Abstract
Background
Prediction of radioresistance of HR-HPV-positive (+) cervical cancer, especially before the course of radiotherapy, is quite beneficial to develop an optimal treatment strategy for individual patients. Unfortunately, the mechanisms responsible for radioresistance of cervical cancer are still largely unexplored. HR-HPV infection leads to a series of changes to normal biophysical process, including miRNAs expression. In this study, we explored the association between miR-375 and radioresistance in HR-HPV (+) cervical cancer.
Material/Methods
qRT-PCR analysis was performed to determine miR-375 expression in HR-HPV-positive (+) cervical cancer patients and in HPV-16-positive SiHa and HPV-18-positive HeLa cervical cancer cell lines. The influence of miR-375 on radiosensitivity and the downstream regulative network were further explored in the cell line models.
Results
The results verified a putative binding site between miR-375 and UBE3A. miR-375 overexpression could significantly reduce UBE3A expression. UBE3A knockdown led to significantly reduced cell survival under radiation treatment. miR-375 promoted radiosensitivity of HR-HPV (+) cancer through decreasing p53 degradation and thereby increasing radiation-induced apoptosis.
Conclusions
The miR-375-UBE3A axis is important in modulating radiosensitivity of HR-HPV (+) cervical cancer.
MeSH Keywords: Human Papillomavirus 16, Human Papillomavirus 18, Uterine Cervical Neoplasms
Background
Cervical cancer is the third most frequent cancer in women [1]. For these patients, radiotherapy is still the most common intervention, either as a primary or an adjuvant therapy [2,3]. Although radiotherapy is used for over 60% of cervical cancers cases, local recurrence is common due to radioresistance [4,5]. Therefore, the prediction of radioresistance, especially before the course of radiotherapy, is quite beneficial to develop an optimal treatment strategy for individual patients. Unfortunately, the mechanisms responsible for radioresistance of cervical cancer are still largely unexplored.
Persistent high-risk HPV (HR-HPV) infection is involved in more than 90% of cervical cancer cases and has been identified as a causal factor in cancer development [6]. Typically, HPV16 and HPV18 cause about 70% of cervical cancer cases [7]. E6 and E7 are 2 primary oncoproteins of HR-HPV. As a pivotal viral oncogene, E6 can combine with cellular protein ubiquitin-protein ligase E3A (UBE3A), also known as E6AP, and initiate proteasomal degradation of p53 [8]. p53 degradation results in reduced p53-mediated apoptosis and p21-mediated cell cycle arrest [9]. In addition, p53 degradation is also involved in the mechanism of radioresistance in several types of tumors [10,11], including cervical cancer [12,13]. However, the upstream regulative network of p53 in radioresistance of HR-HPV-positive (+) cervical cancer is still not clear.
miRNAs are a group of small, conservative, and non-coding RNAs degrading or repressing the translation of target mRNAs through directly binding to the 3′-UTR [14]. Several miRNAs were found involved in radioresistance of cervical cancers, such as miR-630, miR-1246, miR-1290, miR-3138, miR-181, miR-375, and miR-21 [15,16]. However, the downstream regulative network of these miRNAs is not yet fully understood. In this study, we explored the association between miR-375 and radioresistance in HR-HPV (+) cervical cancer. We demonstrated that miR-375 can modulate radiosensitivity of HR-HPV (+) cervical cancer cells by directly downregulating UBE3A expression and thereby promoting p53 regulated cell apoptosis.
Material and Methods
Patient selection and human tissues
We recruited 22 patients from Cangzhou City Central Hospital who were histologically diagnosed as having IA with lymphovascular space invasion (IVSI) or IA2 cervical cancer, confirmed as HPV-16/18 positive and who never received previous chemotherapy. The patients received radiotherapy by standard, pelvic radiation therapy and brachytherapy with a total point A dose of 70 Gy, according to 2013 NCCN Clinical Practice Guideline in Oncology for Cervical Cancer [17]. The radiosensitive and radioresistant cases were assessed by histological examination of residual tumor tissues by colposcopically-directed biopsy 6 months after completion of radiotherapy. Histological assessment was performed by a pathologist without authorship in this study. All of the samples that contained at least 70% tumor cells were used for further studies. The control group consisted of 20 healthy individuals with normal cytology and without HR-HPV infection; specimens were obtained from patients received hysterectomy due to benign gynecologic diseases. Informed consent was obtained from each patient before collecting the specimens.
Cell culture
HEK 293T, HPV-16-positive SiHa and HPV-18-positive HeLa cervical cancer cell lines were obtained from the ATCC and were cultured with Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS; Hyclone Laboratories), 2 mM/L-glutamine, 100U/mL penicillin, and 100 mg/mL streptomycin and were incubated in humidified air with 5% CO2 at 37°C.
qRT-PCR analysis of miR-375 and UBE3A expression
Total RNAs from tissue and cells were extracted using TRIzol reagent (Invitrogen) and from blood were extracted using TRIzol LS reagent (Invitrogen), respectively. cDNAs were synthesized using the PrimeScript RT reagent Kit (TaKaRa). miR-375 expression was quantified by using TaqMan MicroRNA assays (Life Technologies). The 2−ΔΔ Ct method was used to calculate relative miR-375 expression, with RNU6B as a control gene. UBE3A mRNA expression was measured using qRT-PCR with SYBR Green PCR Master Mix (Life Technologies) and UBE3A-specific primers: F, 5′-CCATGGGAAAATGTACATCCA-3′; R, 5′-TTTTTCAGCTGGTTGTGGAGG-3′. GAPDH served as the internal control.
Cell transfection
miR-375 mimics and the negative control, UBE3A siRNA, and the corresponding negative controls were purchased from Ribo Life Science (China). HeLa and SiHa cells were transfected with 75 nM miR-375 mimics or 75 nM UBE3A siRNA, respectively, by using lipofectamine 2000 (Invitrogen). Human UBE3A lentiviral expression vector (Lenti-UBE3A) without 3′-UTR was purchased from GENECHEM. To generate sufficient lentiviral particles for transfection, Lenti-UBE3A and the corresponding packaging mix were cotransfected to HEK-293T cells. Viral supernatant was collected for further experiments at 48 h after transfection. To over-express UBE3A in SiHa and HeLa cells, the cells were treated with the viral supernatants with 8 μg/ml Polybrene (Sigma-Aldrich).
Western blot analysis of UBE3A expression
Total protein from cells was extracted by using RIPA buffer (50 mM TrisHCl, 150 mM NaCl, 2 mM EDTA, 1% NP-40, and 0.1% SDS). Total protein concentration was measured by using BCA protein assay (Pierce, Thermo Scientific) and then separated on 10% SDS PAGE gel and transferred onto nitrocellulose membranes for a conventional Western blot analysis. Antibodies used were Anti-UBE3A (1:2000, ab10488, Abcam), anti-p53 (1:1000, ab131442, Abcam), anti-p21 (1:2000, ab7960, Abcam), anti-survivin (1:1000, ab24479, Abcam), anti-Bax (1:1000, ab7977, Abcam), and anti-active caspase 3 (1:1000, ab2302, Abcam). GAPDH served as loading control and was detected by using ant-GAPDH (1:2500, ab9485, Abcam). Anti-Rabbit IgG (HRP) (1:10000, ab191866, Abcam) was used as a second antibody. Protein signals were detected using SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific).
Colony formation and apoptosis analysis
We seeded 1×105 SiHa or HeLa cells in 6-well plates and prepared for transfection the next day. At 48 h after transfection, the plates were irradiated with 137Cs (Nordion, Ottawa, ON, Canada), with a dose rate of 1.25 Gy/min. The plates were treated with a dose of 0, 2, 4, 6, and 8 Gy in a single fraction. Then, the plates were further incubated in a cell incubator for 10–13 days and then the cells were fixed with 10% paraformaldehyde and stained with 1% crystal violet in 70% ethanol. Colonies (with minimum of 50 cells) were counted. Surviving fraction was defined as the number of colonies/(cells inoculated × plating efficiency) and the radiation survival curve was drawn. Apoptosis of the cells was measured by using the Fluorescein Active Caspase 3 Staining Kit (Abcam, ab65613) according to the manufacturer’s instructions. The proportion of active caspase 3 positive cells was measured by using a flow cytometer (FACSCalibur, BD Biosciences).
MTT assay of cell viability
Cells after different treatments were plated at 5×103 cells/well in 96-well plates. Cells were cultured for 48 h and then cell viability was measured by MTT (Sigma-Aldrich) assay according to the recommended protocol. Absorbance at 490 nm of the solution was read by using a spectrophotometric plate reader. Each test was performed with 3 repeats.
Luciferase microRNA binding assay
The oligonucleotides with wild-type (WT) (with miR-375 binding site) or mutant (MUT) (without miR-375 binding site) sequence of 3′-untranslated region (3′-UTR) of the UBE3A and with flanking PmeI and XbaI restriction enzyme sites were chemically synthesized. The sequence was: WT: F, 5′-aaacGATTTCAGCTACATATATGAACAAATCCTTTATTATTATTt-3′, R, 5′-ctagaAATAATAATAAAGGATTTGTTCATATATGTAGCTGAAATCgttt-3′; MUT: F, 5′-aaacGATTTCAGCTACATATATTCCACCCTCCTTTATTATTATTt-3′; R, 5′-ctagaAATAATAATAAAGGAGGGTGGAATATATGTAGCTGAAATCgttt-3′. The 2 sequences were inserted into the downstream of the firefly luciferase gene in pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega) between PmeI and XbaI sites. The reconstructed vectors were designated as pmirGLO-UBE3A-WT and pmirGLO-UBE3A-MUT, respectively. Insertion was verified by sequencing. HeLa and SiHa cells were co-transfected with 200 ng reporter plasmids and 75 nM miR-375 mimics. At 24 h after transfection, both firefly and Renilla luciferase activities were measured by the Dual-Luciferase Reporter Assay System (Promega) using a Promega GloMax 20/20 luminometer. The firefly luciferase activity was normalized to the Renilla luciferase activity.
Statistical analysis
Data are presented as mean ±SD with at least 3 repeats. Group comparison was performed by unpaired t test. P value <0.05 was considered as significant difference. *, **, and *** donates significance at 0.05, 0.01, and 0.001 level, respectively.
Results
MiR-375 expression is negatively related to radioresistance in HR-HPV (+) cervical cancer
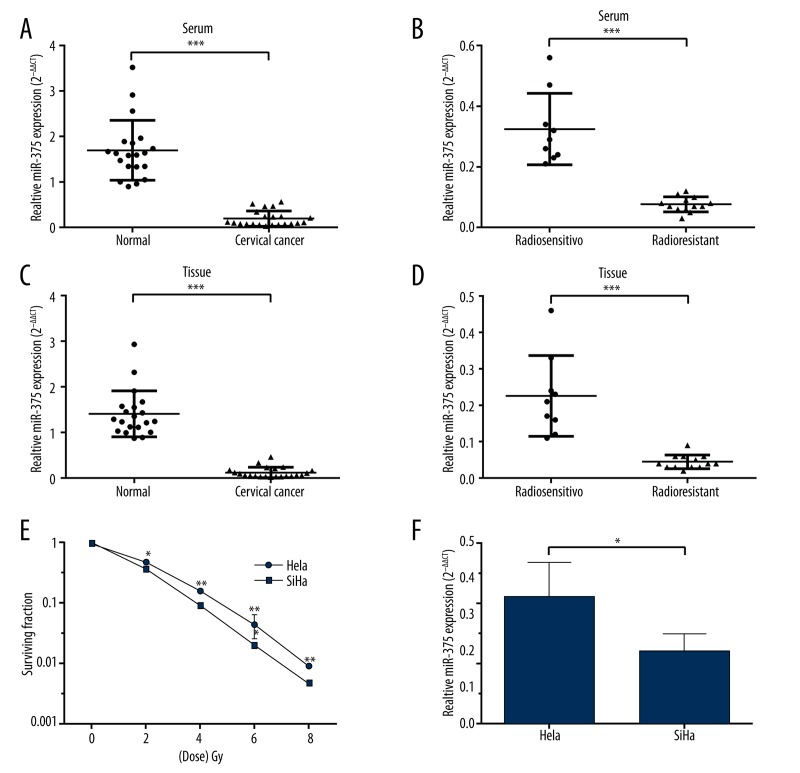
Based on serum and tumor tissue samples from the cervical cancer patients and healthy controls, qRT-PCR results showed that miR-375 expression was significantly lower in the cancer patients than in the controls (Figure 1A, 1C). Its expression was even lower in radioresistant patients than in radiosensitive patients (Figure 1B, 1D). HPV-16-positive SiHa, which had lower response to radiation than HPV-18-positive HeLa cells (Figure 1E), also had lower expression of miR-375 (Figure 1F). These results suggest that miR-375 might be involved in the radioresistance of HPV (+) cervical cancer.
Figure 1.
MiR-375 expression is negatively related to radioresistance in HR-HPV (+) cervical cancer. (A, C) qRT-PCR analysis of relative miR-375 expression in serum (A) and tissue (C) of cervical cancer patients (n=22) and healthy controls (n=20). Relative miR-375 expression was significantly higher in serum (A) and tissue (C) of HR-HPV (+) cervical cancer patients than in healthy controls. (B, D) qRT-PCR analysis showed that miR-375 expression was even lower in both serum (B) and tissue (D) of radioresistant patients (n=13) than in radiosensitive patients (n=9). (E) The surviving fraction of HPV-18-positive HeLa cells was significantly lower than in HPV-16-positive SiHa cells under radiation treatment. (F) qRT-PCR analysis showed that miR-375 expression was significantly lower in SiHa cells than in HeLa cells. Each bar represents the mean ±S.D. of 3 experiments. * P<0.05; ** P<0.01, *** P<0.001.
MiR-375 modulates radiation-induced apoptosis of HR-HPV (+) cervical cancer cells
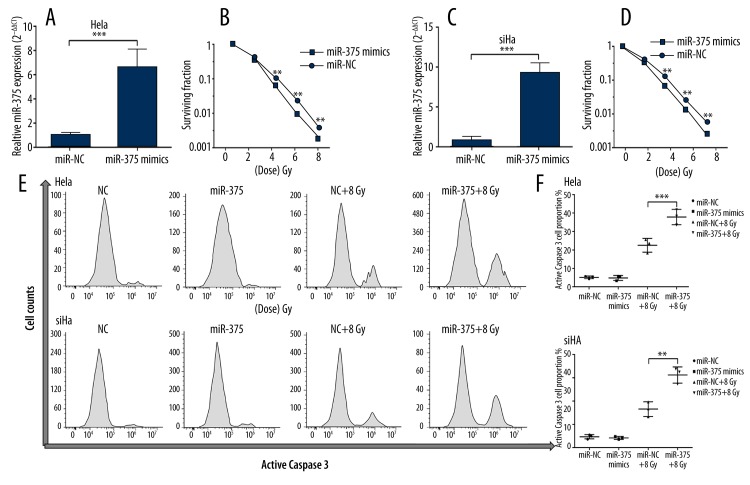
To explore the influence of miR-375 expression on radiation-induced apoptosis, SiHa and HeLa cells were transfected for miR-375 overexpression (Figure 2A, 2C). miR-375 overexpression conferred significantly reduced survival under radiation treatment of both cell lines (Figure 2B, 2D). By measuring the apoptotic marker caspase 3, we observed that the ratios of radiation-induced apoptotic SiHa or HeLa cells with miR-375 overexpression were significantly higher than the corresponding negative controls (Figure 2E, 2F). These results suggest that miR-375 can directly modulate apoptosis of HR-HPV (+) cervical cancer cells induced by radiation.
Figure 2.
MiR-375 modulates radiation-induced apoptosis of HR-HPV (+) cervical cancer cells. (A, C) qRT-PCR analysis confirmed successful transfection of miR-375 into HeLa (A) and SiHa (C) cells. (B, D) Transfection of miR-375 mimics significantly increased radiosensitivity in both HeLa (B) and SiHa (D) cells. (E) Representative images of flow cytometry analysis of apoptotic HeLa and SiHa cells by staining active caspase-3 at 48 h after radiation treatment. (F) (J) Quantification of the apoptotic HeLa and SiHa cells at 48 h after transfection or 48 h after 8Gy treatment. miR-375 overexpression significantly increased the proportion of apoptotic cells treated by radiation. Each bar represents the mean ±S.D. of 3 experiments. * P<0.05; ** P<0.01, *** P<0.001.
MiR-375 directly targets UBE3A and regulates its expression in HR-HPV (+) cervical cancer cells
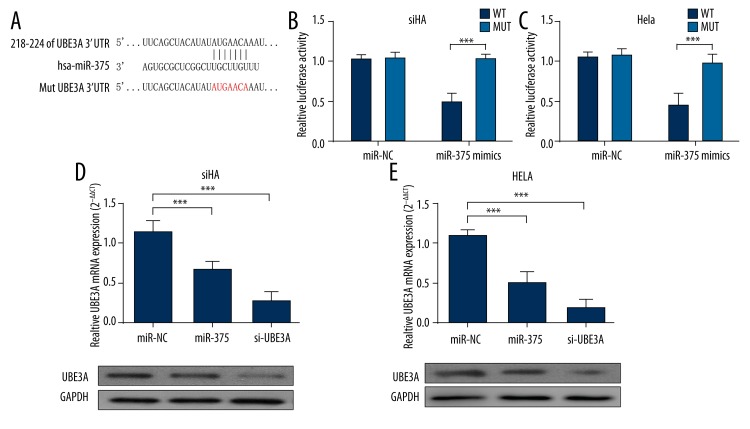
Since miR-375 has a regulative role over radioresistance, we further explored its downstream targets and regulative network in HR-HPV (+) cervical cancer cells. A recent study demonstrated that UBE3A, also known as E6AP, is a direct target of miR-375. In this study, we further verified this target in both SiHa and HeLa cells. miR-375 mimics could inhibit luciferase activity of pmirGLO-UBE3A-WTvector, but had no inhibiting effect over pmirGLO-UBE3A-MUT vector (Figure 3A, 3B). In both of the cell lines, miR-375 overexpression, similar to si-UBE3A expression, effectively interfered with UBE3A expression at both mRNA and protein levels (Figure 3D, 3E). These results suggest that miR-375 can directly target UBE3A and regulate its expression in both HeLa and SiHa cells.
Figure 3.
MiR-375 directly targets UBE3A and regulates its expression in HR-HPV (+) cervical cancer cells. (A) The predicted pairing between miR-375 and 3′-UTR of UBE3A. Designed UBE3A-mutant (MUT) sequence without miR-375 binding sites is shown (B, C). HeLa (B) and SiHa (C) cells were co-transfected with either 75nM miR-375 mimics or NC oligos and 200 ng dual-luciferase reporter plasmids carrying either WT or MUT 3′-UTR of UBE3A. The relative firefly luciferase activity was measured at 24 h after transfection and was normalized with Renilla luciferase activity. miR-375 mimics decreased luciferase activity of WT reporter but not MUT reporter. (D, E) (D) HeLa and SiHa (E) cells were transfected with 75 nM miR-375 mimics or 75 nM UBE3A siRNA, respectively. miR-375 and UBE3A siRNA significantly inhibited UBE3A expression at both mRNA and protein level. Each bar represents the mean ±S.D. of 3 experiments. * P<0.05; ** P<0.01, *** P<0.001.
MiR-375 promotes radiosensitivity through p53 pathway
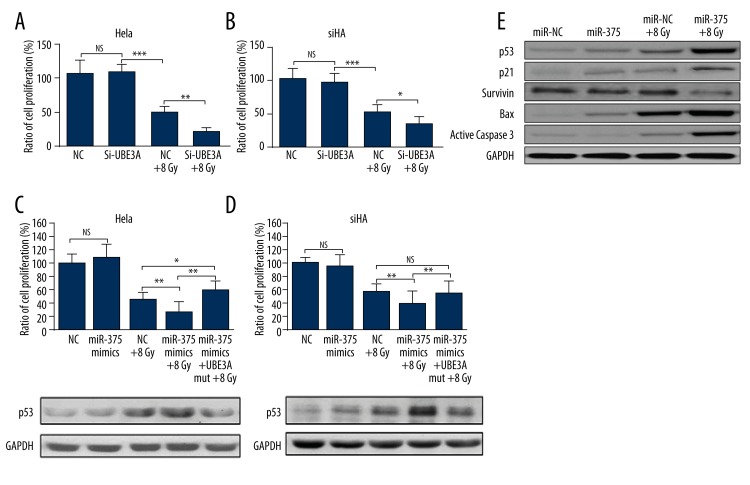
We then further studied the downstream regulation network of miR-375. In both HeLa and SiHa cells, UBE3A knockdown significantly lowered cell viability under radiation treatment (Figure 4A, 4B). Overexpression of UBE3A without the 3′-UTR region rescued miR-375-induced lowered radioresistance in both HeLa and SiHa cells, and also lowered p53 expression (Figure 4C, 4D). The downstream regulation of miR-375 was further explored in HeLa cells. miR-375 overexpression resulted in significantly increased p53 and p21 expression (Figure 4E), both of which are involved in p53-dependent cell cycle G1 phase arrest [18]. miR-375 overexpression also led to significantly higher expression of the apoptotic markers Bax and active caspase 3 under radiation treatment (Figure 4E). In contrast, the expression of survivin, a negative regulation of apoptosis, was remarkably inhibited by miR-375 overexpression (Figure 4E). These results suggest that miR-375 modulates radiation-induced apoptosis of cervical cancer cells at least partially through the p53 pathway.
Figure 4.
MiR-375 promotes radiosensitivity through the p53 pathway. (A, B) MTT analysis of cell viability of HeLa (A) and SiHa (B) cells with UBE3A knockdown 48 h after 8Gy treatment. (C, D) Co-transfection of miR-375 and UBE3A expression vector without 3′-UTR in HeLa (C) and SiHa (D) in response to 8 Gy were analyzed by MTT assays. The expression of p53 under different treatments was detected by Western blot analysis. (E) Western blot analysis of p53, p21, survivin, Bax, and active caspase 3 expression in HeLa cells with/without miR-375 overexpression under radiation treatment. Each bar represents the mean ±S.D. of 3 experiments. * P<0.05; ** P<0.01, *** P<0.001; NS – not significant.
Discussion
HR-HPV infection can alter a series of biophysical process in cervical tissues. In fact, the infection can also affect the expression of multiple miRNAs, thereby affecting the downstream regulations. miR-375 is a miRNA that is significantly downregulated in HR-HPV infection [19,20]. However, how this expression change affects the downstream regulation is still not clearly understood. According to previous studies, miR-375 is involved in a wide range of regulation in different cancers. In gastric cancer, miR-375 is downregulated and can inhibit migration and invasion of cancer cells by targeting Janus kinase 2 (JAK2) [21]. It can also target the p53 gene and regulate cellular response to ionizing radiation and etoposide in gastric cancer cells [22]. In breast cancer, miR-375 directly targets SHOX2 and thereby modulates epithelial-to-mesenchymal transition (EMT) [23]. In cervical cancer, miR-375 is also involved in cancer development processes. For example, in squamous cervical cancer, miR-375 can inhibit cell migration and invasion via targeting transcription factor SP1 [24]. It can also mediate acquired chemo-resistance in cervical cancer by facilitating EMT [25]. Therefore, it is highly possible that miR-375 has a wide regulative network and that its regulation might be different in different cancers. In the current study, we observed that miR-375 was related to radioresistance of HR-HPV (+) cancer. A lower level of miR-375 expression resulted in a higher level of radioresistance. In contrast, miR-375 overexpression in HR-HPV-positive cancer cells significantly promoted radiosensitivity.
Due to the significant association between miR-375 and radioresistance, we decided to explore how it affects radioresistance of HR-HPV (+) cervical cancer. A recent study reported that miR-375 has multiple targets in HPV-associated cancers, including HPV type 16 and 18 transcripts, E6AP and CIP2A [26]. The HPV type 16 and 18 transcripts consist of E6 and E7 protein, the most oncogenic proteins. This study also demonstrated that replenishment of miR-375 in HPV (+) cervical cancer cells significantly reduced the levels of HPV transcripts [26]. In fact, the complex of E6 and UBE3A is quite important for the oncogenic properties of HR-HPVs. It initiates proteasomal degradation of p53 [8] and results in reduced p53-mediated apoptosis and p21-mediated cell cycle arrest. Therefore, we hypothesized that the downregulation of miR-375 might regulate radioresistance through the p53 pathway.
In both SiHa and HeLa cells, we verified the putative binding site between miR-375 and UBE3A. miR-375 overexpression significantly reduced UBE3A expression. UBE3A knockdown led to significantly reduced cell survival under radiation treatment. miR-375 overexpression also had a similar survival-inhibiting effect and caused increased expression of p53. However, this inhibition was rescued by overexpression of UBE3A without binding to miR-375, which was also associated with lower p53 expression. Then, we further explored how radioresistance of these 2 cell lines is modulated in the p53 pathway. The expression of p21, the direct downstream target of p53, which functions as a regulator of cell cycle progression at G1 and S phase [18], increased with miR-375 overexpression. The expression apoptosis regulator BAX and the apoptotic marker active caspase 3 increased significantly in the miR-375 overexpression group. Survivin, an inhibitor of caspase activation and apoptosis [27], significantly decreased with miR-375 overexpression. These results suggest that miR-375 promotes radiosensitivity of HR-HPV (+) cancer through decreasing p53 degradation and thereby increasing radiation-induced apoptosis. Although p53 degradation has already been found in the mechanism of radioresistance in several types of tumors [10,11], our study is the first to demonstrate its upstream regulation in HR-HPV (+) cervical cancer cells.
Conclusions
Based on the evidence obtained above, it is evident that the miR-375-UBE3A axis is important in modulating radiosensitivity of HR-HPV (+) cervical cancer. The modulation occurs at least partly through inhibiting p53 degradation and promoting p53-mediated apoptosis under radiation treatment.
Footnotes
Source of support: Departmental sources
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. Cancer J Clin. 2014;64(1):9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Rogers L, Siu SS, Luesley D, et al. Radiotherapy and chemoradiation after surgery for early cervical cancer. Cochrane Database Syst Rev. 2012;5:CD007583. doi: 10.1002/14651858.CD007583.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Waggoner SE. Cervical cancer. Lancet. 2003;361(9376):2217–25. doi: 10.1016/S0140-6736(03)13778-6. [DOI] [PubMed] [Google Scholar]
- 4.Powell ME. Modern radiotherapy and cervical cancer. Int J Gynecol Cancer. 2010;20(11 Suppl 2):S49–51. doi: 10.1111/igc.0b013e3181f7b241. [DOI] [PubMed] [Google Scholar]
- 5.Frega A, Sopracordevole F, Scirpa P, et al. The re-infection rate of high-risk HPV and the recurrence rate of vulvar intraepithelial neoplasia (VIN) usual type after surgical treatment. Med Sci Monit. 2011;17(9):CR532–35. doi: 10.12659/MSM.881941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunne EF, Park IU. HPV and HPV-associated diseases. Infect Dis Clin North Am. 2013;27(4):765–78. doi: 10.1016/j.idc.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 7.Durst M, Gissmann L, Ikenberg H, zur Hausen H. A papillomavirus DNA from a cervical carcinoma and its prevalence in cancer biopsy samples from different geographic regions. Proc Natl Acad Sci USA. 1983;80(12):3812–15. doi: 10.1073/pnas.80.12.3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scheffner M, Huibregtse JM, Vierstra RD, Howley PM. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75(3):495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 9.Buyru N, Altinisik J, Isin M, Dalay N. p53 codon 72 polymorphism and HPV status in lung cancer. Med Sci Monit. 2008;14(9):CR493–97. [PubMed] [Google Scholar]
- 10.Concin N, Zeillinger C, Stimpfel M, et al. p53-dependent radioresistance in ovarian carcinoma cell lines. Cancer Lett. 2000;150(2):191–99. doi: 10.1016/s0304-3835(99)00393-6. [DOI] [PubMed] [Google Scholar]
- 11.Yang CF, Peng LX, Huang TJ, et al. Cancer stem-like cell characteristics induced by EB virus-encoded LMP1 contribute to radioresistance in nasopharyngeal carcinoma by suppressing the p53-mediated apoptosis pathway. Cancer Lett. 2014;344(2):260–71. doi: 10.1016/j.canlet.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 12.Lindel K, Rieken S, Daffinger S, et al. The transcriptional regulator gene E2 of the Human Papillomavirus (HPV) 16 influences the radiosensitivity of cervical keratinocytes. Radiat Oncol. 2012;7:187. doi: 10.1186/1748-717X-7-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beskow C, Skikuniene J, Holgersson A, et al. Radioresistant cervical cancer shows upregulation of the NHEJ proteins DNA-PKcs, Ku70 and Ku86. Br J Cancer. 2009;101(5):816–21. doi: 10.1038/sj.bjc.6605201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 15.Zhang B, Chen J, Ren Z, et al. A specific miRNA signature promotes radioresistance of human cervical cancer cells. Cancer Cell Int. 2013;13(1):118. doi: 10.1186/1475-2867-13-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ke G, Liang L, Yang JM, et al. MiR-181a confers resistance of cervical cancer to radiation therapy through targeting the pro-apoptotic PRKCD gene. Oncogene. 2013;32(25):3019–27. doi: 10.1038/onc.2012.323. [DOI] [PubMed] [Google Scholar]
- 17.Koh WJ, Greer BE, Abu-Rustum NR, et al. Cervical cancer. J Natl Compr Canc Netw. 2013;11(3):320–43. doi: 10.6004/jnccn.2013.0043. [DOI] [PubMed] [Google Scholar]
- 18.Gartel AL, Radhakrishnan SK. Lost in transcription: p21 repression, mechanisms, and consequences. Cancer Res. 2005;65(10):3980–85. doi: 10.1158/0008-5472.CAN-04-3995. [DOI] [PubMed] [Google Scholar]
- 19.Li Y, Wang F, Xu J, et al. Progressive miRNA expression profiles in cervical carcinogenesis and identification of HPV-related target genes for miR-29. J Pathol. 2011;224(4):484–95. doi: 10.1002/path.2873. [DOI] [PubMed] [Google Scholar]
- 20.Lajer CB, Garnaes E, Friis-Hansen L, et al. The role of miRNAs in human papilloma virus (HPV)-associated cancers: bridging between HPV-related head and neck cancer and cervical cancer. Br J Cancer. 2012;106(9):1526–34. doi: 10.1038/bjc.2012.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu Y, Jin J, Liu Y, et al. Snail-regulated MiR-375 inhibits migration and invasion of gastric cancer cells by targeting JAK2. PloS One. 2014;9(7):e99516. doi: 10.1371/journal.pone.0099516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y, Xing R, Zhang X, et al. miR-375 targets the p53 gene to regulate cellular response to ionizing radiation and etoposide in gastric cancer cells. DNA Repair. 2013;12(9):741–50. doi: 10.1016/j.dnarep.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 23.Hong S, Noh H, Teng Y, et al. SHOX2 is a direct miR-375 target and a novel epithelial-to-mesenchymal transition inducer in breast cancer cells. Neoplasia. 2014;16(4):279–90. e271–75. doi: 10.1016/j.neo.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang F, Li Y, Zhou J, et al. miR-375 is down-regulated in squamous cervical cancer and inhibits cell migration and invasion via targeting transcription factor SP1. Am J Pathol. 2011;179(5):2580–88. doi: 10.1016/j.ajpath.2011.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen Y, Zhou J, Li Y, et al. miR-375 mediated acquired chemo-resistance in cervical cancer by facilitating EMT. PloS One. 2014;9(10):e109299. doi: 10.1371/journal.pone.0109299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jung HM, Phillips BL, Chan EK. miR-375 activates p21 and suppresses telomerase activity by coordinately regulating HPV E6/E7, E6AP, CIP2A, and 14-3-3zeta. Mol Cancer. 2014;13:80. doi: 10.1186/1476-4598-13-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sah NK, Khan Z, Khan GJ, Bisen PS. Structural, functional and therapeutic biology of survivin. Cancer Lett. 2006;244(2):164–71. doi: 10.1016/j.canlet.2006.03.007. [DOI] [PubMed] [Google Scholar]