Abstract
Protozoan mitochondrial tRNAs (mt-tRNAs) are repaired by a process known as 5'-editing. Mt-tRNA sequencing revealed organism-specific patterns of editing G-U base pairs, wherein some species remove G-U base pairs during 5'-editing, while others retain G-U pairs in the edited tRNA. We tested whether 3'-5' polymerases that catalyze the repair step of 5'-editing exhibit organism-specific preferences that explain the treatment of G-U base pairs. Biochemical and kinetic approaches revealed that a 3'-5' polymerase from A. castellanii tolerates G-U wobble pairs in editing substrates much more readily than several other enzymes, consistent with its biological pattern of editing.
Keywords: tRNA editing, mitochondrial tRNA, Thg1-like proteins, Acanthamoeba castellanii, Dictyostelium discoideum, 3'-5' polymerase
A family of 3'-5' polymerases has been discovered, members of which catalyze nucleotide addition in the reverse direction to all known DNA/RNA polymerases (1, 2). Based on sequence similarity, 3'-5' polymerases are further divided into two distinct sub-families: the tRNAHis guanylyltransferases (Thg1) and the Thg1-Like Proteins (TLPs) (3). Thg1 enzymes are strictly eukaryotic and their biological function, addition of an essential 5'-guanosine (G−1) to tRNAHis, is well-established (4-6). In contrast, TLPs, found in a limited number of bacteria but more widespread in archaea and protozoan eukaryotes, are less well-characterized (3, 7-11). However, recent data identified four TLPs as the first candidate mitochondrial-tRNA (mt-tRNA) 5'-editing enzymes (12, 13).
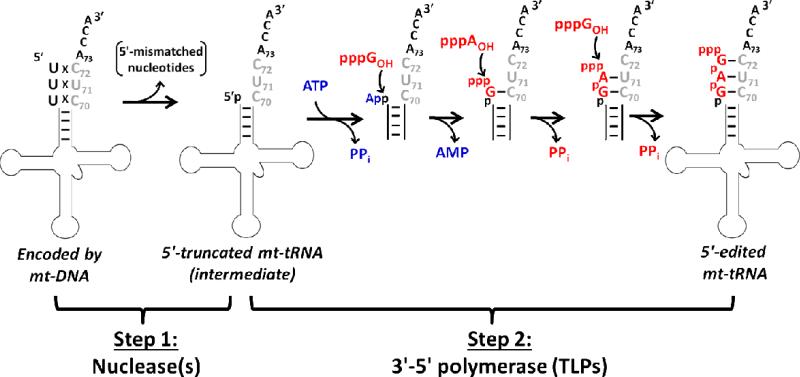
Mitochondrial 5'-editing was discovered in Acanthamoeba castellanii, but has since been demonstrated to occur in many eukaryotic protozoa, including Dictyostelium discoideum (14-20). During editing, mt-tRNA with mismatched nucleotides encoded in their aminoacyl-acceptor stems undergo a 2-step process that first removes 5'-mismatched nucleotides, followed by 5'-end repair of the resulting 5'-truncated tRNA (Figure 1) (3, 21, 22). The second step of the reaction requires a 3'-5' polymerase to extend the 5'-truncated RNA using the 3'-half of the acceptor stem as a template. However, no enzyme with this activity was known until bacterial TLPs were shown to catalyze a similar 5'-end repair activity with model tRNA substrates (9). Subsequently, two D. discoideum TLPs (DdiTLP3 and DdiTLP4) and two A. castellanii TLPs (AcaTLP1 and AcaTLP2) were each demonstrated to repair multiple 5'-truncated mt-tRNA in vitro, suggesting that these enzymes could catalyze 5'-editing (12, 13).
Figure 1. Mechanism of mitochondrial tRNA 5'-editing.
A representative example of mt-tRNA with encoded 5'-mismatches is repaired by 5'-editing. In the first step, mismatched nucleotides are removed from the 5'-end of the aminoacyl-acceptor stem by an unidentified nuclease(s), generating a 5'-truncated mt-tRNA. In Step 2, TLP enzymes repair the truncated 5'-end using their 3'-5' polymerase activity. For the first nucleotide addition to a 5'-monophosphorylated substrate, TLPs activate the 5'-end by adenylylation with ATP (shown in blue). Subsequently, incoming NTPs (shown in red) are selected according to their ability to form WC base pairs with the 3'-end template sequence and added to the growing 5'-end by attack of the NTP 3'-hydroxyl on the activated 5'-end, as indicated. After the first addition, the 5'-triphosphate of the previously added NTP serves as the activated end for nucleotide addition, releasing pyrophosphate as a consequence of the reaction. For intermediates generated during the 3'-5' polymerase reaction, only the aminoacyl-acceptor stem sequence is shown for clarity.
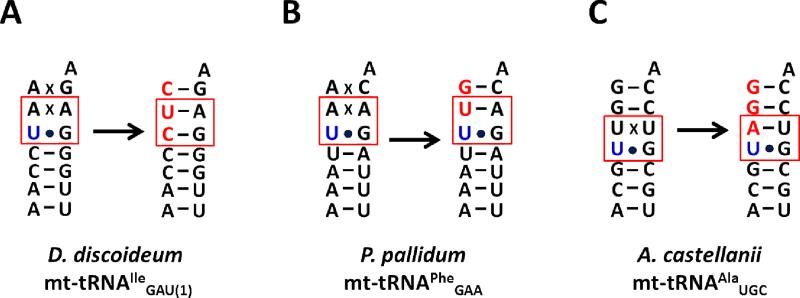
Investigation of mt-tRNA 5'-editing patterns revealed additional complexity in the treatment of G-U wobble base pairs. Direct sequencing of mt-tRNA from 8 different organisms revealed significant differences in the extent of G-U base pair editing (14, 16-19). For example, in D. discoideum a U3-G70 base pair in IleGAU(1) is 100% edited to a Watson-Crick (WC) C3-G70 base pair (Figure 2A) (14). However, editing of a U3-G70 base pair in an identical context in Polysphondylium pallidum is barely detected (in only 6% of sequences) (Figure 2B) (14). Other examples of partially edited or unedited G-U base pairs directly adjacent to edited base pairs are observed in Monoblepharella and Harpochytrium (19). In A. castellanii, editing of a U3-U70 mismatch in mt-tRNAAlaUGC occurs immediately adjacent to a U4-G69 base pair that is retained after editing (Figure 2C) (18). Thus, A. castellanii and D. discoideum exhibit disparate abilities to tolerate U-G base pairs in the context of editing substrates. This observation prompted us to use the four previously characterized TLPs (from A. castellanii and D. discoideum) to test whether organism-specific preferences of TLPs could explain the G-U editing patterns observed in vivo. We hypothesized that if a TLP efficiently extends a substrate mt-tRNA with a G-U base pair at its terminus, then the G-U base pair could remain part of the final edited mt-tRNA. In contrast, if the TLP does not efficiently repair these substrates, then the nucleotide involved in the G-U base pair would necessarily be removed (by the as of yet unidentified nuclease(s) prior to action of the TLP in repairing the 5'-end), resulting in replacement of G-U wobble base pairs with WC base pairs during editing.
Figure 2. Organism-specific outcomes for U-G base pairs during 5'-editing.
Aminoacyl-acceptor stem sequences of (U-G)-containing mt-tRNA that undergo 5'-editing are shown for three different eukaryotes. Genomically-encoded mt-tRNA sequences are shown on the left, and experimentally-determined sequences of the 5'-edited tRNA are shown to the right of each arrow. The U-G base pairs immediately adjacent to mismatches that undergo editing are highlighted by red boxes; nucleotides added by TLPs during 5'-end repair are shown in red. (A) mt-tRNAIleGAU(1) from D. discoideum; the U3-G70 base pair in is 100% edited. (B) mt-tRNAPheGAA from P. pallidum; the U3-G70 base pair is unedited in 94% of sequences, while the adjacent A2-A71 mismatch is fully edited. (C) mt-tRNAAlaUGC from A. castellanii; the U4-G69 base pair adjacent to the edited U3-U70 mismatch is 100% unedited.
To test this, we developed a new assay for 3'-5' polymerase activity, which has significant advantages over previously utilized assays. We measured single-turnover kinetic parameters for single and multiple nucleotide additions to two different 5'-truncated substrates based on D. discoideum mt-tRNAIleGAU(1). The two substrates differ by the identity of their terminal base pair, with one terminating in a WC C-G base pair, while the other terminates in a wobble U-G base pair. Here we show that among the four 3'-5' polymerase repair enzymes, three (DdiTLP3, DdiTLP4 and AcaTLP1) catalyze inefficient repair of the wobble-terminating mt-tRNA relative to the WC-containing substrate. However, one of the A. castellanii TLPs (AcaTLP2) exhibits a substantially improved tolerance for the substrate that terminates with a U-G base pair. This result is consistent with biological patterns of 5'-editing, in which the 5'-end repair reaction leaves a U-G base pair intact adjacent to edited mismatches in A. castellanii, while no examples of this are observed in D. discoideum.
Materials and methods
Preparation of tRNA substrates and 3'-5' polymerases
Sequence-verified plasmids were used as templates for in vitro T7 transcription with a 5'-hammerhead ribozyme engineered to generate the desired 5'-start site for each transcript (C+4 or U+3) (12). Hammerhead-cleaved transcripts were either 5'-end labeled by T4 polynucleotide kinase and γ-32P-ATP or uniformly labeled by inclusion of [α-32P]-ATP during transcription. TLPs from D. discoideum and A. castellanii were expressed as N-terminal His6-tagged proteins in E. coli and purified as previously described (12, 13).
Enzyme titration assay for 3'-5' addition to tRNAIleGAU(1)
5'-truncated mt-tRNAIleGAU(1) (C+4 or U+3-start, as indicated) substrates were incubated at room temperature with five-fold serial dilutions of TLP enzymes (2.7 μM to 0.2 nM), 1 mM CTP and/or UTP and 0.1 mM ATP in reaction buffer (25 mM HEPES, pH 7.5, 10 mM MgCl2, 3 mM DTT, 125 mM NaCl and 0.2 mg/mL BSA). Reactions were quenched after 2 hours by addition of 12.5 mM ethylenediamine tetraacetic acid (EDTA), 5 U/μL RNase T1 (Ambion), 20 mM sodium acetate, pH 5.2 and 100 ng/μL carrier yeast tRNA; RNase T1 cleavage was at 37°C for 30 minutes. An equal volume of formamide loading dye was added and reactions were resolved by 12 % polyacrylamide, 7 M urea, 1X tris-borate-EDTA gel. Dried gels were visualized using a Typhoon imager. For uniformly-labeled substrates, after RNase T1 cleavage, 0.1 U/μL CIAP (Invitrogen) and 1X dephosphorylation buffer were added and reactions were incubated at 37 °C for 30 minutes prior to resolution and visualization as described above.
Single-turnover kinetic measurements of 5'-end repair
TLP enzymes and uniformly-labeled 5'-truncated tRNAIleGAU(1) (initiating with C+4 or U+3) were added to reactions in a ratio of at least 50:1 with varied concentrations for each enzyme (1, 2 or 4μM), the 5'-activating ATP (0.1 or 0.25 mM) and added CTP and/or UTP (0.25, 0.5 and 1 mM). Time courses ranged from 10 sec – 2 hr for U+3-start and from 10 sec – 1 hr for C+4-start. Reactions were resolved by 20 % polyacrylamide, 8 M urea, 1X tris-borate-EDTA gel. Percent product conversion at each time point (Pt) was plotted vs. time and fit to the single exponential rate equation (eq. 1), where ΔP = maximal product formation.
| (eq. 1) |
For very slow reactions, (i.e., DdiTLP3/4 on U+3-start substrate), the method of initial rates was used to calculate kobs from the slope of the line fit to the product formation plot. Rates from three independent measurements at 0.5 mM CTP and/or UTP, as indicated, were averaged and reported errors are standard derivations.
Results
A gel-based assay to detect multiple nucleotide addition to mt-tRNA
Previous characterization of 3'-5' polymerases utilized a phosphatase protection assay developed to study single nucleotide addition catalyzed by S. cerevisiae Thg1 (6). Although the assay has some advantages in resolution and direct quantification of both substrate and product for kinetics, the need for defined standards and sequence incompatibility with some substrates presents challenges for studying multiple nucleotide addition reactions, such as those that occur during 5'-editing.
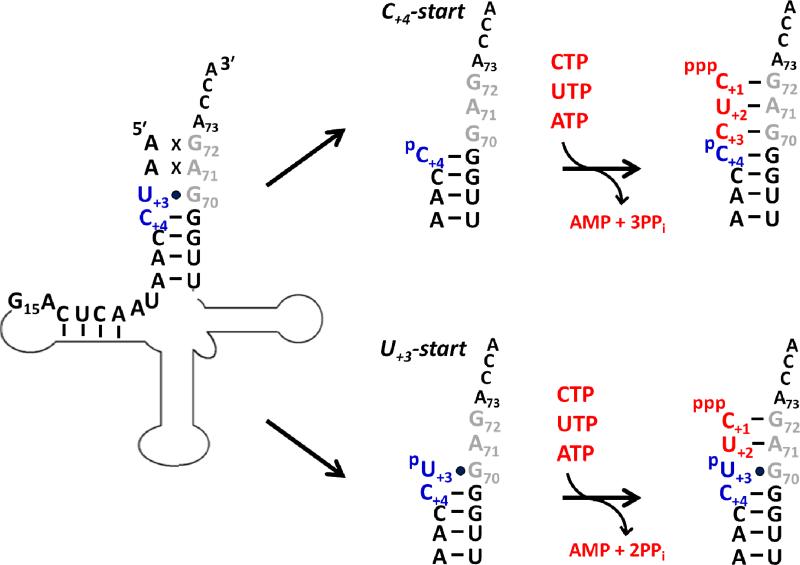
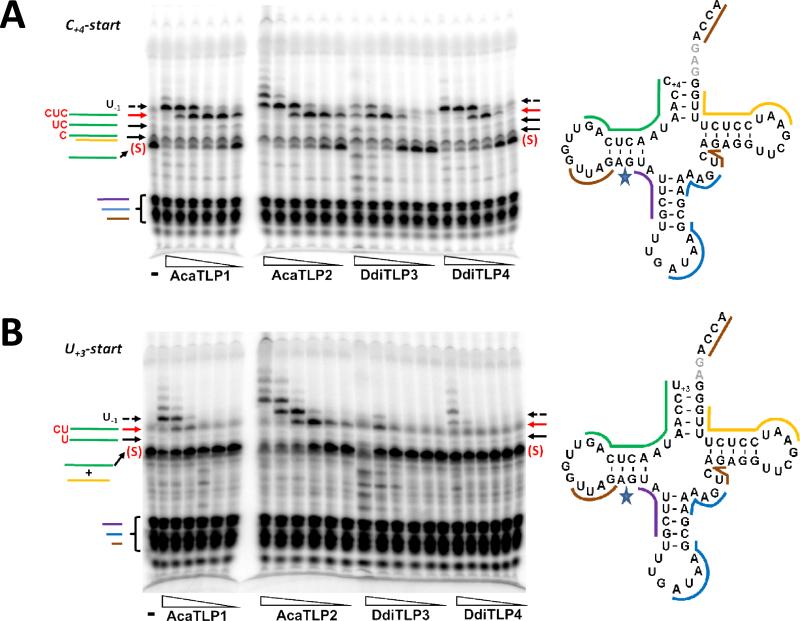
We sought an alternative approach using polyacrylamide gel electrophoretic (PAGE) analysis, with two different substrates based on D. discoideum mt-tRNAIleGAU(1) (Figure 3). The first substrate (C+4-start) mimics the 5'-truncated tRNA after removal of the first three 5'-nucleotides (including the wobble base-paired U+3). The second substrate (U+3-start) mimics a 5'-truncated tRNA resulting from removal of only the first two mismatched A nucleotides, while retaining the terminal wobble base pair. Each 5'-32P labeled substrate was incubated with varied concentrations of purified A. castellanii or D. discoideum TLPs in the presence of ATP (to activate the 5'-monophosphate, as required for the first nucleotide addition, see Figure 1) and CTP and UTP (to be incorporated into the tRNA according to the 3'-end template sequence) (Figure 3). After two hours, reactions were quenched by addition of EDTA and RNase T1, generating a labeled 5'-oligonucleotide from cleavage at G15, and products were resolved by denaturing PAGE (Figure 4).
Figure 3. 5'-end repair substrates based on mt-tRNAIleGAU(1) from D. discoideum.
To evaluate the 5'-repair activity of TLPs, two model tRNA substrates were based on the sequence of mt-tRNAIle. In vitro transcripts initiating with either C+4 (C+4-start) or U+3 (U+3-start) were generated and either 5'-32P-labeled or internally labeled with [α-32P]-ATP, as indicated. The U+3 and C+4 nucleotides are highlighted in blue and nucleotides expected to be added by TLPs to repair the 5'-aminoacyl-acceptor stem according to the 3'-template sequence are marked in red. The position of G15, where RNase T1 cleaves to yield a labeled 5'-fragment to be resolved by PAGE, is indicated.
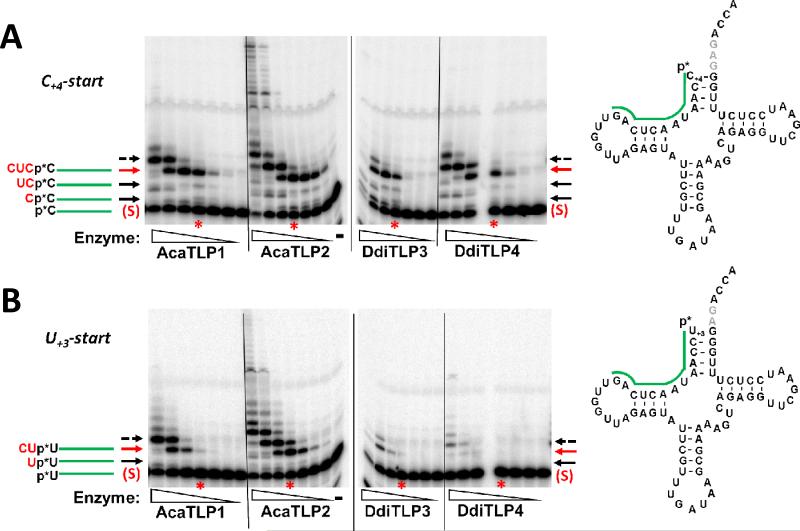
Figure 4. Enzyme titration assay for 5'-end repair with 5'-32P-labeled tRNAIleGAU(1).
The C+4-start (A) or U+3-start (B) substrates were incubated with five-fold serial dilutions (2.7 μM to 0.2 nM) of each of the four TLP enzymes, as indicated, for two hours. Reactions were quenched by addition of EDTA and RNase T1 and the resulting oligonucleotides were resolved by 12% polyacrylamide, 7 M urea PAGE. The tRNA diagrams show the complete tRNA sequence, with the expected labeled oligonucleotide derived from unreacted substrate indicated by the green line; the band corresponding to this oligonucleotide is marked by the red (S) on the gels (as seen in lane -, which is the no enzyme control lane for each substrate). Reaction products corresponding to 3'-5' addition of the indicated NTPs are indicated by arrows, with expected sequences of the added nucleotides shown in red for each band. The red arrows indicate fully-repaired 5'-ends, and the dashed arrow indicates nucleotides added beyond the +1 (full-length) position. Lanes corresponding to reactions performed at 22 nM of the indicated TLP are marked with red stars for comparison between the two substrates.
For the C+4-start tRNA, bands consistent with expected 3'-5' addition products were observed (Figure 4A). For all four enzymes, a major oligonucleotide product (indicated by red arrows) was observed at moderate concentrations of enzyme corresponding to addition of the missing C+3, U+2 and C+1 nucleotides, based on the presence of much lighter, but visible, intermediate bands that migrate in between unreacted substrate and major product bands. At high enzyme concentrations (>100 nM), DdiTLP4 and AcaTLP1 also form a major product that contains one additional 5'-nucleotide (indicated by dashed arrow), consistent with addition of 5'-U−1 to base pair with A73, as reported previously for bacterial TLPs (9). Strikingly, although addition beyond U−1 by AcaTLP1 and DdiTLP4 is detectable, albeit relatively inefficient, AcaTLP2 efficiently adds many additional 5'-nucleotides, apparently corresponding to addition beyond the predicted 3'-end template of the tRNA.
With the U+3-start tRNA, we observed similar 5'-end repair of the missing 2 nucleotides (U+2 and C+1), and also further addition past the +1 position at high enzyme concentrations (Figure 4B). However, three TLPs (DdiTLP3, DdiTLP4 and AcaTLP1) exhibited much less efficient overall activity with the U+3-start tRNA, as judged by significantly weaker intensity of products generated at the same enzyme concentration. In contrast, AcaTLP2 formed similar amounts of product with both RNAs.
Uniformly-labeled tRNA to eliminate migration differences based on differences in 5'-phosphorylation
The chemical nature of the 3'-5' nucleotide addition reaction introduces the possibility of additional complexity, since after each nucleotidyl transfer step, the 5'-triphosphate of the added NTP can remain intact to provide the activated 5'-end for subsequent nucleotide additions (Figure 1) (2). Under some conditions, S. cerevisiae Thg1 hydrolyzes the 5'-triphosphate to a 5'-monophosphate (23), and oligonucleotides bearing different numbers of 5'-phosphates would predictably migrate differently by PAGE. Although 5'-pyrophosphate removal by TLPs has not been tested, we considered the possibility that the slower-migrating reaction products observed at the highest enzyme concentrations (interpreted to correspond to addition of an extra U−1 nucleotide above) could instead correspond to 5'-monophosphorylated versions of the same C+1-containing reaction products produced at intermediate enzyme concentrations.
To test this, we generated the same two in vitro transcripts, instead using α-32P-ATP in the transcription reactions to generate uniformly-labeled tRNA, with only internal phosphodiester bonds 32P-labeled (Supplemental Figure 1) thus allowing treatment with alkaline phosphatase to remove all terminal phosphates from digestion products prior to electrophoresis. PAGE analysis ruled out 5'-end phosphate heterogeneity as the cause of multiple major product bands (Figure 5). Instead, we saw the same pattern of bands observed previously with the 5'-end labeled RNAs, with the majority of products at moderate/low enzyme concentrations corresponding to repair up to addition of the C+1 nucleotide, while at higher enzyme concentrations, addition of further nucleotides (U−1 and beyond) were observed. Moreover, the same enzyme-substrate preferences were observed, with DdiTLP3, DdiTLP4 and AcaTLP1, but not AcaTLP2, exhibiting much less efficient 5'-repair of the U+3-start tRNA than the C+4-start substrate.
Figure 5. 5'-repair assay with uniformly-labeled mt-tRNAIleGAU(1).
Uniformly-labeled C+4-start (A) or U+3-start (B) mt-tRNAIleGAU(1) were generated by in vitro transcription with [α-32P]-ATP. The two RNA substrates were incubated with five-fold serial dilutions (2.7 μM to 1 nM) of the indicated TLP enzymes for two hours. Reactions were quenched by addition of EDTA and RNase T1, and then treated with phosphatase prior to resolution by 12% polyacrylamide, 7 M urea PAGE. The tRNA diagram to the right of each panel shows expected labeled oligonucleotide fragments generated by RNase T1 digestion of the unreacted substrate. Bands were colored according to size: green – 12-mer (C+4-start) or 13-mer (U+3-start); yellow – 11-mer; purple – 6-mer; blue – 5-mer (2 different fragments) and brown – 4-mer (3 different fragments); specific sequences of labeled reaction products are shown in Supplemental Figure 1. Only RNA fragments that contain internal A-nucleotides are visible; for example, the A23-G24 dinucleotide (marked by a star) is not labeled because the 5'-32P of A23 is transferred to G22 as a consequence of RNase T1 digestion, and then removed by phosphatase treatment. Bands corresponding to unreacted substrate are labeled with (S), and bands corresponding to nucleotide addition products are indicated by arrows as in Figure 4, with the red arrow corresponding to the fully-repaired tRNA. On the 12% gels used for these assays, the sequence of the 11-mer (indicated in yellow) causes it to migrate slightly higher than the 12-mer from the 5'-end of unreacted C+4-start and co-migrate with the 13-mer from the 5'-end of unreacted U+3-start; higher percentage gels are required to resolve the 11-mer band, as demonstrated in Figure 6.
Quantification of substrate-specific preferences exhibited by 3'-5' polymerases from D. discoideum vs. A. castellanii
To quantify activities of the four 3'-5' polymerases, we performed a single-turnover kinetic analysis. Reactions with C+4-start or U+3 start uniformly-labeled RNAs were initiated by addition of excess enzyme and resolved by PAGE as above (Figure 6 and Supplemental Figures 2, 3 and 4). Product band intensities were quantified and plotted as a function of time to determine kobs (Supplemental Figure 5). To ensure that maximal concentration-independent rates (with respect to enzyme and NTPs) were measured, measurements were repeated at varied concentrations of each enzyme, the 5'-activating ATP and added CTP and/or UTP. Observed rates were independent of [enzyme] and [ATP] at all concentrations tested (data not shown), and maximal kobs were observed at 0.5 mM CTP and/or UTP (Supplemental figure 6).
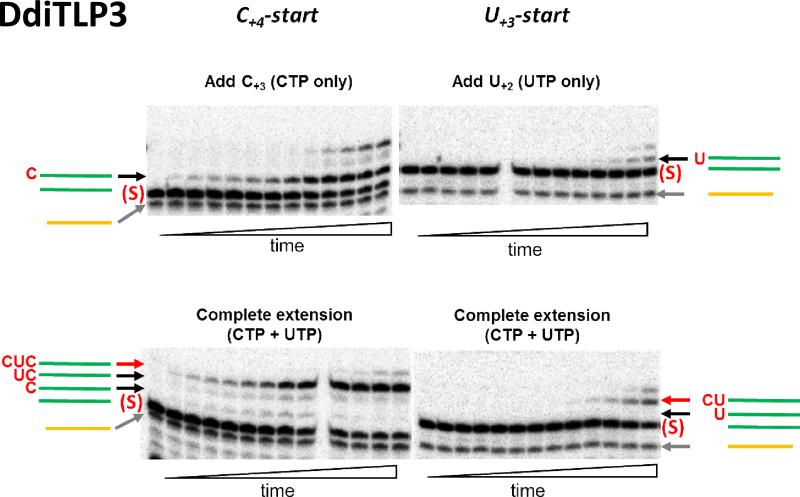
Figure 6. Single-turnover kinetic analysis of DdiTLP3-catalyzed 5'-end repair of uniformly-labeled tRNAIleGAU(1).
DdiTLP3 (2 μM) was used to initiate reactions with uniformly-labeled C+4-start (left) or U+3-start (right) mt-tRNAIleGAU(1) in the presence of 0.1 mM ATP and 0.5 mM NTP (CTP or UTP or both, as indicated). Reactions were quenched at time points ranging from 10 s to 2 h by addition of EDTA and RNase T1, and then treated with phosphatase prior to resolution by 20% polyacrylamide, 8M urea PAGE. Bands corresponding to unreacted substrate (S) and 3'-5' addition products (arrows) are labeled according to the same colors shown in Figure 5 with the identity of the added nucleotide(s) shown in red. Smaller oligonucleotides (4-6-mers) produced from these reactions are not shown for clarity. The 11-mer (yellow labeled band) is well-resolved from unreacted substrate bands on this higher percentage gel and remains constant during the course of the reaction, as expected.
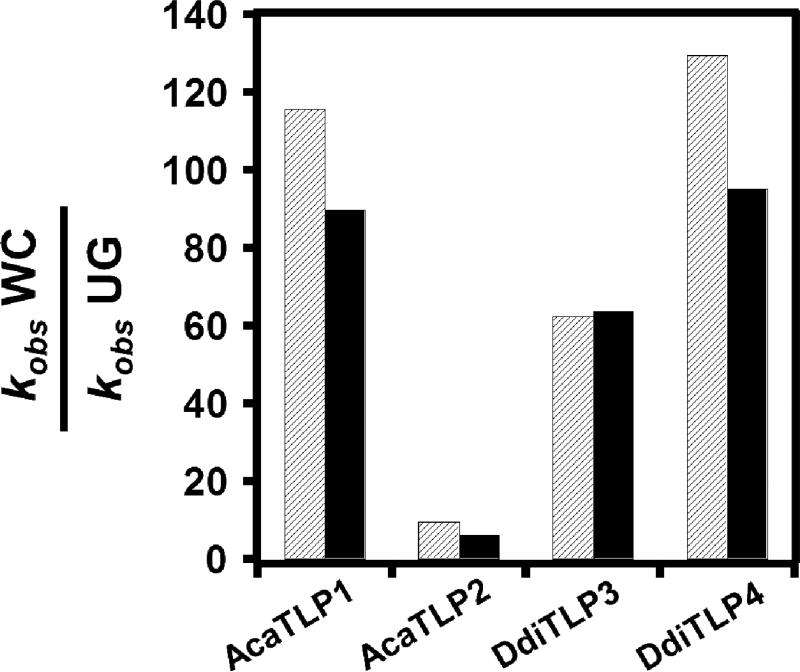
For consistency between reactions involving addition of different numbers of added nucleotides, we first measured kobs for a single nucleotide addition. To do this, we included only the appropriate NTP for the first missing nucleotide from each tRNA (CTP for C+4-start or UTP for U+3 start). Although the single nucleotide addition product was clearly observed in all reactions, we also saw more nucleotides added at longer reaction times, suggesting that incorrect addition can occur (with different efficiencies) in the absence of the correct WC base-pairing NTP (top panels, Figure 6, Supplemental Figures 2, 3 and 4), as observed previously for DdiTLPs (12). Nonetheless, in agreement with the earlier assays, the kobs for single nucleotide addition by DdiTLP3, DdiTLP4 and AcaTLP1 to the WC-terminating C+4 substrate were consistently ~100-fold faster than for addition to the U+3 tRNA (Table 1 and Figure 7). In contrast, the difference in kobs for AcaTLP2 was ≤10-fold.
Table 1.
Single turnover kinetic analysis of 3′-5′ addition catalyzed by TLPs
| kobs (min−1) First Additiona | kobs (min−1) Complete extensiona | |||
|---|---|---|---|---|
| Enzyme | U+3-start | C+4-start | U+3-start | C+4-start |
| AcaTLP1 | 0.022 ± 0.001 | 2.6 ± 0.3 | 0.018 ± 0.011 | 1.6 ± 0.4 |
| AcaTLP2 | 1.2 ± 0.2 | 11 ± 4 | 0.79 ± 0.32 | 4.9 ± 2.2 |
| DdiTLP3 | 0.0084 ± 0.0004b | 0.52 ± 0.18 | 0.0094 ± 0.0014b | 0.60 ± 0.21 |
| DdiTLP4 | 0.0076 ± 0.0014b | 0.98 ± 0.27 | 0.0074 ± 0.0024b | 0.71 ± 0.05 |
Average of three independent measurements performed under saturating conditions with respect to enzyme, ATP and added NTP; errors are standard deviations.
Rates were calculated by the method of linear initial rates because of slow reaction rates.
Figure 7. WC preferences of TLPs from D. discoideum and A. castellanii.
The preference of each of the four TLPs to extend substrates with WC base pair vs. UG base pair termini was determined by calculating the ratio of the average kobs for the C+4-start substrate to the average kobs for the U+3-start substrate. Values for WC preference were calculated for both the first nucleotide addition (hatched bar) and complete extension (solid bar), as indicated for each enzyme.
To rule out effects due to the identity of the added NTP (since the first missing nucleotide is different for each substrate), we also determined kobs for the complete 5'-end repair reaction in the presence of both CTP and UTP (bottom panels, Figure 6, Supplemental Figures 2,3, and 4). Partially repaired intermediates were no longer observed in reactions once both NTPs were included. Bands corresponding to full-length (C+1-containing) and longer nucleotide addition products were quantified to determine kobs. We observed the same pattern of WC preference as for the single nucleotide addition reactions, with DdiTLP3, DdiTLP4 and AcaTLP1 all exhibiting a much faster rate with the WC substrate than with its wobble-base paired counterpart (Table 1 and Figure 7). Interestingly, kobs for complete extension were all strikingly similar to those for the corresponding single nucleotide addition reactions, suggesting that the first addition is the rate-determining step for multiple addition and consistent with a processive mechanism for 3'-5' addition of multiple nucleotides by TLPs.
Discussion
Mitochondrial 5'-editing repairs mismatched base pairs in tRNA aminoacyl-acceptor stems. However, wobble (G-U) base pairs are abundant in tRNA, and comparisons of editing in a variety of organisms was puzzling in that sometimes G-U base pairs in mt-tRNA were “corrected” by editing, while in other cases G-U base pairs were retained or only partially corrected. Here we demonstrate that A. castellanii encodes a 3'-5' polymerase (AcaTLP2) that catalyzes relatively more efficient 3'-5' addition to a 5'-editing substrate that contains a G-U wobble base pair than several other tested TLPs (Table 1, Figure 7). Since 5'-editing in A. castellanii requires repair of mt-tRNAAlaUGC immediately adjacent to the U+4-G69 base pair (Figure 2) (18), AcaTLP2 is a good candidate for the enzyme that catalyzes this repair reaction. In D. discoideum, however, all instances of G-U base pairs immediately adjacent to mismatched bases are edited to WC base pairs (14). This is consistent with the observation that D. discoideum TLPs discriminate against G-U wobble base pairs at the terminus of the 3'-5' polymerase substrate, and suggests that these G-U base pairs must be removed prior to repair by TLPs, consistent with the absence of retained G-U base pairs in D. discoideum mt-tRNA.
It remains to be demonstrated whether only one or both TLPs edit mt-tRNA in vivo in A. castellanii and D. discoideum. In previous in vitro assays, no obvious biochemical differences were observed between the candidate enzymes to imply preferential roles for one or the other in 5'-editing in vivo (12-14). Here, however, we identified distinct biochemical properties of the tested TLPs that are consistent with biological editing patterns. The ability of AcaTLP2 to efficiently repair the wobble-terminating substrate suggests it may be better-suited than AcaTLP1 to edit mt-tRNAAlaUGC in A. castellanii. In light of this proposed function, the extensive 5'-end elongation observed at high concentrations of AcaTLP2 in vitro must also be rationalized. Although the biochemical basis for this unexpected property of AcaTLP2 requires further investigation, it is possible that physiological concentrations of AcaTLP2 are low enough to limit activity to the correct number of nucleotides to repair the mt-tRNA, as observed in Figures 4 and 5. Alternatively, the complete editing enzyme might employ additional factors to counteract addition of extra nucleotides beyond the fully-repaired 5'-end of the mt-tRNA. Further biological characterization of the 5'-editing reaction in A. castellanii will be required to assess these, or other, possibilities. For the D. discoideum TLPs, DdiTLP3 correctly repairs mt-tRNAIleGAU(1) even at high enzyme concentrations, while DdiTLP4 consistently adds one or more nucleotides beyond the correct tRNA 5'-end. This could imply a preferential role for DdiTLP3 in 5'-editing, but regardless is the first observation of any biochemical difference between these two enzymes and may be functionally relevant. Interestingly, DdiTLP3 and AcaTLP2 are predicted to localize to the mitochondria where 5'-editing occurs, but whether these are unique 5'-editing enzymes in either organism awaits further biological characterization.
In this study we developed a new PAGE assay, which can be applied more broadly to study 3'-5' polymerase family enzymes. The decision to use 5'-end labeled vs. uniformly-labeled RNA can be flexible based on similar results of our enzyme titration assays. Although in this study a nuclease was used to generate smaller oligonucleotide reaction products that are better resolved by PAGE than full-length tRNA, nuclease treatment could be omitted, and full-length substrates resolved by PAGE if necessary. The PAGE assay is highly amenable to kinetic characterization of multiple nucleotide addition reactions as demonstrated here, and will be valuable for future investigation of the mechanism of these unusual polymerases. This study provided some kinetic insight into 3'-5' polymerization reactions, suggesting that the first nucleotide addition is rate-limiting. Based on earlier kinetic studies, this may be due to the slower kinetics of the 5'-adenylylation step (which is only required for the first addition to a 5'-monophosphorylated RNA) (24). The PAGE assay opens the door to further investigating enzymatic properties of the unusual 3'-5' reverse polymerase family including substrate specificity, processivity and catalytic mechanism, all of which will advance understanding of the evolutionary connection between canonical and 3'-5' polymerases and the mysterious but versatile roles of these enzymes in biology.
Supplementary Material
Highlights.
Mitochondrial tRNA in many protozoan eukaryotes undergo 5'-editing.
TLPs use 3'-5' polymerase activity to repair 5'-truncated editing substrates.
A. castellanii TLP2 tolerates G-U pairs in editing substrates better than other TLPs.
TLP activities correlate with biological outcomes for G-U base pairs during editing.
Biochemical evidence suggests specialized functions for different eukaryotic TLPs.
Acknowledgments
Funding for this work was provided by NIH grant nos. GM087543 (to J.E.J.). Y.L. was supported by the Pelotonia Graduate Fellowship. The authors are grateful to S. Dodbele, A. Krishnamohan, Q. Liu and K. Patel for valuable comments on the manuscript.
Abbreviations
- Thg1
tRNAHis guanylyltransferase
- TLP
Thg1-like protein
- WC
Watson-Crick
- mt-tRNA
mitochondrial tRNA
- EDTA
ethylenediamine tetraacetic acid
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hyde SJ, Eckenroth BE, Smith BA, Eberley WA, Heintz NH, Jackman JE, Doublie S. tRNA(His) guanylyltransferase (THG1), a unique 3′-5′ nucleotidyl transferase, shares unexpected structural homology with canonical 5′-3′ DNA polymerases. Proc Natl Acad Sci U S A. 2010;107:20305–20310. doi: 10.1073/pnas.1010436107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jackman JE, Phizicky EM. tRNAHis guanylyltransferase catalyzes a 3′-5′ polymerization reaction that is distinct from G-1 addition. Proc Natl Acad Sci U S A. 2006;103:8640–8645. doi: 10.1073/pnas.0603068103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jackman JE, Gott JM, Gray MW. Doing it in reverse: 3′-to-5′ polymerization by the Thg1 superfamily. RNA. 2012;18:886–899. doi: 10.1261/rna.032300.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gu W, Hurto RL, Hopper AK, Grayhack EJ, Phizicky EM. Depletion of Saccharomyces cerevisiae tRNA(His) guanylyltransferase Thg1p leads to uncharged tRNAHis with additional m(5)C. Mol Cell Biol. 2005;25:8191–8201. doi: 10.1128/MCB.25.18.8191-8201.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gu W, Jackman JE, Lohan AJ, Gray MW, Phizicky EM. tRNAHis maturation: an essential yeast protein catalyzes addition of a guanine nucleotide to the 5′ end of tRNAHis. Genes Dev. 2003;17:2889–2901. doi: 10.1101/gad.1148603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jackman JE, Phizicky EM. tRNAHis guanylyltransferase adds G-1 to the 5′ end of tRNAHis by recognition of the anticodon, one of several features unexpectedly shared with tRNA synthetases. RNA. 2006;12:1007–1014. doi: 10.1261/rna.54706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abad MG, Rao BS, Jackman JE. Template-dependent 3′-5′ nucleotide addition is a shared feature of tRNAHis guanylyltransferase enzymes from multiple domains of life. Proc Natl Acad Sci U S A. 2010;107:674–679. doi: 10.1073/pnas.0910961107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hyde SJ, Rao BS, Eckenroth BE, Jackman JE, Doublie S. Structural Studies of a Bacterial tRNA(HIS) Guanylyltransferase (Thg1)-Like Protein, with Nucleotide in the Activation and Nucleotidyl Transfer Sites. PLoS One. 2013;8:e67465. doi: 10.1371/journal.pone.0067465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rao BS, Maris EL, Jackman JE. tRNA 5′-end repair activities of tRNAHis guanylyltransferase (Thg1)-like proteins from Bacteria and Archaea. Nucleic Acids Res. 2011;39:1833–1842. doi: 10.1093/nar/gkq976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heinemann IU, O'Donoghue P, Madinger C, Benner J, Randau L, Noren CJ, Soll D. The appearance of pyrrolysine in tRNAHis guanylyltransferase by neutral evolution. Proc Natl Acad Sci U S A. 2009;106:21103–21108. doi: 10.1073/pnas.0912072106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heinemann IU, Randau L, Tomko RJ, Jr., Soll D. 3′-5′ tRNAHis guanylyltransferase in bacteria. FEBS Lett. 2010;584:3567–3572. doi: 10.1016/j.febslet.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abad MG, Long Y, Willcox A, Gott JM, Gray MW, Jackman JE. A role for tRNA(His) guanylyltransferase (Thg1)-like proteins from Dictyostelium discoideum in mitochondrial 5′-tRNA editing. RNA. 2011;17:613–623. doi: 10.1261/rna.2517111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rao BS, Mohammad F, Gray MW, Jackman JE. Absence of a universal element for tRNAHis identity in Acanthamoeba castellanii. Nucleic Acids Res. 2013;41:1885–1894. doi: 10.1093/nar/gks1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abad MG, Long Y, Kinchen RD, Schindel ET, Gray MW, Jackman JE. Mitochondrial tRNA 5′-editing in Dictyostelium discoideum and Polysphondylium pallidum. J Biol Chem. 2014;289:15155–15165. doi: 10.1074/jbc.M114.561514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gott JM, Somerlot BH, Gray MW. Two forms of RNA editing are required for tRNA maturation in Physarum mitochondria. RNA. 2010;16:482–488. doi: 10.1261/rna.1958810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lonergan KM, Gray MW. Editing of transfer RNAs in Acanthamoeba castellanii mitochondria. Science. 1993;259:812–816. doi: 10.1126/science.8430334. [DOI] [PubMed] [Google Scholar]
- 17.Lonergan KM, Gray MW. Predicted editing of additional transfer RNAs in Acanthamoeba castellanii mitochondria. Nucleic Acids Res. 1993;21:4402. doi: 10.1093/nar/21.18.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Price DH, Gray MW. Confirmation of predicted edits and demonstration of unpredicted edits in Acanthamoeba castellanii mitochondrial tRNAs. Curr Genet. 1999;35:23–29. doi: 10.1007/s002940050428. [DOI] [PubMed] [Google Scholar]
- 19.Laforest MJ, Bullerwell CE, Forget L, Lang BF. Origin, evolution, and mechanism of 5′ tRNA editing in chytridiomycete fungi. RNA. 2004;10:1191–1199. doi: 10.1261/rna.7330504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laforest MJ, Roewer I, Lang BF. Mitochondrial tRNAs in the lower fungus Spizellomyces punctatus: tRNA editing and UAG ‘stop’ codons recognized as leucine. Nucleic Acids Res. 1997;25:626–632. doi: 10.1093/nar/25.3.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bullerwell C, Gray M. In vitro characterization of a tRNA editing activity in the mitochondria of Spizellomyces punctatus, a Chytridiomycete fungus. J Biol Chem. 2005;280:2463–2470. doi: 10.1074/jbc.M411273200. [DOI] [PubMed] [Google Scholar]
- 22.Price DH, Gray MW. A novel nucleotide incorporation activity implicated in the editing of mitochondrial transfer RNAs in Acanthamoeba castellanii. RNA. 1999;5:302–317. doi: 10.1017/s1355838299981840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith BA, Jackman JE. Saccharomyces cerevisiae Thg1 uses 5′-pyrophosphate removal to control addition of nucleotides to tRNA(His.) Biochemistry. 2014;53:1380–1391. doi: 10.1021/bi4014648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith BA, Jackman JE. Kinetic analysis of 3′-5′ nucleotide addition catalyzed by eukaryotic tRNA(His) guanylyltransferase. Biochemistry. 2012;51:453–465. doi: 10.1021/bi201397f. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.