Abstract
It is of great significance to understand the regulatory mechanisms by which plants deal with drought stress. Two EST libraries derived from rapeseed (Brassica napus) leaves in non-stressed and drought stress conditions were analyzed in order to obtain the transcriptomic landscape of drought-exposed B. napus plants, and also to identify and characterize significant drought responsive regulatory genes and microRNAs. The functional ontology analysis revealed a substantial shift in the B. napus transcriptome to govern cellular drought responsiveness via different stress-activated mechanisms. The activity of transcription factor and protein kinase modules generally increased in response to drought stress. The 26 regulatory genes consisting of 17 transcription factor genes, eight protein kinase genes and one protein phosphatase gene were identified showing significant alterations in their expressions in response to drought stress. We also found the six microRNAs which were differentially expressed during drought stress supporting the involvement of a post-transcriptional level of regulation for B. napus drought response. The drought responsive regulatory network shed light on the significance of some regulatory components involved in biosynthesis and signaling of various plant hormones (abscisic acid, auxin and brassinosteroids), ubiquitin proteasome system, and signaling through Reactive Oxygen Species (ROS). Our findings suggested a complex and multi-level regulatory system modulating response to drought stress in B. napus.
Keywords: Brassica napus, EST analysis, Drought stress, Transcription factors, Protein kinases, MicroRNA
Introduction
Growth and development of crop plants are severely affected by several abiotic stresses (Seki et al. 2003; Shinozaki and Yamaguchi-Shinozaki 1996). Among the abiotic stresses, drought has a crucial devastating impact on agricultural production (Farooq et al. 2009; Farooq et al. 2012). Plants recruit complex overlapping mechanisms to reprogram their gene expression to cope with drought stress (Ahuja et al. 2010; Golldack et al. 2011). The orchestrated regulatory networks at transcriptional, post transcriptional and post translational levels are involved in forming the appropriate transcriptome and proteome of stress-exposed plants to show an adaptive response (Duque et al. 2013; Mazzucotelli et al. 2008). Understanding the role of regulatory components will be of great benefit for breeding drought tolerant crops.
Various regulatory proteins including transcription factors, protein kinases and protein phosphatases have been identified to function as the core regulatory elements of stress responses (Ashraf 2010; Hadiarto and Tran 2011; Seki et al. 2007; Shinozaki and Yamaguchi-Shinozaki 1996; Umezawa et al. 2006; Xoconostle-Cazares et al. 2010; Yamaguchi-Shinozaki and Shinozaki 2006). Also, it has been revealed that the expression of some plant microRNAs (miRNAs), as negative regulator of gene expression, alters during stress conditions such as drought, salinity and cold (Khraiwesh et al. 2012; Lu and Huang 2008; Sunkar et al. 2007; Sunkar et al. 2012). Notably, most of these miRNAs target genes encoding transcription factors, which place miRNAs at the center of stress regulatory networks (Lu and Huang 2008; Sunkar et al. 2012).
Expressed sequence tag (EST) analysis is known as a powerful tool to discover and characterize genes involved in different stress responses and developmental cues in several plant species (Deokar et al. 2011; Gao et al. 2008; Gorantla et al. 2007; Gruber et al. 2012; Lata et al. 2010; Pratt et al. 2005; Shamloo-Dashtpagerdi et al. 2013; Zhuang and Zhu 2014). ESTs are also a valuable source for computational identification of miRNAs and study their possible roles in plant stress responses (Guo et al. 2014; Han et al. 2014; Hwang et al. 2011; Panda et al. 2014; Patanun et al. 2013; Ye et al. 2013).
Rapeseed (Brassica napus L.) is globally an important oilseed crop whose quality and quantity is often limited by drought stress in arid and semi-arid regions. Development of rapeseed cultivars with improved tolerance to drought is of great priority in rapeseed breeding programs (Diepenbrock 2000; Raymer 2002; Xie et al. 2007). A high level of homology exists in the protein-coding sequences of rapeseed and Arabidopsis thaliana making possible to use Arabidopsis genome information for functional interpretation in rapeseed (Fourmann et al. 2002; Parkin et al. 2002). EST-based approaches have been frequently employed to elucidate the genetic basis of regulation of rapeseed transcriptome in response to developmental cues, biotic and abiotic stresses, however little is known about the interconnections among different types of regulatory genes in the context of a network system. Generation and analysis of EST sequences from rapeseed developing seeds led to characterize a number of seed specific transcripts which had significant similarities to their Arabidopsis counterparts (Dong et al. 2004). The multiple regulatory factors involved in fatty acid metabolism were identified in rapeseed using EST chip hybridization (Niu et al. 2009). A comparative EST study revealed that the expression pattern of the rapeseed cotyledon transcriptome shifted towards a defense state after damage by flea beetles (Gruber et al. 2012). Indeed, the activity of the transcription factors known to promote or repress genes controlling stress responses, as well as genes involved in primary and secondary metabolism pathways, cell wall synthesis, and transport altered during flea beetle attack (Gruber et al. 2012). Analysis of rapeseed ESTs also discovered some mitogen-activated protein kinases and AP2/ERF transcription factors involved in abiotic stress signaling and regulatory mechanisms (Zhuang and Zhu 2014).
In this study, two EST libraries derived from rapeseed leaves in non-stress and drought stress conditions were mined in order to gain an insight into the transcriptomic landscape changes of drought-exposed rapeseed plants. Afterwards, the differentially expressed regulatory genes encoding transcription factors, protein kinases and protein phosphatases were identified. A computational analysis was also performed to identify and characterize rapeseed drought-responsive miRNAs and their possible target(s). Finally, a regulatory network was depicted which represented a complex and multi-level regulatory system to respond to drought stress in rapeseed.
Materials and methods
Source of B. napus expressed sequence tags data
Sequences of two 5′ EST libraries from leaves of B. napus line DH12075, generated by Agriculture and Agri-food Canada (http://www.agr.gc.ca/), were retrieved from the TIGR Gene Indices of DFCI (http://compbio.dfci.harvard.edu/tgi/). One library (Lib ID: LIBEST 021477; http://www.ncbi.nlm.nih.gov/nucest/?term=LIBEST_021477) contained 6040 sequences from non-stress condition and the other (Lib ID: LIBEST_021491; http://www.ncbi.nlm.nih.gov/nucest/?term=LIBEST_021491) contained 13,063 sequences from drought stress condition. DH12075 is a double haploid line derived from an F1 cross between the French variety Cresor and the Canadian variety Westar (Young et al. 2004). Plants at the four-leaf stage were subjected to drought stress by applying 200 mM mannitol for 4 h and the cDNA libraries were constructed from small to medium sized healthy leaves using SuperScript Plasmid System with Gateway Technology for cDNA Synthesis and Cloning kit (Invitrogen) (http://www.ncbi.nlm.nih.gov/nucest/EV208948). EST sequences were the direct results of Base calling software Phred, and then they were processed using Lucy software. The analysis of these EST data has not been published so far.
Sequence processing, similarity search and annotation
The EST sequences were checked for vector contamination, sequence length and complexity using EGassembler, an online bioinformatics service (http://egassembler.hgc.jp) (Masoudi-Nejad et al. 2006). The vector, repetitive, chloroplast, and mitochondrial sequences were removed and the trimmed sequences were excluded from further analysis when they were less than 100 bp or they had greater than 4 % ambiguous bases (Carlson et al. 2006; Masoudi-Nejad et al. 2006). The remaining high quality ESTs of each library were subsequently clustered and assembled into unigenes (contigs and singletons) using EGassembler with Overlap percent identity cutoff >80 %. All the unigenes were searched against the Arabidopsis protein database (ftp://ftp.arabidopsis.org/) by local BLASTX using CLC Genomics Workbench software (version: 3.6.5) with the cut-off E-value ≤10−5. To categorize the unigenes with significant BLASTX scores into functional modules, modular enrichment analysis (MEA) was performed in each of the non-stress and drought stress conditions using Gene Annotations Co-occurrence Discovery; GeneCodis (http://genecodis.cnb.csic.es/) (Tabas-Madrid et al. 2012). GeneCodis is a web tool for functional enrichment analysis that integrates different information sources (GO, KEGG and Swiss-Prot gene accession databases) to find concurrent annotation of genes and form groups of genes with similar biological functions according to their statistical relevance (Carmona-Saez et al. 2007). In GeneCodis, Arabidopsis was selected as the reference organism and hypergeometric and FDR statistical tests were applied to compute and correct p-values, respectively. Radar charts were used to illustrate the percentage of functional modules.
Identification of drought-responsive regulatory components
In order to identify significant differentially expressed genes between the non-stress and drought EST libraries, all the EST sequences of the two libraries were clustered and assembled together using EGassembler with default parameters. The contigs were subsequently subjected to Audic and Claverie test (Audic and Claverie 1997) with α = 0.05, available at IDEG6 web tool (Romualdi et al. 2003), to check whether the number of ESTs of each library contributing to each contig were significantly different. The total number of ESTs in each library and the number of library specific ESTs in each contig were the inputs to perform Audic and Claverie test.
Of those differentially expressed contigs, the regulatory genes encoding transcription factors, protein kinases and protein phosphatases were specified by two online bioinformatic tools. The contigs were annotated and classified as transcription factors using PlantTFact (http://plantgrn.noble.org/PlantTFcat/) (Dai et al. 2013). The genes encoding protein kinases and protein phosphatases were identified by Plant Transcription factor and Protein Kinase Identifier and Classifier (iTAK) program (http://bioinfo.bti.cornell.edu/cgi-bin/itak/index.cgi).
We also performed a computational analysis to pinpoint and characterize significant differentially expressed miRNAs and their target genes. The Contigs matched with the regulatory genes in the previous step were excluded from the set of differentially expressed contigs. The remaining contigs were searched against all the available mature miRNAs from miRBase (http://www.mirbase.org/) (Griffiths-Jones et al. 2006) using CLC genomics workbench (version: 3.6.5) with expected values ≤10 to raise the hit chance for more potential sequences. The Contig sequences with the maximum four mismatches were subjected to similarity searches against non-redundant protein database using BLASTX (E-value ≤10−5) by CLC Genomics Workbench software (version: 3.6.5). Also, in order to distinguish between potential miRNAs and other small RNAs such as tRNA, rRNA, snRNA and snoRNA, a BLASTN search was carried out against Rfam database (rfam.sanger.ac.uk/). After that, the secondary structures of the remaining contigs were predicted using Mfold (Zuker 2003), The parameters were set as RNA sequence (linear), folding temperature (37 °C), maximum interior/ bulges loop size (30), and all others with default values. Also, the minimal folding free energy index (MFEI) was calculated for the predicted miRNAs (Zhang et al. 2006). The potential target genes of the predicted differentially expressed miRNAs were identified using psRNATarget with default parameters (http://plantgrn.noble.org/psRNATarget/?function=1) (Dai and Zhao 2011). All the differentially expressed genes found in this study were used as the database of psRNATarget. In each EST library, the normalized expression values of the regulatory genes and miRNAs were obtained using IDEG6 web tool as the ratio of the number of ESTs for the gene or miRNA of interest to the total number of ESTs. The fold change of expression for each of the genes and miRNAs was calculated by dividing the expression level in drought stress conditions by that of in non-stress conditions.
The identified regulatory components were networked using Pathway Studio software version 9 based on Fisher test with p-values ≤ 0.01 (Nikitin et al. 2003; Subramanian et al. 2005). The database of software could provide a number of relevant components, which were not identified among the differentially expressed genes, to make the regulatory network more complete.
Results and discussion
Sequence analysis, annotation and functional classification
Pre-processing of 6040 and 13,063 EST sequences derived from B. napus leaves in non-stress and drought stress conditions resulted in 6037 and 13,059 high quality sequences with average length of 863 and 741 bp, respectively. ESTs obtained from the cDNA library of non-stressed plants were clustered and assembled into 4149 unigenes (794 contigs and 3355 singletons), in which the contigs encompassed 2682 (44.43 %) of EST sequences. Also, assembly of ESTs obtained from the cDNA library of drought-stressed plants yielded 5817 unigenes (1945 contigs and 3872 singletons) that 9191 (70.36 %) of ESTs fell into the contigs. The average contig lengths were 898 and 791 bp for the non-stress and drought EST libraries, respectively. The number of ESTs ranged from 2 (465 contigs) to 97 (one contig) in the non-stress contigs and 2 (1140 contigs) to 179 (one contig) in the drought stress contigs representing genes with different levels of expression. Consequently, total lengths of approximately 3395.7 and 3873.5 kb of the non-stress and drought EST libraries were analyzed (Table 1).
Table 1.
Summary statistics of the B. napus ESTs obtained from non-stress and drought stress conditions
| Library | Number of high-quality ESTs | Average length of high-quality ESTs (bp) | Total number of unigenes | Number of contigs | Average length of contigs (bp) | Number of Singletons | Approximate total length of unigenes (Kbp) |
|---|---|---|---|---|---|---|---|
| Non-stress | 6037 | 863 | 4149 | 794 | 898 | 3355 | 3395.7 |
| Drought stress | 13059 | 741 | 5817 | 1945 | 791 | 3872 | 3873.5 |
Three thousand six hundred and four thousand seven hundred sixty-eight unigenes of the non-stress and drought stress B. napus transcriptomes were annotated based on BLASTX homology search against the Arabidopsis protein database. 80.82 and 75.45 % of the unigenes respective to non-stress and drought stress conditions showed significant similarity with genes of known or putative function (Table 2). 5.95 and 6.52 % of the unigenes from the non-stress and drought stress EST libraries were matched to genes with unknown function. The remaining proportion of the unigenes, i.e., 13.23 and 18.03 % corresponding to the non-stress and drought stress libraries respectively, showed no significant match with the Arabidopsis protein database implying that they may be specific genes to B. napus or genes whose biological functions have not yet been reported in Arabidopsis. Therefore, they can be considered as a source for gene discovery.
Table 2.
Summary of BLASTX search for the B. napus unigenes obtained from non-stress and drought stress EST libraries against the Arabidopsis proteins database with expect value cut-off at 10−5
| Library | Unigenes with known function | Unigenes with putative function | Unigenes with unknown function | Unigenes with no significant matches |
|---|---|---|---|---|
| Non-stress | 3236 (78 %) | 117 (2.82 %) | 247 (5.95 %) | 549 (13.23 %) |
| Drought stress | 4165 (71.6 %) | 224 (3.85 %) | 379 (6.52 %) | 1049 (18.03 %) |
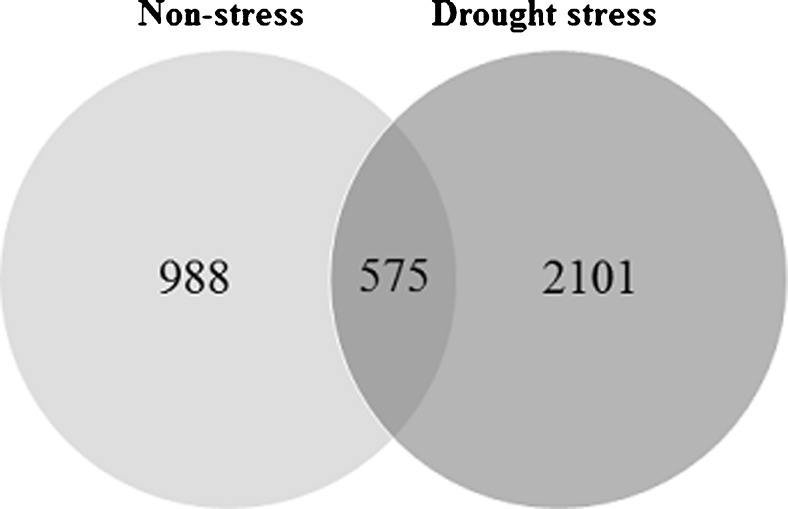
GeneCodis was utilized for functional enrichment of the unigenes with significant hits derived from the two EST libraries in the context of biological process and molecular function ontologies. Based on GeneCodis database, the 1573 and 2676 unigenes of the non-stress and drought stress libraries were ontologically informative and selected for enrichment analysis. Of those, the 988 and 2101 sequences were specific to the non-stress and drought stress EST libraries, respectively, while the 575 unigenes were common between them. The results are shown by Venn diagram in Fig. 1.
Fig. 1.
Venn diagram showing the common and specific informative unigenes obtained from the non-stress and drought EST libraries
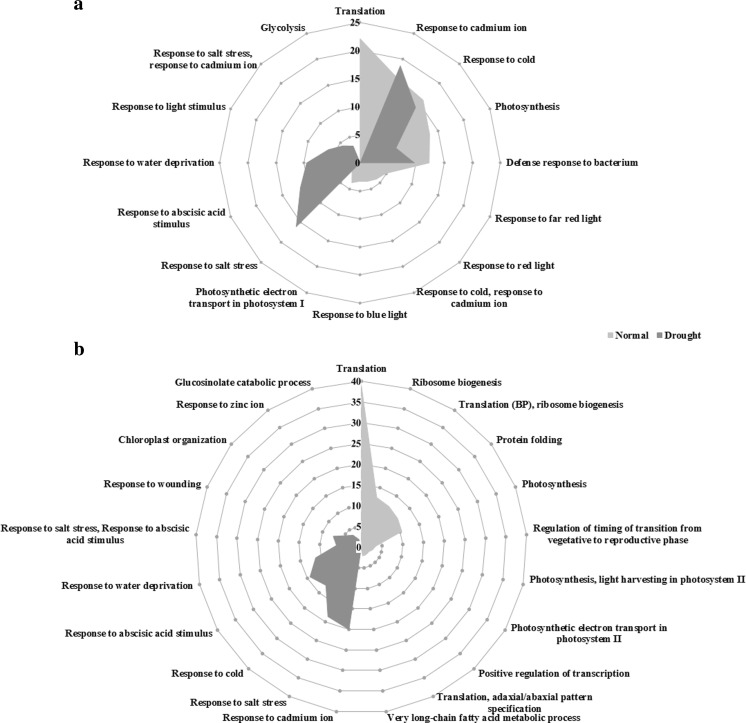
To demonstrate how the genome functions altered in response to drought stress, modular enrichment analysis was conducted for all the informative unigenes obtained from each non-stress and drought stress conditions, and also for the specific informative unigenes of each condition. As a result, all the unigenes of the non-stress and drought stress libraries were enriched in 11 and 10 annotation modules based on biological process ontology (Fig. 2a). The two transcriptomes shared some common annotation modules namely “Photosynthesis”, “Response to cold”, “Response to cadmium ion”, “Defense response to bacterium”, “Response to salt stress” and “electron transport in photosystem Ι”. Over 50 % of the unigenes derived from non-stress conditions were enriched in non-stress responsive groups such as translation and photosynthesis. The results showed that drought stress caused significant changes in the abundance of unigenes assigned to various functional modules. As shown in Fig. 2a, the activity of the enriched annotation modules namely response to “salt stress”, “abscisic acid (ABA)”, “water deprivation”, “light stimulus”, “cadmium ion” and “glycolysis” significantly increased under drought stress conditions, indicating the dramatic change in B. napus genome functions toward response to environmental cues. On the other hand, the percentage of unigenes assigned to “translation”, “response to cold”, “photosynthesis”, “response to bacterium” and “response to far red, red and blue light” modules were significantly underrepresented under drought stress conditions. The results of modular enrichment analysis for the unigenes specific to each condition interestingly revealed that all the unigenes specifically expressed in drought stress fell into the functional modules involved in response to various stresses including cadmium ion, salt, cold, ABA, water deprivation and wounding (Fig. 2b).
Fig. 2.
The percentages of functional modules of (a) all and (b) specific informative unigenes derived from non-stress and drought stress B. napus transcriptomes according to biological process ontology
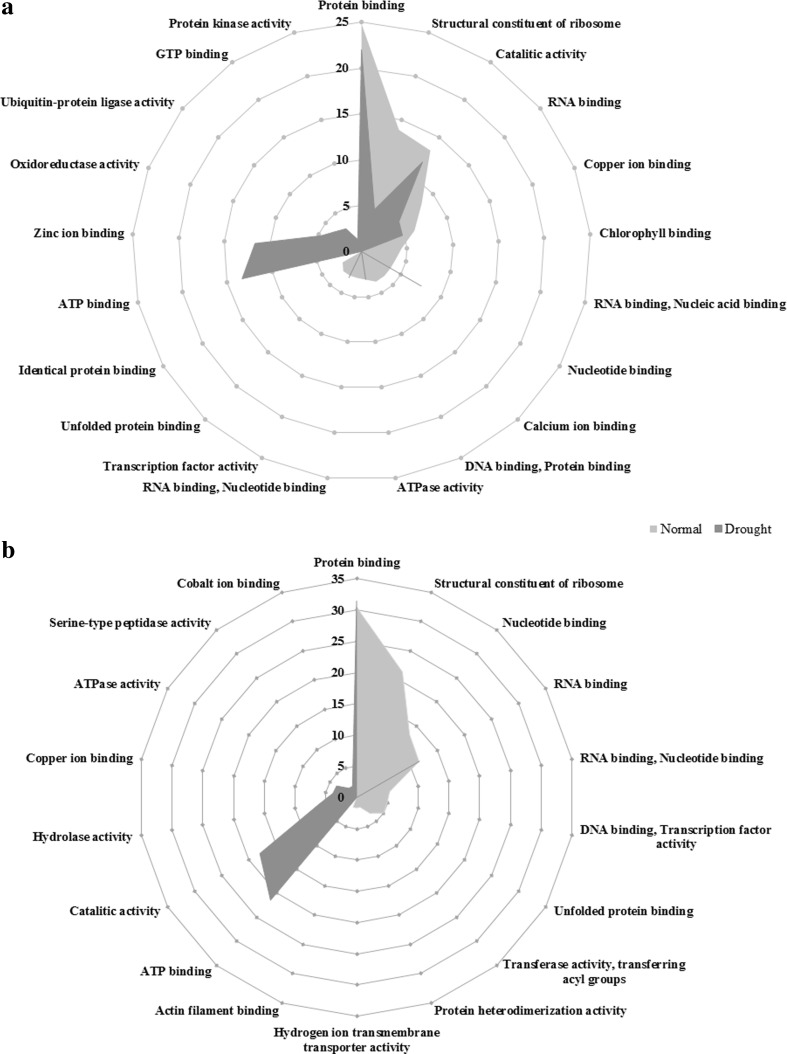
From point of view of molecular function ontology, all the informative unigenes derived from non-stress and drought stress conditions were enriched in 15 and 14 annotation modules, respectively; some of which were common for both conditions but with different abundances of the corresponding unigenes (Fig. 3a). The protein binding module encompassed the highest percentage of the unigenes in non-stress (24.82 %) and drought stress (22 %) conditions. The modules of identical protein binding (2.37 %) and Protein kinase activity (1.37 %) contained the lowest percentage of the unigenes obtained from non-stress and drought stress conditions, respectively. According to molecular function ontology, the activity of ATP binding, zinc ion binding, oxidoreductase, ubiquitin protein ligase, GTP binding, protein kinase, nucleotide binding and transcription factor functional modules enhanced in response to drought. Conversely, the number of transcripts assigned to the functional groups of protein binding, ribosomal constituent, RNA binding, copper ion binding, chlorophyll binding, calcium ion binding and ATPase activity declined under drought stress. Moreover, Fig. 3b illustrated that over 60 % of the unigenes specifically expressed in drought stress were assigned to the three modules including protein binding, ATP binding and catalytic activity. On the other hand, the specific unigenes of non-stress conditions showed different distribution among the functional modules and were divided into more distinct modules; however the module of protein binding still had the maximum percentage (30.45 %) of the unigenes.
Fig. 3.
The percentages of functional modules of (a) all and (b) specific informative unigenes derived from non-stress and drought stress B. napus transcriptomes according to molecular function ontology
The functional ontology analysis revealed a substantial shift in the B. napus transcriptome to govern cellular drought responsiveness via different stress-activated mechanisms including biosynthesis and signaling of various hormones, signaling and neutralization of Reactive Oxygen Species (ROS) and regulation of cellular energy homeostasis. The results were consistent with the fact that plant stress responses cause reduction in photosynthesis and cellular energy thereby plants need to constantly adjust their energy-associated transcriptome (Avin‐Wittenberg et al. 2012). Functional enrichment analysis in Arabidopsis and Rice also demonstrated significant changes in the functional modules related to response and regulation of various plant hormones and photosynthesis under drought and bacterial stresses (Shaik and Ramakrishna 2013).
Identification of the regulatory components
In order to discover the differentially expressed genes between the B. napus transcriptomes in non-stress and drought stress conditions, the total of 19,096 high quality ESTs generated from both EST libraries were assembled and consequently 2345 highly represented genes (contigs) comprising 9534 ESTs (49.93 %) were further analyzed. The number of ESTs in the contigs ranged from 2 (1322 contigs) to 180 (1 contig). The contigs showed the average length of 824 bp.
The 814 contigs with significantly differential expression between non-stress and drought stress conditions were identified using Audic and Claverie test (p-value ≤ 0.05). The contigs are from hereon named as genes. Of those, 441 genes which showed equal or greater than three differences in the number of ESTs between the two EST libraries were used to identify the highly differential regulatory genes and subsequently to infer a simplified plausible drought-responsive regulatory network. As a result, the 26 regulatory genes of the three major groups encoding transcription factors, protein kinases and protein phosphatases were annotated among the differentially expressed genes. The log2 fold change of expression of the genes was calculated and shown in Table 3.
Table 3.
The list of proteins encoded by the regulatory genes with significant differential expressions (p-value ≤ 0.05) between the B. napus transcriptomes derived from non- and drought stress conditions. The proteins were annotated based on homology to Arabidopsis proteins
| Regulatory protein | Family | TAIR ID | Annotation | Fold change (Log2) |
|---|---|---|---|---|
| Transcription factor | C3H | AT4G29190 | Zinc finger C-x8-C-x5-C-x3-H type | 2.54 |
| FHA-SMAD | AT5G67030 | ABA1, zeaxanthin epoxidase (ZEP) | 2.18 | |
| WD40-like | AT1G21680 | DPP6 N-terminal domain-like | 1.87 | |
| CCHC(Zn) | AT2G24590 | RSZ22a | 1.67 | |
| AUX-IAA | AT3G04730 | IAA16, indoleacetic acid-induced protein 16 | 1.45 | |
| C2H2 | AT5G14420 | RGLG2, RING domain ligase2 | 1.44 | |
| GRAS | AT2G37650 | GRAS family transcription factor | 1.44 | |
| Homobox-WOX | AT2G22430 | ATHB6, homeobox protein 6 | 1.44 | |
| MADS-MIKC | AT2G45650 | AGL6, AGAMOUS-like 6 | 1.44 | |
| MYB-HB-like | AT5G05090 | Homeodomain-like | 1.44 | |
| WD40-like | AT1G21680 | DPP6 N-terminal domain-like | 1.44 | |
| bHLH | AT4G36540 | BEE2, BR enhanced expression 2 | −2.86 | |
| C2C2-CO-like | AT5G24930 | ATCOL4, CONSTANS-like 4 | −2.86 | |
| Hap2/NF-YA | AT5G12840 | HAP2A, nuclear factor Y subunit A1 | −2.86 | |
| MADS-MIKC | AT2G45660 | AGL20, SOC1,AGAMOUS-like 20 | −2.86 | |
| Znf-B | AT1G68190 | B-box zinc finger | −2.86 | |
| LIM | AT1G10200 | WLIM1 | −3.38 | |
| Protein kinase | Casein Kinase II | AT3G50000 | CKA2, casein kinase II, alpha chain 2 | 1.87 |
| Leucine Rich Repeat | AT4G33430 | BAK1, BRI1-associated receptor kinase | 1.67 | |
| MAPK | AT2G43790 | MPK6, MAP kinase 6 | 1.44 | |
| SNF1 Related Protein | AT4G30960 | CIPK6, SNRK3.14 | 1.44 | |
| CDC2 Like | AT1G18670 | IBS1, Protein kinase | −2.86 | |
| Receptor Like Kinase I | AT3G56050 | Protein kinase | −2.86 | |
| S Domain (Type 1) | AT1G56145 | Leucine-rich repeat protein kinase | −2.86 | |
| Serine/threonine | AT2G42960 | Protein kinase | −2.86 | |
| Protein phosphatase | Protein phosphatase 2CA | AT4G26080 | ABA insensitive 1; ABI1 | 2.02 |
The 17 transcription factor genes belonging to 15 different families were found indicating that B. napus has evolved a complex regulatory machinery to cope with drought stress. These genes showed different magnitudes and directions of expression changes in response to drought stress (Table 3). The expression of eleven transcription factors significantly increased whereas the six transcription factors showed decreased expression. The gene (AT4G29190) encoding a member of C3H transcription factor family showed the highest level of upregulation in response to drought stress, whilst WLIM1 transcription factor gene had the maximum downregulation following drought stress. In addition, eight protein kinase genes and one protein phosphatase gene were identified showing significant alterations in their expressions during drought stress (Table 3). A significant rise was observed in the expression of the four protein kinase genes (CKA2, BAK1, MPK6 and CIPK6) as well as the protein phosphatase gene, ABI1; while the other four protein kinase genes were sharply downregulated in response to drought stress.
We also found the six miRNAs which were significantly differentially expressed during drought stress revealing the involvement of a post-transcriptional level of regulation for B. napus drought response (Table 4). Five miRNAs were upregulated and one miRNA was downregulated under drought stress conditions. Interestingly, only one miRNAs namely miR396a has already been reported in B. napus and the other five miRNAs are newly reported in this study. There were four miRNAs (miR5658, miR396b, miR1536 and miR1509) with 21 nucleotides in length, while the other two miRNAs (miR2919 and miR435) had the size of 19 and 20 nucleotides, respectively. Further analysis predicted that the seven identified regulatory genes with differential expression between non-stress and drought conditions, including six transcription factor genes and one protein kinase gene were the direct targets of the five differentially expressed miRNAs (Table 4). No target gene was found for miR1509b among the differential regulatory genes. It is noteworthy that miR1536 targeted the three differentially expressed transcription factors (AGL6, IAA16 and RGLG2) showing its importance in regulating drought responses of B. napus.
Table 4.
The miRNAs with significant differential expressions (p-value ≤ 0.05) between the B. napus transcriptomes derived from non- and drought stress conditions. The best matched miRNAs obtained from miRBase database and their potential target genes are also represented
| B. napus mature miRNA sequence | The best matched miRNA | Fold change (Log2) | Target gene | UPE* | Inhibition mechanism |
|---|---|---|---|---|---|
| AUGAUUAUGAUUAUGAUGAGG | ath-miR5658 | 2.37 | BAK1 | 8.098 | Translation |
| AAAGGGGGGGGGGGAAUUU | osa-miR2919 | 0.91 | ABA1 | 22.646 | Cleavage |
| UUCCACAACUUUCUUCAAGUC | Bna-miR396a | 0.91 | B-box zinc finger | 17.6 | Translation |
| UAGAAAAGACAAAUCUGUUUA | gma-miR1536 | 0.91 | AGL6, IAA16, RGLG2 | 13.749 | Cleavage |
| UUUCCCGGUAUUGGACUUGG | osa-miR435 | 0.91 | COL4 | 21.26 | Translation |
| UAAAUCAAGGAAAGAAGGGUU | gma-miR1509b | −2.85 | – | – | – |
* Maximum energy to unpair the target site. The less energy means the more possibility that small RNA is able to contact (and cleave) target mRNA
The regulatory network was predicted among the identified components (Fig. 4). Eleven differentially expressed regulatory genes including seven transcription factors, three protein kinases and one protein phosphatases were placed in the network. Also, two genes namely ABI2 and BRI1 were added to the network by Pathway Studio software. Furthermore, the four identified drought responsive miRNAs (miR5658, miR1536, miR435 and miR2919) were connected to their targets. The network uncovered the relationships among the differentially expressed regulatory genes and the interactions between various regulatory pathways towards an adaptive response to drought in B. napus. These are detailed as follows.
Fig. 4.
The network showing interactions among the drought responsive identified in B. napus
Regulation of ABA biosynthesis
Notably, all the differential regulatory components within the predicted network were, directly or indirectly, connected to the phytohormone ABA, highlighting the significant roles of ABA in B. napus drought responsiveness. Alteration of cellular ABA levels is essential for several developmental and stress responsive processes (Fujita et al. 2011). The results showed that drought stress directly and significantly activated the expression of B. napus ortholog of ABA1 involved in the first step of ABA biosynthesis. The expression level of ABA1 was clearly upregulated by ABA, drought and salt in Arabidopsis shoots (Xiong et al. 2002), whereas its expression was induced by drought in tomato roots but not leaves (Thompson et al. 2000). The regulatory network presented a mutual connection between ABA and ABA1 that is consistent with that ABA synthesis is under a positive feedback regulation as the expression of some genes involved in ABA metabolism are regulated by ABA (Xiong and Zhu 2003). Nevertheless, B. napus ABA1 was known as the potential target of miR2919 which was induced by drought stress. This finding suggested a possible negative regulatory mechanism which antagonistically acts against positive feedback mechanism to precisely regulate ABA1 expression and subsequently ABA level.
Stress signaling through ROS regulation
In this study, we identified a protein phosphatase gene (ABI1), which its expression was significantly enhanced by drought stress. In Arabidopsis, ABI1 and ABI2 are the members of protein phosphatase 2C (PP2C) group and act as the negative regulators of ABA signaling (Leung et al. 1997). As shown in Fig. 4, ABI1 expression is under the direct influence of drought stress, and ABA hormone. ABI1 mediates signal transduction between ABA reception and ROS production under stress conditions (Murata et al. 2001). The Previous studies indicated that MAP kinase signaling is activated following ROS accumulation, as testified to the induction of MPK3 and MPK6 by H2O2 (Jammes et al. 2009; Xing et al. 2008). Interestingly, in this study, the expression of a B. napus homolog of MPK6 was significantly upregulated during drought stress. Arabidopsis MPK6 promotes stomatal closure under osmotic stress (Liu 2012). MPK6 recruits ethylene signaling pathway via phosphorylation and thereby activation of the Arabidopsis transcription factor ERF6 (ETHYLEN RESPONSE FACTOR 6) (Meng et al. 2013). ERF6 specifically binds to the ROS-responsive elements (ROSEs) and interacts with MPK6 to regulate ROS-responsive gene expression at the transcriptional level (Wang et al. 2013). As a result, the identification of the B. napus ABI1 and MPK6 provided evidence to support the role of ROS-mediated-signaling in coping with drought stress.
Response to drought stress via the ubiquitin 26S proteasome proteolytic pathway
The expression of RGLG2, a central component of the ubiquitin 26S proteasome pathway, was significantly downregulated in the drought-stressed B. napus transcriptome. Arabidopsis RGLG2 is a RING domain E3 ligase which mediates ubiquitination of the transcription factor Ethylene Response Factor 53 (ERF53) for proteasome degradation (Cheng et al. 2012). The expression of some drought-responsive transcription factors genes such as DREB2A, CBF4 and RAP2.4 is controlled by ERF53 (Haake et al. 2002; Lin et al. 2008; Sakuma et al. 2006). Therefore, downregulation of RGLG2 expression is required for induction of ERF53 and thereby activation of its downstream genes under drought stress. This study revealed that the B. napus RGLG2 is one of the possible targets of miR1536 which was identified for the first time in the B. napus transcriptome. Similar to ERF53, the involvement of a ubiquitin E3 ligase was known to regulate the abundance of the drought responsive homeobox-leucine zipper transcription factor, AtHB6 (Lechner et al. 2011). AtHB6 is a target of ABI1 and a negative regulator of ABA signaling pathway in Arabidopsis (Himmelbach et al. 2002). The significant upregulation of the B. napus HB6 expression along with the downregulation of the B. napus RGLG2 expression under drought stress resembled the regulatory mechanism, as occurred in Arabidopsis.
Drought-induced Ca2+ signaling and auxin response
The transcript level of B. napus CIPK6 gene was significantly upregulated during drought stress. Arabidopsis CIPK6 encodes a protein kinase involved in sensing Ca2+ signal and induction of Ca2+-dependent ABA signaling (Batistič and Kudla 2012; Halford and Hey 2009; Kim and Maik 2010; Luan 2009; Weinl and Kudla 2009). The overexpression of GhCIPK6 from cotton significantly enhanced tolerance to salt, drought and ABA stresses in transgenic Arabidopsis (He et al. 2013). CIPK6 is also known to function in auxin transport (Tripathi et al. 2009). It has been demonstrated that auxin might participate in positive regulation of drought tolerance through regulation of root architecture, expression of ABA-responsive genes, ROS metabolism and metabolic homeostasis (Shi et al. 2014). In accordance with the activation of B. napus CIPK6 and auxin in response to drought, the results also showed significant downregulation of the B. napus IAA16, a negative regulator of auxin signaling (Park et al. 2011), under drought stress by the increased activity of miR1536. Taken together, these findings uncovered the significance of ABA-mediated-auxin signaling in the modulation of B. napus drought stress responses.
The role of brassinosteroids
The results presented the contribution of brassinosteroids (BRs) to drought response in B. napus. BRs play essential roles in regulating various biological processes including responses to the environment (Wang et al. 2014). BR signaling is induced by binding of BR to a membrane receptor kinase, BRI1. BRI1 makes hetero oligomers with another receptor kinase, BAK1 to shape a phosphorylation string that ultimately causes functional changes of target genes (Nam and Li 2002; Wang et al. 2008). The BRI1 receptor complex induces the redundant genes encoding basic helix-loop-helix (bHLH) namely BEE1, BEE2 and BEE3 (Friedrichsen et al. 2002). The B. napus transcriptome showed a significant increase in the expression of BAK1 under drought stress, although the transcripts of this gene were the potential targets of the drought responsive miR5658 indicating the existence of a negative post-transcriptional regulation for BAK1. On the other hand, the expression of BEE2 was significantly downregulated under drought stress in B. napus leaves. As illustrated in the regulatory network, BEE2 was under the negative regulation of ABA (Friedrichsen et al. 2002). These results showed that ABA and BR signaling pathways act antagonistically in response to drought stress in B. napus. BR and ABA antagonize each other in many biological processes (Wang et al. 2014).
Developmental regulation during drought stress
The expression of the B. napus homologs of the transcription factor genes CONSTANS-like 4 (COL4) and AGAMOUS-LIKE 20 (AGL20) (also known as SOC1) were significantly downregulated under drought stress. The drought responsive regulatory network (Fig. 4) highlighted the induction of miR435 which potentially targets the expression of the B. napus COL4. In agreement with our results, the expression of a number of CONSTANS-like family members was downregulated by drought stress in rice (Shaik and Ramakrishna 2013). The expression of Arabidopsis COL4 was strongly induced by ABA, salt, and osmotic stress (Min et al. 2014). It has been demonstrated that the Arabidopsis SOC1, as a positive regulator of flowering, was upregulated following 4 to 5 days drought treatment indicating that plants reprogram flowering process to shorten the life cycle and promote survival under extreme environments (Su et al. 2013). The apparently contrary responses of these genes to drought may be due to different timing and intensity of stress, and even the existence of various strategies to set time to flowering under stress conditions in plants. Further investigations are required to determine the regulatory roles of B. napus COL4 and SOC1 under stress conditions.
In conclusion, this EST-based transcriptome study provided an insight into how B. napus genome responds to drought stress. The results clearly showed a substantial change in the B. napus genome expression in response to drought in which the corresponding regulatory mechanisms were significantly activated. We identified a number of significant drought responsive regulatory genes and miRNAs with various functions and characterized their interactions using the regulatory network. Our findings indicated that B. napus recruits a complex and overlapping regulatory system to modulate response to drought stress. The results highlighted the importance of biosynthesis and signaling of various plant hormones and their crosstalks to coordinate physiological events under drought stress in B. napus. The obtained drought responsive regulatory network can be utilized to select appropriate genes for transgenic breeding to develop B. napus drought tolerant cultivars.
References
- Ahuja I, de Vos RC, Bones AM, Hall RD. Plant molecular stress responses face climate change. Trends Plant Sci. 2010;15:664–674. doi: 10.1016/j.tplants.2010.08.002. [DOI] [PubMed] [Google Scholar]
- Ashraf M. Inducing drought tolerance in plants: recent advances. Biotechnol Adv. 2010;28:169–183. doi: 10.1016/j.biotechadv.2009.11.005. [DOI] [PubMed] [Google Scholar]
- Audic S, Claverie J-M. The significance of digital gene expression profiles. Genome Res. 1997;7:986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- Avin‐Wittenberg T, Tzin V, Angelovici R, Galili G. Deciphering energy‐associated gene networks operating in the response of Arabidopsis plants to stress and nutritional cues. Plant J. 2012;70:954–966. doi: 10.1111/j.1365-313X.2012.04926.x. [DOI] [PubMed] [Google Scholar]
- Batistič O, Kudla J. Analysis of calcium signaling pathways in plants. Biochim Biophys Acta (BBA) Gen Subj. 2012;1820:1283–1293. doi: 10.1016/j.bbagen.2011.10.012. [DOI] [PubMed] [Google Scholar]
- Carlson JE, et al. EST database for early flower development in California poppy (Eschscholzia californica Cham., Papaveraceae) tags over 6000 genes from a basal eudicot. Plant Mol Biol. 2006;62:351–369. doi: 10.1007/s11103-006-9025-y. [DOI] [PubMed] [Google Scholar]
- Carmona-Saez P, Chagoyen M, Tirado F, Carazo JM, Pascual-Montano A. GENECODIS: a web-based tool for finding significant concurrent annotations in gene lists. Genome Biol. 2007;8:R3. doi: 10.1186/gb-2007-8-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng M-C, Hsieh E-J, Chen J-H, Chen H-Y, Lin T-P. Arabidopsis RGLG2, functioning as a RING E3 ligase, interacts with AtERF53 and negatively regulates the plant drought stress response. Plant Physiol. 2012;158:363–375. doi: 10.1104/pp.111.189738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Zhao PX. psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 2011;39:W155–W159. doi: 10.1093/nar/gkr319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Sinharoy S, Udvardi M, Zhao P. PlantTFcat: an online plant transcription factor and transcriptional regulator categorization and analysis tool. BMC Bioinforma. 2013;14:321. doi: 10.1186/1471-2105-14-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deokar AA, et al. Comparative analysis of expressed sequence tags (ESTs) between drought-tolerant and-susceptible genotypes of chickpea under terminal drought stress. BMC Plant Biol. 2011;11:70. doi: 10.1186/1471-2229-11-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diepenbrock W. Yield analysis of winter oilseed rape (Brassica napus L.): a review. Field Crop Res. 2000;67:35–49. doi: 10.1016/S0378-4290(00)00082-4. [DOI] [Google Scholar]
- Dong J, Keller WA, Yan W, Georges F. Gene expression at early stages of Brassica napus seed development as revealed by transcript profiling of seed-abundant cDNAs. Planta. 2004;218:483–491. doi: 10.1007/s00425-003-1124-2. [DOI] [PubMed] [Google Scholar]
- Duque AS et al. (2013) Abiotic stress responses in plants: unraveling the complexity of genes and networks to survive. INTECH Open Access Publisher
- Farooq M, Wahid A, Kobayashi N. Plant drought stress: effects, mechanisms and management. Agron Sustain Dev. 2009;29:185–212. doi: 10.1051/agro:2008021. [DOI] [Google Scholar]
- Farooq M, Hussain M, Wahid A, Siddique KHM (2012) Drought stress in plants: an overview. In: Aroca R (ed) Plant Responses to Drought Stress. Springer Berlin Heidelberg, pp 1–33. doi:10.1007/978-3-642-32653-0_1
- Fourmann M, Barret P, Froger N, Baron C, Charlot F, Delourme R, Brunel D. From Arabidopsis thaliana to Brassica napus: development of amplified consensus genetic markers (ACGM) for construction of a gene map. Theor Appl Genet. 2002;105:1196–1206. doi: 10.1007/s00122-002-1040-z. [DOI] [PubMed] [Google Scholar]
- Friedrichsen DM, et al. Three redundant brassinosteroid early response genes encode putative bHLH transcription factors required for normal growth. Genetics. 2002;162:1445–1456. doi: 10.1093/genetics/162.3.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Fujita M, Shinozaki K, Yamaguchi-Shinozaki K. ABA-mediated transcriptional regulation in response to osmotic stress in plants. J Plant Res. 2011;124:509–525. doi: 10.1007/s10265-011-0412-3. [DOI] [PubMed] [Google Scholar]
- Gao W-R, et al. Comparative analysis of ESTs in response to drought stress in chickpea (C. arietinum L.) Biochem Biophys Res Commun. 2008;376:578–583. doi: 10.1016/j.bbrc.2008.09.030. [DOI] [PubMed] [Google Scholar]
- Golldack D, Lüking I, Yang O. Plant tolerance to drought and salinity: stress regulating transcription factors and their functional significance in the cellular transcriptional network. Plant Cell Rep. 2011;30:1383–1391. doi: 10.1007/s00299-011-1068-0. [DOI] [PubMed] [Google Scholar]
- Gorantla M, Babu P, Lachagari VR, Reddy A, Wusirika R, Bennetzen JL, Reddy AR. Identification of stress-responsive genes in an indica rice (Oryza sativa L.) using ESTs generated from drought-stressed seedlings. J Exp Bot. 2007;58:253–265. doi: 10.1093/jxb/erl213. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber M, et al. Analysis of expressed sequence tags in Brassica napus cotyledons damaged by crucifer flea beetle feeding. Genome. 2012;55:118–133. doi: 10.1139/g11-083. [DOI] [PubMed] [Google Scholar]
- Guo N et al (2014) Computational identification of novel microRNAs and targets in Glycine max. Mol Biol Rep 1–11 [DOI] [PubMed]
- Haake V, Cook D, Riechmann J, Pineda O, Thomashow MF, Zhang JZ. Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol. 2002;130:639–648. doi: 10.1104/pp.006478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadiarto T, Tran L-SP. Progress studies of drought-responsive genes in rice. Plant Cell Rep. 2011;30:297–310. doi: 10.1007/s00299-010-0956-z. [DOI] [PubMed] [Google Scholar]
- Halford N, Hey S. Snf1-related protein kinases (SnRKs) act within an intricate network that links metabolic and stress signalling in plants. Biochem J. 2009;419:247–259. doi: 10.1042/BJ20082408. [DOI] [PubMed] [Google Scholar]
- Han J, Xie H, Kong M, Sun Q, Li R, Pan J. Computational identification of miRNAs and their targets in Phaseolus vulgaris. Genet Mol Res. 2014;13:310. doi: 10.4238/2014.January.17.16. [DOI] [PubMed] [Google Scholar]
- He L, Yang X, Wang L, Zhu L, Zhou T, Deng J, Zhang X. Molecular cloning and functional characterization of a novel cotton CBL-interacting protein kinase gene (GhCIPK6) reveals its involvement in multiple abiotic stress tolerance in transgenic plants. Biochem Biophys Res Commun. 2013;435:209–215. doi: 10.1016/j.bbrc.2013.04.080. [DOI] [PubMed] [Google Scholar]
- Himmelbach A, Hoffmann T, Leube M, Höhener B, Grill E. Homeodomain protein ATHB6 is a target of the protein phosphatase ABI1 and regulates hormone responses in Arabidopsis. EMBO J. 2002;21:3029–3038. doi: 10.1093/emboj/cdf316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang E-W, Shin S-J, Kwon H-B. Identification of microRNAs and their putative targets that respond to drought stress in Solanum tuberosum. J Korean Soc Appl Biol Chem. 2011;54:317–324. doi: 10.3839/jksabc.2011.051. [DOI] [Google Scholar]
- Jammes F, et al. MAP kinases MPK9 and MPK12 are preferentially expressed in guard cells and positively regulate ROS-mediated ABA signaling. Proc Natl Acad Sci. 2009;106:20520–20525. doi: 10.1073/pnas.0907205106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khraiwesh B, Zhu J-K, Zhu J. Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim Biophys Acta (BBA) Gene Regul Mech. 2012;1819:137–148. doi: 10.1016/j.bbagrm.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T-H, Maik B. Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu Rev Plant Biol. 2010;61:561. doi: 10.1146/annurev-arplant-042809-112226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lata C, Sahu PP, Prasad M. Comparative transcriptome analysis of differentially expressed genes in foxtail millet (Setaria italica L.) during dehydration stress. Biochem Biophys Res Commun. 2010;393:720–727. doi: 10.1016/j.bbrc.2010.02.068. [DOI] [PubMed] [Google Scholar]
- Lechner E, et al. MATH/BTB CRL3 receptors target the homeodomain-leucine zipper ATHB6 to modulate abscisic acid signaling. Dev Cell. 2011;21:1116–1128. doi: 10.1016/j.devcel.2011.10.018. [DOI] [PubMed] [Google Scholar]
- Leung J, Merlot S, Giraudat J. The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell Online. 1997;9:759–771. doi: 10.1105/tpc.9.5.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R-C, Park H-J, Wang H-Y. Role of Arabidopsis RAP2. 4 in regulating light-and ethylene-mediated developmental processes and drought stress tolerance. Mol Plant. 2008;1:42–57. doi: 10.1093/mp/ssm004. [DOI] [PubMed] [Google Scholar]
- Liu Y. Roles of mitogen-activated protein kinase cascades in ABA signaling. Plant Cell Rep. 2012;31:1–12. doi: 10.1007/s00299-011-1130-y. [DOI] [PubMed] [Google Scholar]
- Lu X-Y, Huang X-L. Plant miRNAs and abiotic stress responses. Biochem Biophys Res Commun. 2008;368:458–462. doi: 10.1016/j.bbrc.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Luan S. The CBL–CIPK network in plant calcium signaling. Trends Plant Sci. 2009;14:37–42. doi: 10.1016/j.tplants.2008.10.005. [DOI] [PubMed] [Google Scholar]
- Masoudi-Nejad A, et al. EGassembler: online bioinformatics service for large-scale processing, clustering and assembling ESTs and genomic DNA fragments. Nucleic Acids Res. 2006;34:W459–W462. doi: 10.1093/nar/gkl066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzucotelli E, Mastrangelo AM, Crosatti C, Guerra D, Stanca AM, Cattivelli L. Abiotic stress response in plants: when post-transcriptional and post-translational regulations control transcription. Plant Sci. 2008;174:420–431. doi: 10.1016/j.plantsci.2008.02.005. [DOI] [Google Scholar]
- Meng X, Xu J, He Y, Yang K-Y, Mordorski B, Liu Y, Zhang S. Phosphorylation of an ERF transcription factor by Arabidopsis MPK3/MPK6 regulates plant defense gene induction and fungal resistance. Plant Cell Online. 2013;25:1126–1142. doi: 10.1105/tpc.112.109074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min JH, Chung JS, Lee KH, Kim CS (2014) The CONSTANS‐like 4 transcription factor, AtCOL4, positively regulates abiotic stress tolerance through an abscisic acid‐dependent manner in Arabidopsis. J Integr Plant Biol [DOI] [PubMed]
- Murata Y, Pei Z-M, Mori IC, Schroeder J. Abscisic acid activation of plasma membrane Ca2+ channels in guard cells requires cytosolic NAD (P) H and is differentially disrupted upstream and downstream of reactive oxygen species production in abi1-1 and abi2-1 protein phosphatase 2C mutants. Plant Cell Online. 2001;13:2513–2523. doi: 10.1105/tpc.010210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam KH, Li J. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell. 2002;110:203–212. doi: 10.1016/S0092-8674(02)00814-0. [DOI] [PubMed] [Google Scholar]
- Nikitin A, Egorov S, Daraselia N, Mazo I. Pathway studio—the analysis and navigation of molecular networks. Bioinformatics. 2003;19:2155–2157. doi: 10.1093/bioinformatics/btg290. [DOI] [PubMed] [Google Scholar]
- Niu Y, et al. Global analysis of gene expression profiles in Brassica napus developing seeds reveals a conserved lipid metabolism regulation with Arabidopsis thaliana. Mol Plant. 2009;2:1107–1122. doi: 10.1093/mp/ssp042. [DOI] [PubMed] [Google Scholar]
- Panda D, Dehury B, Sahu J, Barooah M, Sen P, Modi MK. Computational identification and characterization of conserved miRNAs and their target genes in garlic (Allium sativum L.) expressed sequence tags. Gene. 2014;537:333–342. doi: 10.1016/j.gene.2014.01.010. [DOI] [PubMed] [Google Scholar]
- Park J, Lee N, Kim W, Lim S, Choi G. ABI3 and PIL5 collaboratively activate the expression of SOMNUS by directly binding to its promoter in imbibed Arabidopsis seeds. Plant Cell Online. 2011;23:1404–1415. doi: 10.1105/tpc.110.080721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin IA, Lydiate D, Trick M. Assessing the level of collinearity between Arabidopsis thaliana and Brassica napus for A. thaliana chromosome 5. Genome. 2002;45:356–366. doi: 10.1139/g01-160. [DOI] [PubMed] [Google Scholar]
- Patanun O, Lertpanyasampatha M, Sojikul P, Viboonjun U, Narangajavana J. Computational identification of microRNAs and their targets in cassava (Manihot esculenta Crantz.) Mol Biotechnol. 2013;53:257–269. doi: 10.1007/s12033-012-9521-z. [DOI] [PubMed] [Google Scholar]
- Pratt LH, et al. Sorghum expressed sequence tags identify signature genes for drought, pathogenesis, and skotomorphogenesis from a milestone set of 16,801 unique transcripts. Plant Physiol. 2005;139:869–884. doi: 10.1104/pp.105.066134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymer PL. Canola: an emerging oilseed crop. Trends New Crops New Uses. 2002;1:122–126. [Google Scholar]
- Romualdi C, Bortoluzzi S, d’Alessi F, Danieli GA. IDEG6: a web tool for detection of differentially expressed genes in multiple tag sampling experiments. Physiol Genomics. 2003;12:159–162. doi: 10.1152/physiolgenomics.00096.2002. [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Maruyama K, Osakabe Y, Qin F, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell Online. 2006;18:1292–1309. doi: 10.1105/tpc.105.035881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Kamei A, Yamaguchi-Shinozaki K, Shinozaki K. Molecular responses to drought, salinity and frost: common and different paths for plant protection. Curr Opin Biotechnol. 2003;14:194–199. doi: 10.1016/S0958-1669(03)00030-2. [DOI] [PubMed] [Google Scholar]
- Seki M, Umezawa T, Urano K, Shinozaki K. Regulatory metabolic networks in drought stress responses. Curr Opin Plant Biol. 2007;10:296–302. doi: 10.1016/j.pbi.2007.04.014. [DOI] [PubMed] [Google Scholar]
- Shaik R, Ramakrishna W. Genes and co-expression modules common to drought and bacterial stress responses in Arabidopsis and rice. PLoS One. 2013;8:e77261. doi: 10.1371/journal.pone.0077261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamloo-Dashtpagerdi R, Razi H, Lindlöf A, Niazi A, Dadkhodaie A, Ebrahimie E. Comparative analysis of expressed sequence tags (ESTs) from Triticum monococcum shoot apical meristem at vegetative and reproductive stages. Genes Genomics. 2013;35:365–375. doi: 10.1007/s13258-013-0091-7. [DOI] [Google Scholar]
- Shi H, Chen L, Ye T, Liu X, Ding K, Chan Z. Modulation of auxin content in Arabidopsis confers improved drought stress resistance. Plant Physiol Biochem. 2014;82:209–217. doi: 10.1016/j.plaphy.2014.06.008. [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. Molecular responses to drought and cold stress. Curr Opin Biotechnol. 1996;7:161–167. doi: 10.1016/S0958-1669(96)80007-3. [DOI] [PubMed] [Google Scholar]
- Su Z, Ma X, Guo H, Sukiran NL, Guo B, Assmann SM, Ma H. Flower development under drought stress: morphological and transcriptomic analyses reveal acute responses and long-term acclimation in Arabidopsis. Plant Cell Online. 2013;25:3785–3807. doi: 10.1105/tpc.113.115428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Chinnusamy V, Zhu J, Zhu J-K. Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci. 2007;12:301–309. doi: 10.1016/j.tplants.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Sunkar R, Li Y-F, Jagadeeswaran G. Functions of microRNAs in plant stress responses. Trends Plant Sci. 2012;17:196–203. doi: 10.1016/j.tplants.2012.01.010. [DOI] [PubMed] [Google Scholar]
- Tabas-Madrid D, Nogales-Cadenas R, Pascual-Montano A. GeneCodis3: a non-redundant and modular enrichment analysis tool for functional genomics. Nucleic Acids Res. 2012;40:W478–W483. doi: 10.1093/nar/gks402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AJ, Jackson AC, Parker RA, Morpeth DR, Burbidge A, Taylor IB (2000) Abscisic acid biosynthesis in tomato: regulation of zeaxanthin epoxidase and 9-cis-epoxycarotenoid dioxygenase mRNAs by light/dark cycles, water stress and abscisic acid. Plant Mol Biol 42:833–845 [DOI] [PubMed]
- Tripathi V, Parasuraman B, Laxmi A, Chattopadhyay D. CIPK6, a CBL‐interacting protein kinase is required for development and salt tolerance in plants. Plant J. 2009;58:778–790. doi: 10.1111/j.1365-313X.2009.03812.x. [DOI] [PubMed] [Google Scholar]
- Umezawa T, Fujita M, Fujita Y, Yamaguchi-Shinozaki K, Shinozaki K. Engineering drought tolerance in plants: discovering and tailoring genes to unlock the future. Curr Opin Biotechnol. 2006;17:113–122. doi: 10.1016/j.copbio.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Wang X, et al. Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev Cell. 2008;15:220–235. doi: 10.1016/j.devcel.2008.06.011. [DOI] [PubMed] [Google Scholar]
- Wang P, Du Y, Zhao X, Miao Y, Song C-P. The MPK6-ERF6-ROS-responsive cis-acting Element7/GCC box complex modulates oxidative gene transcription and the oxidative response in Arabidopsis. Plant Physiol. 2013;161:1392–1408. doi: 10.1104/pp.112.210724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Bai M-Y, Wang Z-Y. The brassinosteroid signaling network—a paradigm of signal integration. Curr Opin Plant Biol. 2014;21:147–153. doi: 10.1016/j.pbi.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinl S, Kudla J. The CBL–CIPK Ca2+−decoding signaling network: function and perspectives. New Phytol. 2009;184:517–528. doi: 10.1111/j.1469-8137.2009.02938.x. [DOI] [PubMed] [Google Scholar]
- Xie FL, Huang SQ, Guo K, Xiang AL, Zhu YY, Nie L, Yang ZM. Computational identification of novel microRNAs and targets in Brassica napus. FEBS Lett. 2007;581:1464–1474. doi: 10.1016/j.febslet.2007.02.074. [DOI] [PubMed] [Google Scholar]
- Xing Y, Jia W, Zhang J. AtMKK1 mediates ABA‐induced CAT1 expression and H2O2 production via AtMPK6‐coupled signaling in Arabidopsis. Plant J. 2008;54:440–451. doi: 10.1111/j.1365-313X.2008.03433.x. [DOI] [PubMed] [Google Scholar]
- Xiong L, Zhu J-K. Regulation of abscisic acid biosynthesis. Plant Physiol. 2003;133:29–36. doi: 10.1104/pp.103.025395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Lee H, Ishitani M, Zhu J-K. Regulation of osmotic stress-responsive gene expression by theLOS6/ABA1 locus in Arabidopsis. J Biol Chem. 2002;277:8588–8596. doi: 10.1074/jbc.M109275200. [DOI] [PubMed] [Google Scholar]
- Xoconostle-Cazares B, Ramirez-Ortega FA, Flores-Elenes L, Ruiz-Medrano R. Drought tolerance in crop plants. Am J Plant Physiol. 2010;5:241–256. doi: 10.3923/ajpp.2010.241.256. [DOI] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- Ye K, Chen Y, Hu X, Guo J. Computational identification of microRNAs and their targets in apple. Genes Genomics. 2013;35:377–385. doi: 10.1007/s13258-013-0070-z. [DOI] [Google Scholar]
- Young LW, Wilen RW, Bonham‐Smith PC. High temperature stress of Brassica napus during flowering reduces micro‐and megagametophyte fertility, induces fruit abortion, and disrupts seed production. J Exp Bot. 2004;55:485–495. doi: 10.1093/jxb/erh038. [DOI] [PubMed] [Google Scholar]
- Zhang B, Pan X, Cox S, Cobb G, Anderson T. Evidence that miRNAs are different from other RNAs. Cell Mol Life Sci. 2006;63:246–254. doi: 10.1007/s00018-005-5467-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang J, Zhu B. Analysis of Brassica napus ESTs: gene discovery and expression patterns of AP2/ERF-family transcription factors. Mol Biol Rep. 2014;41:45–56. doi: 10.1007/s11033-013-2836-4. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]