Abstract
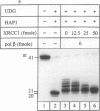
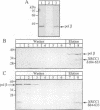
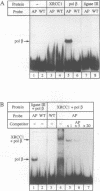
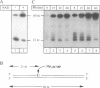
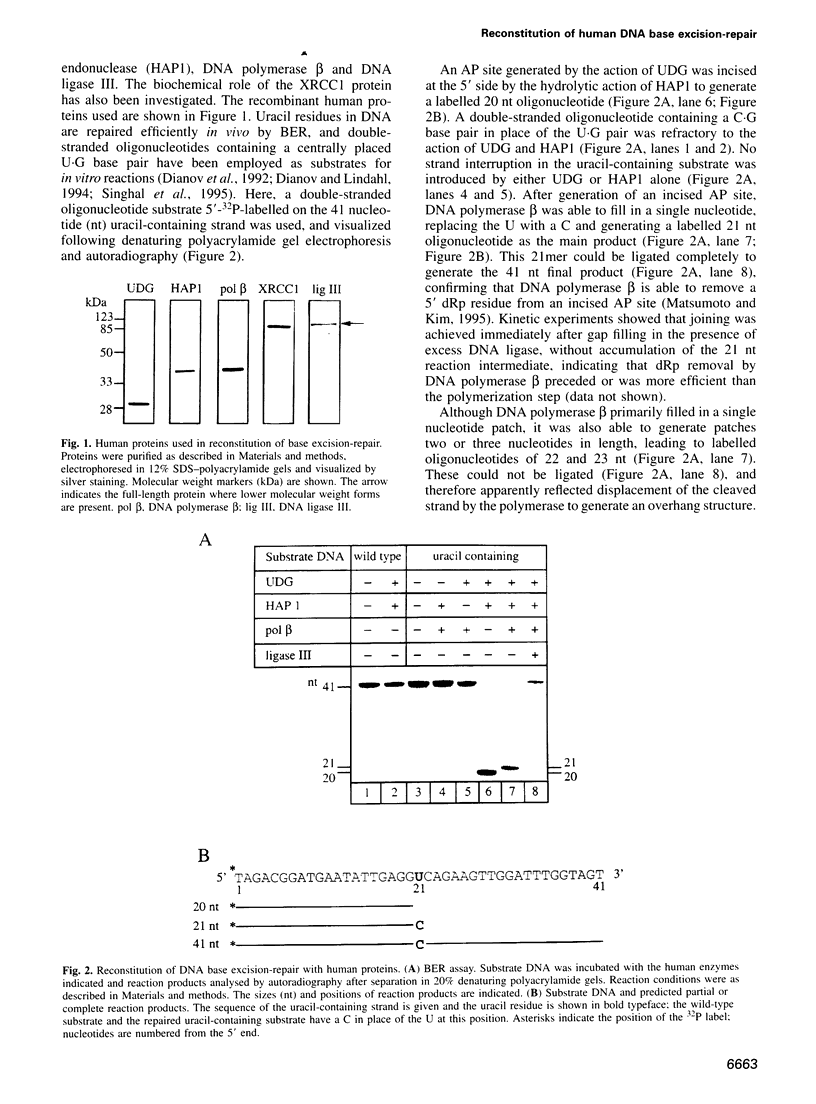
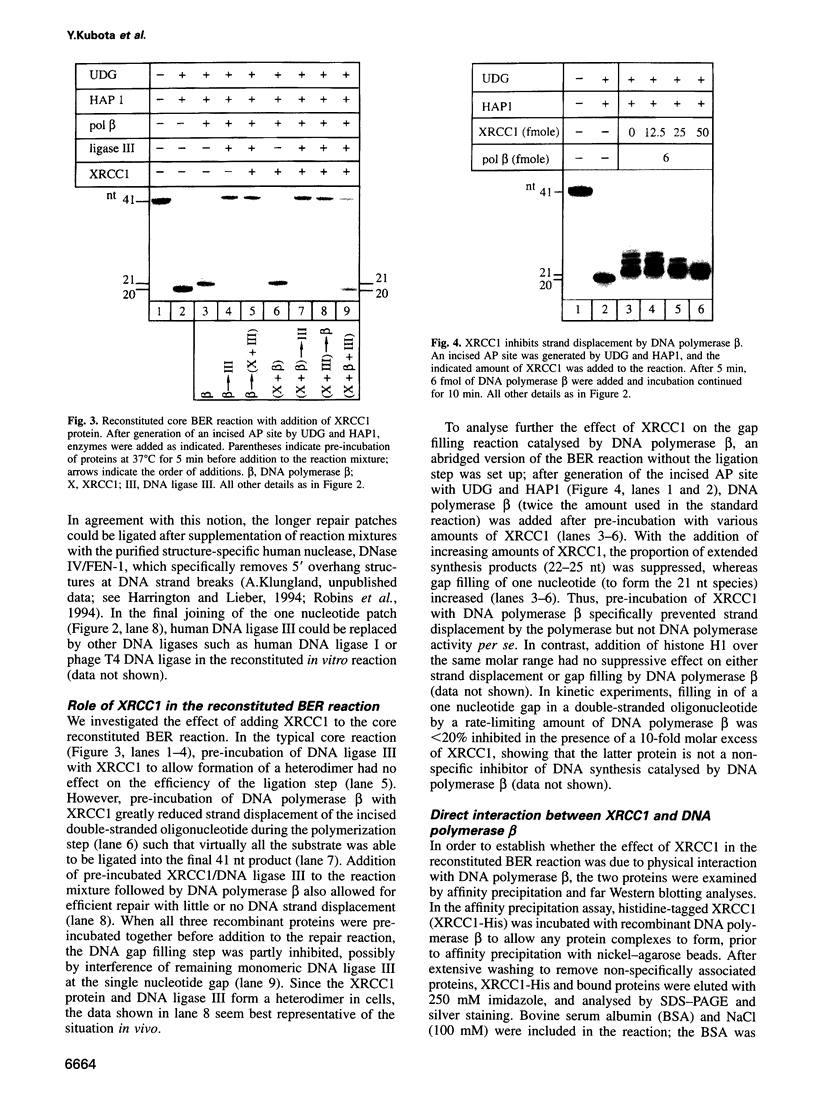
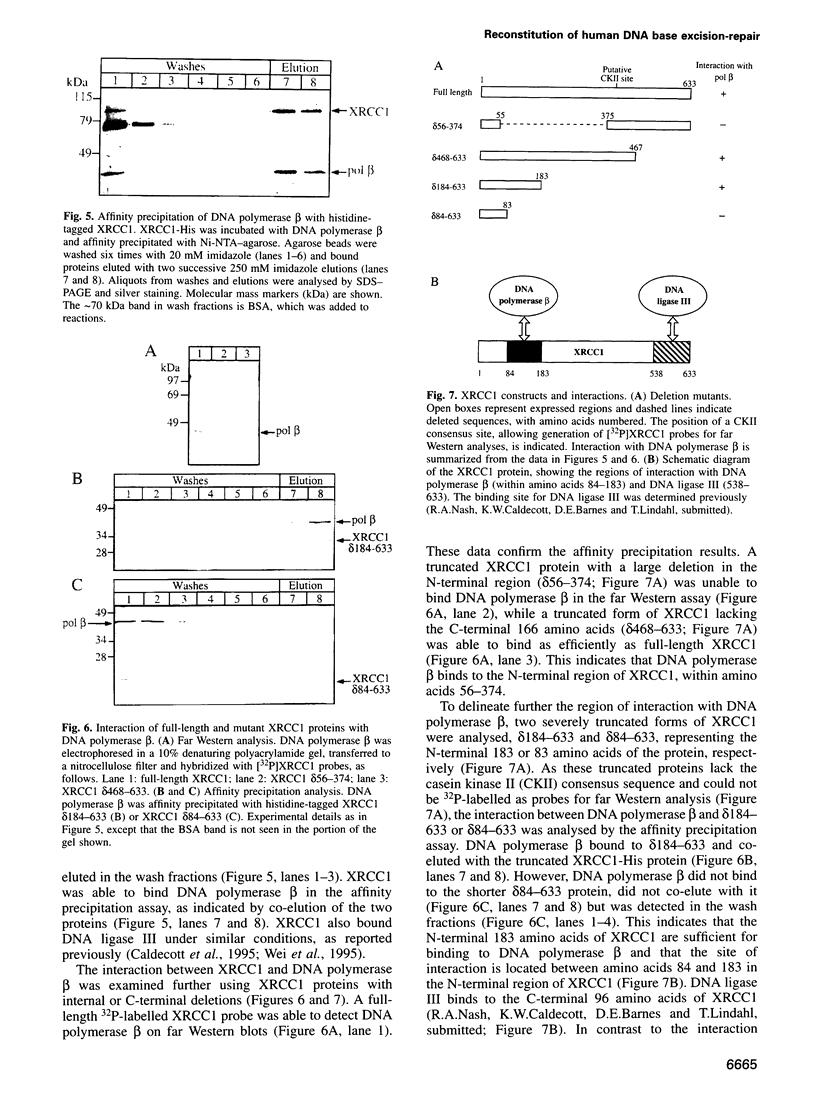
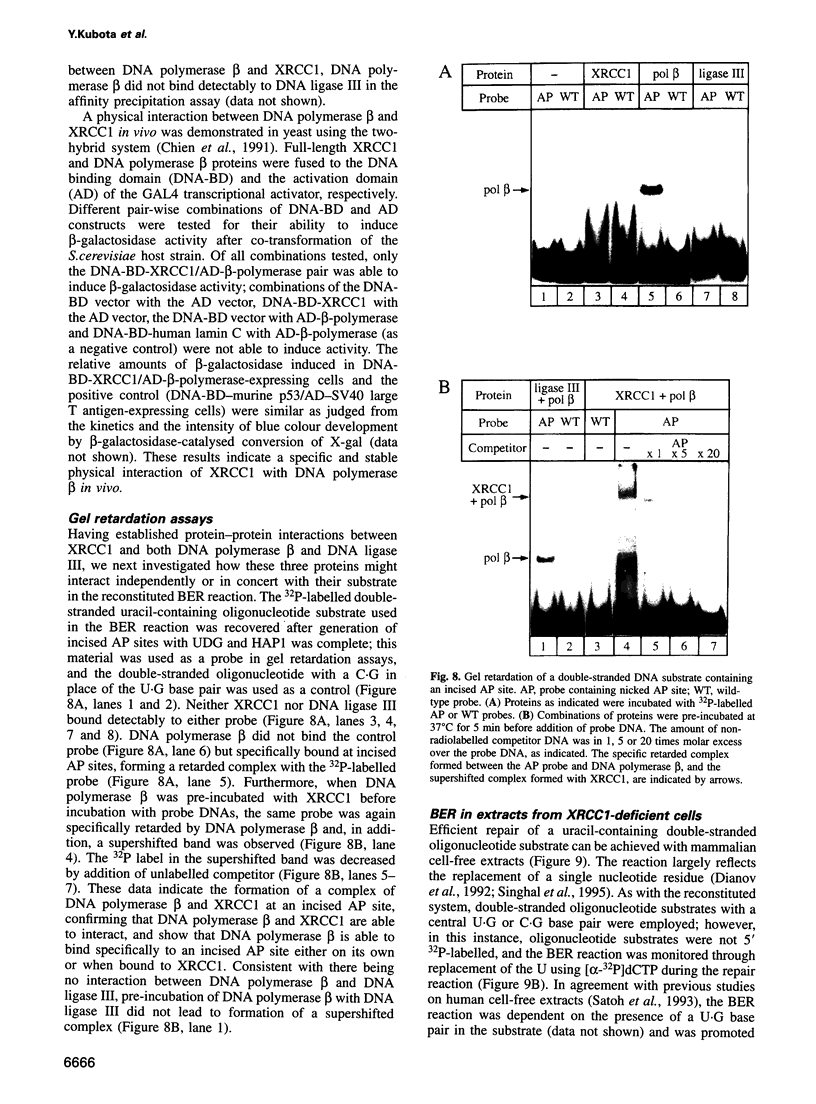
Repair of a uracil-guanine base pair in DNA has been reconstituted with the recombinant human proteins uracil-DNA glycosylase, apurinic/apyrimidinic endonuclease, DNA polymerase beta and DNA ligase III. The XRCC1 protein, which is known to bind DNA ligase III, is not absolutely required for the reaction but suppresses strand displacement by DNA polymerase beta, allowing for more efficient ligation after filling of a single nucleotide patch. We show that XRCC1 interacts directly with DNA polymerase beta using far Western blotting, affinity precipitation and yeast two-hybrid analyses. In addition, a complex formed between DNA polymerase beta and a double-stranded oligonucleotide containing an incised abasic site was supershifted by XRCC1 in a gel retardation assay. The region of interaction with DNA polymerase beta is located within residues 84-183 in the N-terminal half of the XRCC1 protein, whereas the C-terminal region of XRCC1 is involved in binding DNA ligase III. These data indicate that XRCC1, which has no known catalytic activity, might serve as a scaffold protein during base excision-repair. DNA strand displacement and excessive gap filling during DNA repair were observed in cell-free extracts of an XRCC1-deficient mutant cell line, in agreement with the results from the reconstituted system.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abbotts J., SenGupta D. N., Zmudzka B., Widen S. G., Notario V., Wilson S. H. Expression of human DNA polymerase beta in Escherichia coli and characterization of the recombinant enzyme. Biochemistry. 1988 Feb 9;27(3):901–909. doi: 10.1021/bi00403a010. [DOI] [PubMed] [Google Scholar]
- Aboussekhra A., Biggerstaff M., Shivji M. K., Vilpo J. A., Moncollin V., Podust V. N., Protić M., Hübscher U., Egly J. M., Wood R. D. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell. 1995 Mar 24;80(6):859–868. doi: 10.1016/0092-8674(95)90289-9. [DOI] [PubMed] [Google Scholar]
- Blank A., Kim B., Loeb L. A. DNA polymerase delta is required for base excision repair of DNA methylation damage in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1994 Sep 13;91(19):9047–9051. doi: 10.1073/pnas.91.19.9047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldecott K. W., McKeown C. K., Tucker J. D., Ljungquist S., Thompson L. H. An interaction between the mammalian DNA repair protein XRCC1 and DNA ligase III. Mol Cell Biol. 1994 Jan;14(1):68–76. doi: 10.1128/mcb.14.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldecott K. W., Tucker J. D., Stanker L. H., Thompson L. H. Characterization of the XRCC1-DNA ligase III complex in vitro and its absence from mutant hamster cells. Nucleic Acids Res. 1995 Dec 11;23(23):4836–4843. doi: 10.1093/nar/23.23.4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien C. T., Bartel P. L., Sternglanz R., Fields S. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci U S A. 1991 Nov 1;88(21):9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demple B., Harrison L. Repair of oxidative damage to DNA: enzymology and biology. Annu Rev Biochem. 1994;63:915–948. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- Dianov G., Lindahl T. Reconstitution of the DNA base excision-repair pathway. Curr Biol. 1994 Dec 1;4(12):1069–1076. doi: 10.1016/s0960-9822(00)00245-1. [DOI] [PubMed] [Google Scholar]
- Dianov G., Price A., Lindahl T. Generation of single-nucleotide repair patches following excision of uracil residues from DNA. Mol Cell Biol. 1992 Apr;12(4):1605–1612. doi: 10.1128/mcb.12.4.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dianov G., Sedgwick B., Daly G., Olsson M., Lovett S., Lindahl T. Release of 5'-terminal deoxyribose-phosphate residues from incised abasic sites in DNA by the Escherichia coli RecJ protein. Nucleic Acids Res. 1994 Mar 25;22(6):993–998. doi: 10.1093/nar/22.6.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frosina G., Fortini P., Rossi O., Carrozzino F., Raspaglio G., Cox L. S., Lane D. P., Abbondandolo A., Dogliotti E. Two pathways for base excision repair in mammalian cells. J Biol Chem. 1996 Apr 19;271(16):9573–9578. doi: 10.1074/jbc.271.16.9573. [DOI] [PubMed] [Google Scholar]
- Gietz D., St Jean A., Woods R. A., Schiestl R. H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992 Mar 25;20(6):1425–1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper J. W., Adami G. R., Wei N., Keyomarsi K., Elledge S. J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993 Nov 19;75(4):805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- Harrington J. J., Lieber M. R. The characterization of a mammalian DNA structure-specific endonuclease. EMBO J. 1994 Mar 1;13(5):1235–1246. doi: 10.1002/j.1460-2075.1994.tb06373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T. New class of enzymes acting on damaged DNA. Nature. 1976 Jan 1;259(5538):64–66. doi: 10.1038/259064a0. [DOI] [PubMed] [Google Scholar]
- Manley J. L., Fire A., Samuels M., Sharp P. A. In vitro transcription: whole-cell extract. Methods Enzymol. 1983;101:568–582. doi: 10.1016/0076-6879(83)01038-1. [DOI] [PubMed] [Google Scholar]
- Matsumoto Y., Kim K., Bogenhagen D. F. Proliferating cell nuclear antigen-dependent abasic site repair in Xenopus laevis oocytes: an alternative pathway of base excision DNA repair. Mol Cell Biol. 1994 Sep;14(9):6187–6197. doi: 10.1128/mcb.14.9.6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto Y., Kim K. Excision of deoxyribose phosphate residues by DNA polymerase beta during DNA repair. Science. 1995 Aug 4;269(5224):699–702. doi: 10.1126/science.7624801. [DOI] [PubMed] [Google Scholar]
- Montecucco A., Savini E., Weighardt F., Rossi R., Ciarrocchi G., Villa A., Biamonti G. The N-terminal domain of human DNA ligase I contains the nuclear localization signal and directs the enzyme to sites of DNA replication. EMBO J. 1995 Nov 1;14(21):5379–5386. doi: 10.1002/j.1460-2075.1995.tb00222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neddermann P., Gallinari P., Lettieri T., Schmid D., Truong O., Hsuan J. J., Wiebauer K., Jiricny J. Cloning and expression of human G/T mismatch-specific thymine-DNA glycosylase. J Biol Chem. 1996 May 31;271(22):12767–12774. doi: 10.1074/jbc.271.22.12767. [DOI] [PubMed] [Google Scholar]
- O'Regan N. E., Branch P., Macpherson P., Karran P. hMSH2-independent DNA mismatch recognition by human proteins. J Biol Chem. 1996 Jan 19;271(3):1789–1796. doi: 10.1074/jbc.271.3.1789. [DOI] [PubMed] [Google Scholar]
- Piersen C. E., Prasad R., Wilson S. H., Lloyd R. S. Evidence for an imino intermediate in the DNA polymerase beta deoxyribose phosphate excision reaction. J Biol Chem. 1996 Jul 26;271(30):17811–17815. doi: 10.1074/jbc.271.30.17811. [DOI] [PubMed] [Google Scholar]
- Prasad R., Singhal R. K., Srivastava D. K., Molina J. T., Tomkinson A. E., Wilson S. H. Specific interaction of DNA polymerase beta and DNA ligase I in a multiprotein base excision repair complex from bovine testis. J Biol Chem. 1996 Jul 5;271(27):16000–16007. doi: 10.1074/jbc.271.27.16000. [DOI] [PubMed] [Google Scholar]
- Robins P., Pappin D. J., Wood R. D., Lindahl T. Structural and functional homology between mammalian DNase IV and the 5'-nuclease domain of Escherichia coli DNA polymerase I. J Biol Chem. 1994 Nov 18;269(46):28535–28538. [PubMed] [Google Scholar]
- Satoh M. S., Poirier G. G., Lindahl T. NAD(+)-dependent repair of damaged DNA by human cell extracts. J Biol Chem. 1993 Mar 15;268(8):5480–5487. [PubMed] [Google Scholar]
- Shivji M. K., Podust V. N., Hübscher U., Wood R. D. Nucleotide excision repair DNA synthesis by DNA polymerase epsilon in the presence of PCNA, RFC, and RPA. Biochemistry. 1995 Apr 18;34(15):5011–5017. doi: 10.1021/bi00015a012. [DOI] [PubMed] [Google Scholar]
- Singhal R. K., Prasad R., Wilson S. H. DNA polymerase beta conducts the gap-filling step in uracil-initiated base excision repair in a bovine testis nuclear extract. J Biol Chem. 1995 Jan 13;270(2):949–957. doi: 10.1074/jbc.270.2.949. [DOI] [PubMed] [Google Scholar]
- Slupphaug G., Eftedal I., Kavli B., Bharati S., Helle N. M., Haug T., Levine D. W., Krokan H. E. Properties of a recombinant human uracil-DNA glycosylase from the UNG gene and evidence that UNG encodes the major uracil-DNA glycosylase. Biochemistry. 1995 Jan 10;34(1):128–138. doi: 10.1021/bi00001a016. [DOI] [PubMed] [Google Scholar]
- Sobol R. W., Horton J. K., Kühn R., Gu H., Singhal R. K., Prasad R., Rajewsky K., Wilson S. H. Requirement of mammalian DNA polymerase-beta in base-excision repair. Nature. 1996 Jan 11;379(6561):183–186. doi: 10.1038/379183a0. [DOI] [PubMed] [Google Scholar]
- Stephenson C., Karran P. Selective binding to DNA base pair mismatches by proteins from human cells. J Biol Chem. 1989 Dec 15;264(35):21177–21182. [PubMed] [Google Scholar]
- Thompson L. H., Brookman K. W., Dillehay L. E., Carrano A. V., Mazrimas J. A., Mooney C. L., Minkler J. L. A CHO-cell strain having hypersensitivity to mutagens, a defect in DNA strand-break repair, and an extraordinary baseline frequency of sister-chromatid exchange. Mutat Res. 1982 Aug;95(2-3):427–440. doi: 10.1016/0027-5107(82)90276-7. [DOI] [PubMed] [Google Scholar]
- Thompson L. H., Brookman K. W., Jones N. J., Allen S. A., Carrano A. V. Molecular cloning of the human XRCC1 gene, which corrects defective DNA strand break repair and sister chromatid exchange. Mol Cell Biol. 1990 Dec;10(12):6160–6171. doi: 10.1128/mcb.10.12.6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Wu X., Friedberg E. C. DNA repair synthesis during base excision repair in vitro is catalyzed by DNA polymerase epsilon and is influenced by DNA polymerases alpha and delta in Saccharomyces cerevisiae. Mol Cell Biol. 1993 Feb;13(2):1051–1058. doi: 10.1128/mcb.13.2.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y. F., Robins P., Carter K., Caldecott K., Pappin D. J., Yu G. L., Wang R. P., Shell B. K., Nash R. A., Schär P. Molecular cloning and expression of human cDNAs encoding a novel DNA ligase IV and DNA ligase III, an enzyme active in DNA repair and recombination. Mol Cell Biol. 1995 Jun;15(6):3206–3216. doi: 10.1128/mcb.15.6.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiebauer K., Jiricny J. Mismatch-specific thymine DNA glycosylase and DNA polymerase beta mediate the correction of G.T mispairs in nuclear extracts from human cells. Proc Natl Acad Sci U S A. 1990 Aug;87(15):5842–5845. doi: 10.1073/pnas.87.15.5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood R. D., Robins P., Lindahl T. Complementation of the xeroderma pigmentosum DNA repair defect in cell-free extracts. Cell. 1988 Apr 8;53(1):97–106. doi: 10.1016/0092-8674(88)90491-6. [DOI] [PubMed] [Google Scholar]
- Xanthoudakis S., Smeyne R. J., Wallace J. D., Curran T. The redox/DNA repair protein, Ref-1, is essential for early embryonic development in mice. Proc Natl Acad Sci U S A. 1996 Aug 20;93(17):8919–8923. doi: 10.1073/pnas.93.17.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahradka P., Ebisuzaki K. A shuttle mechanism for DNA-protein interactions. The regulation of poly(ADP-ribose) polymerase. Eur J Biochem. 1982 Oct;127(3):579–585. [PubMed] [Google Scholar]