Abstract
Quest for Orthologs (QfO) is a community effort with the goal to improve and benchmark orthology predictions. As quality assessment assumes prior knowledge on species phylogenies, we investigated the congruency between existing species trees by comparing the relationships of 147 QfO reference organisms from six Tree of Life (ToL)/species tree projects: The National Center for Biotechnology Information (NCBI) taxonomy, Opentree of Life, the sequenced species/species ToL, the 16S ribosomal RNA (rRNA) database, and trees published by Ciccarelli et al. (Ciccarelli FD, et al. 2006. Toward automatic reconstruction of a highly resolved tree of life. Science 311:1283–1287) and by Huerta-Cepas et al. (Huerta-Cepas J, Marcet-Houben M, Gabaldon T. 2014. A nested phylogenetic reconstruction approach provides scalable resolution in the eukaryotic Tree Of Life. PeerJ PrePrints 2:223) Our study reveals that each species tree suggests a different phylogeny: 87 of the 146 (60%) possible splits of a dichotomous and rooted tree are congruent, while all other splits are incongruent in at least one of the species trees. Topological differences are observed not only at deep speciation events, but also within younger clades, such as Hominidae, Rodentia, Laurasiatheria, or rosids. The evolutionary relationships of 27 archaea and bacteria are highly inconsistent. By assessing 458,108 gene trees from 65 genomes, we show that consistent species topologies are more often supported by gene phylogenies than contradicting ones. The largest concordant species tree includes 77 of the QfO reference organisms at the most. Results are summarized in the form of a consensus ToL (http://swisstree.vital-it.ch/species_tree) that can serve different benchmarking purposes.
Keywords: Tree of Life, species tree, gene tree support
Introduction
Because important applications in the Life Sciences rely on orthology inference, scientists—among whom many authors of orthology databases—joined a community effort named “Quest for Orthologs (QfO),” one of whose goals is to compare and improve orthology predictions (Gabaldon et al. 2009; Dessimoz et al. 2012; Sonnhammer et al. 2014). The evaluation of such inference depends on the thorough understanding of the evolution of gene families and taxa. To generate reference gene trees (Trachana et al. 2011, 2014; Sonnhammer et al. 2014), it is crucial to find a core set of species whose relative histories are known to the best of our knowledge. After realizing the incongruence of Trees of Life (ToL) at the third Quest for Orthologs (QfO3) conference in Lausanne (Switzerland) in 2013
(http://questfororthologs.org/meetings#quest_for_orthologs_3, last accessed July 7, 2015), a QfO species tree working group was initiated to survey the status of species phylogenies as well as to establish contacts between experts of the ToL community. Within the same year, exchanges were initiated between the two research communities at the Biodiversity Information Standards conference (Taxonomic Databases Working Group) in 2013 in Florence, Italy (http://www.slideshare.net/suzi.lewis/q4-o-at-tdwg-2013, last accessed July 7, 2015).
ToL links inferred species histories on a global level. Although there is an ongoing debate on tree or network-based evolution—particularly in prokaryotes where horizontal gene transfer (HGT) can be massive (Koonin et al. 2001; Treangen and Rocha 2011)—the concept of a ToL is an important pillar in evolutionary biology. In the context of quality assessment for orthology predictions, species trees are key in reconciling species and gene histories and decoding discordance between the two trees. Yet, understanding to which extent species trees are robust recapitulations of the evolution of life and to which extent they depend on method and data set contingencies is challenging. Initially, morphological and anatomical observations led to the assumption that species are related to each other, and species were grouped and classified accordingly. Molecular phylogenetics revolutionized the inference of species phylogenies, thanks to its ability to trace the evolution of proteins, genes, and genomes. Be it for predicting species phylogenies or understanding the evolutionary relationships of genes, the underlying analyses are alike (Whelan et al. 2001). As a first step, genes related to each other (homologs) are identified, followed by in-depth analysis on how the genes are related, that is, whether they diverged following a speciation event (ortholog), gene duplication (paralog), or were laterally transferred between organisms (xenolog). In other words, the prediction of gene relationships is based on knowledge of the species tree, and the species tree can be inferred from the study of gene relationships. Hence, there is a risk of circularity when inferring orthology predictions from ortholog-derived species trees.
In this study, we address the question of the current knowledge and reliability of species relationships by assessing the congruence among the most inclusive ToLs and species trees. To do this, we compared tree topologies for 147 species that were selected for the QfO reference data set 2013: 120 eukaryotes, 7 archaea, and 20 bacteria (hereafter named QfO reference organisms; taxa are listed in supplementary table S1, Supplementary Material online). The prediction of their evolutionary relationship differs in many ways which are all able to influence the tree topology (compare supplementary table S2, Supplementary Material online). To begin with, the size of the six analyzed species phylogenies ranges from 191 to 2,227,481 taxa. Taxon sampling has a strong impact on tree reconstruction. As an example, high species coverage has been shown to avoid systematic bias and long branch attraction (LBA) in phylogenetic analysis of molecular sequences (Philippe et al. 2000; Heath et al. 2008). Of importance is also the type of data: In contrast to noncoding genes, coding genes can be analyzed at the level of both nucleotide sequences and amino acid sequences, which can lead to complementary resolution at different levels in the tree space. The analysis of a single gene presumes that the gene history reflects that of the species, as opposed to the study of multiple marker genes. The latter allows the comparison of all gene tree topologies with each other, which helps to identify topological incongruence such as nonvertical gene traffic, which is especially relevant to the study of prokaryotic relationships. Phylogenetic signals can also be obtained from domain occurrence in proteomes, and the analysis of the binary matrix constitutes a fast tree-building approach for regular revision. Another fundamental difference is that not all species trees are built from scratch: Some ToLs use an existing species classification as the initial tree, to which phylogenetic information is added or to guide a phylogenetic analysis. In this case, species trees are not independent, and are thus likely to share higher topological congruence. Last but not the least, some species trees are rooted, while others are not, thus influencing the analysis as well as the interpretation of the tree topology.
The six species trees under comparison are the following: 1) The NCBI taxonomy is probably the most widely used species classification in molecular phylogenetics because it is the repository for standardized nomenclature and taxonomic identifiers for international sequence databases (Federhen 2011). As such, it is often implicitly used as the guide for classification. However, the NCBI taxonomy database is not an authoritative source for nomenclature or classification (http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Root, last accessed July 7, 2015). It currently stores data from 971,052 species and 1,254,947 taxonomic nodes (NCBI taxonomy browser; www.ncbi.nlm.nih.gov/Taxonomy/taxonomyhome.html/index.cgi?chapter=statistics, February 19, 2015). The species classification, based on expert knowledge, includes a large number of multifurcating nodes that represent yet unknown species phylogenies. 2) The Open Tree of Life (Opentree) strives to capture the spectrum of known biodiversity. Initially based on the NCBI taxonomy, published species phylogenies are stepwise mapped on the species classification (http://biorxiv.org/content/early/2014/12/15/012260, last accessed July 7, 2015). With over 2.3 million taxa, it is the largest of the six species trees. 3) The sequenced species Tree of Life (sToL) is an NCBI taxonomy-guided, bifurcating likelihood tree based on the protein domain composition predicted for cellular organisms with complete proteomes (Fang et al. 2013). 4) Huerta-Cepas, Marcet-Houben, et al. (2014) developed a nested, hierarchical approach to maximize phylogenetic signals from the sequence data used for tree reconstruction (hereafter named Tree-HC). The species tree thus analyzed includes 216 eukaryotic species and no prokaryotes. 5) Cicarelli et al. (2006) published a species tree based on 31 concatenated universal proteins from 191 species, of which 23 are eukaryotes, 18 archaea, and 150 bacteria (hereafter named ToL-C). Although this tree has been updated, thanks to an improved HGT detection methodology (Creevey et al. 2011) and the possibility of careful data selection from a considerably increased collection of completely sequenced genomes, we decided to oppose this early version of a species tree—constructed by an automated procedure—to more recent and larger ToLs. 6) The 16S rRNA project is probably the greatest attempt for the detailed analysis of a gene history (Munoz et al. 2011; Yilmaz et al. 2013). In 1977, the study of this gene by Woese and Fox (1977) resulted in the identification of Archaea as a third domain of life. Today, the expert-curated multiple sequence alignment of ribosomal RNA sequences consists of data from over 10,000 taxa, and the 16S rRNA has become an important marker gene for metagenomics. However, more than one copy of this gene has been observed in genomes (Case et al. 2007). As the gene could be subject to HGT, the gene tree can differ from the actual species phylogeny. Consequently, alternative markers have been proposed to determine bacterial biodiversity (Case et al. 2007; Lang et al. 2013; Mende et al. 2013).
In this study, we show that—for the six species trees—the tree topology of the 147 QfO reference organisms is congruent at 87 of the 146 possible internal nodes of a rooted binary tree; all other nodes are incongruent in at least one of the species trees. In addition, we assessed the discordance of the different species trees with individual gene trees by using available phylomes for QfO species available at PhylomeDB (Huerta-Cepas, Capella-Gutierrez, et al. 2014). For benchmarking in the context of the QfO activities, we propose the use of an annotated consensus tree (ToLc-147) whose internal branches can be multifurcated at different levels of consistency. We provide a first version here.
Material and Methods
Terms
The terms to describe tree relationships and properties have been used in different ways in publications. In this document, the general notion congruency (antonym: incongruency) refers to identical relationships of taxa in independent gene phylogenies or species phylogenies, as well as for splits obtained from these trees. The term concordance (antonym: discordance) is used to describe congruence between a gene tree and a species tree, likewise when trees were pruned to sets of common taxa. A tree topology—or part of it—is consistent, if identical relationships of taxa or clades have been reconstructed based on different data sets.
Species Tree Comparison
The NCBI taxonomic classification tree was generated at http://www.ncbi.nlm.nih.gov/Taxonomy/CommonTree/wwwcmt.cgi (July 14, 2014) based on the taxonomic identifiers for the species of the QfO reference proteomes. The sToL tree was constructed at http://supfam.org/SUPERFAMILY/cgi-bin/genome_names.cgi (July 8, 2014). Twenty-five species differed in their strain and could not be mapped based on the taxonomic identifiers. These species were mapped based on the species names (“relaxed species mapping”). Opentree provided a draft of the species tree (April 16, 2014) and the corresponding species mapping to taxonomic identifiers. The 216 eukaryotic species tree published by Huerta-Cepas (peerj.com/preprints/223/) was provided, including NCBI taxonomic identifiers and annotated branch support values (aLRT SH). Seventy-one species were mapped by taxonomic identifiers, 27 by species name. The ToL by Ciccarelli et al. (ToL-C) was copied from http://itol.embl.de/ (July 14, 2014). Forty-four QfO reference organisms were identified by their scientific names. The 16S rRNA-based species phylogeny of the all-species living tree project was obtained from http://www.arb-silva.de/projects/ (LTPs115). Twenty-two archaea and bacteria were mapped to the QfO data set by the species name. Species not matching the QfO reference proteomes were pruned from the trees using the Newick utilities (Junier and Zdobnov 2010); species names were standardized using Perl scripts. Robinson–Foulds (RF) distances between pairs of species trees—except for the two complementing species trees that have no taxa in common—were calculated with ETE v2.2 (Huerta-Cepas et al. 2010).
The species consensus tree (ToLc-147) was constructed manually and consistency levels annotated for multifurcation at different levels of topological congruence. As a first approach, consistency values were assigned for each node and each species tree, according to the following instances: +1, node is congruent with the ToLc-147 topology; +2, same as +1, with significant branch support; 0, multifurcating node; −1, alternative topology (incongruent with ToLc-147); −2, alternative topology with significant branch support (supplementary table S3, Supplementary Material online). The average consistency value was calculated for each node, and results classified into four levels: Level L90 (>1), level L70 (0.75–1), level L50 (>0 to <0.75), and level L10 (≤0). The assigned levels correspond to the annotated consistency values (90, 70, 50, 10) in ToLc-147, so nodes can be multifurcated at different levels with existing tree visualization tools. Further evidence for the evolutionary relationship of species was obtained from the literature, and levels adapted according to findings in a similar way as described above; for instance, when published clade-specific analyses significantly supported nodes of the consensus tree. Finally, a fifth level was introduced for practical reasons, for nodes with conflicting results which await further classification (L30).
Assessment of Gene Tree Support for Species Topologies
For each phylome reconstructed for the QfO project (phyIDs 500–542), the trees were downloaded from phylogenomic database phylomeDB (Huerta-Cepas, Capella-Gutierrez, et al. 2014). These gene trees were constructed with a methodology similar to the one used to infer the species phylogeny of Tree-HC, although based on a different set of proteomes. Because gene tree reconstruction was not guided by a species tree and because this experiment is not a quality assessment, this collection of gene phylogenies is suitable to estimate gene tree support for our consensus tree and alternative topologies. Each tree in each phylome was first compared with a given species tree. If they overlapped in less than three species, the tree was discarded. Trees were then pruned so that they contained only species that were present in the species tree. Trees were then rooted by the species placed closest to the root according to the species tree. The tree was then split into orthologous trees following the methodology explained previously (Marcet-Houben and Gabaldon 2011) as implemented in ETE v2.2. Briefly, all duplication nodes were detected in the tree by using a species overlap algorithm (Huerta-Cepas et al. 2007). All the possible combinations of the duplicated parts of the tree were done and then the tree was pruned as many times as combinations of duplicated nodes were found. These pruned trees are called orthologous trees as all the leaves in them are orthologous to each other. Only trees with less than 100 associated orthologous trees were considered to reduce computing time. A support measure was then calculated as follows: Species trees and orthologous trees were pruned so that they contained exactly the same set of species. Then, for each node in the remaining species tree, we searched each orthologous tree for the presence of the node. The number of trees that contained the node was divided by the total amount of trees that contained the species derived from the node, whether they were monophyletic or not. If a tree resulted in more than one orthologous tree, then each orthologous tree was checked for the presence of the node and then divided by the number of orthologous trees that contained the species derived from the node. This was done to ensure that each tree contributed equally to the final result.
Results and Discussion
Species Supernetworks: Survey on the Topological Congruence of Species Trees
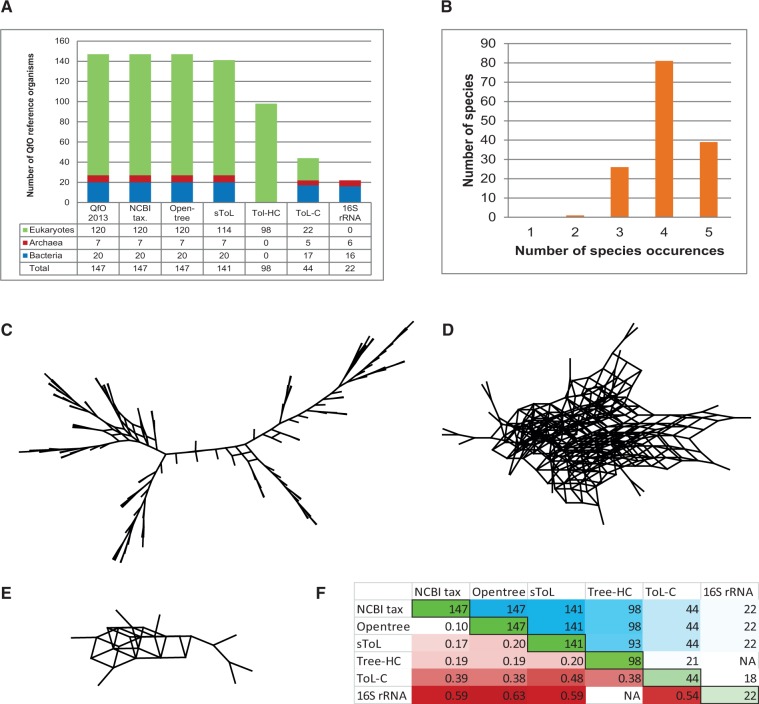
Six well-known broad species phylogenies were compared: The NCBI taxonomic classification, the Opentree of Life, the sequenced sToL, the 16S rRNA-based species tree, and the species phylogenies published by Ciccarelli et al (ToL-C) and Huerta-Cepas et al (Tree-HC). All these trees were generated by applying different methodologies and using different databases, data sets, and sources of phylogenetic signals (supplementary table S1, Supplementary Material online), which makes the search for a consensus species tree especially meaningful. Nonrelevant taxa were pruned from all trees, resulting in cladograms of up to 147 QfO reference organisms (fig. 1A and supplementary file S1, Supplementary Material online). Four of the trees cover species from all the three domains of life (ToLs), and the two complementing species trees consist of prokaryotes and eukaryotes, respectively. Thus, each species can be found in the analyzed data sets—at the most five times—and the average frequency was 4.1 (fig. 1B).
Fig. 1.—
Comparison of the six species trees. (A) Coverage of QfO species in the analyzed ToLs/species trees: Stacked bar chart of species from the Quest for Orthologs reference proteome set 2013 mapped to the species trees, color-coded by domains of life. The far left column presents the QfO reference organisms. (B) Frequency of QfO reference organisms in the analyzed ToLs/species trees. On average, each QfO reference organism occurred in the data set about 4.1 times; represented only twice are the amoeba Polysphondylium_pallidum (NCBI TaxId: 13642), the fungi Rhizopus delemar (TaxId: 246409) and Batrachochytrium_dendrobatidis (NCBI TaxId: 684364). Supernetwork of the eukaryote (C), bacterial (D), and archaeal (E) clade visualize topological congruence and incongruence between ToLs/species trees. (F) RF distances between ToLs/species trees: For each tree, the table shows the number of species in common with the species of the QfO reference data set (green cells), the number of QfO reference organisms shared by two trees (blue), and the average RF distances per node between trees (red).
At first, the degree of incongruence between the predicted species phylogenies was explored through supernetworks for each of the three domains of life (fig. 1C–E). In a phylogenetic supernetwork, identical topologies are merged into a tree structure and alternative topologies are combined into a network (Huson et al. 2004). Considerably higher topological congruence is observed in the eukaryotic clade compared with the two prokaryotic clades. One known major factor disturbing phylogenetic reconstruction in prokaryotes is the pervasiveness of HGT in these domains (Bapteste et al. 2004). Rates of HGT have been estimated to lie in the range of 1.6–32.6% (Koonin et al. 2001) and higher (McDaniel et al. 2010), and a constant rate of interspecies gene traffic was reported for universally single copy genes (Trachana et al. 2014).
Quantification of Topological Congruence between Species Trees
There are basically two approaches for the quantification of differences in tree topologies: 1) Tree incompatibility, which is calculated from contradicting splits and does not take into account multifurcation, and 2) tree dissimilarity, which is determined from all nonidentical splits, thus measuring incongruent branching patterns including multifurcation. At first, the former sounds more suitable when aiming to identify a consensus, however it does not take into account unresolved (multifurcating) nodes, and thus equates a star-like tree with the perfect tree. This is why we decided to measure the dissimilarity between species phylogenies by applying the RF metric (Robinson and Foulds 1981) averaged over the number of nodes common to a tree pair (fig. 1F). The resulting normalized RF distances between pairs of species trees range from 10% to 63%, thus confirming that none of the species phylogenies are congruent. Distances are shortest between trees which include a high fraction of eukaryotic species, while they increase according to the fraction of prokaryotic species. This result indicates once again high dissimilarity in bacterial and archaeal phylogenies and is thus in agreement with the above described supernetworks. The smallest distance between ToLs was calculated for the NCBI taxonomy and Opentree. This is not surprising, as Opentree is based on the NCBI taxonomic classification. Both trees still share a relatively large number of unresolved nodes, but Opentree synthesizes the taxonomic hierarchy with published species phylogenies, which results in a higher resolution in Homininae or early diverging metazoans (Amphimedon, Trichoplax, Bilateria) for instance. The RF distance is the most commonly used dissimilarity measure for pairs of tree topologies, regardless of its high susceptibility to certain topological differences which can result in maximum distance values, even for the interchange of a single leaf (Lin et al. 2011). Thus one needs to exercise caution and avoid overinterpreting the results. It should also be recalled that—at this stage—it is not possible to assess the quality of inferred species trees for many reasons, most important of all the lack of knowledge on the true tree. Thus, low distances do not indicate high accuracy nor do high distances indicate failure. The goal of this study is to identify topological incongruence between species phylogenies and, to this end, we summarize node consistency based on the comparison of all six species phylogenies: 87 of the 146 possible splits of a bifurcating, rooted tree are congruent in the compared species trees; all other splits are incongruent in at least one of the six trees (supplementary table S3, Supplementary Material online). Noteworthy, NCBI reflects well topological incongruence in the form of multifurcating nodes, but misses some of the highly supported nodes, such as the consistent node for Diptera/Bombyx mori, Metazoa/Monosiga, or Opisthokonta.
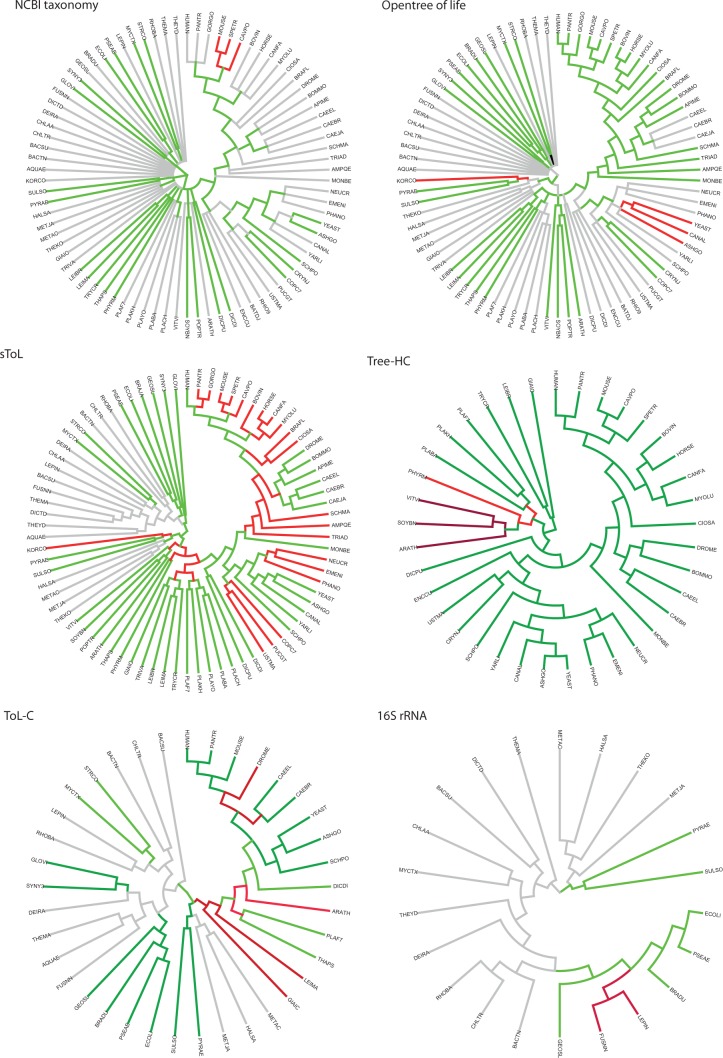
In eukaryotes, 67% (80/119) of the internal nodes of a rooted binary tree are congruent, 79% (94/119) are congruent or compatible (multifurcating), and alternative phylogenies are suggested for 21% (25/119) of the internal nodes. Various reasons can be given for the topological differences within the eukaryotic clade. Tree reconstruction artifacts are frequently explained by the lack of a phylogenetic signal or the failing to discriminate a phylogenetic signal from noise. The former is commonly observed in closely related species, species which diverged in a short interval from a common lineage (short common branch length) or in genes under strong structural and functional constraints, thus evolving at a low rate and lacking shared traits. When homoplasy prevails synapomorphy in genes of fast-evolving species, taxa tend to be grouped by mistake—an artifact known as LBA (Schulmeister 2004). For the 147 QfO reference organisms, such knowledge can, for instance, help explain topological incongruence observed for discrete representatives of invertebrate clades sampled along the human lineage as well as for species which diverged early from major clades such as ecdysozoans. Differing phylogenies are not only observed at deep nodes, but also at recent speciation events (figs. 2 and 3A). The phylogeny of Homininae is an example: The topology is unresolved in the NCBI classification, and a monophyletic origin of chimp and gorilla is suggested by sToL. Opentree implemented a resolved but differing species history—human being more closely related to chimp than to gorilla—based on relevant published phylogenies. One of the underlying studies is a recent phylogenetic analysis of complete mitochondrial primate genomes that provides significant support for a common ancestor of human and chimp after the divergence of gorilla, which is in agreement with many other clade-specific analyses (Pozzi et al. 2014). Within the mammalian branch, we note two further clades with contradicting phylogenies: Glires and Laurasiatheria. Within the first group, the rodents Cavia porcellus and Spermophilus tridecemlineatus show an interchanged divergence order, and each of the two topologies is suggested by two of the four species trees that include the relevant QfO reference organisms. Tree-HC provides significant branch support for a monophyletic origin of Murinae and C. porcellus, and Opentree has implemented supporting results for the same topology. For Laurasiatheria, none of the species trees suggest the same phylogeny. For the same clades, incongruence is also observed in various published phylogenies, thus indicating that phylogenetic signals which are derived from the different data sets are ambiguous.
Fig. 2.—
Overview of critical spots in reconstructed species phylogenies. For lack of space, all species trees were pruned to include only species which illustrate yet unresolved phylogenies and contradicting topologies. Color codes: Light green = topologies supporting the consensus tree (fig. 3); dark green = topologies supporting the consensus tree with significant support; red = topologies differ from the consensus tree; dark red = topologies with significant support differ from the consensus tree; light gray = unresolved and/or unknown topologies.
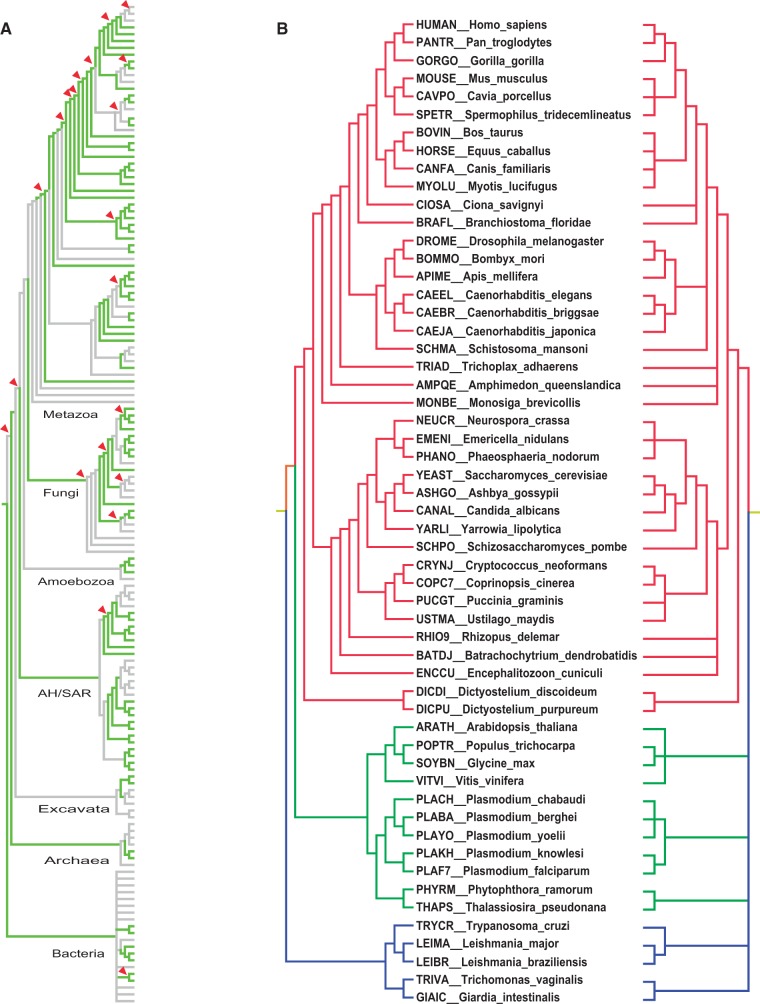
Fig. 3.—
Consensus tree. (A) Consensus phylogeny of the 147 QfO reference organisms. Green branches highlight congruent and bifurcating topologies, grey branches indicate topologies that are either multifurcating or incongruent in at least one of the species trees. Red triangles mark nodes that are supported by at least 75% of the gene trees (see also supplementary file S4, Supplementary Material online). (B) Eukaryotic clade of the consensus tree at highest (bifurcating, left handed) and lowest (L90, right handed) resolution, pruned to the species set as in figure 2. Bifurcation is not yet possible for most internal nodes in the archaeal and bacterial clades; the topologies are thus identical for both trees.
In nonvertebrate eukaryotes, differing bifurcating tree topologies are observed within chordates for species of the genus Ciona and Branchiostoma floridae, for the platyhelminth Schistosoma mansoni, for early divergence groups within ecdysozoans, for the metazoans Trichoplax adhaerens and Amphimedon queenslandica; within fungi for the ascomycetes Ashbya gossypii, Candida albicans, and Phaeosphaeria nodorum, and for the basidiomycete Puccinia graminis; within Viridiplantae for the rosids Arabidopsis thaliana and Vitis vinifera, and within the genus Plasmodium for Plasmodium berghei. At deep nodes, incongruent phylogenies concern the groups Halvaria, AH/SAR, the recently suggested clade Neozoa (Unikonta and AH/SAR; He et al. 2014), and Excavata.
As for prokaryotes, 27% (7/26, archaea: 50% [3/6], bacteria: 21% [4/19]) of the internal nodes are congruent, 38% (10/26, archaea: 50% [3/6], bacteria: 37% [7/19]) are congruent or compatible (multifurcating), and 62% (16/26, archaea: 50% [3/6], bacteria 63% [12/19]) are incongruent. At the deepest nodes in the ToL, all relevant species trees agree on the monophyly of bacteria, and that of archaea when rooted with bacteria. The representative group of archaeal QfO reference organisms include both crenarchaeotes and euryarchaeotes, and their monophyly is still disputed (Wolf et al. 2001). Noteworthy, the tree topology for five archaeal species common to Tol-C and the 16S rRNA species tree is congruent and in agreement with a recently published archaeal phylogeny based on over 200 marker genes (Petitjean et al. 2015). Consistent clades are observed within Archaea for the class Thermoprotei (Pyrobaculum aerophilum, Sulfolobus solfataricus) and the phylum Euryarchaeota (Halobacterium salinarum, Methanosarcina acetivorans, Methanocaldococcus jannaschii, Thermococcus kodakaraensis), within bacteria for the phylum Cyanobacteria (Gloeobacter violaceus, Synechocystis sp.), the order Actinomycetales (Mycobacterium tuberculosis, Streptomyces coelicolor), the class Gammaproteobacteria (Escherichia coli, Pseudomonas aeruginosa), and the phylum Proteobacteria (Bradyrhizobium diazoefficiens, Geobacter sulfurreducens, and Gammaproteobacteria). Other nodes within prokaryotes are unresolved or conflicting.
In summary, for the QfO reference organisms the topological congruence between species trees is more than 3-fold higher in the eukaryotic clade than in the bacterial clade. Discrepancies can be explained with tree reconstruction artifacts and differences in genome evolution. As HGT is prevalent in prokaryotes, inclusion of species or genes prominently subjected to it will heavily affect phylogeny (Trachana et al. 2014).
Consequences for Orthology Prediction and Benchmarking
Needless to say, a resolved evolutionary species history would be preferable for orthology prediction and benchmarking. The largest nonconflicting and bifurcating tree generated from the six species trees would, however, only include 77 species at the most (ToLc-77; supplementary file S2, Supplementary Material online). Thus, we constructed a consensus tree composed of all 147 QfO reference organisms (ToLc-147) taking into consideration both the degree of topological incongruence and the branch support, if available. A large fraction of consistent nodes of the eukaryotic clade present in Tree-HC was significantly supported. For yet unresolved and inconsistent nodes, high quality phylogenetic studies were conducted from the literature, because clade-specific analyses commonly possess a higher species density, more marker genes, and less ambiguous or missing characters that originate from distantly related species. So far, all studies that have been taken into account concern eukaryotic clades: Primates (Fabre et al. 2009; Pozzi et al. 2014), laurasiatherians (Zhou et al. 2011; Pozzi et al. 2014), chordates (Delsuc et al. 2006; Putnam et al. 2008), nonchordate metazoans (Halanych et al. 1995; Telford et al. 2005; Delsuc et al. 2006; Dunn et al. 2008; Srivastava et al. 2008; Nosenko et al. 2013; Misof et al. 2014), choanozoans (Shalchian-Tabrizi et al. 2008), fungi (Ebersberger et al. 2011; Medina et al. 2011; Capella-Gutierrez et al. 2012), amoebozoans (Lahr et al. 2011; Fiz-Palacios et al. 2013), plants (Wang et al. 2009; Burleigh et al. 2010; Zeng et al. 2014), halvarians (Simpson et al. 2006; Krief et al. 2010; Burki et al. 2012), and deep branches in eukaryotes (Simpson et al. 2006; Hampl et al. 2009; Burleigh et al. 2010). In contrast to eukaryotes, the predicted evolutionary relationships for bacteria and archaea are too divergent to generate a possible bifurcating tree. Consequently, for the time being we retain the polytomy of the NCBI taxonomic classification. As a result, orthology prediction for these domains of life has to be complemented with non–tree-based methods for HGT prediction such as the analysis of codon usage or oligonucleotide profiles. The proposed species consensus tree ToLc-147 is presented in figure 3 and available at http://swisstree.vital-it.ch/species_tree (last accessed July 7, 2015). The assigned consistency values are annotated and can be used to multifurcate nodes at different levels of topological congruence between species trees.
However, even if there were a bifurcating, resolved species tree, there could be a risk in oversimplifying the view on genome evolution. Assuming that the main gene stream—inherited genes with full-length orthology—reflects species evolution, each genome is likely to contain a fraction of genes which is discordant with any species tree. This can be because of horizontal/endosymbiont gene transfer and pseudo-orthologs in accurate gene trees, but it can also be because of the lack of phylogenetic signal and noise resulting from incomplete lineage sorting, gene conversion, changes in domain architectures, wrong gene models, sequence errors, and so on. As an illustration, a recent analysis of the evolution of 48 bird species and the genes encoded in their genomes revealed that no single gene tree was fully congruent with the reconstructed species phylogeny (Jarvis et al. 2014). In a group of closely related vertebrate species where the role of HGT is negligible, such a result indicates that both incomplete lineage sorting and stochastic noise can create widespread discordance between gene trees and species trees. The intentional exploration of this nonsupporting gene fraction might provide advanced insight into genome and species evolution. Consequently, it is worthwhile reflecting the reliability of predicted gene relationships by means of confidence scores which allow experiment-specific data selection by database users. In addition, confidence scores could be used for improved orthology benchmarking, accounting for the sensitivity/specificity trade-off.
Assessment of Gene Tree Support for Species Tree Topologies
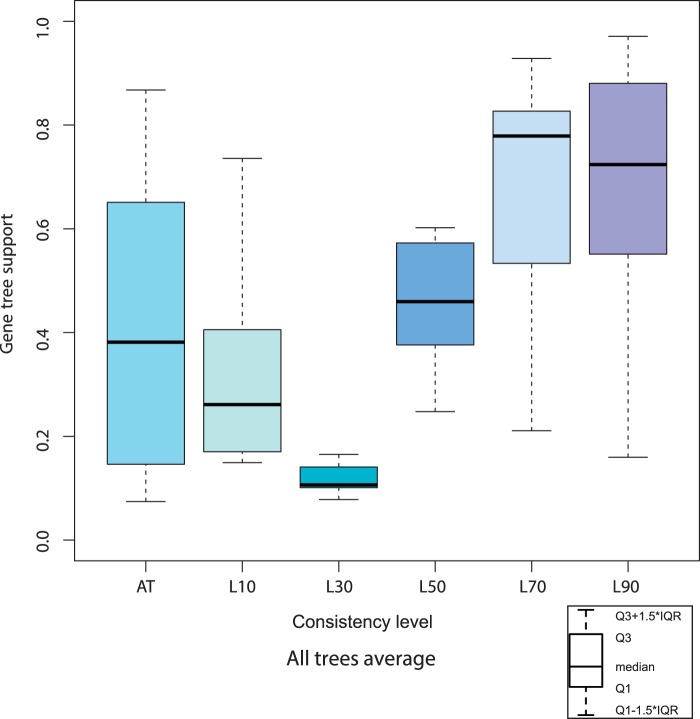
To assess the level of discordance between the different species trees and the individual gene trees, we compared each species tree node with 458,108 gene phylogenies built for proteins encoded in 65 QfO reference species (Huerta-Cepas, Capella-Gutierrez, et al. 2014). This analysis was performed on ToLc-147 as well as on each species tree. By analyzing results according to the assigned consistency levels, we show that gene tree topologies coincide more often with consistent nodes (consistency levels L90 and L70) in species trees than with conflicting ones (consistency levels L10, L30, and L50) (fig. 4). This trend is also observed in the individual box plots for each project (supplementary file S3, Supplementary Material online). Interestingly, species trees which differ from the consensus tree (hereafter named “alternative species topologies” and assigned category “AT” for practical reasons) occur as the most dispersed group. The range of its gene tree support values corresponds to that determined for incongruent nodes. In fact, category AT includes many prokaryotic speciation nodes which are still polytomous in ToLc-147 because of incongruent topologies in the species trees, thus explaining the comparatively tall box plot. Even when assuming ToLc-147 to present the true tree, gene trees congruent with alternative species topologies can be correct, for instance, when containing xenologs or pseudo-orthologs. ToLc-147 with annotated gene tree support is presented in supplementary file S4, Supplementary Material online.
Fig. 4.—
Box plot of gene tree fractions supporting species tree topologies at different consistency levels. Consistent species tree topologies with (L90) and without (L70) significant branch support are generally in compliance with the analyzed gene trees. The fraction of supporting gene trees drops considerably when species tree topologies are incongruent, once or more, between the species trees (L10, L30, L50). Consistency categories “L30” and AT were assigned for practical reasons. Level L30 is the default value for conflicting nodes prior to evaluation, and the two remaining nodes (Excavata, Proteobacteria) show on the one hand conflicting species topologies, on the other hand significant branch support in at least one of the species trees. Only a low fraction of our gene trees supports these speciation nodes. Category AT indicates alternative topologies suggested by the species trees, and results cover the range of conflicting levels (L10, L50); this makes sense because alternative topologies are incongruent with the consensus tree and between species trees. For each box plot, bottom of the box is the first quartile (Q1), top of the box is the third quartile (Q3), the middle bar is the median, whiskers represent the 1.5 interquartile range (IQR).
Conclusions
This study sheds light on the current resolution of the species phylogeny for the 147 QfO reference proteomes. Open questions concern not only the true species tree, but likewise which type of phylogenetic signal is most informative for tree reconstruction at a particular level of depth and taxonomic range. As for sequence-based phylogenetic analysis, the selection of good marker genes and a higher and balanced species density in the tree space may help to stabilize the tree topology. A comparison of species trees obtained from the analysis of different types of characters, including morphological ones, can help to identify strengths and weakness of each approach in order to optimize species tree inference. Other issues worthwhile discussing between expert communities include taxon sampling and visualization of annotated evolutionary changes which result in the characteristics of extant species at any level of organization. We hope that this work will contribute to a lively exchange between the QfO and ToL communities.
For orthology benchmarking, we propose a species consensus tree with conservation values associated to each node for multifurcation at the desired consistency level. We provide here the first version of one such reference tree covering the QfO reference species.
Supplementary Material
Supplementary tables S1–S3 and supplementary files S1–S4 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors wish to thank Vivienne Baillie Gerritsen for proofreading. This work was supported by the Swiss Federal Government through the Federal Office of Education and Science (B.B.). Funding for open access charge: Swiss Institute of Bioinformatics. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. T.G. group research is funded in part by a grant from the Spanish ministry of Economy and Competitiveness (BIO2012-37161), a Grant from the Qatar National Research Fund (NPRP 5-298-3-086), and a grant from the European Research Council under the European Union’s Seventh Framework Programme (FP/2007-2013)/ERC (grant agreement no. ERC-2012-StG-310325). M.M. acknowledges support from the Wellcome Trust (grant number WT095908) and the European Molecular Biology Laboratory.
Literature Cited
- Bapteste E, Boucher Y, Leigh J, Doolittle WF. 2004. Phylogenetic reconstruction and lateral gene transfer. Trends Microbiol. 12:406–411. [DOI] [PubMed] [Google Scholar]
- Burki F, Okamoto N, Pombert JF, Keeling PJ. 2012. The evolutionary history of haptophytes and cryptophytes: phylogenomic evidence for separate origins. Proc Biol Sci. 279:2246–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burleigh JG, et al. 2010. Genome-scale phylogenetics: inferring the plant tree of life from 18,896 gene trees. Syst Biol. 60:117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutierrez S, Marcet-Houben M, Gabaldon T. 2012. Phylogenomics supports microsporidia as the earliest diverging clade of sequenced fungi. BMC Biol. 10:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case RJ, et al. 2007. Use of 16S rRNA and rpoB genes as molecular markers for microbial ecology studies. Appl Environ Microbiol. 73:278–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccarelli FD, et al. 2006. Toward automatic reconstruction of a highly resolved tree of life. Science 311:1283–1287. [DOI] [PubMed] [Google Scholar]
- Creevey CJ, Doerks T, Fitzpatrick DA, Raes J, Bork P. 2011. Universally distributed single-copy genes indicate a constant rate of horizontal transfer. PLoS One 6:e22099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delsuc F, Brinkmann H, Chourrout D, Philippe H. 2006. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature 439:965–968. [DOI] [PubMed] [Google Scholar]
- Dessimoz C, Gabaldon T, Roos DS, Sonnhammer EL, Herrero J. 2012. Toward community standards in the quest for orthologs. Bioinformatics 28:900–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn CW, et al. 2008. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature 452:745–749. [DOI] [PubMed] [Google Scholar]
- Ebersberger I, et al. 2011. A consistent phylogenetic backbone for the fungi. Mol Biol Evol. 29:1319–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre PH, Rodrigues A, Douzery EJ. 2009. Patterns of macroevolution among Primates inferred from a supermatrix of mitochondrial and nuclear DNA. Mol Phylogenet Evol. 53:808–825. [DOI] [PubMed] [Google Scholar]
- Fang H, et al. 2013. A daily-updated tree of (sequenced) life as a reference for genome research. Sci Rep. 3:2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federhen S. 2011. The NCBI taxonomy database. Nucleic Acids Res. 40:D136–D143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiz-Palacios O, et al. 2013. Did terrestrial diversification of amoebas (amoebozoa) occur in synchrony with land plants? PLoS One 8:e74374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabaldon T, et al. 2009. Joining forces in the quest for orthologs. Genome Biol. 10:403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halanych KM, et al. 1995. Evidence from 18S ribosomal DNA that the lophophorates are protostome animals. Science 267:1641–1643. [DOI] [PubMed] [Google Scholar]
- Hampl V, et al. 2009. Phylogenomic analyses support the monophyly of Excavata and resolve relationships among eukaryotic “supergroups”. Proc Natl Acad Sci U S A. 106:3859–3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He D, et al. 2014. An alternative root for the eukaryote tree of life. Curr Biol. 24:465–470. [DOI] [PubMed] [Google Scholar]
- Heath TA, Hedtke SM, Hillis DM. 2008. Taxon sampling and the accuracy of phylogenetic analyses. J Syst Evol. 46:239–257. [Google Scholar]
- Huerta-Cepas J, Capella-Gutierrez S, Pryszcz LP, Marcet-Houben M, Gabaldon T. 2014. PhylomeDB v4: zooming into the plurality of evolutionary histories of a genome. Nucleic Acids Res. 42:D897–D902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J, Dopazo H, Dopazo J, Gabaldon T. 2007. The human phylome. Genome Biol. 8:R109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J, Dopazo J, Gabaldon T. 2010. ETE: a python Environment for Tree Exploration. BMC Bioinformatics 11:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J, Marcet-Houben M, Gabaldon T. 2014. A nested phylogenetic reconstruction approach provides scalable resolution in the eukaryotic Tree Of Life. PeerJ PrePrints 2:223. [Google Scholar]
- Huson DH, Dezulian T, Klopper T, Steel MA. 2004. Phylogenetic super-networks from partial trees. IEEE/ACM Trans Comput Biol Bioinform 1:151–158. [DOI] [PubMed] [Google Scholar]
- Jarvis ED, et al. 2014. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science 346:1320–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junier T, Zdobnov EM. 2010. The Newick utilities: high-throughput phylogenetic tree processing in the UNIX shell. Bioinformatics 26:1669–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Makarova KS, Aravind L. 2001. Horizontal gene transfer in prokaryotes: quantification and classification. Annu Rev Microbiol. 55:709–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krief S, et al. 2010. On the diversity of malaria parasites in African apes and the origin of Plasmodium falciparum from Bonobos. PLoS Pathog. 6:e1000765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahr DJ, Grant J, Nguyen T, Lin JH, Katz LA. 2011. Comprehensive phylogenetic reconstruction of amoebozoa based on concatenated analyses of SSU-rDNA and actin genes. PLoS One 6:e22780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang JM, Darling AE, Eisen JA. 2013. Phylogeny of bacterial and archaeal genomes using conserved genes: supertrees and supermatrices. PLoS One 8:e62510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Rajan V, Moret BM. 2011. A metric for phylogenetic trees based on matching. IEEE/ACM Trans Comput Biol Bioinform. 9:1014–1022. [DOI] [PubMed] [Google Scholar]
- Marcet-Houben M, Gabaldon T. 2011. TreeKO: a duplication-aware algorithm for the comparison of phylogenetic trees. Nucleic Acids Res. 39:e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel LD, et al. 2010. High frequency of horizontal gene transfer in the oceans. Science 330:50. [DOI] [PubMed] [Google Scholar]
- Medina EM, Jones GW, Fitzpatrick DA. 2011. Reconstructing the fungal tree of life using phylogenomics and a preliminary investigation of the distribution of yeast prion-like proteins in the fungal kingdom. J Mol Evol. 73:116–133. [DOI] [PubMed] [Google Scholar]
- Mende DR, Sunagawa S, Zeller G, Bork P. 2013. Accurate and universal delineation of prokaryotic species. Nat Methods. 10:881–884. [DOI] [PubMed] [Google Scholar]
- Misof B, et al. 2014. Phylogenomics resolves the timing and pattern of insect evolution. Science 346:763–767. [DOI] [PubMed] [Google Scholar]
- Munoz R, et al. 2011. Release LTPs104 of the All-Species Living Tree. Syst Appl Microbiol. 34:169–170. [DOI] [PubMed] [Google Scholar]
- Nosenko T, et al. 2013. Deep metazoan phylogeny: when different genes tell different stories. Mol Phylogenet Evol. 67:223–233. [DOI] [PubMed] [Google Scholar]
- Petitjean C, Deschamps P, Lopez-Garcia P, Moreira D, Brochier-Armanet C. 2015. Extending the conserved phylogenetic core of archaea disentangles the evolution of the third domain of life. Mol Biol Evol. 32:1242–54. [DOI] [PubMed] [Google Scholar]
- Philippe H, et al. 2000. Early-branching or fast-evolving eukaryotes? An answer based on slowly evolving positions. Proc Biol Sci. 267:1213–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozzi L, et al. 2014. Primate phylogenetic relationships and divergence dates inferred from complete mitochondrial genomes. Mol Phylogenet Evol. 75:165–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam NH, et al. 2008. The amphioxus genome and the evolution of the chordate karyotype. Nature 453:1064–1071. [DOI] [PubMed] [Google Scholar]
- Robinson DR, Foulds LR. 1981. Comparison of phylogenetic trees. Math Biosci. 53:131–147. [Google Scholar]
- Schulmeister S. 2004. Inconsistency of maximum parsimony revisited. Syst Biol. 53:521–528. [DOI] [PubMed] [Google Scholar]
- Shalchian-Tabrizi K, et al. 2008. Multigene phylogeny of choanozoa and the origin of animals. PLoS One 3:e2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson AG, Inagaki Y, Roger AJ. 2006. Comprehensive multigene phylogenies of excavate protists reveal the evolutionary positions of “primitive” eukaryotes. Mol Biol Evol. 23:615–625. [DOI] [PubMed] [Google Scholar]
- Sonnhammer EL, et al. 2014. Big data and other challenges in the quest for orthologs. Bioinformatics 30:2993–2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M, et al. 2008. The Trichoplax genome and the nature of placozoans. Nature 454:955–960. [DOI] [PubMed] [Google Scholar]
- Telford MJ, Wise MJ, Gowri-Shankar V. 2005. Consideration of RNA secondary structure significantly improves likelihood-based estimates of phylogeny: examples from the bilateria. Mol Biol Evol. 22:1129–1136. [DOI] [PubMed] [Google Scholar]
- Trachana K, et al. 2011. Orthology prediction methods: a quality assessment using curated protein families. Bioessays 33:769–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachana K, et al. 2014. A phylogeny-based benchmarking test for orthology inference reveals the limitations of function-based validation. PLoS One 9:e111122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treangen TJ, Rocha EP. 2011. Horizontal transfer, not duplication, drives the expansion of protein families in prokaryotes. PLoS Genet. 7: e1001284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, et al. 2009. Rosid radiation and the rapid rise of angiosperm-dominated forests. Proc Natl Acad Sci U S A. 106:3853–3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan S, Lio P, Goldman N. 2001. Molecular phylogenetics: state-of-the-art methods for looking into the past. Trends Genet. 17:262–272. [DOI] [PubMed] [Google Scholar]
- Woese CR, Fox GE. 1977. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci U S A. 74:5088–5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf YI, Rogozin IB, Grishin NV, Tatusov RL, Koonin EV. 2001. Genome trees constructed using five different approaches suggest new major bacterial clades. BMC Evol Biol. 1:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz P, et al. 2013. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 42:D643–D648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, et al. 2014. Resolution of deep angiosperm phylogeny using conserved nuclear genes and estimates of early divergence times. Nat Commun. 5:4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, et al. 2011. Phylogenomic analysis resolves the interordinal relationships and rapid diversification of the laurasiatherian mammals. Syst Biol. 61:150–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.