Abstract
Rice blast fungus, Magnaporthe oryzae, is the most destructive pathogen in the rice-growing area. This fungus has a biotrophic phase early in infection and later switches to a necrotrophic lifestyle. During the biotrophic phase, the fungus competes with its host for nutrients and oxygen. Continuous uptake of oxygen is essential for successful establishment of blast disease of this pathogen. Here, we report transcriptional responses of the fungus to oxygen limitation. Transcriptome analysis using RNA-Seq identified that 1,047 genes were up-regulated in response to hypoxia. Those genes are involved in mycelial development, sterol biosynthesis, and metal ion transport based on hierarchical GO terms, and are well-conserved among three fungal species. In addition, null mutants of two hypoxia-responsive genes were generated and their roles in fungal development and pathogenicity tested. The mutant for the sterol regulatory element-binding protein gene, MoSRE1, exhibited increased sensitivity to a hypoxia-mimicking agent, increased conidiation, and delayed invasive growth within host cells, which is suggestive of important roles in fungal development. However, such defects did not cause any significant decrease in disease severity. The other null mutant, for the alcohol dehydrogenase gene MoADH1, showed no defect in the hypoxia-mimicking condition (using cobalt chloride) and fungal development. Taken together, this comprehensive transcriptional profiling in response to a hypoxic condition with experimental validations would provide new insights into fungal development and pathogenicity in plant pathogenic fungi.
Introduction
Oxygen (O2) is an essential element for most eukaryotes due to its roles in the maintenance of many physiological processes such as the biosynthesis of heme, sterols, and fatty acids, and also as a terminal electron acceptor in oxidative phosphorylation. Oxygen concentration ranges from 0% to 20.9% in the environment. Habitats lacking oxygen are known as anoxic, while those at the higher end of the oxygen range are known as aerobic [1]. Microbes living in those niches can be exposed to diverse oxygenic environments including hypoxia (a reduced oxygen level associated with physiological and pathophysiological processes) [2], and have adapted to microbial growth in their hosts. The exposure to oxygen depletion; i.e., hypoxia, dramatically influences various physiological processes [3–5] and global transcriptional regulation in eukaryotic cells [6, 7]. Thus, microbes have evolved a set of cellular and molecular mechanisms to survive in low-oxygen environments.
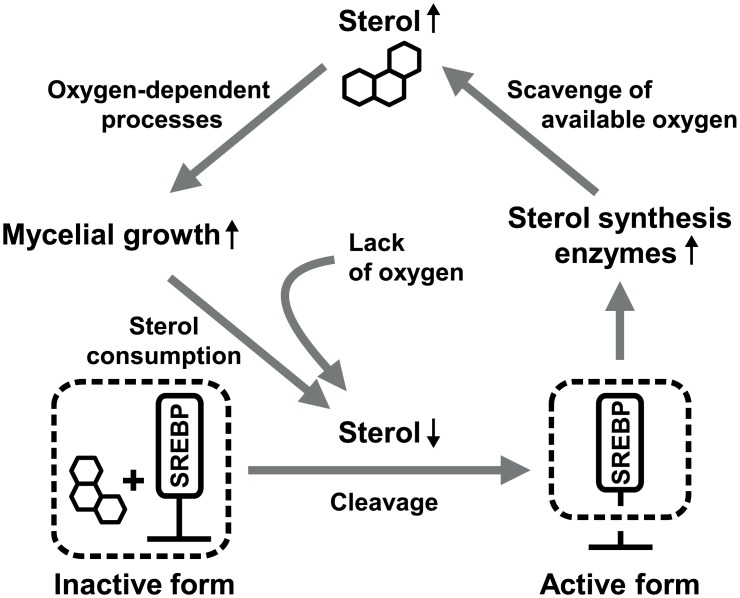
In healthy humans, oxygen levels vary depending on the location in the body, ranging from 2.5% in the kidneys to 9% in the lungs [1, 8]. Human fungal pathogens, therefore, encounter changes in oxygen concentration and also can cause oxygen depletion (hypoxia) when they invade host tissue. For example, Aspergillus fumigatus infects the lungs of mice where it is exposed to the hypoxic microenvironment, and adapts its physiological processes to overcome this. One of the adaptation processes is to change energy metabolism to a mechanism such as ethanol fermentation, resulting in accumulation of ethanol [9]. A null mutant for the alcohol dehydrogenase gene caused increased host inflammatory responses while still causing aspergillosis to the same degree as the wild-type [9]. Another example of fungal response to hypoxia is the sterol regulatory element-binding protein (SREBP) pathway involved in regulation of sterol biosynthesis [10]. As shown in Fig 1, membrane-bound SREBP is activated when sterols are depleted due to hypoxia. Active SREBP turns on the expression of sterol synthesis enzymes and other oxygen-dependent proteins. SREBP is essential for virulence and growth under hypoxic conditions in Cryptococcus neoformans and A. fumigatus [11–13], suggesting that adaptation to hypoxia is important for fungal virulence in mammals. However, little is known about hypoxia responses of plant pathogenic fungi and the importance of such responses in disease establishment within plants.
Fig 1. Schematic diagram of SREBP regulation for adaptation to hypoxia.
Sterols control activation of SREBP: it is inactive in the presence of and active in the absence of sterols. Growth in limited oxygen conditions inevitably resulted in a lack of sterols, activating SREBP. Increases in enzymes for sterol biosynthesis and other oxygen-dependent pathways produce more sterols, which can be used for mycelial growth.
In plant tissues, active metabolic reactions result in low internal oxygen concentrations even under normal oxygen (normoxia) conditions [14]. For example, a decrease of 1–12% in internal oxygen levels has been reported in petioles, root meristems, phloems, and seeds of various plants [5, 15–17]. However, less has been reported about the internal oxygen concentration in leaves, where oxygen is produced. When infected with biotrophic plant pathogens (obligate parasites), hosts and pathogens must compete for oxygen as well as nutrients. Such competition might lead to decreased availability of oxygen. In particular, in darkness, plants use oxygen through respiration without photosynthesis or production of oxygen, and the internal oxygen level in the leaves decreases. Thus, plant-invading biotrophic agents who encounter oxygen-depleted environments are able to adapt to hypoxic conditions for successful infection.
Rice blast caused by Magnaporthe oryzae is a socioeconomically important disease that results in enormous yield loss [18]. The Oryza sativa-M. oryzae pathosystem has been studied as a model system of plant-fungal pathogen interactions. This fungus establishes a biotrophic interaction within the host early in the infection process and then switches to a necrotrophic lifestyle [19]. Dramatic developmental changes occur in the fungus during establishment of infection in the host. Firstly, a dome-shaped appressorium is developed at the tip of a germ tube. The appressorium matures as glycerol accumulates within it, generating high osmotic pressure. [20]. A penetration peg from the mature appressorium pierces the plant cell wall with mechanical force and forms a thin primary hypha down into the plant cell [21]. Secondly, the primary hypha differentiates into thick bulbous infectious hyphae that fill the first-invaded cell, and then moves to neighboring cells. Later in infection, the fungus switches lifestyle to a necrotrophic phase in which it kills neighboring host cells and gains nutrients from the dead cells.
We hypothesized that plant pathogens are exposed to hypoxic microenvironments during a biotrophic phase in host cells and their ability to overcome this challenge is essential for the successful establishment of disease. High-throughput RNA sequencing (RNA-Seq) was employed to analyze transcriptional responses of the fungus to hypoxic environments and to compare our results to those of other fungal pathogens and to in planta transcriptomes. Additionally, null mutants for the specific hypoxia-responsive genes were generated and tested for their development and pathogenicity on rice.
Materials and Methods
Sampling under hypoxia culture conditions
Magnaporthe oryzae strain KJ201 was incubated in liquid complete medium (LCM; 0.6% yeast extract, 1% sucrose, and 0.6% tryptone) at room temperature for three days. Homogenized mycelia were filtered through two layers of cheesecloth. Mycelia were harvested using Miracloth (Calbiochem, CA, USA) after three washes with 0.9% NaCl. The harvested mycelia were resuspended in 20 ml of 0.9% NaCl and 500 μl of the suspension were inoculated on V8 juice agar plates (8% V8 juice and 1.5% agar, pH 6.7) layered with a 0.45-μm pore cellulose nitrate membrane filter (Whatman, Maidstone, England). All 20 plates were pre-incubated for three hours at 25°C. Ten of these plates were then incubated in the hypoxia chamber (Coy Lab Products, MI, USA), at 99% N2 and 1% O2. The other 10 were incubated outside of the chamber (normoxia control). After 12 hours of incubation, mycelia were harvested from the plates. The harvested fungal tissues were frozen immediately in liquid nitrogen and stored at -80°C for RNA isolation.
RNA isolation and RNA-Seq
Total RNAs were extracted using TRIzol reagent (Life Technologies, MD, USA). The integrity of RNA was estimated using Bioanalyzer and Pico RNA chip, following the eukaryotic total RNA assay protocol [22]. The TruSeq RNA sample preparation kit (Illumina, CA, USA) was used, following the manufacturer’s instructions. Briefly, poly A tailed transcript RNA was isolated by oligo(dT) selection using streptavidin-coated magnetic beads and fragmented randomly by Mg2+ ion treatment. The fragmented mRNA was subjected to cDNA synthesis using Superscript II reverse transcriptase and random hexamers (Invitrogen, CA, USA). RNAs with a length of 300 bp to 400 bp were selected and sequenced on the HiSeq 2000 system (Illumina, CA, USA).
Read filtering, mapping, and expression quantification
Paired-end reads were filtered as follows: 1) Adaptor sequences were trimmed from the reads using the NGS QC toolkit (ver. 2.3) [23]. 2) Reads with less than 70% of ‘Q > 20’ bases were removed. 3) Only paired reads were selected for further analyses. TopHat (ver. 2.0.8b) [24] was used to map the filtered reads against the reference genome (M. oryzae strain 70–15; http://www.broadinstitute.org). The minimum and maximum intron lengths were adjusted as five and 2,120 bp based on the exon information of M. oryzae. The other parameters were set as default values. The Cufflinks (ver. 2.1.1) tool calculated FPKM (fragments per kilobase of transcript per million mapped reads) values using only uniquely mapped reads. A false discovery rate (FDR, P < 0.05) was applied for correction. Fold changes were calculated using a modified function, ([FPKMHYPOXIA +1] / [FPKMNORMOXIA +1]).
Gene ontology
Weight algorithm with Fisher’s exact tests implemented in the R package ‘topGO’ version 2.12.0 was employed to calculate GO term significance for an enrichment analysis of gene sets and visualization of a GO hierarchical structure [25]. GO terms for genes of M. oryzae were retrieved from Comparative Fungal Genomics Platform (http://cfgp.snu.ac.kr) [26].
Fungal transformation
Transformation was performed as described previously [27]. Briefly, the MoADH1 and MoSRE1 genes were replaced with the constructs having a hygromycin B phosphotranferase gene cassette (HPH) with flanking regions of two genes [28]. The primers used for the PCR amplification are listed in S1 Table. After PEG-mediated transformation, mutant candidates were chosen on the selective media (20% sucrose, 1% glucose, 0.3% yeast extract, 0.3% casamino acids, 0.8% agar and 200 ppm of hygromycin). The candidates for MoADH1 and MoSRE1 mutants were confirmed by PCR and Southern blot analysis. For genetic complementation of the targeted gene deletion mutant, the fragment containing native promoter region and the full-length ORF of the MoSRE1 gene was amplified by PCR and used to transform the ΔMosre1 mutant with a geneticin resistance gene cassette originated from pII99 [29]. All strains used in this study have been deposited in the Center for Fungal Genetic Resources (http://genebank.snu.ac.kr).
Southern hybridization assay
Genomic DNA for Southern blotting was prepared by the ‘quick & safe’ method [30] or standard protocols [31]. Restriction enzyme digestion, gel electrophoresis, and Southern hybridization were performed by standard procedures [32]. DNA probes for hybridization were labeled with 32P by using the Rediprime II Random Prime Labeling System kit following the manufacturer’s protocol (Amersham Pharmacia Biotech, NJ, USA).
Assays for fungal development
Growth rate was estimated by measuring diameter of mycelia on the modified complete medium (CM; 0.2% peptone, 1% glucose, 1% casamino acid, 0.1% yeast extract, 0.15% KH2PO4, 0.05% KCl, 0.6% NaNO3, 0.05% MgSO4, 0.1% trace element (v/v) and 0.1% vitamin supplement (v/v), pH 6.5) and minimal medium (0.15% KH2PO4, 0.05% KCl, 0.6% NaNO3, 1% glucose, 0.05% MgSO4, 0.1% trace element and 0.1% vitamin supplement, pH 6.5) as previously described [33]. The number of asexual spores (conidia) was counted with a hemocytometer under a microscope. Observation of conidiophore development was performed on oatmeal agar media (5% oatmeal (w/v) and 2.5% agar (w/v)) at 16 hours after incubation under a microscope [34]. Conidia were harvested with sterilized distilled water from V8 juice agar media at seven days after incubation and filtered with double-layered Miracloth (CalBiochem, CA, USA). To measure frequencies of germination and appressorium formation, conidial suspensions adjusted to 2 × 104 conidia/ml were spotted onto hydrophobic coverslips with three replicates and incubated in a humid container. Germination of conidia was estimated by the observation of ~100 conidia at four hours after incubation. Appressorium formation rate was determined by the number of formed appressoria to the total number of germinated spores at 16 hours after incubation.
Pathogenicity and sheath infiltration assays
To evaluate pathogenicity on rice, 10 ml of conidial suspension (105 conidia/ml) were prepared with sterilized distilled water supplemented with Tween 20 (250 ppm final concentration) and sprayed onto rice seedlings (Oryza sativa cv. Nakdongbyeo) of three- to four-leaf stage. Rice seedlings were stored in a dew chamber for 24 hours in the dark and then moved into a growth chamber at 25°C, 80% humidity with a photoperiod of 16 hours of light [35]. To monitor lesion development, the pathogenicity assay was also examined by using drop inoculation of conidial suspensions. The 30 μl of conidia suspension adjusted to 105 conidia/ml were dropped onto detached rice leaves and incubated in humid box at room temperature. Rice leaf sheaths were used to observe invasive growth in the rice cells. Conidial suspensions adjusted to 2 × 104 conidia/ml were syringed into the excised sheath cells and incubated in a humid chamber at 25°C [36]. The layers of epidermal cells were observed under the microscope after clipping out chlorophyll-enriched area at 48 hours after inoculation.
Extracellular oxidative stress sensitivity test
To determine the sensitivity to extracellular oxidative stress, mycelial growth rates were measured on solid CM supplemented H2O2 (2.5 and 5 mM), 3 mM methyl viologen (paraquat, Aldrich, 856177) and 200 ppm Congo Red (Sigma, C6277). This experiment was performed with three biological replicates.
Real-time quantitative reverse transcription PCR (qRT-PCR)
cDNA was prepared using ImProm-II Reverse Transcription System (Promega, Madison, WI, USA) according to manufacturer’s recommendation. qRT-PCR reactions were carried out in the mixture of cDNA template (25 ng/ 2 μl), reverse and forward primers(100 nM each in total 3 μl) and Power SYBR Green PCR Master Mix (5 μl, Applied Biosystems, Foster City, CA, USA). The AB7500 Real-Time PCR system was used for amplification and detection (Applied Biosystems, Foster City, CA, USA). PCR condition consists of 40 cycles of 15 s at 95°C, 30 s at 60°C, and 30 s at 72°C after initial denaturation.
Results and Discussion
Mapping and quantifying fungal transcriptomes under hypoxia
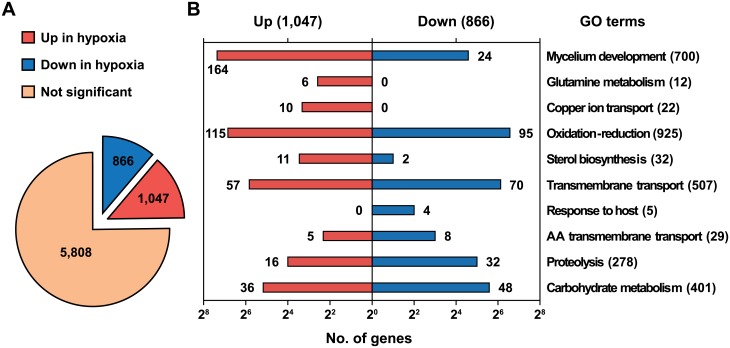
To examine transcriptomic changes of the rice blast fungus in response to hypoxia, we incubated each of 10 plates separately under hypoxia (1% O2) and normoxia (20.9% O2) for 12 hours after pre-culture on complete medium (CM) for three days. Those 10 plates were used as biological replicates for each conditions. Total RNAs sampled from both conditions were pooled (one lane each) and sequenced using next-generation sequencing technology with three technical repeats (multiplexing with fluorescence dyes; e.g. Hypoxia_0, Hypoxia_1 and Hypoxia_2 in S2 Table). In total, 139 and 167 million reads were generated from hypoxia and normoxia samples, respectively (S2 Table). The original sequencing datasets have been deposited to a public data repository (http://www.ncbi.nlm.nih.gov/geo, accession number: GSE51597). We selected 113 and 140 million reads from hypoxia and normoxia, respectively, as valuable sequences after in-house filtering processes (S2 Table). The TopHat software [24] was employed for large-scale mapping of the filtered reads. Seventy six percent of the filtered reads were mapped onto the genome of M. oryzae (http://www.broadinstitute.org) with 87% coverage of all transcripts (11,293 out of 12,991, S3 Table). Expression levels of the mapped transcripts were estimated using FPKM (fragments per kilobase of exon per million fragments mapped), which was calculated by the Cufflink software [37]. After applying FDR corrections (P < 0.05), we selected a total of 7,720 transcripts (Fig 2A and S3 Table), of which 1,047 transcripts were significantly up-regulated and 866 transcripts were down-regulated in response to hypoxia (two-fold, Fig 2A and S3 Table).
Fig 2. Transcriptional response of M. oryzae to hypoxic conditions.
(A) Expression of annotated transcripts in hypoxia. A total of 7,720 transcripts were selected by the FDR correction (P < 0.05). A twofold threshold was applied for significantly regulated genes. (B) Enrichment of specific gene ontology terms under hypoxia. Numbers represent the counts of transcripts supporting the GO terms.
Genome-wide transcriptomic analysis for hypoxia-responsive genes
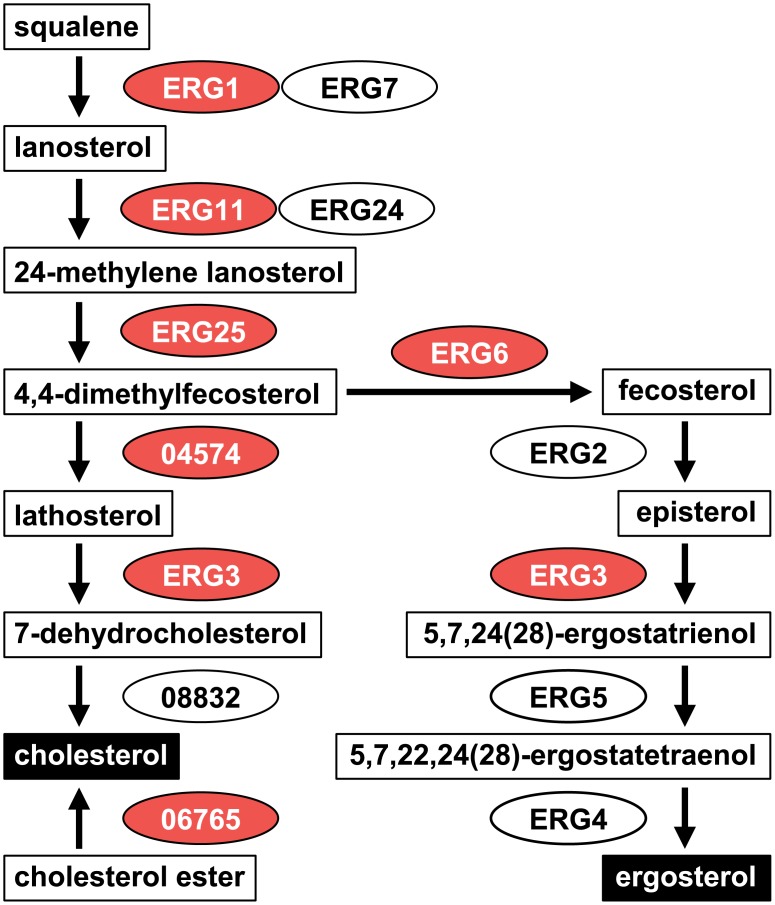
Gene Ontology (GO) enrichment analysis was conducted to understand functions of genes influenced by hypoxia. The R package ‘topGO’ was used to identify GO terms enriched in the differentially expressed gene sets, giving 16 and 12 enriched GO terms in the up- and down-regulated gene sets, respectively (P < 0.05, Fisher's exact test, Table 1 and Fig 2B). The up-regulated genes contained terms involved in mycelial growth: mycelium development (GO:0043581), energy metabolism (GO:0006123 and GO:0015671), and biosynthesis of amino acids (GO:0006541, GO:0009082, GO:0009084, GO:0009450, and GO:0042026), in addition to lipid (GO:0016126 and GO:0072330), phosphate (GO:0070409), and NAD (GO:0034354). As summarized in Fig 1, ‘mycelium development,’ ‘sterol biosynthesis’, and ‘oxygen transport’ terms are closely related in the sterol biosynthesis process. We also examined the steroid biosynthesis pathway in the KEGG database (http://www.genome.jp/kegg-bin/show_pathway?mgr00100). Most genes in this pathway were highly induced under the hypoxic condition and identified as having the ‘sterol biosynthesis’ or ‘oxidation-reduction process’ terms (Fig 3). In the term ‘oxidation-reduction process’, oxygen-consuming enzymes like catalase (MGG_06442) and laccases (MGG_08523 and MGG_11608) were up-regulated, suggesting that lack of oxygen seems to activate oxygen-dependent processes to maintain normal levels of growth in the oxygen-limited environment. An increase in transcripts of catalase and laccase indicates that reactive oxygen species (ROS) might have accumulated in the cell under hypoxia. In addition, genes in the ‘copper ion transport’ term (MGG_02774, MGG_02283, MGG_00930, etc.) are involved in responses to ROS as well as copper homeostasis, supporting ROS accumulation.
Table 1. Significant GO terms in response to hypoxia.
| GO IDs | Terms | GO term counts | |||
|---|---|---|---|---|---|
| Annotated | Observed | Expected | Significance | ||
| <Up-regulated in hypoxia > | |||||
| GO:0043581 | mycelium development | 700 | 164 | 68.9 | 0.000 |
| GO:0006541 | glutamine metabolic process | 12 | 6 | 1.2 | 0.000 |
| GO:0006825 | copper ion transport | 14 | 6 | 1.4 | 0.001 |
| GO:0055114 | oxidation-reduction process | 925 | 115 | 91.1 | 0.003 |
| GO:0016126 | sterol biosynthetic process | 14 | 5 | 1.4 | 0.009 |
| GO:0007623 | circadian rhythm | 2 | 2 | 0.2 | 0.010 |
| GO:0009450 | gamma-aminobutyric acid catabolic process | 2 | 2 | 0.2 | 0.010 |
| GO:0015671 | oxygen transport | 2 | 2 | 0.2 | 0.010 |
| GO:0052372 | modulation by symbiont of entry into host | 2 | 2 | 0.2 | 0.010 |
| GO:0070409 | carbamoyl phosphate biosynthetic process | 2 | 2 | 0.2 | 0.010 |
| GO:0009082 | branched chain family amino acid biosynthetic process | 15 | 5 | 1.5 | 0.012 |
| GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen | 3 | 2 | 0.3 | 0.027 |
| GO:0034354 | de novo NAD biosynthetic process from tryptophan | 3 | 2 | 0.3 | 0.027 |
| GO:0042026 | protein refolding | 3 | 2 | 0.3 | 0.027 |
| GO:0072330 | monocarboxylic acid biosynthetic process | 27 | 6 | 2.7 | 0.044 |
| GO:0009084 | glutamine family amino acid biosynthetic process | 21 | 5 | 2.1 | 0.049 |
| <Down-regulated in hypoxia > | |||||
| GO:0055085 | transmembrane transport | 507 | 70 | 38.7 | 0.000 |
| GO:0075136 | response to host | 5 | 4 | 0.4 | 0.000 |
| GO:0055114 | oxidation-reduction process | 925 | 95 | 70.6 | 0.001 |
| GO:0003333 | amino acid transmembrane transport | 29 | 8 | 2.2 | 0.001 |
| GO:0005975 | carbohydrate metabolic process | 401 | 48 | 30.6 | 0.001 |
| GO:0015847 | putrescine transport | 2 | 2 | 0.2 | 0.006 |
| GO:0015848 | spermidine transport | 2 | 2 | 0.2 | 0.006 |
| GO:0008643 | carbohydrate transport | 39 | 8 | 3.0 | 0.008 |
| GO:0015851 | nucleobase transport | 7 | 3 | 0.5 | 0.012 |
| GO:0006508 | proteolysis | 278 | 32 | 21.2 | 0.012 |
| GO:0043419 | urea catabolic process | 3 | 2 | 0.2 | 0.017 |
| GO:0006071 | glycerol metabolic process | 8 | 3 | 0.6 | 0.019 |
Fig 3. The steroid biosynthesis pathway in M. oryzae.
Intermediates are boxed in white and final products are boxed in black. Genes involved in the pathway are circled. Numbers in the gene names are gene IDs that start with ‘MGG.’ Up-regulated genes are colored in red and down-regulated genes in blue. ‘No significant regulation’ is colored in white.
In the down-regulated genes, the ‘transport’-related terms such as ‘transmembrane transport’, ‘transport of amino acid, carbohydrates, nucleobase, spermidine, and putrescine’, were enriched, indicating that intra- and extracellular transport was inactive or delayed due to low oxygen (Table 1). The term ‘oxidation-reduction process’ was enriched in the down-regulated genes as well as up-regulated genes, suggesting that the genes belonging to the term might play pivotal roles in adaptation to different oxygen concentrations (Fig 2B). In this term, alcohol oxidases (MGG_ 09072 and MGG_02127) and alcohol dehydrogenases (MGG_04556 and MGG_05519) were down-regulated. Those enzymes participate in the conversion of alcohol to aldehyde using oxygen. Thus, oxygen availability in this fungus would be improved by decreasing expression of those genes. The ‘response to host’ term consists of five genes and four of them (MGG_02863, MGG_05871, MGG_12476, and MGG_10315 (MPG1)) were found in the down-regulated transcripts under the hypoxic condition. MPG1 is known as a hydrophobin that functions as a surface sensor when infection-related morphological development begins [38]. Therefore, appressorium formation could be suppressed with the reduced expression of MPG1. Given that the genes belonging to the ‘mycelium development’ term were induced under hypoxia, decrease in the MPG1 transcripts suggests that fungal cells under hypoxia might abandon energy-consuming morphological development and expand the mycelium by simple radial growth to save energy and increase the chance of oxygen uptake. Taken together, the evidence indicated that the rice blast fungus responds to oxygen deprivation mainly by increasing oxygen availability and by shutting down energy-consuming processes such as appressorium formation.
Comparative analysis of hypoxia-responsive pathways in fungi
We investigated the literature on the genome-wide transcriptional analyses of hypoxia-responsive genes in other fungi. Several microarray datasets whereby wild-type strains were exposed to hypoxia had been generated for Candida albicans [39], Cryptococcus neoformans [11, 12], Aspergillus nidulans [40], A. fumigatus [41], and Blastocladiella emersonii [42]. Conditions used for hypoxia were varied in the studies, including oxygen concentrations (0–1%), composition of other gases (N2 and CO2), incubation times (1–6 h), and the culture media. However, genes encoding ergosterol biosynthesis, glycolysis, fermentation, fatty acid metabolism, stress response, and hyphal growth were commonly identified in all experiments with different fungi, showing consistency with our GO enrichment results. In addition, we carried out genome-wide comparison of our data with those in C. albicans [39] and C. neoformans [11], where 111 and 178 genes were significantly up-regulated under hypoxic conditions, respectively. We identified that 95 and 137 homologous genes of C. albicans and C. neoformans exist in the M. oryzae genome, respectively (BLASTP, e-value cutoff < e-10). Among them, 42 (45%) and 37 (25%) were included in the up-regulated genes of M. oryzae under hypoxia, respectively (S4 Table). Eight genes were commonly induced in all three fungi; these encoded C4-methyl sterol oxidase (ERG25), C5 sterol desaturase (ERG3), fatty acid hydroxylase (SCS7), delta-9 fatty acid desaturase, alcohol dehydrogenase (ADH1), mitochondrial hypoxia responsive domain-containing protein, carboxypeptidase Y, and heat shock protein 70. Considering the total gene numbers of the three fungi, the probability that this happened by chance is extremely low (P = 1.4 × e-11). Interestingly, the three genes, ERG25, ERG3, and SCS7, had also been identified as SREBP-dependent proteins in Schizosaccharomyces pombe [10]. In addition to SREBP (MGG_11534), SREBP cleavage activating protein (SCAP, MGG_08135) and Insig (MGG_06428) are known as regulators for SREBP function and they were highly up-regulated by 5.2- and 3.9-fold in our expression data, respectively. Thus, SREBP-mediated transcriptional responses to hypoxia are well-conserved among these fungi.
We further compared our data to in planta transcriptomes which had been analyzed with biotrophic phases of M. oryzae [43, 44]. The hypoxia-induced genes shared 25 genes (11.7%) with the in planta transcriptome (samples harvested at 24 hpi, 213 up-regulated genes in total) and 5 genes (11.1%) in the in planta transcriptome (at 36 hpi, 45 genes), respectively (Table 2). This comparative analysis revealed that about ten percent of the in planta expressed genes overlapped with the hypoxia-induced genes. And they included genes encoding oxidoreductase, minor extracellular protease, isotrichodermin C-15 hydroxylase which is one of cytochrome P450 proteins (CYPs), laccase, alpha-amylase, amino acid transport system protein, and integral membrane protein. This result suggests that the hypoxia would be one of the environments that the rice blast fungus encountered during infection.
Table 2. The in planta expressed genes in the hypoxia-induced transcriptome.
| Genes | Protein names | Fold change in hypoxia | References |
|---|---|---|---|
| MGG_03374 | beta-1,6-galactanase | 4.1 | [43] |
| MGG_03671 | hypothetical protein | 2.1 | [43] |
| MGG_04015 | mannan endo-1,6-alpha-mannosidase DCW1 | 3.3 | [43] |
| MGG_07709 | hypothetical protein | 2.5 | [43] |
| MGG_10710 | oxidoreductase | 11.8 | [43] |
| MGG_00050 | hypothetical protein | 2.3 | [44] |
| MGG_00099 | hypothetical protein | 3.7 | [44] |
| MGG_00703 | MAS3 protein | 2.6 | [44] |
| MGG_00715 | glucose-repressible protein | 4.4 | [44] |
| MGG_01094 | hypothetical protein | 3.2 | [44] |
| MGG_02329 | isotrichodermin C-15 hydroxylase | 2.0 | [44] |
| MGG_03988 | hypothetical protein | 2.5 | [44] |
| MGG_04378 | integral membrane protein | 5.9 | [44] |
| MGG_04994 | plasma membrane H+-ATPase | 2.7 | [44] |
| MGG_05164 | hypothetical protein | 2.6 | [44] |
| MGG_05638 | hypothetical protein | 8.3 | [44] |
| MGG_06542 | N amino acid transport system protein | 8.3 | [44] |
| MGG_06578 | hypothetical protein | 3.1 | [44] |
| MGG_07200 | plasma membrane ATPase | 3.7 | [44] |
| MGG_07341 | hypothetical protein | 2.4 | [44] |
| MGG_08158 | hypothetical protein | 2.6 | [44] |
| MGG_08523 | laccase-1 | 31.8 | [44] |
| MGG_08535 | hypothetical protein | 11.9 | [44] |
| MGG_08811 | hypothetical protein | 2.3 | [44] |
| MGG_09321 | hypothetical protein | 6.0 | [44] |
| MGG_09640 | alpha-amylase 1 | 3.6 | [44] |
| MGG_09763 | hypothetical protein | 2.5 | [44] |
| MGG_12247 | hypothetical protein | 7.2 | [44] |
| MGG_12655 | hypothetical protein | 6.1 | [44] |
| MGG_13063 | hypothetical protein | 3.8 | [44] |
| Sum | 30 |
Growth of null mutants for hypoxia-responsive genes
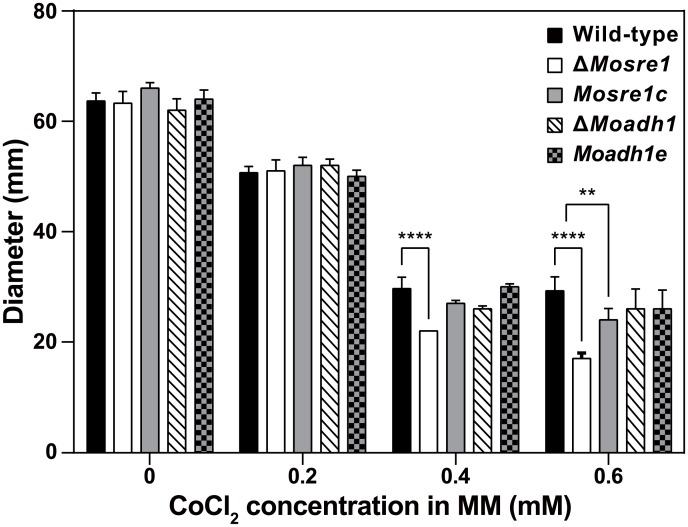
We chose two hypoxia-responsive genes—SREBP (MoSRE1, MGG_11534) and alcohol dehydrogenase (MoADH1, MGG_03880) for further characterization. They were up-regulated in our RNA-seq data (2.6- and 4.1-fold, respectively); up-regulation was confirmed by qRT-PCR. For those genes, deletion mutants were generated by gene replacement via homologous recombination. Correct integration of deletion constructs into the M. oryzae genome was demonstrated by PCR and Southern hybridization (S1 Fig). We investigated whether those genes are required for overcoming conditions of limited oxygen. As CoCl2 (cobalt(II) chloride) has been reported as a chemical hypoxia-mimicking agent in eukaryotic cells [45], the wild-type, mutants and their complementary strains were grown on minimal media (MM) with or without CoCl2 (Sigma-Aldrich, 255599). Neither the wild-type nor any of the mutant strains showed any significant differences in mycelial growth on MM (Fig 4). When grown on MM containing CoCl2 (0.2 mM to 0.6 mM final concentration), growth of the ΔMosre1 mutant was barely detectable at 0.4 mM and 0.6 mM of CoCl2, compared to the wild-type and ΔMoadh1 (Fig 4). That is, growth of the ΔMosre1 mutant was more sensitive to hypoxia-mimicking conditions than the others, although there were no differences in radial growth between this mutant and the wild-type when grown on 0.2 mM CoCl2-supplemented MM. Interestingly, the M. oryzae wild-type strain was more sensitive to CoCl2 than the other fungi. In C. neoformans [12] and A. fumigatus [46], the mutants in which MoSRE1-homologous genes had been deleted could not grow on the medium containing 0.6 mM CoCl2, while the wild-type strains of both species exhibited normal growth on this medium. On the other hand, the ΔMosre1 mutant exhibited a significant growth delay on complete medium (CM) without CoCl2 (Table 3). When treated with 0.6 mM, growth of the mutant was unaffected by the addition of CoCl2, while the wild-type and complementary strains showed a similar growth defect. Thus, deletion of MoSRE1 in the wild-type results in a similar effect on fungal growth, as the addition of 0.6 mM CoCl2 disturbs normal mycelial growth (mimicry of hypoxia). This observation is also consistent with the suggested role of CoCl2 in directly disturbing several oxygen-requiring steps in ergosterol biosynthesis [45]. Taken together, these data suggest that the SREBP-mediated pathway plays a key role in ergosterol biosynthesis, which correlates with hyphal growth in both plant and human pathogens.
Fig 4. Mycelial growth of M. oryzae on CoCl2-containing medium.
Wild-type and two deletion mutants were inoculated on MM with or without CoCl2. Plates were incubated at room temperature for 9 days. MM: minimal medium. Two-way ANOVA was performed with Dunnett’s multiple comparison.
Table 3. Developmental phenotypes of the transformants for MoADH1 and MoSRE1 genes in M. oryzae.
| Strain | Mycelial growth (mm) | Conidiation (x104/ml) | Germination (%)* | Appressorium formation (%)** | |
|---|---|---|---|---|---|
| CM | CM+CoCl2 | ||||
| Wild-type | 62.0±1.0A *** | 39.7±0.6A | 33.6±4.7A | 95.7±2.3A | 93.0±1.7A |
| ΔMoadh1 | 59.3±2.1A | 42.0±2.6A | 33.4±4.6A | 95.0±2.6A | 94.7±2.1A |
| Moadh1e | 61.0±2.0A | 40.7±3.8A | 32.1±2.7A | 94.3±1.5A | 94.0±1.0A |
| ΔMosre1 | 43.3±4.7B | 40.7±2.5A | 81.1±6.7B | 94.0±2.6A | 94.7±2.9A |
| Mosre1c | 61.3±0.6A | 39.7±0.6A | 49.9±9.5C | 94.7±1.2A | 94.0±0.0A |
*Germination rate was observed four hours after incubation.
**Appressorium formation rate was observed 16 hours after incubation.
***Two-way ANOVA was performed with Dunnett’s multiple comparison.
Roles of MoSRE1 and MoADH1 in fungal development
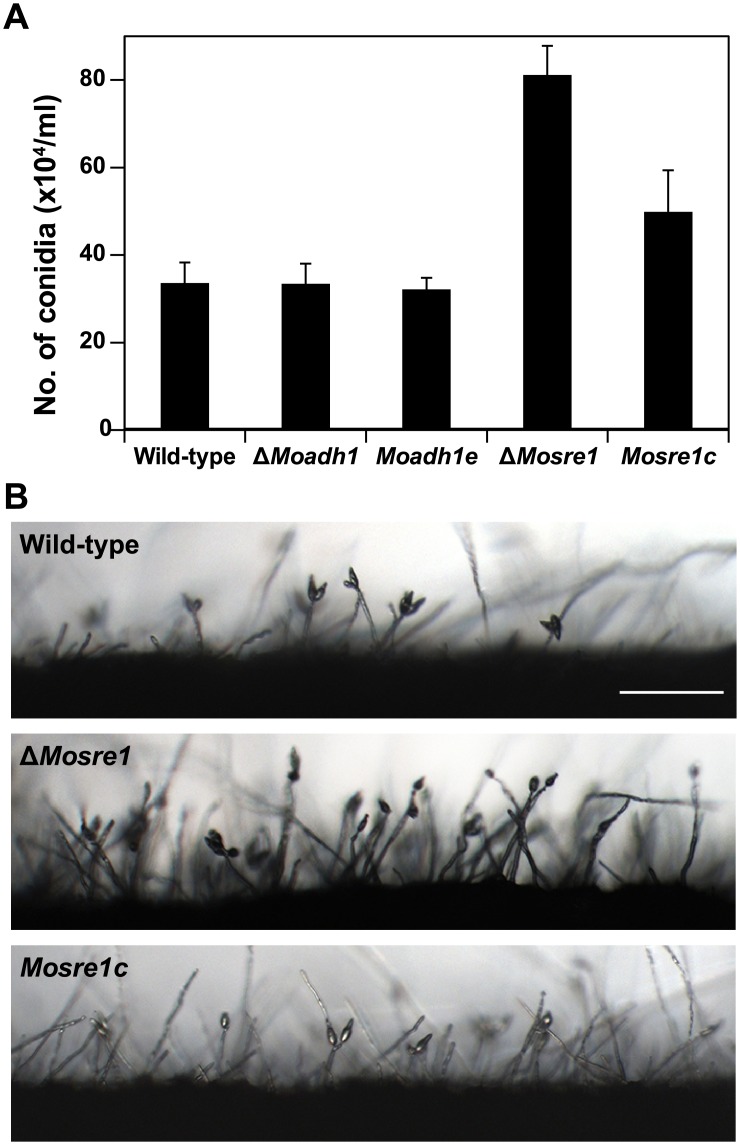
To further investigate the roles of the MoSRE1 and MoADH1 genes in fungal development and pathogenicity of M. oryzae, we tested mutants for phenotypes such as conidiation, conidial germination, and appressorium formation. No significant difference was observed in conidial germination (4 h) and appressorium formation (16 h) between the wild-type and either of the two deletion mutants (Table 3). Interestingly, the ΔMosre1 mutant produced twice as many conidia than the other strains (Fig 5A and Table 3, P < 0.05). Densely developed conidiophores were observed in the ΔMosre1 mutant compared to the wild-type and the complemented strain, Mosre1c (Fig 5B), suggesting that MoSRE1 has a negative function in conidiation. Because aeration due to unsealing plates generally causes initiation of conidiation in M. oryzae, the increased conidia in the ΔMosre1 mutant was contrary to our expectations. We expected that the oxygen concentration would be decreased in the sealed plates and would stimulate expression of MoSRE1 as shown in the RNA-seq data. Alternatively, null mutation of MoSRE1 might promote conidia production in an independent manner. The hypoxia effect via deletion of the MoSRE1 gene (discussed in Fig 4) could be recognized as a signal that the fungus encountered in an unfavorable environment. It then produced more spores to increase its survival rate under this adverse condition. In addition, the GO enrichment analysis revealed the circadian rhythm term (GO:0007623) was enriched in up-regulated genes (Table 1). The MoFRQ1 gene (MGG_17344) belongs to this term and its homolog in Neurospora crassa has been known as a circadian clock regulator, showing variation in conidiation [47]. In M. oryzae, the MoFRQ1 mutant exhibited short conidiophores with dense conidia [48].
Fig 5. Conidiation of the wild-type, ΔMoadh1 and ΔMosre1.
(A) Conidia were collected from 7-day-old cultures on V8 juice agar plates with 5-ml water. The number of conidia was counted using a hemocytometer under the microscope. (B) Conidiogenesis was monitored under a microscope 16 hours after incubation. Strains were grown on oatmeal agar plates and scraped for inducing conidiogenesis at the same time. Scale bar indicates 100 μm.
Roles of MoSRE1 and MoADH1 in fungal pathogenicity
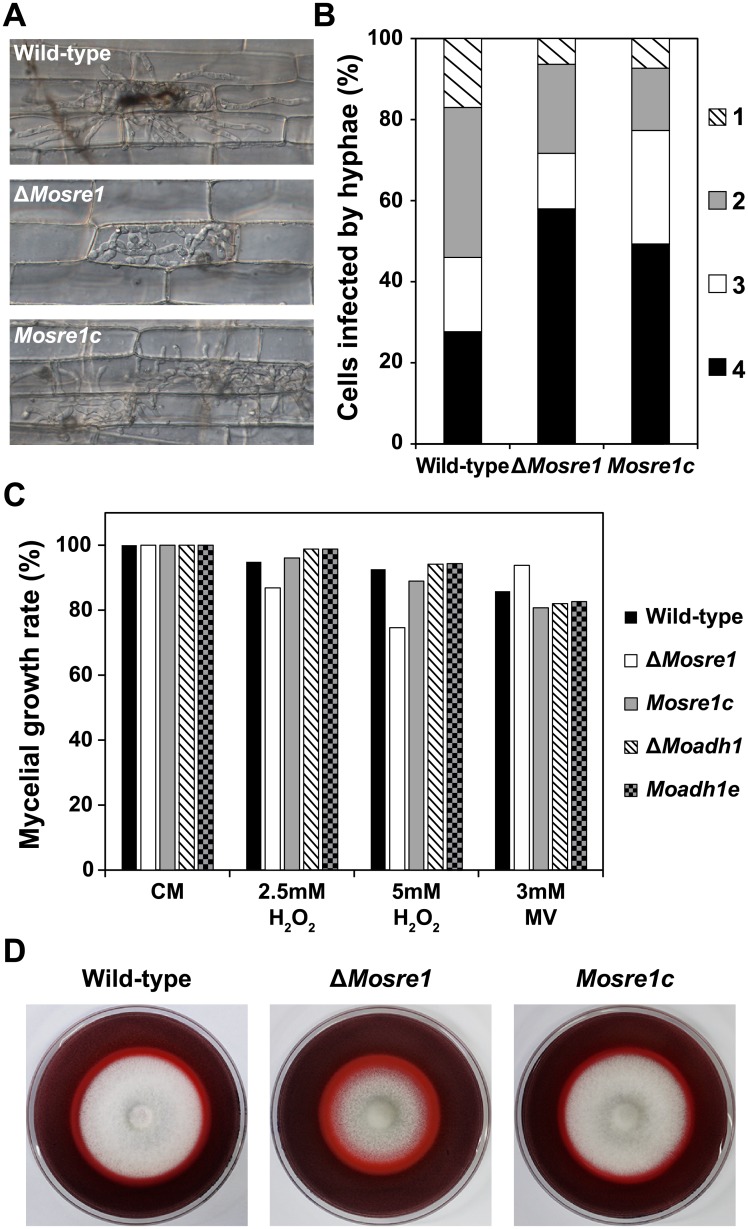
When a conidial suspension was injected into the rice sheath cells, infectious hyphae of the wild-type and both deletion mutants extended over the adjacent cells after filling inside of the first-infected cell (Fig 6A and 6B). However, penetration and infectious growth of the ΔMosre1 mutant were much delayed than those of the wild-type and other mutants. More than 50% of the ΔMosre1 conidia produced only appressorium and no penetration peg was observed in them, while less than 30% of the wild-type conidia were marked as the same category, “No penetration” (Fig 6B). Delay in penetration might be explained by the down-regulated genes belonging to the ‘glycerol metabolism’ term (GO:0006071) (Table 1). Because glycerol accumulation increases osmotic pressure within an appressorium [49], a delay in glycerol metabolism led to the penetration defects in the ΔMosre1 cells. The proportions of “Primary hyphae”, “One cell filled”, and “Multiple cells” in the mutant were less than those in the wild-type (Fig 6B), supporting overall delay in invasive growth.
Fig 6. Invasive growth, oxidative stress sensitivity and enzyme activity of the wild-type, ΔMoadh1 and ΔMosre1.
(A) Infectious growth was observed in rice sheath cells. A conidial suspension (2 × 104 conidia/ml) was inoculated into the excised rice sheath. Photographs were taken 48 hours after incubation. Scale bar indicates 20 μm. (B) Frequency of infected rice cells was determined by counting at least 100 appressorium-mediated penetration pegs with three replicates. Invasive growth was observed as described above. 1, Move to adjacent cell; 2, One cell filled; 3, Primary hyphae; 4, No penetration. (C) Extracellular oxidative stress sensitivity of the wild-type and two deletion mutants were examined. Wild-type and two deletion mutants were inoculated on CM and CM including 2.5 or 5 mM H2O2 and 3 mM methyl viologen (MV). (D) Wild-type, ΔMosre1 and Mosre1c were inoculated on CM containing 200 ppm Congo Red. Discoloration (halo) of Congo Red was observed at 9 days after incubation.
In addition, we examined responses to oxidative stresses because hypoxia-induced transcripts such as laccase, catalase, and copper-transport genes might be caused by ROS accumulation in the fungal cell (GO:0055114 and GO:0006825 in Table 1). When the growth on CM was set as 100%, the wild-type showed 95% and 93% of mycelial growth on the CM containing 2.5 mM and 5 mM H2O2, respectively (Fig 6C). The ΔMosre1 mutant showed 87% and 75% of mycelial growth on the media, indicating that the mutant is sensitive to H2O2. However, the mutant showed opposite trend to the oxidative stress of methyl viologen, a chemical inducer of superoxide free radical. The ΔMosre1 mutant was not vulnerable (94%) to methyl viologen while the wild-type and other strains showed slight reduction in response to it (81~86%, Fig 6C). In addition, we tested a degrading activity of Congo Red (a secondary diazo dye) to see extracellular peroxidase or laccase activity of the fungus. Interestingly, the ΔMosre1 mutant showed apparent halo compared to the wild-type (Fig 6D). Image analysis revealed that the relative ratio of halo size to mycelial size was higher in the mutant (1.61) than the wild-type (1.40) although total degraded area was almost similar between them (Fig 6D). This indicates that the ΔMosre1 mutant generated more extracellular peroxidase or laccase. These results may also explain one of the reasons why the ΔMosre1 mutant showed the delayed growth in the sheath inoculation assay (Fig 6A), suggesting that fungal SREBP might be involved in recognition and responses of ROS within host cells. Thus, given that the growth of the ΔMosre1 mutant was delayed under hypoxia-mimicking conditions using CoCl2 (Fig 4), our observation is the first indirect evidence that hypoxia occurs within plant leaves.
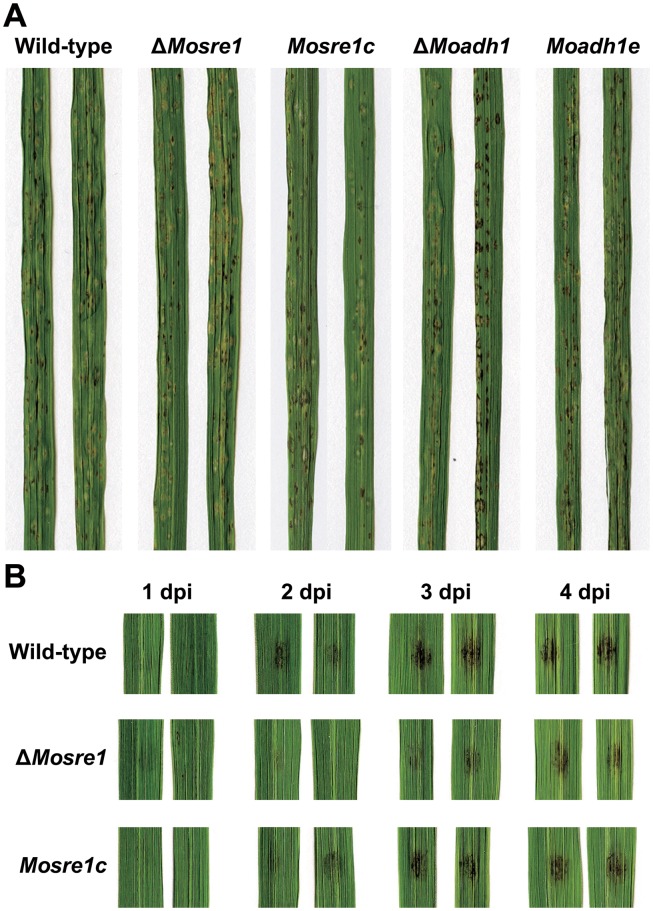
We then performed spray inoculation assays to determine the effects of the MoSRE1 and MoADH1 genes on fungal pathogenicity. Rice leaves were spray-inoculated using a conidial suspension and were observed at seven days post inoculation (dpi). Both mutants exhibited similar disease symptoms compared to the wild-type, Moadh1e and Mosre1c strains (Fig 7A). To determine when this mutant overcome the delayed growth within host cells, lesion development was monitored using rice leaves by dropping conidial suspensions. Dark lesion areas began to be shown at two days post inoculation with the wild-type conidial suspension while similar patterns of lesions were developed at three days post inoculation in the ΔMosre1 mutant (Fig 7B). The delay in ΔMosre1 was almost recovered four days post inoculation. These results suggest that ΔMosre1 seems to overcome the hypoxic environment in the host cells, which is not the case for SREBP-null mutants of human fungal pathogens within their hosts [13]. It might be explained by the fact that M. oryzae, unlike human fungal pathogens, switches from a biotrophic to a necrotrophic lifestyle in the middle of the infection.
Fig 7. Pathogenicity of wild-type, ΔMoadh1 and ΔMosre1.
(A) The pathogenicity assay was performed by spraying a conidia suspension (5 × 104 conidia/ml) of each strain onto susceptible rice seedlings. Photographs were taken 7 days after inoculation. (B) The pathogenicity assay via drop inoculation was examined. Conidia suspension (105 conidia/ml) was dropped onto detached rice leaves and incubated at room temperature. dpi, days post inoculation.
Conclusion
Changes in transcriptomes and physiological observations in this study suggest that the hemibiotrophic rice blast fungus might encounter hypoxia in a living host cell during infection. Under hypoxic conditions, this fungus altered transcript levels of diverse sets of genes including those involved in mycelial development, sterol biosynthesis and redox processes. These transcriptional alterations were also observed in human fungal pathogens whose interactions with hosts have been well studied. Especially, ergosterol biosynthesis-related genes (ERG25, ERG3 and SCS7) which are associated with the SREBP pathway were induced in M. oryzae and in all the fungi used for comparison. The SREBP-mediated pathway, in response to a given oxygen concentration, induces ergosterol biosynthesis and hyphal growth in order to scavenge more oxygen in human fungal pathogens. In M. oryzae, a function for SREBP as an oxygen sensor against hypoxia could be extrapolated from the results that the ΔMosre1 mutant exhibited delayed growth in the plant cells and hypoxia-mimicking environment. In addition, SREBP also functions as a negative regulator for conidiation. Our data provides a new platform to decipher molecular machinery underpinning surveillance of hypoxic conditions during interactions between plant hosts and fungal pathogens.
Supporting Information
(A) ΔMoadh1. Wild-type and the ΔMoadh1 mutant had a 1.8-kb and 3.9-kb SalI fragment, respectively. (B) ΔMosre1. Wild-type and the ΔMosre1 mutant had a 5.3-kb and 3.5-kb XbaI fragment, respectively.
(PPTX)
(DOCX)
(DOCX)
(XLSX)
(XLSX)
Data Availability
All relevant data are available in the manuscript, its Supporting Information files, and via the NCBI GEO database (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE51597.
Funding Statement
This work was supported by National Research Foundation of Korea grants funded by the Korean government (2014R1A2A1A10051434 and 2013-010928), and the Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ011154) Rural Development Administration, Republic of Korea. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Ernst JF, Tielker D. Responses to hypoxia in fungal pathogens. Cell Microbiol. 2009;11(2):183–90. 10.1111/j.1462-5822.2008.01259.x [DOI] [PubMed] [Google Scholar]
- 2. Semenza GL. Life with oxygen. Science. 2007;318(5847):62–4. [DOI] [PubMed] [Google Scholar]
- 3. Packer L, Fuehr K. Low oxygen concentration extends lifespan of cultured human diploid cells. Nature. 1977;267(5610):423–5. [DOI] [PubMed] [Google Scholar]
- 4. Jacobson MD, Raff MC. Programmed cell death and Bcl-2 protection in very low oxygen. Nature. 1995;374(6525):814–6. [DOI] [PubMed] [Google Scholar]
- 5. van Dongen JT, Schurr U, Pfister M, Geigenberger P. Phloem metabolism and function have to cope with low internal oxygen. Plant Physiol. 2003;131(4):1529–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zitomer RS, Carrico P, Deckert J. Regulation of hypoxic gene expression in yeast. Kidney Int. 1997;51(2):507–13. [DOI] [PubMed] [Google Scholar]
- 7. Igwe E, Essler S, Al-Furoukh N, Dehne N, Brune B. Hypoxic transcription gene profiles under the modulation of nitric oxide in nuclear run on-microarray and proteomics. BMC Genomics. 2009;10(1):408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Erecinska M, Silver IA. Tissue oxygen tension and brain sensitivity to hypoxia. Respir Physiol. 2001;128(3):263–76. [DOI] [PubMed] [Google Scholar]
- 9. Grahl N, Puttikamonkul S, Macdonald JM, Gamcsik MP, Ngo LY, Hohl TM, et al. In vivo hypoxia and a fungal alcohol dehydrogenase influence the pathogenesis of invasive pulmonary aspergillosis. PLoS Path. 2011;7(7):e1002145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hughes AL, Todd BL, Espenshade PJ. SREBP pathway responds to sterols and functions as an oxygen sensor in fission yeast. Cell. 2005;120(6):831–42. [DOI] [PubMed] [Google Scholar]
- 11. Chun CD, Liu OW, Madhani HD. A link between virulence and homeostatic responses to hypoxia during infection by the human fungal pathogen Cryptococcus neoformans . PLoS Path. 2007;3(2):e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chang YC, Bien CM, Lee H, Espenshade PJ, Kwon-Chung KJ. Sre1p, a regulator of oxygen sensing and sterol homeostasis, is required for virulence in Cryptococcus neoformans . Mol Microbiol. 2007;64(3):614–29. [DOI] [PubMed] [Google Scholar]
- 13. Willger SD, Puttikamonkul S, Kim KH, Burritt JB, Grahl N, Metzler LJ, et al. A sterol-regulatory element binding protein is required for cell polarity, hypoxia adaptation, azole drug resistance, and virulence in Aspergillus fumigatus . PLoS Path. 2008;4(11):e1000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Geigenberger P. Response of plant metabolism to too little oxygen. Curr Opin Plant Biol. 2003;6(3):247–56. [DOI] [PubMed] [Google Scholar]
- 15. Ober ES, Sharp RE. A microsensor for direct measurement of O2 partial pressure within plant tissues. J Exp Bot. 1996;47(296):447–54. [Google Scholar]
- 16. Rolletschek H, Borisjuk L, Koschorreck M, Wobus U, Weber H. Legume embryos develop in a hypoxic environment. J Exp Bot. 2002;53(371):1099–107. [DOI] [PubMed] [Google Scholar]
- 17. Rijnders JGHM, Armstrong W, Darwent MJ, Blom CWPM, Voesenek LACJ. The role of oxygen in submergence-induced petiole elongation in Rumex palustris: in situ measurements of oxygen in petioles of intact plants using micro-electrodes. New Phytol. 2000;147(3):497–504. [DOI] [PubMed] [Google Scholar]
- 18. Khush G, Jena KK. Current status and future prospects for research on blast resistance in rice (Oryza sativa L.) In: Wang G-L, Valent B, editors. Advances in Genetics, Genomics and Control of Rice Blast Disease: Springer; 2009. p. 1–10. [Google Scholar]
- 19. Kankanala P, Czymmek K, Valent B. Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. Plant Cell. 2007;19(2):706–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Howard RJ, Valent B. Breaking and entering: Host penetration by the fungal rice blast pathogen Magnaporthe grisea . Annu Rev Microbiol. 1996;50:491–512. [DOI] [PubMed] [Google Scholar]
- 21. Khang CH, Berruyer R, Giraldo MC, Kankanala P, Park SY, Czymmek K, et al. Translocation of Magnaporthe oryzae effectors into rice cells and their subsequent cell-to-cell movement. Plant Cell. 2010;22(4):1388–403. 10.1105/tpc.109.069666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, et al. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7:3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Patel RK, Jain M. NGS QC toolkit: a toolkit for quality control of next generation sequencing data. PLoS ONE. 2012;7(2):e30619 10.1371/journal.pone.0030619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–11. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Alexa A, Rahnenfuhrer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006;22(13):1600–7. [DOI] [PubMed] [Google Scholar]
- 26. Choi J, Cheong K, Jung K, Jeon J, Lee GW, Kang S, et al. CFGP 2.0: a versatile web-based platform for supporting comparative and evolutionary genomics of fungi and Oomycetes. Nucleic Acids Res. 2013;41(D1):D714–D9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Park J, Kong S, Kim S, Kang S, Lee Y-H. Roles of forkhead-box transcription factors in controlling development, pathogenicity, and stress response in Magnaporthe oryzae . Plant Pathol J. 2014;30(2):136–50. 10.5423/PPJ.OA.02.2014.0018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Choi J, Kim Y, Kim S, Park J, Lee YH. MoCRZ1, a gene encoding a calcineurin-responsive transcription factor, regulates fungal growth and pathogenicity of Magnaporthe oryzae . Fungal Genet Biol. 2009;46(3):243–54. 10.1016/j.fgb.2008.11.010 [DOI] [PubMed] [Google Scholar]
- 29. Yi M, Chi MH, Khang CH, Park SY, Kang S, Valent B, et al. The ER chaperone LHS1 is involved in asexual development and rice infection by the blast fungus Magnaporthe oryzae . Plant Cell. 2009;21(2):681–95. 10.1105/tpc.107.055988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chi MH, Park SY, Lee YH. A quick and safe method for fungal DNA extraction. Plant Pathol J. 2009;25(1):108–11. [Google Scholar]
- 31. Choi J, Park J, Jeon J, Chi MH, Goh J, Yoo SY, et al. Genome-wide analysis of T-DNA integration into the chromosomes of Magnaporthe oryzae . Mol Microbiol. 2007;66(2):371–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sambrook J, Russel DW. Molecular Cloning: A Laboratory Manual. 3rd ed. ed. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 33. Talbot NJ, Ebbole DJ, Hamer JE. Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea . Plant Cell. 1993;5(11):1575–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lau GW, Hamer JE. Acropetal: A genetic locus required for conidiophore architecture and pathogenicity in the rice blast fungus. Fungal Genet Biol. 1998;24(1–2):228–39. [DOI] [PubMed] [Google Scholar]
- 35. Valent B, Farrall L, Chumley FG. Magnaporthe grisea genes for pathogenicity and virulence identified through a series of backcrosses. Genetics. 1991;127(1):87–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Koga H, Dohi K, Nakayachi O, Mori M. A novel inoculation method of Magnaporthe grisea for cytological observation of the infection process using intact leaf sheaths of rice plants. Physiol Mol Plant Pathol. 2004;64(2):67–72. [Google Scholar]
- 37. Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–7. 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Talbot NJ, Kershaw MJ, Wakley GE, deVries OMH, Wessels JGH, Hamer JE. MPG1 encodes a fungal hydrophobin involved in surface interactions during infection-related development of Magnaporthe grisea . Plant Cell. 1996;8(6):985–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Setiadi ER, Doedt T, Cottier F, Noffz C, Ernst JF. Transcriptional response Candida albicans to hypoxia: Linkage of oxygen sensing and Efg1p-regulatory networks. J Mol Biol. 2006;361(3):399–411. [DOI] [PubMed] [Google Scholar]
- 40. Masuo S, Terabayashi Y, Shimizu M, Fujii T, Kitazume T, Takaya N. Global gene expression analysis of Aspergillus nidulans reveals metabolic shift and transcription suppression under hypoxia. Mol Genet Genomics. 2010;284(6):415–24. 10.1007/s00438-010-0576-x [DOI] [PubMed] [Google Scholar]
- 41. Barker BM, Kroll K, Vodisch M, Mazurie A, Kniemeyer O, Cramer RA. Transcriptomic and proteomic analyses of the Aspergillus fumigatus hypoxia response using an oxygen-controlled fermenter. BMC Genomics. 2012;13(1):62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Camilo CM, Gomes SL. Transcriptional response to hypoxia in the aquatic fungus Blastocladiella emersonii . Eukaryot Cell. 2010;9(6):915–25. 10.1128/EC.00047-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mosquera G, Giraldo MC, Khang CH, Coughlan S, Valent B. Interaction transcriptome analysis identifies Magnaporthe oryzae BAS1-4 as biotrophy-associated secreted proteins in rice blast disease. Plant Cell. 2009;21: 1273–1290. 10.1105/tpc.107.055228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kawahara Y, Oono Y, Kanamori H, Matsumoto T, Itoh T, Minami E. Simultaneous RNA-seq analysis of a mixed transcriptome of rice and blast fungus interaction. PLoS One. 2012;7: e49423 10.1371/journal.pone.0049423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lee H, Bien CM, Hughes AL, Espenshade PJ, Kwon-Chung KJ, Chang YC. Cobalt chloride, a hypoxia-mimicking agent, targets sterol synthesis in the pathogenic fungus Cryptococcus neoformans . Mol Microbiol. 2007;65(4):1018–33. [DOI] [PubMed] [Google Scholar]
- 46. Blatzer M, Barker BM, Willger SD, Beckmann N, Blosser SJ, Cornish EJ, et al. SREBP coordinates iron and ergosterol homeostasis to mediate triazole drug and hypoxia responses in the human fungal pathogen Aspergillus fumigatus . PLoS Genet. 2011;7(12):e1002374 10.1371/journal.pgen.1002374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. McClung CR, Fox BA, Dunlap JC. The Neurospora clock gene frequency shares a sequence element with the Drosophila clock gene period . Nature. 1989;339(6225):558–62. [DOI] [PubMed] [Google Scholar]
- 48. Park J, Lee Y-H. Bidirectional-genetics platform, a dual-purpose mutagenesis strategy for filamentous fungi. Eukaryot Cell. 2013;12(11):1547–53. 10.1128/EC.00234-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. de Jong JC, McCormack BJ, Smirnoff N, Talbot NJ. Glycerol generates turgor in rice blast. Nature. 1997;389(6648):244–5. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) ΔMoadh1. Wild-type and the ΔMoadh1 mutant had a 1.8-kb and 3.9-kb SalI fragment, respectively. (B) ΔMosre1. Wild-type and the ΔMosre1 mutant had a 5.3-kb and 3.5-kb XbaI fragment, respectively.
(PPTX)
(DOCX)
(DOCX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are available in the manuscript, its Supporting Information files, and via the NCBI GEO database (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE51597.