Abstract
The DEAD-box RNA helicases are involved in almost every aspect of RNA metabolism, associated with diverse cellular functions including plant growth and development, and their importance in response to biotic and abiotic stresses is only beginning to emerge. However, none of DEAD-box genes was well characterized in tomato so far. In this study, we reported on the identification and characterization of two putative DEAD-box RNA helicase genes, SlDEAD30 and SlDEAD31 from tomato, which were classified into stress-related DEAD-box proteins by phylogenetic analysis. Expression analysis indicated that SlDEAD30 was highly expressed in roots and mature leaves, while SlDEAD31 was constantly expressed in various tissues. Furthermore, the expression of both genes was induced mainly in roots under NaCl stress, and SlDEAD31 mRNA was also increased by heat, cold, and dehydration. In stress assays, transgenic tomato plants overexpressing SlDEAD31 exhibited dramatically enhanced salt tolerance and slightly improved drought resistance, which were simultaneously demonstrated by significantly enhanced expression of multiple biotic and abiotic stress-related genes, higher survival rate, relative water content (RWC) and chlorophyll content, and lower water loss rate and malondialdehyde (MDA) production compared to wild-type plants. Collectively, these results provide a preliminary characterization of SlDEAD30 and SlDEAD31 genes in tomato, and suggest that stress-responsive SlDEAD31 is essential for salt and drought tolerance and stress-related gene regulation in plants.
Introduction
Plants, as sessile organisms, are continuously exposed to various environmental stresses such as salinity, drought, low temperature, and UV light, which can cause damage to lipids, proteins and DNA, reducing plants genome stability, growth, and productivity [1, 2]. Plants respond to these stresses via the cascades of molecular networks and changes in expression profiles of diverse classes of genes, including those involved in nucleic acid metabolism, which are induced in response to the DNA damage by activating a network of DNA repair enzymes including helicases [3–5]. RNA helicases, which catalyze the unwinding of energetically stable duplex RNA secondary structures in an ATP-dependent manner, thereby have been implicated in every aspect of RNA metabolism, including nuclear transcription, pre-mRNA splicing, ribosome biogenesis, RNA transport, translation initiation, RNA degradation, and organelle gene expression [6, 7]. Whereas DNA helicases unwind duplex DNA and are involved in replication, repair, recombination, and transcription [8].
The majority of RNA helicases belong to the superfamily 2 (SF2) subclass, characterized by sequence homology within the helicase domain that contains eight or nine conserved motifs. The SF2 consists of three subfamilies, termed DEAD, DEAH and DExH/D, based on variations within motif II of the proteins [9, 10]. The DEAD-box RNA helicases are by far the largest family of RNA helicases, and they are found in all of the organisms from prokaryotes to eukaryotes, and have been shown to be involved in almost every aspect of RNA metabolism [11, 12]. In addition, DEAD-box RNA helicases are also prominent candidates for RNA chaperones because they can use energy derived from ATP hydrolysis to disrupt misfolded RNA structures and promote correct folding, potentially playing roles in diverse cellular functions [9]. So far, biochemical activities characteristic of RNA helicases, namely RNA-dependent RNA unwinding and ATPase were only demonstrated for a small proportion of the RNA helicase. For example, the redox-regulated cyanobacterial helicase, CrhR, catalyzes annealing of complementary ssRNA (single-stranded RNA) into dsRNA (double-stranded RNA) and combines the helicase and annealing activities to promote RNA-strand exchange [13].
In plants, the comprehensive analysis and classification of the RNA helicase gene family in Arabidopsis and rice, which contain 113 and 115 RNA helicase genes, respectively, has been reported recently [14]. Presently, limited reports have emerged indicating that DEAD-box RNA helicase expression or activity is regulated not only with respect to participation in those housekeeping processes mentioned above, but also in plant growth and development, and in response to biotic and abiotic stresses [5, 15–17]. For example, the tobacco VDL gene encodes a plastid DEAD-box protein involved in chloroplast differentiation and plant morphogenesis as its recessive mutant vdl showed variegated leaves and abnormal roots and flowers [18]. In rice, the OsBIRH1 gene encodes a DEAD-box RNA helicase, ectopic expression of OsBIRH1 in Arabidopsis showed increased tolerance to pathogen and oxidative stresses [17]. The Arabidopsis LOS4 RNA helicase was showed to be a cold stress-regulated gene, which was proved as an early regulator of CBF transcription factor expression in response to chilling [19, 20]. In addition, overexpression of AtRH25 results in enhanced freezing tolerance in transgenic Arabidopsis [21]. On the contrary, mutations in STRS1 (STRESS RESPONSE SUPPRESSOR 1) and STRS2 which encode DEAD-box RNA helicases, cause increased expression of stress-responsive transcriptional activators and abiotic stress tolerance, suggesting that both STRS1 and STRS2 function as negative regulators [16]. Despite the involvement of DEAD-box RNA helicases in many diverse biological processes, the actual biological, biochemical, and molecular function of most plant DEAD-box proteins still remain to be further elucidated.
Tomato (Solanum lycopersicum) is one of the most widely produced and versatile vegetable crops, and it is also considered as one of the best-characterized plant systems employed in genetic and developmental studies whose genome has been fully sequenced [22, 23]. Although a genome-wide analysis of the RNA helicase gene family has identified 42 DEAD-box genes, 36 DEAH-box genes, and 52 DExD/H-box genes in tomato [24], none of the DEAD-box RNA helicase genes was well characterized in tomato so far. To explore the exact roles of tomato DEAD-box helicases, in the present study, two putative DEAD-box RNA helicase genes, termed SlDEAD30 and SlDEAD31, were cloned and characterized from tomato. The overexpression of SlDEAD31 led to enhanced salt and drought tolerance, and upregulated expression of stress-related genes. Our results suggest that SlDEAD31 acts as a positive regulator in defence responses against abiotic stress, and may hold promising applications in the engineering of salt- and drought-tolerant plants.
Results
Sequence Analysis of SlDEAD30 and SlDEAD31 Genes
Sequence analysis showed that SlDEAD30 and SlDEAD31 encode polypeptides consisting of 488 and 439 amino acids, and share 50.6% and 42.9% identity at the nucleotide level and amino acid level, respectively. In addition, two typical conserved domains: the DEAD-box helicase superfamily domain and the helicase C-terminal domain, as well as nine conserved helicase motifs, which are common characteristics of DEAD-box RNA helicases, are all present in these two SlDEAD proteins (S1 and S2 Figs). Generally, these motifs are functional in RNA substrate binding and ATP binding or exhibit ATPase activity [10]. To further analyze the conserved helicase motifs of SlDEAD30 and SlDEAD31 proteins, the amino acid sequences of two previously reported DEAD-box proteins, AtRH9 and AtRH36 from Arabidopsis, which show a high sequence similarity with SlDEAD30 and SlDEAD31, were analyzed for alignment. SlDEAD30 protein shares a close relationship with AtRH36 (66.4% identity, S1 Table), which is essential for Arabidopsis female gametogenesis [25], whereas SlDEAD31 is much closer related to AtRH9 (79.9% identity, S1 Table), which has an important role in cold tolerance in Arabidopsis [21]. The results show that all the nine conserved helicase motifs are present in the four DEAD-box proteins, Q-motif and motif IV are relatively diverse, while the other motifs are highly conserved (Fig 1).
Fig 1. Multiple sequence alignment of the amino acid sequences of SlDEAD30, SlDEAD31, AtRH9, and AtRH36 proteins.
Identical or similar amino acids are shaded in black and grey, respectively. The locations of the nine conserved helicase motifs are indicated on top of the sequences and DEAD domains are shown in the box. The conserved phenylalanine residue is indicated by an inverted triangle. SlDEAD30 (KJ739798), SlDEAD31 (KJ713393), AtRH9 (NM_125492), and AtRH36 (NM_101494).
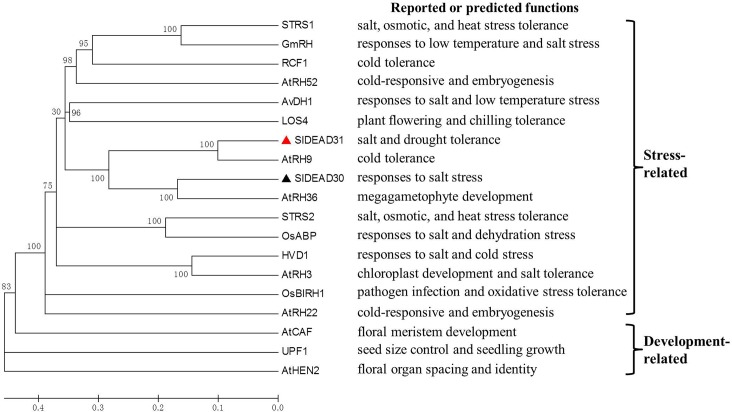
To investigate the divergence of SlDEAD30 and SlDEAD31 proteins with other plant DEAD-box proteins during evolution, a phylogenetic tree was constructed using the amino acid sequences of SlDEAD30, SlDEAD31, and 17 previously published plant DEAD-box proteins with known functions. These proteins are putatively divided into two distinct subgroups, a stress-related subgroup and a development-related subgroup based on their relatives’ function and the phylogenetic tree. SlDEAD30 and SlDEAD31 proteins are present in the stress-related subgroup (Fig 2). In addition, the phylogenetic analysis also revealed that SlDEAD30 and SlDEAD31 proteins share a close relationship with the AtRH36 and AtRH9, respectively, suggesting that they may carry out similar function. Besides, the possible functions of SlDEAD30, SlDEAD31, and the other previously reported plant DEAD-box RNA helicases are also presented (Fig 2).
Fig 2. Phylogenetic tree and reported or predicted functions of SlDEAD30, SlDEAD31, and other known plant DEAD-box proteins.
The phylogenetic tree analysis was constructed with MEGA 5.2 software using the neighbor-joining method and the following parameters: bootstrap analysis of 1000 replicates, poisson model and pairwise deletion. The numbers at the nodes indicate the bootstrap values. SlDEAD30 and SlDEAD31 are marked with a black triangle and a red triangle, respectively. Accession numbers and corresponding references for the proteins used are as follows: Solanum lycopersicum: SlDEAD30, KJ739798; SlDEAD31, KJ713393. Arabidopsis thaliana: AtRH3, NM_001036866 [28]; AtRH9, NM_125492 [21]; AtRH22, NM_104691 [26]; AtRH36, NM_101494 [25]; AtRH52, NM_115719 [26]; STRS1, AY080680 [16]; STRS2, AY035114 [16]; LOS4, BT002444 [19]; RCF1, BT002030 [54]; AtCAF, AF187317 [27]; AtHEN2, AY050658 [55]. UPF1, AF484122 [56]. Oryza sativa: OsBIRH1, Q0DVX2 [17]; OsABP, LOC_Os06g33520 [57]; Hordeum vulgare: HVD1, AB164680 [58]; Glycine max: GmRH, FJ462142 [59]. Apocynum venetum: AvDH1, EU145588 [15].
Organ-Specific Expression of SlDEAD30 and SlDEAD31 Genes in Wild-Type Tomato
Tissue specificity may be associated with specific biological functions. Previous reports have demonstrated that a range of DEAD-box RNA helicases are involved in regulating the essential steps in plant growth and development, such as morphogenesis and embryogenesis [18, 26], floral meristems [27], and chloroplast development [28]. The results showed that the expression of SlDEAD30 was relatively higher in roots and mature leaves, whereas moderate to weak signals of its expression were detected in other tissues such as stems, flowers, and fruits. SlDEAD31 mRNAs were constantly expressed in all the organs tested (Fig 3).
Fig 3. Expression profiles of SlDEAD30 and SlDEAD31 genes in different tissues of wild-type tomato.
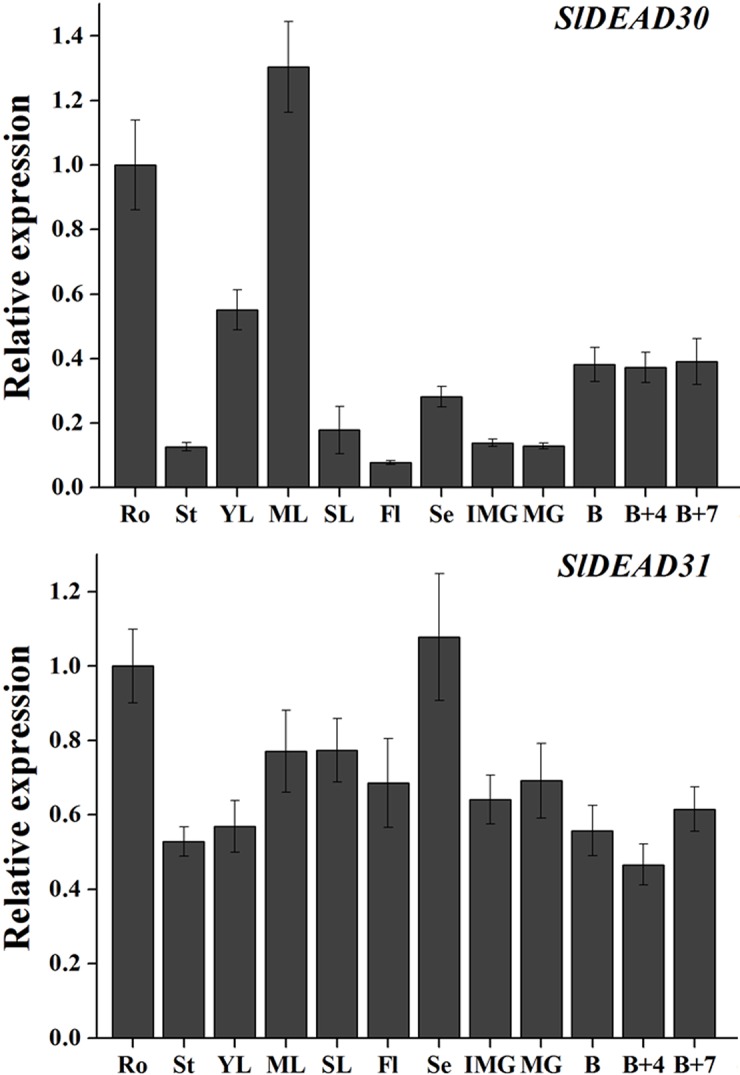
Ro, roots; St, stems; Yl, young leaves; Ml, mature leaves; Sl, senescent leaves; Fl, flowers; Se, sepals; IMG, immature green fruits; MG, mature green fruits; B, breaker fruits; B+4, 4 day after breaker stage; B+7, 7 day after breaker stage. The relative expression levels were normalized to 1 in the roots. Bars represent mean relative expression values ± SE (n = 3).
The Expression of SlDEAD30 and SlDEAD31 Genes in Response to Various Abiotic Stresses
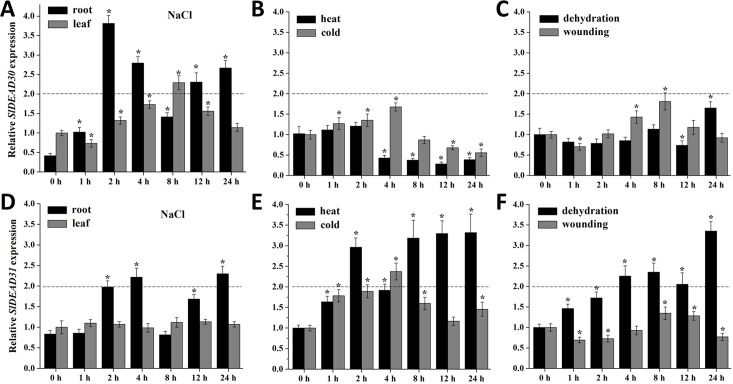
To speculate on the physiological and functional relevance of SlDEAD30 and SlDEAD31 genes, we detected their expression profiles under various abiotic stresses by quantitative RT-PCR. We found that their expression was not affected by circadian rhythm under the normal growth condition (S3 Fig). SlDEAD30 transcripts were induced apparently in roots after NaCl stress compared with controls, and to a lesser degree in leaves (Fig 4A). SlDEAD31 expression was also induced by NaCl in roots, whereas remained unchanged in leaves (Fig 4D). The results indicate that the expression of both SlDEAD30 and SlDEAD31 was induced mainly in roots but not in leaves under NaCl stress. When treated with heat and cold, SlDEAD30 mRNA was not increased by cold, while down-regulated by heat (Fig 4B). In contrast, SlDEAD31 expression was induced by both stresses (Fig 4E). In addition, the expression of SlDEAD30 was not induced by dehydration and wounding, while SlDEAD31 mRNA was induced by dehydration almost by a gradual increase (Fig 4C and 4F). The induction of SlDEAD30 and SlDEAD31 genes by abiotic stress prompted us to check their promoter sequences by searching against the promoter database PLACE. The results show that their promoter sequences indeed contain putative stress response-related cis-elements, such as MYB, MYC, and WRKY recognition sites (S2 Table).
Fig 4. Expression patterns of SlDEAD30 and SlDEAD31 genes under various stress treatments including NaCl (200 mM, A, D), heat (40°C) and cold (4°C, B, E), dehydration, and wounding (C, F).
Gene expression was detected by RT-PCR using total RNA from leaves or roots of 35-day-old tomato plants. Bars represent mean relative expression values ± SE (n = 3). Values are presented relative to untreated plants (0 h). Asterisks indicate a significant difference (P<0.05) between untreated and treated plants. The twofold threshold is indicated by a dotted line.
Overexpression of SlDEAD31 in Tomato Plants Significantly Improves Salt Tolerance
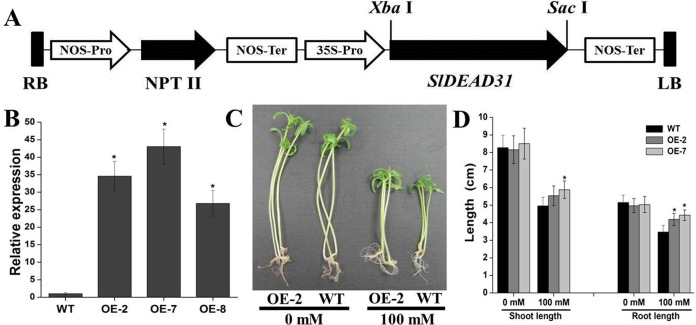
Taking into account that only SlDEAD31 expression was induced by various stresses, therefore, to further analyze its biological function, we generated transgenic tomato plants overexpressing SlDEAD31 (Fig 5A). A total of 7 independent SlDEAD31-overexpressing transgenic lines were obtained. Besides, we also attempted to silence SlDEAD31 gene in stably transformed tomato plants by RNAi (RNA interference) using the constitutive 35S promoter, but this failed three times. Therefore, only SlDEAD31-overexpressing transgenic lines were used for further studies. Quantitative RT-PCR analysis showed that the relative mRNA level of SlDEAD31 in three overexpression transgenic lines (OE-2, OE-7, and OE-8) increased by 34.6, 43.1, and 26.8 times compared with WT plants, respectively (Fig 5B).
Fig 5. Salt sensitivity of SlDEAD31 overexpressing plants at postgermination stage.
(A) SlDEAD31-overexpressing vector. RB, right border; Pro, promoter; Ter, terminator; LB, left border. (B) Quantitative RT-PCR analysis of SlDEAD31 expression in the leaves of WT and transgenic lines OE-2, OE-7, and OE-8. Values represent the means ± SE (n = 3). Asterisks indicate a significant difference from WT (p<0.05). (C) and (D) Representative growth performance (C) and shoot and root length (D) of WT and SlDEAD31 overexpressing plants in MS medium containing 0 and 100 mM NaCl at postgermination stage. The shoot and root length of seedlings (n≥20 each) were counted after growth for 10 d. Values represent the means ± SE (n = 3). Asterisks indicate a significant difference from WT (p<0.05).
Salt sensitivity in tomato is most evident during seed germination and subsequent seedling growth [29]. As described above, the induction of SlDEAD31 expression by NaCl suggested that it might be involved in the tolerance to salt stress (Fig 4D). To confirm this, salt tolerance performance of the transgenic tomato seedlings was tested at the post-germination stage. WT and transgenic tomato seeds of consistent germination were transferred to medium containing 0 and 100 mM NaCl. In the presence of 100 mM NaCl, plant growth was inhibited apparently in both WT and overexpressing tomato seedlings, whereas the average length of roots of WT seedlings were shorter than that of transgenic plants, while the difference of shoot length was not that significant. No discernible difference was observed for seedlings grown in the control medium (Fig 5C and 5D), indicating that the salt-caused retarded growth on WT plants was more severe than on transgenic plants.
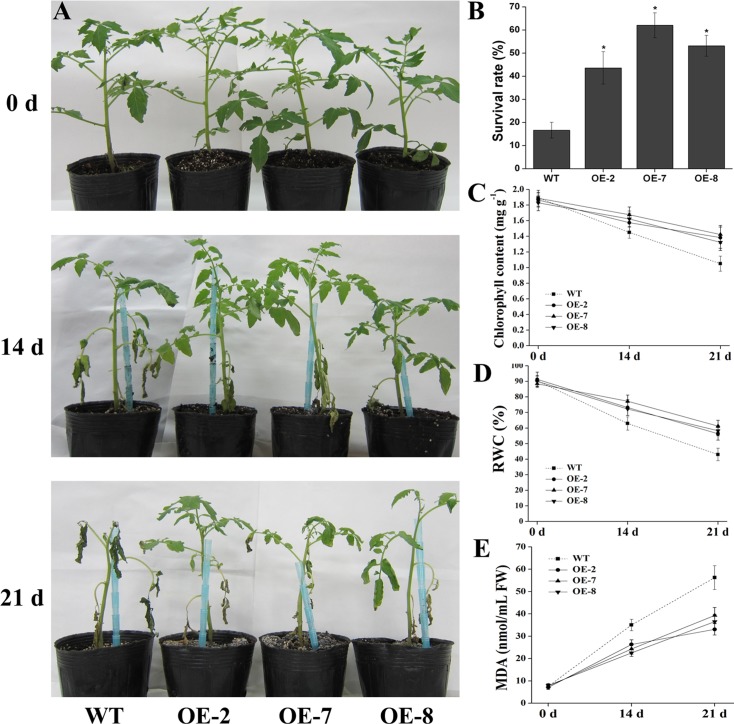
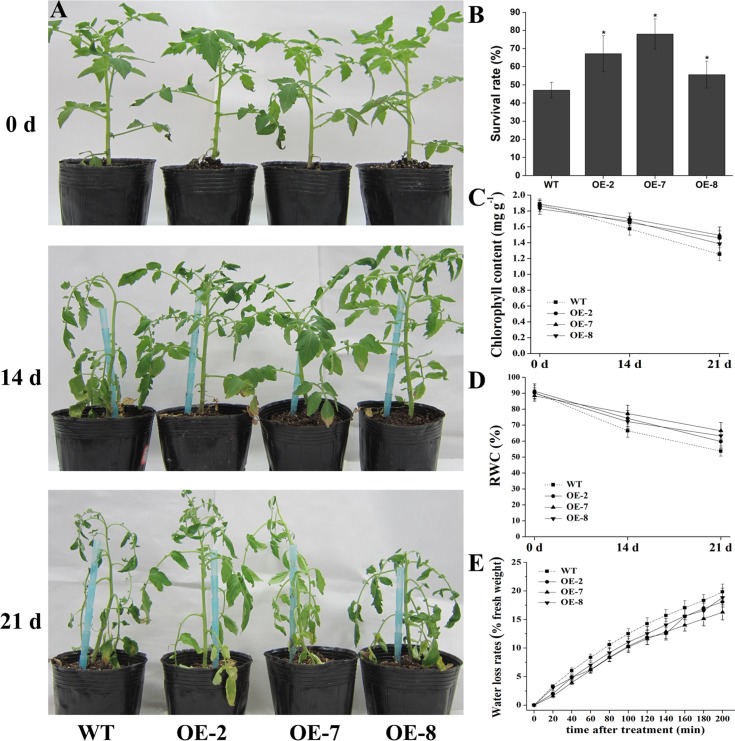
To further evaluate the performance of SlDEAD31-overexpressing plants under salt stress in soil, 8-week-old plants of WT and transgenic lines were irrigated with water containing 400 mM NaCl (200 mL) every 72 h. Under normal growth conditions, the transgenic plants showed no abnormal morphological phenotype compared with WT plants (Fig 6A). However, WT plants showed more notorious and quick damage signs during all the time course of the salt assay. NaCl-induced symptoms were observable 14 d after salt stress with chlorosis and wilting of lower leaves in both WT and transgenic plants, whereas the transgenic lines displayed apparently less wilting and necrosis, and most of the leaves remained vigorous. After 21 d, all the leaves of WT plants showed severe necrosis and wilting, and some were dead, whereas the upper leaves of transgenic plants remained green and vigorous (Fig 6A). Besides, most of the WT plants were dead 5 d after stopping the treatment, whereas 43–62% of the transgenic plants survived (Fig 6B).
Fig 6. Salt stress tolerance of SlDEAD31-overexpressing plants.
(A) Growth characteristics of WT and transgenic tomato plants at 0, 14 and 21 d after salt stress. 8-week-old plants were irrigated with 200 mL 400 mM NaCl every 72 h from the bottom of the pots. Representative plants are shown. (B) Survival rate of the plants shown after 21 d of salt treatment and 5 d of recovery. Each bar represents an average of eight plants ± SE (n = 3). Asterisks indicate a significant difference between WT and transgenic lines (P<0.05). (C)-(E) Comparisons of chlorophyll content (C), relative water content (RWC, D), and malondialdehyde (MDA) content (E) of WT and transgenic plants at 0, 14 and 21 d after salt stress. Data represent the means ± SE (n = 3).
Salt damage was evaluated by measuring chlorophyll content, relative water content (RWC) and malondialdehyde (MDA) content, which are typical physiological parameters for evaluating abiotic stress tolerance in crop plants. Under normal conditions, these physiological parameters were similar for transgenic and WT plants (Fig 6C–6E). Upon exposure to salt stress, there were obvious reductions in chlorophyll content in the leaves of both WT and transgenic tomato plants. WT plants lost about 23 and 43% of the chlorophyll at 14 and 21 d after salt assay, respectively. However, in SlDEAD31-overexpressing plants this reduction was about 11–16 and 24–29%, respectively (Fig 6C). RWC in transgenic plants decreased to 72–77% of the normal levels at 14 d, and to 56–61% at 21 d post-treatment. Whereas this parameter in WT plants went down to about 63 and 43% at 14 and 21 d post-treatment, respectively (Fig 6D). The MDA content, which is an indicator of lipid peroxidation, was increased by the salt treatment, but the degree of increase in transgenic plants was lower than in WT plants (Fig 6E), suggesting that overexpression of SlDEAD31 induces potential antioxidative processes preventing significant membrane damage.
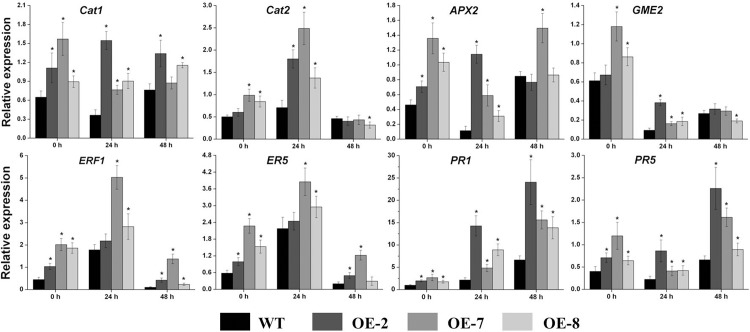
Stress-Related Genes Are Upregulated in SlDEAD31-Overexpressing Tomato Plants
To gain further information concerning the increased salt tolerance phenotype observed in SlDEAD31-overexpressing tomato plants, transcripts of previously reported stress-related genes were utilized for comparative analysis between WT and transgenic tomato roots under normal and salt-stressed conditions. The key ascorbic acid (AsA) synthetase gene GME2 [30]; two catalase (CAT) genes, Cat1 and Cat2 [31]; an ascorbate peroxidase (APX) gene, APX2 [32]; an ethylene-responsive LEA gene (ER5) [33]; the ethylene-responsive factor ERF1 [34], and two pathogenesis-related (PR) genes, PR1 and PR5 [35] were selected (Fig 7). Based on the reported literature, these stress tolerance genes are involved in the regulation of biotic and abiotic stress responses (such as PR1, PR5, ER5, and ERF1), production of osmolytes (such as GME2), detoxification and redox homeostasis (such as Cat1, Cat2, and APX2). Quantitative RT-PCR analysis showed that the transcripts of Cat1, ERF1, ER5, PR1, and PR5 were significantly elevated in almost all the overexpression lines under both normal and salt-stressed conditions, although the fold changes showed some variation for some genes among the three transgenic lines (Fig 7). Relatively higher expression levels of Cat2, APX2, and GME2 were detected mainly under normal conditions and at 24 h following salt stress in overexpression lines. The transcripts of these stress-related genes were also analyzed in the leaves of WT and transgenic plants, the results showed that their induction levels were less significant than in roots, as expected (taking into account that the expression of SlDEAD31 was only induced in roots by NaCl stress, Fig 4D, S4 Fig).
Fig 7. The relative expression levels of biotic and abiotic stress-related genes of WT and SlDEAD31-overexpressing plants under normal and salt stress conditions.
8-week-old plants were irrigated with water containing 400 mm NaCl (200 mL) from the bottoms of the pots, then root samples were harvested after 24 and 48 h. Seedlings harvested before the salt stress were used as controls. Bars represent mean relative expression values ± SE (n = 3). Asterisks indicate a significant difference (P<0.05) between WT and transgenic plants.
Overexpression of SlDEAD31 in Tomato Plants Slightly Improves Drought Resistance
To further test the regulatory effects of SlDEAD31 on drought resistance, a drought assay was performed by withholding water for 21 d in soil and then physiological parameters were measured. The leaves of WT plants started to show typical drought-induced symptoms after 14 d of drought stress, such as leaf rolling and wilting with a concomitant loss of chlorophyll (Fig 8A). Whereas the transgenic plants showed delayed leaf-rolling compared with the WT, only slight symptoms of drought-induced damage were observed, and most of the leaves remained vigorous. Only slight differences were observed on the 21th day after drought stress, almost all the leaves of WT plants displayed severe wilting and curling, while slightly less withered and rolled leaves were observed in transgenic plants (Fig 8A). In addition, after rewatering the plants for 5 d, 55–78% of SlDEAD31 transgenic plants recovered from the drought stress, while about 47% of the WT plants recovered (Fig 8B).
Fig 8. Drought tolerance testing of SlDEAD31-overexpressing plants.
A water withholding assay was performed with 8-week-old plants for up to 21 d followed by a recovery period of 5 d. (A) Representative phenotypes of wild-type and SlDEAD31 overexpression plants at 0, 14 and 21 d after initiation of drought assay. (B) Survival rates of WT and transgenic plants after rewatering for 5 d following 21 d of drought assay. Each bar represents an average of eight plants ± SE. Asterisks indicate a significant difference from WT (p < 0.05). (C) and (D) Comparisons of chlorophyll content (C) and relative water content (RWC, D) of WT and transgenic plants at 0, 14 and 21 d after drought treatment. Values represent the means ± SE (n = 3). (E) Water loss rates of WT and transgenic plants. Similar leaves were excised and weighed at the indicated times. Water loss was calculated as the percentage of initial fresh weight. At least 15 leaves of similar developmental stages were excised and weighed at different times after the detachment. The data represent the means ± SE (n = 3).
To further verify the drought tolerance phenotype, the chlorophyll and RWC values of WT and transgenic plants were monitored in leaves during the drought assay. Apparent chlorophyll degradation was observed in both WT and transgenic plants under drought stress, while the reduction in WT was higher than that in transgenic plants both at 14 and 21 d of the drought assay (Fig 8C). RWC in WT plants decreased to about 67 and 54% at 14 and 21 d post-treatment, respectively. Whereas in transgenic lines, this parameter went down to about 73–79% at 14 d and to 60–67% at 21 d following the same drought stress, respectively (Fig 8D). In addition, the water loss rate of detached leaves, which is also an important physiological parameter to investigate plants’ drought tolerance, was slightly lower in SlDEAD31-overexpressing plants compared to the WT at each dehydration time point (Fig 8E).
Discussion
Agricultural productivity is subjected to increasing environmental constraints, particularly to drought and salt stress due to their high magnitude of impact and wide distribution. The Solanaceae family, which includes tomato, potato, pepper, and tobacco, represents one of the most valuable families of vegetable crops. Cultivated tomatoes are susceptible to a wide range of environmental stresses, for example, most tomato cultivars being moderately sensitive to drought and salt stress during seed germination, seedling emergence, vegetative growth, and reproduction, thus fail to produce high yields in a fragile ecosystem [36]. Bioengineering stress-signaling pathways to develop stress-tolerant crops is one of the major goals of agricultural research. The involvement and significance of RNA helicases in response to biotic and abiotic stresses have only recently begun to emerge, and Vashisht and Tuteja (5] suggested that stress-responsive DEAD-box helicases have uncovered a new pathway to engineer plant stress tolerance. In the present study, we characterized two tomato DEAD-box genes, SlDEAD30 and SlDEAD31, which each encodes a putative DEAD-box RNA helicase with all the conserved domains and motifs that are characteristic of DEAD-box proteins. Our phylogenetic analysis showed that they cluster together with stress-related DEAD-box proteins, which was further supported by the induction of SlDEAD30 and SlDEAD31 by multiple abiotic stresses (Figs 2 and 4), and that overexpression of SlDEAD31 significantly enhanced salt tolerance and slightly improved drought resistance. Besides, MYB, MYC, and WRKY transcription factors have been suggested to be involved in responses and adaptation to biotic and/or abiotic stress in plants [37, 38]. Thus, the presence of these stress responsive cis-acting elements also suggests critical roles of these two genes in stress tolerance. Moreover, phylogenetic analyse indicated that a cross-talk may exist between stress response and plant development in DEAD-box RNA helicases. For example, in the stress-related subgroup, the Arabidopsis LOS4 plays a role in chilling tolerance linked with other developmental processes such as flowering or vernalization [19, 20]. Arabidopsis RH3 gene plays a critical role in chloroplast development, while rh3 mutant was more sensitive to NaCl stress [28]. Similarly, the high or constant expression of SlDEAD30 and SlDEAD31 in specific tissues suggest that they may also have a specialized function in plant growth and/or reproductive development, while further studies are needed.
Evidence that SlDEAD31 plays important roles in salt and drought tolerance, as demonstrated by its inducible expression by multiple abiotic stress treatments, including NaCl and dehydration, as well as by phenotypes, physiological indices, and significantly enhanced expression of multiple stress-related genes in the SlDEAD31-overexpressing tomato plants, was presented. Similarly, there are several examples in which similar effects have been demonstrated through the overexpression of stress-responsive DEAD-box helicase genes that can improve the stress tolerance of transgenic plants. For example, OsBIRH1-overexpressing transgenic rice showed an enhanced disease resistance and oxidative stress tolerance [17]. PDH45 was proven to be induced by various abiotic stresses including NaCl, and its overexpression in rice confers salinity tolerance and improved physiological traits [39].
The establishment of seedlings at early growth stages, which is one of the most important determinants for crop production, is severely affected by soil salinity. Therefore, vigorous early growth under salty soils is preferred. In our present study, we observed a better growth of SlDEAD31-transgenic lines compared to WT plants (Fig 5C and 5D), indicating enhanced salt tolerance during this sensitive and critical stage. In addition, environmental stresses often cause physiological changes in plants. Physiological indices, including chlorophyll content, RWC and MDA, are typical and effective physiological parameters for evaluating abiotic stress tolerance and resistance in crop plants. Chlorophyll content, expressing the retention of chlorophyll, is taken as an index of the damage done to the photosynthetic apparatus. Leaf RWC, expressing the relative amount of water present in the plant tissues, is a useful indicator of plant water balance. Adverse stress conditions often generate reactive oxygen species (ROS), thus inducing oxidative damage to macromolecules such as membrane lipids. The content of MDA can serve as an indicator of oxidative damage in the cell [40]. Our results showed that both the RWC and chlorophyll content of transgenic lines were higher than in WT plants under salt- and drought-stress conditions (Figs 6 and 8), suggesting that SlDEAD31-overexpressing plants had more robust photosynthetic capabilities and better water balance than the WT. In addition, transgenic tomato plants showed lower MDA content than the WT plants under salt stress (Fig 6E), which may result from an enhanced capacity for scavenging ROS. Water loss rate is an important parameter of plants under water-deficit conditions and has been proposed as an indicator of water status [41]. Therefore, the slightly enhanced drought tolerance of transgenic plants could be attributed, at least in part, to their lower transpiration rates (Fig 8E). In addition, SlDEAD31-overexpressing plants showed significantly higher survival rates than WT plants under both stresses (Figs 6B and 8B), further supporting the usefulness of SlDEAD31 in improving salt and drought resistance in tomato.
The expression of multiple abiotic stress-related genes was significantly upregulated in the SlDEAD31-overexpressing tomato plants (Fig 7), something considered to be advantageous for the resistance to abiotic stress, also indicating the protective role of SlDEAD31 against environmental stresses. The expression of stress-related genes results in the accumulation of metabolites and the alteration of biochemical and physiological pathways that are vital for the plant adaptation to adverse stress conditions. Our results are in a good agreement with previous report, in which enhanced tolerance to various stresses is mainly attributed to increased expression of biotic and abiotic stress-related genes in the OsBIRH1-overexpressing plants [17]. In plants, biotic and abiotic stresses can trigger the generation of ROS, which include hydrogen peroxide (H2O2) that can oxidize and damage cellular components [42]. The ROS defense mechanisms involve the synthesis of antioxidant compounds (such as AsA) and antioxidant enzymes (such as CAT1/2 and APX2). An increase in AsA content can confer tolerance to various stresses including salt, ozone, and chilling [30]. The upregulated expression of GME2 was detected in the transgenic plants suggesting an elevated AsA content, and thus conferring tolerance to salt stress in SlDEAD31-overexpressing plants. In this context, CAT and APX can detoxify ROS by converting H2O2 to H2O in plant cells under oxidative stress [31, 32]. Therefore, the enhanced tolerance observed in transgenic tomato plants might be due to the upregulation of APX2 and Cat1/2, and thus an increased protection against ROS-induced damages. In addition, a significant upregulation in ERF1 expression was observed in transgenic plants. It has been shown that overexpression of ERF1 provides improved drought tolerance in transgenic tomato [34]. We have reported previously that overexpression of SlERF5 in tomato promotes adaptation to drought and salt tolerance [43]. Furthermore, ER5, an ethylene-responsive LEA gene confers desiccation tolerance [33], was also increased in transgenic plants, indicating a less damage of the transgenic lines under abiotic stress. Interestingly, transcripts encoding pathogenesis-related proteins, PR1 and PR5, were also upregulated in transgenic lines. Similarly, they were also elevated in the OsBIRH1-overexpressing Arabidopsis plants [17]. Thus our results imply that SlDEAD31, besides having a role in plant adaptation to abiotic stress, may also integrates signals derived from both biotic and abiotic stresses in tomato, although this remains to be elucidated in detail. Collectively, our data indicate that biotic or abiotic stresses can induce the expression of SlDEAD31, leading to the upregulation of transcripts encoding functional and regulatory proteins involved in antistress metabolism. Therefore, their consistent upregulated expression likely increases the expression of downstream genes, probably enhancing integrative tolerance to multiple stresses due to cross-talk between various environmental stresses, especially salt and drought [44, 45].
In conclusion, the present study provides a preliminary characterization of SlDEAD30 and SlDEAD31 genes in tomato, and suggests that stress-responsive SlDEAD31 functions in positive modulation of abiotic stress tolerance. Presently, the functions of RNA helicases are still poorly understood in plants. Clearly, understanding the role of SlDEAD31 will not only extend knowledge of the biological function of the DEAD-box RNA helicases, but will also provide insight into the significance of DEAD-box RNA helicase-mediated metabolism in regulation of plant defence responses. Although the detailed mechanisms underlying the functions of SlDEAD31 remain to be completely revealed, for example, what role SlDEAD31 may play in plant growth and development, how SlDEAD31 protein regulates the expression of stress-related genes, and how SlDEAD31 operates different stress signaling pathways in tomato, the data we present here indicate that overexpression of SlDEAD31 confers enhanced salt and drought stress tolerance of tomato. Thus, SlDEAD31 may hold a promising application in the production of stress-tolerant plants.
Materials and Methods
Identification and Isolation of SlDEAD Genes
To identify DEAD-box genes in tomato, the protein sequences of AtRHs (RNA helicase genes) from Arabidopsis were used as driver sequences to search the database of Sol Genomics Network (SGN, http://solgenomics.net/), which is a clade-oriented database containing biological data for the Solanaceae. Different AtRHs were used to interrogate the database, and the returned results were compared to each other. Only the sequences that contain the DEAD-box helicases superfamily domains and all the nine conserved RNA helicase motifs were selected and used for further analyse. For the two gene predictions selected eventually, SlDEAD30 and SlDEAD31, gene-specific primers (S3 Table) were designed for PCR to isolate the cDNA, and their nucleotide and deduced amino acid sequence data have been deposited in GenBank under the accession numbers KJ739798 and KJ713393, respectively.
DNA and Protein Sequence Analysis
The translation of SlDEAD genes using the ExPASy translate tool, grand average of hydropathicity (GRAVY) and theoretical molecular weight (Mw) were calculated using the ExPASy ProtParam tool (http://www.expasy.org/). Conserved structure domains were annotated according to the ScanProsite (http://prosite.expasy.org/scanprosite/). Multiple sequences alignment and phylogenetic tree were conducted using the DNAMAN 5.2.2 program and MEGA 5.2 software, respectively. To analyze putative cis-elements in the promoter regions of SlDEAD30 and SlDEAD31 genes, promoter sequences (1000 bp regions upstream the 5’ end of the predicted ORFs) of both genes were extracted from SGN database and searched against the promoter database PLACE (http://www.dna.affrc.go.jp/PLACE/index.html) [46].
Plant Materials for Organ-Specific Expression
The wild-type (WT) tomato (Solanum lycopersicum Mill. cv. Ailsa Craig) seeds were germinated and the seedlings were grown in a greenhouse under sodium lights timed for 16 h days (27°C) and 8 h nights (19°C). For organ-specific expression profiling of SlDEAD30 and SlDEAD31 genes, the roots, stems, flowers, sepals, fruits, and leaves of similar age and position at different stages were collected. Flowers were tagged at anthesis and fruit development was recorded as days post-anthesis (DPA). The fruit ripening stages were divided into IMG (immature green, 28 DPA), MG (mature green, 35 DPA, full fruit expansion but no visible color change), B (breaker, fruit showing the first signs of ripening-associated color change from green to yellow), B+4 (4 d after breaker), and B+7 (7 d after breaker). All plant samples for the preparation of total RNA were taken at the same time each day, frozen in liquid nitrogen and stored at -80°C until required.
Plant Stress Treatments
To verify the expression profiles of SlDEAD30 and SlDEAD31 genes under abiotic stress treatments, WT tomato seeds were germinated and the seedlings were grown in greenhouses. All the plant treatments were performed using potted 35 day-old tomato seedlings. In each case, individual plants were used for each treatment with three biological replicates. Untreated seedlings were used as controls, and all leaf or root samples were harvested at 1, 2, 4, 8, 12, 24 h after each treatment. Heat and cold treatments were conducted by incubating the seedlings at 4 and 40°C, respectively. Salinity treatment was applied by submerging the roots of seedlings in a solution with 200 mM NaCl, then root and leaf samples were collected. For dehydration, whole tomato seedlings were gently pulled out, washed carefully with water to remove the soil, and left on a piece of dry filter paper. For wounding, the leaves of tomato seedlings were cut with a razor blade into small pieces and left on wetted filter paper in sealed pots at 25 ±1°C [43, 47]. All harvested samples were immediately frozen in liquid nitrogen and stored at -80°C until RNA extraction.
RNA Extraction and Quantitative RT-PCR Analysis
Total RNA was isolated from different plant tissues accordingly to the protocol of the Trizol reagent (Invitrogen, Shanghai, China). 2 μg of RNA were predigested with Dnase-I and reverse-transcribed using M-MLV reverse transcriptase (Promega, Beijing, China) with oligo(dT)18 primer. The quantitative RT-PCR reaction consisted of 5 μL 2× GoTaq qPCR Master Mix (Promega, Beijing, China), 0.25 μL 10 mM each primer, 1 μL cDNA and distilled water to a final volume of 10 μL. Quantitative RT-PCR was performed using the CFX96 Real-Time System (Bio-Rad, USA) using a two-step method: 95°C 2 min, followed by 40 cycles of 95°C 15 s, 60°C 1 min and then a melting curve was generated and analyzed. The tomato CAC gene was selected as internal standard for organ-specific expression [48], and the tomato EF1α gene was used as internal control under abiotic stress [49]. The comparative 2-ΔΔCT method was used to analyze the relative gene expression levels [50]. Gene-specific primers used for quantitative RT-PCR are listed in S3 Table, and a standard curve by serial dilution was analyzed for each specific gene using WT tomato cDNA.
Overexpression Vector Construction and Tomato Transformation
The coding region of SlDEAD31 gene was amplified from WT tomato cDNA using the primers Ov-SlDEAD31-F/R (S3 Table), with PrimeSTAR HS DNA polymerase (TaKaRa, Dalian, China) under the following PCR procedure: 98°C for 5 min followed by 35 cycles of 30 s at 98°C, 15 s at 56°C and 3 min at 72°C, the PCR products were confirmed by DNA sequencing. Then SlDEAD31 was cloned into the plant expression vector pBIN121 between the Xba I and Sac I sites, under the control of the cauliflower mosaic virus 35S promoter. Afterwards, the resultant construct pBIN121-SlDEAD31 was transformed into tomato cv Ailsa Craig by Agrobacterium tumefaciens (strain LBA4404) by the freeze-thaw method [51]. Positive transgenic lines were screened for kanamycin (50 mg L−1) resistance. Genomic DNA of transgenic tomato plants was extracted according to the manufacturer's protocol of Genomic DNA Extraction Kit (Invitrogen, Shanghai, China), and was used for the detection of the presence of T-DNA by PCR with the primers NPTII-F/R (S3 Table). T2 seeds from the selected transgenic lines were germinated on a medium with kanamycin (50 mg L−1) and homozygous plants were selected for further study.
Salinity and Drought Stress Tolerance Assays
Seeds of WT and homozygous T2 SlDEAD31-overexpressing tomato plants were surface-sterilized and sown on the MS medium. Then WT and transgenic tomato seeds of consistent germination were selected and transferred to MS medium containing 0 and 100 mM NaCl. Culture vessels containing the seeds were incubated in a growth chamber for 10 d. Then the seedlings were scanned and used for shoot and root growth measurements by ImageJ software. For high-salinity stress in soil, WT and transgenic seedlings were pre-grown under normal conditions for 15 d and transferred to a greenhouse. Then 8-week-old plants of uniform size were used for the salt tolerance assay. The plants were irrigated from the bottom of the pots with water containing 400 mm NaCl (200 mL) every 72 h for 21 d, followed by irrigating normal water for 5 d. Then the plants whose upper leaves were alive were considered to survive. Pictures were taken to record the phenotypes.
For the drought tolerance assay, 8-week-old WT and homozygous T2 transgenic tomato plants were used. The drought treatment consisted of withholding water for up to 21 d, then plants were rewatered for 5 d, and the plants that could recover were considered to survive. Pictures were taken to record the phenotypes.
Evaluation of Stress Tolerance
The measurement of RWC was performed as in our previous report [43]. For chlorophyll (Chl) measurement, each leaf sample was randomly selected from three individual plants, then weighed, ground in liquid nitrogen, and extracted with 10 mL 80% aqueous acetone (v/v). The extract was centrifuged at 4000 g for 5 min and the absorbance of the supernatants at 663 and 646 nm was recorded using a lambda 900 scanning spectrophotometer (PerkinElmer). Total chlorophyll content was calculated using the formulas according to the method of Wellburn [52]: Total Chl (μg mL-1) = 20.29A646+8.02A663.
To measure the water loss rate under dehydration conditions, leaves of a similar size, age and position were detached from WT and transgenic plants, and placed on dry filter papers. The relative humidity was ~45% at room temperature during the dehydration period. The leaves were weighed every 20 min for 200 min.
The level of lipid peroxidation was measured in terms of malondialdehyde (MDA) content, a product of lipid peroxidation. Fresh tissue (0.2 g) was ground in liquid nitrogen and homogenised in 4 mL of 0.1% trichloroacetic acid (TCA). The homogenate was centrifuged at 15,000 g for 5 min. 4.0 mL of 20% TCA containing 0.5% thiobarbituric acid (TBA) was added to 1.0 mL aliquot of the supernatant. The mixture was incubated at 95°C in a hot water bath for 30 min and then the reaction was terminated in an ice bath. After centrifugation at 10,000 g for 10 min, the absorbance of the supernatant was recorded at 532 and 600 nm for the correction of non-specific turbidity. The MDA equivalent was calculated as follows [53]: MDA (nmol/mL FW) = ((A532-A600)/155, 000)×106.
Statistical Analysis
When appropriate, data were subjected to one-way analysis of variance (ANOVA) and differential expression data were regarded as statistically significant when passing the Dunnett’s test at P<0.05 level. In addition, we adopted an experiential threshold of twofold for analyzing stress induction or repression, and threefold for organ-specific expression. The expression levels were designated as ‘induced’, ‘repressed’ or ‘different’ only if such differences met the above criteria.
Supporting Information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We thank all laboratory members for help, advice and discussion.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the National Natural Science Foundation of China (31171968), the Natural Science Foundation of Chongqing of China (CSTC2013jcyjA0001), and the Fundamental Research Funds for the Central Universities (No. 106112015CDJZR235504). ZH received the funding. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Tuteja N, Ahmad P, Panda BB, Tuteja R. Genotoxic stress in plants: shedding light on DNA damage, repair and DNA repair helicases. Mutat Res. 2009;681(2):134–49. [DOI] [PubMed] [Google Scholar]
- 2. Schützendübel A, Polle A. Plant responses to abiotic stresses: heavy metal-induced oxidative stress and protection by mycorrhization. J Exp Bot. 2002;53(372):1351–65. [PubMed] [Google Scholar]
- 3. Zhu J, Dong C-H, Zhu J-K. Interplay between cold-responsive gene regulation, metabolism and RNA processing during plant cold acclimation. Curr Opin Plant Biol. 2007;10(3):290–5. [DOI] [PubMed] [Google Scholar]
- 4. Kazan K. Alternative splicing and proteome diversity in plants: the tip of the iceberg has just emerged. Trends Plant Sci. 2003;8(10):468–71. [DOI] [PubMed] [Google Scholar]
- 5. Vashisht AA, Tuteja N. Stress responsive DEAD-box helicases: a new pathway to engineer plant stress tolerance. J Photochem Photobiol B: Biol. 2006;84(2):150–60. [DOI] [PubMed] [Google Scholar]
- 6. Rocak S, Linder P. DEAD-box proteins: the driving forces behind RNA metabolism. Nat Rev Mol Cell Bio. 2004;5(3):232–41. [DOI] [PubMed] [Google Scholar]
- 7. Linder P. Dead-box proteins: a family affair—active and passive players in RNP-remodeling. Nucleic Acids Res. 2006;34(15):4168–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tuteja N. Plant DNA helicases: the long unwinding road. J Exp Bot. 2003;54(391):2201–14. [DOI] [PubMed] [Google Scholar]
- 9. Tanner NK, Linder P. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol Cell. 2001;8(2):251–62. [DOI] [PubMed] [Google Scholar]
- 10. Linder P, Owttrim GW. Plant RNA helicases: linking aberrant and silencing RNA. Trends Plant Sci. 2009;14(6):344–52. 10.1016/j.tplants.2009.03.007 [DOI] [PubMed] [Google Scholar]
- 11. Boudet N, Aubourg S, Toffano-Nioche C, Kreis M, Lecharny A. Evolution of intron/exon structure of DEAD helicase family genes in Arabidopsis, Caenorhabditis, and Drosophila . Genome Res. 2001;11(12):2101–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. [DOI] [PubMed] [Google Scholar]
- 13. Owttrim GW. RNA helicases and abiotic stress. Nucleic Acids Res. 2006;34(11):3220–30. 10.1093/nar/gkl408 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Umate P, Tuteja R, Tuteja N. Genome-wide analysis of helicase gene family from rice and Arabidopsis: a comparison with yeast and human. Plant Mol Biol. 2010;73(4–5):449–65. 10.1007/s11103-010-9632-5 [DOI] [PubMed] [Google Scholar]
- 15. Liu H, Liu J, Fan S, Song M, Han X, Liu F, et al. Molecular cloning and characterization of a salinity stress-induced gene encoding DEAD-box helicase from the halophyte Apocynum venetum . J Exp Bot. 2008;59(3):633–44. 10.1093/jxb/erm355 [DOI] [PubMed] [Google Scholar]
- 16. Kant P, Kant S, Gordon M, Shaked R, Barak S. STRESS RESPONSE SUPPRESSOR1 and STRESS RESPONSE SUPPRESSOR2, two DEAD-box RNA helicases that attenuate Arabidopsis responses to multiple abiotic stresses. Plant Physiol. 2007;145(3):814–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li D, Liu H, Zhang H, Wang X, Song F. OsBIRH1, a DEAD-box RNA helicase with functions in modulating defence responses against pathogen infection and oxidative stress. J Exp Bot. 2008;59(8):2133–46. 10.1093/jxb/ern072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang Y, Duby G, Purnelle B, Boutry M. Tobacco VDL gene encodes a plastid DEAD box RNA helicase and is involved in chloroplast differentiation and plant morphogenesis. Plant Cell. 2000;12(11):2129–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gong Z, Lee H, Xiong L, Jagendorf A, Stevenson B, Zhu J-K. RNA helicase-like protein as an early regulator of transcription factors for plant chilling and freezing tolerance. P Natl Acad Sci USA. 2002;99(17):11507–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gong Z, Dong C-H, Lee H, Zhu J, Xiong L, Gong D, et al. A DEAD box RNA helicase is essential for mRNA export and important for development and stress responses in Arabidopsis . Plant Cell. 2005;17(1):256–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim JS, Kim KA, Oh TR, Park CM, Kang H. Functional characterization of DEAD-box RNA helicases in Arabidopsis thaliana under abiotic stress conditions. Plant Cell Physiol. 2008;49(10):1563–71. 10.1093/pcp/pcn125 . [DOI] [PubMed] [Google Scholar]
- 22. Zouine M, Latché A, Rousseau C, Regad F, Pech J-C, Philippot M, et al. The tomato genome sequence provides insights into fleshy fruit evolution. Nature. 2012;485:635–41. 10.1038/nature11119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Spooner DM, Peralta IE, Knapp S. Comparison of AFLPs with other markers for phylogenetic inference in wild tomatoes [Solanum L. section Lycopersicon (Mill.) Wettst.]. Taxon. 2005:43–61. [Google Scholar]
- 24. Xu R, Zhang S, Lu L, Cao H, Zheng C. A genome-wide analysis of the RNA helicase gene family in Solanum lycopersicum . Gene. 2012;513(1):128–40. 10.1016/j.gene.2012.10.053 [DOI] [PubMed] [Google Scholar]
- 25. Liu M, Shi DQ, Yuan L, Liu J, Yang WC. SLOW WALKER3, encoding a putative DEAD-box RNA helicase, is essential for female gametogenesis in Arabidopsis . Journal of Integrative Plant Biology. 2010;52(9):817–28. 10.1111/j.1744-7909.2010.00972.x [DOI] [PubMed] [Google Scholar]
- 26. Tripurani SK, Nakaminami K, Thompson KB, Crowell SV, Guy CL, Karlson DT. Spatial and temporal expression of cold-responsive DEAD-box RNA helicases reveals their functional roles during embryogenesis in Arabidopsis thaliana . Plant Mol Biol Rep. 2011;29(4):761–8. [Google Scholar]
- 27. Jacobsen SE, Running MP, Meyerowitz EM. Disruption of an RNA helicase/RNAse III gene in Arabidopsis causes unregulated cell division in floral meristems. Development. 1999;126(23):5231–43. [DOI] [PubMed] [Google Scholar]
- 28. Lee KH, Park J, Williams DS, Xiong Y, Hwang I, Kang BH. Defective chloroplast development inhibits maintenance of normal levels of abscisic acid in a mutant of the Arabidopsis RH3 DEAD-box protein during early post-germination growth. Plant J. 2013;73(5):720–32. 10.1111/tpj.12055 [DOI] [PubMed] [Google Scholar]
- 29. Kaveh H, Nemati H, Farsi M, Jartoodeh S. How salinity affect germination and emergence of tomato lines. J Biol Environ Sci. 2011;5:159–63. [Google Scholar]
- 30. Zhang C, Liu J, Zhang Y, Cai X, Gong P, Zhang J, et al. Overexpression of SlGMEs leads to ascorbate accumulation with enhanced oxidative stress, cold, and salt tolerance in tomato. Plant Cell Rep. 2011;30(3):389–98. 10.1007/s00299-010-0939-0 [DOI] [PubMed] [Google Scholar]
- 31. Bussink HJ, Oliver R. Identification of two highly divergent catalase genes in the fungal tomato pathogen, Cladosporium fulvum . Eur J Biochem. 2001;268(1):15–24. [DOI] [PubMed] [Google Scholar]
- 32. Najami N, Janda T, Barriah W, Kayam G, Tal M, Guy M, et al. Ascorbate peroxidase gene family in tomato: its identification and characterization. Mol Genet Genomics. 2008;279(2):171–82. [DOI] [PubMed] [Google Scholar]
- 33.Bernadac A, Audran C, Bouzayen M. ER5, a tomato LEA-like gene induced by ethylene and ABA confers desiccation tolerance to seeds. In: Vendrell M, Klee H, Pech JC, Romojaro F, editors. Biology and Biotechnology of the Plant Hormone Ethylene Iii. Nato Science Series, Sub-Series I: Life and Behavioural Sciences. 3492003. p. 118–9.
- 34. Lu C, Li Y, Chen A, Li L, Zuo J, Tian H, et al. LeERF1 improves tolerance to drought stress in tomato (Lycopersicon esculentum) and activates downstream stress-responsive genes. Afr J Biotechnol. 2013;9(38):6294–300. [Google Scholar]
- 35. Lim CJ, Kim WB, Lee B-S, Lee HY, Kwon T-H, Park JM, et al. Silencing of SlFTR-c, the catalytic subunit of ferredoxin: thioredoxin reductase, induces pathogenesis-related genes and pathogen resistance in tomato plants. Biochem Biophys Res Commun. 2010;399(4):750–4. 10.1016/j.bbrc.2010.08.016 [DOI] [PubMed] [Google Scholar]
- 36. Foolad MR. Current status of breeding tomatoes for salt and drought tolerance Advances in molecular breeding toward drought and salt tolerant crops: Springer; 2007. p. 669–700. [Google Scholar]
- 37. Eulgem T, Rushton PJ, Robatzek S, Somssich IE. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000;5(5):199–206. [DOI] [PubMed] [Google Scholar]
- 38. Yamaguchi-Shinozaki K, Shinozaki K. Organization of cis-acting regulatory elements in osmotic-and cold-stress-responsive promoters. Trends Plant Sci. 2005;10(2):88–94. [DOI] [PubMed] [Google Scholar]
- 39. Amin M, Elias SM, Hossain A, Ferdousi A, Rahman MS, Tuteja N, et al. Over-expression of a DEAD-box helicase, PDH45, confers both seedling and reproductive stage salinity tolerance to rice (Oryza sativa L.). Mol Breed. 2012;30(1):345–54. [Google Scholar]
- 40. Mittler R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002;7(9):405–10. [DOI] [PubMed] [Google Scholar]
- 41. Dhanda S, Sethi G. Inheritance of excised-leaf water loss and relative water content in bread wheat (Triticum aestivum). Euphytica. 1998;104(1):39–47. [Google Scholar]
- 42. Zhang W, Yang H, You S, Xu Y, Ran K, Fan S. Cloning, characterization and functional analysis of the role MhNCED3, a gene encoding 9-cis-epoxycarotenoid dioxygenase in Malus hupehensis Rehd., plays in plant tolerance to osmotic and Cd2+ stresses. Plant Soil. 2014;381(1–2):143–60. [Google Scholar]
- 43. Pan Y, Seymour GB, Lu C, Hu Z, Chen X, Chen G. An ethylene response factor (ERF5) promoting adaptation to drought and salt tolerance in tomato. Plant Cell Rep. 2012;31(2):349–60. 10.1007/s00299-011-1170-3 [DOI] [PubMed] [Google Scholar]
- 44. Knight H, Knight MR. Abiotic stress signalling pathways: specificity and cross-talk. Trends Plant Sci. 2001;6(6):262–7. [DOI] [PubMed] [Google Scholar]
- 45. Zhu JK. Salt and drought stress signal transduction in plants. Annu Rev Plant Biol. 2002;53:247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999;27(1):297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tang Y, Liu M, Gao S, Zhang Z, Zhao X, Zhao C, et al. Molecular characterization of novel TaNAC genes in wheat and overexpression of TaNAC2a confers drought tolerance in tobacco. Physiol Plant. 2012;144(3):210–24. 10.1111/j.1399-3054.2011.01539.x [DOI] [PubMed] [Google Scholar]
- 48. Expósito-Rodríguez M, Borges AA, Borges-Pérez A, Pérez JA. Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biology. 2008;8(1):131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nicot N, Hausman J-F, Hoffmann L, Evers D. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J Exp Bot. 2005;56(421):2907–14. [DOI] [PubMed] [Google Scholar]
- 50. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using Real-Time puantitative PCR and the 2-ΔΔCT Method. Methods. 2001;25(4):402–8. [DOI] [PubMed] [Google Scholar]
- 51. An G. Binary ti vectors for plant transformation and promoter analysis. Methods Enzymol. 1987;153:292–305. [Google Scholar]
- 52. Wellburn AR. The spectral determination of chlorophylls a and b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. J Plant Physiol. 1994;144(3):307–13. [Google Scholar]
- 53. Heath RL, Packer L. Photoperoxidation in isolated chloroplasts: I. Kinetics and stoichiometry of fatty acid peroxidation. Arch Biochem Biophys. 1968;125(1):189–98. [DOI] [PubMed] [Google Scholar]
- 54. Guan Q, Wu J, Zhang Y, Jiang C, Liu R, Chai C, et al. A dead box RNA helicase is critical for pre-mRNA splicing, cold-responsive gene regulation, and cold tolerance in Arabidopsis . Plant Cell. 2013;25(1):342–56. 10.1105/tpc.112.108340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Western TL, Cheng Y, Liu J, Chen X. HUA ENHANCER2, a putative DExH-box RNA helicase, maintains homeotic B and C gene expression in Arabidopsis . Development. 2002;129(7):1569–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yoine M, Nishii T, Nakamura K. Arabidopsis UPF1 RNA helicase for nonsense-mediated mRNA decay is involved in seed size control and is essential for growth. Plant Cell Physiol. 2006;47(5):572–80. [DOI] [PubMed] [Google Scholar]
- 57. Macovei A, Vaid N, Tula S, Tuteja N. A new DEAD-box helicase ATP-binding protein (OsABP) from rice is responsive to abiotic stress. Plant Signaling & Behavior. 2012;7(9):1138–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nakamura T, Muramoto Y, Yokota S, Ueda A, Takabe T. Structural and transcriptional characterization of a salt-responsive gene encoding putative ATP-dependent RNA helicase in barley. Plant Sci. 2004;167(1):63–70. [Google Scholar]
- 59. Chung E, Cho CW, Yun BH, Choi HK, So HA, Lee SW, et al. Molecular cloning and characterization of the soybean DEAD-box RNA helicase gene induced by low temperature and high salinity stress. Gene. 2009;443(1):91–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.