Abstract
Objective
E26 transformation specific sequence 1 (ETS-1) belongs to the ETS family of transcription factors that regulate the expression of various immune-related genes. Increasing evidence indicates that ETS-1 could contribute to the pathogenesis of autoimmune disease. Recent research has provided evidence that ETS-1 might correlate with rheumatoid arthritis (RA), but it's not clearly defined. In this study, we aimed to identify whether polymorphisms of ETS-1 play a role in Rheumatoid arthritis (RA) susceptibility and development in Chinese Han population.
Methods
Four single nucleotide polymorphisms (SNPs) within ETS-1 were selected based on HapMap data and previous associated studies. Whole blood and serum samples were obtained from 158 patients with RA and 192 healthy subjects. Genotyping was performed with polymerase chain reaction-high resolution melting (PCR-HRM) assay and the data was analyzed using SPSS17.0.
Results
A significantly positive correlation was observed between the SNP rs73013527 of ETS-1 and RA susceptibility, DAS28 and CRP (P<0.001, P = 0.001, and P = 0.028, respectively). Carriers of the haplotype CCT or TCT for rs4937333, rs11221332 and rs73013527 were associated with decreased risk of RA as compared to controls. No statistical significant difference was observed in the distribution of rs10893872, rs4937333 and rs11221332 genotypes between RA patients and controls.
Conclusions
Our data further supports that ETS-1 has a relevant role in the pathogenesis and development of RA. Allele T of rs73013527 plays a protective role in occurrence of RA but a risk factor in the high disease activity. Rs10893872, rs11221332 and rs4937333 are not associated with RA susceptibility and clinical features.
Introduction
Rheumatoid arthritis (RA) is a complicated autoimmune disease, whose clinical course ranges from mild joint swelling to severe poly-arthritis with progressive destruction of cartilage and bone. It is a serious pathological condition that can lead to incapacitation and decreased life expectancy compared with the normal population [1]. People suffering from RA weigh around 1% of world’s population [2]. In China, the prevalence of RA in the overall population is 0.32%-0.36%. What’s more, according to the data published by Ministry of Health of China, RA is one of the major chronic diseases in rural area of China. So far, the etiology of RA has yet to be clarified, but it has been established that genetic factors play a role in susceptibility to this condition [3], and the genetic variants may account for 50%–60% of the etiology [4].
To date, many genes have been identified to be associated with RA and the main genetic factor is the HLA-DRB1 gene, which accounts for one-third of the genetic liability to the disease [5]. Besides HLA-DRB1, non-HLA genes (e.g. PTPN22, NFAIP3, STAT4, TRAF1/C5, IL2RB, AFF3, CD40, CTLA4, MMEL1 and PADI4) have also been implicated in disease susceptibility, but many other genes still remain to be discovered [6]. Heo SH and their colleagues had confirmed that E26 transformation specific sequence 1 (ETS-1) was involved in the regulation of matrix metalloproteinases (MMPs) [7] which played a direct and important role in tissue destruction[8]. Recently, studies suggested that ETS-1 might be a crucial factor in the cytokine-mediated inflammatory and destructive cascade characteristic of RA [9, 10].
ETS-1 belongs to the ETS family of transcription factors. It mainly expresses in lymphoid cells and plays multiple roles in the lymphocyte development, function, apoptosis and inflammation [11–14]. Many viral and cellular genes, including those encoding growth factor genes and proto-oncogenes, are activated by ETS-1. At the same time, the regulation genes of some important immune functional proteins are activated (TCRα, Cd3δ, IL-5, Lck, c-fms, mb1, Igh, csf2) or repressed (TCRβ, IL-2) by ETS-1 as well [15, 16]. Recently, studies demonstrated that ETS-1 is a factor in causing the SLE in East Asian [17, 18]. Meanwhile, some single nucleotide polymorphisms (SNPs) of ETS-1 were identified as the susceptible variants of some immune diseases. For example, rs10893872, rs4937333 of ETS-1 were found as significant variants associated with SLE susceptibility [17, 19]. Rs10893872 may affect the genetic predisposition to pediatric uveitis [20] and rs11221332 is related with celiac disease and RA in Caucasians [10, 21]. Also, recent Genome-Wide Association studies (GWAS) revealed that rs73013527 associated with RA in Europeans [22]. All these diseases mentioned above are characterized by excessive activation of the immune system.
Therefore, ETS-1 is a very promising research field for understanding diseases of immune deficiency and autoimmunity. The aim of the present study is to identify the role of the ETS1 polymorphisms rs73013527, rs10893872, rs4937333 and rs11221332 in RA in Chinese Han population.
Materials and Methods
Participants
Our study recruited 350 Chinese Han subjects, including 158 RA patients and 192 healthy controls, from September 2012 to September 2014 in West China Hospital. All the subjects to be enrolled in the case-control study signed informed consents. All patients fulfilled rheumatoid arthritis criteria 2010 released by ACR/EULAR [23]. Controls were randomly selected by inviting people who lived in the same area as the patients to take part in the study. Individuals were considered healthy if their medical history did not reveal any chronic diseases, endemic infectious diseases or autoimmune diseases and their physical examination and blood tests prompt normal. Otherwise, any individual with one or more of these conditions was excluded from the control group. The study was approved by the Ethics Committee of West China Hospital.
Patient information
The following information was gathered from the patients’ medical records and collated on a form specifically prepared for the purpose: age, gender, disease duration (DD), age at onset (AO), painful joint and swollen joint. Moreover, many tests were done to get more information including rheumatoid factor (RF), anti-cyclic citrullinated peptide antibody (anti-CCP), erythrocyte sedimentation rate (ESR) and C-reactive protein (CRP) during assessment. Disease activity was determined according to the disease activity score for 28 painful/swollen joints (DAS28) [24].
ETS-1 polymorphisms genotyping
All the four SNPs (rs10893872, rs4937333, rs11221332, rs73013527) were genotyped using polymerase chain reaction-high resolution melting (PCR-HRM) and analysis performed on Light Cycler 480 (Roche Diagnostics, Penzberg, Bavaria, Germany). Genomic DNA kit (Biotake Corporation, Beijing, China) was used to extract the free circulating DNA from the blood sample and the concentration was measured by Nanodrop 2000c spectrophotometer (Thermo Scientific, DE). SNP genotyping was performed in a 20μL reaction system contains 10μL Roche Master Mix (Roche Applied Science) which comprises FastStart Taq DNA Polymerase and the High Resolution Melting Dye in a reaction buffer, 2.4μL 25 mM MgCl2, 0.2μL 10 mmol/L Forward Primer and 0.2μL 10 mmol/L Reverse Primer, 6.2μL deionized water and finally 1μL DNA sample as recommended by the manufacturer. The whole genotyping process encompasses four programmes, namely, pre-denaturation, amplification, high resolution melting and cooling. It was performed under the following conditions: an initial denaturation step at 95°C for 10 min, then continued with 50 cycles of 95°C for 15 s, touchdown cycling (decreasing 1°C/cycle), annealing in the range of 65–55°C for 10 s, and 72°C for 10 s. After the amplification phase, PCR products were denatured at 95°C for 1 min and cooled to 40°C for 1 min to form double-stranded DNA. Then the HRM analyses were performed by gradually increasing the temperature from 65 to 95°C at a rate of 0.01°C/s. After the melting process, the instrument was cooled down to 40°C. When finished, the results were analyzed by the corresponding Gene Scanning Software v1.2 (Roche Diagnostic) primarily based on the shape of the melting curve.
Laboratory assays
Serology markers of RA were analyzed using the following methods: RF and CRP were tested using Beckman Coulter IMMAGE 800 immunoassay (Beckman Coulter,Inc, CA, USA). Anti-CCP was analyzed by Elecsys Modular E170 immunoassay (Roche Diagnostics, GmbH, Mannheim, Germany). Anti-keratin antibody (AKA) was tested by indirect immunofluorescence. All the tests were conducted in accordance with manufacturers’ instruction.
Statistical analysis
Hardy-Weinberg equilibrium was independently appraised for each polymorphism. Demographic and clinical data between groups were compared by Student’s t test or Mann-Whitney U test for continuous variables, as appropriate. Haploview software was used to explore whether ETS-1 polymorphisms were in strong linkage disequilibrium (LD) or they independently contribute to the susceptibility of RA, and can capture additional significant variants since it’s more sensitive than the single SNP analysis. Pearson’s chi-square test or Fisher’s exact test were used to analyze the allele case-control comparisons. Association of SNPs with development as well as susceptibility of RA was estimated by figuring out the odds ratio (OR) and 95% Confidence Interval (CI). When comparing the two groups of subjects (case and control), several analytic methods were used: allele frequency distribution of the two groups (allele A versus allele B, A as the major allele, B as the minor allele, this applied to the following methods); dominant model (AB+BB versus AA); recessive model (AA+AB versus BB). All statistical analyses were performed using the Statistical Package for the Social Sciences (SPSS, SPSS Inc., Chicago, IL, USA), version 17.0. A two-sided P value <0.05 was deemed as statistically significant.
Results
Characteristics of the study population
A total of 158 RA patients and 192 healthy controls were included in the study. Their main demographic and clinical characteristics were showen in Table 1 and S1 Table. Subjects were adequately matched for age and sex between RA patients and controls (P = 0.953 and P = 0.731, respectively). The mean age in RA patients and controls were 54.4 and 55.1 years. The age at onset of case group was 43.9±14.4 years and disease duration is 10.5±9.9 years. The mean of erythrocyte sedimentation rate for patients is 70.5 mm/h (SD±34.6) and mean of DAS28 score is 6.32 (SD±1.62). Furthermore, 84.81% of Patients were rheumatoid factor (RF) positive and 83.54% were anti-cyclic citrullinated peptide (anti-CCP) positive. The positive percentage of c-reactive protein (CRP) and anti-keratin antibody (AKA) in RA patients is 81.01% and 45.95%, respectively.
Table 1. The main demographic and clinical characteristics of patients and controls.
| Characteristics | Case | Control | P value |
|---|---|---|---|
| Age, mean±SD (years) | 54.4±12.3 | 55.1±9.8 | 0.953 |
| Female(%)/Male(%) | 76.6/23.4 | 75.0/25.0 | 0.731 |
| Age at onset, mean±SD (years) | 43.9±14.4 | - | - |
| Disease duration, mean±SD (years) | 10.5±9.9 | - | - |
| DAS28, mean±SD | 6.32±1.62 | - | - |
| RF-positive (%) | 84.81 | - | - |
| AntiCCP-positive (%) | 83.54 | - | - |
| AKA-positive (%) | 45.95 | - | - |
| CRP,mean±SD (%) | 34.8±43.5 | - | - |
| ESR, mean±SD (mm/h) | 70.5±34.6 | - | - |
Note: a) Data were expressed as median ± SD or median (interquartile range).
b)-: data were not available.
Abbreviations: DAS28, disease activity score for 28 painful/swollen joints; RF, rheumatoid factor; AntiCCP, anti-cyclic citrullinated peptide; CRP, c-reactive protein; AKA, anti-keratin antibody; ESR, erythrocyte sedimentation rate.
Genotyping and LD evaluation
In this study, all subjects were clearly genotyped using PCR-HRM methods for the four SNPs and the correctness of assays was verified by direct sequencing for PCR products of randomly selected samples. Sequencing results were in complete accord with all the corresponding genotypes. No significant deviation was found from Hardy-Weinberg equilibrium (HWE), as determined at the 0.05 significance level.
Using Haploview to conduct linkage disequilibrium evaluation, two SNPs (rs10893872 and rs4937333) in ETS-1 were in complete linkage disequilibrium (Fig 1). Therefore, the analyses were only carried out on the rs4937333 for its higher minor allele frequency (MAF = 0.44 and 0.42 for rs4937333 and rs 10893872, respectively).
Fig 1. Linkage disequilibrium for four SNPs in 350 individuals.
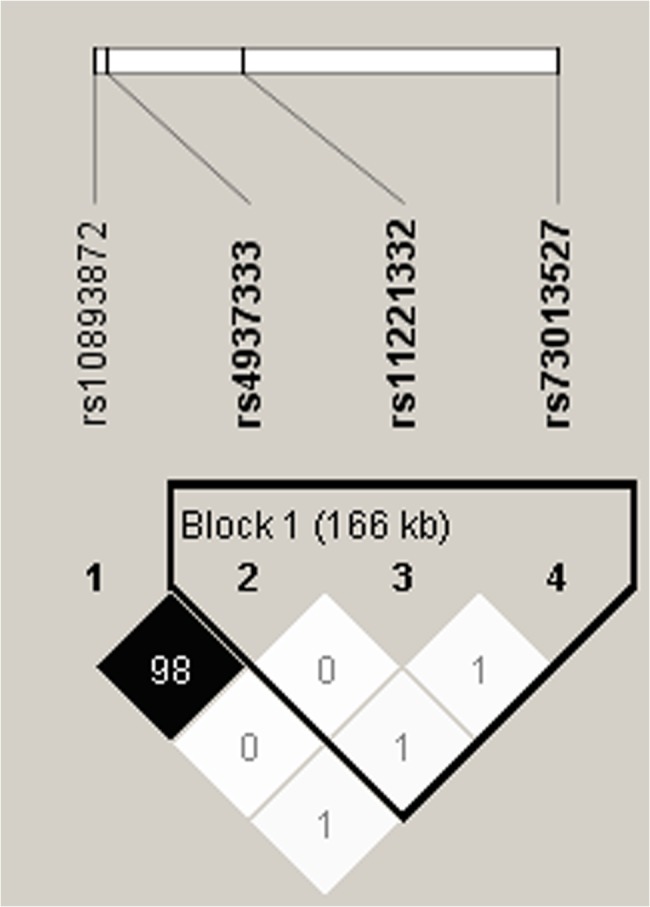
The linkage disequilibrium plot shows r² values between each pair of SNPs. rs10893872 and rs4937333 were in strong LD (black squares). D’ = 1.000, r2 = 0.980.
Association between ETS-1 SNPs and the risk of RA
Genotype distributions of rs73013527 were found to be statistically different between RA patients and the controls. The frequencies of the two groups differed significantly (dominant model, OR = 0.43, 95%CI = 0.28–0.66, P<0.001; recessive model, OR = 0.44, 95%CI = 0.23–0.84, P = 0.012; allelic model, OR = 0.51, 95%CI = 0.37–0.70, P<0.001) (Table 2).
Table 2. Genotype distributions of ETS-1 in RA patients and Controls in Chinese Han population.
| SNPs | Model | Genotype | RA (n = 158) | Controls(n = 192) | OR(95%CI) | P value | ||
|---|---|---|---|---|---|---|---|---|
| N | % | N | % | |||||
| rs73013527 | Dominant | CC | 87 | 55.1 | 66 | 34.4 | 1.00 | |
| CT+TT | 71 | 44.9 | 126 | 65.6 | 0.43(0.28–0.66) | <0.001 | ||
| Recessive | TT | 14 | 8.9 | 35 | 18.2 | 1.00 | ||
| CC+CT | 144 | 91.1 | 157 | 81.8 | 0.44(0.23–0.84) | 0.012 | ||
| Allele | C | 231 | 73.1 | 223 | 58.1 | 1.00 | ||
| T | 85 | 26.9 | 161 | 41.9 | 0.51(0.37–0.70) | <0.001 | ||
| rs4937333 | Dominant | CC | 53 | 33.5 | 61 | 31.8 | 1.00 | |
| CT+TT | 105 | 66.5 | 131 | 68.2 | 0.92(0.59–1.45) | 0.725 | ||
| Recessive | TT | 19 | 12.0 | 30 | 15.6 | 1.00 | ||
| CC+CT | 139 | 88.0 | 162 | 84.4 | 0.74(0.40–1.37) | 0.334 | ||
| Allele | C | 192 | 60.8 | 223 | 58.1 | 1.00 | ||
| T | 124 | 39.2 | 161 | 41.9 | 0.90(0.66–1.21) | 0.472 | ||
| rs11221332 | Dominant | CC | 140 | 88.6 | 166 | 86.5 | 1.00 | |
| CT+TT | 18 | 11.4 | 26 | 13.5 | 0.82(0.43–1.56) | 0.546 | ||
| Recessive | TT | 0 | 0.0 | 2 | 1.0 | - | ||
| CC+CT | 158 | 100.0 | 190 | 99.0 | - | 0.503 | ||
| Allele | C | 298 | 94.3 | 356 | 92.7 | 1.00 | ||
| T | 18 | 5.7 | 28 | 7.3 | 0.77(0.42–1.42) | 0.397 | ||
Abbreviation: OR, odds ratio; CI, confidence interval.
Note: a) Data were presented as number and percentage for every group.
b) Significant p-values (<0.05) are highlighted in bold.
However, none of rs4937333 and rs11221332 polymorphisms achieved a significant difference in the genotype distributions between cases and controls (dominant model, OR = 0.92, 95%CI = 0.59–1.45, P = 0.725 and OR = 0.82, 95%CI = 0.43–1.56, P = 0.546, respectively) (Table 2).
Haplotype analysis of ETS-1 with RA susceptibility
Haplotypes were constructed on the basis of 3 SNPs (rs4937333, rs11221332 and rs73013527) and four common haplotypes were inferred out (frequency>5%). The most common haplotype was CCC, whose frequency was 0.342. Meanwhile, the other three haplotypes were TCC, CCT and TCT according to the frequency order (Table 3).
Table 3. Haplotype analysis of 3 ETS-1 polymorphisms in Chinese Han population.
| Haplotype* | All(freq)** | Case(freq) | Control(freq) | OR(95%CI) | P value |
|---|---|---|---|---|---|
| CCC | 239.3(0.342) | 127.3(0.403) | 112.0(0.292) | 1.000 | |
| TCC | 195.2(0.279) | 96.6(0.306) | 98.6(0.257) | 0.864(0.592–1.261) | 0.449 |
| CCT | 139.8(0.200) | 51.2(0.162) | 88.6(0.231) | 0.505(0.329–0.775) | 0.002 |
| TCT | 79.7(0.114) | 22.9(0.072) | 56.8(0.148) | 0.356(0.206–0.615) | <0.001 |
Abbreviation: OR, odds ratio; CI, confidence interval.
Note: a) Data were presented as number (percentage) for every group. A two-sided p value < 0.05 was considered as statistically significant.
b) * All frequencies <0.03 had been ignored in analysis. Loci chosen for hap-analysis were in this order: rs4937333, rs11221332, rs73013527.
c) **Data exhibited haplotype frequencies (percentage) in the whole 350 subjects.
Moreover, the results of haplotype analysis showed that haplotype block CCT and TCT significantly correlated with reduced risk of RA susceptibility (CCT: OR = 0.505, 95%CI = 0.329–0.775, P = 0.002; TCT: OR = 0.356, 95%CI = 0.206–0.615, P<0.001) (Table 3), which was in line with the results of genotype analyses.
Association of ETS-1 polymorphisms with clinical characteristics in RA patients
Table 4 summarizes the influence analysis of ETS-1 polymorphisms on clinical features of RA patients which shows that rs73013527 was associated with DAS28 and CRP (P = 0.001 and P = 0.028, respectively). These two indexes of different genotype group of rs73013527 differed significantly (DAS28: recessive model, P<0.001; CRP: dominant model, P = 0.022; recessive model, P = 0.034). However, for RF and anti-CCP, not so conspicuous difference as DAS28/CRP was observed between the genotypes of rs73013527 (RF: P = 0.821; anti-CCP: P = 0.351). No significant association was detected between rs4937333/rs11221332 and DAS28, RF, CRP and anti-CCP (DAS28: P = 0.127 and P = 0.423; RF: P = 0.313 and P = 0.874; CRP: P = 0.983 and P = 0.860; anti-CCP: P = 0.777 and P = 0.262, respectively). (Table 4).
Table 4. Association of ETS-1 polymorphisms with clinical characteristics in RA patients.
| SNPs | Model | Genotype | DAS28 | RF(IU/L) | CRP(mg/L) | Anti-CCP(U/L) | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Md±IQR | P Value | Md±IQR | P Value | Md±IQR | P Value | Md±IQR | P Value | |||
| rs73013527 | CC | 6.50±1.53 | 344.00±1527.45 | 11.05±38.67 | 307.15±398.05 | |||||
| CT | 6.62±2.21 | 0.001 | 323.00±1123.8 | 0.821 | 18.70±55.53 | 0.028 | 371.40±257.45 | 0.351 | ||
| TT | 7.81±1.01 | 465.00±939.5 | 49.5±42.68 | 369.80±235.82 | ||||||
| Dominant | CC | 6.50±1.53 | 344.00±1527.45 | 11.05±38.67 | 307.15±398.05 | |||||
| CT+TT | 6.80±1.84 | 0.162 | 366.00±1077.80 | 0.952 | 21.60±57.72 | 0.022 | 371.40±249.90 | 0.15 | ||
| Recessive | TT | 7.81±1.01 | 465.00±939.5 | 49.5±42.68 | 369.80±235.82 | |||||
| CC+CT | 6.54±1.62 | <0.001 | 342.00±1140.45 | 0.572 | 15.60±41.99 | 0.034 | 351.10±382.70 | 0.509 | ||
| rs4937333 | CC | 6.67±1.69 | 342.00±1535.25 | 16.10±49.36 | 328.60±403.54 | |||||
| CT | 6.52±1.98 | 0.127 | 310.00±923.55 | 0.313 | 18.50±44.75 | 0.983 | 363.40±376.00 | 0.777 | ||
| TT | 6.71±1.66 | 635.00±1407.5 | 10.7±55.9 | 350.10±220.5 | ||||||
| Dominant | CC | 6.67±1.69 | 342.00±1535.25 | 16.10±49.36 | 328.60±403.54 | |||||
| CT+TT | 6.58±1.65 | 0.354 | 410.00±986.62 | 0.592 | 17.05±44.21 | 0.859 | 361.65±327.32 | 0.585 | ||
| Recessive | TT | 6.71±1.66 | 635.00±1407.5 | 10.7±55.9 | 350.10±220.5 | |||||
| CC+CT | 6.56±1.70 | 0.134 | 329.00±1129.12 | 0.216 | 17.65±46.32 | 0.926 | 361.65±383.3 | 0.559 | ||
| rs11221332 | CC | 6.60±1.65 | 342.00±1188.85 | 16.7±47.32 | 340.10±379.7 | |||||
| CT | 6.73±1.37 | 0.423 | 431.00±1015.2 | 0.874 | 17.20±48.73 | 0.860 | 416.65±242.5 | 0.262 | ||
| Dominant | CC | 6.60±1.65 | 342.00±1188.85 | 16.7±47.32 | 340.10±379.7 | |||||
| CT+TT | 6.73±1.37 | 0.423 | 431.00±1015.2 | 0.874 | 17.20±48.73 | 0.860 | 416.65±242.5 | 0.262 | ||
Abbreviation: Md±IQR: median±quartile interval.
Note: Significant p-values (<0.05) are highlighted in bold.
Discussion
RA is a complex heterogeneous chronic autoimmune disease, whereby both environmental and genetic factors contribute to the etiology and/or clinical severity. The exact immunopathogenesis of RA has been one of the hottest research topics all the time. In this study, we have demonstrated that ETS-1 is associated with RA in Chinese Han population.
We identified a SNP, rs73013527 of ETS-1, which was significantly associated with RA in Chinese Han population, with minor allele T being correlated with a reduced risk of RA susceptibility. To our knowledge, rs73013527 wasn’t found to be correlated with other autoimmune disease and this is the first report demonstrating an association of this SNP of ETS-1 and RA in Han Chinese. In previous reports, some SNPs of ETS-1 were found to be significantly with many autoimmune diseases such as SLE, pediatric uveitis and celiac disease. In agreement with these autoimmune diseases, we observed that ETS-1 was also a susceptibility gene for RA at least in southern Han Chinese. This finding provides a support for that ETS-1 is a susceptibility gene for many autoimmune diseases. As we know, ETS-1 is a transcription factor and a member of the ETS family that activate transcription by binding to cis-regulatory elements in target genes [25]. It was initially discovered as the proto-oncogene corresponding to v-ets of the avian erythroblastosis virus (E26), which contains a conserved DNA-binding domain mediating specific DNA binding to the GGAA/T motif [26, 27]. Concerning the important roles of ETS-1, some researches indicated it might play a crucial role in RA. For example, ETS-1 was reported to be overexpressed in RA synovial membrane and to be involved in the destructive pathway of RA [28], but it was not clearly defined. In this study, the decreased frequency of the rs73013527 TT genotype in patients suggests that ETS-1 may be a predisposing factor in RA. What’s more, recent research has demonstrated that SNPs at miRNA binding sites likely affect the expression of the miRNA target genes and thus, may contribute to susceptibility to autoimmune diseases [29]. Since rs73013527 is located upstream of the ETS-1 gene and would not affect the sequence of the ETS-1 mRNA and hence could not affect miRNA binding. Therefore, we presumed that the SNP might influence the activity of an upstream enhancer of ETS-1 or might be in linkage disequilibrium with another genetic alteration that influences expression or activity of ETS-1. However, the detailed mechanism of this functional relationship requires further investigation.
Meanwhile, a significant correlation was found between rs73013527 and DAS28 as well as CRP, with minor allele T associated with an increase risk of disease activity. The TT genotype might influence the development of RA by a lasting high disease activity. It is interesting that minor allele T of rs73013527 plays a protective role in occurrence of RA but as a risk factor of high disease activity. A similar phenomenon was found in other researches. For example, Systemic lupus erythematosus (SLE) is a sexually dimorphic autoimmune disease which is more common in women, but male patients often experience a more severe disease state. Amr Sawalha speculated that men require a higher cumulative genetic load than women to develop the disease [30, 31]. The dimorphism of rs73013527 in RA is not clearly defined and it may correlate with gene-gene interaction and complex immune microenviroment, which worth to be studied further.
Although other studies showed that ETS-1 SNP rs11221332 has a positive association with RA susceptibility in Caucasians [10], it seemed not to be correlated with RA susceptibility in Chinese Han population in our study. It may be explained by that the risk variants are often population-specific and the difference can be truly originate from ethnic disparity since the degree of genetic variations differs among individuals of different ethnicities [32]. Besides, no significant correlation was found in the genotype, allele or haplotype frequencies of rs4937333 between RA patients and controls in our study. Considering that rs4937333 was significantly associated with SLE in East Asian populations, there were probably different target genes and pathway of regulation in these two similar diseases. Moreover, our study didn’t find the association between rs11221332 or rs4937333 and the clinical features such as DAS28, CRP, RF and anti-CCP.
As a crucial transcription factor, ETS-1 widely expressed in lymphocytes, vascular endothelial and various invasion tumor cells. It regulates the development, senescence and death of many immune cells, and also plays a role in both innate and adaptive immune response [25, 33]. Accumulating evidence points to an important role for ETS1 in regulating the differentiation of immune cells such as T-cell differentiation into a helper population, terminal differentiation of B cells, development of natural killer (NK) cells and NK T cells and the expression of cytokine and chemokine genes in a wide variety of different cell lineages [16, 25, 34–38]. What’s more, animal experiments showed that autoimmune disease developed in ETS-1 knockout mice, as investigated by the production of high titers of autoantibodies, and immune cell infiltration into organs accounted for aberrations in lymphocyte differentiation [36]. Since RA is a disorder of T cell dysfunction and systemic inflammation, ETS-1 might play a role in occurrence and development of RA. We will investigate this hypothesis in next study.
In summary, our study identified that the common variant (rs73013527) of ETS-1 confer susceptibility to RA. This SNP of ETS-1 also associated with the development of RA. This study might provide further evidence to improve our understanding of the exact function of ETS-1 in the pathogenesis of autoimmune diseases. It is worthwhile to mention that there are several limitations in our present study. The sample of patients in our study is relatively small and only Han Chinese cohorts in west China are included. Therefore, further studies in a large sample size and other ethnic populations are needed to confirm the results observed in this study.
Supporting Information
(DOC)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the Natural Science Foundation of China 81202354 to ZCH and the Natural Science Foundation of China 81301496 to BY. (http://npd.nsfc.gov.cn/fundingProjectSearchAction.action) These funders supported our work including study design, data collection and decision to publish.
References
- 1. Peterson JC, Paget SA, Lachs MS, Reid MC, Charlson ME. The risk of comorbidity. Ann Rheum Dis. 2012;71:635–637. 10.1136/annrheumdis-2011-200473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tobon GJ, Youinou P, Saraux A. The environment, geo-epidemiology, and autoimmune disease: Rheumatoid arthritis. J Autoimmun. 2010;35:10–14. 10.1016/j.jaut.2009.12.009 [DOI] [PubMed] [Google Scholar]
- 3. Yarwood A, Huizinga TW, Worthington J. The genetics of rheumatoid arthritis: risk and protection in different stages of the evolution of RA. Rheumatology. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Viatte S, Plant D, Raychaudhuri S. Genetics and epigenetics of rheumatoid arthritis. Nat Rev Rheumatol. 2013;9:141–153. 10.1038/nrrheum.2012.237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Reynolds RJ, Ahmed AF, Danila MI, Hughes LB, Gregersen PK, Raychaudhuri S, et al. HLA-DRB1-associated rheumatoid arthritis risk at multiple levels in African Americans: hierarchical classification systems, amino acid positions, and residues. Arthritis Rheumatol. 2014;66:3274–3282. 10.1002/art.38855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barton A, Worthington J. Genetic susceptibility to rheumatoid arthritis: an emerging picture. Arthritis Rheum. 2009;61:1441–1446. 10.1002/art.24672 [DOI] [PubMed] [Google Scholar]
- 7. Heo SH, Cho JY. ELK3 suppresses angiogenesis by inhibiting the transcriptional activity of ETS-1 on MT1-MMP. Int J Biol Sci. 2014;10:438–447. 10.7150/ijbs.8095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kothari P, Pestana R, Mesraoua R, Elchaki R, Khan KM, Dannenberg AJ, et al. IL-6-mediated induction of matrix metalloproteinase-9 is modulated by JAK-dependent IL-10 expression in macrophages. J Immunol. 2014;192:349–357. 10.4049/jimmunol.1301906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Song KS, Yoon JH, Kim KS, Ahn DW. c-Ets1 inhibits the interaction of NF-kappaB and CREB, and downregulates IL-1beta-induced MUC5AC overproduction during airway inflammation. Mucosal Immunol. 2012;5:207–215. 10.1038/mi.2011.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chatzikyriakidou A, Voulgari PV, Georgiou I, Drosos AA. Altered sequence of the ETS1 transcription factor may predispose to rheumatoid arthritis susceptibility. Scand J Rheumatol. 2013;42:11–14. 10.3109/03009742.2012.711367 [DOI] [PubMed] [Google Scholar]
- 11. Sharrocks AD. The ETS-domain transcription factor family. Nat Rev Mol Cell Biol. 2001;2:827–837. [DOI] [PubMed] [Google Scholar]
- 12. Tsao HW, Tai TS, Tseng W, Chang HH, Grenningloh R, Miaw SC, et al. Ets-1 facilitates nuclear entry of NFAT proteins and their recruitment to the IL-2 promoter. Proc Natl Acad Sci U S A. 2013;110:15776–15781. 10.1073/pnas.1304343110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Van Beek JP, Kennedy L, Rockel JS, Bernier SM, Leask A. The induction of CCN2 by TGFbeta1 involves Ets-1. Arthritis Res Ther. 2006;8:R36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang W, Zhao J, Lee JF, Gartung A, Jawadi H, Lambiv WL, et al. ETS-1-mediated transcriptional up-regulation of CD44 is required for sphingosine-1-phosphate receptor subtype 3-stimulated chemotaxis. J Biol Chem. 2013;288:32126–32137. 10.1074/jbc.M113.495218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Robinson L, Panayiotakis A, Papas TS, Kola I, Seth A. ETS target genes: identification of egr1 as a target by RNA differential display and whole genome PCR techniques. Proc Natl Acad Sci U S A. 1997;94:7170–7175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. John S, Russell L, Chin SS, Luo W, Oshima R, Garrett-Sinha LA. Transcription factor Ets1, but not the closely related factor Ets2, inhibits antibody-secreting cell differentiation. Mol Cell Biol. 2014;34:522–532. 10.1128/MCB.00612-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yang W, Shen N, Ye DQ, Liu Q, Zhang Y, Qian XX, et al. Genome-wide association study in Asian populations identifies variants in ETS1 and WDFY4 associated with systemic lupus erythematosus. PLoS Genet. 2010;6:e1000841 10.1371/journal.pgen.1000841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dang J, Shan S, Li J, Zhao H, Xin Q, Liu Y, et al. Gene-gene interactions of IRF5, STAT4, IKZF1 and ETS1 in systemic lupus erythematosus. Tissue Antigens. 2014;83:401–408. 10.1111/tan.12349 [DOI] [PubMed] [Google Scholar]
- 19. Han JW, Zheng HF, Cui Y, Sun LD, Ye DQ, Hu Z, et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat Genet. 2009;41:1234–1237. 10.1038/ng.472 [DOI] [PubMed] [Google Scholar]
- 20. Wei L, Zhou Q, Hou S, Bai L, Liu Y, Qi J, et al. MicroRNA-146a and Ets-1 gene polymorphisms are associated with pediatric uveitis. PLoS One. 2014;9:e91199 10.1371/journal.pone.0091199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dubois PC, Trynka G, Franke L, Hunt KA, Romanos J, Curtotti A, et al. Multiple common variants for celiac disease influencing immune gene expression. Nat Genet. 2010;42:295–302. 10.1038/ng.543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Okada Y, Wu D, Trynka G, Raj T, Terao C, Ikari K, et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506:376–381. 10.1038/nature12873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Biliavska I, Stamm TA, Martinez-Avila J, Huizinga TW, Landewe RB, Steiner G, et al. Application of the 2010 ACR/EULAR classification criteria in patients with very early inflammatory arthritis: analysis of sensitivity, specificity and predictive values in the SAVE study cohort. Ann Rheum Dis. 2013;72:1335–1341. 10.1136/annrheumdis-2012-201909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wells G, Becker JC, Teng J, Dougados M, Schiff M, Smolen J, et al. Validation of the 28-joint Disease Activity Score (DAS28) and European League Against Rheumatism response criteria based on C-reactive protein against disease progression in patients with rheumatoid arthritis, and comparison with the DAS28 based on erythrocyte sedimentation rate. Ann Rheum Dis. 2009;68:954–960. 10.1136/ard.2007.084459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Garrett-Sinha LA. Review of Ets1 structure, function, and roles in immunity. Cell Mol Life Sci. 2013;70:3375–3390. 10.1007/s00018-012-1243-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wei GH, Badis G, Berger MF, Kivioja T, Palin K, Enge M, et al. Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. EMBO J. 2010;29:2147–2160. 10.1038/emboj.2010.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Babayeva ND, Wilder PJ, Shiina M, Mino K, Desler M, Ogata K, et al. Structural basis of Ets1 cooperative binding to palindromic sequences on stromelysin-1 promoter DNA. Cell Cycle. 2010;9:3054–3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Redlich K, Kiener HP, Schett G, Tohidast-Akrad M, Selzer E, Radda I, et al. Overexpression of transcription factor Ets-1 in rheumatoid arthritis synovial membrane: regulation of expression and activation by interleukin-1 and tumor necrosis factor alpha. Arthritis Rheum. 2001;44:266–274. [DOI] [PubMed] [Google Scholar]
- 29. Hikami K, Kawasaki A, Ito I, Koga M, Ito S, Hayashi T, et al. Association of a functional polymorphism in the 3'-untranslated region of SPI1 with systemic lupus erythematosus. Arthritis Rheum. 2011;63:755–763. 10.1002/art.30188 [DOI] [PubMed] [Google Scholar]
- 30. Gaudreau MC, Johnson BM, Gudi R, Al-Gadban MM, Vasu C. Gender bias in lupus: Does immune response initiated in the gut mucosa have a role? Clin Exp Immunol. 2015;180:393–407. 10.1111/cei.12587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hughes T, Adler A, Merrill JT, Kelly JA, Kaufman KM, Williams A, et al. Analysis of autosomal genes reveals gene-sex interactions and higher total genetic risk in men with systemic lupus erythematosus. Ann Rheum Dis. 2012;71:694–699. 10.1136/annrheumdis-2011-200385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fu J, Festen EA, Wijmenga C. Multi-ethnic studies in complex traits. Hum Mol Genet. 2011;20:R206–213. 10.1093/hmg/ddr386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sasaki I, Kaisho T. Transcriptional control of dendritic cell differentiation. Curr Top Microbiol Immunol. 2014;381:257–278. 10.1007/82_2014_378 [DOI] [PubMed] [Google Scholar]
- 34. Maier H, Colbert J, Fitzsimmons D, Clark DR, Hagman J. Activation of the early B-cell-specific mb-1 (Ig-alpha) gene by Pax-5 is dependent on an unmethylated Ets binding site. Mol Cell Biol. 2003;23:1946–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shah S, Schrader KA, Waanders E, Timms AE, Vijai J, Miething C, et al. A recurrent germline PAX5 mutation confers susceptibility to pre-B cell acute lymphoblastic leukemia. Nat Genet. 2013;45:1226–1231. 10.1038/ng.2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Russell L, Garrett-Sinha LA. Transcription factor Ets-1 in cytokine and chemokine gene regulation. Cytokine. 2010;51:217–226. 10.1016/j.cyto.2010.03.006 [DOI] [PubMed] [Google Scholar]
- 37. Mouly E, Chemin K, Nguyen HV, Chopin M, Mesnard L, Leite-de-Moraes M, et al. The Ets-1 transcription factor controls the development and function of natural regulatory T cells. J Exp Med. 2010;207:2113–2125. 10.1084/jem.20092153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ramirez K, Chandler KJ, Spaulding C, Zandi S, Sigvardsson M, Graves BJ, et al. Gene deregulation and chronic activation in natural killer cells deficient in the transcription factor ETS1. Immunity. 2012;36:921–932. 10.1016/j.immuni.2012.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.


