Abstract
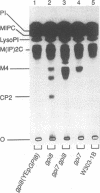
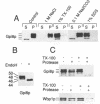
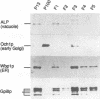
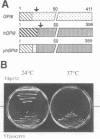
Glycosylphosphatidylinositol (GPI) anchors are added onto newly synthesized proteins in the ER. Thereby a putative transamidase removes a C-terminal peptide and attaches the truncated protein to the free amino group of the preformed GPI. The yeast mutant gpi8-1 is deficient in this addition of GPIs to proteins. GPI8 encodes for an essential 47 kDa type I membrane glycoprotein residing on the luminal side of the ER membrane. GPI8 shows significant homology to a novel family of vacuolar plant endopeptidases one of which is supposed to catalyse a transamidation step in the maturation of concanavalin A and acts as a transamidase in vitro. Humans have a gene which is highly homologous to GPI8 and can functionally replace it.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abe Y., Shirane K., Yokosawa H., Matsushita H., Mitta M., Kato I., Ishii S. Asparaginyl endopeptidase of jack bean seeds. Purification, characterization, and high utility in protein sequence analysis. J Biol Chem. 1993 Feb 15;268(5):3525–3529. [PubMed] [Google Scholar]
- Alonso J. M., Granell A. A putative vacuolar processing protease is regulated by ethylene and also during fruit ripening in Citrus fruit. Plant Physiol. 1995 Oct;109(2):541–547. doi: 10.1104/pp.109.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bause E., Hettkamp H. Primary structural requirements for N-glycosylation of peptides in rat liver. FEBS Lett. 1979 Dec 15;108(2):341–344. doi: 10.1016/0014-5793(79)80559-1. [DOI] [PubMed] [Google Scholar]
- Bause E., Lehle L. Enzymatic N-glycosylation and O-glycosylation of synthetic peptide acceptors by dolichol-linked sugar derivatives in yeast. Eur J Biochem. 1979 Nov;101(2):531–540. doi: 10.1111/j.1432-1033.1979.tb19748.x. [DOI] [PubMed] [Google Scholar]
- Becker C., Shutov A. D., Nong V. H., Senyuk V. I., Jung R., Horstmann C., Fischer J., Nielsen N. C., Müntz K. Purification, cDNA cloning and characterization of proteinase B, an asparagine-specific endopeptidase from germinating vetch (Vicia sativa L.) seeds. Eur J Biochem. 1995 Mar 1;228(2):456–462. [PubMed] [Google Scholar]
- Becker D. M., Guarente L. High-efficiency transformation of yeast by electroporation. Methods Enzymol. 1991;194:182–187. doi: 10.1016/0076-6879(91)94015-5. [DOI] [PubMed] [Google Scholar]
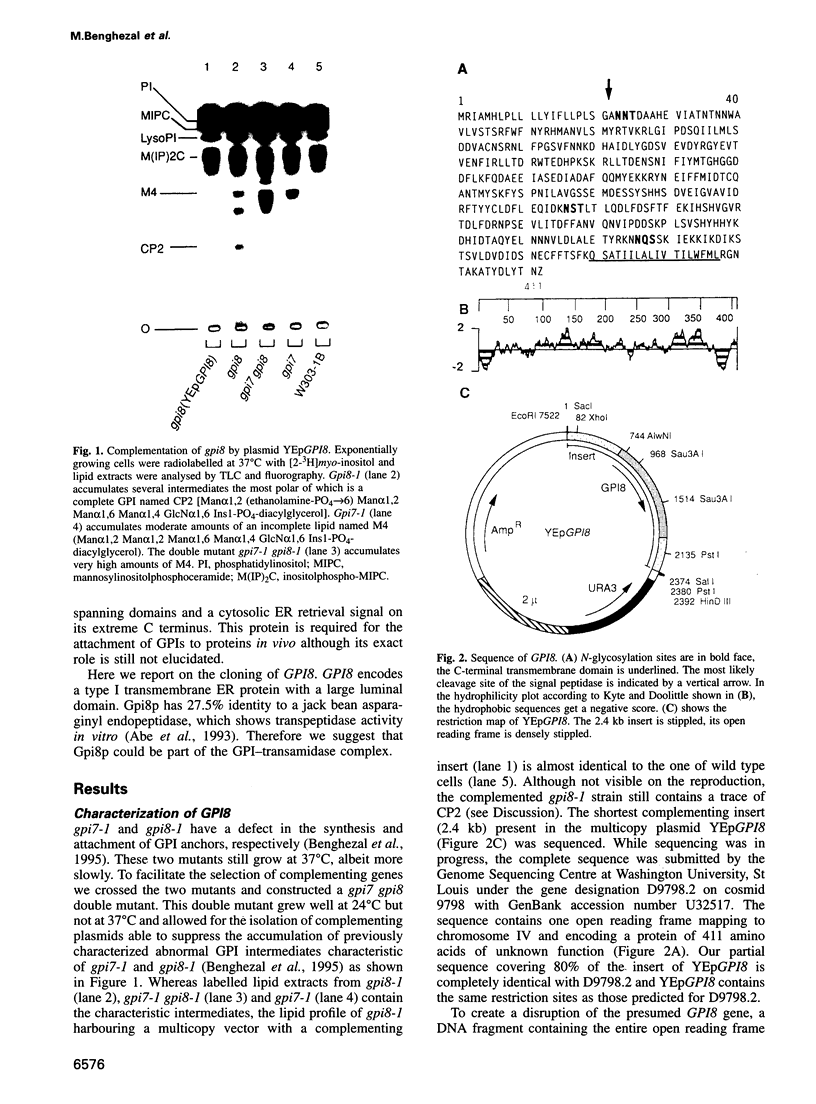
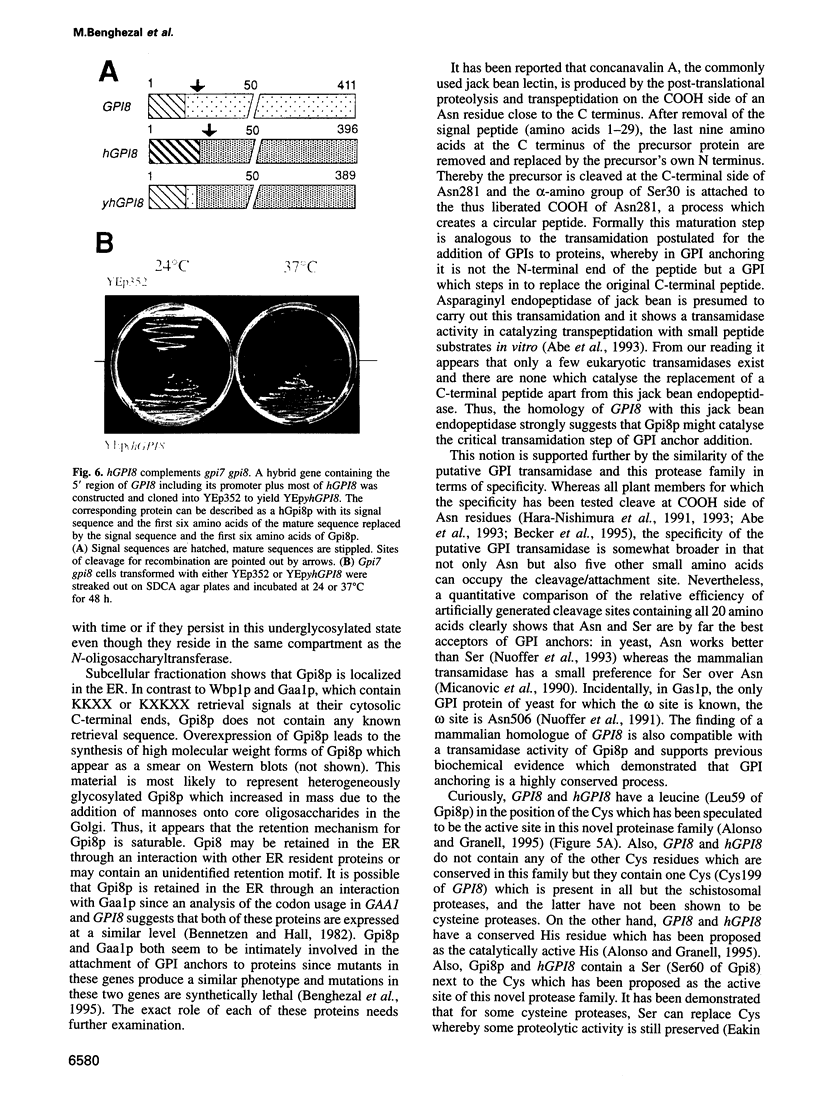
- Benghezal M., Lipke P. N., Conzelmann A. Identification of six complementation classes involved in the biosynthesis of glycosylphosphatidylinositol anchors in Saccharomyces cerevisiae. J Cell Biol. 1995 Sep;130(6):1333–1344. doi: 10.1083/jcb.130.6.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. Codon selection in yeast. J Biol Chem. 1982 Mar 25;257(6):3026–3031. [PubMed] [Google Scholar]
- Caras I. W., Weddell G. N., Davitz M. A., Nussenzweig V., Martin D. W., Jr Signal for attachment of a phospholipid membrane anchor in decay accelerating factor. Science. 1987 Nov 27;238(4831):1280–1283. doi: 10.1126/science.2446389. [DOI] [PubMed] [Google Scholar]
- Conzelmann A., Puoti A., Lester R. L., Desponds C. Two different types of lipid moieties are present in glycophosphoinositol-anchored membrane proteins of Saccharomyces cerevisiae. EMBO J. 1992 Feb;11(2):457–466. doi: 10.1002/j.1460-2075.1992.tb05075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conzelmann A., Spiazzi A., Bron C., Hyman R. No glycolipid anchors are added to Thy-1 glycoprotein in Thy-1-negative mutant thymoma cells of four different complementation classes. Mol Cell Biol. 1988 Feb;8(2):674–678. doi: 10.1128/mcb.8.2.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conzelmann A., Spiazzi A., Hyman R., Bron C. Anchoring of membrane proteins via phosphatidylinositol is deficient in two classes of Thy-1 negative mutant lymphoma cells. EMBO J. 1986 Dec 1;5(12):3291–3296. doi: 10.1002/j.1460-2075.1986.tb04642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cysteine or serine proteinase? Nature. 1989 Nov 9;342(6246):132–132. doi: 10.1038/342132b0. [DOI] [PubMed] [Google Scholar]
- Davis A. H., Nanduri J., Watson D. C. Cloning and gene expression of Schistosoma mansoni protease. J Biol Chem. 1987 Sep 15;262(26):12851–12855. [PubMed] [Google Scholar]
- Englund P. T. The structure and biosynthesis of glycosyl phosphatidylinositol protein anchors. Annu Rev Biochem. 1993;62:121–138. doi: 10.1146/annurev.bi.62.070193.001005. [DOI] [PubMed] [Google Scholar]
- Feldheim D., Schekman R. Sec72p contributes to the selective recognition of signal peptides by the secretory polypeptide translocation complex. J Cell Biol. 1994 Aug;126(4):935–943. doi: 10.1083/jcb.126.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor E. C., te Heesen S., Graham T. R., Aebi M., Emr S. D. Signal-mediated retrieval of a membrane protein from the Golgi to the ER in yeast. J Cell Biol. 1994 Nov;127(3):653–665. doi: 10.1083/jcb.127.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber L. D., Kodukula K., Udenfriend S. Phosphatidylinositol glycan (PI-G) anchored membrane proteins. Amino acid requirements adjacent to the site of cleavage and PI-G attachment in the COOH-terminal signal peptide. J Biol Chem. 1992 Jun 15;267(17):12168–12173. [PubMed] [Google Scholar]
- Gorbalenya A. E., Donchenko A. P., Blinov V. M., Koonin E. V. Cysteine proteases of positive strand RNA viruses and chymotrypsin-like serine proteases. A distinct protein superfamily with a common structural fold. FEBS Lett. 1989 Jan 30;243(2):103–114. doi: 10.1016/0014-5793(89)80109-7. [DOI] [PubMed] [Google Scholar]
- Goud B., Salminen A., Walworth N. C., Novick P. J. A GTP-binding protein required for secretion rapidly associates with secretory vesicles and the plasma membrane in yeast. Cell. 1988 Jun 3;53(5):753–768. doi: 10.1016/0092-8674(88)90093-1. [DOI] [PubMed] [Google Scholar]
- Hamburger D., Egerton M., Riezman H. Yeast Gaa1p is required for attachment of a completed GPI anchor onto proteins. J Cell Biol. 1995 May;129(3):629–639. doi: 10.1083/jcb.129.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara-Nishimura I., Inoue K., Nishimura M. A unique vacuolar processing enzyme responsible for conversion of several proprotein precursors into the mature forms. FEBS Lett. 1991 Dec 2;294(1-2):89–93. doi: 10.1016/0014-5793(91)81349-d. [DOI] [PubMed] [Google Scholar]
- Hara-Nishimura I., Takeuchi Y., Nishimura M. Molecular characterization of a vacuolar processing enzyme related to a putative cysteine proteinase of Schistosoma mansoni. Plant Cell. 1993 Nov;5(11):1651–1659. doi: 10.1105/tpc.5.11.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higaki J. N., Evnin L. B., Craik C. S. Introduction of a cysteine protease active site into trypsin. Biochemistry. 1989 Nov 28;28(24):9256–9263. doi: 10.1021/bi00450a004. [DOI] [PubMed] [Google Scholar]
- Higgins D. G., McConnell D. J., Sharp P. M. Malarial proteinase? Nature. 1989 Aug 24;340(6235):604–604. doi: 10.1038/340604a0. [DOI] [PubMed] [Google Scholar]
- Hill J. E., Myers A. M., Koerner T. J., Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986 Sep;2(3):163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- Kinoshita T., Nishimura M., Hara-Nishimura I. Homologues of a vacuolar processing enzyme that are expressed in different organs in Arabidopsis thaliana. Plant Mol Biol. 1995 Oct;29(1):81–89. doi: 10.1007/BF00019120. [DOI] [PubMed] [Google Scholar]
- Kodukula K., Amthauer R., Cines D., Yeh E. T., Brink L., Thomas L. J., Udenfriend S. Biosynthesis of phosphatidylinositol-glycan (PI-G)-anchored membrane proteins in cell-free systems: PI-G is an obligatory cosubstrate for COOH-terminal processing of nascent proteins. Proc Natl Acad Sci U S A. 1992 Jun 1;89(11):4982–4985. doi: 10.1073/pnas.89.11.4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodukula K., Gerber L. D., Amthauer R., Brink L., Udenfriend S. Biosynthesis of glycosylphosphatidylinositol (GPI)-anchored membrane proteins in intact cells: specific amino acid requirements adjacent to the site of cleavage and GPI attachment. J Cell Biol. 1993 Feb;120(3):657–664. doi: 10.1083/jcb.120.3.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee N. H., Weinstock K. G., Kirkness E. F., Earle-Hughes J. A., Fuldner R. A., Marmaros S., Glodek A., Gocayne J. D., Adams M. D., Kerlavage A. R. Comparative expressed-sequence-tag analysis of differential gene expression profiles in PC-12 cells before and after nerve growth factor treatment. Proc Natl Acad Sci U S A. 1995 Aug 29;92(18):8303–8307. doi: 10.1073/pnas.92.18.8303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidich S. D., Drapp D. A., Orlean P. A conditionally lethal yeast mutant blocked at the first step in glycosyl phosphatidylinositol anchor synthesis. J Biol Chem. 1994 Apr 8;269(14):10193–10196. [PubMed] [Google Scholar]
- Leidich S. D., Kostova Z., Latek R. R., Costello L. C., Drapp D. A., Gray W., Fassler J. S., Orlean P. Temperature-sensitive yeast GPI anchoring mutants gpi2 and gpi3 are defective in the synthesis of N-acetylglucosaminyl phosphatidylinositol. Cloning of the GPI2 gene. J Biol Chem. 1995 Jun 2;270(22):13029–13035. doi: 10.1074/jbc.270.22.13029. [DOI] [PubMed] [Google Scholar]
- Maxwell S. E., Ramalingam S., Gerber L. D., Brink L., Udenfriend S. An active carbonyl formed during glycosylphosphatidylinositol addition to a protein is evidence of catalysis by a transamidase. J Biol Chem. 1995 Aug 18;270(33):19576–19582. doi: 10.1074/jbc.270.33.19576. [DOI] [PubMed] [Google Scholar]
- McConville M. J., Ferguson M. A. The structure, biosynthesis and function of glycosylated phosphatidylinositols in the parasitic protozoa and higher eukaryotes. Biochem J. 1993 Sep 1;294(Pt 2):305–324. doi: 10.1042/bj2940305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merckelbach A., Hasse S., Dell R., Eschlbeck A., Ruppel A. cDNA sequences of Schistosoma japonicum coding for two cathepsin B-like proteins and Sj32. Trop Med Parasitol. 1994 Sep;45(3):193–198. [PubMed] [Google Scholar]
- Micanovic R., Gerber L. D., Berger J., Kodukula K., Udenfriend S. Selectivity of the cleavage/attachment site of phosphatidylinositol-glycan-anchored membrane proteins determined by site-specific mutagenesis at Asp-484 of placental alkaline phosphatase. Proc Natl Acad Sci U S A. 1990 Jan;87(1):157–161. doi: 10.1073/pnas.87.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohney R. P., Knez J. J., Ravi L., Sevlever D., Rosenberry T. L., Hirose S., Medof M. E. Glycoinositol phospholipid anchor-defective K562 mutants with biochemical lesions distinct from those in Thy-1- murine lymphoma mutants. J Biol Chem. 1994 Mar 4;269(9):6536–6542. [PubMed] [Google Scholar]
- Moran P., Caras I. W. A nonfunctional sequence converted to a signal for glycophosphatidylinositol membrane anchor attachment. J Cell Biol. 1991 Oct;115(2):329–336. doi: 10.1083/jcb.115.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran P., Caras I. W. Fusion of sequence elements from non-anchored proteins to generate a fully functional signal for glycophosphatidylinositol membrane anchor attachment. J Cell Biol. 1991 Dec;115(6):1595–1600. doi: 10.1083/jcb.115.6.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran P., Caras I. W. Requirements for glycosylphosphatidylinositol attachment are similar but not identical in mammalian cells and parasitic protozoa. J Cell Biol. 1994 Apr;125(2):333–343. doi: 10.1083/jcb.125.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran P., Raab H., Kohr W. J., Caras I. W. Glycophospholipid membrane anchor attachment. Molecular analysis of the cleavage/attachment site. J Biol Chem. 1991 Jan 15;266(2):1250–1257. [PubMed] [Google Scholar]
- Muramatsu M., Fukazawa C. A high-order structure of plant storage proprotein allows its second conversion by an asparagine-specific cysteine protease, a novel proteolytic enzyme. Eur J Biochem. 1993 Jul 1;215(1):123–132. doi: 10.1111/j.1432-1033.1993.tb18014.x. [DOI] [PubMed] [Google Scholar]
- Nuoffer C., Horvath A., Riezman H. Analysis of the sequence requirements for glycosylphosphatidylinositol anchoring of Saccharomyces cerevisiae Gas1 protein. J Biol Chem. 1993 May 15;268(14):10558–10563. [PubMed] [Google Scholar]
- Nuoffer C., Jenö P., Conzelmann A., Riezman H. Determinants for glycophospholipid anchoring of the Saccharomyces cerevisiae GAS1 protein to the plasma membrane. Mol Cell Biol. 1991 Jan;11(1):27–37. doi: 10.1128/mcb.11.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B., Casadio R., Fariselli P., Sander C. Transmembrane helices predicted at 95% accuracy. Protein Sci. 1995 Mar;4(3):521–533. doi: 10.1002/pro.5560040318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönbächler M., Horvath A., Fassler J., Riezman H. The yeast spt14 gene is homologous to the human PIG-A gene and is required for GPI anchor synthesis. EMBO J. 1995 Apr 18;14(8):1637–1645. doi: 10.1002/j.1460-2075.1995.tb07152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T., Hiraiwa N., Nishimura M., Hara-Nishimura I. Vacuolar processing enzyme of soybean that converts proproteins to the corresponding mature forms. Plant Cell Physiol. 1994 Jun;35(4):713–718. doi: 10.1093/oxfordjournals.pcp.a078648. [DOI] [PubMed] [Google Scholar]
- Stagljar I., te Heesen S., Aebi M. New phenotype of mutations deficient in glucosylation of the lipid-linked oligosaccharide: cloning of the ALG8 locus. Proc Natl Acad Sci U S A. 1994 Jun 21;91(13):5977–5981. doi: 10.1073/pnas.91.13.5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathern J. N., Higgins D. R. Recovery of plasmids from yeast into Escherichia coli: shuttle vectors. Methods Enzymol. 1991;194:319–329. doi: 10.1016/0076-6879(91)94024-7. [DOI] [PubMed] [Google Scholar]
- Takeda J., Kinoshita T. GPI-anchor biosynthesis. Trends Biochem Sci. 1995 Sep;20(9):367–371. doi: 10.1016/s0968-0004(00)89078-7. [DOI] [PubMed] [Google Scholar]
- Takeda O., Miura Y., Mitta M., Matsushita H., Kato I., Abe Y., Yokosawa H., Ishii S. Isolation and analysis of cDNA encoding a precursor of Canavalia ensiformis asparaginyl endopeptidase (legumain). J Biochem. 1994 Sep;116(3):541–546. doi: 10.1093/oxfordjournals.jbchem.a124559. [DOI] [PubMed] [Google Scholar]
- Vossen J. H., Ram A. F., Klis F. M. Identification of SPT14/CWH6 as the yeast homologue of hPIG-A, a gene involved in the biosynthesis of GPI anchors. Biochim Biophys Acta. 1995 Apr 13;1243(3):549–551. doi: 10.1016/0304-4165(95)00002-s. [DOI] [PubMed] [Google Scholar]
- Wach A., Brachat A., Pöhlmann R., Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994 Dec;10(13):1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Wickerham L. J. A Critical Evaluation of the Nitrogen Assimilation Tests Commonly Used in the Classification of Yeasts. J Bacteriol. 1946 Sep;52(3):293–301. [PMC free article] [PubMed] [Google Scholar]
- te Heesen S., Rauhut R., Aebersold R., Abelson J., Aebi M., Clark M. W. An essential 45 kDa yeast transmembrane protein reacts with anti-nuclear pore antibodies: purification of the protein, immunolocalization and cloning of the gene. Eur J Cell Biol. 1991 Oct;56(1):8–18. [PubMed] [Google Scholar]
- von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986 Jun 11;14(11):4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]