Abstract
As space travel is expanding to include private tourism and travel beyond low-Earth orbit, so is the risk of exposure to space radiation. Galactic cosmic rays and solar particle events have the potential to expose space travelers to significant doses of radiation that can lead to increased cancer risk and other adverse health consequences. Metabolomics has the potential to assess an individual’s risk by exploring the metabolic perturbations in a biofluid or tissue. In this study, C57BL/6 mice were exposed to 0.5 and 2 Gy of 1 GeV/nucleon of protons and the levels of metabolites were evaluated in urine at 4 h after radiation exposure through liquid chromatography coupled to time-of-flight mass spectrometry. Significant differences were identified in metabolites that map to the tricarboxylic acid (TCA) cycle and fatty acid metabolism, suggesting that energy metabolism is severely impacted after exposure to protons. Additionally, various pathways of amino acid metabolism (tryptophan, tyrosine, arginine and proline and phenylalanine) were affected with potential implications for DNA damage repair and cognitive impairment. Finally, presence of products of purine and pyrimidine metabolism points to direct DNA damage or increased apoptosis. Comparison of these metabolomic data to previously published data from our laboratory with gamma radiation strongly suggests a more pronounced effect on metabolism with protons. This is the first metabolomics study with space radiation in an easily accessible biofluid such as urine that further investigates and exemplifies the biological differences at early time points after exposure to different radiation qualities.
INTRODUCTION
Space exploration, while still primarily restricted to astronauts, is expanding to include future commercial travel through private companies and travel beyond low-Earth orbit to asteroids and Mars. One of the primary concerns for space travel is exposure to space radiation. Radiation exposure and more significantly space radiation have been connected with increased carcinogenesis, cataract development, neurological health risks of the central nervous system and cardiovascular disease, as well as with short-term adverse effects that could compromise mission success (1–6). Radiation can also leads to perturbations of metabolic processes and inflammatory processes, and these changes can be reflected in biofluids, such as urine and blood, that could provide a snap shot of an individual’s current physiological state. Metabolomics, the qualitative and quantitative assessment of small molecules, for example, can provide such a rapid identification of metabolites that would allow for the monitoring of radiation exposure. More importantly, metabolomics has the potential to provide individual assessment of disease risks based on levels of metabolic markers in easily accessible biofluids.
The main radiation exposure of astronauts is attributed to galactic cosmic rays (GCRs) and solar particle events (SPEs). GCRs originate from deep space, of which about 90% consist of very high-energy protons and helium, and to lesser extent heavy ions (HZE), such as 56Fe, 16O, 28Si and 12C. SPEs on the other hand consist of very high flux midenergy (~100 MeV) protons, helium and HZE ions with protons as >80% of the particles. While both types have the potential to expose astronauts to high levels of radiation, SPEs are particularly worrisome during extra vehicular activities as astronauts have little time to seek adequate protection in areas with more shielding. SPEs and GCRs have alternating patterns of activity. GCRs are highest during solar minimum, with significant increases in solar activity approximately every 11 years. While space exploration in low-Earth orbit, e.g. International Space Station, provides a certain protection from direct space radiation due to Earth’s magnetosphere, exploration beyond that point will certainly lead to higher levels of radiation exposure for astronauts.
Previous radiation metabolomic studies from our laboratory have focused on external gamma exposure, as in situations that may be encountered during radiological and nuclear events, and our extensive efforts have established the field of radiation metabolomics (7–10). Here we extend the studies to external proton irradiation, as may be encountered by astronauts in space. As protons, with a relative biological effectiveness (RBE) of 1.1, constitute the majority of GCRs and SPEs, it is imperative to assess their biological significance and determine the differences and similarities between the two radiation qualities (11) and more importantly the overall metabolic responses.
In the current study, mouse urine was analyzed through ultra performance liquid chromatography coupled to time-of-flight mass spectrometry (UPLC-TOFMS) at 4 h post exposure to 0.5–2 Gy of 1 GeV/nucleon of protons. Results were compared to previously published gamma-radiation urinary results for generation of comparative metabolomic biosignatures and additional biomarkers for protons exposure were mapped to biochemical pathways. Observed metabolic perturbations included altered amino acid excretion, purine and pyrimidine metabolism pathway molecules and TCA cycle products. To the best of our knowledge, this is the first metabolomics study in urine from mice exposed to radiation qualities that may be encountered in space and the first study to assess metabolic changes of space radiation at an early time point. We propose that metabolomics, through the generation of biosignatures in easily accessible biofluids, can provide a rapid method for monitoring radiation exposure. This may be implemented in future space exploration to not only assess biologic effects of exposure but also future health effects.
MATERIALS AND METHODS
Mice, Radiation Exposure and Sample Collection
Eight-week-old male C57BL/6 mice were bred at Georgetown University according to Georgetown University Institutional Animal Care and Use Committee (GUACUC) protocols (#12–035) and shipped to Brookhaven National Laboratory (BNL) one week prior to radiation exposure. Radiation exposure to protons (1 GeV/nucleon, LET 0.25 keV/µm) was performed at the NASA Space Radiation Laboratory (NSRL), as previously described. Mice were either sham irradiated (n = 8), or exposed to 0.5 Gy (n = 8) or 2 Gy (n = 7) of protons. Spot urine samples were collected at 4 h postirradiation either just before (spontaneous bladder voiding) or directly after euthanasia with CO2 (direct needle aspiration of the bladder) and stored at−80°C until further processing.
Sample Processing and Data Acquisition
Water and acetonitrile were of analytical grade or higher purity. Debrisoquine sulfate and nitrobenzoic acid were obtained from Sigma- Aldrich (St. Louis, MO) and utilized as internal standards. Urine samples were processed as previously described (7) and analyzed with a BEH C18 Column, 130 Å, 1.7 µM, 2.1 × 50 mm on an Acquity UPLC H-Class (Waters Corporation, MA) coupled to a Xevo G2 mass spectrometer (Waters Corporation, UK) (UPLC-TOFMS). The capillary voltage was set at 3.0 kV and the source temperature at 120°C. Injections consisted of 2 µl with a column temperature of 40°C and flow rate of 0.5 mL/min. Mobile phase A consisted of water + 0.1% formic acid and Mobile phase B consisted of acetonitrile + 0.1% formic acid. The gradient was as follows: 0–0.5 min consisted of 95% A; 0.5–4 min from 95% A to 80% A; 4–8 min from 80% A to 5% A; 8–9 min maintain A at 5%; 9–9.1 min from 5% to 95% A; and 9.1– 10.5 min maintain 95% A. The Xevo G2 was operated in both positive and negative electrospray ionization modes with a range of 50–1,200 m/z and mass spectrometry data was acquired in centroid mode.
Data Processing and Analysis
Data processing was conducted using Progenesis QI Informatics (Nonlinear Dynamics, Newcastle, UK). Normalization was performed to all compounds. Data analysis was conducted with the statistical software package MetaboLyzer (12). Statistically significant ions were identified through Welch’s t test (complete presence ions) and Fisher’s exact test (partial presence ions) with a P < 0.05 considered significant. Putative identities were assigned according to matches with databases incorporated in MetaboLyzer [Human Metabolome Database (HMDB), Kyoto Encyclopedia of Genes and Genomes (KEGG) and Lipid Maps] with a ppm error of <10. Multivariate data analysis was conducted with the software SIMCA-P+ version 13.0 and patterns of global metabolite changes in each sample according to their relative concentrations were represented as heatmaps with the Gene Expressions Dynamics Inspector (GEDI) software, utilizing Euclidean distance. GEDI utilizes self-organizing maps (SOM) and have been successfully employed to represent radiation changes through metabolomics in previous studies (9, 13). Groups of ions with similar patterns are clustered together in a tile or in the neighboring vicinity of the grid. Sample integrity is preserved in each analysis and each tile in the same analysis across samples represents the same ions, hence allowing for the direct interpretation of the results as a dose response. More information regarding GEDI can be found on the website (http://bit.ly/1yN3c0P).
Validation of Putative Metabolites Through Tandem Mass Spectrometry
Verification of the identity of the putative metabolites was performed through tandem mass spectrometry (MS/MS) against pure chemicals. Pyridoxine was obtained from Acros Organics (Fair Lawn, NJ) and hexanoylglycine was obtained from ONBIO, Inc. (Ontario, Canada). All other chemicals were obtained directly from Sigma- Aldrich (St. Louis, MO). Pure chemicals were diluted in 50:50 acetonitrile:water and fragmented with ramping collision energy between 10 and 50 eV. Fragmentation patterns of the pure chemicals were compared to fragmentation patterns of putative metabolites in urine samples. As an additional confirmatory step, the fragmentation patterns were compared to those on the online database METLIN (http://metlin.scripps.edu/index.php).
RESULTS
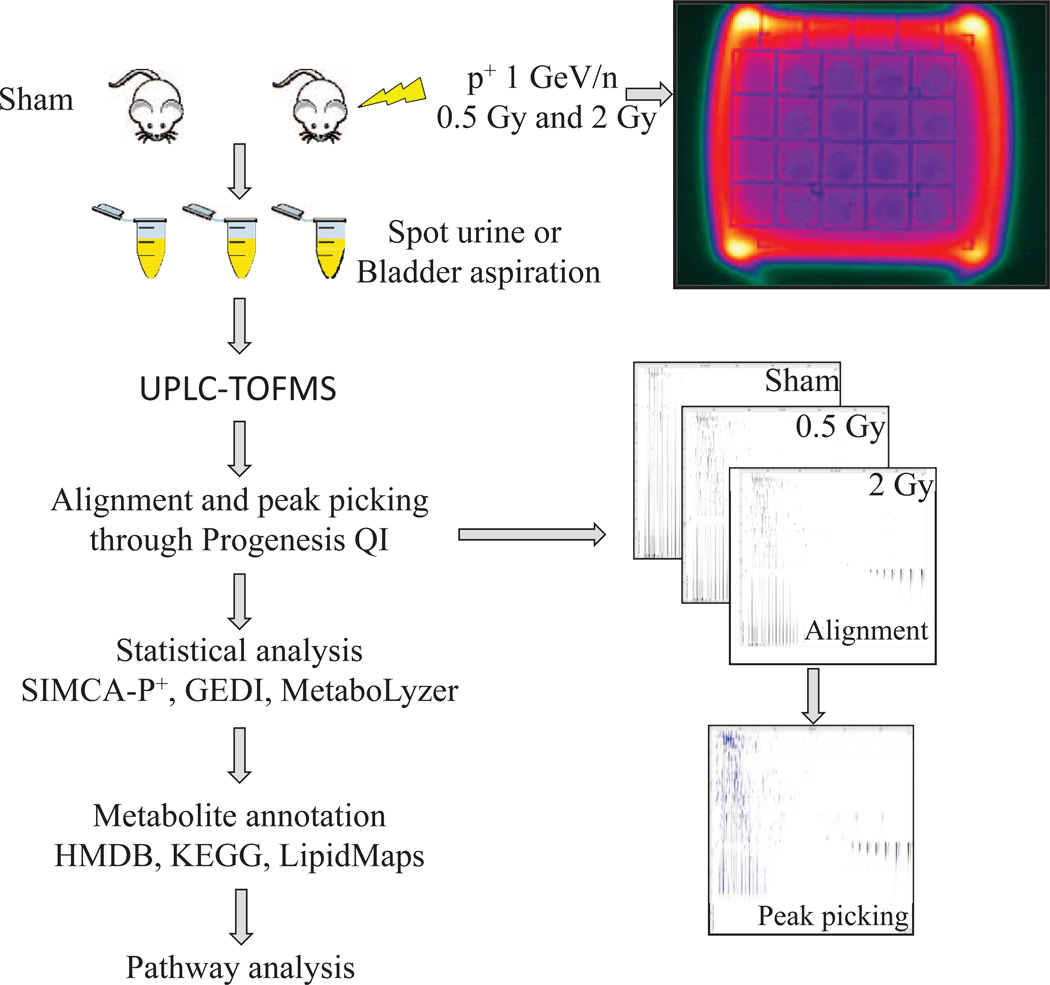
Previous metabolomic studies from our laboratory have focused on creating a urinary metabolomic signature specific to gamma-ray exposure. The current study concentrated on the urinary metabolomics of protons irradiation and compared the results to those published with gamma rays. A graphical depiction of the experimental method is shown in Fig. 1. Following peak alignment, peak picking, and deconvolution with the software Progenesis QI, the levels of creatinine were analyzed as a potential normalization method, as has been used in past studies of urinary metabolomics (7–10). However, given that the samples acquired were spot urines and therefore not a true representation of the rate of creatinine excretion, creatinine levels were not comparable between irradiated and nonirradiated groups (Supplementary Fig. S1; http://dx.doi.org/10.1667/RR13967.1.S1). Therefore, a different normalization method was employed, normalizing to all compounds, through Progenesis QI.
FIG. 1.
Study design and workflow for metabolomic analysis. C57BL/6 mice were irradiated with 0.5 or 2 Gy of protons (p+) 1 GeV/n or sham irradiated at the NSRL facility at BNL. Urine was collected at 4 h postirradiation either through spontaneous bladder voiding or direct needle aspiration of the bladder after euthanasia with CO2. Metabolomic profiling was performed with a Waters UPLC-TOFMS. Raw chromatographic data was aligned and peaks were picked with the software Progenesis QI. Following statistical analysis, markers were assigned putative identities through online database searches and pathway assignment was conducted through KEGG searches.
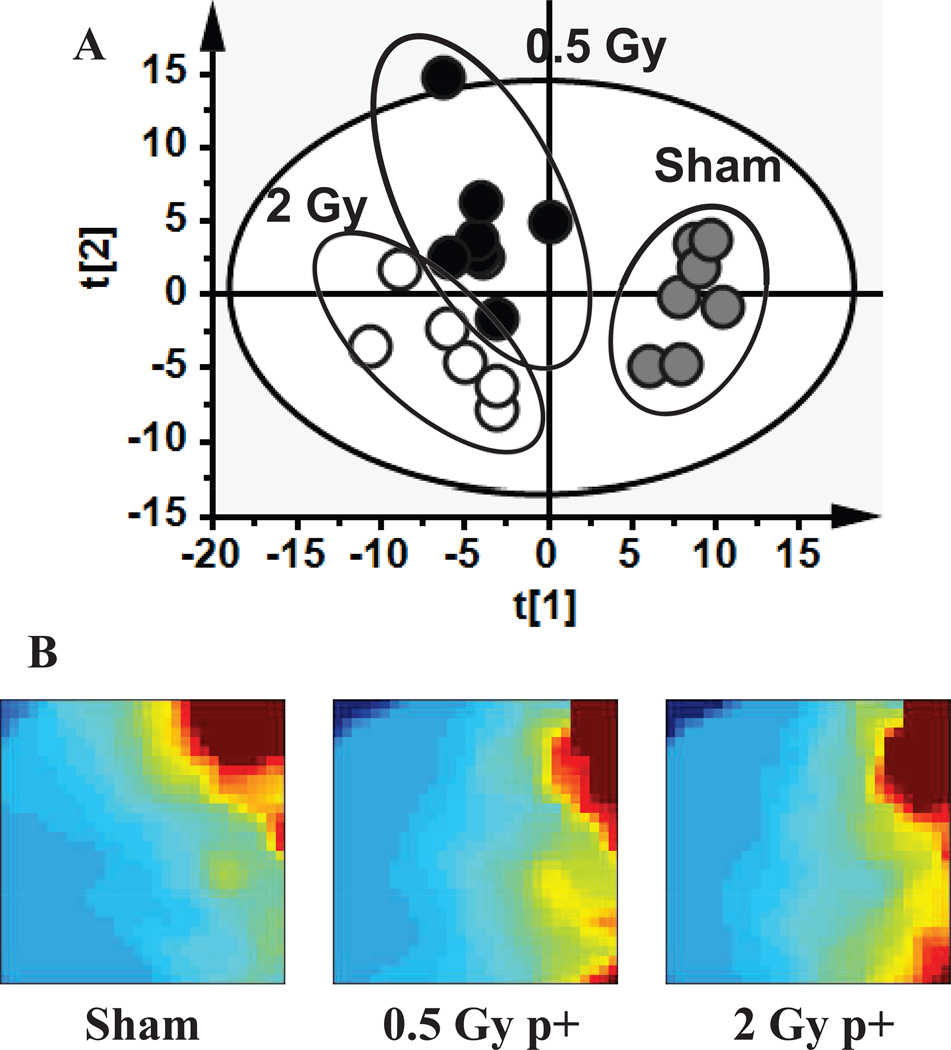
A principal component analysis through SIMCA-P+ utilizing the ESI+ data showed distinct clustering of the samples in their respective experimental groups (Fig. 2A), with R2X(cum)=0.221 and Q2(cum)=0.0936 for the first component and R2X(cum) = 0.316 and Q2(cum) = 0.0455 for the second component. Overall, separation between the groups based on their global metabolomic differences was concentrated on principal component 1. Analysis with the statistical features Distance to Model X and Hotelling’s T2Range did not reveal any outliers in the sample set, so all samples were included in further analysis. Global metabolomic profiling with the software GEDI utilizing self-organizing maps revealed patterns of increased and decreased levels of metabolites, as shown in Fig. 2B. As evident, there is a dose-response alteration in levels of metabolite excretion, signifying significant early perturbations of metabolic pathways after exposure to high-energy protons.
FIG. 2.
Principal component analysis (panel A) of metabolomics data from the positive ionization mode reveals distinct clustering of urine samples in their respective experimental groups. Self-organizing maps (panel B) show a dose response of biomarkers with increasing changes in patterns of upregulated regions.
Analysis through MetaboLyzer identified statistically significant ions (P < 0.05) with both Welch’s t test and Fisher’s exact test. Since analysis with MetaboLyzer is binary, four different runs were conducted, all utilizing sham irradiated as the controls. The number of statistically significant ions per analysis is summarized in Supplementary Table S1 (http://dx.doi.org/10.1667/RR13967.1.S2). Putative identifications through databases (KEGG, HMDB and Lipid maps) allowed for pathway analysis and comparison with previously reported urinary metabolites of gamma-radiation exposure. In particular, as depicted in Table 1, similar excretion patterns were identified for uric acid, N-acetylspermidine, hexanoylglycine, 2′-deoxyuridine and creatine between external gamma exposure and protons. While common pathways perturbed by the two different types of ionizing radiation were identified, differences were observed for citric acid, oxoglutaric acid, hippuric acid and choline, allowing for distinction between the two types of radiation exposure.
TABLE 1.
Extrapolation of Previously Published Urinary Radiation Markers (Gamma or X-ray Radiation) to Urine from Mice Exposure to Protons (p+)
| Metabolite identity | Published markers of external single dose gamma or X-ray exposure |
Single dose protons p+ |
Publication |
|---|---|---|---|
| Citric acid | ↓ | Dose specific | a |
| Uric acid | ↑ | ↑ | b |
| Oxoglutaric acid | ↓ | Dose specific | a |
| N-acetylspermidine | ↑ | ↑ | c |
| Hippuric acid | No change on day 1 | ↓ | a |
| Hexanoylglycine | ↑ | ↑ | d |
| 2′-Deoxyuridine* | ↑ | ↑ | c,e |
| Creatine | ↑ | ↑ | a |
| Choline* | ↑ | ↓ | a |
Chen, C., Brenner, D.J., Brown, T.R. Identification of urinary biomarkers from X-irradiated mice using NMR spectroscopy. Radiat Res 2011; 175:622–30.
Laiakis, E.C., Hyduke, D.R., Fornace, A.J. Comparison of mouse urinary metabolic profiles after exposure to the inflammatory stressors gamma radiation and lipopolysaccharide. Radiat Res 2012; 177:187–99.
Manna, S.K., Krausz, K.W., Bonzo, J.A., Idle, J.R., Gonzalez, F.J. Metabolomics reveals aging-associated attenuation of noninvasive radiation biomarkers in mice: potential role of polyamine catabolism and incoherent DNA damage-repair. J Proteome Res 2013; 12:2269–81.
Tyburski, J.B., Patterson, A.D., Krausz, K.W., Slavik, J., Fornace, A.J.J., Gonzalez, F.J. et al. Radiation metabolomics. 1. Identification of minimally invasive urine biomarkers for gamma-radiation exposure in mice. Radiat Res 2008; 170:1–14.
Tyburski, J.B., Patterson, A.D., Krausz, K.W., Slavik, J., Fornace, A.J.J., Gonzalez, F.J. et al. Radiation metabolomics. 2. Dose- and time-dependent urinary excretion of deaminated purines and pyrimidines after sublethal gamma-radiation exposure in mice. Radiat Res 2009; 172:42–57.
Designates putative metabolities in p+ experiments.
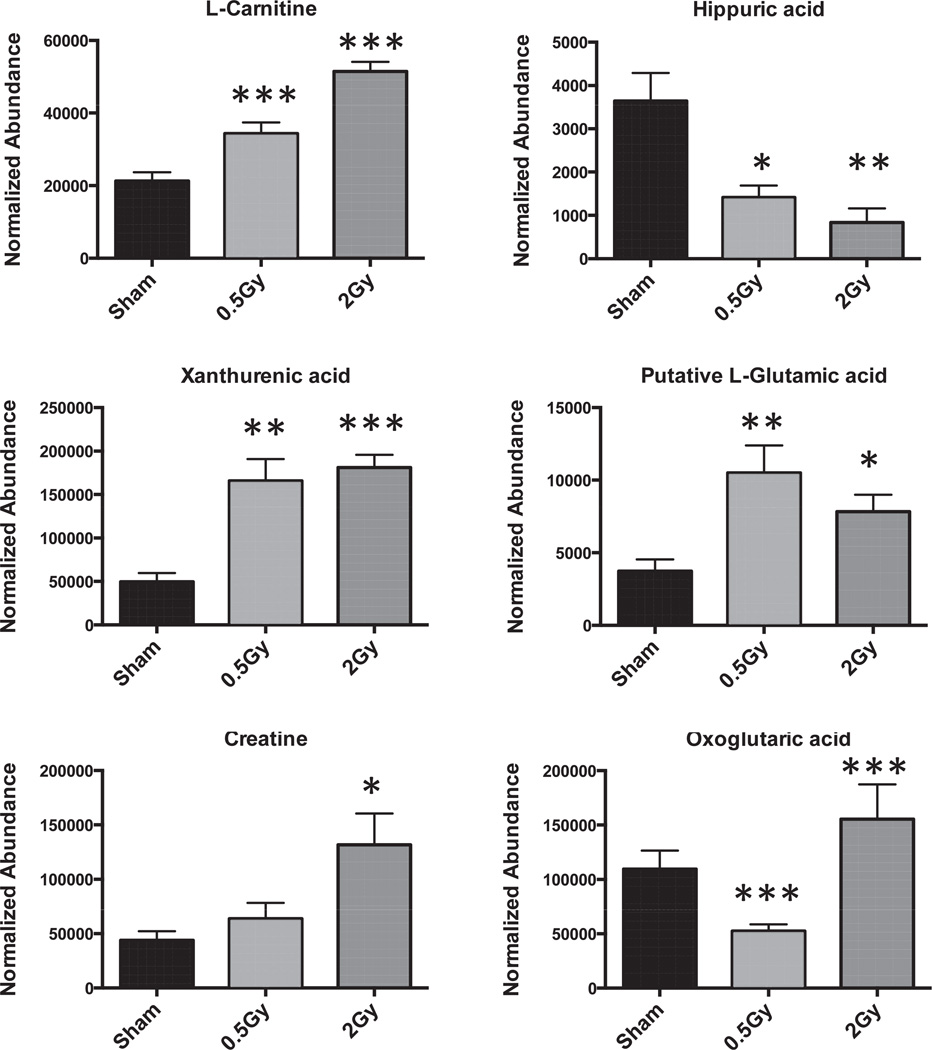
Select metabolites were validated through tandem mass spectrometry and fragmentation patterns were matched with those generated from pure standards and verified with the online database METLIN. Those results are shown in Table 2, where the validated metabolites are marked with an asterisk. The fold changes and P values for each of the metabolites (validated and putative) are also shown in Table 2. The highest fold changes are observed with dihydrouracil (8.34 for 0.5 Gy and 5.2 for 2 Gy), methylmalonic acid (12.34 for 0.5 Gy and 6.44 for 2 Gy) and 2′-deoxyuridine (138.48 for 0.5 Gy and 339.38 for 2 Gy). For other metabolites dose specific changes were observed, with examples of different patterns of excretion highlighted in Fig. 3. Regarding specific identities of altered metabolite excretion post proton exposure, pathway analysis revealed a significant number of putative ions mapping to the purine and pyrimidine pathways, the TCA cycle, fatty acid metabolism and various amino acids (Table 3).
TABLE 2.
Fold Changes and P Values of Putative Markers in Urine of Mice Exposed to Protons
| Fold change | P value | |||
|---|---|---|---|---|
| Metabolite | 0.5 Gy | 2 Gy | Sham vs. 0.5 Gy |
Sham vs. 2 Gy |
| Dihydrouracil | 8.34 | 5.20 | 0.0004 | 0.0008 |
| L-carnitine* | 1.62 | 2.42 | 0.005 | <0.0001 |
| Phenylpyruvic acid | 0.58 | 0.37 | 0.0441 | 0.0055 |
| Xanthurenic acid* | 3.33 | 3.64 | 0.002 | <0.0001 |
| Epinephrine | 0.31 | 0.41 | 0.0071 | 0.0152 |
| Norepinephrine | 0.20 | 0.16 | 0.0071 | 0.0058 |
| N-acetylornithine | 0.89 | 1.41 | 0.6917 | 0.2212 |
| N-acetyl-L-alanine | 2.36 | 2.35 | <0.0001 | <0.0001 |
| Aminoadipic acid | 2.47 | 2.28 | <0.0001 | 0.0008 |
| Fumaric acid | 3.09 | 2.75 | 0.0019 | 0.0028 |
| Citric acid* | 0.58 | 1.05 | 0.0005 | 0.6258 |
| Leucine/isoleucine | 2.78 | 1.69 | 0.0071 | 0.1228 |
| L-proline | 0.47 | 0.49 | 0.0198 | 0.031 |
| L-glutamic acid | 2.82 | 2.10 | 0.0083 | 0.0143 |
| L-phenylalanine | 0.44 | 0.26 | 0.002 | 0.0003 |
| Methylmalonic acid | 12.34 | 6.44 | 0.0028 | 0.1242 |
| SDMA | 1.49 | 1.83 | 0.0107 | 0.0003 |
| Pantothenic acid* | 1.23 | 1.67 | 0.1023 | 0.0004 |
| Kynurenic acid* | 1.45 | 2.01 | 0.1066 | 0.0006 |
| N-acetylspermidine* | 1.82 | 2.77 | 0.0171 | 0.005 |
| Uric acid* | 2.91 | 3.31 | 0.0019 | 0.012 |
| Hippuric acid* | 0.39 | 0.23 | 0.0102 | 0.0028 |
| Hexanoylglycine* | 2.03 | 3.63 | 0.0016 | 0.0034 |
| Creatine* | 1.45 | 3.00 | 0.2571 | 0.0216 |
| Oxoglutaric acid* | 0.48 | 1.42 | 0.0005 | 0.0007 |
| Choline | 0.68 | 0.15 | 0.4864 | 0.041 |
| 2′-deoxyruridine | 138.48 | 339.38 | 0.01 | 0.0014 |
Designates metabolites that were validated through MS/MS by matching to fragmentation patterns of pure chemicals and/or through the online database METLIN.
FIG. 3.
Representative bar graphs of validated metabolites following exposure to p+. Differential patterns are observed, with dose difference for select markers (L-carnitine, L-glutamic acid, creatine, oxoglutaric acid). However, some markers (hippuric acid and xanthurenic acid) exhibit responses, independent of dose. Results are represented as mean 6 SEM. Statistical significance was calculated with Welch’s t test between control (sham) and treated (0.5 or 2 Gy). P values are represented with asterisks, where *<0.05, **<0.01 and ***<0.001.
TABLE 3.
Urinary Markers Detected after Exposure to p+
| No. | Adduct | Found m/z | ppm error | Metabolite | Elemental composition |
Biofunction according to HMDB |
|---|---|---|---|---|---|---|
| 1 | [M+H]+ | 115.0509 | 5.85 | Dihydrouracil | C4H6N2O2 | Pyrimidine metabolism; pantothenate and CoA biosynthesis; beta-Alanine metabolism |
| 2 | [M+H]+ | 162.1123 | 0.92 | L-carnitine | C7H15NO3 | Fatty acid metabolism; alanine and aspartate metabolism |
| 3 | [M+H]+ | 165.0537 | 5.87 | Phenylpyruvic acid | C9H8O3 | Phenylalanine, tyrosine, tryptophan metabolism |
| 4 | [M+H]+ | 206.044 | 3.75 | Xanthurenic acid | C10H7NO4 | Tryptophan metabolism |
| 5 | [M+Na]+ | 206.0786 | 0.98 | Epinephrine | C9H13NO3 | Tyrosine metabolism; Neuroactive ligand-receptor interaction |
| 6 | [M+Na]+ | 192.0624 | 3.86 | Norepinephrine | C8H11NO3 | Tyrosine metabolism; Neuroactive ligand-receptor interaction; Precursor of epinephrine |
| 7 | [M+H]+ | 175.1063 | 8.06 | N-acetylornithine | C7H14N2O3 | Arginine and proline metabolism |
| 8 | [M−H]− | 130.0503 | 4.87 | N-acetyl-L-alanine | C5H9NO3 | Protein synthesis and amino acid biosynthesis |
| 9 | [M−H]− | 160.0611 | 2.6 | Aminoadipic acid | C6H11NO4 | Lysine metabolism |
| 10 | [M−H]− | 115.0031 | 5.01 | Fumaric acid | C4H4O4 | TCA cycle |
| 11 | [M−H]− | 191.0187 | 5.45 | Citric acid | C6H8O7 | TCA cycle |
| 12 | [M−H]− | 130.0873 | 0.15 | Leucine/isoleucine | C6H13NO2 | Branched chain amino acids |
| 13 | [M−H2O−H] | 96.0452 | 2.62 | L-proline | C5H9NO2 | Arginine and proline metabolism |
| 14 | [M−H]− | 146.0453 | 3.75 | L-glutamic acid | C5H9NO4 | Amino acid, various metabolic pathways |
| 15 | [M−H2O−H]− | 146.0606 | 0.14 | L-phenylalanine | C9H11NO2 | Phenylalanine, tyrosine, tryptophan metabolism |
| 16 | [M−H2O−H]− | 99.0083 | 0.61 | Methylmalonic acid | C4H6O4 | Pyrimidine metabolism; Valine, leucine and isoleucine metabolism |
| 17 | [M−H]− | 273.0916 | 9.4 | SDMA | C8H18N4O2 | Endogenous inhibitor of nitric oxide synthase |
| 18 | [M−H]− | 218.1029 | 2.2 | Pantothenic acid | C9H17NO5 | Pantothenate and CoA biosynthesis; beta-Alanine metabolism |
| 19 | [M−H]− | 188.0349 | 2.33 | Kynurenic acid | C10H7NO3 | Tryptophan metabolism |
| 20 | [M+H]+ | 188.175 | 3.71 | N-acetylspermidine | C9H21N3O | Polyamine |
| 21 | [M−H]− | 167.0204 | 3.86 | Uric acid | C5H4N4O3 | Purine metabolism |
| 22 | [M+H]+ | 180.0637 | 10 | Hippuric acid | C9H9NO3 | Phenylalanine metabolism |
| 23 | [M−H]− | 172.0979 | 5 | Hexanoylglycine | C8H15NO3 | Metabolite of fatty acids |
| 24 | [M+H]+ | 132.0764 | 2.63 | Creatine | C4H9N3O2 | Glycine, serine, threonine metabolism; Arginine and proline metabolism |
| 25 | [M−H]− | 145.0137 | 3 | Oxoglutaric acid | C5H6O5 | TCA cycle |
| 26 | [M+Na]+ | 127.0968 | 3.15 | Choline | C5H14NO | Glycine, serine, threonine metabolism; Glycerophospholipid metabolism; Bile secretion |
| 27 | [M−H]− | 227.0676 | 0.4 | 2′-deoxyruridine | C9H12N2O5 | Pyrimidine nucleosides and analogues |
DISCUSSION
We previously reported on the metabolomics of urine from mice exposed to gamma rays. In this study, we extended the urinary metabolomics to identify markers of exposure to protons. As part of GCRs, high-energy protons can affect space missions and satellites and expose astronauts to significant doses of radiation. Mid-energy protons constitute part of solar particle events, encountered primarily during solar maximum. In this study we investigated the effects of high-energy protons in the mouse metabolome, as reflected in the urine at 4 h postirradiation.
While creatinine levels are primarily utilized for normalization of urinary metabolomic data to account for changes in glomerular filtration rates, they are most informative when urine is collected over a predetermined period of time, i.e. 24 h, when indication of kidney injury can be inferred by increased levels of creatinine excretion. Spot urine samples, while giving a snapshot of the metabolism at a given moment, can often times be misleading regarding the creatinine levels and kidney function. As such, different normalization methods offer a more robust method for biomarker discovery. This issue was encountered in previous studies and different normalization methods were discussed (8). One such method, normalization to all compounds, was utilized in this study in the post-processing step, to account for the differences in urinary excretion and express the level of each ion as a fraction of total metabolites.
As has been reported with gamma radiation, a dose specific change in the excretion of urinary metabolites is evident after proton exposure as well. Previous gamma- and X-ray studies (Table 1) have utilized sublethal but also lethal (LD50/30 and above) doses, primarily within the first 24 h postirradiation (over a 24 h time period or conducting a time course study). The overall metabolomic profiles in this study show an increase in excretion of metabolite levels with the higher dose of 2 Gy as early as 4 h postirradiation. This is consistent with time studies with gamma radiation, where some biomarkers exhibited increased excreted levels between 6 and 8 h after exposure (10). It could be inferred therefore that on the urinary metabolomic level, the global metabolomic responses between different radiation types with similar RBEs show similar patterns, at least at early time points after exposure.
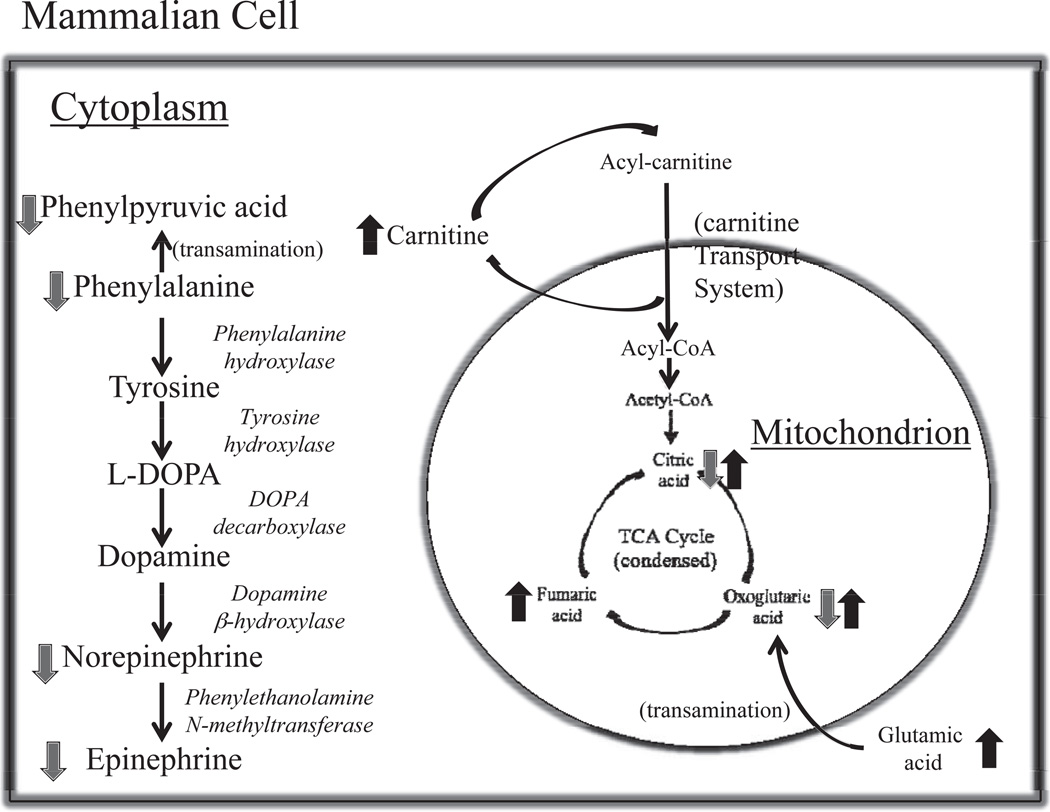
Although the similarities exist on a global level, significant differences between the two types of radiation began to emerge when we investigated individual metabolites. Based on previous identified metabolites of external gamma exposure, we compared and contrasted these metabolites with this study. As was shown in Table 1, markers such as citric acid, oxoglutaric acid, hippuric acid, and choline show either a trend opposite of those previously identified with gamma radiation or a dose difference that was not observed after gamma exposure. Further analysis of the pathway perturbations indicates that the TCA cycle displays important perturbations in the first steps, resulting in affected levels of citric acid, oxoglutaric acid and fumaric acid, as seen in Fig. 4. Previous studies with cells have shown that exposure to low doses of protons decreases the activity of certain enzymes involved in the TCA cycle in the progeny of bystander cells such as aconitase, without an effect at higher doses such as 2 Gy (14). In our studies this can translate into urinary markers, as excretion of citric acid and the downstream oxoglutaric acid are reduced after a 0.5 Gy dose and increased after exposure to the higher dose of 2 Gy. Fumaric acid levels on the other hand showed increased excretion with both doses, however the fold change was more prominent with 0.5 Gy. This might suggest that with increasing dose of protons, the TCA cycle is progressively unable to maintain a balance for sufficient energy production. As levels of proteins can be directly affected by oxidative stress due to increased levels of reactive oxygen species (ROS) after proton irradiation (11), so the intermediary metabolite levels can be dysregulated as a direct effect of insufficient metabolism. The TCA cycle’s primary role is to generate energy that can be eventually fed into the oxidative phosphorylation pathway to produce ATP and its dysregulation can have a direct effect on downstream pathways and processes, such as in apoptosis and DNA repair, which both have been found to be substantially impacted after proton exposure (11).
FIG. 4.
Pathway illustrations of some of the most important metabolic perturbations observed with proton irradiation. The TCA cycle is severely impacted, hence overall energy metabolism, while changes in the tyrosine pathway and particularly the catecholamines may impact cognitive abilities.
The TCA cycle is further impacted by evidence of insufficient β-oxidation of fatty acids. The primary component for transportation of fatty acids into the mitochondria, L-carnitine, is significantly excreted in both low and higher doses of protons. This urinary marker is novel and specific to external proton exposure compared to gamma exposure. Impairment of entry of fatty acids to the mitochondria to be metabolized to acetyl-CoA, a precursor of the TCA cycle, leads to energy metabolism defects. To further demonstrate that radiation exposure has a generalized effect on energy metabolism, hexanoylglycine, a marker of gamma radiation exposure (9) and, in this study, of proton exposure, shows increased levels in urine. This marker is generally associated with medium chain fatty acid (MCFA) metabolism impairment; MCFA enter the mitochondria in a process other than the carnitine transport for preferential oxidation (15).
Additional important perturbations with subsequent potential generalized effects involve the metabolism of various amino acids. In particular tryptophan, tyrosine, arginine and proline, and phenylalanine metabolism all displayed significant disorganization in urine from proton-irradiated mice compared to the sham irradiated. This generalized alteration of amino acid metabolism, as evident in urine, has not been previously observed or reported with external gamma radiation and can therefore be attributed to direct exposure to high-energy protons. Perturbations of amino acid levels can have a potential impact on both protein levels and therefore DNA damage repair. Interestingly, part of the tyrosine pathway that shows significant changes is associated with the production of norepinephrine and epinephrine, with the pathway and the changes shown in Fig. 4. Decreases in catecholamines and more specifically norepinephrine have been associated with cognitive impairment (16–19) after exposure to space radiation. Alternations in cognitive abilities remains a primary area of research for NASA and other space agencies, e.g. ESA, and models of risk assessments are being developed as more scientific evidence regarding the effects of space radiation of the central nervous system are published.
A final theme that emerges from our analysis is the generalized perturbations of purine and pyrimidine metabolism. We have previously described metabolomic markers of external gamma radiation exposure that map to the catabolism of purines (8) or pyrimidines (9, 10) and elevated excretion was observed in a dose specific manner as early as 4 h postirradiation (10). Early excretion of similar markers was also observed in this proton study, consistent with the damage associated with ionizing radiation. It remains however to determine whether these markers are direct products of clustered DNA damage and/or increased apoptosis, both of which are more extensive with exposure to protons versus photons (11, 20–22).
As long term effects of exposure to space radiation persist and can lead to carcinogenesis and central nervous system effects, as has been previously demonstrated in mouse models (4, 5, 16, 17, 23–25), it is imperative to understand the early effects of radiation exposure and the metabolic changes that can contribute to genetic alterations through persistent inflammatory or oxidative responses. Urinary metabolomics can rapidly provide a snapshot of the metabolic state of an individual and as shown this first study of exposure to protons identify a significant number of metabolites that are involved in important biochemical processes. Understanding and monitoring the metabolic changes due to radiation exposure can potentially lead to development of effective protective countermeasures against space radiation.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank Ms. Juijung Chung, MS for invaluable help with sample preparation for LC-MS analysis and Ms. Pelagie Ake for breeding of the mice. This work was funded by the National Space Biomedical Research Institute (NSBRI) grant PF019004 (P.I., DT) under NASA Cooperative Agreement #NCC 9–58, and NASA Grant NNX09AU95G (P.I., AJF Jr). The authors acknowledge the Lombardi Comprehensive Cancer Proteomics and Metabolomics Shared Resource (PMSR), which are in part supported by Award Number P30CA051008 (P.I., LW) from the National Cancer Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
Footnotes
The online version of this article (DOI: 10.1667/RR13967.1) contains supplementary information that is available to all authorized users.
SUPPLEMENTARY INFORMATION
Fig. S1. Creatinine levels at 4 h postirradiation. Since levels of exposed group show statistically significant differences compared to the sham control, this marker is not a reliable method for normalization.
Table S1. Number of statistically significant ions for each ionization mode as determined through analysis with the software MetaboLyzer.
REFERENCES
- 1.Chancellor JC, Scott GB, Sutton JP. Space radiation: the number one risk to astronaut health beyond low earth orbit. Life (Basel) 2014;4:491–510. doi: 10.3390/life4030491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kennedy AR. Biological effects of space radiation and development of effective countermeasures. Life Sci Space Res (Amst) 2014;1:10–43. doi: 10.1016/j.lssr.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parihar VK, Pasha J, Tran KK, Craver BM, Acharya MM, Limoli CL. Persistent changes in neuronal structure and synaptic plasticity caused by proton irradiation. Brain Struct Funct. 2014 doi: 10.1007/s00429-014-0709-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trani D, Datta K, Doiron K, Kallakury B, Fornace AJ., Jr Enhanced intestinal tumor multiplicity and grade in vivo after HZE exposure: mouse models for space radiation risk estimates. Radiat Environ Biophys. 2010;49:389–396. doi: 10.1007/s00411-010-0292-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trani D, Nelson SA, Moon BH, Swedlow JJ, Williams EM, Strawn SJ, et al. High-energy particle-induced tumorigenesis throughout the gastrointestinal tract. Radiat Res. 2014;181:162–171. doi: 10.1667/RR13502.1. [DOI] [PubMed] [Google Scholar]
- 6.Yan X, Sasi SP, Gee H, Lee J, Yang Y, Mehrzad R, et al. Cardiovascular risks associated with low dose ionizing particle radiation. PLoS One. 2014;9:e110269. doi: 10.1371/journal.pone.0110269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laiakis EC, Hyduke DR, Fornace AJ., Jr Comparison of mouse urinary metabolic profiles after exposure to the inflammatory stressors gamma radiation and lipopolysaccharide. Radiat Res. 2012;177:187–199. doi: 10.1667/rr2771.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laiakis EC, Mak TD, Anizan S, Amundson SA, Barker CA, Wolden SL, et al. Development of a metabolomic radiation signature in urine from patients undergoing total body irradiation. Radiat Res. 2014;181:350–361. doi: 10.1667/RR13567.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tyburski JB, Patterson AD, Krausz KW, Slavik J, Fornace AJ, Jr, Gonzalez FJ, et al. Radiation metabolomics. 1. Identification of minimally invasive urine biomarkers for gamma-radiation exposure in mice. Radiat Res. 2008;170:1–14. doi: 10.1667/RR1265.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tyburski JB, Patterson AD, Krausz KW, Slavik J, Fornace AJ, Jr, Gonzalez FJ, et al. Radiation metabolomics. 2. Dose- and time-dependent urinary excretion of deaminated purines and pyrimidines after sublethal gamma-radiation exposure in mice. Radiat Res. 2009;172:42–57. doi: 10.1667/RR1703.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Girdhani S, Sachs R, Hlatky L. Biological effects of proton radiation: what we know and don’t know. Radiat Res. 2013;179:257–272. doi: 10.1667/RR2839.1. [DOI] [PubMed] [Google Scholar]
- 12.Mak TD, Laiakis EC, Goudarzi M, Fornace AJ., Jr MetaboLyzer: a novel statistical workflow for analyzing Postprocessed LC-MS metabolomics data. Anal Chem. 2014;86:506–513. doi: 10.1021/ac402477z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patterson AD, Li H, Eichler GS, Krausz KW, Weinstein JN, Fornace AJ, Jr, et al. UPLC-ESI-TOFMS-based metabolomics and gene expression dynamics inspector self-organizing metabolomic maps as tools for understanding the cellular response to ionizing radiation. Anal Chem. 2008;80:665–674. doi: 10.1021/ac701807v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buonanno M, De Toledo SM, Pain D, Azzam EI. Long-term consequences of radiation-induced bystander effects depend on radiation quality and dose and correlate with oxidative stress. Radiat Res. 2011;175:405–415. doi: 10.1667/RR2461.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Papamandjaris AA, Macdougall DE, Jones PJ. Medium chain fatty acid metabolism and energy expenditure: obesity treatment implications. Life Sci. 1998;62:1203–1215. doi: 10.1016/s0024-3205(97)01143-0. [DOI] [PubMed] [Google Scholar]
- 16.Cucinotta FA, Kim MH, Chappell LJ, Huff JL. How safe is safe enough? Radiation risk for a human mission to Mars. PLoS One. 2013;8:e74988. doi: 10.1371/journal.pone.0074988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davis CM, Decicco-Skinner KL, Roma PG, Hienz RD. Individual differences in attentional deficits and dopaminergic protein levels following exposure to proton radiation. Radiat Res. 2014;181:258–271. doi: 10.1667/RR13359.1. [DOI] [PubMed] [Google Scholar]
- 18.Hyland K. Inherited disorders affecting dopamine and serotonin: critical neurotransmitters derived from aromatic amino acids. J Nutr. 2007;137:1568S–1572S. doi: 10.1093/jn/137.6.1568S. discussion 1573S. [DOI] [PubMed] [Google Scholar]
- 19.Moore TL, Schettler SP, Killiany RJ, Herndon JG, Luebke JI, Moss MB, et al. Cognitive impairment in aged rhesus monkeys associated with monoamine receptors in the prefrontal cortex. Behav Brain Res. 2005;160:208–221. doi: 10.1016/j.bbr.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 20.Di Pietro C, Piro S, Tabbi G, Ragusa M, Di Pietro V, Zimmitti V, et al. Cellular and molecular effects of protons: apoptosis induction and potential implications for cancer therapy. Apoptosis. 2006;11:57–66. doi: 10.1007/s10495-005-3346-1. [DOI] [PubMed] [Google Scholar]
- 21.Gerelchuluun A, Hong Z, Sun L, Suzuki K, Terunuma T, Yasuoka K, et al. Induction of in situ DNA double-strand breaks and apoptosis by 200 MeV protons and 10 MV X-rays in human tumour cell lines. Int J Radiat Biol. 2011;87:57–70. doi: 10.3109/09553002.2010.518201. [DOI] [PubMed] [Google Scholar]
- 22.Ristic-Fira AM, Todorovic DV, Koricanac LB, Petrovic IM, Valastro LM, Cirrone PG, et al. Response of a human melanoma cell line to low and high ionizing radiation. Ann N Y Acad Sci. 2007;1095:165–174. doi: 10.1196/annals.1397.020. [DOI] [PubMed] [Google Scholar]
- 23.Cheema AK, Suman S, Kaur P, Singh R, Fornace AJ, Jr, Datta K. Long-term differential changes in mouse intestinal metabolomics after gamma and heavy ion radiation exposure. PLoS One. 2014;9:e87079. doi: 10.1371/journal.pone.0087079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Datta K, Suman S, Kallakury BV, Fornace AJ., Jr Exposure to heavy ion radiation induces persistent oxidative stress in mouse intestine. PLoS One. 2012;7:e42224. doi: 10.1371/journal.pone.0042224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Datta K, Suman S, Kallakury BV, Fornace AJ., Jr Heavy ion radiation exposure triggered higher intestinal tumor frequency and greater beta-catenin activation than gamma radiation in APC(Min/+) mice. PLoS One. 2013;8:e59295. doi: 10.1371/journal.pone.0059295. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.