Abstract
The order Trichosporonales (Tremellomycotina, Basidiomycota) includes various species that have clinical, agricultural and biotechnological value. Thus, understanding why and how evolutionary diversification occurred within this order is extremely important. This study clarified the phylogenetic relationships among Tricosporonales species. To select genes suitable for phylogenetic analysis, we determined the draft genomes of 17 Trichosporonales species and extracted 30 protein-coding DNA sequences (CDSs) from genomic data. The CDS regions of Trichosporon asahii and T. faecale were identified by referring to mRNA sequence data since the intron positions of the respective genes differed from those of Cryptococcus neoformans (outgroup) and are not conserved within this order. A multiple alignment of the respective gene was first constructed using the CDSs of T. asahii, T. faecale and C. neoformans, and those of other species were added and aligned based on codons. The phylogenetic trees were constructed based on each gene and a concatenated alignment. Resolution of the maximum-likelihood trees estimated from the concatenated dataset based on both nucleotide (72,531) and amino acid (24,173) sequences were greater than in previous reports. In addition, we found that several genes, such as phosphatidylinositol 3-kinase TOR1 and glutamate synthase (NADH), had good resolution in this group (even when used alone). Our study proposes a set of genes suitable for constructing a phylogenetic tree with high resolution to examine evolutionary diversification in Trichosporonales. These can also be used for epidemiological and biogeographical studies, and may also serve as the basis for a comprehensive reclassification of pleomorphic fungi.
Introduction
The order Trichosporonales was proposed by Fell et al. [1] and, in “The Yeasts, A Taxonomic Study” 5th ed., it includes the genera Trichosporon (37 species) [2] and Cryptotrichosporon (1 species) [3], as well as some species of highly polyphyletic genera, Bullera (3 of 41 species listed) [4] and Cryptococcus (10 of 70 species listed) [5]. Since the type species of the genera Bullera and Cryptococcus were assigned to the Tremellales, species within these two genera in the Trichosporonales are misleading taxonomically.
The genus Trichosporon, as it is defined at this time, is polyphyletic and divided into five clades; namely, Brassicae, Cutaneum, Gracile, Ovoides and Porosum. Each is traditionally characterized based on a combination of serological properties (serogroup) and the major ubiquinone (as the phenotype), as shown in Fig 1a. In the Cryptococcus species mentioned above, Weiss et al. [6] recently reported the amended concept of the genus Vanrija [7] by following several discussions on this group [2, 3, 5], and proposed the use of Vanrija as the genus name for Cryptococcus humicola and phylogenetically related species.
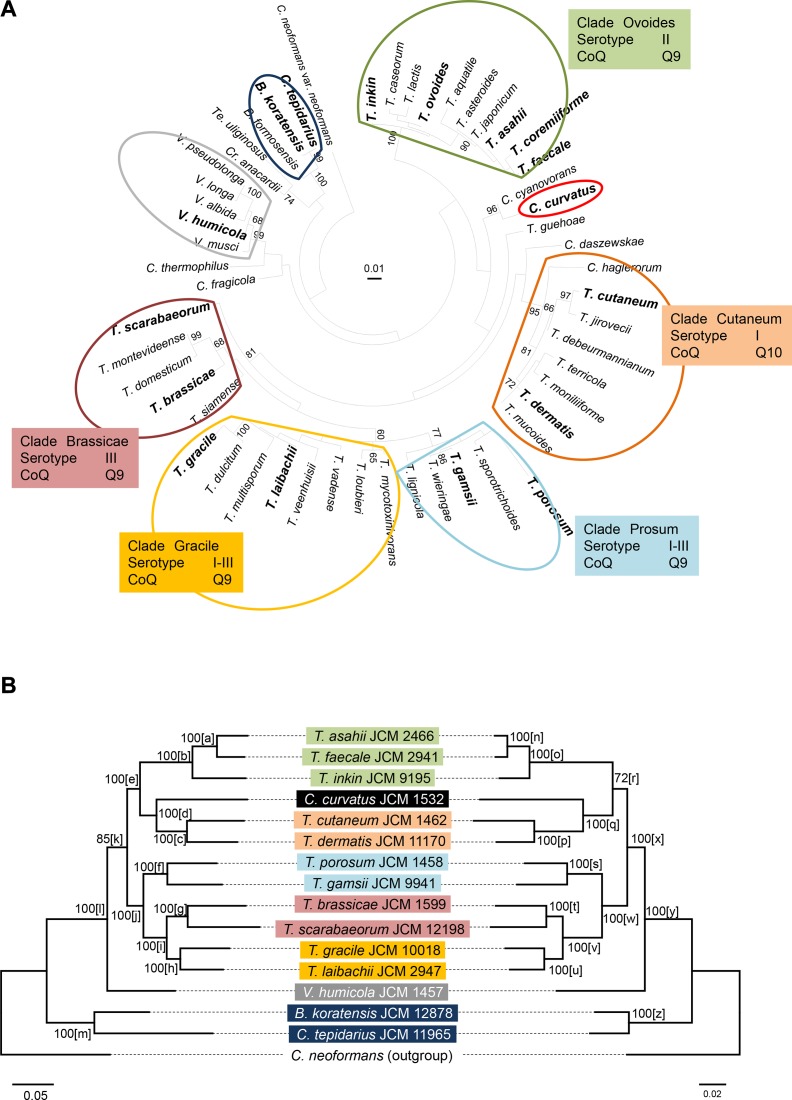
Fig 1. Phylogenetic trees of Trichosporon species and related taxa based on (a) D1/D2 LSU rRNA gene sequences, (b) nucleotide sequences of 30 concatenated genes, and (c) amino acid sequences of 30 concatenated genes.
The tree in (a) was constructed using the maximum likelihood method based on the Tamura-Nei model [44]. The names of clades are based on those in Fig. 161.1 in The Yeasts, A Taxonomic Study, 5th ed. [2]. Boldface indicates that the species was used for the 30-gene concatenation analysis. The trees in (b) and (c) were constructed using the maximum likelihood method based on the Tamura-Nei model [44] for (b) and a JTT matrix-based model [45] for (c). Numerals on each node represent the percentages from 100 replicating bootstrap samplings (frequencies of less than 60% are not shown) [46]. Letters in brackets after the bootstrap values in (b) and (c) indicate the node name (see Table 3). Positions used: (a) 604 nucleotides, (b) 72,531 nucleotides, (c) 24,173 amino acids. Highest log likelihood: (a) -3894.3736, (b) -611997.4313, (c) -216445.1306. B., Bullera; C., Cryptococcus; Cr., Cryptotrichosporon; T., Trichosporon; Te., Tetragoniomyces; V., Vanrija.
Clinically, biotechnologically and agriculturally important species are included in this order: approximately one-third of species in the genus Trichosporon are associated with infections or allergies in humans [2]; Trichosporon asahii (Ovoides clade) is the causative agent of trichosporonosis and summer-type hypersensitivity pneumonitis (SHP) [8]. Trichosporon mycotoxinivorans (Gracile clade) detoxifies mycotoxins, such as ochratoxin A and zearalenone [9], and Trichosporon porosum (Porosum clade) is thought to function in the global carbon cycle because it possesses CbhI genes for cellobiohydrolase [10] and oleaginous activity [11]. Additional oleaginous species have been reported among the Trichosporonales, including Cryptococcus curvatus [12], Vanrija albida [13], Vanrija musci [13], Trichosporon cacaoliposimilis (Gracile clade) [14] and Trichosporon oleaginosus (Cutaneum clade) [14]. Some species show cellulase activity, and most species can assimilate various compounds including aromatic and aliphatic compounds [2]. Since the expression (as shown above) and implied potential activities have been characterized in this order, understanding why and how this evolutionary diversification occurred is important. We have performed various studies on the genus Trichosporon and related species, including intra- and inter-species diversity; however, the phylogenetic relationships and evolutionary processes in this order remain unclear [2].
This study was performed to clarify the phylogenetic relationships among Trichosporonales species, especially basal species and clusters within this lineage. The draft genomes of 17 Trichosporonales species were determined, 30 orthologous protein-coding DNA sequences (CDSs) were selected, and phylogenetic trees were constructed based on the respective genes and a concatenated alignment. When selecting CDSs suitable for this analysis, the draft transcriptomes of T. asahii and T. faecale were used to identify coding regions for these species. Each CDS of T. asahii and T. faecale was aligned with that of Cryptococcus neoformans (outgroup), and those of other species were added and aligned based on codons, because the intron position of the respective genes is not conserved in this order.
Results and Discussion
Gene selection
A summary of the draft genome sequences is shown in Table 1. The obtained genomic size of T. asahii was similar to other recently published data (ALBS01000000 and AMBO01000000). The range of genomic sizes of Trichosporonales species, estimated based on the total length of contigs in this study, ranged from 18.2 Mb (C. curvatus) to 42.2 Mb (Trichosporon coremiiforme). We did not observe a trend between genome size and phylogenetic position or morphology in this study.
Table 1. Strains used in this study.
| Scientific name | Strain no. | Source*1 | # Contigs | Total length(bases) | N50(bases) | Maximum(bases) | Average (bases) |
|---|---|---|---|---|---|---|---|
| Trichosporon asahii | JCM 2466 | nail of a psoriasis patient, Fukuoka, Japan | 1,262 | 24,495,558 | 77,499 | 335,598 | 19,410 |
| Trichosporon brassicae | JCM 1599 | cabbage, T. Nakase, Tokyo, Japan | 988 | 23,545,754 | 46,311 | 203,527 | 23,832 |
| Trichosporon coremiiforme | JCM 2938 | lesion on the head of a farmer, Costa Rica | 3,232 | 42,172,195 | 26,704 | 150,431 | 13,048 |
| Trichosporon cutaneum | JCM 1462 | skin lesion | 3,076 | 22,615,927 | 14,018 | 146,985 | 7,352 |
| Trichosporon dermatis | JCM 11170 | infected human skin, Tübingen, Germany | 1,016 | 23,295,971 | 46,612 | 233,244 | 22,929 |
| Trichosporon faecale | JCM 2941 | human feces, A.C. Batista | 551 | 24,482,466 | 94,928 | 366,476 | 44,433 |
| Trichosporon gamsii | JCM 9941 | moist humus around roots, W. Gams, Sierra Nevada de Santa Marta, Colombia | 7,995 | 25,860,364 | 17,022 | 114,205 | 3,235 |
| Trichosporon gracile | JCM 10018 | sour milk, Germany | 1,672 | 24,039,830 | 29,596 | 175,298 | 14,378 |
| Trichosporon inkin | JCM 9195 | human skin with tinea cruris caused by Trichophyton, L. do Carmo-Sousa | 3,680 | 19,793,539 | 17,312 | 173,610 | 5,379 |
| Trichosporon laibachii | JCM 2947 | soil | 3,380 | 30,595,110 | 21,469 | 138,973 | 9,052 |
| Trichosporon ovoides | JCM 9940 | hair of a white piedra patient, A. Lasagni | 3,336 | 39,800,440 | 45,378 | 382,552 | 11,931 |
| Trichosporon porosum | JCM 1458 | exudate of English yew (Taxus baccata), Hamburg, Germany | 1,838 | 25,943,715 | 30,685 | 146,128 | 14,115 |
| Trichosporon scarabaeorum | JCM 12198 | gut of a larval scarab beetle, South Africa | 2,755 | 30,549,951 | 30,687 | 133,129 | 11,089 |
| Bullera koratensis | JCM 12878 | leaf of plant, Thailand | 1,443 | 25,088,877 | 33,905 | 163,727 | 17,387 |
| Cryptococcus curvatus | JCM 1532 | sputum of a patient with tuberculosis | 531 | 18,156,997 | 81,043 | 240,381 | 34,194 |
| Cryptococcus tepidarius | JCM 11965 | water in stream, Owakudani, Hakone, Japan | 3,328 | 22,106,340 | 11,260 | 65,094 | 6,643 |
| Vanrija humicola | JCM 1457 | heath soil in Switzerland | 3,111 | 22,380,125 | 15,011 | 154,669 | 7,194 |
*1 data from [2, 5] and http://jcm.brc.riken.jp/en/catalogue_e.
The candidate genes for phylogenetic analysis selected in this study are shown in Table 2. These 30 genes were selected as described in the Materials and Methods without consideration of function. They are orthologous genes shown in the Microbial Genome Database for Comparative Analysis (MBGD) (http://mbgd.nibb.ac.jp). Notably, we found that the genomes of T. coremiiforme and T. ovoides occasionally include two paralogs for some of the studied genes, whereas those of other species contained only one (Table 2). Even including the two paralogs from T. ovoides and T. coremiiforme, all phylogenetic trees based on a single gene showed that the former constituted a subclade with T. inkin and the latter with T. asahii and T. faecale (data not shown). This topology was the same as reported previously [2], indicating that the paralogous genes were not transferred from phylogenetically distant species. Therefore, these two species were excluded from the robustness analyses of the tree topology.
Table 2. Genes used in this study.
| Product | Homolog to: (locus_tag of C. neoformans) | Function category*1 | Sequence accession numbers for 17 species | Number of paralogs detected in † | |
|---|---|---|---|---|---|
| T. coremiiforme | T. ovoides | ||||
| methylenetetrahydrofolate reductase (NADPH) | CNL04820 | Amino acid biosynthesis [1.2] | AB920057-AB920077 | 2 | 2 |
| argininosuccinate lyase | CNC04420 | Amino acid biosynthesis [1.4] | AB919708-AB919727 | 2 | 1 |
| glutamate synthase (NADH) | CNJ02910 | Amino acid biosynthesis [1.4] | AB920036-AB920056 | 2 | 1 |
| orotidine monophosphate pyrophosphorylase (URA5) | CNG03730 | Purines, pyrimidines, nucleosides, and nucleotides [2.4] | AB919901-AB919919 | 2 | 2 |
| pyruvate carboxylase | CNF00650 | Fatty acid, phospholipid and sterol metabolism [3.1], Transport and binding proteins [7.4], Other categories [14] | AB919845-AB919864 | 1 | 2 |
| actin | CNA04650 | Central intermediary metabolism [5.2] | AB919611-AB919629 | 2 | 2 |
| adenylyl-sulfate kinase | CNE03380 | Central intermediary metabolism [5.5] | AB919806-AB919824 | 2 | 2 |
| translation elongation factor 1-α | CNM01300 | Central intermediary metabolism [5.5], Translation [10.4] | AB920098-AB920117 | 1 | 2 |
| adenosylhomocysteinase | CND00240 | Central intermediary metabolism [5] | AB919748-AB919768 | 2 | 2 |
| 1,3-beta-glucan synthase | CNN02320 | Central intermediary metabolism [5] | AB920158-AB920176 | 2 | 2 |
| citrate synthase | CNA00510 | Energy metabolism [6.13] | AB919588-AB919610 | 2 | 2 |
| vacuolar ATP synthase | CNI01180 | Energy metabolism [6.4] | AB919979-AB919997 | 2 | 2 |
| phosphoenolpyruvate carboxykinase | CNI03590 | Energy metabolism [6.8] | AB919998-AB920016 | 2 | 1 |
| malate synthase | CNH02910 | Energy metabolism [6] | AB919920-AB919940 | 2 | 2 |
| isocitrate lyase | CNH03280 | Energy metabolism [6] | AB919941-AB919961 | 1 | 2 |
| plasma membrane H(+)-ATPase 1 | CNN01260 | Transport and binding proteins [7.4] | AB920136-AB920157 | 2 | 2 |
| ATP-binding cassette (ABC) transporter | CND00300 | Transport and binding proteins [7.6] | AB919769-AB919786 | 2 | 1 |
| ATP dependent DNA helicase | CNB05360 | DNA replication, restriction, modification, recombination, and repair [8.2] | AB919670-AB919688 | 2 | 2 |
| DNA replication licensing factor cdc19 (cell division control protein 19) | CNG02380 | DNA replication, restriction, modification, recombination, and repair [8.2] | AB919883-AB919900 | 1 | 2 |
| DNA-dependent RNA polymerase II RPB140 (RPB2) | CND03540 | Transcription [9.2] | AB919787-AB919805 | 1 | 2 |
| DNA-directed RNA polymerase ii largest subunit, putative (RPB1) | CNE03720 | Transcription [9.2] | AB919825-AB919844 | 2 | 2 |
| DNA-directed RNA polymerase | CNI00420 | Transcription [9.2] | AB919962-AB919978 | 1 | 1 |
| RAS2 | CNJ01920 | Translation [10.5] | AB920017-AB920035 | 1 | 2 |
| glycogen synthase kinase 3 | CNB00720 | Regulatory functions [11.1] | AB919630-AB919650 | 2 | 2 |
| mitogen-activated protein kinase | CNC06590 | Regulatory functions [11.1] | AB919728-AB919747 | 2 | 2 |
| phosphatidylinositol 3-kinase TOR1 | CNF03740 | Regulatory functions [11.1] | AB919865-AB919882 | 1 | 2 |
| chaperone | CNM01520 | Cellular processes [13.3] | AB920118-AB920135 | 1 | 2 |
| alpha tubulin | CNB02810 | Cellular processes and signaling; Cytoskeleton (COG)*2 | AB919651-AB919669 | 2 | 2 |
| beta1-tubulin, putative | CNC03260 | Cellular processes and signaling; Cytoskeleton (COG)*2 | AB919689-AB919707 | 2 | 2 |
| eukaryotic translation initiation factor 3 subunit 6 | CNL05160 | Unknown (KEGG)*2 | AB920078-AB920097 | 2 | 2 |
*1 Data from Microbial Genome Database for Comparative Analysis (MBGD) (http://mbgd.nibb.ac.jp). Numerals in brackets show the detailed category in MBGD.
*2 Since function category was not shown in MBGD, data was obtained from the databases, COG and KEGG.
† For other species, only one homolog was detected.
Phylogeny of Trichosporonales
First, we obtained sequence data for three regions (RNA polymerase II subunits I (RPB1) and II (RPB2) and translation elongation factor 1-α (TEF1-α)) that have been used for the phylogenetic analyses in Ascomycotina and Basidiomycotina [15–19]. The sequences of these regions of 12 ascomycetous yeasts (S1 Table) were obtained from the database and aligned to extract the corresponding regions from our data. The conserved domains of genes predicted by Microbial Genome Annotation Pipeline (MiGAP) (http://www.migap.org/) showed good alignment based on the codons, and we found that Trichosporonales species possessed spliceosomal introns and that their position was strain-specific. Intron positions also differed from those in C. neoformans, which was used as a model species for gene prediction using HMM-based gene finder AUGUSTUS [20] (Fig 2 and S2 Table). While the intron positions of Agaricomycetes species are sufficiently conserved to distinguish and characterize the higher taxa [16], those of Trichosporonales species are not conserved. Therefore, we identified and aligned CDS regions of T. asahii and T. faecale by referring to their mRNA data and then added genes from other species, as indicated in the Materials and Methods. The sequences used in this study covered almost the complete CDS region; 72,531 nucleotides (24,173 residues) among 30 concatenated genes. Gene alignments are available upon request.
Fig 2. Amino acid sequences and intron positions of CDS homologs of translation elongation factor 1-α (TEF1-α), DNA-dependent RNA polymerase II RPB140 (RPB2), and DNA-directed RNA polymerase II largest subunit (RPB1) from Trichosporon asahii JCM 2466.
Numbers refer to intron positions.
Phylogenetic trees for the 16 species based on nucleotide and amino acid sequences using the 30 concatenated genes are shown in Fig 1b and 1c, respectively. The trees based on a single gene are presented in (S3 Table). Trees inferred from the concatenated alignments based on nucleotide (Fig 1b) and amino acid (Fig 1c) sequences had the same topology and increased reliability compared with the tree based on the D1/D2 region of the LSU rRNA gene (Fig 1a) and other single-gene trees. A phylogenetic tree inferred from the amino acid sequence alignment including consensus sequences of T. coremiiforme and T. voides (see Materials and Methods) showed an identical topology (S1 Fig). Thus, they currently represent the best estimate of the phylogenetic relationship in Trichosporonales.
To evaluate the robustness of the topology of the tree inferred from the concatenated alignments, we identified all nodes recovered during the single-gene analyses and recorded bootstrap values when the nodes were present in a tree inferred from the concatenated alignment (Table 3). Although T. ovoides should be included in the phylogenetic analyses of the Trichosporon genus since this is the type species of the genus, we excluded this species from the analysis, as mentioned above. Nonetheless, we continue to use the name of the clade, Ovoides, in this report.
Table 3. Comparison of topology between the concatenated tree and single trees.
| CDS product | Homolog to: (locus_tag of C. neoformans) | Based on nucleotide sequences | ||||||||||||||||
| Positions used | Highest log likelihood | Bootstrap value of each node*1 | # nodes contained in concatenated tree*2 | Divergence within Trichosporonales*3 | ||||||||||||||
| a | b | c | d | e | f | g | h | i | j | k | l | m | ||||||
| phosphatidylinositol 3-kinase TOR1 | CNF03740 | 7167 | -71807.7691 | 100 | 100 | 100 | (57) | 96 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 12/13 | 0.2765973 |
| glutamate synthase (NADH) | CNJ02910 | 6525 | -58590.6785 | 100 | 100 | - | 100 | 100 | 100 | 75 | 100 | 100 | 93 | 95 | 100 | 96 | 12/13 | 0.2325986 |
| malate synthase | CNH02910 | 1641 | -13875.1721 | 100 | 100 | 100 | - | (53) | 100 | 100 | 72 | 100 | 100 | 84 | 100 | 90 | 11/13 | 0.2328670 |
| phosphoenolpyruvate carboxykinase | CNI03590 | 1668 | -10299.0077 | 100 | 100 | 81 | 100 | 81 | 75 | 92 | - | 76 | 99 | - | 100 | 97 | 11/13 | 0.1416784 |
| ATP-binding cassette (ABC) transporter | CND00300 | 1932 | -14912.2764 | 100 | 100 | 100 | 81 | - | 100 | 91 | (38) | 100 | 100 | - | 100 | 98 | 10/13 | 0.1990241 |
| glycogen synthase kinase 3 | CNB00720 | 1272 | -9228.1996 | 100 | 100 | 100 | - | - | 99 | 87 | - | 100 | 73 | - | 100 | 100 | 9/13 | 0.1819497 |
| DNA-directed RNA polymerase ii largest subunit, putative (RPB1) | CNE03720 | 5535 | -54837.8043 | 100 | 100 | 100 | - | - | 100 | 98 | - | 99 | 100 | - | 100 | 100 | 9/13 | 0.2865913 |
| DNA replication licensing factor cdc19 (cell division control protein 19) | CNG02380 | 2928 | -29876.1716 | 100 | 100 | 100 | 73 | - | 100 | - | 100 | - | 92 | - | 100 | 94 | 9/13 | 0.3063356 |
| isocitrate lyase | CNH03280 | 1698 | -15133.6988 | 100 | 100 | 100 | - | - | 87 | 80 | 71 | 98 | - | - | 100 | 100 | 9/13 | 0.2597505 |
| DNA-directed RNA polymerase | CNI00420 | 3414 | -31208.3767 | 87 | 100 | 100 | (48) | - | 93 | (63) | 99 | 76 | 76 | (55) | 100 | 100 | 9/13 | 0.2359366 |
| methylenetetrahydrofolate reductase (NADPH) | CNL04820 | 1929 | -16548.0025 | 100 | 100 | 100 | (53) | 85 | 100 | - | 99 | 99 | 99 | - | 100 | - | 9/13 | 0.2302595 |
| ATP dependent DNA helicase | CNB05360 | 2235 | -22405.5341 | 100 | 100 | 100 | - | (24) | 100 | 72 | 79 | 82 | (45) | (2) | 100 | - | 8/13 | 0.2732160 |
| adenosylhomocysteinase | CND00240 | 1347 | -8971.2698 | 98 | 100 | 100 | - | - | 82 | 90 | 76 | - | - | - | 98 | 99 | 8/13 | 0.1546073 |
| DNA-dependent RNA polymerase II RPB140 (RPB2) | CND03540 | 3849 | -33545.1855 | 100 | 100 | 100 | - | - | 100 | 99 | 100 | (62) | - | - | 100 | 96 | 8/13 | 0.2237954 |
| pyruvate carboxylase | CNF00650 | 3729 | -28326.3212 | 100 | 100 | 100 | - | (37) | 92 | (65) | 100 | 100 | - | - | 100 | 100 | 8/13 | 0.1847746 |
| Eukaryotic translation initiation factor 3 subunit 6 | CNL05160 | 1920 | -16255.6032 | 100 | 100 | 100 | - | - | 100 | - | 100 | 100 | - | - | 100 | 99 | 8/13 | 0.2257396 |
| plasma membrane H(+)-ATPase 1 | CNN01260 | 3135 | -20529.9850 | 100 | 100 | 99 | - | - | 100 | 93 | (62) | 100 | 77 | - | 100 | - | 8/13 | 0.1515828 |
| 1,3-beta-glucan synthase | CNN02320 | 5511 | -47328.6072 | 100 | 100 | 100 | - | - | 100 | 100 | 100 | 100 | (46) | - | 100 | - | 8/13 | 0.2236312 |
| mitogen-activated protein kinase | CNC06590 | 1107 | -8324.0754 | 100 | 99 | 97 | - | - | 81 | 99 | (62) | 83 | (43) | - | 98 | (68) | 7/13 | 0.1666310 |
| orotidine monophosphate pyrophosphorylase (URA5) | CNG03730 | 687 | -6763.7318 | 100 | 100 | 100 | 98 | - | 91 | - | - | (53) | (63) | - | 100 | 99 | 7/13 | 0.3037264 |
| vacuolar ATP synthase | CNI01180 | 1587 | -11933.2227 | 100 | 100 | 89 | - | - | (52) | 75 | (37) | 99 | - | - | 100 | 97 | 7/13 | 0.1767209 |
| Ras2 | CNJ01920 | 1101 | -6180.5535 | 99 | 100 | 99 | - | - | 97 | - | 88 | - | - | - | 100 | 99 | 7/13 | 0.1254647 |
| chaperone | CNM01520 | 2139 | -13392.3250 | 100 | 100 | 100 | - | (39) | 100 | - | 96 | - | - | - | 100 | 100 | 7/13 | 0.1363423 |
| citrate synthase | CNA00510 | 1416 | -9234.5688 | 100 | 100 | - | - | (57) | 89 | (52) | - | 93 | 98 | (69) | 100 | - | 6/13 | 0.1478656 |
| actin | CNA04650 | 1161 | -5631.8845 | 98 | 100 | 73 | - | - | - | 74 | (63) | 85 | - | - | 99 | (62) | 6/13 | 0.0915010 |
| alpha tubulin | CNB02810 | 1371 | -7641.2544 | 100 | 97 | 98 | - | (54) | 89 | - | - | - | - | (65) | 100 | 94 | 6/13 | 0.1094242 |
| argininosuccinate lyase | CNC04420 | 1416 | -11523.0058 | 100 | 100 | 88 | 100 | - | - | (31) | - | - | 73 | - | 100 | - | 6/13 | 0.2090371 |
| beta tubulin | CNC03260 | 1377 | -7299.8992 | 100 | 100 | 100 | - | - | 90 | - | - | - | - | - | 92 | - | 5/13 | 0.1023791 |
| adenylyl-sulfate kinase | CNE03380 | 641 | -5714.2900 | 100 | - | 96 | - | - | (57) | 92 | - | (41) | - | - | 100 | 98 | 5/13 | 0.2429367 |
| translation elongation factor 1-α | CNM01300 | 1389 | -7333.6338 | - | 100 | (60) | - | - | 100 | 98 | (39) | - | - | - | 96 | - | 4/13 | 0.0985092 |
| # nodes contained in concatenated tree*2 | 29/30 | 29/30 | 27/30 | 6/30 | 4/30 | 26/30 | 18/30 | 15/30 | 19/30 | 13/30 | 3/30 | 30/30 | 20/30 | |||||
| CDS product | Homolog to: (locus_tag of C. neoformans) | Based on amino acid sequences | ||||||||||||||||
| Positions used | Highest log likelihood | Bootstrap value of each node*1 | # nodes contained in concatenated tree*2 | Divergence within Trichosporonales*4 | ||||||||||||||
| n | o | p | q | r | s | t | u | v | w | x | y | z | ||||||
| phosphatidylinositol 3-kinase TOR1 | CNF03740 | 2388 | -20501.3857 | 100 | 100 | 100 | 100 | 98 | 100 | 93 | 100 | 100 | 100 | 99 | 100 | 100 | 13/13 | 0.1414040 |
| DNA-directed RNA polymerase ii largest subunit, putative (RPB1) | CNE03720 | 1844 | -19055.5663 | 100 | 100 | 100 | 100 | - | 100 | 80 | - | 100 | 85 | 84 | 91 | 100 | 11/13 | 0.1861724 |
| glutamate synthase (NADH) | CNJ02910 | 2174 | -23206.1602 | 100 | 100 | 100 | 100 | (42) | 100 | 80 | 91 | 100 | 100 | (38) | 100 | 100 | 11/13 | 0.1956882 |
| malate synthase | CNH02910 | 546 | -6042.9704 | 100 | 100 | 100 | - | (41) | 97 | 96 | (63) | 91 | 97 | 73 | 100 | 98 | 10/13 | 0.2240399 |
| methylenetetrahydrofolate reductase (NADPH) | CNL04820 | 633 | -5915.2895 | 100 | 100 | 100 | 80 | (59) | 100 | 75 | 100 | 98 | 90 | - | - | 99 | 10/13 | 0.1540453 |
| ATP-binding cassette (ABC) transporter | CND00300 | 643 | -5438.2601 | 97 | 100 | 100 | (40) | - | 99 | 90 | (49) | 100 | 97 | (53) | 98 | 100 | 9/13 | 0.1424313 |
| DNA-directed RNA polymerase | CNI00420 | 1137 | -8524.1458 | 86 | 100 | 100 | (65) | - | 96 | 78 | - | 100 | 96 | - | 99 | 100 | 9/13 | 0.1097242 |
| 1,3-beta-glucan synthase | CNN02320 | 1836 | -17034.1009 | 97 | 100 | 100 | - | - | 100 | 99 | 100 | 100 | (24) | (51) | 100 | 100 | 9/13 | 0.1593036 |
| pyruvate carboxylase | CNF00650 | 1242 | -10659.4494 | 99 | 100 | 100 | - | (42) | (60) | 87 | 100 | 100 | - | - | 100 | 100 | 8/13 | 0.1286247 |
| DNA replication licensing factor cdc19 (cell division control protein 19) | CNG02380 | 975 | -11547.8857 | 97 | 100 | 100 | (60) | - | 96 | - | 100 | 71 | (43) | - | 96 | 100 | 8/13 | 0.2288008 |
| isocitrate lyase | CNH03280 | 565 | -6484.3719 | 99 | 100 | 100 | (64) | - | 94 | - | 82 | 93 | - | - | 98 | 100 | 8/13 | 0.2222622 |
| vacuolar ATP synthase | CNI01180 | 528 | -3637.3220 | 96 | 99 | - | (63) | - | 95 | 94 | (53) | 79 | 70 | - | 85 | 100 | 8/13 | 0.0735907 |
| phosphoenolpyruvate carboxykinase | CNI03590 | 555 | -3829.0758 | 97 | 100 | 92 | 85 | - | (44) | - | - | 89 | 97 | (56) | 97 | 100 | 8/13 | 0.0931894 |
| plasma membrane H(+)-ATPase 1 | CNN01260 | 1044 | -8907.8908 | 100 | 100 | 100 | - | (47) | 100 | 77 | (54) | 100 | (62) | - | 98 | 100 | 8/13 | 0.1269335 |
| ATP dependent DNA helicase | CNB05360 | 744 | -7343.5154 | 100 | 100 | 100 | (36) | - | 99 | - | (29) | 95 | (50) | - | 74 | 100 | 7/13 | 0.1617158 |
| adenosylhomocysteinase | CND00240 | 447 | -4136.0745 | (24) | 100 | 91 | - | - | 72 | 93 | 71 | - | - | - | 77 | 100 | 7/13 | 0.1579080 |
| DNA-dependent RNA polymerase II RPB140 (RPB2) | CND03540 | 1280 | -10617.4474 | 99 | 100 | 99 | - | - | 100 | - | (63) | 100 | - | - | 96 | 100 | 7/13 | 0.1122744 |
| orotidine monophosphate pyrophosphorylase (URA5) | CNG03730 | 228 | -2840.2696 | 100 | 99 | 97 | 96 | - | 86 | - | - | (69) | (33) | - | 100 | 100 | 7/13 | 0.2999401 |
| Ras2 | CNJ01920 | 366 | -2531.3792 | 97 | 100 | 94 | (55) | - | 95 | - | 90 | - | - | - | 100 | 100 | 7/13 | 0.0971751 |
| Eukaryotic translation initiation factor 3 subunit 6 | CNL05160 | 639 | -6146.9708 | 97 | 100 | 95 | - | - | 100 | (61) | 76 | 100 | - | - | - | 100 | 7/13 | 0.1704422 |
| chaperone | CNM01520 | 712 | -5502.4940 | 100 | 100 | 99 | - | - | 92 | (59) | 86 | 70 | - | - | (61) | 100 | 7/13 | 0.1150690 |
| glycogen synthase kinase 3 | CNB00720 | 421 | -2686.2647 | 88 | 76 | 95 | - | - | (52) | - | (62) | 100 | - | - | 98 | 100 | 6/13 | 0.0792372 |
| argininosuccinate lyase | CNC04420 | 471 | -4055.3207 | 91 | 100 | 95 | 99 | - | 96 | - | (58) | - | - | (59) | - | 100 | 6/13 | 0.1375764 |
| actin | CNA04650 | 386 | -1520.7293 | 82 | 93 | 84 | - | - | - | - | (69) | 96 | - | - | - | 99 | 5/13 | 0.0254971 |
| beta tubulin | CNC03260 | 458 | -2074.0608 | 93 | 100 | 98 | - | - | (61) | - | 90 | - | - | - | (50) | 100 | 5/13 | 0.0307975 |
| mitogen-activated protein kinase | CNC06590 | 368 | -1562.5243 | 97 | 81 | 100 | (68) | - | 89 | - | - | (63) | (42) | - | (54) | 74 | 5/13 | 0.0313906 |
| adenylyl-sulfate kinase | CNE03380 | 212 | -2039.7504 | 98 | 80 | 77 | - | - | (62) | - | 74 | (49) | - | - | (59) | 97 | 5/13 | 0.1536762 |
| citrate synthase | CNA00510 | 471 | -4170.0870 | 100 | 100 | (67) | - | - | (31) | - | (19) | (49) | (57) | - | 100 | 71 | 4/13 | 0.1482440 |
| translation elongation factor 1-α | CNM01300 | 462 | -2674.2830 | (56) | 100 | - | - | - | 94 | 99 | (44) | - | - | - | - | 97 | 4/13 | 0.0626686 |
| alpha tubulin | CNB02810 | 456 | -2114.6945 | 100 | - | - | - | (48) | 83 | - | - | - | - | (21) | (39) | 89 | 3/13 | 0.0321153 |
| # nodes contained in concatenated tree*2 | 28/30 | 29/30 | 26/30 | 7/30 | 1/30 | 23/30 | 13/30 | 13/30 | 20/30 | 9/30 | 3/30 | 20/30 | 30/30 | |||||
*1, Node names are shown in Fig 1b and 1c. Numerals show the bootstrap value of each node which corresponded to that of the concatenated tree [boldface, more than 70%; (), less than 70%].—indicates that the node on the concatenated tree is not shown in respective CDS tree.
*2, Nodes showing more than 70% bootstrap value (boldface) were treated as "positive".
*3, The number of base substitutions per site from averaging over all sequence pairs are shown. Analyses were conducted using the Kimura 2-parameter model [47]. All ambiguous positions were removed for each sequence pair.
*4, The number of amino acid substitutions per site from averaging over all sequence pairs are shown. Analyses were conducted using the JTT matrix-based model [45]. All ambiguous positions were removed for each sequence pair.
In the Ovoides clade, T. asahii, T. faecale and T. inkin formed a subclade, as shown by the high bootstrap values in the single-gene trees for the majority of genes [nodes a (29/30) and b (29/30) in Fig 1b and n (28/30) and o (29/30) in Fig 1c and Table 3, and hereafter]. Regarding the Cutaneum clade, the two species examined (T. cutaneum and T. dermatis) formed a cluster in trees inferred from the concatenated alignment, which was also observed in single-gene trees [nodes c (27/30) and p (26/30)]. The Ovoides and Cutaneum clades formed a monophyletic group, which was supported with 100% and 72% bootstrap values for nucleotide and amino acid sequences, respectively. The tree based on the D1/D2 region of the LSU rRNA gene did not support this cluster (Fig 1a), and single-gene trees showing more than 70% bootstrap values were 4 of 30 [nodes e] and 1 of 30 [node r] based on nucleotide and amino acid sequences, respectively.
The Porosum and Gracile clades formed a cluster, and the connection of the Brassicae clade was shown in the tree based on the D1/D2 region of the LSU rRNA gene (Fig 1a). In trees inferred from the concatenated alignment, the Brassicae and Gracile clades were clustered and connected to the Porosum clade with 100% bootstrap values for both nucleotide and amino acid sequences. The bootstrap values for single-gene trees were similar to those for trees inferred from the concatenated alignment: two-thirds supported the connection of the Brassicae and Gracile clades [nodes i (19/30) and v (20/30)], and one-third supported their connection to the Porosum clade [nodes j (13/30) and w (9/30)].
Cryptococcus curvatus is in an adjunct position in the Cutaneum clade with 100% bootstrap values for both nucleotide and amino acid sequences (Fig 1b and 1c). We previously evaluated the phylogenetic position of the species based on rRNA genes and spacer regions, but were unable to obtain good resolution [21–22]. In this study, the single-gene trees did not frequently support the node [more than 70% bootstrap support nodes for d (6/30) and q (7/30)].
Vanrija humicola was located outside of the Trichosporon species, which has been reported previously [2, 5, 6, 21, 23]. As described above, Weiss et al. [6] proposed the use of genus Vanrijia as the genus name, and we follow their suggestion in this report. The genus Vanrija sensu Moore [7] is polyphyletic; thus the emendation and subsequent reclassification including new combinations is required. We believe that our genes will contribute to the reclassification; namely, delineation for the species at the basal position between Vanrija and other genera.
The Bullera koratensis and Cryptococcus tepidarius clades occurred at remote positions in the Trichosporonales. In addition, the intron positions were closer to those of C. neoformans than to those of other Trichosporon species (S2 Table). Okoli et al. [3] proposed the genus Cryptotrichosporon based on the D1/D2 region of the LSU rRNA plus ITS regions and the 18S rRNA gene, and noted that B. koratensis and C. tepidarius were phylogenetically closely related to their species. These species might be pivotal in clarifying the phylogenetic relationships between Tremellales and Trichosporonales, since they obtain intermediate positions.
Phenotypic properties and phylogeny
Trichosporon has been divided into five clades based on a combination of serological properties and major ubiquinone as described above [2, 24]. Based on morphological aspects, species in the clades Brassicae, Cutaneum, Gracile, and Ovoides produce abundant arthroconidia, whereas those in the Porosum clade rarely produce arthroconidia or not at all [2]. Fig 1b and 1c showed clustering of the Brassicae and Gracile clades and a connection to the Porosum clade with 100% bootstrap values for both nucleotide and amino acid sequences, indicating that this morphological characteristic supports the linkage between Brassicae and Gracile clades. Recently, Nagy et al. [25] reported that Zn-finger transcription factor families are related to switches between yeast and filamentous forms. Since the production of arthroconidium has been reported in the genera Guehomyces and Tausonia (Cystofilobasidiales) and species in the family Dipodascaceae (Saccharomycetales), genes associated with arthroconidium production may be found when the genomic data are accumulated.
Regarding serological properties, five clades; namely, Cutaneum, Ovoides, Brassicae, Gracile and Porosum, are characterized by serogroup I, II, III, I-III and I-III (reacting with both factors I and III), respectively [26]. The clustering topology of clades Brassicae (serogroup I) and Gracile (I-III), and the connection to clade Porosum (I-III), indicated that these three clades can be characterized by positive reactions with III as common antisera (S2 Fig). This grouping, serogroups I, II and III, was proposed previously [27] and confirmed in this study. The epitopes of serological group II and III have been reported [28–29]; thus, this characteristic would be a useful property for differentiation.
For the major ubiquinone in ascomycetous yeasts, ubiquinone isoprene chain length ranged from 6 to 10 (namely Q-6 to Q-10) and showed a strong relationship with phylogeny of higher taxa with some exceptions, and was used as a phenotypic characteristic [15, 30]. In basidiomycetous yeasts, most species contain Q-10 and few genera are characterized by different chain lengths: Mrakia and Cystofilobasidium (both in Cystofilobasidiales) are characterized by Q-8, and Q-9 was observed in Trichosporon (excluding the Cutaneum clade), Vanrija, Guehomyces, and Itersonilia in Agaricomycotina. In this study, Q-10 species C. curvatus clustered with the Cutaneum clade, which has been characterized by Q-10 (S2 Fig), and this characteristic supports this linkage. Although the taxonomic significance of ubiquinone isoprene chain length remains unclear due to the occurrence of both Q-9 and Q-10 in the same genus; e.g., in Leucosporidium [31], this could be used as a distinctive characteristic with serological properties when the Cutaneum clade together with C. curvatus is proposed as a separate genus. We believe that further studies will clarify how the isoprene chain length is controlled by prenyltransferase and related mechanisms.
Clarification of the topology of basal species in a lineage by obtaining a highly resolved tree
Recently, the number of fungal phylogenetic trees based on genomic data have increased due to the greater amount of genomic data available e.g., [32–36]. However, studies on fungi are still fewer compared with bacteria and archaea, mainly because of the amount of available genomic data. Based on the NCBI BioProject (http://www.ncbi.nlm.nih.gov/bioproject/), only 84 and 230 species were available for yeasts and filamentous fungi, respectively, as of May 15th, 2014. With the available fungal genomic data, Rokas et al. [32] demonstrated that a minimum of 20 concatenated genes in an analysis were sufficient to represent the phylogeny based on all genomic data for Saccharomyces species. Kuramae et al. [33] showed that 40–45 concatenated proteins would be required to resolve the tree of life of the fungal kingdom by cophenetic correlation analysis. Based on these reports, we believe that concatenated analyses of genes could be used for robust studies on the fungal tree of life, although the number of genes required would depend on the range of the target groups.
Rokas et al. [32] used 106 genes and compared a total length of 127,026 nucleotides (42,342 residues), which corresponded to roughly 1% of the genomic sequences and 2% of the predicted genes. According to this estimation, the use of “a concatenated analysis of 20 or more genes”, corresponding to roughly 0.2% of the genomic sequences, is sufficient to construct an accurate phylogeny. Our data based on 72,531 nucleotides and corresponding to 0.24% (for Trichosporon laibachii, genome size 30.6 Mb) to 0.40% (for C. curvatus, genome size 18.2 Mb) of the total genomic sequences of Trichosporonales support the results of Rokas et al. [32].
While the number of genes required to represent a tree with a resolution comparable to the resolution of a genome tree has been discussed [32, 34], an example of such a set of genes has not been reported. In this study, we propose a set of genes suitable for constructing a phylogenetic tree with high resolution to examine the evolutionary diversification in Trichosporonales. Table 3 indicates that the use of more genes does not improve the resolution of the tree of this taxonomic group. In addition, among the nucleotide sequences used in this study, those for four genes (phosphatidylinositol 3-kinase TOR1, glutamate synthase (NADH), malate synthase and phosphoenolpyruvate carboxykinase) showed better resolution than those typically used for multigene analyses, such as TEF1-α, RPB1 and RPB2. For the tree based on amino acid sequences of phosphatidylinositol-3-kinase TOR1 (even if used alone), the same topology was obtained as for the tree inferred from the concatenated alignment. The tree inferred from the concatenated alignment based on RPB1, RPB2 and TEF1-α (regions corresponding to those used by Kurtzman and Robnett [15]) revealed the existence of five clades in Trichosporon (nodes b, c, f, g and h) with 100% bootstrap values and a clear relationship among the Brassicae, Gracile and Porosum clades (nodes i and j; S3 Fig). This tree had better resolution than the respective single-gene trees, as shown in (S4 Table), but did not clarify the relationship between the Ovoides clade and the other four Trichosporon clades (nodes e and r), or the positions of V. humicola (nodes k and x) and C. curvatus (node d). Obviously, the use of these three genes is not enough to clarify the phylogenetic relationship of the Trichosporonales. This was our original motivation for the present study.
In yeasts, resolution of the phylogenetic relationships has particularly been increased for the ascomycetous teleomorphic genera using partial sequences of TEF1-α and cytochrome oxidase II, in addition to rRNA genes. The later addition of RPB1 and RPB2 improved the resolution for the Saccharomycetales yeast species [15]. Recently, Koufopanou et al. [37] reported primer sets for 14 genes to clarify the topology of basal species among the Saccharomycetales. Of our 30 genes, glutamate synthase and adenosyl homocysteinase were used for multiple gene analysis by Koufopanou et al. [37], and CDC19 was selected as a candidate during the first screening stage but was not used in the published analysis. Gene duplication in the Tor pathway was also reported in some fungi by Shertz et al. [38]. Careful attention must be paid when conducting multigene analyses in other taxa using the Tor gene; however, this gene can be used for Trichosporonales, since we did not detect a Tor paralog in our experiments.
Conclusions
In this study, phylogenetic trees inferred from concatenated alignments (nucleotide and amino acid sequences) of 30 genes extracted from draft genome data showed greater resolution than in previous reports, indicating that they currently represent the best estimate of the phylogenetic relationship in Trichosporonales. In addition, we suggest that the genes identified in this study are good candidates for constructing a multi-locus sequence analysis system for both inter- and intra-species analyses. Specific primers are required to adopt these newly discovered promising genes. These genes can also be used for epidemiological and biogeographical studies. A set of genes suitable for constructing a phylogenetic tree to examine evolutionary diversification. The relationship between the closely related Trichosporonales and Tremellales in the Agaricomycotina might be clarified using these genes [6, 17, 39]. Moreover, our study provides a platform for future studies on a comprehensive reclassification of pleomorphic fungi.
Materials and Methods
Genome sequencing
Strains and DNA and RNA extraction
Strains used in this study were received from the Japan Collection of Microorganisms at the RIKEN Bioresource Center (Table 1). Cultures grown on YM agar (Difco, Detroit, MI; BD, Franklin Lakes, NJ, USA) were collected and freeze-dried, and DNA was prepared according to a previous method [40]. The integrity and quality of the extracted genomic DNA were examined by agarose gel electrophoresis using a NanoDrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA). The concentration of DNA was determined using a Quant-iT PicoGreen dsDNA Assay Kit (Life Technologies, Gaithersburg, MD, USA). Total RNA was prepared from T. asahii and T. faecale cells cultivated at 37°C or 28°C using an RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions after disruption by glass beads. The integrity of the RNA was verified by agarose gel electrophoresis. The RNA concentration was determined using a NanoDrop 2000 spectrophotometer.
Genome sequencing
Draft genomes of 17 selected species in the Trichosporonales were determined using a 454 GS FLX (Roche Diagnostics, Mannheim, Germany) and/or Illumina (Illumina Inc., San Diego, CA, USA) next-generation sequencer. Since karyotyping data is not available for the Trichosporonales, we were unable to estimate the genome sizes of these species before the beginning of this study. Thus, the genome size of C. neoformans (approximately 20 Mb) was used as an initial estimate.
DNA library preparation and sequencing were performed according to the manufacturer’s instructions. Briefly, a paired-end DNA library was prepared from 1 μg genomic DNA, following DNA fragmentation into approximately 350-bp average sized fragments using an S2 ultrasonicator (Covaris, Woburn, MA, USA) and then sequenced using a HiSeq 2000 sequencer (Illumina Inc.; S5 Table). The T. asahii genome was also sequenced using the GS FLX Titanium platform (Roche Diagnostics). A DNA library was prepared from 0.6 μg T. asahii genomic DNA, followed by fragmentation into approximately 900-bp average sized fragments using a Covaris S2 ultrasonicator and then sequencing using GS FLX pyrosequencing technologies. De novo assembly was performed using the CLC Genomics Workbench (version 4.5 or later) and CLC Genomics Server (version 3.6 or later) (CLC Bio Co., Aarhus, Denmark) with the default settings, including the “Update contigs” option, which edited contig sequences based on the reads mapped to the contig. The de novo assembly produced contigs with an average N50 value of 37,614. Key attributes of the draft genome sequences are summarized in Table 1. Library generation, sequencing and de novo assembly were performed at the Genome Network Analysis Support Facility, RIKEN CLST (Yokohama, Japan).
Transcriptomic sequencing
An RNA-Seq library was prepared as described previously [41]. Briefly, polyA(+) RNA was extracted from 10 μg of total RNA using Dynabeads Oligo(dT)25 (Life Technologies), according to the manufacturer’s protocols, and was fragmented by heating at 70°C for 3.5 min in 0.5× fragmentation buffer (Life Technologies). Fragmented RNA was purified using the RNeasy MinElute Kit (Qiagen) and then dephosphorylated using Antarctic phosphatase (New England Biolabs, Beverly, MA, USA). The dephosphorylated RNA was phosphorylated using T4 polynucleotide kinase (New England Biolabs) with 10 mM ATP and was purified again using the RNeasy MinElute Kit. Purified RNA was concentrated and ligated at both ends with 3’ and 5’ adapters from the TruSeq Small RNA Sample Prep Kit (Illumina Inc.) using T4 RNA ligase 2 and T4 RNA ligase 1 (New England Biolabs), respectively. Adapted RNA was reverse transcribed using PrimeScript Reverse Transcriptase (Takara Bio, Kyoto, Japan) and amplified using Phusion High-Fidelity DNA Polymerase (New England Biolabs), as well as forward and reverse primers from the TruSeq Small RNA Sample Prep Kit. cDNA libraries with a size range of 160–320 bp were selected by removing PCR primers using 1.2 volumes of AMPure XP beads (Beckman Coulter, Miami, FL, USA) and subsequent gel extraction using Pippin Prep (Sage Science, Beverly, MA, USA). The four resulting cDNA libraries were pooled and then subjected to 101-bp single-read sequencing on a HiSeq 2000 sequencer. The sequencing produced 97 million and 88 million reads for T. asahii cultivated at 37°C and 28°C, respectively, and 80 million and 117 million reads for T. faecale cultivated at 37°C and 28°C, respectively. The reads were trimmed of adapter, low-quality and ambiguous sequences using the CLC Genomics Workbench with default parameters and were then assembled de novo with the CLC Genomics Workbench and CLC Genomics Server. The draft transcriptome assemblies of T. asahii at 37°C and 28°C and of T. faecale at 37°C and 28°C consisted of 26,569 (N50 = 1,307), 26,769 (N50 = 1,382), 24,105 (N50 = 1,362) and 25,349 (N50 = 1,389) transcripts, respectively. Library generation, sequencing, and de novo assembly were performed at the Genome Network Analysis Support Facility, RIKEN CLST.
Auto-annotation
After the de novo assembly, auto-annotation was performed using MiGAP (http://www.migap.org/), a publicly available annotation pipeline system of microbial and fungal genome draft sequences operated by the National Institute of Genetics. Briefly, MiGAP provides consecutive gene searching and annotation functions and consists of prediction programs. Exons and introns were predicted using the HMM software AUGUSTUS 2.5.5 [20]. The HMM model species used for this prediction is cryptococcus_neoformans_neoformans_JEC21. After gene prediction, each ORF was subjected to a homology search against the following protein sequence databases using NCBI BLAST: the fungi division of RefSeq, whole entries of TrEMBL and whole entries of NR.
Alignment and phylogenetic analyses
Gene selection and alignment
For selection of genes, we used genomic data from C. neoformans (AE017341) and C. gattii (CP000286) as references. Since our target genes were those that can be used for genus-level discrimination, they were required to be conserved in the same genus. A total of 1,234 CDSs were selected from C. neoformans, C. gattii and T. asahii genomic data under the conditions of identity = 60 and coverage = 60, using in-silico molecular cloning (IMC; In Silico Biology, Inc., Yokohama, Japan). CDS homologs showing more than 97% similarity between C. neoformans and C. gattii were listed. Candidate T. asahii genes were selected using IMC under the criteria that they include approximately 300 or more amino acids and do not include many introns, since our purpose was to develop signature/marker sequences. CDS homologs of other species were then selected using IMC. Genes used in this study are listed in Table 2 with DDBJ/GenBank/EMBL accession numbers.
The coding regions of genes from T. asahii and T. faecale were determined using a BLAST search against the respective mRNA data. Multiple alignment of respective genes was first performed using the CDSs of T. asahii and T. faecale using those of C. neoformans as an outgroup. Subsequently those of other species were added and aligned based on codons. In some cases, part of a gene could be found in one contig and the other parts in different contigs. In this case, manual alignment was performed.
When making an alignment file, the following treatment was performed for two CDSs beforehand: An obvious insertion (1,557 nucleotides) in CND03540 (DNA-dependent RNA polymerase II (RPB2)) homolog was found in T. asahii but not in other species, and the region was eliminated from the analyses. Since the same was observed in the published T. asahii genome data (for both ALBS01000000 and AMBO01000000), this insertion was assumed to be distinct to this species or these strains in Trichosporonales. For CNF03740 (phosphatidylinositol-3-kinase TOR1) homolog, the length of CDSs of 9 species studied, viz. Trichosporon brassicae, T. cutaneum, T. dermatis, Trichosporon scarabaeorum, B. koratensis, C. curvatus, C. tepidarius, V. humicola and C. neofomans (outgroup) were 7,167 nucleotides (2,388 amino acids), whereas those of other species were found to be longer and varied. Since they aligned well from the 5’ end to position 7,167 by codon, 7,167 nucleotides (2,388 amino acids) were used for the analyses; the downstream sequences were not used. Since CDSs were orthologous genes shown in the Microbial Genome Database for Comparative Analysis (MBGD) (http://mbgd.nibb.ac.jp), CDSs were concatenated using SeaView [42].
Alignment including the consensus amino acid sequence of T. coremiiforme and T. ovoides
As shown in Table 2, we found paralogous genes in T. ovoides and T. coremiiforme. In this case, we created a consensus sequence from paralogs (from a multiple alignment) with X indicating a non-identical amino acid. When a partial sequence of gene was found only in one of the two sequences, this partial sequence was included in the consensus sequence.
Alignment of frequently used regions of RPB1, RPB2 and TEF1-α in fungi
Sequence alignments of RPB1, RPB2, and TEF1-α that are often used for the phylogenetic analyses of fungi were generated by referring to sequences of 12 ascomycetous yeasts [15]. The reference sequences used for this extraction are shown in (S1 Table).
Alignment of the D1/D2 region of LSU rRNA gene
All sequences of the D1/D2 region of LSU rRNA gene were obtained from GenBank/EMBL/DDBJ database (S1 Table). A multiple alignment was performed using MEGA6 [43] and edited manually.
Phylogenetic analyses
Trees (a phylogenetic tree based on each gene and a tree inferred from the concatenated alignment) were constructed using the maximum likelihood method in MEGA6 [43]. When a tree was constructed based on nucleotide sequences, the Tamura-Nei model [44] was used for the analyses with options Rates and Patterns = uniform rates. Initial trees for the heuristic search were obtained automatically by applying the neighbor-joining and BIONJ algorithms to a matrix of pairwise distances estimated using the maximum composite likelihood (MCL) approach and then selecting the topology with the highest log likelihood value. When a tree was inferred based on amino acid sequences, the JTT matrix-based model [45] was used for analyses with option Rates and Patters = uniform dates. Initial trees for the heuristic search were obtained automatically by applying the neighbor-joining and BIONJ algorithms to a matrix of pairwise distances estimated using a JTT model and then selecting the topology with the highest log likelihood value. All sites were used in all analysis with 100 times bootstrap analyses [46].
Average base substitutions per site and average amino acid substitutions per site for each gene were conducted using the Kimura 2-parameter model [47] and the JTT matrix-based model [45], respectively.
Supporting Information
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
The amino acid consensus sequences for T. coremiiforme and T. ovoides were created (see Materials and Methods) and labeled “consensus” in parenthesis following the species name. The tree was constructed using the maximum likelihood method based on a JTT matrix-based model [45]. Numerals on each node represent the percentages from 100 replicating bootstrap samplings [46]. There were a total of 24,173 amino acids in the dataset, and the tree with the highest log likelihood (-219231.8349) is shown. B., Bullera; C., Cryptococcus; T., Trichosporon; V., Vanrija.
(TIF)
Phylogenetic tree in Fig 1b is shown with phenotypic properties [2]. B., Bullera; C., Cryptococcus; T., Trichosporon; V., Vanrija.
(TIF)
(a) Nucleotide sequences and (b) amino acid sequences. The trees in (a) and (b) were constructed using the maximum likelihood method based on the Tamura-Nei model [44] for (a) and a JTT matrix-based model [45] for (b). Numerals on each node represent the percentages from 100 replicating bootstrap samplings (frequencies of less than 60% are not shown) [46]. Letters in brackets after bootstrap values indicate the node name (see Table 3). Positions used: (a) 5,934 nucleotides, (b) 1,975 amino acids. Highest log likelihood: (a) -51394.7872, (b) -16428.1661. B., Bullera; C., Cryptococcus; T., Trichosporon; V., Vanrija.
(TIF)
Acknowledgments
The authors thank Prof. Hideaki Sugawara of the National Institute of Genetics in Japan for valuable suggestions and for supporting the study.
Data Availability
All sequence data used in this study are available from the DDBJ/GenBank/EMBL database (AB919588-AB920176).
Funding Statement
This study was supported primarily by a grant from the Institute of Fermentation, Osaka, awarded to MT. RM was supported by a research grant from the Japanese Ministry of Education, Culture, Sports, Science and Technology (MEXT) to the RIKEN Center for Life Science Technologies (the RIKEN Omics Science Center ceased to exist on 1 April 2013 owing to the reorganization of RIKEN) and by a grant of the Innovative Cell Biology by Innovative Technology (the Cell Innovation Program) from MEXT, Japan to Yoshihide Hayashizaki. In silico biology, inc. provided support in the form of salaries for author AO but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of this author is articulated in the ‘author contributions’ section.'
References
- 1. Fell JW, Boekhout T, Fonseca Á, Scorzetti G, Statzell-Tallman A. Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. Int J Syst Evol Microbiol. 2000;50: 1351–1371. [DOI] [PubMed] [Google Scholar]
- 2. Sugita T. Trichosporon Behrend (1890) In: Kurtzman CP, Fell JW, Boekhout T, editors. The Yeasts, A Taxonomic Study, 5th edn Amsterdam: Elsevier; 2011. pp. 2015–2061. [Google Scholar]
- 3. Okoli I, Oyeka CA, Kwon-Chung KJ, Theelen B, Robert V, Groenewald JZ, et al. Cryptotrichosporon anacardii gen. nov., sp. nov., a new trichosporonoid capsulate basidiomycetous yeast from Nigeria that is able to form melanin on niger seed agar. FEMS Yeast Res. 2007;7: 339–350. [DOI] [PubMed] [Google Scholar]
- 4. Boekhout T, Bai FY, Nakase T. Bullera Derx (1930) In: Kurtzman CP, Fell JW, Boekhout T, editors. The Yeasts, A Taxonomic Study, 5th edn Amsterdam: Elsevier: 2011. pp. 1623–1659. [Google Scholar]
- 5. Fonseca Á, Boekhout T, Fell JW. Cryptococcus Vuillemin (1901) In: Kurtzman CP, Fell JW, Boekhout T, editors. The Yeasts, A Taxonomic Study, 5th edn Amsterdam: Elsevier; 2011. pp. 1661–1737. [Google Scholar]
- 6. Weiss M, Bauer R, Sampaio JP, Oberwinkler F. Tremellomycetes and related groups In: McLaughlin DJ, Spatafora JW, editors. The Mycota VII Part A, Systematic and Evolution, 2nd edn Berlin Heiderberg: Springer-Verlag; 2014. pp. 331–355. [Google Scholar]
- 7. Moore RT. Taxonomic proposals for the classification of marine yeasts and other yeast-like fungi including smuts. Botanica Marina 1980;23: 361–373. [Google Scholar]
- 8. Colombo AL, Padovan AC, Chaves GM. Current knowledge of Trichosporon spp. and trichosporonosis. Clin Microbiol Rev. 2011;24: 682–700. 10.1128/CMR.00003-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Molnar O, Schatzmayr G, Fuchs E, Prillinger H. Trichosporon mycotoxinivorans sp. nov., a new yeast species useful in biological detoxification of various mycotoxins. Syst Appl Microbiol. 2004;27: 661–671. [DOI] [PubMed] [Google Scholar]
- 10. Baldrian P, Kolařík M, Stursová M, Kopecký J, Valášková V, Větrovský T, et al. Active and total microbial communities in forest soil are largely different and highly stratified during decomposition. ISME J. 2012;6: 248–258. 10.1038/ismej.2011.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schulze I, Hansen S, Großhans S, Rudszuck T, Ochsenreither K, Syldatk C, et al. Characterization of newly isolated oleaginous yeasts—Cryptococcus podzolicus, Trichosporon porosum and Pichia segobiensis . AMB Express. 2014;4: 24 10.1186/s13568-014-0024-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hassan M, Blanc PJ, Pareilleux A, Goma G. Production of cocoa butter equivalents from prickly-pear juice fermentation by an unsaturated fatty acid auxotroph of Cryptococcus curvatus grown in batch culture. Process Biochem. 1995;30: 629–634. [Google Scholar]
- 13. Tanimura A, Takashima M, Sugita T, Endoh R, Kikukawa M, Yamaguchi S, et al. Selection of oleaginous yeasts with high lipid productivity for practical biodiesel production. Bioresour Technol. 2014;153: 230–235. 10.1016/j.biortech.2013.11.086 [DOI] [PubMed] [Google Scholar]
- 14. Gujjari P, Suh SO, Coumes K, Zhou JJ. Characterization of oleaginous yeasts revealed two novel species: Trichosporon cacaoliposimilis sp. nov. and Trichosporon oleaginosus sp. nov. Mycologia 2011;103: 1110–1118. 10.3852/10-403 [DOI] [PubMed] [Google Scholar]
- 15. Kurtzman CP, Robnett CJ. Relationships among genera of the Saccharomycotina (Ascomycota) from multigene phylogenetic analysis of type species. FEMS Yeast Res. 2013;13: 23–33. 10.1111/1567-1364.12006 [DOI] [PubMed] [Google Scholar]
- 16. Matheny PB, Wang Z, Binder M, Curtis JM, Lim YW, Nilsson RH, et al. Contributions of rpb2 and tef1 to the phylogeny of mushrooms and allies (Basidiomycota, Fungi). Mol Phylogenet Evol. 2007;43: 430–451. [DOI] [PubMed] [Google Scholar]
- 17. Hibbett DS, Binder M, Bischoff JF, Blackwell M, Cannon PF, Eriksson OE, et al. A higher-level phylogenetic classification of the Fungi. Mycol Res. 2007;111: 509–547. [DOI] [PubMed] [Google Scholar]
- 18. Findley K, Rodriguez-Carres M, Metin B, Kroiss J, Fonseca Á, Vilgalys R, et al. Phylogeny and phenotypic characterization of pathogenic Cryptococcus species and closely related saprobic taxa in the Tremellales. Eukaryot Cell 2009;8: 353–361. 10.1128/EC.00373-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guerreiro MA, Springer DJ, Rodrigues JA, Rusche LN, Findley K, Heitman J, et al. Molecular and genetic evidence for a tetrapolar mating system in the basidiomycetous yeast Kwoniella mangrovensis and two novel sibling species. Eukaryot Cell 2013;12: 746–760. 10.1128/EC.00065-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stanke M, Schöffmann O, Morgenstern B, Waack S. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics 2006;7: 62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sugita T, Takashima M, Ikeda R, Nakase T, Shinoda T. Phylogenetic and taxonomic heterogeneity of Cryptococcus humicolus by analysis of the sequences of the internal transcribed spacer regions and 18S rDNA, and the phylogenetic relationships of C. humicolus, C. curvatus, and the genus Trichosporon . Microbiol Immunol. 2000;44: 455–461. [DOI] [PubMed] [Google Scholar]
- 22. Takashima M, Nakase T. Molecular phylogeny of the genus Cryptococcus and related species based on the sequences of SSU rDNA and internal transcribed spacer regions. Microbiol Cult Coll. 1999;15: 35–47. [Google Scholar]
- 23. Takashima M, Sugita T, Shinoda T, Nakase T. Reclassification of the Cryptococcus humicola complex. Int J Syst Evol Microbiol. 2001;51: 2199–2210. [DOI] [PubMed] [Google Scholar]
- 24. Sugita T, Kikuchi K, Makimura K, Urata K, Someya T, Kamei K, et al. Trichosporon species isolated from guano samples obtained from bat-inhabited caves in Japan. Appl Environ Microbiol. 2005;71: 7626–7629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nagy LG, Ohm RA, Kovács GM, Floudas D, Riley R, Gácser A, et al. Latent homology and convergent regulatory evolution underlies the repeated emergence of yeasts. Nat Commun. 2014;5: 4471 10.1038/ncomms5471 [DOI] [PubMed] [Google Scholar]
- 26. Ikeda R, Yokota M, Shinoda T. Serological characterization of Trichosporon cutaneum and related species. Microbiol Immunol. 1996;40: 813–819. [DOI] [PubMed] [Google Scholar]
- 27. Sugita T, Nakase T. Molecular phylogenetic study of the basidiomycetous anamorphic yeast genus Trichosporon and related taxa based on small subunit ribosomal DNA sequences. Mycoscience 1998;39: 7–13. [Google Scholar]
- 28. Ichikawa T, Nishikawa A, Ikeda R, Shinoda T. Structural studies of a cell wall polysaccharide of Trichosporon asahii containing antigen II. Eur J Biochem. 2001;268: 5098–5106. [DOI] [PubMed] [Google Scholar]
- 29. Ichikawa T, Nishikawa A, Wada H, Ikeda R, Shinoda T. Structural studies of the antigen III cell wall polysaccharide of Trichosporon domesticum . Carbohydr Res. 2001;330: 495–503. [DOI] [PubMed] [Google Scholar]
- 30. Daniel HM, Lachance MA, Kurtzman CP. On the reclassification of species assigned to Candida and other anamorphic ascomycetous yeast genera based on phylogenetic circumscription. Antonie van Leeuwenhoek 2014;106: 67–84. 10.1007/s10482-014-0170-z [DOI] [PubMed] [Google Scholar]
- 31. Sampaio JP. Leucosporidium Fell, Statzell I.L. Hunter & Phaff (1969) In: Kurtzman CP, Fell JW, Boekhout T, editors. The Yeasts, A Taxonomic Study, 5th edn Amsterdam: Elsevier; 2011. pp. 1485–1494. [Google Scholar]
- 32. Rokas A, Williams BL, King N, Carroll SB. Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature 2003;425: 798–804. [DOI] [PubMed] [Google Scholar]
- 33. Kuramae EE, Robert V, Echavarri-Erasun C, Boekhout T. Cophenetic correlation analysis as a strategy to select phylogenetically informative proteins: an example from the fungal kingdom. BMC Evol Biol. 2007;7: 134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kuramae EE, Robert V, Snel B, Weiss M, Boekhout T. Phylogenomics reveal a robust fungal tree of life. FEMS Yeast Res. 2006;6: 1213–1220. [DOI] [PubMed] [Google Scholar]
- 35. Fitzpatrick DA, Logue ME, Stajich JE, Butler G. A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis. BMC Evol Biol. 2006;6: 99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang H, Xu Z, Gao L, Hao B. A fungal phylogeny based on 82 complete genomes using the composition vector method. BMC Evol Biol. 2009;9: 195 10.1186/1471-2148-9-195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Koufopanou V, Swire J, Lomas S, Burt A. Primers for fourteen protein-coding genes and the deep phylogeny of the true yeasts. FEMS Yeast Res. 2013;13: 574–584. 10.1111/1567-1364.12059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shertz CA, Bastidas RJ, Li W, Heitman J, Cardenas ME. Conservation, duplication, and loss of the Tor signaling pathway in the fungal kingdom. BMC Genomics 2010;11: 510 10.1186/1471-2164-11-510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Millanes AM, Diederich P, Ekman S, Wedin M. Phylogeny and character evolution in the jelly fungi (Tremellomycetes, Basidiomycota, Fungi). Mol Phylogenet Evol. 2011;61: 12–28. 10.1016/j.ympev.2011.05.014 [DOI] [PubMed] [Google Scholar]
- 40. Takashima M, Nakase T. Four new species of the genus Sporobolomyces isolated from leaves in Thailand. Mycoscience 2000;41: 357–369. [Google Scholar]
- 41. Kawaji H, Lizio M, Itoh M, Kanamori-Katayama M, Kaiho A, Nishiyori-Sueki H, et al. Comparison of CAGE and RNA-seq transcriptome profiling using a clonally amplified and single molecule next generation sequencing. Genome Res. 2014;24: 708–717. 10.1101/gr.156232.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gouy M, Guindon S, Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27: 221–224. 10.1093/molbev/msp259 [DOI] [PubMed] [Google Scholar]
- 43. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10: 512–526. [DOI] [PubMed] [Google Scholar]
- 45. Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8: 275–282. [DOI] [PubMed] [Google Scholar]
- 46. Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 1985;39: 783–791. [DOI] [PubMed] [Google Scholar]
- 47. Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16: 111–120. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
The amino acid consensus sequences for T. coremiiforme and T. ovoides were created (see Materials and Methods) and labeled “consensus” in parenthesis following the species name. The tree was constructed using the maximum likelihood method based on a JTT matrix-based model [45]. Numerals on each node represent the percentages from 100 replicating bootstrap samplings [46]. There were a total of 24,173 amino acids in the dataset, and the tree with the highest log likelihood (-219231.8349) is shown. B., Bullera; C., Cryptococcus; T., Trichosporon; V., Vanrija.
(TIF)
Phylogenetic tree in Fig 1b is shown with phenotypic properties [2]. B., Bullera; C., Cryptococcus; T., Trichosporon; V., Vanrija.
(TIF)
(a) Nucleotide sequences and (b) amino acid sequences. The trees in (a) and (b) were constructed using the maximum likelihood method based on the Tamura-Nei model [44] for (a) and a JTT matrix-based model [45] for (b). Numerals on each node represent the percentages from 100 replicating bootstrap samplings (frequencies of less than 60% are not shown) [46]. Letters in brackets after bootstrap values indicate the node name (see Table 3). Positions used: (a) 5,934 nucleotides, (b) 1,975 amino acids. Highest log likelihood: (a) -51394.7872, (b) -16428.1661. B., Bullera; C., Cryptococcus; T., Trichosporon; V., Vanrija.
(TIF)
Data Availability Statement
All sequence data used in this study are available from the DDBJ/GenBank/EMBL database (AB919588-AB920176).