing pace thanks to the development of new genomic resources in nonmodel organisms (Janes et al. 2008), such as the recent release of several chelonian (turtle) genomes: The western painted turtle Chrysemys picta (CPI) (Shaffer et al. 2013), the Chinese softshell turtle Pelodiscus sinensis, and the sea turtle Chelonia mydas (Wang et al. 2013). These turtle genomes complement recent sequencing efforts on other major reptilian groups including lizards (Alfoldi et al. 2011), crocodilians (St John et al. 2012; Green et al. 2014), and snakes (Castoe et al. 2013; Vonk et al. 2013). Phylogenetic analyses enabled by these chelonian genomes strongly support the hypothesis that turtles are sister to Archosaurs (birds and crocodilians) (Chiari et al. 2012; Crawford et al. 2012; Deakin et al. 2013; Shaffer et al. 2013), a result of paramount importance as the accurate placement of turtles in the tree of life is essential for the reconstruction of the evolutionary history of vertebrate traits and genomes.
A major aspect of genome organization influencing genome function and evolution is its compartmentalization into chromosomes, because changes in the synteny of genes and gene blocks alter their regulatory environment (Ahituv et al. 2005), affecting transcription (De et al. 2009) and adaptation (Kirkpatrick and Barton 2006; Hoffmann and Rieseberg 2008; Loxdale 2010). Genome organization varies among taxa and coevolves with other traits: An example is the coevolution of chromosome number and sex determination in turtles (Valenzuela and Adams 2011), or the location and types of repeat elements and evolutionary breakpoints of chromosomes prone to rearrangements (Flint et al. 1994; Azzalin et al. 2001; Ruiz-Herrera et al. 2005). Additionally, karyological evolution is linked to lineage diversification in a variety of organisms, including reptiles (Olmo et al. 2002; Ayala and Coluzzi 2005; Olmo 2005; Hoffmann and Rieseberg 2008). Thus, evolutionary and functional genomics benefit not only from sequence data but also from cytogenetic information that places DNA sequences in their physical and phylogenetic context to enable evolutionary inferences across species.
In particular, comparative cytogenetic and sequence analyses have illuminated many aspects of vertebrate genome evolution (Deakin and Ezaz 2014) although much remains to be learned. For instance, the sequencing and physical mapping of the chicken genome revealed the homology between bird and human chromosomes (Nanda et al. 2000; Schmid et al. 2000), and the high conservation of the avian genome previously attributed to the scarcity of repeat elements (Backstrom et al. 2008) was later confirmed by additional genome analyses (Dalloul et al. 2010; but see Griffin et al. 2007). Sequencing of outgroup genomes is also important for phylogenomics. For example, the opossum and platypus genomes revealed shared and unique genomic components in monotremes, birds, and therian mammals (Mikkelsen et al. 2007; Warren et al. 2008), whereas genome evolution in teleosts and gnathostomes is anchored by the coelacanth and lamprey genomes (Kasahara et al. 2007; Amemiya et al. 2013; Smith et al. 2013). Comparative approaches have also permitted the reconstruction of ancestral karyotypes in lineages such as primates, marsupials, amniotes, tetrapods, and vertebrates (De Leo et al. 1999; Richard et al. 2003; Kemkemer et al. 2006, 2009; Kohn et al. 2006; Nakatani et al. 2007; Stanyon et al. 2008; Uno et al. 2012; Deakin et al. 2013; Romanov et al. 2014), among others. Although sequence comparisons between the recently sequenced turtle genomes and those of other vertebrates revealed a less prominent GC-rich isochore structure in turtles than in mammals and birds (Shaffer et al. 2013), we know less about the chromosomal rearrangements that have accrued during chelonian evolution.
Turtles are a reptile group reported to have highly conserved karyotypes when compared with lizards and snakes in terms of the number, morphology, and G-banding pattern of their chromosomes (Bickham 1981; Olmo 2008). Within turtles, this conservation is greater in the suborder Criptodira—to which all newly sequenced turtles belong—relative to the suborder Pleurodira. Previous studies have consistently identified highly conserved homology between some turtle chromosomes and those of other vertebrates, most notably between the six largest turtle and chicken chromosomes (Matsuda et al. 2005), including CHICKEN-Z and P. sinensis turtle chromosome 6 (PELODISCUS-6) (Matsuda et al. 2005; Kawai et al. 2007); PELODISCUS-Z/W and CHICKEN-15 (Kawagoshi et al. 2009), and PELODISCUS-6 and Elaphe quadrivirgata snake chromosome 2 (ELAPHE-2) (Matsuda et al. 2005). Turtles resemble birds and lizards in the presence of microchromosomes, some of which may also represent ancient syntenies conserved since the rise of vertebrates 400 Ma (Burt 2002), but which are notably absent in mammals and crocodilians. Thus, more extensive analyses encompassing a larger portion of the turtle karyotypes are still needed to gain a comprehensive understanding of genome evolution in turtles and vertebrates.
Here, we present an improved genome assembly and the first physical BAC mapping of the painted turtle (CPI) genome, the first of any vertebrate with temperature-dependent sex determination (TSD), and a comparison with other vertebrates where information is available (mainly chicken and human). Importantly, we found evidence dispelling the full conservation of several purported syntenies while supporting the conservation of some significant vertebrate gene blocks and the occurrence of numerous chromosomal rearrangements over more than 300 Myr of vertebrate evolution. Specifically, we complement previous basic cytogenetic data for CPI (Killebrew 1977; De Smet 1978) with detailed G- and C-banded karyotypic and molecular cytogenetic information otherwise lacking for this emerging model species (Valenzuela 2009), including 1) the distribution of repeat elements, 18S rDNA and telomeres; 2) the first banded ideogram for this species (diagrammatic representation of the haploid chromosome set); and 3) the mapping of 61 sequenced BACs (some containing genes involved in sexual development). Additionally, using a new set of bioinformatic algorithms we obtained an improved genome assembly with fewer and larger scaffolds (see Supplementary Information) than the original released CPI genome (Shaffer et al. 2013). Our molecular cytogenetic data permit the refinement of the painted turtle genome assembly by anchoring scaffolds to chromosomes and therefore provide the most detailed picture yet of the structure of turtle chromosomes.
Materials and Methods
Cell Culture, Chromosome Preparation, and Chromosome Banding
Primary fibroblast cell cultures for cytogenetic analyses were established using limb tissue from one male and one female CPI 6-month-old hatchling as well as the adult female whose genome was sequenced and reported in Shaffer et al. (2013). The sex of all individuals was assessed by gonadal inspection. Briefly, fibroblast cell cultures were established from collagenase (Sigma) digests and cultured using a medium which was composed of 50% RPMI 1640 and 50% Leibowitz media supplemented with 15% fetal bovine serum, 2 mM l-glutamine, and 1% antibiotic-antimycotic solution (Sigma). Cultures were incubated at 30 °C with no CO2 supplementation (Badenhorst et al. 2013). Four hours prior to harvesting 10 μg/ml colcemid (Roche) was added to the cultures. Metaphase chromosomes were harvested after colcemid arrest (KaryoMAX; Invitrogen), hypotonic exposure, and fixed in 3:1 methanol:acetic acid following standard procedures (Ezaz et al. 2006; Martinez et al. 2008; Badenhorst et al. 2013). G- and C-banding followed conventional protocols (Seabright 1971; Sumner 1972). The distribution of NORs (the genomic region containing the genes for the 18S, 5.8S, and 28S ribosomal subunits) was investigated by silver staining (Ag-NOR) (Goodpasture and Bloom 1975), and by fluorescent in-situ hybrydization (FISH) using a turtle-specific 18S DNA fragment labeled by nick-translation and coprecipitated with salmon sperm DNA (Badenhorst et al. 2013). A telomeric probe containing the repeat motif (TTAGGG)n was generated and labeled by polymerase chain reaction, starting with (TTAGGG)4 and (CCCTAA)4 primers in the absence of template DNA (Ijdo et al. 1991).
BAC Clone Sequencing and Hybridization
Sets of randomly chosen clones from a CPI BAC library (library VMRC CHY3 produced by the Joint Genome Institute) were sequenced (through Illumina) as part of the turtle genome sequencing project (Shaffer et al. 2013), and others (12 BACs) were screened for the putative presence of genes in the turtle and vertebrate sex determination network (Valenzuela 2010; Valenzuela et al. 2013) and sequenced independently in full (454 platform) or in part (Sanger sequencing) to confirm the presence of genes of interest. These sequenced BACs (81 in total) were used to link cytogenetic and DNA sequence data, to refine the genome assembly, and to establish the syntenic relationships (relative genome position) of functional genes. BAC DNA (∼1 µg) was extracted and labeled by standard nick-translation (Abbott Molecular) using either biotin or digoxingenin dUTPs (Roche) and coprecipitated with human cot-1 DNA and turtle cot-1 DNA. FISH was carried out using BAC, telomere, and 18S probes, by dehydrating the slides through an ethanol series followed by denaturing the chromosome preparations together with the probe-mix on a hot plate at 65 °C for 2 min, and hybridization took place overnight (two nights for 18S rDNA and telomere probes) in a humid chamber at 37 °C. Posthybridization washes were comprised of a first wash in 0.4 × Saline-Sodium Citrate (SSC)/0.3% Tween 20 for 2 min at 60°C, followed by a second wash in 2 × SSC/0.1% Tween 20 for 1 min at room temperature. Fluorochrome detection was performed with 4XT/relevant antibody in a 200-μl final volume at 37°C for 45 min. Slides were subsequently washed thrice in 4XT at 37 °C, counterstained with DAPI (6 μl DAPI 2 mg/ml in 50 ml 2 × SSC), and mounted using an antifade solution (Vectashield). Signals were assigned to specific chromosomes according to their morphology, size, and DAPI-banding. FISH was repeated after G-banding to improve anchoring of BAC sequences to chromosomal regions, and two to four BAC FISH was used to determine relative position within chromosomes. Images were taken with a Photometrics CoolSnap ES2 Digital Monochrome camera attached to an Olympus BX41 fluorescent microscope, and analyzed using CytoVision cytogenetic analysis system (Applied Imaging/Genetix).
BAC Bioinformatics for Cytogenetic Analysis
All 61 sequenced BACs were mapped to CPI’s reference genome assembly 3.0.3 using Geneious v.6.1.6 (Kearse et al. 2012), as follows. First, Megablast was used to search for the complete BAC sequence within the annotated CPI genome. The annotations in the CPI genome were used to identify putative protein-coding genes that co-occur on the same genomic scaffold as the mapped BAC (61 in total). Genomic scaffolding was improved in this study (assembly 3.0.3) with respect to the original genome assembly 3.0.1 (Shaffer et al. 2013) (see Supplementary information). The genomic location of those genes (1,425 genes in total) in the human and chicken genomes was determined from a direct search in the NCBI (National Center for Biotechnology Information) Database (Assemblies: homSapGRCh38, galGal4), and information in other vertebrates was obtained from the published literature.
Results and Discussion
Cytogenetic Data and Ideogram
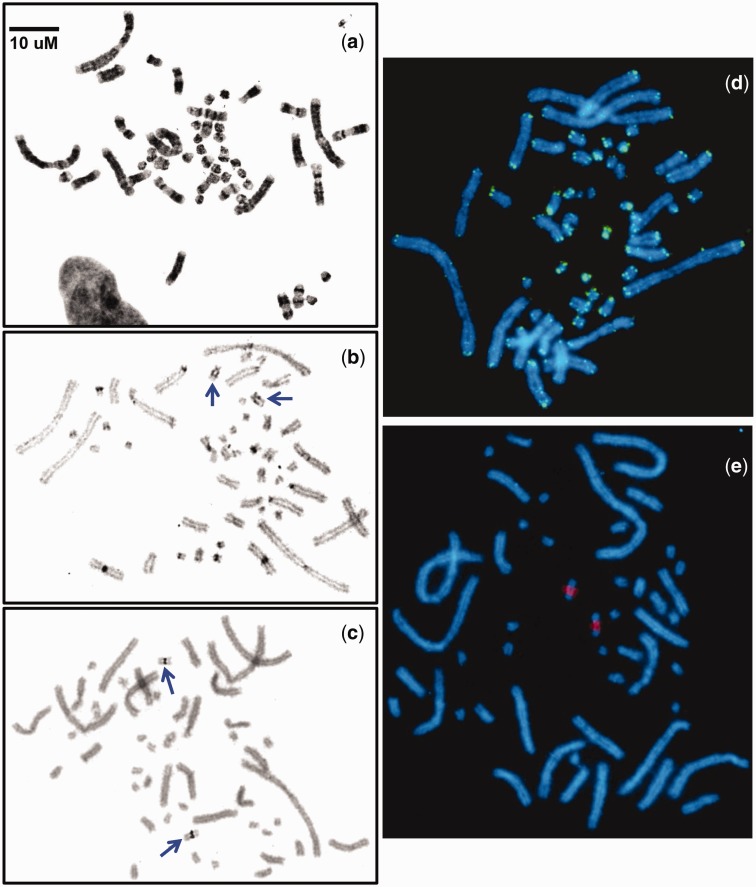
A combination of classic and molecular cytogenetics allowed a deeper characterization of the CPI karyotype than was previously reported for this species (Killebrew 1977; De Smet 1978), and included G- and C-banding, Ag-NOR (nucleolar-organizing region), 18S rRNA, and telomere DNA mapping (figs. 1 and 2). At least ten cells per individual were analyzed for all cytogenetic procedures. The G-banded ideogram of the haploid genome of CPI presented here depicts 189 defined G-bands (dark AT-rich heterochromatin and light CG-rich euchromatic bands; fig. 2). The smaller microchromosomes of CPI were nearly indistinguishable from each other by shape and were ordered by approximate size and G-banding pattern where possible. Ours is the first banded ideogram developed for any turtle and for any vertebrate with TSD and lacking sex chromosomes (Valenzuela et al. 2014).
Fig. 1.
— G-banded (a), C-banded (b), and Ag-NOR stained (c) metaphase chromosomes of CPI, and the distribution of telomeric DNA (d) and 18S rDNA repeats (e) on CPI metaphase spreads. Arrows indicate C-positive interstitial bands (b) and NOR localization (c).
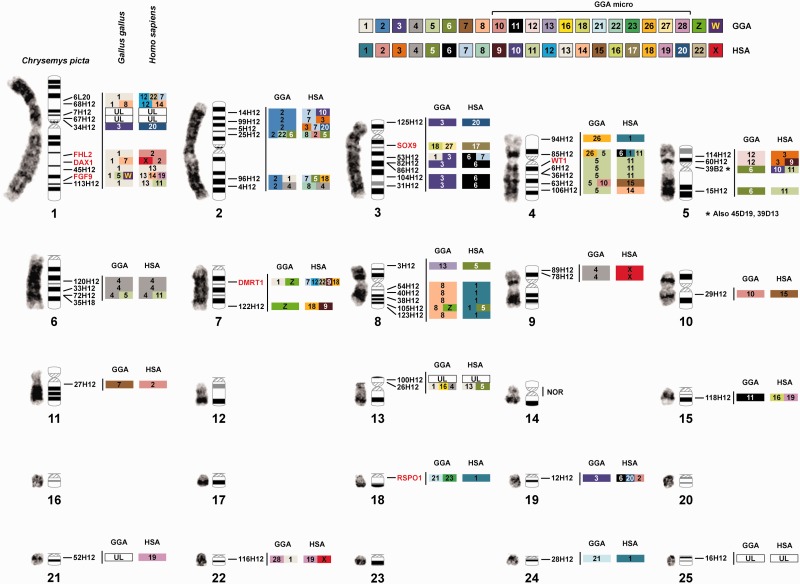
Fig. 2.
— CPI GTG-banded karyotype and ideogram showing the physical location of FISH-anchored BAC clones, with sex-linked BACs in red, along with the homology of gene blocks to chicken (GGA) and human (HSA) chromosomes, depicted as colored blocks, and with the NOR location on CPI 14. “UL” indicates uncharacterized loci, that is, CPI BACs that did not map to annotated scaffolds and thus homology to GGA and HSA was precluded. Note that BACs 45D19, 35H18, 39D13 colocate with 39B2 in CPI-5, and BAC 6L20 shows some hybridization to this region as well, likely due to the presence of shared repeat sequences (see supplementary table S2, Supplementary Material online).
All three individuals examined had 2N = 50 chromosomes with 26 macrochromosomes and 24 microchromosomes (4 metacentric, 18 submetacentric, 28 acrocentric), a result that differs slightly from the 24 macrochromosomes and 20 metacentric/submetacentric chromosomes described by De Smet (1978), but in agreement with Killebrew (1977). The subtle difference from the report by De Smet (1978) could be due to individual or population differences, but is more likely due to the difficulty of unambiguously identifying the morphology of turtle microchromosomes, because they may appear telocentric, making the position of the centromere difficult to discern (De Smet 1978).
The C-positive material representing constitutive heterochromatin is largely restricted to the centromeric regions with clearly observable blocks only visible in some chromosome pairs. Variation in the abundance of heterochromatin in the centromeric regions among chromosomes is observed by C-banding in a variety of taxa including humans, and reflects differences in the types and number of repeat elements and satellite DNA (Sumner 2003). There is, however, interstitial heterochromatin visible in chromosome pair 14 (fig. 1b) that corresponds to the NOR (containing genes encoding the three major ribosomal RNA subunits—18S, 5.8S, and 28S [Shaw and McKeown 2011]). Indeed, a single interstitial NOR was detected on chromosome pair 14 by both silver staining, which detects active NORs, and 18S rDNA FISH, which detects active and inactive NORs (fig. 1c and e), corresponding to this C-positive band. These results agree with a previous report using silver staining only (Bickham and Rogers 1985). Interstitial NORs are not ubiquitous in turtles; only 12 of 28 investigated turtles display interstitial NOR sites, but they seem widespread in the subfamily Emydinae, to which CPI belongs (Bickham and Rogers 1985). C-positive blocks are also found in reptilian sex chromosomes (Kawai et al. 2007; Singh 2011; Badenhorst et al. 2013; Matsubara et al. 2014; Rojo et al. 2014) and may colocalize with the NOR in the W chromosome of some turtles (Kawai et al. 2007; Badenhorst et al. 2013) and lizards (Matsubara et al. 2014; Rojo et al. 2014). However, our BAC mapping data rule out the hypothesis that CPI-14 is homologous to any of these reptilian sex chromosomes, and instead suggest that the NOR has undergone translocations to multiple chromosomal locations independently in various reptilian lineages.
Telomeric repeats localized exclusively to the end of the chromosome arms (fig. 1d), as expected given that telomeres play a key role in maintaining chromosome stability [(Bolzán and Bianchi 2006)]. Interstitial telomeres are indicative of past chromosomal rearrangements and evolutionary unstable genome regions (Ruiz-Herrera et al. 2008). The absence of interstitial telomeric sequences in CPI corresponds with previous reports in other turtles which also lack them (e.g., Trachemys dorbigni and Chelonoidis donosobarrosi [Martinez et al. 2009]), as well as tuatara (O’Meally et al. 2009). However, these data contrast with lizards where they are present, for example, in microchromosomes in Pogona vitticeps (Young et al 2013), macrochromosomes in Iberolacerta monticola (Rojo et al. 2014) or both in Leiolepis lizards (Srikulnath et al. 2011). However, additional data on the chromosomal location of telomeric sequences in a larger subset of turtle taxa are needed to test whether such contrasting patterns among these major reptilian lineages are generalizable.
BAC-Mapping and Bioinformatic Analysis
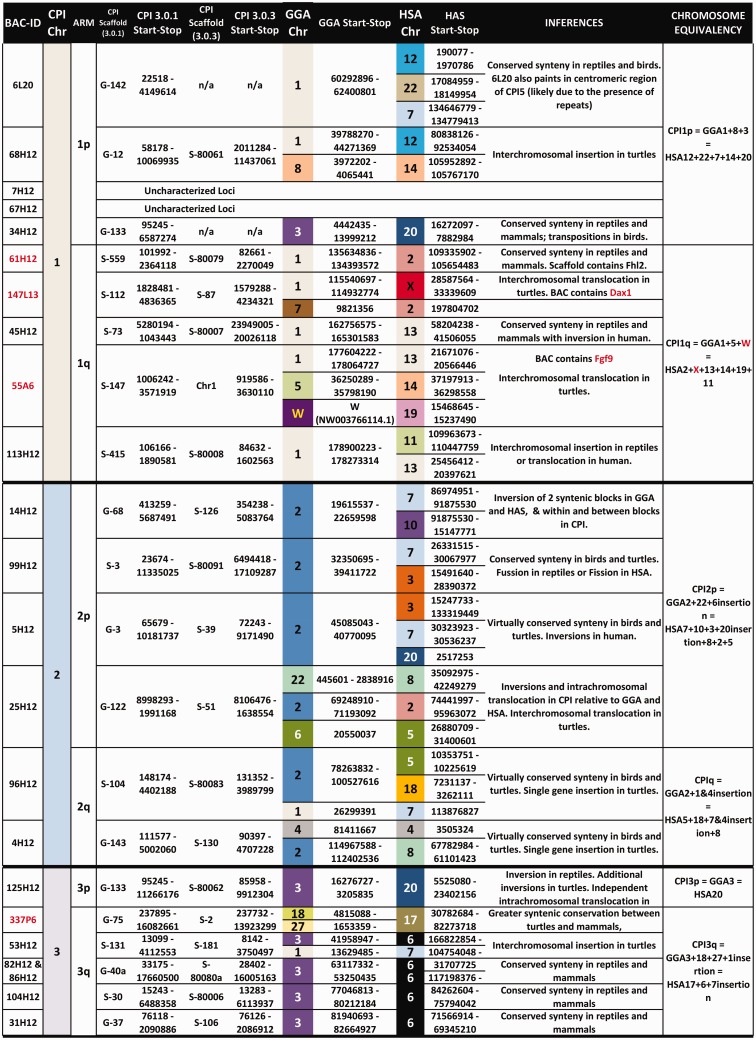
A total of 61 of the 81 fully sequenced BAC clones were successfully assigned to a unique location in the CPI ideogram (figs. 2 and 3). BAC clones were assigned to 12 of the 13 macrochromosomes and 7 of the 12 microchromosomes. Multicolor FISH was used to anchor BACs to microchromosomes and to macrochromosomes of similar size and shape. Thus, 19 of the 25 chromosome pairs have BAC markers assigned to them, and chromosome pair 14 is distinguishable by its C-positive block and by the localization of the NOR detectable by silver staining and/or 18S-FISH (fig. 1). This cytogenetic BAC mapping information was combined with bioinformatics analyses to refine the painted turtle genome assembly (see supplementary information, Supplementary Material online) and resulted in improved ultrascaffolds and chromosomal information. For instance, chromosomal AGPs were created and centromeres were positioned using the BAC maps, which localized 461 Mb of genomic DNA to 18 chromosomes. AGPs are “A Golden Path” description files of the components of each chromosome. This is the first chromosomal AGP produced for a turtle and the second for nonavian reptiles (Alfoldi et al. 2011). The improved genome sequence of CPI 3.0.3 was deposited in the DDBJ/EMBL/GenBank database (accession number AHGY00000000.2). There were a few mapped BACs containing DNA sequences with no annotation and are referred to as “uncharacterized loci” in the figures and tables.
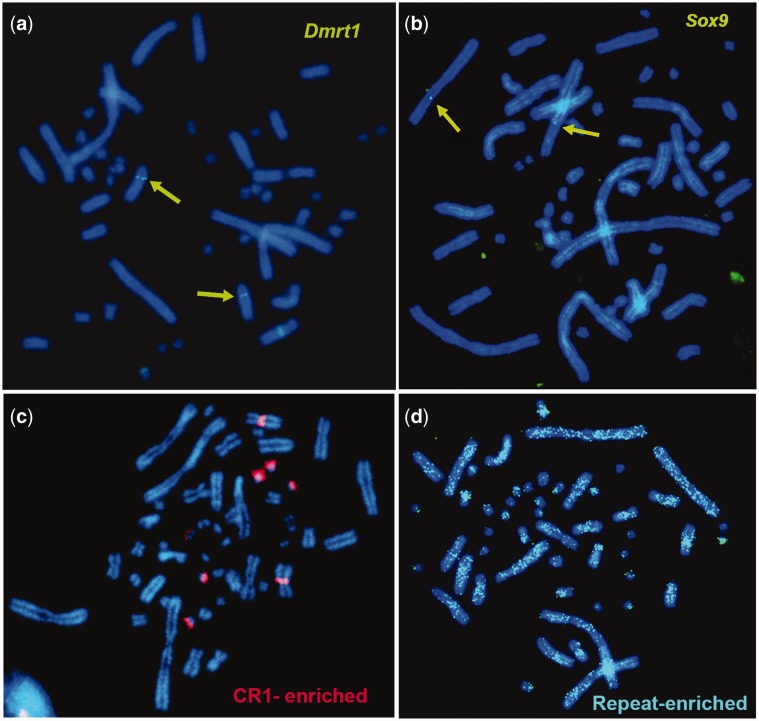
Fig. 3.
— Examples of BAC FISH mapping showing the hybridization pattern of BACs containing genes related to sexual development (a, b), CR1 and Gypsy-enriched BACs (c), and simpler repeat-enriched BACs (d).
An additional subset of 18 BAC clones produced too much background during FISH to permit accurate localization (fig. 3). Interestingly, these problematic BACs contained a large proportion of various repeat elements indicating that repeats are abundant and widespread throughout the turtle genome (supplementary table S2, Supplementary Material online; fig. 3d), consistent with the sequenced genome analysis (Shaffer et al. 2013). In contrast, the hybridization signal from four BACs enriched for CR1-like and Gypsy repeat sequences exhibited a clustered pattern in the centromeric region of five chromosome pairs, including a macrochromosome (CPI-5) and four microchromosomes (fig. 3c). These repeat elements are also shared by four additional BACs that map uniquely to the same region in CPI-5. These results indicate that unlike other simpler repeat types, these transposable elements are not randomly distributed in the CPI genome but instead predominate on five chromosome pairs. This is the first indication that turtle centromeric and pericentromeric regions are not uniform in their composition, similar to what is observed in chicken (Shang et al. 2010), humans, and other metazoans (Maddox et al. 2012; Fukagawa and Earnshaw 2014). These results are also important because these repeats may affect the evolution and regulation of these genomic regions disproportionately (Kudla et al. 2006), and transposable elements have played a significant role in the evolution of other vertebrate genomes, such as in mammals (Mikkelsen et al. 2007).
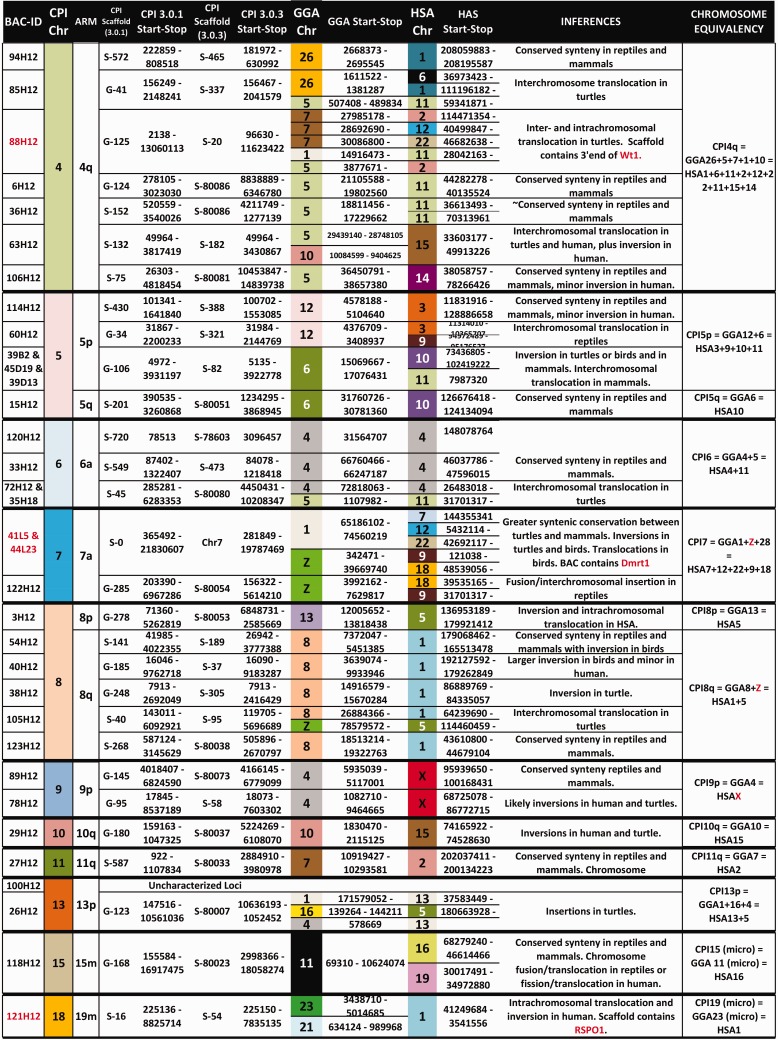
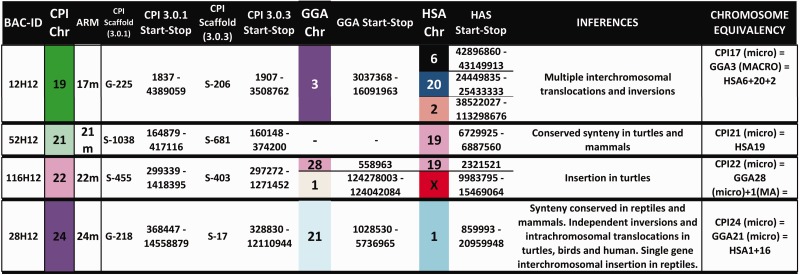
Bioinformatic analyses of the BAC sequences and the CPI genome scaffolds to which they map permitted the first assessment of homology between the painted turtle chromosomes and those of chicken (Gallus gallus [CHICKEN]) and human (Homo sapiens [HUMAN]), as well as a few other reptiles where partial genome or cytogenetic information was available (fig. 4, table 1), specifically the turtle P. sinensis (PELODISCUS), the snake E. quadrivirgata (ELAPHE), and several lizards (Varanus salvator [VARANUS], Leiolepsis reevesii, (LEIOLEPSIS) Po. vitticeps [POGONA]) (Matsuda et al. 2005; Matsubara et al. 2006; Srikulnath et al. 2013; Young et al. 2013). In general, our data challenge the previously reported conservation of macrochromosomes between birds and turtles (Matsuda et al. 2005; Kasai et al. 2012). Specifically, by using a much larger gene data set from our BAC clones and the scaffolds to which they map in the CPI genome (1,425 genes) we identified numerous putative chromosomal rearrangements that passed undetected when using fewer markers in other turtles (e.g., 57 genes in Matsuda et al. [2005]). Additionally, our data set allowed inferences of homologies for over 70% of the turtle chromosome pairs (18 of 25) for the first time, including intermediate sized and microchromosomes. Specifically, we found that CPI and chicken macrochromosomes 1, 2, and 3 represent the highest conserved synteny. However, this is not a fully conserved synteny because they contain regions orthologous to at least four and six chicken chromosomes; this number is a conservative estimate as our BAC coverage is not complete on the CPI genome. Furthermore, CPI-4 contains larger gene block regions that are orthologous to at least five chicken chromosomes compared with the smaller blocks that interrupt the synteny of CPI-1, CPI-2 and CPI-3 and CHICKEN-1, CHICKEN-2 and CHICKEN-3, respectively. The gene blocks identified in CPI-1 to CPI-4 are orthologous to numerous human chromosomes including HUMAN-X (figs. 2 and 4, supplementary table S1, Supplementary Material online). Homology to three chicken chromosomes was also detected in CPI-8 and CPI-13, and homology to two chicken chromosomes was identified in CPI-5, CPI-6, CPI-7, and CPI-18. All these CPI chromosomes exhibited homology to between two and five human macrochromosomes. The contrast of gene blocks among species permitted the detection of interchromosomal translocations and inversions in turtle alone, some only in chicken, and others in both turtle and chicken and thus possibly shared across turtles and archosaurs, although tests in crocodilians are needed to confirm this hypothesis (fig. 4).
Fig. 4.
— Chromosomal homology, synteny, and rearrangements identified between CPI turtles and the chicken and human genomes. Multiple gene blocks may be encompassed by the start and stop positions listed within each chromosome and are detailed in the supplementary table S1, Supplementary Material online, along with their gene content.
Table 1.
Partial Homology of Painted Turtle Chromosomes to Those of Other Reptiles
| Gene | CPI | VSA | LRE | EQU | PSI | PVI | SCR | STR | Reference |
|---|---|---|---|---|---|---|---|---|---|
| EPB41L3 | 2 | 3p | 1 | ||||||
| TOP2B | 2 | 4q | 2 | ||||||
| TAX1BP1 | 2 | Zp | 6p | 1, 3 | |||||
| RAB5A | 2 | Zp, Wq | 6p | 1, 3 | |||||
| CTNNB1 | 2 | 4q | 6q | Zp, Zcen, Wcen | 6p | 1, 2, 3 | |||
| KAT2B | 2 | 6p | 3 | ||||||
| ARG1 | 3 | 3q | 4 | ||||||
| WT1 | 4 | 2q | 1q | 1q | X, Y | 2, 5 | |||
| COQ6 | 4 | 5q | 4 | ||||||
| EIF2B2 | 4 | 5q | 4 | ||||||
| CTBP2 | 5 | 3q | 3 | ||||||
| DMRT1 | 7 | X, Y | 6 | ||||||
| DCTN4 | 8 | 2q | 1 | ||||||
| RUFY1 | 8 | 1q | 2q | 2q | 2 | ||||
| TPR | 8 | 3p | 1 | ||||||
| RPE65 | 8 | 8p | 2 | ||||||
| SPARC | 8 | Micro 13 | 4 | ||||||
| BRD7 | 15 | Micro | Micro | Micro | 2 | ||||
| ENO1 | 24 | Micro | Micro | 2 |
Note.—VSA, Varanus salvator macromaculatus; LRE, Leiolepsis reevesii rubritaeniata; EQU, Elaphe quadrivirgata; PSI, Pelodiscus sinensis; PVI, Pogona vitticeps; SCR, Siebenrockiella crassicollis; STR, Staurotypus triporcatus; cen, centromeric; Micro, microchromosome. References: 1, Matsubara et al. (2006); 2, Srikulnath et al. (2013); 3, Young et al. (2013); 4, Matsuda et al. (2005); 5, Kawagoshi et al. (2012); 6, Kawagoshi et al. (2014). Shaded cells denote a split region in turtle with respect to snakes.
Our data also revealed syntenic blocks between painted turtle autosomes and amniote sex chromosomes, and the correspondence is not always one to one. For instance, macrochromosomes CPI-7 and CPI-8 harbor gene blocks that are syntenic in CHICKEN-Z, whereas macrochromosomes CPI-1, CPI-9, and microchromosome CPI-22 contain gene blocks orthologous to HUMAN-X (fig. 4). In contrast, CPI-2 contains genes that mapped to snake ELAPHE-Z and ELAPHE-W (table 1). Although available data are scarce for other reptiles, other regions of homology and rearrangements were also detected, involving autosomes and sex chromosomes. Namely, macrochromosome CPI-2 contains a gene block homologous to VARANUS-4, POGONA-6, and ELAPHE-Z (table 1). CPI-2 also shows partial homology to ELAPHE-3, whereas ELAPHE-3 contains another gene block located in CPI-8, and CPI-8 harbors a different gene block that maps to ELAPHE-2, revealing several chromosomal rearrangements between snakes and turtles (table 1). A CPI-4 region appears homologous to PELODISCUS-5 (both macrochromosomes), whereas a gene in CPI-8 (macrochromosome) maps to microchromosome PELODISCUS-13. Some microchromosomes appear to be syntenic across reptiles, as genes in CPI-15 and CPI-24 map to VARANUS, LEIOLEPIS, and ELAPHE microchromosomes as well (table 1).
Of the 61 BACs that mapped successfully to a single location, seven contained genes or mapped to scaffolds containing genes in the sex determination network of turtles and vertebrates (FgF9, Dax1, Sox9, Dmrt1, Fhl2, Wt1, and Rspo1) (Valenzuela 2008a; Badenhorst et al. 2013). The relative position of these genes revealed additional chromosomal rearrangements among amniotes and regions of homology between CPI autosomes and sex chromosomes in other taxa. Namely, BACs containing Fhl2, FgF9 and Dax1 mapped to CPI-1q and to CHICKEN-1, whereas this gene block is split in human as they map to chromosome HUMAN-2 (Chan et al. 1998), HUMAN-13 (Mattei et al. 1995), and HUMAN-X (Zanaria et al. 1994), respectively. Additionally, Sox9 mapped to CPI-3q whereas it is located on the CHICKEN-18 microchromosome (Kuroiwa et al. 2002) and on HUMAN-17 (Foster et al. 1994). G-banding (fig. 2) and whole-chromosome-specific painting using Trachemys scripta (TRACHEMYS) probes indicate that CPI-3 is orthologous to TRACHEMYS-3 (Badenhorst D, Montiel Jiménez EE, Stanyon R, Ferguson-Smith MA, O'Brien PCM, Valenzuela N, unpublished data), which in turn appears homologous to CHICKEN-3 (Kasai et al. 2012) (i.e., CPI-3 = TRACHEMYS-3 = CHICKEN-3). Therefore, our results suggest the transposition of Sox9 chromosomal location between macrochromosomes in turtles (Sox9 = CPI-3 = TRACHEMYS-3) and a chicken microchromosome (Sox9 = CHICKEN-18). On the other hand, Dmrt1 mapped to CPI-7 (our study, fig. 3a), and it is located in Gekko hokouensis lizards GEKKO-Z (Kawai et al. 2009), and in PELODISCUS-6 which is homologous to CHICKEN-Z (Kawai et al. 2007). CHICKEN-Z in turn is homologous to Staurotypus triporcatus turtles STAUROTYPUS-X/Y (Kawagoshi et al. 2014). As our chromosome-specific painting shows CPI-7 to be homologous to TRACHEMYS-6 (Badenhorst D, Montiel Jiménez EE, Stanyon R, Ferguson-Smith MA, O'Brien PCM, Valenzuela N, unpublished data), then CPI-7 appears to be homologous to PELODISCUS-6 as well. The apparent conserved autosomal synteny of Dmrt1 (a strong candidate for avian sex-determining gene [Smith et al. 2009]) across turtles is of interest, because CPI exhibits TSD and lacks sex chromosomes (Valenzuela et al. 2014), whereas P. sinensis displays a ZZ/ZW sex-determining system (Kawai et al. 2007), and PELODISCUS-Z is homologous to CHICKEN-15 (Kawagoshi et al. 2009). Furthermore, Wt1, a candidate gene for a role as a TSD master gene in CPI based on transcriptional profiling (Valenzuela 2008b; Valenzuela et al. 2013), maps to CPI-4 and this region shows homology to CHICKEN-5 and HUMAN-11, and Siebenrockiella crassicollis turtles SIEBENROCKIELLA-X/Y (Kawagoshi et al. 2012). These observations combined with the homology of CPI-7 to CHICKEN-Z and GEKKO-Z, and of CPI-2 to ELAPHE-Z and POGONA-6 (table 1) of Po. vitticeps, a lizard with ZZ/ZW micro sex chromosomes (Ezaz et al. 2005), all support the notion that the mechanisms of sex-determination have evolved independently between birds and turtles (Kawai et al. 2007), as well as among turtles, snakes, and lizards (Ezaz et al. 2009). Otherwise, all reptilian sex chromosomes would have shown homology to a single CPI chromosome.
Conclusion
In summary, our study extends the currently available cytogenetic and DNA sequence (Shaffer et al. 2013) data for painted turtles, an emerging model for ecology and evolution (Valenzuela 2009). Importantly, the improved assembly and physical mapping presented here advance our understanding of the evolution of amniote genomes. For instance, our data reveal that macrochromosome synteny is not fully retained between birds and turtles for the six largest chromosomes as previously reported between turtles and archosaurs (birds and crocodilians) (Matsuda et al. 2005; Kasai et al. 2012). Indeed, rearrangements were identified involving both these and other macro and microchromosomes. Our results also support the notion that sex-determining mechanisms have evolved independently multiple times in birds, turtles, and squamates. Indeed, regions in seven different CPI chromosomes show homology to sex chromosomes of other turtles, birds, squamates, and human, supporting the idea that not one but multiple chromosomes were recruited as sex chromosomes in different vertebrate lineages. We hope that this study, the first of its kind in turtles and TSD vertebrates, fosters further research into the fascinating evolution of vertebrate genomes.
Supplementary Material
Supplementary information and tables S1–S3 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgment
This work was funded in part by grants NSF MCB 0815354 to N.V. and S.V.E, and MCB 1244355 to N.V. from the National Science Foundation of the United States.
Literature Cited
- Ahituv N, et al. 2005. Mapping cis-regulatory domains in the human genome using multi-species conservation of synteny. Hum Mol Genet. 14:3057-3063. [DOI] [PubMed] [Google Scholar]
- Alfoldi J, et al. 2011. The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature 477:587-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amemiya CT, et al. 2013. The African coelacanth genome provides insights into tetrapod evolution. Nature 496:311-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala FJ, Coluzzi M. 2005. Chromosome speciation: humans, Drosophila, and mosquitoes. Proc Natl Acad Sci U S A. 102:6535-6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzalin CM, et al. 2001. Human intrachromosomal telomeric-like repeats: sequence organization and mechanisms of origin. Chromosoma 110:75-82. [DOI] [PubMed] [Google Scholar]
- Backstrom N, et al. 2008. A gene-based genetic linkage map of the collared flycatcher (Ficedula albicollis) reveals extensive synteny and gene-order conservation during 100 million years of avian evolution. Genetics 179:1479-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badenhorst D, et al. 2013. A ZZ/ZW microchromosome system in the spiny softshell turtle, Apalone spinifera, reveals an intriguing sex chromosome conservation in Trionychidae. Chromosome Res. 21:137-147. [DOI] [PubMed] [Google Scholar]
- Bickham JW. 1981. 200,000,000-year-old chromosomes—deceleration of the rate of karyotypic evolution in turtles. Science 212:1291-1293. [DOI] [PubMed] [Google Scholar]
- Bickham JW, Rogers DS. 1985. Structure and variation of the nucleolus organizer region in turtles. Genetica 67:171-184. [Google Scholar]
- Bolzán AD, Bianchi MS. 2006. Telomeres, interstitial telomeric repeat sequences, and chromosomal aberrations. Mutat Res - Rev Mut Res. 612:189-214. [DOI] [PubMed] [Google Scholar]
- Burt DW. 2002. Origin and evolution of avian microchromosomes. Cytogenet Genome Res. 96:97-112. [DOI] [PubMed] [Google Scholar]
- Castoe TA, et al. 2013. The Burmese python genome reveals the molecular basis for extreme adaptation in snakes. Proc Natl Acad Sci U S A. 110:20645-20650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KK, et al. 1998. Molecular cloning and characterization of FHL2, a novel LIM domain protein preferentially expressed in human heart. Gene 210:345-350. [DOI] [PubMed] [Google Scholar]
- Chiari Y, et al. 2012. Phylogenomic analyses support the position of turtles as the sister group of birds and crocodiles (Archosauria). BMC Biol. 10:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford NG, et al. 2012. More than 1000 ultraconserved elements provide evidence that turtles are the sister group of archosaurs. Biol Lett. 8:783-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalloul RA, et al. 2010. Multi-platform next-generation sequencing of the domestic turkey (Meleagris gallopavo): genome assembly and analysis. PLoS Biol. 8:e1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Leo AA, et al. 1999. Comparative chromosome painting between marsupial orders: relationships with a 2n=14 ancestral marsupial karyotype. Chromosome Res. 7:509-517. [DOI] [PubMed] [Google Scholar]
- De S, et al. 2009. The impact of genomic neighborhood on the evolution of human and chimpanzee transcriptome. Genome Res. 19:785-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet WHO. 1978. Chromosomes of 22 species of Chelonia (Reptilia). Acta Zool Pathol Antverp. 70:15-34. [Google Scholar]
- Deakin JE, et al. 2013. Reconstruction of the ancestral marsupial karyotype from comparative gene maps. BMC Evol Biol. 13:1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deakin JE, Ezaz T. 2014. Tracing the evolution of amniote chromosomes. Chromosoma 123:201-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezaz T, et al. 2005. The dragon lizard Pogona vitticeps has ZZ/ZW micro-sex chromosomes. Chromosome Res. 13:763-776. [DOI] [PubMed] [Google Scholar]
- Ezaz T, et al. 2006. An XX/XY sex microchromosome system in a freshwater turtle, Chelodina longicollis (Testudines : Chelidae) with genetic sex determination. Chromosome Res. 14:139-150. [DOI] [PubMed] [Google Scholar]
- Ezaz T, et al. 2009. The ZW sex microchromosomes of an Australian dragon lizard share no homology with those of other reptiles or birds. Chromosome Res. 17:965-973. [DOI] [PubMed] [Google Scholar]
- Flint J, et al. 1994. Healing of broken human chromosomes by the addition of telomeric repeats. Am J Hum Genet. 55:505-512. [PMC free article] [PubMed] [Google Scholar]
- Foster JW, et al. 1994. Campomelic dysplasia and autosomal sex reversal caused by mutations in an Sry-related gene. Nature 372:525-530. [DOI] [PubMed] [Google Scholar]
- Fukagawa T, Earnshaw WC. 2014. The centromere: chromatin foundation for the kinetochore machinery. Dev Cell. 30:497-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodpasture C, Bloom SE. 1975. Visualization of nucleolar organizer regions im mammalian chromosomes using silver staining. Chromosoma 53:37-50. [DOI] [PubMed] [Google Scholar]
- Green RE, et al. 2014. Three crocodilian genomes reveal ancestral patterns of evolution among archosaurs. Science 346 :1254449-1–1254449-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin DK, et al. 2007. The evolution of the avian genome as revealed by comparative molecular cytogenetics. Cytogenet Genome Res. 117:64-77. [DOI] [PubMed] [Google Scholar]
- Hoffmann AA, Rieseberg LH. 2008. Revisiting the impact of inversions in evolution: from population genetic markers to drivers of adaptive shifts and speciation? Annu Rev Ecol Evol Syst. 39:21-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ijdo JW, et al. 1991. Improved telomere detection using a telomere repeat probe (TTAGGG)n generated by PCR. Nucleic Acids Res. 19:4780-4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janes DE, et al. 2008. New resources inform study of genome size, content and organization in non-avian reptiles. Integr Comp Biol. 48:447-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara M, et al. 2007. The medaka draft genome and insights into vertebrate genome evolution. Nature 447:714-719. [DOI] [PubMed] [Google Scholar]
- Kasai F, et al. 2012. Extensive homology of chicken macrochromosomes in the karyotypes of Trachemys scripta elegans and Crocodylus niloticus revealed by chromosome painting despite long divergence times. Cytogenet Genome Res. 136:303-307. [DOI] [PubMed] [Google Scholar]
- Kawagoshi T, et al. 2009. The ZW micro-sex chromosomes of the Chinese soft-shelled turtle (Pelodiscus sinensis, Trionychidae, Testudines) have the same origin as chicken chromosome 15. Cytogenet Genome Res. 125:125-131. [DOI] [PubMed] [Google Scholar]
- Kawagoshi T, et al. 2012. The origin and differentiation process of X and Y chromosomes of the black marsh turtle (Siebenrockiella crassicollis, Geoemydidae, Testudines). Chromosome Res. 20:95-110. [DOI] [PubMed] [Google Scholar]
- Kawagoshi T, et al. 2014. The Staurotypus turtles and Aves share the same origin of sex chromosomes but evolved different types of heterogametic sex determination. PLoS One 9:e105315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai A, et al. 2007. Different origins of bird and reptile sex chromosomes inferred from comparative mapping of chicken Z-linked genes. Cytogenet Genome Res. 117:92-102. [DOI] [PubMed] [Google Scholar]
- Kawai A, et al. 2009. The ZW sex chromosomes of Gekko hokouensis (Gekkonidae, Squamata) represent highly conserved homology with those of avian species. Chromosoma 118:43-51. [DOI] [PubMed] [Google Scholar]
- Kearse M, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemkemer C, et al. 2006. Reconstruction of the ancestral ferungulate karyotype by electronic chromosome painting (E-painting). Chromosome Res. 14:899-907. [DOI] [PubMed] [Google Scholar]
- Kemkemer C, et al. 2009. Gene synteny comparisons between different vertebrates provide new insights into breakage and fusion events during mammalian karyotype evolution. BMC Evol Biol. 9:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killebrew FC. 1977. Mitotic chromosomes of turtles. IV. Emydidae. Texas J Sci. 29:245-253. [Google Scholar]
- Kirkpatrick M, Barton N. 2006. Chromosome inversions, local adaptation and speciation. Genetics 173:419-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn M, et al. 2006. Reconstruction of a 450-My-old ancestral vertebrate protokaryotype. Trends Genet. 22:203-210. [DOI] [PubMed] [Google Scholar]
- Kudla G, et al. 2006. High guanine and cytosine content increases mRNA levels in mammalian cells. PLoS Biol. 4:933-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroiwa A, et al. 2002. Chromosome assignment of eight SOX family genes in chicken. Cytogenet Genome Res. 98:189-193. [DOI] [PubMed] [Google Scholar]
- Loxdale HD. 2010. Rapid genetic changes in natural insect populations. Ecol Entomol. 35:155-164. [Google Scholar]
- Maddox PS, et al. 2012. Structure, assembly and reading of centromeric chromatin. Curr Opin Genet Dev. 22:139-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez P, et al. 2008. An XX/XY heteromorphic sex chromosome system in the Australian chelid turtle Emydura macquarii, a new piece in the puzzle of sex chromosome evolution in turtles. Chromosome Res. 16:815-825. [DOI] [PubMed] [Google Scholar]
- Martinez PA, et al. 2009. Karyotypic characterization of Trachemys dorbigni (Testudines: Emydidae) and Chelonoidis (Geochelone) donosobarrosi (Testudines: Testudinidae), two species of Cryptodiran turtles from Argentina. Genetica 137:277-283. [DOI] [PubMed] [Google Scholar]
- Matsubara K, et al. 2006. Evidence for different origin of sex chromosomes in snakes, birds, and mammals and step-wise differentiation of snake sex chromosomes. Proc Natl Acad Sci U S A. 103:18190-18195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara K, et al. 2014. Highly differentiated ZW sex microchromosomes in the Australian Varanus species evolved through rapid amplification of repetitive sequences. PLoS One. 9:e95226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda Y, et al. 2005. Highly conserved linkage homology between birds and turtles: bird and turtle chromosomes are precise counterparts of each other. Chromosome Res. 13:601-615. [DOI] [PubMed] [Google Scholar]
- Mattei MG, et al. 1995. The human fgf9 gene maps to chromosomal region 13q11-q12. Genomics 29:811-812. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, et al. 2007. Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature 447:167-177. [DOI] [PubMed] [Google Scholar]
- Nakatani Y, et al. 2007. Reconstruction of the vertebrate ancestral genome reveals dynamic genome reorganization in early vertebrates. Genome Res. 17:1254-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda I, et al. 2000. Conserved synteny between the chicken Z sex chromosome and human chromosome 9 includes the male regulatory gene DMRT1: a comparative (re)view on avian sex determination. Cytogenet Cell Genet. 89:67-78. [DOI] [PubMed] [Google Scholar]
- Olmo E. 2005. Rate of chromosome changes and speciation in reptiles. Genetica 125:185-203. [DOI] [PubMed] [Google Scholar]
- Olmo E. 2008. Trends in the evolution of reptilian chromosomes. Integr Comp Biol. 48:486-493. [DOI] [PubMed] [Google Scholar]
- Olmo E, et al. 2002. Different genomic evolutionary rates in the various reptile lineages. Gene 295:317-321. [DOI] [PubMed] [Google Scholar]
- O’Meally D, et al. 2009. The first cytogenetic map of the tuatara, Sphenodon punctatus. Cytogenet Genome Res. 127:213-223. [DOI] [PubMed] [Google Scholar]
- Richard F, et al. 2003. Reconstruction of the ancestral karyotype of eutherian mammals. Chromosome Res. 11:605-618. [DOI] [PubMed] [Google Scholar]
- Rojo V, et al. 2014. Karyological characterization of the endemic Iberian rock lizard, Iberolacerta monticola (Squamata, Lacertidae): insights into sex chromosome evolution. Cytogenet Genome Res. 142:28-39. [DOI] [PubMed] [Google Scholar]
- Romanov MN, et al. 2014. Reconstruction of gross avian genome structure, organization and evolution suggests that the chicken lineage most closely resembles the dinosaur avian ancestor. BMC Genomics 15. 1060:1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Herrera A, et al. 2005. Evolutionary breakpoints are co-localized with fragile sites and intrachromosomal telomeric sequences in primates. Cytogenet Genome Res. 108:234-247. [DOI] [PubMed] [Google Scholar]
- Ruiz-Herrera A, et al. 2008. Telomeric repeats far from the ends: mechanisms of origin and role in evolution. Cytogenet Genome Res. 122:219-228. [DOI] [PubMed] [Google Scholar]
- Schmid M, et al. 2000. First report on chicken genes and chromosomes. Cytogenet Cell Genet. 90:171-218. [DOI] [PubMed] [Google Scholar]
- Seabright M. 1971. A rapid banding technique for human chromosomes. Lancet 2:971-972. [DOI] [PubMed] [Google Scholar]
- Shaffer HB, et al. 2013. The western painted turtle genome, a model for the evolution of extreme physiological adaptations in a slowly evolving lineage. Genom Biol. 14:R28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang WH, et al. 2010. Chickens possess centromeres with both extended tandem repeats and short non-tandem-repetitive sequences. Genome Res. 20:1219-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw PJ, McKeown PC. 2011. The structure of rDNA chromatin. In: Olson MOJ, editor. The nucleolus. New York: Springer; p. 43-55. [Google Scholar]
- Singh L. 2011. The charms of sex chromosomes in snakes. J Biosci (Bangalore). 36:17-21. [DOI] [PubMed] [Google Scholar]
- Smith CA, et al. 2009. The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature (London) 461:267-271. [DOI] [PubMed] [Google Scholar]
- Smith JJ, et al. 2013. Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nat Genet. 45:415-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikulnath K, et al. 2011. Chromosomal localization of the 18S-28S and 5S rRNA genes and (TTAGGG)n sequences of butterfly lizards (Leiolepis belliana belliana and Leiolepis boehmei, Agamidae, Squamata). Genet Mol Biol. 34:583-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikulnath K, et al. 2013. Karyotype evolution in monitor lizards: cross-species chromosome mapping of cDNA reveals highly conserved synteny and gene order in the Toxicofera clade. Chromosome Res. 21:805-819. [DOI] [PubMed] [Google Scholar]
- St John JA, et al. 2012. Sequencing three crocodilian genomes to illuminate the evolution of archosaurs and amniotes. Genom Biol. 13:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanyon R, et al. 2008. Primate chromosome evolution: ancestral karyotypes, marker order and neocentromeres. Chromosome Res. 16:17-39. [DOI] [PubMed] [Google Scholar]
- Sumner AT. 1972. Simple technique for demonstrating centromeric heterochromatin. Exp Cell Res. 75:304. [DOI] [PubMed] [Google Scholar]
- Sumner AT. 2003. Chromosomes: organization and function. Oxford (United Kingdom): Blackwell Publishing; p. 287. [Google Scholar]
- Uno Y, et al. 2012. Inference of the protokaryotypes of amniotes and tetrapods and the evolutionary processes of microchromosomes from comparative gene mapping. PLoS One 7:e53027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela N. 2008a. Evolution of the gene network underlying gonadogenesis in turtles with temperature-dependent and genotypic sex determination. Integr Comp Biol. 48:476-485. [DOI] [PubMed] [Google Scholar]
- Valenzuela N. 2008b. Relic thermosensitive gene expression in genotypically-sex-determined turtles. Evolution 62:234-240. [DOI] [PubMed] [Google Scholar]
- Valenzuela N. 2009. The painted turtle, Chrysemys picta: a model system for vertebrate evolution, ecology, and human health. Cold Spring Harb Protoc. 410.1101/pdb.emo124: 1-9. [DOI] [PubMed] [Google Scholar]
- Valenzuela N. 2010. Multivariate expression analysis of the gene network underlying sexual development in turtle embryos with temperature-dependent and genotypic sex determination. Sex Dev. 4:39-49. [DOI] [PubMed] [Google Scholar]
- Valenzuela N, Adams DC. 2011. Chromosome number and sex determination co-evolve in turtles. Evolution 65:1808-1813. [DOI] [PubMed] [Google Scholar]
- Valenzuela N, et al. 2013. Transcriptional evolution underlying vertebrate sexual development. Dev Dyn. 242:307-319. [DOI] [PubMed] [Google Scholar]
- Valenzuela N, et al. 2014. Molecular cytogenetic search for cryptic sex chromosomes in painted turtles Chrysemys picta. Cytogenet Genome Res. 144:39-46 [DOI] [PubMed] [Google Scholar]
- Vonk FJ, et al. 2013. The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system. Proc Natl Acad Sci U S A. 110:20651-20656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, et al. 2013. The draft genomes of soft-shell turtle and green sea turtle yield insights into the development and evolution of the turtle-specific body plan. Nat Genet. 45:701-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren WC, et al. 2008. Genome analysis of the platypus reveals unique signatures of evolution. Nature 453:175-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MJ, et al. 2013. Molecular cytogenetic map of the central bearded dragon, Pogona vitticeps (Squamata: Agamidae). Chromosome Res. 21:361-374. [DOI] [PubMed] [Google Scholar]
- Zanaria E, et al. 1994. An unusual member of the nuclear hormone-receptor superfamily responsible for X-linked adrenal hypoplasia congenita. Nature 372:635-641 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.