Abstract
One of the main challenges in functional diffuse optical tomography (DOT) is to accurately recover the depth of brain activation, which is even more essential when differentiating true brain signals from task-evoked artifacts in the scalp. Recently, we developed a depth-compensated algorithm (DCA) to minimize the depth localization error in DOT. However, the semi-infinite model that was used in DCA deviated significantly from the realistic human head anatomy. In the present work, we incorporated depth-compensated DOT (DC-DOT) with a standard anatomical atlas of human head. Computer simulations and human measurements of sensorimotor activation were conducted to examine and prove the depth specificity and quantification accuracy of brain atlas-based DC-DOT. In addition, node-wise statistical analysis based on the general linear model (GLM) was also implemented and performed in this study, showing the robustness of DC-DOT that can accurately identify brain activation at the correct depth for functional brain imaging, even when co-existing with superficial artifacts.
Keywords: Diffuse optical tomography, Near infrared spectroscopy, Depth compensation, Superficial artifacts, Anatomical atlas of human head, General linear model
1. Introduction
Diffuse optical tomography (DOT) is an emerging neuroimaging technology that uses low-power near infrared light (650 to 950 nm) to measure the changes of cerebral blood flow and oxygenation associated with neuronal activity (Villringer and Chance, 1997; Boas et al., 2004a). Compared with other neuroimaging modalities, such as functional magnetic resonance imaging (fMRI), DOT has the advantage of being portable and cost effective, with excellent temporal resolution. The near infrared spectroscopy (NIRS) methods used to measure the hemodynamic response to brain activation were first described by several groups in 1993 (Villringer et al., 1993; Hoshi and Tamura, 1993; Chance et al., 1993). Early studies used a sparse array of light sources and detectors; the spatial resolution was comparable to the source-detector separation, namely, in a few centimeters. In the past decade, the technology has been advanced to high-density DOT (Boas et al., 2004b; Zeff et al., 2007), which records data with a high-density array of light sources and detectors and reconstructs volumetric images of brain hemodynamics by solving the forward and inverse problems of light propagation in tissue. High-density DOT significantly improves the spatial resolution and positional accuracy of optical brain imaging (White and Culver, 2010; Eggebrecht et al., 2012; Zhan et al., 2012).
One of the major problems needing attention in DOT development is its severe sensitivity decay along depth. It is known that optical sensitivity in DOT has an approximately exponential decay with increased depth (Lee et al., 2005; Dehghani et al., 2009a), which makes DOT measurements hypersensitive to hemodynamic fluctuations in the scalp rather than to the more pertinent signals from the brain. The ill-posed sensitivity matrix causes positional errors in image reconstruction since the severe sensitivity decay biases the reconstructed brain activation towards the superficial layer. A variety of approaches have been developed to minimize depth error in reconstructed images. A widely-used approach is to apply a spatially variant regularization (SVR) parameter to regular DOT reconstruction (i.e., SVR-DOT) (Pogue et al., 1999; Culver et al., 2003; Dehghani et al., 2009a). Another approach is a depth-compensated DOT (DC-DOT) (Niu et al., 2010), which modifies the depth-variant sensitivity matrix directly rather than modifying the regularization parameter. While these two approaches are mathematically similar, DC-DOT apparently has a wider effective range in depth and can adapt for both sparse and dense geometries (Kavuri et al., 2012). One concern about DC-DOT is that the true quantity of hemodynamic changes is lost due to necessary modification on the sensitivity matrix. To solve this problem, a scaling factor can be applied on either the modified sensitivity matrix or the reconstructed image. Tian et al. (2010) demonstrated that by applying a scaling factor on the reconstructed image, DC-DOT actually improves quantification accuracy.
1.1 Depth-compensated DOT to discriminate brain activities from superficial artifacts
DC-DOT also improves depth specificity of brain activation in the existence of non-activation physiological interferences. It is well known that optical signals from the brain contain several physiological fluctuations that originate from cardiac pulsation, respiration, change of blood pressure, and so on. These physiological fluctuations are systemic and contribute greater signal changes than local brain activities. Tian et al. (2011) have reported that DC-DOT is able to recover brain activation at correct depth even when systemic fluctuations coexist. The image quality can be further enhanced by adaptive removal (Zhang et al., 2007a; 2007b; Zhang et al., 2009; Tian et al., 2011) or linear regression (Saager and Berger 2005; Saager et al., 2011) of the systemic fluctuations that are sampled at a short source-detector separation (typically within 1.3 cm). Besides the systemic physiological fluctuations, currently there is increasing attention on the task-evoked superficial artifacts which can lead to false positive findings in functional brain activities (Kirilina et al., 2012 ; Takahashi et al., 2011). These superficial artifacts arise from local vascular oscillations in the scalp although the physiological origin is still controversial and might be varied. One goal of the present study was to investigate whether DC-DOT has the capability to discriminate brain activation from the superficial artifacts according to their depths.
1.2 Integration of DC-DOT with a standard anatomical atlas of human head
Previous studies in DC-DOT (Niu et al., 2010; Tian et al., 2010; 2011 ; Kavuri et al., 2012) have used a homogeneous, semi-infinite model of diffusion theory to recover volumetric images of brain activation. While this model is computationally efficient because of the capability to create analytic solutions, it significantly deviates from realistic human head anatomy. In order to localize and quantify brain activation accurately, it is necessary to use a computational model derived from real head anatomy. This requires acquisition of MRI images of each subject’s head with optode positions marked, followed by generation of a subject-specific head model, which enables investigators to solve the forward and inverse diffusion equation more accurately (Gibson et al., 2003; Dehghani et al., 2009a; Eggebrecht et al., 2012; Zhan et al., 2012). However, this approach reduces the convenience and feasibility of optical brain imaging as a stand-alone technology. An alternative approach is to fit a standard MRI head template (atlas) for all subjects via affine registration (Custo et al., 2010; Cooper et al., 2012); this approach has proved to be consistent with and comparable to the subject-specific approach. Another goal of the present study, therefore, was to incorporate DC-DOT with a finite element model derived from an anatomical MRI atlas of human head. Based on this model, the depth specificity and quantification accuracy of DC-DOT can be assessed through computer simulations and human measurements under a selected functional stimulation.
1.3 Analysis of volumetric DC-DOT image series with general linear model
In addition, the present study also implemented model-based analysis according to the general linear model (GLM). GLM is a statistical linear expression that models measured signals as a linear combination of predicted responses to independent stimulation variables plus an error term. GLM can be used to analyze a variety of experimental data acquired by many measurement modalities. As an example, fMRI has popularly used GLM to model blood oxygen level-dependent (BOLD) signal changes. For time-dependent data series, such as those seen in fMRI or functional NIRS, GLM-based analysis matches both the temporal pattern and the magnitude of signals, thereby providing us with a robust tool to quantitatively characterize functional brain responses.
While GLM has been used as a standard data analysis method in fMRI (Beckmann et al, 2003; Friston et al., 1995; Bullmore et al., 1996), it can be helpful for functional NIRS data analysis especially in cases of severe light attenuation. Schroeter et al. (2004) first proposed the application of GLM as a standard data analysis strategy in functional NIRS. Later, Plichta et al. (2007) showed that model-based GLM provided a powerful test of visual cortex activation in a rapid event-related paradigm. It would be beneficial to compare and interpret both types of neuroimaging data if both functional NIRS and fMRI images can be studied using the same GLM framework. Towards this direction, Ye et al. (2009) have developed a software package (known as NIRS-SPM) for functional NIRS, based on a statistical parametric mapping (SPM) toolbox that is widely used in the field of fMRI. However, all of these studies have implemented GLM-based analysis that can be applied only to channel-wise NIRS data. Post-GLM topographic images are generated by interpolating data between adjacent channels and thus result in limited spatial resolution. To date, very few studies have applied GLM to volumetric image series achieved through DOT. Thus, the final goal of this study was to design, implement, and demonstrate a node-wise GLM approach to analyze the volumetric image series generated from brain atlas-based DC-DOT.
In order to demonstrate the overall improvement in DOT image quality by the three imaging and data analysis methods developed in this study, we utilized human sensorimotor activations evoked by a finger-tapping task to generate volumetric DC-DOT images. The primary sensorimotor regions of the human brain are optically accessible and have been intensively studied by researchers in this field as reviewed by Leff et al. (2011). Thus, the corresponding brain activations in response to a finger tapping task are well understood. While the emphasis of the present study was on advances and integration of three DOT-based imaging and data analysis methods, we measured sensorimotor activities during a well-known finger-tapping task among human subjects for a demonstrative purpose, without utilizing fMRI for validation.
2. Materials and Methods
2.1 Anatomical atlas of human head
We used a standard MRI atlas of human head, known as ICBM 152 nonlinear asymmetric template, with a spatial resolution of 1×1×1 mm3 (http://www.bic.mni.mcgill.ca/ServicesAtlases/ICBM152NLin2009) (Fonov et al., 2009; 2011). This template was generated as an unbiased non-linear average among normal population in a broad range of ages (18.5 to 43.5 years); the anatomical structures were originally segmented using the ANIMAL+INSECT algorithm (Collins et al., 1999). In this study, a total of five head/brain tissues were defined: the scalp, skull, cerebrospinal fluid (CSF), gray matter and white matter. Optical properties of the segmented tissues at two wavelengths, 750 nm and 850 nm, were set according to current literature (Eggebrecht et al., 2012; Zhan et al., 2012), which are summarized in Table 1.
Table 1.
Absorption coefficient μa, reduced scattering coefficient μs′ and refractive index n of the segmented head tissues
| 750nm |
850 nm |
Refractive index n |
|||
|---|---|---|---|---|---|
| μa (cm−1) | μs′ (cm−1) | μa (cm−1) | μs′ (cm−1) | ||
| Scalp | 0.17 | 7.40 | 0.19 | 6.40 | 1.4 |
|
| |||||
| Skull | 0.12 | 9.40 | 0.14 | 8.40 | 1.4 |
|
| |||||
| CSF | 0.04 | 3.00 | 0.04 | 3.00 | 1.4 |
|
| |||||
| Gray matter | 0.18 | 8.36 | 0.19 | 6.73 | 1.4 |
|
| |||||
| White matter | 0.17 | 11.91 | 0.21 | 10.11 | 1.4 |
2.2 Probe geometry, spatial registration, and head atlas-based meshing
A rectangle probe was used to image the sensorimotor cortex on the left hemisphere in both simulation and human head measurements. The probe was composed of 21 sources and 21 detectors that were arranged alternatively, as shown in Fig. 1(a). The probe provided a total of 71 channels at the first nearest source-detector separation of 1.6 cm (or 1st nearest neighbors, 1st NN) and a total of 98 channels at the second nearest source-detector separation of 3.6 cm (or 2nd nearest neighbors, 2nd NN). Other larger source-detector separations were not considered because their signals were too weak in reality.
Fig. 1.
Probe configuration and spatial registration: (a) Optical probe configuration. The probe was composed of 21 sources and 21 detectors that were arranged alternatively, providing a total of 71 channels at the 1st nearest source-detector separation and a total of 98 channels at the 2nd nearest source-detector separation. (b) Projected probe location on the head mesh for human Subject 1. The head mesh and projected probe location were also used in the computer simulations.
The probe was placed along a para-coronal line between the left and right preauricular points. The center of the probe was located approximately at C3 according to the international 10-20 system. In human head measurements, once the probe was set in place, the positions of optodes along with five cranial landmarks (the nasion, inion, left and right preauricular points, and vertex) were measured using a PATRIOT motion tracking system (Polhemus, Colchester, Vermont, USA.). The cranial landmarks served as mediators for converting the standard Montreal Neurological Institute (MNI) coordinates used in the standard head atlas into the real-world stereotaxic coordinates based on affine transformation (Singh et al., 2005). Following the methods given by Custo et al. (2010), the standard head atlas was registered into the real-world space to match each subject’s head. Then a finite element mesh (FEM) model was generated from the registered atlas using the iso2mesh software package (Fang and Boas, 2009). At this step, the maximum volume of the tetrahedral element was set to be 2 mm3. The resultant FEM mesh of the whole head consisted of approximately 90,000 nodes and 500,000 tetrahedral elements. After the probe was projected onto the FEM mesh, the forward and inverse problems of light propagation could be solved based on diffusion theory in order to reconstruct a volumetric image of brain activation, which will be described in details in Section 2.6. The spatial registration and head meshing were performed for each individual subject. As an example, Fig. 1(b) shows the head mesh and the projected probe position on the mesh for Subject 1.
2.3 Simulative experiments
In the first part of this study, simulative experiments were conducted to justify the performance of brain atlas-based DC-DOT. In simulations, the initial cerebrovascular conditions and task-evoked changes were all known, so we were able to obtain comprehensive and quantitative evaluations on DC-DOT, which otherwise were not accessible for validation in human measurements. Two types of simulation were conducted sequentially: the first was to assess the capability of DC-DOT to recover a static absorber at different depths, which mimicked the scenario where a time-averaged change in local absorption was imaged without considering any temporal alternation. In the second type of simulation, a set of time-dependent cerebral hemodynamics were created to mimic resultant fluctuations due to 1) functional brain activations evoked by a finger tapping paradigm, 2) systemic physiological noises, and 3) superficial artifacts from the scalp vasculature. The reconstructed time-dependent volumetric image series by DC-DOT were analyzed based on GLM for each node. The registered probe position shown in Fig. 1(b), which was taken from Subject 1, was used in both types of simulations. In particular, we generated two sets of head mesh: one mesh (mesh #1) was used to simulate the optical signals measured by the detectors, and the other one (mesh #2) was used to compute the sensitivity matrix and to reconstruct the volumetric image. The use of two separated head meshes in simulation was to avoid the so-called ‘inverse crime’, which denotes the act of employing the same computational model to generate and to invert synthetic data (Wirgin, 2004). Under every simulated cerebrovascular condition, the optical signals measured by the detectors (based on mesh #1) and the sensitivity matrix (based on mesh #2) were computed using NIRFAST (Dehghani et al., 2009b), a widely-used FEM-based software package for modeling propagation of near infrared light in biological tissues.
2.3.1 Simulating a static absorber at different depths
A spherical absorber, 0.6 cm in diameter, was embedded in the region of sensorimotor cortex to mimic a local absorption change from the baseline. The absorber had a higher absorption coefficient (μa) than that of background at both 750 nm and 850 nm, while keeping the same reduced scattering coefficient (μs′) as the background. At both wavelengths, the absorber induced an identical absorption perturbation of Δμa = 0.05 cm−1 from the background tissues. To comprehensively assess the performance of DC-DOT, five separated trials were simulated by placing the absorber at a different depth, namely 0.5, 1.0, 1.5, 2.0 or 2.5 cm from the simulated head surface. The localization and quantification accuracies of DC-DOT were evaluated at each depth and also compared with those using conventional DOT without any spatially variant regularization or depth compensation (Con-DOT), SVR-DOT and DC-DOT.
2.3.2 Simulating time-dependent cerebral hemodynamics in a finger tapping paradigm
The simulated paradigm was a common blocked-design paradigm composed of 12 repetitive blocks with 10 seconds of finger tapping and 20 seconds of rest per block. There was an initial baseline of 20-second rest prior to the first block. Data were simulated at a sampling rate of 10 Hz. The scattering properties of the head were assumed to be stable throughout the paradigm. The task-evoked functional brain activation and physiological interferences were assumed to be pure absorption changes.
In a previous study (Tian et al., 2011), we performed human measurements using a similar paradigm. Prior knowledge obtained from that study directed us to characterize the distinct hemodynamic evolutions of functional activation and physiological interferences separately. In the last step of this simulative experiment, we combined the hemodynamic evolutions of functional activation and physiological interferences to simulate the overall time-dependent hemodynamic measurement from the head. The following sub-sections describe the steps performed to generate different time-dependent hemodynamic components.
2.3.2.1 Task-evoked functional activation
The task-evoked functional activation was characterized by a hemodynamic fluctuation localized within a spherical volume on the sensorimotor cortex. The spherical volume had a diameter of 1.0 cm and was embedded 2.2 cm below the scalp surface. The hemodynamic fluctuation of functional activation, f(t), was modeled as the convolution of a stimulation function s(t) [s(t) = 1 for finger tapping and 0 for rest] and the normalized hemodynamic response function (HRF) h(t), namely, f(t) = s(t) ⊗ h(t). The hemodynamic response function h(t) was defined as (Cohen, 1997):
| (1) |
where c was adjusted to give unit amplitude at equilibrium and t is time in second. While eq. (1) was introduced for fMRI signals, it was also reasonable to approximately represent the time-dependent hemodynamic responses measured by functional NIRS. This is because both fMRI and NIRS detect signals that stem from the same origin: the hemodynamic changes in the brain, while they may have somewhat different sensitivities toward different cerebrovascular compartments. For example, it is known that fMRI is more sensitive to venous blood (Huettel et al., 2009) whereas NIRS is more sensitive to capillary bed (Liu et al., 1995). Indeed, an optical imaging method that was used to quantify stimulation-evoked changes in absolute HbO and Hb concentrations in an animal brain served as a cornerstone for validation of fMRI (Huettel et al., 2009). Although it would be ideal to find an NIRS-derived HRF to simulate functional activation and to perform consequent GLM analysis, such HRF is often not available since it requires careful studies with well-designed experimental paradigms. In contrast, fMRI-derived HRFs have been intensively studied and are available in literature; thus, it is scientifically appropriate to utilize such a HRF in our simulative experiments and in GLM-based DC-DOT image analysis.
2.3.2.2 Systemic fluctuations
The optical signals propagating through the brain contain several spontaneous fluctuations that originate from cardiac pulsation, respiration and change of blood pressure (also referred to as the Mayer waves). These fluctuations cause systemic hemodynamic changes in the entire head except in the CSF. In particular, the cardiac pulsation generates fast-oscillating waves, which are distinct from task-evoked functional activation and can be efficiently filtered out. Therefore, in the simulations, we considered the slow systemic fluctuations only, including Mayer waves and respiration waves. The magnitudes, frequencies, and coherence of these two types of fluctuations were comprehensively investigated previously (Tian et al., 2011). According to the prior knowledge gained (Tian et al., 2011), these two fluctuations were determined to have the following characteristics:
Mayer waves
The Mayer waves oscillate along the change of arterial blood pressure and have a frequency about 0.1 Hz. However, the exact mechanism of Mayer waves is not well understood. Additionally, there were consistent reports on a phase shift between the oxy-hemoglobin (HbO2) related and deoxy-hemoglobin (Hb) related Mayer waves (Obrig et al., 2000; Tian et al., 2011), with Hb-related Mayer waves always taking the lead. Therefore, we used a random time series to simulate the initial origins of Mayer waves, followed by band-pass filtering between 0.06 and 0.12 Hz to generate continuous HbO2-related Mayer waves, namely m(t). We generated Hb-related Mayer waves separately, as m(t+t0), where t0 was set to be 3 seconds.
Respiration waves
The respiration waves usually have a frequency range of 0.2 to 0.4 Hz, and the phase shift between Hb-related and HbO2-related components is negligible. In this study, a continuous oscillation, generated by band-pass filtering between 0.2 and 0.4 Hz of a random time series, r(t), was used to simulate both the Hb-related and HbO2-related respiration waves.
2.3.2.3 Task-evoked superficial artifacts
The task-evoked superficial artifacts were simulated by hemodynamic fluctuations in a spherical volume in the scalp. The spherical volume had a diameter of 0.6 cm and was embedded 0.5 cm below the scalp surface. According to Kirilina et al. (2012), the hemodynamic fluctuation of superficial artifacts, a(t), was characterized by a block-locked cosine function: a(t) = cos(2πfartifactt). The frequency fartifact was the same as the repetitive frequency of task blocks, i.e., fartifact = 0.033 Hz.
It is noted that the temporal patterns of the superficial artifacts given in literature are variable and might depend on specific tasks (Takahashi et al., 2011; Kirilina et al., 2012). Here we chose a cosine function because it was distinct from the functional brain activation; in this way, the superficial artifacts and functional brain activation could be easily distinguished in the reconstructed image series. In real applications, however, there is a possibility that the superficial artifacts have a temporal pattern similar to functional activation, which will be discussed by the end of this paper.
2.3.2.4 Overall time-dependent cerebral hemodynamics
The overall hemodynamic changes in the head were a combination of task-evoked functional activation, systemic fluctuations, and task-evoked superficial artifacts with different absolute magnitudes. We assumed that each type of systemic fluctuation had identical magnitude in the scalp, skull and brain. We also assumed that there was no systematic fluctuation in CSF. The task-evoked functional activation and task-evoked superficial artifacts were localized in two localized spherical regions, namely Ractivation and Rartifact. In summary, the overall hemodynamic changes in the ith non-CSF node of the head model were:
| (2) |
| (3) |
where ΔCi denotes the concentration change of each hemoglobin species in the ith non-CSF node; Af, Am, Ar and Aa denote the magnitudes of functional activation, Mayer waves, respiration and superficial artifacts in each hemoglobin species, respectively. These magnitudes were determined as follows:
Functional activation
A finger tapping task can evoke about 1 μM increase of HbO2 concentration and a much smaller decrease of Hb concentration as measured at a source-detector separation of 3 cm (Tian et al., 2011). For a channel-wise measurement, it is well known that a partial volume effect exists and causes under-estimation of the focal hemoglobin changes in the brain; namely, the focal hemoglobin changes in the brain are “averaged” to the whole volume of tissues probed by a source-detector channel (Boas et al., 2001; Strangman et al., 2003). To minimize the partial volume effect, the measured channel-wise hemoglobin changes should be corrected by a sensitivity correction factor, SCF, which is defined as a ratio between the summed optical sensitivity of a source-detector channel to brain tissues without superficial layers (i.e., scalp and skull) and the summed sensitivity of this channel to all of the head tissues including the superficial layers. For a source-detector separation of 3.0 cm, a prior calculation using aforementioned mesh #2 and NIRFAST found that SCF was about 1/30. So, we estimated the magnitudes of task-evoked brain activation to be a 30-μM increase in HbO2 concentration and a 6-μM decrease in Hb concentration.
Mayer waves
It is estimated that Mayer waves introduce about 1 μM changes of HbO2 concentration in the head (Tian et al., 2011). Also, it is known that Mayer waves originate from the arterial blood flow with high oxygen saturation. Therefore, we utilized 0.1 μM to represent the magnitude of Hb concentration changes induced by Mayer waves.
Respiration waves
It is also assumed that the respiration waves introduce about 0.3 μM changes of HbO2 concentration in the entire head (Tian et al., 2011). It has been demonstrated that respiration waves are mainly related to venous flow with oxygen saturation values around 60-70% (Franceschini et al., 2002). Thus, we estimated 0.2 μM to be the magnitude of Hb concentration changes induced by respiration waves.
Superficial artifacts
According to relevant literature, task-evoked superficial artifacts cause about 0.3 μM decreases in HbO2 concentration and much smaller changes in Hb concentration, which are measured at a source-detector separation of 3 cm (Kirilina et al., 2012). These artifacts originate from the superficial blood vessels that are relatively small in size. Similar to the functional activation case, a sensitivity correction factor is also needed to estimate the actual hemoglobin changes in the superficial blood vessels from the measured signals. Under the assumption that the artifacts come from a spherical volume in the scalp with a diameter of 0.6 cm, we calculated SCF for superficial artifacts using aforementioned mesh #2 and NIRFAST and found that the corresponding SCF was about 1/50 at source-detector separation of 3.0 cm. Therefore, we estimated the magnitudes of the superficial artifacts to represent a 15-μM decrease of HbO2 concentration and a 3-μM increase of Hb concentration, which may need to be confirmed in future studies.
Figure 2 shows the time-dependent Hb and HbO2 concentration fluctuations induced by simulated functional activation, Mayer waves, respiration waves, and superficial artifacts, as given in four respective panels. The magnitudes, frequency ranges, and time lag between Hb and HbO2 for all four time-dependent hemodynamic components are summarized in Table 2.
Fig. 2.
Time-dependent Hb and HbO2 fluctuations (unit: μM) of the simulated (a) functional brain activation, (b) Mayer waves, (c) respiration waves and (d) superficial artifacts. The dash line in top panel marks the starting time of each block.
Table 2.
Magnitudes, frequency ranges, and time lags between Hb and HbO2 of the task-evoked functional activation, slow systemic fluctuations (Mayer waves and respiration waves) and task-evoked superficial artifacts used in the simulation
| Magnitude (μM) | Frequency (Hz) | THb→HbO2 (s)* | ||
|---|---|---|---|---|
| HbO2 | Hb | |||
| Functional activation | 30 | −6 | 0 | |
| Mayer waves | 1 | 0.1 | 0.06 - 0.12 | 3 |
| Respiration | 0.3 | 0.2 | 0.2 - 0.4 | 0 |
| Superficial artifacts | −15 | 3 | 0.033 | 0 |
Time lag from Hb-related oscillation to HbO2-related oscillation
The cerebral hemodynamic changes resulted in absorption changes in the head model at both wavelengths (750 nm and 850 nm), which were computed as:
| (4) |
| (5) |
where , , and denote the molar absorption coefficients of each hemoglobin species at 750 nm and 850 nm, respectively.
2.3.3 Calculating the optical signals received by the detectors
Once the absorption and scattering coefficients were selected under every simulated state, the propagation of near infrared light in the head mesh was modeled using the NIRFAST software package. It provided light intensity received by each detector (based on aforementioned mesh #1) from every specific source, similar to real data collection on a human subject’s head. Details of light propagation modeling have been reported by Dehghani et al. (2009b).
Additionally, electronic noises always exist in real data collection. To make the simulated data closer to the reality, in the second type of simulation (time-dependent cerebral hemodynamics), we added white noise (i.e., a random noise with a flat power spectral density) into the time-dependent light intensities output from NIRFAST. In general, a shorter source-detector separation results in a bigger received light intensity and better signal-to-noise ratio (SNR). Therefore, we added 1% white noises to all the 1st NN source-detector channels relative to their baseline intensities and 3% white noises to all the 2nd NN source-detector channels. In the first type of simulation with static absorbers, we did not add any random noise because of the fact that the white noise in the time-averaged data could be reduced significantly.
2.4. Human head measurements
In the second part of this study, human measurements were conducted to compare and confirm the performance of brain atlas-based DC-DOT using the same finger tapping paradigm as described in Section 2.3.2. Subjects were recruited from the local community of the University of Texas at Arlington. The study protocol was approved by the University of Texas at Arlington Institutional Review Board. Written informed consent was obtained from every subject prior to the experiment.
2.4.1 Subjects
Five healthy subjects (all males, age range 22-39 years) were included in the study. The subjects were all right-handed. Subjects were screened and excluded for a past or current neurological disorder, a presently unstable medical condition, or current intake of any medication. Only eligible subjects who passed screening underwent the experiment.
2.4.2 Instrument
A high-density DOT system (Cephalogics LLC., Boston, MA) (Zeff et al., 2007) was used to acquire data from human subjects using the probe shown in Fig. 1(a), with a sampling rate of 10.8 Hz. In particular, the probe was made with low-weight fibers purchased separately from TechEn Inc. (Boston, MA). The low-weight fibers and flexible probe assembly ensured subjects’ comfort during the experiments.
2.4.3 Task
Each subject participated in one session of finger tapping experiment using the same paradigm described in Section 2.3.2. Briefly, it was a blocked-design paradigm composed of 12 repetitive blocks, each of which had 10 seconds of finger tapping and 20 seconds of rest. An initial baseline of 20-second rest was taken prior to the first block. Each subject sat on a chair stably throughout the experiment. During the task period, each subject was instructed to tap his index and middle fingers against the thumb at a comfortable rhythm, which was about 2 to 3 Hz (Tian et al., 2011). Each subject was instructed to stay still and relaxed during the rest period. Ambient light was blocked during the experiment to ensure that it did not contaminate the optical signals.
2.5 Data preprocessing
2.5.1 Screening of channel-wise data acquired from the human subjects
In human measurements, data from some channels might have very low SNR due to the poor contact of optodes on the scalp. The data with bad quality or low SNR could degrade the quality of reconstructed images significantly. Therefore, for the human data, in the pre-processing stage, we excluded all the channels that had high-frequency (>0.4 Hz) noise in large amplitudes, that is, larger than 15% of the baseline intensities. For the qualified channels, we further excluded some blocks that showed significant data discontinuities (with a signal swing of 15% or larger from the raw baseline intensities) resulting from motion artifacts.
2.5.2 Filtering of time-dependent data in both simulation and human measurements
After data screening, the time-dependent hemodynamic signals from both simulation and human measurements were filtered in a common frequency range. First, the raw data in light intensity were low-pass filtered at a cut-off frequency of 0.4 Hz to reduce the electronic noise. The low-pass filter also removed the fast-oscillating cardiac waves. Then, changes in optical signals relative to the baseline were computed as changes in optical density, ΔOD, at all respective times. At last, the computed ΔOD value for each channel was further high-pass filtered at a cut-off frequency of 0.01 Hz to remove the baseline drift during the experiment.
2.6 Tomography
2.6.1 Forward problem
Given the fact that a biological medium has a much greater light scattering coefficient than the absorption coefficient in the near infrared range, light propagation in tissue can be modeled by the diffusion approximation of the Boltzmann transport equation (Arridge, 1999; Durduran et al., 2010). The diffusion approximation is given by:
| (6) |
where v is the speed of light in the medium, S(r, t) is the isotropic source providing the number of photons emitted at position r and time t, and Φ(r, t) is the photon fluence rate that is proportional to the photon number density U(r, t), namely, Φ(r, t) = vU(r, t). D(r) is the diffusion coefficient defined as D=v/[3(μa + μs′)], where μa is the absorption coefficient and μs′ is the reduced scattering coefficient. The latter, μs′, is defined as μs′ = (1 - <cosθ>) μs, where θ is the scattering angle and <cosθ> is the mean of cosine θ.
The implementation of DOT for functional brain imaging is to reconstruct the absorption perturbation from a known background, for which the forward solution of equation (6) can be expressed by Rytov approximation (O’Leary, 1996):
| (7) |
where Δμa is the relative absorption change, detected between a source at position rs and a detector at position rd, between two measurement states, Φ0 and Φ. L is the effective optical path length of light propagating through the medium. Φ0(rd, rs) ≡ Φ(rd, rs, t0) is the measurement taken at baseline and Φ(rd, rs, t) is the measurement taken when the absorption is changed in the medium. The left hand side of equation (7) is the definition of optical density change for the given source-detector pair, i.e., ΔOD. The right hand side of equation (7) is known as the modified Beer-Lambert law (Delpy et al., 1988), which can be generalized for a set of discrete elements (i.e., nodes) with potentially different absorption changes:
| (8) |
where Lj is the effective optical path length of a given source-detector pair in the jth node of the medium, and Nnode represents the number of nodes along the optical path between this set of source and detector. For multiple source-detector measurements, equation (8) can be written in a matrix form as:
| (9) |
where y is a column vector of channel-wise measurements in optical density changes written as y = (ΔOD1, ΔOD2, …ΔODNmeas)T with Nmeas being the number of measurements, and x is also a column vector that represents node-wise absorption changes in the 3D image space, x = (Δμa1, Δμa2, …, ΔμaNnode)T. A is a Nmeas × Nnode sensitivity matrix of Li,j, where Li,j is the effective path length between the ith source-detector pair passing through the jth node. Note that index i covers from 1 to the total number of measurements (source-detector pairs), Nmeas, on a 2D measurement surface, while index j runs from 1 to the total number of nodes, Nnode, in a 3D image space. Matrix A is also called the Jacobian matrix or weight matrix.
2.6.2 Inverse problem
Con-DOT: Due to the limited numbers of sources and detectors used in DOT, in equation (9), the number of measurements is much fewer than the number of nodes to be reconstructed, namely, Nmeas << Nnode. As a result, the equation is highly ill-determined and the solution is not unique. Therefore, regularization to the solutions is required. The conventional way is to use the Tikhonov regularization method (Tikhonov, 1963), which reconstructs the image from the following equation:
| (10) |
where λ = α · smax (A) is the regularization parameter and smax (A) denotes the maximum singular value of A matrix, ||x||2 is the l2 norm of x defined as .
However, A matrix is also ill-posed since it has an approximately exponential decay as depth increases due to severe attenuation of biological tissues. The nodes from the influence of the superficial layer always contribute much greater weights to the measured signals than the nodes in deep layers (e.g., the brain). Thus, the superficial layer eventually biases the reconstructed brain activation towards the head surface with a significant depth error. As reviewed in the beginning of this paper, two approaches have been used to solve this problem:
SVR-DOT
The scheme of SVR is to regularize the depth-dependent sensitivities with variable λ-values, namely, bigger regularization values to suppress the hyper sensitivities in the superficial layer and thus to achieve a more homogenous spatial distribution of the sensitivity. According to Zhan et al. (2012), the SVR approach replaces the original A matrix in Equation (10) with a spatially variant form:
| (11) |
where χ is the spatial regularization factor. According to the literature (Dehghani et al., 2009b; Eggebrecht et al., 2012; Zhan et al., 2012), χ = 0.01 was chosen in our SVR-DOT calculations.
DC-DOT
The DC-DOT approach induces a depth-variant modification function to modify the ill-posed A matrix directly. The key idea behind DCA was to find or compose a weight matrix M, which has a pseudo exponential increase in magnitude with depth so as to counterbalance the severe loss of sensitivity of A in depth. Originally, matrix M was developed by Niu et al. (2010) for a perfectly-layered homogeneous model; thus, M can be noted hereafter by Mlayer. In this study, since the heterogeneous head mesh does not have a perfectly-layered structure, we modified the compensation weight function by using a direct exponential term, as defined below:
| (12) |
where mj (j = 1, 2, … Nnode) is a depth-compensation weight for the jth node, dj (j = 1, 2, … Nnode) is the depth of the jth node in centimeter below the scalp surface, and η is a constant which controls the compensation power and should have an optimal value to the specific probe configuration and tissue optical properties. Equation (12) forms a node-wise, depth-compensation matrix, noted hereafter by Mnode, which is a modified or simplified version from its original form of Mlayer, given by Niu et al. (2010). While the compositions of Mnode and Mlayer are not exactly the same, both of them offer a similar trend of exponential increase in depth-compensation weight as the depth increases. More details on the derivation and discussion on Mnode and η are given in the Supplementary Material.
Following the same logic and steps used in DCA (Niu et al., 2010) with Mlayer, we generated a node-wise, depth-compensated sensitivity matrix, A#=AMnode, for DC-DOT image reconstruction on the heterogeneous head mesh. Because mj is always larger than 1 [see eq. (12) and the Appendix], every element in A# is greater than its uncompensated element in A. This mismatch in amplitude between A# and A gives rise to the inability to recover correct hemodynamic perturbation in the reconstructed DC-DOT images. To fix this problem, an additional scaling factor k is introduced in Eq. (13) to keep equal quantity between matrixes A# and A within the brain region:
| (13) |
where j runs 1, 2, … Nnode to cover all the nodes within the head mesh, while i covers all the measurement channels, Nmeas, or pairs of sources and detectors. It is noted that the scaling factor k is determined at each wavelength. By keeping the quantities consistent between A# and A at each wavelength, factor k adds an extra constraint that may help minimize the potential crosstalk when calculating the changes of HbO2 and Hb concentrations. Our results to be shown in Section 3 (i.e., section 3.1.2) will provide evidence to support this statement.
After matrix A# = AMnode is generated and scaling factor k is determined, the DC-DOT approach is formed to reconstruct DC-DOT images as (Niu et al., 2010; Tian et al., 2011):
| (14) |
where λ = α · smax (A#).
In the present study, the NIRFAST software package (Dehghani et al., 2009b) was used to solve the forward problem and to derive the sensitivity matrix A based on the optical properties given in Table 1. To determine the depth of each node inside the head mesh, dj, we first defined the surface of the head as 1) a cohort of the surface nodes between the air and the scalp (maximum intermodal distance was 2 mm on the head surface) and together with 2) a cohort of the centroids of the surface triangles. Then for each node inside the head mesh, we computed its distance to all of the identified surface nodes and centroids of surface triangles; then we selected the minimum distance as its depth beneath the identified head surface. After the depth of each node was determined, the inverse problem was solved under the region interrogated by the probe within the depth ranging from 0 to 3.5 cm, which included about 15,000 nodes. An identical regularization parameter α = 0.1 was used for all of the inverse calculations, namely, for the conventional DOT, SVR-DOT and DC-DOT.
Specifically for our DC-DOT image reconstructions, an optimal compensation power η = 3 based on the heterogeneous head mesh was determined following the method described by Niu et al. (2010); its derivation and confirmation are given in the Appendix. Figure 3 shows a comparison of the normalized, depth-dependent sensitivities before and after applying depth compensation with η = 3. Finally, the reconstructed absorption images at two wavelengths (750 nm and 850 nm) were converted to the HbO and Hb images by inversely solving equations (4) and (5).
Fig. 3.
Normalized depth-dependent sensitivity of optical measurements: (a) Without depth compensation and (b) With depth compensation. The depth-dependent sensitivity was computed as a mean of node-wise sensitivities in a depth step of 2 mm for each source-detector separation. Without depth compensation, in panel (a), both the 1st NN and 2nd NN measurements were more sensitive to the superficial tissues. With depth compensation, in panel (b), the 1st NN measurements were more sensitive to the superficial tissues and the 2nd NN measurements were more sensitive to the brain.
2.7 Node-wise model-based analysis using GLM
After the time-dependent, volumetric images were reconstructed using the approaches given in Sections 2.5 and 2.6, GLM analysis was performed on the node-wise data series. As mentioned in Section 1.3, GLM models the measured brain response as a linear combination of predicted responses due to variable stimulations plus an error term. It can be used to analyze different types of experimental data acquired by a variety of measurement modalities. For example, in fMRI, a hemodynamic response function is used to serve as a model to predict the blood oxygen level-dependent (BOLD) signals due to variable stimulations, followed by data-driven analysis using t-statistics for final formation of fMRI images. The characteristic and advantage of model-based GLM analysis are the ability to integrate model-driven and data-driven analysis approaches smoothly with a reliable statistical power. This is why it has been popularly utilized in fMRI (Beckmann et al, 2003; Friston et al., 1995; Bullmore et al., 1996).
Since the reconstructed images in DOT were concentrations of HbO2 and Hb, we utilized the similar GLM approach to select statistically meaningful nodes by t-statistics so as to form 3D DC-DOT images. Specifically, for each node, our GLM expression was formulated as:
| (15) |
where z(t) is the reconstructed time series of hemodynamic signals (e.g., HbO2 and Hb) at each node, f(t) is the predicted stimulation-evoked response which is computed as the hemodynamic response function, h(t), convoluted with the stimulation function, s(t), (refer to Section 2.3.2.1), β is the estimated amplitudes of HbO2 or Hb, and ε is the error term. The hemodynamic response function given in eq. (1) was also used in our GLM analysis. By fitting Eq. (14) to the reconstructed time series of HbO2 and Hb at each node, we would be able to obtain the estimated amplitude β and its variance, and thus a statistical t-value representing the statistical power of brain activation at the respective node.
In our analysis, the error term ε in GLM was assumed to be spatially uncorrelated and normally distributed. Furthermore, it is known that the slow systemic fluctuations (Mayer waves and respiration waves) produce structured “noise” to the volumetric data (Plichta et al. 2007). In this study, we used linear regression to reduce the systemic fluctuations from the reconstructed volumetric image series. For this step, a reference of the systemic fluctuations was obtained by averaging the data from the superficial nodes within a depth less than 1 cm. After linear regression, the image series from both simulation and human measurements were down sampled to 1 Hz, followed by node-wise GLM analysis. Both β-map and t-statistics map (or t-map) were derived. Both maps were thresholded at the corresponding half maxima or half minima to identify the region of activation.
3. Results
3.1 Simulative experiments
3.1.1 A static absorber at different depths
By positioning a static absorber at different depths, the localization and quantification accuracies of DC-DOT were assessed and compared with those from Con-DOT and SVR-DOT. Here we only show the results at 850 nm. In Fig. 4, the left column shows the reconstructed images of this absorber at five different depths using Con-DOT, in a coronal view of the brain that was along the middle line of optode array (as marked by the vertical dashed lines in both panels of Fig. 1). In each figure, the small circle indicates the actual location of the absorber, with an absorption perturbation of Δμa = 0.05 cm−1 relative to the background. It is seen that conventional DOT reconstructs images of the absorber mainly in a shallow region under the scalp surface, independent of the actual depth of the absorber. The recovered absorption perturbations of the absorber were also much smaller than the true value (Δμa = 0.05 cm−1), with the maximum recovered perturbation being 15% of the true value occurring at an absorber depth of 0.5 cm. The middle column of Fig. 4 shows the reconstructed images using SVR-DOT, which does improve the depth accuracy in the reconstructed images, especially before the absorber reaches 2.0 cm in depth. The right column of Fig. 4 shows the reconstructed images using DC-DOT. From actual absorber depths of 0.5 cm to 2.0 cm, the depth localization errors in the reconstructed images are small, within a couple of millimeters. The algorithm begins to break down at actual absorber depth of 2.5 cm. Compared with Con-DOT and SVR-DOT, DC-DOT shows improved depth accuracy when the absorber is located between 0.5 cm and 2.5 cm in depth within simulated brain tissues. Moreover, DC-DOT also provides us with better quantification accuracy. The maximum recovered absorption perturbation by DC-DOT, occurring also at the actual absorber depth of 0.5 cm, is about 60% of the true value.
Fig. 4.
Reconstructed images of a static absorber at depths of 0.5 cm, 1.0 cm, 1.5 cm, 2.0 cm and 2.5 cm: the first, second, and third columns counting from left show the reconstructed images using conventional DOT, SVR-DOT, and DC-DOT, respectively. All of the images were normalized by the true absorption perturbation (Δμa = 0.05 cm−1) and cut off at the corresponding half maxima. The color scale in each image represents a recovery rate, which is a ratio between the recovered absorption perturbation and the true absorption perturbation.
3.1.2 Time-dependent cerebral hemodynamics in a finger tapping paradigm
It is known that time-dependent optical signals measured by functional NIRS or DOT can be affected by three factors: 1) task-evoked brain functional activation, 2) systemic fluctuations caused by physiological noises, and 3) superficial hemodynamic artifacts from the scalp vasculature. To evaluate the performance of DC-DOT in response to the simulated finger tapping task, we studied two separate cases: in the first case, we considered the situation of factors 1) and 2) as overlapped; namely, the simulated optical signals included both task-evoked brain activation signals and systemic physiological fluctuations. In the second case, we assumed that all three factors coexisted. In both cases, node-wise GLM analysis was performed on the volumetric image series reconstructed with DC-DOT, followed by formation of the magnitude (β-map) and statistical power (t-map) images of time-dependent functional activation. A comparison between these two cases proves that DC-DOT is able to discriminate task-evoked brain activation from the systemic fluctuations and superficial artifacts, as presented in details below.
3.1.2.1 Task-evoked functional activation overlapped with systemic fluctuations
Before looking into the reconstructed images, we first compared the time-dependent channel-wise data and node-wise data. As reviewed in the beginning of this paper, previous studies (Schroeter et al., 2004; Plichta et al., 2007; Ye et al., 2009) had performed GLM analysis on the channel-wise data directly; post-GLM topographic images were generated by interpolating data between adjacent channels. Therefore, the magnitude and statistical power of brain activation in these studies were determined by the channel-wise data. In the present study, a series of volumetric images were generated first through DC-DOT, followed by GLM analysis on the node-wise data, leading to volumetric magnitude and statistical power maps of brain activation. So, a comparison between the channel-wise and node-wise data would reveal the difference of GLM-based results between the previous studies and present study.
For the comparison, we selected a special 2nd NN channel which was the most sensitive channel to the spherical absorbing volume of simulated brain activation. We also randomly selected a node that was within the spherical absorbing volume of simulated brain activation. In Fig. 5, panels (a) and (b) show the real-time and block-averaged hemodynamic signals detected by the selected channel, respectively. Although this channel is supposed to be the most sensitive to functional brain activation, it is clearly seen that in panels of Figs. 5(a) and 5(b), the task-evoked brain activation patterns are hardly recognized due to strong overlap or interference of the systemic physiological fluctuations. Panels (c) and (d) of Fig. 5 display the real-time and block-averaged HbO2 and Hb profiles, respectively, from the selected node after DC-DOT image reconstruction (before the systemic fluctuations were removed by linear regression). The task-evoked functional activation patterns are clearly seen in both of these panels. This comparison demonstrates that DC-DOT has the ability to isolate or separate true brain activation from systemic fluctuations, greatly improving SNR in the region of brain activation.
Fig. 5.
Comparison of the channel-wise data and reconstructed node-wise HbO2 and Hb profiles from the simulated task-evoked brain activation with systematic fluctuations: (a) Time-dependent channel-wise data from a specific 2nd NN channel that was most sensitive to the brain activation. (b) Block-averaged data from the same channel. (c) Time-dependent node-wise HbO2 and Hb profiles from a node inside the volume of brain activation. (d) Block-averaged data from the same node. In panels (a) and (c), the dash line indicates the starting time of each block.
In Fig. 6, panels (a) and (b) show the HbO2-based and Hb-based β-maps derived from node-wise GLM, respectively. In both maps, the functional brain activation is identified at approximately the correct location and depth. The recovered maximum magnitudes of functional activation are 2.5 μM in HbO2 and −0.5 μM in Hb; both are about 8% of the corresponding true values (30 μM in HbO2 and −6 μM in Hb, see Table 2). In addition, the ratio between the recovered HbO2 magnitude and the recovered Hb magnitude (i.e., 2.5 μM vs. −0.5 μM) is identical to the ratio between the corresponding true values (i.e., 30 μM vs. −6 μM), indicating that DC-DOT induces a minimal crosstalk between the two hemoglobin species.
Fig. 6.
Identified brain activation locations after excluding the systematic fluctuations by voxel-wise GLM analysis: (a) HbO2-based β-map, (b) Hb-based β-map, (c) HbO2-based t-map, and (d) Hb-based t-map. Each image was thresholded at the corresponding half maxima or half minima. The small circle in each panel indicates the real location and size of the simulated brain activation. The color scales in (a) and (b) represent the real β-amplitude in μM. Images (c) and (d) were normalized by their corresponding maximum or minimum t-values, and thus have unit amplitude.
Panels (c) and (d) in Fig. 6 show the normalized t-maps derived from node-wise GLM for HbO2 and Hb, respectively. In panel (c), the identified region of activation from HbO2-based t-map shows a good match with its actual location. In panel (d), however, the identified region of activation from Hb-based t-map is a little deeper than its actual location. More discussion on the activation region being in a “deeper” depth is given in Section 4.4 and Supplementary Material.
3.1.2.2 Task-evoked functional activation coexisted with superficial artifacts and systemic fluctuations
It has been recently reported that some stimulation tasks can evoke hemodynamic changes in the scalp vasculature besides the functional activation in the brain (Kirilina et al., 2012 ; Takahashi et al., 2011). The task-evoked superficial artifacts can induce a significant bias or false positive reading to functional brain activation because both fluctuations have similar task-dependent frequencies. However, DC-DOT may have the capability to separate these two fluctuations in the reconstructed images because they originate from different depths. Following this rationale, we performed our node-wise GLM analysis on the simulated hemodynamic signals that included the functional activation, systemic fluctuations, and task-evoked superficial artifacts. The corresponding β-maps of HbO2 and Hb are shown in Figs. 7(a) and 7(b). In both panels, we can see that DC-DOT is able to localize the functional brain activation and superficial artifacts at correct depths. The recovered maximum magnitudes of functional brain activation are about 2.5 μM in HbO2 and −0.5 μM in Hb; both are about 8% of the corresponding true values. Panels (c) and (d) in Fig. 7 show the normalized t-maps derived from the same node-wise GLM analysis when both the functional brain activation and superficial artifacts co-existed. In both panels, the regions of brain activation are identified at the right locations, although they are ‘pushed’ deeper into the brain by the superficial artifacts. More discussion on the reconstructed activation region being in a “deeper” depth is Section 4.4 and Supplementary Material.
Fig. 7.
Identified brain activation and superficial artifacts from the systematic fluctuations based on voxel-wise GLM analysis: (a) HbO2-based β-map, (b) Hb-based β-map, (c) HbO2-based t-map, and (d) Hb-based t-map. Each image was thresholded at the corresponding half maxima and half minima. The bigger circle in each image indicates the actual location and size of the simulated brain activation; the smaller circle in each image indicates the actual location and size of the simulated superficial artifacts. The color scales in (a) and (b) represent the real β-amplitude in μM. Images (c) and (d) were normalized by their corresponding maximum or minimum t-values.
Based on channel-wise data, Kirilina et al. (2012) recently reported that brain activation cannot be identified when task-evoked superficial artifacts are predominant. The simulation-based results shown in this paper demonstrate that the combination of DC-DOT with node-wise GLM analysis is capable of differentiating task-evoked brain activation from the task-evoked superficial artifacts. Further improvement in our algorithm will continue in order to reconstruct more accurately the depth of brain activation.
3.2 Human measurements
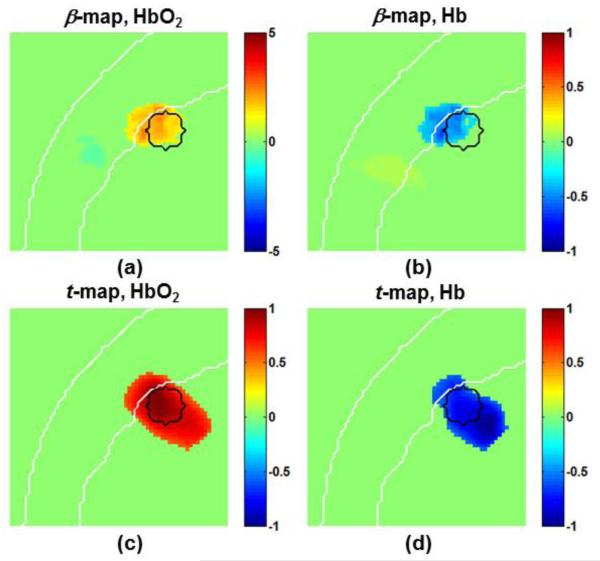
To confirm and support the simulative results given in Section 3.1, human measurements were performed using a well-studied finger-tapping protocol. In the actual human measurements, the optical probe covered a broad cortical region including partial frontal, parietal, and temporal lobes (see Fig. 1). In this study we restricted our focus on activation around the primary sensorimotor area, contralateral to the tapping hand. The results from DC-DOT with node-wise GLM included β-maps and t-maps of both HbO2 and Hb. These results were reviewed subject by subject. As an example, Fig. 8 shows the results from Subject 1 in a coronal view of the primary sensorimotor area, along the middle line of optode array (as marked by the vertical dashed lines in both panels of Fig. 1) in response to the finger tapping protocol. Panels (a) and (b) are the derived β-maps for HbO2 and Hb, respectively; panels (c) and (d) are the derived t-maps for HbO2 and Hb, respectively. While the regions of brain activation identified by t-maps include those seen in β-maps, the t-maps outline the activation region deeper into the brain, as seen in Figs. 6 and 7. Again, further discussion on this issue will be given Section 4.4 and Supplementary Material. Note that this subject does not show obvious superficial artifacts in any of those maps.
Fig. 8.
Identified sensorimotor activation of human Subject 1 based on voxel-wise GLM analysis: (a) HbO2-based β-map, (b) Hb-based β-map, (c) HbO2-based t-map, and (d) Hb-based t-map. Each image was thresholded at the corresponding half maxima or half minima. The dash arc in panel (a) indicates the selected region for surface imaging of the sensorimotor activation. The color scales in (a) and (b) represent the real β-amplitude in μM. Images (c) and (d) were normalized by their corresponding maximum or minimum t-values.
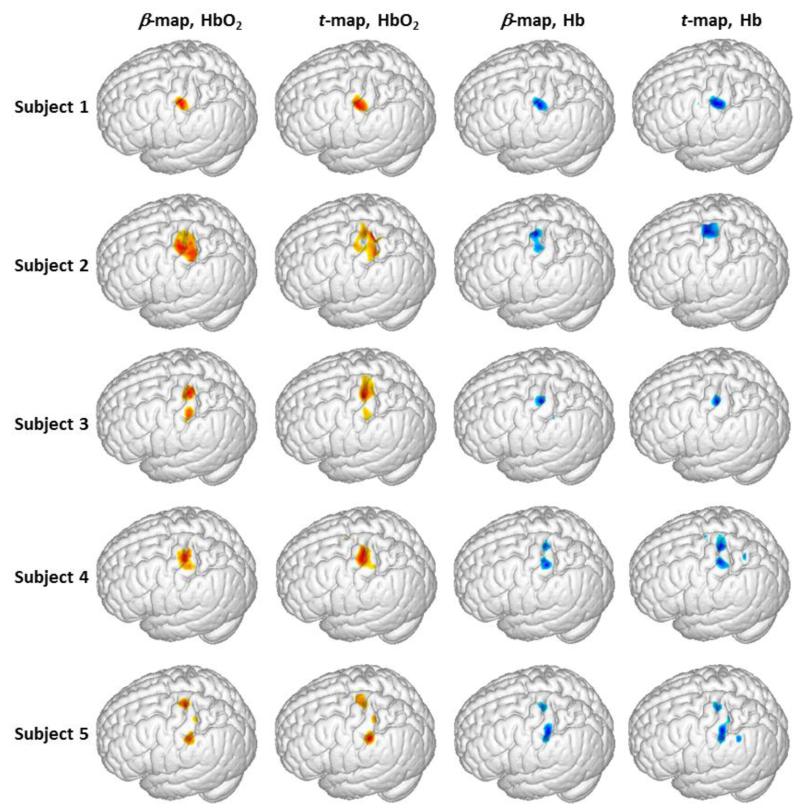
In order to characterize or visualize coverage of brain activation on the cortical surface, we selected an arc region/volume on or near the brain surface with depths ranging from 1.5 cm to 2.5 cm below the scalp surface, as indicated in Fig. 8(a). The β-values and t-values within the arc region/volume were averaged across the depth to generate a 2D image of brain activation and then projected on the surface of the brain in a way similar to our previous study (Tian et al., 2012). Using this projection method, we generated 3D rendered, activation-evoked brain images from all of the five subjects, as shown in Fig. 9. This figure clearly illustrates that the sensorimotor cortical region was consistently activated during the finger tapping task across all the subjects. The β-maps and t-maps are also in good agreement with each other in both HbO2 and Hb cases.
Fig. 9.
Activation images of 3D rendered sensorimotor cortex evoked by the finger-tapping task from all five subjects. The images were generated by averaging the β-values or t-values across the depth of the selected arc region, as marked in Fig. 8(a). The HbO2-based β-maps and t-maps were thresholded at the corresponding half maxima, in which the warm colors represent values from half maximum to full maxima. The Hb-based β-maps and t-maps were thresholded at the corresponding half minima, in which the cool colors represent values from half minima to full minima.
4. Discussion
4.1 Localization and quantification accuracies of DC-DOT
In the Introduction section, we mentioned that one of the goals for the present study was to incorporate DC-DOT with a finite element model derived from a standard brain atlas. Throughout the texts in the Methods and Results sections, we have shown in details how DC-DOT is incorporated with a standard brain MRI atlas for volumetric imaging of human sensorimotor activation. Improved depth localization and quantification accuracies by the brain atlas-based DC-DOT have been clearly demonstrated through computer simulations and human measurements. For depth localization accuracy, DC-DOT can recover both superficial and cortical absorption perturbations at their correct depths, which is expected and consistent with our previous reports based on a homogeneous, semi-infinite model (Niu et al., 2010). For quantification accuracy, DC-DOT has improved the recovery rate of absorption perturbation (e.g., 60% using DC-DOT, as compared to 15% and 14% using Con-DOT and SVR-DOT seen in Fig. 4 at an absorber depth of 0.5 cm) at each wavelength by inducing a scaling factor in reconstruction, which is also consistent with our previous reports (Tian et al., 2010). However, we acknowledge that the improvement in quantification accuracy becomes less meaningful in larger depths. For example, in the time-dependent hemodynamic simulations, the recovered maximum magnitudes of functional activation (actual depth was 2.2 cm) are about 8% of the true values. Importantly, when quantifying hemoglobin concentrations based on at least two wavelengths, the scaling factors should follow the spectral constraint. Otherwise, a crosstalk between the reconstructed concentrations of Hb and HbO2 could occur due to inappropriate scaling factors at different wavelengths. In the example of simulated time-dependent cerebral hemodynamics, we did not find any noticeable crosstalk between the derived Hb and HbO2 concentrations in either node-wise time courses or reconstructed images. Thus, the scaling factors we used are expected to be correct across the two wavelengths.
It is noted that although DC-DOT provides a mathematic approach to improve depth specificity, the eventual image quality of DC-DOT still relies on the arrangement of optode array and dynamic range of the instrument, which mandate the appropriate source-detector separations that can be used. It has been well documented in literature (Dehghani et al., 2009a) that the inclusion of larger source-detector separations leads to improved depth profiling into the deep brain region. However, the SNR of optical signals falls off exponentially as the source-detector separation increases. In this study, we only used the 1st NN and 2nd NN source-detector separations in the probe, which had acceptable SNR in our human measurements. Because of this limitation, we speculate that DC-DOT used in this study may be able to accurately recover functional activation only on or shallowly below the cortical surface (i.e., 2.5 cm or less from the scalp surface). The main objective of this paper is to prove that DC-DOT provides reliable images of the cortex by using a simple experimental setup that can be commonly achieved in human studies. We expect that incorporation of DC-DOT with larger source-detector separations will further improve the quality of deeper brain imaging, with future validation.
4.2 Discriminating brain activation from task-evoked superficial artifacts
It was also mentioned in the introduction that we sought to discover if DC-DOT can be used to discriminate brain activation from superficial artifacts. By using the time-dependent simulation throughout the text, we have demonstrated the capability of DC-DOT to discriminate the brain activation from the task-evoked superficial artifacts, which is otherwise difficult to achieve. We chose distinct temporal patterns for the superficial artifacts and functional brain activation (cosine function vs. gamma function) so that they could be easily distinguished from one another in the reconstructed image. In real applications, however, it is possible that the superficial artifacts and functional brain activations have similar temporal patterns. In this case, the influences from both fluctuations will be overlapped in the transition region, making it difficult to separate the two fluctuations in reconstructed images. The ability of DC-DOT to isolate the influence of superficial artifacts from true brain activation when they have similar temporal patterns remains to be further studied and validated.
The capability of DC-DOT to discriminate brain activation from task-evoked superficial artifacts has many implications. Historically, functional NIRS used single source-detector separation (typically 2.5 to 4 centimeters) to detect brain activities. Then, data were processed in a channel-wise format, which did not allow components from the brain and scalp vasculature to be differentiated. Because the superficial artifacts are also task-evoked, they can lead to false positive findings in optical brain imaging. Adaptive filtering (Zhang et al., 2007a; 2007b; Zhang et al., 2009; Tian et al., 2011) and linear regression (Saager and Berger 2005; Saager et al., 2011), which are designed to remove the systemic fluctuations, may be used to reduce the task-evoked superficial artifacts. However, these methods have limitations. For example, if the superficial artifacts are from regional scalp vasculature, an adaptive filter or linear regression with a distant short-separation reference may not be able to remove them efficiently. Another scenario is that task-evoked superficial artifacts have a similar temporal frequency to that of brain activation (Takahashi et al., 2011). Then, the use of adaptive filtering or linear regression will remove not only the superficial artifacts but also the brain activation as well. This scenario may also occur when recording the brain activities in response to an external electrical or magnetic stimulation, such as repetitive transcranial magnetic stimulation (Näsi et al., 2012). In all of the above-mentioned scenarios, DC-DOT may provide an alternative solution, namely, to be able to discriminate the brain activation from the superficial artifacts according to their depths.
4.3 Node-wise GLM analysis used with DC-DOT
For the first time, the present study has implemented and performed node-wise GLM analysis on the time-dependent volumetric image series of DC-DOT. In both computer simulations and human measurements, the GLM-derived β-map in magnitude and t-map in statistical power agreed with each other, and both provided reasonable images of sensorimotor activations. In particular, linear regression of systemic fluctuations prior to GLM analysis was conducted by averaging the shallow nodes (depth < 1 cm) as a reference. This operation was different from the procedure given in the literature that used the 1st NN channels as a reference (Saager and Berger 2005; Saager et al., 2011). The reason is that the 1st NN source-detector separation in the current probe configuration was 1.6 cm, which was large enough to detect certain brain activities. Therefore, the use of 1st NN channels as a reference in this study would reduce the activation-related fluctuations significantly. We confirmed that using an average of shallow nodes as a reference helped decrease the systemic fluctuations efficiently while retaining the activation-related brain fluctuations.
Compared with several previous studies that used channel-wise GLM analysis, DC-DOT in combination with the node-wise GLM analysis has several advantages. First, DC-DOT improves the SNR of node-wise data in the region of activation as compared with the channel-wise data used in topography, as we demonstrated in time-dependent simulation (see Fig. 5). Therefore, it is expected that DC-DOT combined with node-wise GLM analysis will lead to a greater statistical power than channel-wise GLM analysis even though both methods may share the same raw dataset. Second, in channel-wise GLM analysis, post-GLM topographic images are often generated by interpolating data between adjacent channels, and thus the spatial resolution is limited to the distance between the adjacent channels. On the other hand, DOT itself has the ability to reach a high spatial resolution, surely higher than the source-detector separation, according to several published studies (Eggebrecht et al., 2012; Habermehl et al. 2012). Thus, the node-wise GLM analysis based on DC-DOT is expected to provide a higher spatial resolution while a validation study seems to be the next step. Finally, spatial comparison and co-registration between a 2D topographic image from channel-wise-based fNIRS and a 3D volumetric image from fMRI is indirect and difficult to perform. In contrast, the integration and correlation between images of DC-DOT and fMRI are straightforward because both of the modalities offer volumetric images. Therefore, DC-DOT in combination with node-wise GLM analysis, in a similar way to what fMRI does, will ease and benefit the direct comparison between these two modalities in future.
It is also noted that there are several differences between the fMRI-based GLM and DC-DOT-based GLM. The main difference is that while fMRI images each voxel directly, DC-DOT generates a volumetric image through a complex reconstruction procedure, where each voxel is interrogated by multiple source-detector channels possibly having different noise levels. This means that DC-DOT-based GLM analysis could deliver quite different outcomes, depending on the pre-processing and reconstruction procedures used to form time-dependent volumetric DC-DOT image series. Specifically, we have noticed several aspects that may affect the quality of reconstructed images and node-wise GLM analysis: first, a single source-detector channel with a poor SNR would severely affect a group of nodes/voxels along its optical pathway. To avoid degrading the image quality, data screening prior to DC-DOT reconstruction is necessary so as to identify and eliminate all removable noise sources among the qualified channels. Second, GLM analysis has to be combined with DC-DOT for its best operation. In a relevant study, Habermehl et al. (2012) performed classical t-test analysis on the volumetric image series measured by a high-density DOT system. Without using any depth compensation approach, however, they found that the identified regions of activation were mis-located in the superficial layer about 1.5 cm away from the brain. Their results highlight the importance of depth compensation in DOT prior to statistical analysis.
4.4 Limitations of the present study
At last, it is important to note that there are several limitations in the present study: First, it is known that the general brain atlas-based reconstruction leads to a greater localization error than the subject-specific image reconstruction due to a combination of imperfect optode registration, anatomical differences between the generalized brain atlas and individual brain, and so on (Cooper et al., 2012). In our human results, it is expected that the localization errors of cortical activation could be 1-2 centimeters, large enough to misidentify the primary motor and primary sensory activations.
Second, the present GLM analysis used in this study was based on a hemodynamic response function derived from fMRI. Because the two image modalities have different sensitivities to cerebrovascular components in their measurements, their hemodynamic response functions evoked by given tasks are not necessarily the same. Having an NIRS-specific hemodynamic response function would provide more accurate node-wise GLM analysis.
Third, we acknowledge that great improvement in depth localization of brain activation has been achieved by developing brain-atlas-guided DC-DOT combined with GLM analysis. Further refinement and advances are needed since the errors in depth localization of brain activation are still non-trivial and need to be reduced significantly for any meaningful clinical application. For example, we have consistently observed that the identified region of activation from the GLM-derived t-maps is deeper than the actual location as well as than the region resulting from the GLM-derived β-maps (see Figs. 6 to 8). One particular question worthy of further investigation is whether the node-wise, depth-compensation matrix, Mnode, is sufficient enough to achieve high-quality or high-SNR data so as to reconstruct localized images (or t-maps) at deep depths. Based on the discussion given in the Supplementary Material, we expect that eq. (12) needs to be refined in order to improve the spatial resolution along depth. One possible and feasible solution is to resume layer-wise DC-DOT by appropriately finding and segmenting specific layers along human head curvatures. The expectation that layer-wise DC-DOT may be able to improve the stretched t-maps in the “deeper” brain region, as seen in Figs. 6-8, is reasonable and needs to be tested in our future studies.
At last, the improvement on quantification accuracy has been good only for static absorbers at a shallow depth, not for time-dependent hemodynamic changes at deeper regions. Also, the spatial resolution of DC-DOT in lateral dimension was not studied. We noticed that a few methods (Kavuri et al., 2012; Shimokawa et al., 2012) were developed recently to improve the in lateral resolution of DOT. The combination of these methods with DC-DOT may further advance the image reconstruction algorithm and improve its 3D spatial resolution, but it is beyond the scope of the present work and may be a possible direction for future studies.
5. Conclusions
We have incorporated depth-compensated DOT (DC-DOT) with a standard brain atlas. Improved depth specificity and quantitative accuracy of brain atlas-based DC-DOT were demonstrated through simulation and human measurements of sensorimotor activation. For the first time, node-wise GLM analysis was implemented and performed on the volumetric image series of DC-DOT, showing the robustness of DC-DOT and its ability to accurately identify the brain activation at the correct depth, even in existence of task-evoked superficial artifacts.
Supplementary Material
Highlights.
Development of brain atlas-based, depth-compensated diffuse optical tomography (DC-DOT) for accurate volumetric functional brain imaging
Validation and demonstration through computer simulations to show that DC-DOT is able to specify the brain activation from the task-evoked superficial artifacts.
For the first time, robustness of node-wise GLM analysis in combination with DC-DOT is demonstrated and able to clearly identify the human sensorimotor activation.
Acknowledgements
This work was supported in part by NIH Grant R01 (5R33CA101098). The authors would like to thank Dr. Qianqian Fang at Department of Radiology, Harvard Medical School for his timely support in facilitating the use of iso2mesh software. The authors would also like to thank the NIRFAST group at Dartmouth College for their training on NIRFAST software.
Abbreviations
- 3D
three-dimensional
- BOLD
blood oxygen level dependent
- Con-DOT
conventional DOT without any spatially variant regularization or depth compensation
- DC
depth compensation
- DC-DOT
depth-compensated DOT
- DCA
depth-compensated algorithm
- DOT
diffuse optical tomography
- fMRI
functional magnetic resonance imaging
- FEM
finite element mesh
- GLM
general linear model
- Hb
deoxy-hemoglobin concentration
- HbO2
oxy-hemoglobin concentration
- HRF
hemodynamic response function
- MNI
Montreal Neurological Institute coordinates
- MRI
magnetic resonance imaging
- n
refractive index
- NIRFAST
a FEM-based MATLAB package for modeling propagation of near infrared light in biological tissues
- NIRS
near infrared spectroscopy
- NIRS-SPM
a SPM-based software package for functional NIRS data analysis
- OD
optical density
- SCF
sensitivity correction factor
- SNR
signal-to-noise ratio
- SPM
statistical parametric mapping
- SVR
spatially variant regularization
- SVR-DOT
apply spatially variant regularization to regular DOT reconstruction
- μa
absorption coefficient
- μs′
reduced scattering coefficient
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arridge SR. Optical tomography in medical imaging. Inverse Problems. 1999;15(2):R41. [Google Scholar]
- Beckmann CF, Jenkinson M, Smith SM. General multilevel linear modeling for group analysis in FMRI. NeuroImage. 2003;20:1052–1063. doi: 10.1016/S1053-8119(03)00435-X. [DOI] [PubMed] [Google Scholar]
- Boas DA, Gaudette T, Strangman G, Cheng X, Marota JJ, Mandeville JB. The accuracy of near infrared spectroscopy and imaging during focal changes in cerebral hemodynamics. NeuroImage. 2001;13(1):76–90. doi: 10.1006/nimg.2000.0674. [DOI] [PubMed] [Google Scholar]
- Boas DA, Dale AM, Franceschini MA. Diffuse optical imaging of brain activation: approaches to optimizing image sensitivity, resolution, and accuracy. NeuroImage. 2004a;23(Suppl. 1):S275–S288. doi: 10.1016/j.neuroimage.2004.07.011. [DOI] [PubMed] [Google Scholar]
- Boas DA, Chen K, Grebert D, Franceschini MA. Improving the diffuse optical imaging spatial resolution of the cerebral hemodynamic response to brain activation in humans. Optics Letters. 2004b;29:1506–1509. doi: 10.1364/ol.29.001506. [DOI] [PubMed] [Google Scholar]
- Bullmore E, Brammer M, Williams SC, Rabe Hesketh S, Janot N, David A, Mellers J, Howard R, Sham P. Statistical methods of estimation and inference for functional MR image analysis. Magn. Reson. Med. 1996;35(2):261–277. doi: 10.1002/mrm.1910350219. [DOI] [PubMed] [Google Scholar]
- Chance B, Zhuang Z, UnAh C, Alter C, Lipton L. Cognition activated low frequency modulation of light absorption in human brain. Proc. Natl. Acad. Sci. USA. 1993;90:2660–2774. doi: 10.1073/pnas.90.8.3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen MS. Parametric analysis of fMRI data using linear systems methods. NeuroImage. 1997;6(2):93–103. doi: 10.1006/nimg.1997.0278. [DOI] [PubMed] [Google Scholar]
- Collins DL, Zijdenbos AP, Baaré WFC, Evans AC. ANIMAL+INSECT: Improved Cortical Structure Segmentation. IPMI Lecture Notes in Computer Science. 1999;1613:210–223. [Google Scholar]
- Cooper RJ, Caffini M, Dubb J, Fang Q, Custo A, Tsuzuki D, Fischl B, Wells W, 3rd, Dan I, Boas DA. Validating atlas-guided DOT: a comparison of diffuse optical tomography informed by atlas and subject-specific anatomies. NeuroImage. 2012;62(3):1999–2006. doi: 10.1016/j.neuroimage.2012.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culver JP, Durduran T, Furuya D, Cheung C, Greenberg JH, Yodh AG. Diffuse optical tomography of cerebral blood flow, oxygenation, and metabolism in rat during focal ischemia. J. Cereb. Blood Flow Metab. 2003;23:911–924. doi: 10.1097/01.WCB.0000076703.71231.BB. [DOI] [PubMed] [Google Scholar]
- Custo A, Boas DA, Tsuzuki D, Dan I, Mesquita R, Fischl B, Grimson WE, Wells W., 3rd Anatomical atlas-guided diffuse optical tomography of brain activation. NeuroImage. 2010;49(1):561–567. doi: 10.1016/j.neuroimage.2009.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehghani H, White BR, Zeff BW, Tizzard A, Culver JP. Depth sensitivity and image reconstruction analysis of dense imaging arrays for mapping brain function with diffuse optical tomography. Appl. Opt. 2009a;48(10):D137–D143. doi: 10.1364/ao.48.00d137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehghani H, Eames ME, Yalavarthy PK, Davis SC, Srinivasan S, Carpenter CM, Pogue BW, Paulsen KD. Near infrared optical tomography using NIRFAST: Algorithm for numerical model and image reconstruction. Communications in Numerical Methods in Engineering. 2009b;25:711–732. doi: 10.1002/cnm.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delpy DT, Cope M, van der Zee P, Arridge S, Wray S, Wyatt J. Estimation of optical pathlength through tissue from direct time of flight measurement. Physics in Medicine and Biology. 1988;33(12):1433. doi: 10.1088/0031-9155/33/12/008. [DOI] [PubMed] [Google Scholar]
- Durduran T, Choe R, Baker WB, Yodh AG. Diffuse optics for tissue monitoring and tomography. Rep. Prog. Phys. 2010;73:076701. doi: 10.1088/0034-4885/73/7/076701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggebrecht AT, White BR, Ferradal SL, Chen C, Zhan Y, Snyder AZ, Dehghani H, Culver JP. A quantitative spatial comparison of high-density diffuse optical tomography and fMRI cortical mapping. NeuroImage. 2012;61(4):1120–1128. doi: 10.1016/j.neuroimage.2012.01.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Q, Boas DA. Tetrahedral mesh generation from volumetric binary and gray-scale images. Proceedings of IEEE International Symposium on Biomedical Imaging. 20092009:1142–1145. [Google Scholar]
- Franceschini MA, Boas DA, Zourabian A, Diamond SG, Nadgir S, Lin DW, Moore JB, Fantini S. Near-infrared spiroximetry: noninvasive measurements of venous saturation in piglets and human subjects. J Appl Physiol. 2002;92(1):372–384. doi: 10.1152/jappl.2002.92.1.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston KJ, Holmes A, Worsley K, Poline J. Statistical parametric maps in functional imaging: a general linear approach. Hum. Brain Mapp. 1995;2:189–210. [Google Scholar]
- Fonov VS, Evans AC, McKinstry RC, Almli CR, Collins DL. Unbiased nonlinear average age-appropriate brain templates from birth to adulthood. NeuroImage. 2009;47:S102–S102. [Google Scholar]
- Fonov VS, Evans AC, Botteron K, Almli CR, McKinstry RC, Collins DL, Brain Development Cooperative Group Unbiased average age-appropriate atlases for pediatric studies. NeuroImage. 2011;54(1):313–327. doi: 10.1016/j.neuroimage.2010.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson AP, Riley J, Schweiger M, Hebden JC, Arridge SR, Delpy DT. A method for generating patient-specific finite element meshes for head modelling. Phys Med Biol. 2003;48(4):481–495. doi: 10.1088/0031-9155/48/4/305. [DOI] [PubMed] [Google Scholar]
- Habermehl C, Holtze S, Steinbrink J, Koch SP, Obrig H, Mehnert J, Schmitz CH. Somatosensory activation of two fingers can be discriminated with ultrahigh-density diffuse optical tomography. NeuroImage. 2012;59(4):3201–3211. doi: 10.1016/j.neuroimage.2011.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshi Y, Tamura M. Detection of dynamic changes in cerebral oxygenation coupled to neuronal function during mental work in man. Neurosci. Lett. 1993;150(1):5–8. doi: 10.1016/0304-3940(93)90094-2. [DOI] [PubMed] [Google Scholar]
- Huettel SA, Song AW, McCarthy G. Functional magnetic resonance imaging. 2nd edition Sinauer Associates, Inc; Sunderland, MA, USA: 2009. [Google Scholar]
- Kavuri VC, Lin ZJ, Tian F, Liu H. Sparsity enhanced spatial resolution and depth localization in diffuse optical tomography. Biomedical Optics Express. 2012;3(5):943–957. doi: 10.1364/BOE.3.000943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirilina E, Jelzow A, Heine A, Niessing M, Wabnitz H, Brühl R, Ittermann B, Jacobs AM, Tachtsidis I. The physiological origin of task-evoked systemic artefacts in functional near infrared spectroscopy. NeuroImage. 2012;61(1):70–81. doi: 10.1016/j.neuroimage.2012.02.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Sun C, Lee P, Lee H, Yang C, Jiang C, Tong Y, Yeh T, Hsieh J. Study of photon migration with various source-detector separations in near-infrared spectroscopic brain imaging based on three-dimensional Monte Carlo modeling. Opt. Express. 2005;13:8339–8348. doi: 10.1364/opex.13.008339. [DOI] [PubMed] [Google Scholar]
- Leff DR, Orihuela-Espina F, Elwell CE, Athanasiou T, Delpy DT, Darzi AW, Yang GZ. Assessment of the cerebral cortex during motor task behaviours in adults: a systematic review of functional near infrared spectroscopy (fNIRS) studies. Neuroimage. 2011;54(4):2922–2936. doi: 10.1016/j.neuroimage.2010.10.058. [DOI] [PubMed] [Google Scholar]
- Liu H, Hielscher AH, Tittel FK, Jacques SL, Chance B. Influence of blood vessels on the measurement of hemoglobin oxygenation as determined by time-resolved reflectance spectroscopy. Medical Physics. 1995;22:1209–1217. doi: 10.1118/1.597520. [DOI] [PubMed] [Google Scholar]
- Näsi T, Mäki H, Kotilahti K, Nissilä I, Haapalahti P, Ilmoniemi RJ. Magnetic-stimulation-related physiological artifacts in hemodynamic near-infrared spectroscopy signals. PLoS One. 2011;6(8):e24002. doi: 10.1371/journal.pone.0024002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H, Tian F, Lin ZJ, Liu H. Development of a compensation algorithm for accurate depth localization in diffuse optical tomography. Opt. Lett. 2010;35:429–431. doi: 10.1364/OL.35.000429. [DOI] [PubMed] [Google Scholar]
- Obrig H, Neufang M, Wenzel R, Kohl M, Steinbrink J, Einhäupl K, Villringer A. Spontaneous low frequency oscillations of cerebral hemodynamics and metabolism in human adults. NeuroImage. 2000;12(6):623–639. doi: 10.1006/nimg.2000.0657. [DOI] [PubMed] [Google Scholar]
- O’Leary MA. Ph.D. dissertation. University of Pennsylvania; United States - Pennsylvania: 1996. Imaging with diffuse photon density waves. [Google Scholar]
- Plichta MM, Heinzel S, Ehlis A-C, Pauli P, Fallgatter AJ. Model-based analysis of rapid event-related functional near-infrared spectroscopy (NIRS) data: A parametric validation study. NeuroImage. 2007;35(2):625–634. doi: 10.1016/j.neuroimage.2006.11.028. [DOI] [PubMed] [Google Scholar]
- Pogue BW, McBride TO, Prewitt J, Österberg UL, Paulsen KD. Spatially variant regularization improves diffuse optical tomography. Appl. Opt. 1999;38:2950–2961. doi: 10.1364/ao.38.002950. [DOI] [PubMed] [Google Scholar]
- Saager RB, Berger AJ. Direct characterization and removal of interfering absorption trends in two-layer turbid media. J. Opt. Soc. Am. A. 2005;22(9):1874–1882. doi: 10.1364/josaa.22.001874. [DOI] [PubMed] [Google Scholar]
- Saager RB, Telleri NL, Berger AJ. Two-detector Corrected Near Infrared Spectroscopy (C-NIRS) detects hemodynamic activation responses more robustly than single-detector NIRS. NeuroImage. 2011;55(4):1679–1685. doi: 10.1016/j.neuroimage.2011.01.043. [DOI] [PubMed] [Google Scholar]
- Schroeter ML, Bucheler MM, Muller K, Uludag K, Obrig H, Lohmann G, Tittgemeyer M, Villringer A, von Cramon DY. Towards a standard analysis for functional near-infrared imaging. NeuroImage. 2004;21(1):283–290. doi: 10.1016/j.neuroimage.2003.09.054. [DOI] [PubMed] [Google Scholar]
- Shimokawa T, Kosaka T, Yamashita O, Hiroe N, Amita T, Inoue Y, Sato M. Hierarchical Bayesian estimation improves depth accuracy and spatial resolution of diffuse optical tomography. Optics Express. 2012;20(18):20427–20446. doi: 10.1364/OE.20.020427. [DOI] [PubMed] [Google Scholar]
- Singh AK, Okamoto M, Dan H, Jurcak V, Dan I. Spatial registration of multichannel multi-subject fNIRS data to MNI space without MRI. NeuroImage. 2005;27(4):842–851. doi: 10.1016/j.neuroimage.2005.05.019. [DOI] [PubMed] [Google Scholar]
- Strangman G, Franceschini MA, Boas DA. Factors affecting the accuracy of near-infrared spectroscopy concentration calculations for focal changes in oxygenation parameters. NeuroImage. 2003;18:865–879. doi: 10.1016/s1053-8119(03)00021-1. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Takikawa Y, Kawagoe R, Shibuya S, Iwano T, Kitazawa S. Influence of skin blood flow on near-infrared spectroscopy signals measured on the forehead during a verbal fluency task. NeuroImage. 2011;57(3):991–1002. doi: 10.1016/j.neuroimage.2011.05.012. [DOI] [PubMed] [Google Scholar]
- Tian F, Niu H, Khadka S, Lin ZJ, Liu H. Algorithmic depth compensation improves quantification and noise suppression in functional diffuse optical tomography. Biomed. Opt. Express. 2010;1:441–452. doi: 10.1364/BOE.1.000441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian F, Khan B, Niu H, Behbehani K, Alexandrakis G, Liu H. Enhanced Functional Brain Imaging by Using Adaptive Filtering and a Depth Compensation Algorithm in Diffuse Optical Tomography. IEEE Trans. Med. Imaging. 2011;30(6):1239–1251. doi: 10.1109/TMI.2011.2111459. [DOI] [PubMed] [Google Scholar]
- Tian F, Kozel A, Yennu A, Croarkin PE, McClintock SM, Mapes KS, Husain MM, Liu H. Test-retest assessment of cortical activation induced by repetitive transcranial magnetic stimulation with brain atlas-guided optical topography. J. Biomed. Opt. 2012;17(11):116020. doi: 10.1117/1.JBO.17.11.116020. [DOI] [PubMed] [Google Scholar]
- Tikhonov A. Solution of incorrectly formulated problems and the regularization method. Soviet Math. Doklady. 1963;4:1035–1038. [Google Scholar]
- Villringer A, Planck J, Hock C, Schleinkofer L, Dirnagl U. Near infrared spectroscopy (NIRS): a new tool to study hemodynamic changes during activation of brain function in human adults. Neurosci. Lett. 1993;154(1-2):101–104. doi: 10.1016/0304-3940(93)90181-j. [DOI] [PubMed] [Google Scholar]
- Villringer A, Chance B. Non-invasive optical spectroscopy and imaging of human brain function. Trends Neurosci. 1997;20:435–442. doi: 10.1016/s0166-2236(97)01132-6. [DOI] [PubMed] [Google Scholar]
- Wirgin A. The inverse crime. 2004. Arxiv preprint math-ph, 0401050. [Google Scholar]
- White BR, Culver JP. Quantitative evaluation of high-density diffuse optical tomography: in vivo resolution and mapping performance. J. Biomed. Opt. 2010;15:026006. doi: 10.1117/1.3368999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye JC, Tak S, Jang KE, Jung J, Jang J. NIRS-SPM: statistical parametric mapping for near-infrared spectroscopy. NeuroImage. 2009;44(2):428–447. doi: 10.1016/j.neuroimage.2008.08.036. [DOI] [PubMed] [Google Scholar]
- Zeff BW, White BR, Dehghani H, Schlaggar BL, Culver JP. Retinotopic mapping of adult human visual cortex with high-density diffuse optical tomography. Proc. Natl. Acad. Sci. USA. 2007;104(29):12169–12174. doi: 10.1073/pnas.0611266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan Y, Eggebrecht AT, Culver JP, Dehghani H. Image quality analysis of high-density diffuse optical tomography incorporating a subject-specific head model. Front Neuroenergetics. 2012;4:6. doi: 10.3389/fnene.2012.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Brown E, Strangman G. Adaptive filtering for global interference cancellation and real-time recovery of evoked brain activity: a Monte Carlo simulation study. J. Biomed. Opt. 2007a;12(4):044014. doi: 10.1117/1.2754714. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Brown E, Strangman G. Adaptive filtering to reduce global interference in evoked brain activity detection: a human subject case study. J. Biomed. Opt. 2007b;12(6):064009. doi: 10.1117/1.2804706. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Strangman G, Ganis G. Adaptive filtering to reduce global interference in non-invasive NIRS measures of brain activation: How well and when does it work? NeuroImage. 2009;45(3):788–794. doi: 10.1016/j.neuroimage.2008.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.