Abstract
More than 2 decades ago, experiments on the antiviral mechanisms of interferons led to the discovery of Janus kinases (JAK) and their downstream effectors, the signal transducer of activation (STAT) proteins. This pathway has since become a paradigm for membrane to nucleus signaling and has come to explain how a broad range of soluble factors, including cytokines and hormones, mediate their diverse functions. Jak/STAT research has not only impacted basic science, particularly in the context of intercellular communication and cell-extrinsic control of gene expression, but has also become a prototype for transition from bench to bedside, culminating in the development and clinical implementation of pathway-specific therapeutics. This brief review will synthesize current understanding of Jak/STAT biology while taking stock of the lessons learned and the challenges that lie ahead.
Introduction
Metazoans, and even unicellular aggregates, must coordinate the diverse activities of cellular hordes to achieve collective goals. This imperative underlies two fundamental questions in biology - how do individual cells sense external cues and, in turn, how are these external cues interpreted to program behavior? Evolution has provided a number of elegant solutions and, among these, the Jak/STAT pathway has emerged a paradigm, as evidenced by its conservation across phylogeny from vertebrates to lower organisms like Dictyostelium (1). Not surprisingly, the study of this ancient pathway has yielded fundamental insights about cellular communication and the role of membrane to nucleus signaling in controlling gene expression. It has also shaped our understanding of the mammalian immune system, perhaps the most intricate of all cellular networks, and provided both the impetus and rationale for pathway-specific therapeutics. This brief review will discuss current theories on how Jak/STAT signaling works at the cellular, molecular and genomic levels, with particular emphasis on human disease. Our aim is not to be expansive but, rather, to highlight emerging ideas and to present a modern view of Jak/STAT research as it strides towards into its third decade.
Specificity and redundancy in Jak/STAT signaling
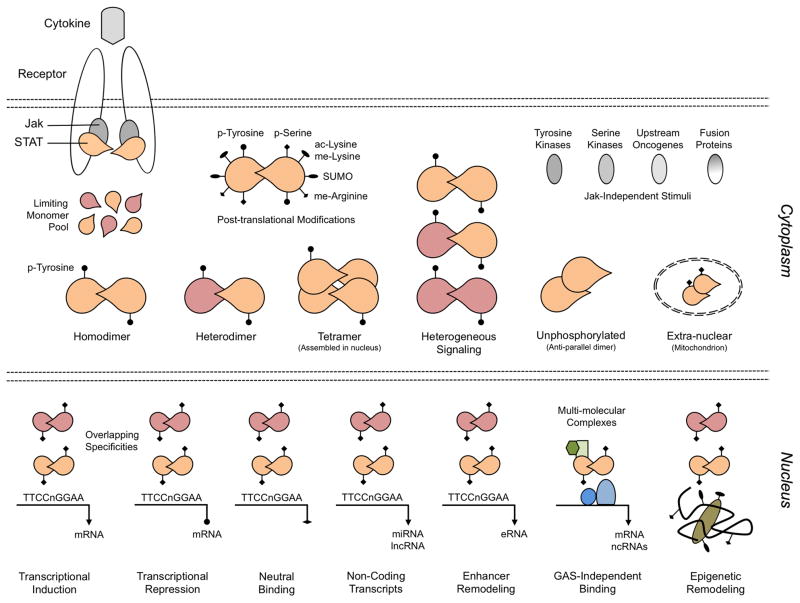
The biochemistry of Jak/STAT signaling has been studied extensively (2). Briefly, signaling begins with extracellular association of cytokines or growth factors with their corresponding transmembrane receptors. This facilitates trans-activation of receptor-bound Janus kinases (Jaks) by putting them in spacial proximity and by prompting conformational changes that distance their kinase domains from inhibitory pseudokinase domains (3). Activated Jaks then phosphorylate latent STAT monomers, leading to dimerization, nuclear translocation and DNA binding (Figure 1). In mammals, 4 Jaks (JAK1, JAK2, JAK3, TYK2) and 7 STATs (STAT1, STAT2, STAT3, STAT4, STAT5a, STAT5b, STAT6) are employed by more than 50 cytokines and growth factors, raising the question of how specificity is achieved with so few building blocks. One explanation is that specificity is lineage-dependent, meaning that cytokines which activate the same STATs may operate on different cell types (or states), each with distinct panels of STAT-sensitive genes. However, this does not explain why cytokines that ‘look’ similar in terms of STAT signaling cascades can have different outcomes within the same cell type or state. A classic example of this dichotomy is the case of IL-6 and IL-10; both are potent STAT3 activators in myeloid cells, but the former exerts mostly pro-inflammatory effects while the latter is mostly anti-inflammatory. There is evidence that qualitative differences in the duration and or intensity of STAT3 signaling underlie the disparity, but this may not be the whole story (4). Another point to consider is that, while each cytokine is traditionally associated with a particular STAT, almost all engage more than one family member to varying degrees (i.e. heterogeneous signaling; Figure 1). For example, type I interferons are prototypical STAT1 activators but also activate STAT3 and STAT4 (5). In some instances, cytokines engage multiple STATs with comparable potency, as with IL-27, a strong activator of both STAT1 and STAT3 (6), while in others there is clear hierarchy, as with IFN-γ, which elicits a strong STAT1 response coupled to a relatively weak STAT3 response (7). This heterogeneity is philosophically satisfying because graded combinations of STATs may contribute to cytokine specificity, though it is also unlikely that such quantitative differences account for all of it.
Figure 1.
Canonical Jak/STAT signaling begins with the association of cytokines and their corresponding transmembrane receptors. This brings Jaks in proximity, leading to phosphorylation of both the Jaks themselves and the cytoplasmic tails of the receptors, creating requisite docking sites for latent STAT monomers. STAT tyrosine phosphorylation (p-Tyr) then proceeds as the major activating event, leading to dimerization, translocation, DNA binding and target gene induction. Non-canonical deviations from this model should also be considered. First, p-Tyr can occur via Jak-independent pathways and, in fact, is not required for some activities (unphosphorylated STATs). Additional post-translational modifications (e.g. serine phosphorylation) can also influence STATs. Next, although each cytokine/receptor is typically associated with a particular STAT, most engage more than one family member (heterogeneous signaling), leading to the formation of not only homodimers but also heterodimers and higher order tetramers. These migrate to the nucleus where they bind to DNA not only through consensus GAS elements, but also through GAS-independent mechanisms, leading to induction, repression or neutral binding of protein-coding (mRNAs) or non-coding (miRNAs and lncRNAs) target genes, typically as part of multi-molecular complexes. Due to a shared affinity for GAS motifs, multiple STATs may bind to the same target sites (overlapping specificities). They can also bind to and promote remodeling of gene enhancers and can regulate the epigenetic status of target genes. Lastly, extra-nuclear functions have increasingly been recognized, most notably in the mitochondrion.
It is well established that STATs can bind directly to DNA, thus acting as classical transcription factors (TFs). Exactly where and how they bind are matters of ongoing debate. Traditionally, these questions have been addressed using low-throughput, candidate-driven approaches but, with the advent of next-generation sequencing, it is now possible to tackle them on a genome-wide scale. Most notably, deep sequencing can be coupled with chromatin immunoprecipitation (ChIP-seq) to produce comprehensive, unbiased maps of STAT-DNA binding which can then be integrated with transcriptomics and loss-of-function studies (i.e. ‘knock-out’ mice, siRNA) to catalogue target genes subject to both STAT occupancy and STAT-dependent transcriptional regulation (8). This methodology was first applied to STAT1 and has since been used to interrogate each family member (10). The emerging theme is that STATs disperse throughout the genome and regulate transcription of both protein-coding and non-coding genes (9)(Figure 1).
Genome-wide analyses have yielded a number of surprises about STAT biology. First is the pervasive nature of STAT binding. In helper T cells, thousands of binding sites have been mapped for each family member, sometimes proximal to transcriptional start sites, as would have been predicted, but more often distal, typically associated with enhancers or other cis regulatory elements (9). Many of these binding events are not associated with consensus DNA motifs (i.e. GAS elements), which suggests either degeneracy in the recognition code or alternative targeting strategies (10), and many do not correlate to changes in gene expression (Figure 1)(11, 12). This latter phenomenon, sometimes referred to as ‘neutral’ or ‘opportunistic’ binding, is typically under-emphasized in genome-wide profiles. Another unexpected finding is the high degree of overlap between different STATs (i.e. overlapping specificities; Figure 1)(13). It is now apparent that many sites (and or genes) can be bound by multiple family members, though it should also be noted that each STAT does have non-redundant functions, which implies that unique targets are still highly relevant.
It has long been known that all STATs recognize the same DNA sequence, known as the GAS motif. There is some selectivity in the core nucleotides preferred by each family member - and STAT6 prefers an extra spacer nucleotide – but all STATs can engage each other’s ‘optimal’ binding sites, albeit with variable affinities (7, 14). This provides a tidy explanation for the genomic overlap between individual STATs and, in turn, for why some genes are regulated by multiple family members. A singular consensus motif has also led to the idea that STATs may antagonize one another by competing for access to the same genomic locales. This concept has been validated for STAT3 and STAT5 - first in T cells, where STAT3-driven IL-17 transcription is blocked by STAT5, and later in dendritic cells and cancer cell lines (15–17). Further studies are needed to determine whether other STATs engage in this type of head-to-head competition and the question of how STAT-mediated transcriptional inhibition works, whether it reflects direct regulation of the loci themselves or induction of secondary agents (i.e. inhibitory TFs, miRNAs), is also pressing.
It is undeniable that STATs are critical determinants of cytokine specificity and function. However, STAT-independent signals emanating from cytokine receptors and or parallel stimuli also contribute, in part, by influencing STAT responses. Recent examples include ITAM-bearing receptors, which modulate STAT1 signaling, and TRAF-activating receptors, which modulate STAT3 signaling (18, 19).
STATs set the stage for transcription
An important application of ChIP-seq is to correlate TF binding with epigenetics. These two phenomena are intimately linked because, on one hand, TFs are beholden to the epigenetic structure of their target genes while, on the other, TFs can elicit toggling between ‘open’ and ‘closed’ states, which suggests that they can also be upstream of epigenetic changes (20, 21). Echoing work from other model systems, genome-wide analyses in T cells have shown that STATs are both regulators of and regulated by the epigenetic landscape (Figure 1). These studies have not only affirmed that STAT signaling can drive widespread epigenetic changes - which had previously been demonstrated for only a limited set loci (e.g. Ifng, Il4/Il13) - but have also provided an epigenetic basis for T cell lineage commitment and plasticity (8, 9, 21). It is also notable that STAT may differ in the way that they influence the epigenetic landscape. For instance, while STAT4 and STAT6 each create ‘open’ chromatin at target loci, the former does so mainly by promoting permissive marks and the latter by limiting repressive marks (11). The exact mechanism by which STATs elicit these changes, whether by recruitment and or transcriptional regulation of the epigenetic machinery, are currently under investigation. However, a direct link between STATs and repressive marks has recently been described in B cells where STAT5 was shown to recruit the methyltransferase, EZH2, thereby guiding deposition of H3K27me3 at silenced loci (22).
As with epigenetic marks, STAT binding profiles can be integrated with those of other TFs to identify regulatory networks. The key idea emerging from these studies is that STATs are core elements within multi-molecular complexes that congregate at transcriptionally relevant DNA elements, such as promoters or enhancers (Figure 1). Recent examples come from the field of stem cell biology, where STAT3 was shown to be involved in a pluripotency network together with Oct4, Sox2 and Smad1 (23), and from immunology where STAT3 was shown to be involved in a network together with IRF4, BATF and RORγt (24). Broadly speaking, STATs participate in at least 3 types of networks in immune cells: 1) those involved in the general transcriptional or epigenetic machinery, with members including RNA polymerase, p300, methyltransferases and or acetyltransferases (25, 26), 2) those involved in general inflammatory functions, with members including AP-1, IRF and or NF-κB family TFs (24, 25, 27), and 3) those involved in lineage commitment, with members including ‘master’ TFs that are critical for lineage specification. This latter group has been studied extensively in T cells where it is known that STATs bind near (and sometimes physically associate with) ‘master’ TFs, like T-bet, GATA-3, ROR_t and FoxP3. An outstanding question is whether STATs are the pioneers in these multi-molecular complexes or just essential components. The answer is likely to be context dependent, varying from gene-to-gene and across cell types, but one study argues that, in helper T cells, IRF4 precedes and is required for STAT3 binding to target genes, making the former a pioneer and the latter a secondary recruit (28).
STATs and long distance relationships
It has long been known that cis regulatory elements can have profound influence on gene expression. However, only with the advent of next-generation sequencing technologies has it become possible to enumerate these on a genome-wide scale. Among the most widely studied regulatory elements are enhancers, which are critical for expression of distally associated genes, and can be mapped using a combination of epigenetic (e.g. K27ac) and TF (e.g. p300) binding profiles (20). This approach has revealed that global distribution of active enhancers is extensively remodeled during cellular differentiation, thus presenting a defining characteristic, or fingerprint, of lineage commitment (29). Moreover, regions with dense enhancer clusters, termed ‘super’ or ‘stretch’ enhancers, tend to decorate lineage-defining genes, thereby making areas of the genome that determine cell identity and which, therefore, must be tightly controlled at the transcriptional level (30, 31). The role of STATs in assembling T cell enhancers has now been investigated on a genome-wide scale. These studies confirm the long-standing idea that lineage-specific loci, including Ifng, Il4/Il13 and Il17a/Il17f, are regulated by STAT-bound enhancers, and further demonstrate that STAT-dependent enhancer remodeling is pervasive, affecting thousands of sites throughout the genome (Figure 1). In many cases, these remodeling events were shown to be independent of ‘master’ TFs, thus positioning STATs as upstream mediators of lineage commitment and cell identity (32).
Inherent to next generation sequencing is the ability to probe genomic regions that are ignored by predictive approaches, including ‘gene deserts’ far from protein coding genes. This capacity has enabled identification of hundreds of cytokine-responsive microRNAs (miRNAs) and long non-coding RNAs (lncRNAs), many of which can be traced back to STAT-bound loci (Figure 1)(33, 34). In fact, recent work has shown that STATs directly control transcription of lncRNAs (33). One high-profile example is NEST (Tmevpg1), a STAT4-regulated lncRNA that impacts T cell responses (35, 36). It is also noteworthy that STATs can be the targets of miRNAs and lncRNAs. Several miRNAs, including miR-17 and miR-145, and at least one lncRNA, lnc-DC, are known to directly regulate STATs (37, 38).
Non-canonical aspects of Jak/STAT signaling
Although Jak/STAT signaling is typically presented as a simple pathway, decades of research have revealed it to be full of intricacy. For instance, though it is widely held that STATs act only as homodimers, they are also known to form heterodimers and higher order tetramers (Figure 1). In fact, classic studies demonstrated that a key difference between type I and type II interferons is that the former elicits STAT1/STAT2 heterodimers (ISGF3 complex), while the latter elicits mostly STAT1 homodimers (GAF complex)(2). Other examples have subsequently been reported, such as STAT1/STAT3 and STAT1/STAT4 heterodimers, but functional relevance remains largely unexplored (7, 39). STAT tetramers are also poorly understood. Unlike dimers, which are assembled in the cytoplasm, the bulk of experimental evidence suggests that tetramerization occurs in the nucleus subsequent to the binding of 2 STAT dimers to adjacent (or tandem) DNA elements. In terms of function, those involving STAT5 have been most extensively studied and are thought to be critical for optimal transcriptional activity (40). STAT1 tetramerization has also been investigated, with recent work demonstrating that it is important for type II interferons but dispensable for type I interferons (41). Another common misconception is that the cytoplasmic pool of latent STATs is kept at saturating density. In reality, expression of STAT proteins is tightly regulated and oscillating concentrations have important consequences, as seen in NK cells, where relative levels of STAT1 and STAT4 determine the response to type I interferons (Figure 1)(42). Consistent with this latter point, STAT loci harbor ‘super’ or ‘stretch’ enhancers, which suggests a high degree of transcriptional control (30).
The canonical model of Jak/STAT signaling posits that STATs are triggered by Jak-dependent tyrosine phosphorylation (p-Tyr). This is clearly the dominant route downstream of cytokine receptors but the obligatory role of Jaks has been challenged in other settings. Jak-independent p-Tyr has been linked to the STAT hyperactivity seen in many cancers (discussed bellow) and is also thought to play a role in normal physiological processes (Figure 1). For instance, some receptor tyrosine kinases (e.g. Flt-3 receptor) can elicit STAT5 activation without involving Jaks (43). Additionally, the nucleic acid sensor, STING, has been shown to invoke Jak-independent p-Tyr of STAT6, thereby eliciting host protective antiviral responses, and the pyruvate kinase M2 has been shown to invoke Jak-independent p-Tyr of STAT3 in response to metabolic cues (44, 45). The central role of p-Tyr itself, whether Jak-dependent or not, has also been scrutinized and a growing number of functions have been ascribed to ‘unphosphorylated’ STATs (Figure 1)(2). These are truly unconventional in that they do not require p-Tyr for nuclear translocation and or transcriptional activity, and because they form distinct anti-parallel dimers (2).
Beyond p-Tyr, several post-translational modifications can influence STAT function (Figure 1). The first to be recognized was serine phosphorylation (p-Ser), which regulates a variety of STAT activities, including DNA binding and interaction with accessory proteins. All STATs are phosphorylated on at least one serine residue, often without concurrent p-Tyr, and numerous external signals can induce p-Ser. These, in turn, dictate the identity of the upstream kinases, which include Map Kinases, cyclin dependent kinases and IKK, among others (2, 46, 47). In vitro studies have established that p-Ser is important for transcriptional responses, as seen with STAT4-driven IFN-γ in T cells (48), while in vivo models have shown its impact on immune function and hematopoietic transformation (49–51).
Additional chemical modifications of STATs include acetylation, methylation and sumoylation (Figure 1)(2). These can be broadly divided into two categories: those that promote STAT function and those that limit STAT function. Lysine acetylation and arginine methylation can be placed in the first category; the former having been shown to promote STAT function through effects on dimerization and transcriptional activity, and the latter having been shown to block association with PIAS proteins, a well known class of STAT inhibitors. However, it should also be noted that the mechanistic basis and biological significance of these findings has been disputed (52, 53). It is generally agreed that lysine methylation plays an important role in STAT function, although there is some discord about whether it is a positive or negative regulatory event (54, 55). To date, sumoylation has been exclusively regarded as a negative regulator, having been shown to limit the function of STAT1 and STAT5 (56, 57).
Another dogma that has been challenged is that STATs are active only in the nucleus. Recent studies have shown that STAT3 can localize to the mitochondrion, where it promotes oxidative phosphorylation and membrane permeability (Figure 1). This effect is dependent on serine but not tyrosine phosphorylation and is thought to be relevant in settings where cellular respiration is altered, such as cellular stress and cancer (58). All other STATs (save STAT4) have now been detected in the mitochondrion but their function within this organelle has not been rigorously assessed (58). Another intriguing ‘geographical’ deviation involves Jak2, which has been shown to mediate p-Tyr of nuclear histones (59).
Jaks and STATs in immunity and disease
The Jak/STAT pathway is critical for meeting the diverse challenges faced by the immune system, from resisting infection to maintaining immune tolerance, enforcing barrier functions and guarding against cancer. However, this capacity comes at a steep price. Errant immune responses can inflict great bodily harm and, not surprisingly, exaggerated or protracted Jak/STAT signaling has been implicated in just about every type of autoimmune disease. Therefore, a delicate balance must be reached to allow for both efficient induction of the Jak/STAT pathway when the immune system is called to action, and proper diminution when the instigating threat subsides. The following is a brief overview of how Jak/STAT dysfunction impacts human disease. For a more extensive discussion of this topic, along with tables summarizing the role of each Jak/STAT component, we refer to our previous reviews (8, 60).
The broad in vivo relevance of the Jak/STAT pathway first became apparent with the identification monogenic diseases resulting from germline mutations of Jak/STAT signaling components and associated cytokines/receptors. These rare ‘experiments of nature’ typically exhibit Mendelian inheritance patterns and dramatic phenotypes, including immunodeficiency, susceptibility to infection and autoimmunity, thus bringing them to the attention of geneticists and immunologists alike (61).
A striking example of monogenic disease is the ‘bubble boy’ syndrome (X-linked severe combined immunodeficiency). These patients suffer from extreme pathogen susceptibility due to a lack of T and NK cells caused by loss-of-function variants of the common γ chain (62). Based on in vitro studies demonstrating a selective role for Jak3 in common γ chain signal transduction, it was discovered that a subset of SCID patients harbor mutations in this kinase, thus providing the first demonstration of non-redundant in vivo functions for any Jak/STAT component (63, 64). Analogous phenotypes in IL-7/IL-7R-deficient humans and STAT5 deficient mice have since confirmed the involvement of the Jak3/STAT5 axis, though it also bears noting that humans with mutations of STAT5B are not immunodeficient but, instead, develop autoimmunity due, in large part, to a defect in T regulatory cells (61).
Following the discovery of Jak3-SCID, a number of additional Mendelian disorders involving Jak/Stat components have been reported. These findings emphasize the central role of Jak/STAT signaling in resistance to infection and, in fact, heritable susceptibility has been key to identifying these mutant alleles. All of the known STAT loss-of-function mutations can be associated with particular types of pathogens. Patients with STAT1 mutations are prone to mycobacterial and viral infections, those with STAT2 mutations are prone to viral infections and those with STAT3 mutations are prone to fungal infections (60). Gain-of-function STAT1 mutations are also prone to fungal infections but, in this case, hyperactive STAT1 appears to limit STAT3-driven anti-fungal responses (61). In general, STAT deficiency phenotypes are mirrored in patients with mutations of upstream/downstream mediators, though at least one exception should be considered; those harboring loss-of-function alleles of IL-10/IL-10R, which is upstream of STAT3, are not susceptible to fungi but, instead, present with inflammatory bowel disease (IBD)(61). They also do not develop many of the clinical symptoms seen in patients with hypomorphic STAT3 mutations, such as hyper IgE syndrome and developmental abnormalities (i.e. Job’s Syndrome). A similar dichotomy is evident in mice; IL-10- and IL-10R-deficient animals develop colitis while those bearing a hypomorphic STAT3 allele common in HIES patients do not (65). Adding further complexity, the recent identification of patients with gain-of-function STAT3 mutations highlights the central role of this pathway in promoting autoimmune disease. These individuals develop a spectrum of clinical symptoms, most notably type 1 diabetes, which is consistent with numerous studies demonstrating that STAT3 can drive pathological inflammation in mice (66).
Next-generation sequencing has led to an explosion of genome-wide association studies (GWAS) where disease phenotypes are linked to single nucleotide polymorphisms (SNPs) in affected populations (67). Not surprisingly, GWAS have implicated SNPs within STAT genes in numerous common diseases. Salient examples of this growing list include STAT3 SNPs associated with Crohn’s disease, psoriasis and ankylosing spondylitis, STAT4 SNPs associated with rheumatoid arthritis, Crohn’s disease, systemic lupus erythematosus and Sjogren’s syndrome, and STAT6 SNPs associated with asthma (8, 68). As with monogenic diseases, SNPs found in Jaks and upstream cytokines/receptors support the central role of STATs in many of these disorders.
Over the past 20 years, the link between Jak/STAT signaling and oncogenesis has become a major thread in cancer biology. It has long been known that STAT hyperactivity can drive cellular transformation downstream of classic oncogenic signals like BCR-ABL, Ras and Src (69, 70). This hyperactivity, which typically involves STAT3 and or STAT5, is now considered a defining characteristic of most solid and blood cancers (69). In addition, somatic mutations of the Jak kinases themselves have increasingly been recognized as primary oncogenic lesions. One landmark discovery was the finding that gain-of-function JAK2 mutations underlie myeloproliferative malignancies (71). This work has been informative not only for disease etiology but also in the context of Jak structure - many of the mutations are found in the pseudo-kinase domain, which has recently been shown to have catalytic activity and to limit ‘auto-activation’ of Jaks prior to receptor oligomerization (72). JAK2 fusion proteins have also been implicated in hematological malignancies, as have JAK3 mutations (60, 71). Despite notable exceptions, few mutant STAT alleles are associated with carcinogenesis, which suggests that their oncogenic potential is largely secondary to upstream events.
Aside from cell intrinsic effects, JAK/STAT signals can influence the tumor microenvironment via extrinsic pathways. Again, STAT3 appear to be a principal culprit, with numerous studies showing that it can limit anti-tumor immunity. Among its subversive activities are the ability to promote myeloid-derived suppressor cells and the ability to mediate the signaling of IL-23, a cytokine that has recently been established as an important pro-cancer agent. On the other end of the spectrum is STAT1, which is both a critical cell-intrinsic anti-cancer signal and an important cell extrinsic mediator of immunosurveillance (70).
Therapeutic targeting of STAT signaling – that’s a fact Jak!
Given the data implicating Jak/STAT in autoimmune disease and malignancy, it is not surprising that this pathway has become an attractive target for pharmaceuticals. Ruxolitinib, a JAK1 and JAK2 inhibitor with efficacy for the treatment of polycythemia vera and myelofibrosis, was the first FDA-approved Jak inhibitor, or Jakinib. The second was Tofacitinib, which blocks JAK3, and to a lesser extent JAK1 and Jak2 (73). First approved for rheumatoid arthritis, it is now in clinical trials for a range of maladies including psoriasis, IBD, transplant rejection, juvenile arthritis and spondyloarthropathy, among others (60)(https://www.clinicaltrials.gov/ct2/results?term=Tofacitinib&Search=Search). A third Jak inhibitor, Oclacitinib, is now approved for dermatitis in dogs (74), and a number of additional Jakinibs, including ‘second generation’ versions with improved potency and selectivity, are currently under development.
Unlike Jaks, whose kinase domains are a clear pharmacological target, STATs do not have catalytic activity and, thus, present a more challenging objective. Oligonucleotide-based STAT inhibitors, which presumably sequester STATs away from ‘dangerous’ loci, are the most well-developed approach and are currently undergoing human testing for a number of malignancies (75). Alternative strategies include small molecule inhibitors, which are also in clinical trials (https://clinicaltrials.gov/ct2/results?term=STAT3&Search=Search), and STAT-binding ‘intrabodies’, which have been investigated only in animal models (76). STATs can also be impacted by drugs that are not pathway-specific, such as pyrimethamine, curcumin and platinum-based drugs, though it should be noted that their effects are likely to be indirect, perhaps reflecting reductions availability of STAT stimuli rather than biochemical effects on the STATs themselves.
Conclusions
The Jak/STAT pathway is a fascinating case study of how basic science can be parlayed into clinical gains, first by strengthening our understanding of human disease, then by informing the development of targeted therapeutics. Beyond these pragmatic ends, the study of Jak/STAT signaling continues to yield transformative insights about the nature of cellular communication and the regulation of gene expression. Befitting its ascendant status, the impact of Jak/STAT is now felt beyond immunology and onto a number of emerging fields, including stem cell biology and metabolomics, thus setting the stage for the next phase of research to meet the lofty standard set in the past 2 decades.
Acknowledgments
We regret that, due to space constraints, many relevant publications were omitted from this review. J.J.O’S. and the NIH hold patents related to therapeutic targeting of JAKs and have a Collaborative Research Agreement and Development Award with Pfizer.
References
- 1.Wang Y, Levy DE. Comparative evolutionary genomics of the STAT family of transcription factors. JAKSTAT. 2012;1:23–33. doi: 10.4161/jkst.19418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stark GR, Darnell JE., Jr The JAK-STAT pathway at twenty. Immunity. 2012;36:503–514. doi: 10.1016/j.immuni.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brooks AJ, Dai W, O’Mara ML, Abankwa D, Chhabra Y, Pelekanos RA, Gardon O, Tunny KA, Blucher KM, Morton CJ, Parker MW, Sierecki E, Gambin Y, Gomez GA, Alexandrov K, Wilson IA, Doxastakis M, Mark AE, Waters MJ. Mechanism of Activation of Protein Kinase JAK2 by the Growth Hormone Receptor. Science. 2014:344. doi: 10.1126/science.1249783. [DOI] [PubMed] [Google Scholar]
- 4.O’Shea JJ, Murray PJ. Cytokine Signaling Modules in Inflammatory Responses. Immunity. 2008;28:477–487. doi: 10.1016/j.immuni.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho SS, Bacon CM, Sudarshan C, Rees RC, Finbloom D, Pine R, O’Shea JJ. Activation of STAT4 by IL-12 and IFN-alpha: evidence for the involvement of ligand-induced tyrosine and serine phosphorylation. J Immunol. 1996;157:4781–4789. [PubMed] [Google Scholar]
- 6.Villarino AV, Hibbert L, Lieberman L, Wilson E, Mak T, Yoshida H, Kastelein RA, Saris C, Hunter CA. The IL-27R (WSX-1) is required to suppress T cell hyperactivity during infection. Immunity. 2003;19:645–655. doi: 10.1016/s1074-7613(03)00300-5. [DOI] [PubMed] [Google Scholar]
- 7.Seidel HM, Milocco LH, Lamb P, Darnell JE, Jr, Stein RB, Rosen J. Spacing of palindromic half sites as a determinant of selective STAT (signal transducers and activators of transcription) DNA binding and transcriptional activity. PNAS. 1995;92:3041–3045. doi: 10.1073/pnas.92.7.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Shea JJ, Lahesmaa R, Vahedi G, Laurence A, Kanno Y. Genomic views of STAT function in CD4+ T helper cell differentiation. Nature Rev Immunol. 2011;11:239–250. doi: 10.1038/nri2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanno Y, Vahedi G, Hirahara K, Singleton K, O’Shea JJ. Transcriptional and Epigenetic Control of T Helper Cell Specification: Molecular Mechanisms Underlying Commitment and Plasticity. Annu Rev Immunol. 2012;30:707–731. doi: 10.1146/annurev-immunol-020711-075058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Langlais D, Couture C, Balsalobre A, Drouin J. The Stat3/GR Interaction Code: Predictive Value of Direct/Indirect DNA Recruitment for Transcription Outcome. Mol Cell. 2012:1–12. doi: 10.1016/j.molcel.2012.04.021. [DOI] [PubMed] [Google Scholar]
- 11.Wei L, Vahedi G, Sun HW, Watford WT, Takatori H, Ramos HL, Takahashi H, Liang J, Gutierrez-Cruz G, Zang C, Peng W, O’Shea JJ, Kanno Y. Discrete Roles of STAT4 and STAT6 Transcription Factors in Tuning Epigenetic Modifications and Transcription during T Helper Cell Differentiation. Immunity. 2010;32:840–851. doi: 10.1016/j.immuni.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu BM, Kang K, Yu JH, Chen W, Smith HE, Lee D, Sun HW, Wei L, Hennighausen L. Genome-wide analyses reveal the extent of opportunistic STAT5 binding that does not yield transcriptional activation of neighboring genes. Nucleic Acids Res. 2012;40:4461–4472. doi: 10.1093/nar/gks056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang K, Robinson GW, Hennighausen L. Comprehensive meta-analysis of Signal Transducers and Activators of Transcription (STAT) genomic binding patterns discerns cell-specific cis-regulatory modules. BMC Genomics. 2013;14:4. doi: 10.1186/1471-2164-14-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Decker T, Kovarik P, Meinke A. GAS elements: a few nucleotides with a major impact on cytokine-induced gene expression. J Interferon Cyt Res. 1997;17:121–134. doi: 10.1089/jir.1997.17.121. [DOI] [PubMed] [Google Scholar]
- 15.Walker SR, Nelson EA, Yeh JE, Pinello L, Yuan GC, Frank DA. STAT5 Outcompetes STAT3 To Regulate the Expression of the Oncogenic Transcriptional Modulator BCL6. Mol Cell Biol. 2013;33:2879–2890. doi: 10.1128/MCB.01620-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wan CK, Oh J, Li P, West EE, Wong EA, Andraski AB, Spolski R, Yu ZX, He J, Kelsall BL, Leonard WJ. The Cytokines IL-21 and GM-CSF Have Opposing Regulatory Roles in the Apoptosis of Conventional Dendritic Cells. Immunity. 2013;38:514–527. doi: 10.1016/j.immuni.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang XP, Ghoreschi K, Steward-Tharp SM, Rodriguez-Canales J, Zhu J, Grainger JR, Hirahara K, Sun HW, Wei L, Vahedi G, Kanno Y, O’Shea JJ, Laurence A. Opposing regulation of the locus encoding IL-17 through direct, reciprocal actions of STAT3 and STAT5. Nature Immunol. 2011:1–9. doi: 10.1038/ni.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bezbradica JS, Rosenstein RK, Demarco RA, Brodsky I, Medzhitov R. A role for the ITAM signaling module in specifying cytokine-receptor functions. Nature Immunol. 2014;15:333–342. doi: 10.1038/ni.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagashima H, Okuyama Y, Asao A, Kawabe T, Yamaki S, Nakano H, Croft M, Ishii N, So T. The adaptor TRAF5 limits the differentiation of inflammatory CD4(+) T cells by antagonizing signaling via the receptor for IL-6. Nature Immunol. 2014;15:449–456. doi: 10.1038/ni.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nature Rev Genetics. 2010;12:7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 21.Wilson CB, Rowell E, Sekimata M. Epigenetic control of T-helper-cell differentiation. Nature Rev Immunol. 2009;9:91–105. doi: 10.1038/nri2487. [DOI] [PubMed] [Google Scholar]
- 22.Mandal M, Powers SE, Maienschein-Cline M, Bartom ET, Hamel KM, Kee BL, Dinner AR, Clark MR. Epigenetic repression of the Igk locus by STAT5-mediated recruitment of the histone methyltransferase Ezh2. Nature Immunol. 2011;12:1212–1220. doi: 10.1038/ni.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, Loh YH, Yeo HC, Yeo ZX, Narang V, Govindarajan KR, Leong B, Shahab A, Ruan Y, Bourque G, Sung WK, Clarke ND, Wei CL, Ng HH. Integration of External Signaling Pathways with the Core Transcriptional Network in Embryonic Stem Cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 24.Ciofani M, Madar A, Galan C, Sellars M, Mace K, Pauli F, Agarwal A, Huang W, Parkurst CN, Muratet M, Newberry KM, Meadows S, Greenfield A, Yang Y, Jain P, Kirigin FK, Birchmeier C, Wagner EF, Murphy KM, Myers RM, Bonneau R, Littman DR. A Validated Regulatory Network for Th17 Cell Specification. Cell. 2012;151:289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farlik M, Reutterer B, Schindler C, Greten F, Vogl C, Müller M, Decker T. Nonconventional Initiation Complex Assembly by STAT and NF-κB Transcription Factors Regulates Nitric Oxide Synthase Expression. Immunity. 2010;33:25–34. doi: 10.1016/j.immuni.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Korzus E, Torchia J, Rose DW, Xu L, Kurokawa R, McInerney EM, Mullen TM, Glass CK, Rosenfeld MG. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 27.Qiao Y, Giannopoulou EG, Chan CH, Park SH, Gong S, Chen J, Hu X, Elemento O, Ivashkiv LB. Synergistic Activation of Inflammatory Cytokine Genes by Interferon-gamma-Induced Chromatin Remodeling and Toll-like Receptor Signaling. Immunity. 2013;39:454–469. doi: 10.1016/j.immuni.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwon H, Thierry-Mieg D, Thierry-Mieg J, Kim HP, Oh J, Tunyaplin C, Carotta S, Donovan CE, Goldman ML, Tailor P, Ozato K, Levy DE, Nutt SL, Calame K, Leonard WJ. Analysis of Interleukin-21-Induced Prdm1 Gene Regulation Reveals Functional Cooperation of STAT3 and IRF4 Transcription Factors. Immunity. 2009;31:941–952. doi: 10.1016/j.immuni.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Natoli G. Maintaining Cell Identity through Global Control of Genomic Organization. Immunity. 2010;33:12–24. doi: 10.1016/j.immuni.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 30.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, Hoke HA, Young RA. Super-Enhancers in the Control of Cell Identity and Disease. Cell. 2013:1–28. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parker SCJ, Stitzel ML, Taylor DL, Orozco JM, Erdos MR, Akiyama JA, van Bueren KL, Chines PS, Narisu N, Black BL, Visel A, Pennacchio LA, Collins FS. Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. PNAS. 2013;110:17921–17926. doi: 10.1073/pnas.1317023110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vahedi G, Takahashi H, Nakayamada S, Sun HW, Sartorelli V, Kanno Y, O’Shea JJ. STATs Shape the Active Enhancer Landscape of T Cell Populations. Cell. 2012;151:981–993. doi: 10.1016/j.cell.2012.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu G, Tang Q, Sharma S, Yu F, Escobar TM, Muljo SA, Zhu J, Zhao K. Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation. Nature Immunol. 2013 doi: 10.1038/ni.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuchen S, Resch W, Yamane A, Kuo N, Li Z, Chakraborty T, Wei L, Laurence A, Yasuda T, Peng S, Hu-Li J, Lu K, Dubois W, Kitamura Y, Charles N, Sun HW, Muljo S, Schwartzberg PL, Paul WE, O’Shea JJ, Rajewsky K, Casellas R. Regulation of MicroRNA Expression and Abundance during Lymphopoiesis. Immunity. 2010;32:828–839. doi: 10.1016/j.immuni.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Collier SP, Collins PL, Williams CL, Boothby MR, Aune TM. Cutting Edge: Influence of Tmevpg1, a Long Intergenic Noncoding RNA, on the Expression of Ifng by Th1 Cells. J Immunol. 2012;189:2084–2088. doi: 10.4049/jimmunol.1200774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gomez JA, Wapinski OL, Yang YW, Bureau JF, Gopinath S, Monack DM, Chang HY, Brahic M, Kirkegaard K. The NeST Long ncRNA Controls Microbial Susceptibility and Epigenetic Activation of the Interferon-gamma Locus. Cell. 2013;152:743–754. doi: 10.1016/j.cell.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kohanbash G, Okada H. MicroRNAs and STAT interplay. Sem Cancer Biol. 2011:1–6. doi: 10.1016/j.semcancer.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, Jiang Z, Xu J, Liu Q, Cao X. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. 2014;344:310–313. doi: 10.1126/science.1251456. [DOI] [PubMed] [Google Scholar]
- 39.Collison LW, Delgoffe GM, Guy CS, Vignali KM, Chaturvedi V, Fairweather D, Satoskar AR, Garcia KC, Hunter CA, Drake CG, Murray PJ, Vignali DA. The composition and signaling of the IL-35 receptor are unconventional. Nature Immunol. 2012;13:290–299. doi: 10.1038/ni.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin JX, Li P, Liu D, Jin HT, He J, Rasheed MAU, Rochman Y, Wang L, Cui K, Liu C, Kelsall BL, Ahmed R, Leonard WJ. Critical Role of STAT5 Transcription Factor Tetramerization for Cytokine Responses and Normal Immune Function. Immunity. 2012;36:586–599. doi: 10.1016/j.immuni.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Begitt A, Droescher M, Meyer T, Schmid CD, Baker M, Antunes F, Knobeloch KP, Owen MR, Naumann R, Decker T, Vinkemeier U. STAT1-cooperative DNA binding distinguishes type 1 from type 2 interferon signaling. Nature Immunol. 2014;15:168–176. doi: 10.1038/ni.2794. [DOI] [PubMed] [Google Scholar]
- 42.Miyagi T, Gil MP, Wang X, Louten J, Chu WM, Biron CA. High basal STAT4 balanced by STAT1 induction to control type 1 interferon effects in natural killer cells. J Exp Med. 2007;204:2383–2396. doi: 10.1084/jem.20070401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang S, Fukuda S, Lee Y, Hangoc G, Cooper S, Spolski R, Leonard WJ, Broxmeyer HE. Essential role of signal transducer and activator of transcription (Stat)5a but not Stat5b for Flt3-dependent signaling. J Exp Med. 2000;192:719–728. doi: 10.1084/jem.192.5.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao X, Wang H, Yang JJ, Liu X, Liu ZR. Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Mol Cell. 2012;45:598–609. doi: 10.1016/j.molcel.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen H, Sun H, You F, Sun W, Zhou X, Chen L, Yang J, Wang Y, Tang H, Guan Y, Xia W, Gu J, Ishikawa H, Gutman D, Barber G, Qin Z, Jiang Z. Activation of STAT6 by STING is critical for antiviral innate immunity. Cell. 2011;147:436–446. doi: 10.1016/j.cell.2011.09.022. [DOI] [PubMed] [Google Scholar]
- 46.Bancerek J, Poss ZC, Steinparzer I, Sedlyarov V, Pfaffenwimmer T, Mikulic I, Dölken L, Strobl B, Müller M, Taatjes DJ, Kovarik P. CDK8 Kinase Phosphorylates Transcription Factor STAT1 to Selectively Regulate the Interferon Response. Immunity. 2013 doi: 10.1016/j.immuni.2012.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Decker T, Kovarik P. Serine phosphorylation of STATs. Oncogene. 2000;19:2628–2637. doi: 10.1038/sj.onc.1203481. [DOI] [PubMed] [Google Scholar]
- 48.Morinobu A, Gadina M, Strober W, Visconti R, Fornace A, Montagna C, Feldman GM, Nishikomori R, O’Shea JJ. STAT4 serine phosphorylation is critical for IL-12-induced IFN-gamma production but not for cell proliferation. PNAS. 2002;99:12281–12286. doi: 10.1073/pnas.182618999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shen Y, Schlessinger K, Zhu X, Meffre E, Quimby F, Levy DE, Darnell JE., Jr Essential role of STAT3 in postnatal survival and growth revealed by mice lacking STAT3 serine 727 phosphorylation. Mol Cell Biol. 2004;24:407–419. doi: 10.1128/MCB.24.1.407-419.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Varinou L, Ramsauer K, Karaghiosoff M, Kolbe T, Pfeffer K, Müller M, Decker T. Phosphorylation of the Stat1 transactivation domain is required for full-fledged IFN-gamma-dependent innate immunity. Immunity. 2003;19:793–802. doi: 10.1016/s1074-7613(03)00322-4. [DOI] [PubMed] [Google Scholar]
- 51.Friedbichler K, Kerenyi MA, Kovacic B, Li G, Hoelbl A, Yahiaoui S, Sexl V, Mullner EW, Fajmann S, Cerny-Reiterer S, Valent P, Beug H, Gouilleux F, Bunting KD, Moriggl R. Stat5a serine 725 and 779 phosphorylation is a prerequisite for hematopoietic transformation. Blood. 2010;116:1548–1558. doi: 10.1182/blood-2009-12-258913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meissner T, Krause E, Lödige I, Vinkemeier U. Arginine methylation of STAT1: a reassessment. Cell. 2004;119:587–9. doi: 10.1016/j.cell.2004.11.024. discussion 589–590. [DOI] [PubMed] [Google Scholar]
- 53.O’Shea JJ, Kanno Y, Chen X, Levy DE. Stat acetylation - a key facet of cytokine signaling? Science. 2005;307:217–218. doi: 10.1126/science.1108164. [DOI] [PubMed] [Google Scholar]
- 54.Kim E, Kim M, Woo DH, Shin Y, Shin J, Chang N, Oh YT, Kim H, Rheey J, Nakano I, Lee C, Joo KM, Rich JN, Nam DH, Lee J. Phosphorylation of EZH2 Activates STAT3 Signaling via STAT3 Methylation and Promotes Tumorigenicity of Glioblastoma Stem-like Cells. Cancer Cell. 2013;23:839–852. doi: 10.1016/j.ccr.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang J, Huang J, Dasgupta M, Sears N, Miyagi M, Wang B, Chance MR, Chen X, Du Y, Wang Y, An L, Wang Q, Lu T, Zhang X, Wang Z, Stark GR. Reversible methylation of promoter-bound STAT3 by histone-modifying enzymes. PNAS. 2010;107:21499–21504. doi: 10.1073/pnas.1016147107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Begitt A, Droescher M, Knobeloch KP, Vinkemeier U. SUMO conjugation of STAT1 protects cells from hyperresponsiveness to IFN. Blood. 2011;118:1002–1007. doi: 10.1182/blood-2011-04-347930. [DOI] [PubMed] [Google Scholar]
- 57.Van Nguyen T, Angkasekwinai P, Dou H, Lin F-M, Lu L-S, Cheng J, Chin YE, Dong C, Yeh ETH. SUMO-specific protease 1 is critical for early lymphoid development through regulation of STAT5 activation. Mol Cell. 2012;45:210–221. doi: 10.1016/j.molcel.2011.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meier JA, Larner AC. Toward a new STATe: The role of STATs in mitochondrial function. Sem Immunol. 2014:1–9. doi: 10.1016/j.smim.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dawson MA, Bannister AJ, Göttgens B, Foster SD, Bartke T, Green AR, Kouzarides T. JAK2 phosphorylates histone H3Y41 and excludes HP1alpha from chromatin. Nature. 2009;461:819–822. doi: 10.1038/nature08448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.O’Shea JJ, Holland SM, Staudt LM. JAKs and STATs in Immunity, Immunodeficiency, and Cancer. N Engl J Med. 2013;368:161–170. doi: 10.1056/NEJMra1202117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Casanova JL, Holland SM, Notarangelo LD. Inborn Errors of Human JAKs and STATs. Immunity. 2012;36:515–528. doi: 10.1016/j.immuni.2012.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Noguchi M, Yi H, Rosenblatt HM, Filipovich AH, Adelstein S, Modi WS, McBride OW, Leonard WJ. Interleukin-2 receptor gamma chain mutation results in X-linked severe combined immunodeficiency in humans. Cell. 1993;73:147–157. doi: 10.1016/0092-8674(93)90167-o. [DOI] [PubMed] [Google Scholar]
- 63.Macchi P, Villa A, Giliani S, Sacco MG, Frattini A, Porta F, Ugazio AG, Johnston JA, Candotti F, O’Shea JJ. Mutations of Jak-3 gene in patients with autosomal severe combined immune deficiency (SCID) Nature. 1995;377:65–68. doi: 10.1038/377065a0. [DOI] [PubMed] [Google Scholar]
- 64.Russell SM, Tayebi N, Nakajima H, Riedy MC, Roberts JL, Aman MJ, Migone TS, Noguchi M, Markert ML, Buckley RH, O’Shea JJ, Leonard WJ. Mutation of Jak3 in a patient with SCID: essential role of Jak3 in lymphoid development. Science. 1995;270:797–800. doi: 10.1126/science.270.5237.797. [DOI] [PubMed] [Google Scholar]
- 65.Steward-Tharp SM, Laurence A, Kanno Y, Kotlyar A, Villarino AV, Sciumè G, Kuchen S, Resch W, Wohlfert EA, Jiang K, Hirahara K, Vahedi G, Sun HW, Feigenbaum L, Milner JD, Holland SM, Casellas R, Powrie F, O’Shea JJ. A mouse model of HIES reveals pro- and anti-inflammatory functions of STAT3. Blood. 2014;123:2978–2987. doi: 10.1182/blood-2013-09-523167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Flanagan SE, Haapaniemi E, Russell MA, Caswell R, Lango Allen H, De Franco E, McDonald TJ, Rajala H, Ramelius A, Barton J, Heiskanen K, Heiskanen-Kosma T, Kajosaari M, Murphy NP, Milenkovic T, Seppänen M, Lernmark Å, Mustjoki S, Otonkoski T, Kere J, Morgan NG, Ellard S, Hattersley AT. Activating germline mutations in STAT3 cause early-onset multi-organ autoimmune disease. Nature Genetics. 2014;46:812–814. doi: 10.1038/ng.3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Manolio TA. Bringing genome-wide association findings into clinical use. Nature Rev Genetics. 2013;14:549–558. doi: 10.1038/nrg3523. [DOI] [PubMed] [Google Scholar]
- 68.Vercelli D. Discovering susceptibility genes for asthma and allergy. Nature Rev Immunol. 2008;8:169–182. doi: 10.1038/nri2257. [DOI] [PubMed] [Google Scholar]
- 69.Bromberg J. Stat proteins and oncogenesis. J Clin Invest. 2002;109:1139–1142. doi: 10.1172/JCI15617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Levine RL, Pardanani A, Tefferi A, Gilliland DG. Role of JAK2 in the pathogenesis and therapy of myeloproliferative disorders. Nat Rev Cancer. 2007;7:673–683. doi: 10.1038/nrc2210. [DOI] [PubMed] [Google Scholar]
- 72.Ungureanu D, Wu J, Pekkala T, Niranjan Y, Young C, Jensen ON, Xu CF, Neubert TA, Skoda RC, Hubbard SR, Silvennoinen O. The pseudokinase domain of JAK2 is a dual-specificity protein kinase that negatively regulates cytokine signaling. Nature Struct Mol Biol. 2011;18:971–976. doi: 10.1038/nsmb.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ghoreschi K, Laurence A, O’Shea JJ. Selectivity and therapeutic inhibition of kinases: to be or not to be? Nature Immunol. 2009;10:356–360. doi: 10.1038/ni.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cosgrove SB, Wren JA, Cleaver DM, Walsh KF, Follis SI, King VI, Tena JKS, Stegemann MR. A blinded, randomized, placebo-controlled trial of the efficacy and safety of the Janus kinase inhibitor oclacitinib (Apoquel®) in client-owned dogs with atopic dermatitis. Vet Dermatol. 2013;24:587–97. e141–2. doi: 10.1111/vde.12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sen M, Grandis JR. Nucleic acid-based approaches to STAT inhibition. JAKSTAT. 2012;1:285–291. doi: 10.4161/jkst.22312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Koo MY, Park J, Lim JM, Joo SY, Shin SP, Shim HB, Chung J, Kang D, Woo HA, Rhee SG. Selective inhibition of the function of tyrosine-phosphorylated STAT3 with a phosphorylation site-specific intrabody. PNAS. 2014;111:6269–6274. doi: 10.1073/pnas.1316815111. [DOI] [PMC free article] [PubMed] [Google Scholar]