Abstract
Porcine epidemic diarrhea virus (PEDV), an Alphacoronavirus in the family Coronaviridae, causes acute diarrhea, vomiting, dehydration, and high mortality rates in neonatal piglets. PEDV can also cause diarrhea, agalactia, and abnormal reproductive cycles in pregnant sows. Although PEDV was first identified in Europe, it has resulted in significant economic losses in many Asian swine-raising countries, including Korea, China, Japan, Vietnam, and the Philippines. However, from April 2013 to the present, major outbreaks of PEDV have been reported in the United States, Canada, and Mexico. Moreover, intercontinental transmission of PEDV has increased mortality rates in seronegative neonatal piglets, resulting in 10% loss of the US pig population. The emergence and re-emergence of PEDV indicates that the virus is able to evade current vaccine strategies. Continuous emergence of multiple mutant strains from several regions has aggravated porcine epidemic diarrhea endemic conditions and highlighted the need for new vaccines based on the current circulating PEDV. Epidemic PEDV strains tend to be more pathogenic and cause increased death in pigs, thereby causing substantial financial losses for swine producers. In this review, we described the epidemiology of PEDV in several countries and present molecular characterization of current strains. We also discuss PEDV vaccines and related issues.
Keywords: Porcine epidemic diarrhea, Vaccine, Genetics, Epidemiology
Introduction
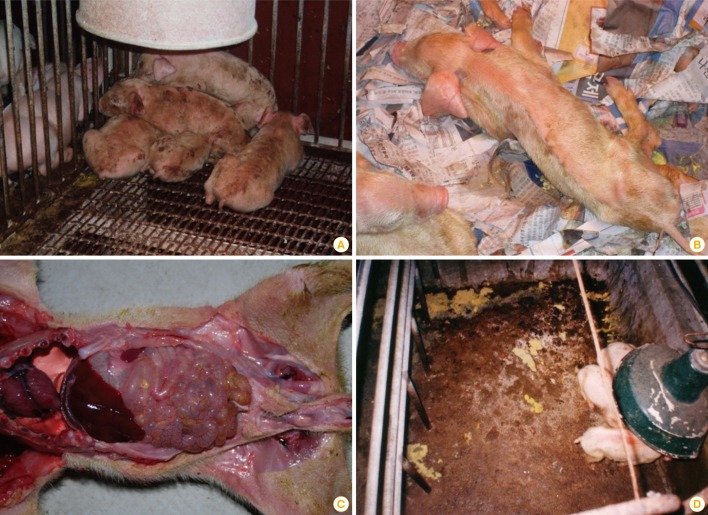
Porcine epidemic diarrhea (PED) is a common type of viral enteritis in pigs that is caused by PED virus (PEDV). Consistent with the name of the disease, diarrhea is the major symptom of PED. Additionally, PED presents with various other clinical signs, including vomiting, anorexia, dehydration, and weight loss (Fig. 1) [1]. PEDV can infect pigs of any age, from neonates to sows or boars; however, the severity of PED in pigs differs according to age [2]. Importantly, PEDV infection in neonatal pigs commonly induces death from watery diarrhea and dehydration. Indeed, in a previous study, researchers stated that over 1,000,000 piglets have died from PEDV infection, with a death rate of 80%-100% [3]. Such high death rates are associated with huge economic losses.
Fig. 1.
Photographic records of porcine epidemic diarrhea virus (PEDV) outbreaks. (A, B) During a 2006 outbreak on a commercial farm in Gimpo. South Korean, piglets <1 week of age died from severe watery diarrhoea after showing signs of dehydration. After the acute outbreak, piglets were anorectic, depressed, vomiting, and producing water faeces that did not contain any signs of blood. (C) Necropsies of deceased piglets from the Gimpo outbreak uncovered gross lesions in the small intestines, which were typically fluidic, distended, and yellow, containing a mass of curdled, undigested milk. Atrophy of the villi caused the walls of the small intestines to become thin and almost transparent. (D) Yellowish watery diarrhea in sucling piglets after acute infection of PEDV.
PED was first observed in Europe in 1971 [4]. The coronavirus-like strain CV777 was isolated from pigs exhibiting diarrhea in a PED outbreak in Belgium in 1976 [1]. This coronavirus-like strain CV777 was designated PEDV and was classified in the genus Alphacoronavirus, family Coronaviridae. During the 1970s and 1980s, the virus spread throughout Europe. However, during the 1980s and 1990s, the number of PED outbreaks decreased markedly in the region. Only a few severe outbreaks have been reported since the 1980s in Europe. However, PED has become an endemic disease in Asian pig farming countries, such as Korea, China, Vietnam, Japan, the Philippines, Taiwan, and Thailand [5,6,7,8,9].
Until 2013, PED was thought to have been restricted to Asian countries. However, an outbreak of PEDV infection occurred in the United States in Iowa in April 2013, and within 1 year, PEDV had spread to Canada and Mexico [10], which share borders with the United States. Additionally, PED outbreaks occurred in Korea and Japan, across the Pacific Ocean, also within 1 year of the US outbreak [11,12]. The PEDV strain isolated in the United States was genetically related to the Chinese PEDV strain reported in 2012 [13]. Interestingly, the Korean and Taiwanese PEDV strains isolated after the US outbreak were genetically related to the US PEDV strain [11].
After spring 2013, PED was no longer found only in Asia. US scientists who had not researched PEDV began to study this disease, which had previously not been a problem in American. From this, a PED vaccine reflecting the genetic characteristics of the PEDV strain isolated during the US outbreak was commercialized [14]. Moreover, many veterinary scientists have focused on the development of more effective PED vaccines because of the major economic losses associated with PED outbreaks. After the US outbreak, sporadic PED outbreaks occurred in Germany. Therefore, European countries were not isolated from the spread of the US PEDV [15]. Thus, the US outbreak was important turning point in PED research, and PED research can be said to be divided into two eras: before and after 2013.
This review covers the vaccine, epidemiology, genetic structure, and characteristics of PED/PEDV after 2013. This report may improve our understanding of this disease, which is currently the most fatal disease in pigs and one of the most costly health issues in animals. Furthermore, this review may provide insight into important topics for investigation in PED research.
Genetic Structure and Characteristics of PEDV
The genome of PEDV is positive-sense, single-stranded RNA. The size of PEDV genomic RNA is about 28 kb. The organization of the PEDV genome is presented in Table 1. From the 5' cap to the 3' poly A tail, PED genomic RNA contains seven open reading frames (ORFs) encoding viral proteins. ORF1a and ORF1b encode the viral polymerase. ORF3 encodes a nonstructural protein with an unknown function; this protein is thought to be related to viral pathogenicity [16]. Additionally, the other ORFs have specific names according to the proteins encoded in these regions, i.e., spike (S), envelope (E), matrix (M), and nucleocapsid (N) proteins [7,17]. Table 1 presents the characteristics of the PEDV proteins as described in previous articles.
Table 1.
Summary of the genomic organization and its functions of PEDV
| ORF | Functions and remarks |
|---|---|
| ORF1a/b | These called polymerase (Pol), collectively. ORF 1 divided into two ORF, 1a and 1b by ribosomal frameshift mechanism. ORF 1a contains proteases and ORF 1b contains RNA dependent RNA polymerase, exonuclease, endoribonuclease, and methyltransferase. |
| S | This is type I glycoprotein with 1,383 amino acids containing neutralizing epitopes. This protein is comprised of 2 subdomains, S1 and S2. S protein has the important role in interaction with host cell receptor and entry. Especially S1 domain is crucial for viral neutralizing and genetic diversity. |
| ORF3 | ORF3 is thought to be related with virulence of PEDV. |
| E | E protein is responsible for the assembly of viron in conjunction with M protein. And it plays a role of ion channel affecting the release of virus. |
| M | It may induce the alpha interferon after infection. M protein is also related with neutralizing and viral assembly. |
| N | N protein is a phosphoprotein associated with viral genome. This protein is useful for the diagnosis of PEDV infection. |
PEDV, porcine epidemic diarrhea virus; ORF, open reading frame; S, spike; E, envelope; M, matrix; N, nucleocapsid.
Of the PEDV proteins, the S protein is considered the most antigenic. As summarized in Table 1, the S protein is responsible for the interaction with host cellular receptor molecules [18,19]. This interaction is crucial for the entry of the virus and is related to induction of neutralizing antibodies against the virus [18,20,21]. Additionally, these critical characteristics of the S protein are used for analysis of the molecular epidemiology of PEDV.
Thus, PEDV researchers have started to analyze the genotypes of PEDV using the S gene [22]. While other genes, such as the gene encoding the M protein and the gene encoded in ORF3 [23,24,25] have been used for phylogeny or molecular epidemiological studies, genetic diversity of the S gene is the focus of this review. One of the most interesting characteristics of the S gene is the diversity in this gene that has occurred from the end of 1970s to the present. Genotypes in the S gene of PEDV could be important because this gene may affect the pathogenicity of novel PED outbreaks based on variations in the S gene [3,10,11,12,16,26,27,28,29]. Moreover, the Chinese PED outbreak in 2011 involved the presence of new variants based on the nucleotide similarities in the S gene. This variant PEDV exhibited 93% similarity with the CV777 prototype PEDV strain in the S gene [29]. Moreover, the PEDV isolated after the US outbreak also exhibited 89%-92% similarity with the S protein of CV777 [26,27]. Phylogenetic analyses of PEDV RNA have been reported in many previous studies [7,16,26,27,30].
Whether the 10% nucleotide difference often observed in the S gene can affect the pathogenesis of PEDV has not been clarified. However, the similarities in the S gene, which is responsible for the host interaction and normalization, the S1 region in particular could be important in vaccine efficacy and development strategies [7,17]. Neutralizing epitopes of the PEDV S protein have been investigated [20]. In particular, the CO-26K equivalent (COE) is a neutralizing epitope within the CO-26K collagenase fragment in transmissible gastroenteritis virus (TGEV) [31,32]. In the prototype PEDV (CV777) and vaccine strains, certain COEs are different from field isolates that were isolated after 2011. The different COEs are 549T and 594G in the prototype and vaccine strains, respectively (Table 2); however, other COEs that differ from that of CV777 have been shown to be identical to the vaccine strains 83P-5 100th or attenuated DR13 [33]. In PEDV isolates acquired after 2011 in China, these amino acids were changed to serine; similar observations were made for the PEDV strain from the US outbreak [29]. The new variant PEDV strains have similar mutational patterns for the neutralizing epitope. In contrast, PEDV strains isolated from 2002 to 2009 exhibit different patterns in 549T and 594G compared to strains isolated after 2011. Interestingly, at amino acid position 549, some strains exhibit no changes; however, in most strains, amino acid 549 is changed to arginine (R). Additionally, most strains exhibit a change from G to S at amino acid position 594, similar to current strains [22].
Table 2.
Comparison of the COE region of PEDV S gene
| Strains (year/country) | Accession number or reference | Amino acid COE in CV777 (AF353511) | |
|---|---|---|---|
| 549T | 594G | ||
| 83P-5 100th (1985/Japan) | AB548621 | - | - |
| SM98 (1998/Korea) | GU937797 | - | - |
| Chinju99 (1999/Korea) | AY167285 | R | - |
| Attenuated DR13 (2001/Korea) | JQ023162 | - | - |
| DBI1825 (2002/Korea) | (19) | - | S |
| BI1482 (2003/Korea) | (19) | - | S |
| E1695 (2003/Korea) | (19) | - | S |
| BI1166 (2003/Korea) | (19) | S | S |
| JS-2004-2 (2004/China) | AY653204 | S | S |
| M2503 (2005/Korea) | (19) | R | S |
| E2540 (2005/Korea) | (19) | R | S |
| KNU-0802 (2008/Korea) | GU180143 | R | S |
| MF3809 (2008/Korea) | KF779469 | S | S |
| KNU-0904 (2009/Korea) | GU180147 | R | - |
| KNU-0905 (2009/Korea) | GU180148 | R | S |
| CHGD-01 (2011/China) | JX261936 | S | S |
| CH-GDHY-2011 (2011/China) | JX145339 | S | S |
| USA/Colorado/2013 (2013/USA) | KF272920 | S | S |
COE, CO-26K equivalent; PEDV, porcine epidemic diarrhea virus.
In order to elucidate the positive correlations between changes in the S protein and virulence or vaccine efficacy, further studies involving animal models are needed.
In addition to the changes in the S protein described above, more variations have been discovered. For example, Park et al. [34] reported large deletion in the S protein from amino acid 713 to amino acid 916, within the S1 and S2 domains. This virus was detected in 2008, and other regions of the S protein share 92.8%-96.9% amino acid identity in the partial S1 domain with other strains isolated in Korean in 2008 and 2009. In the United States, a deletion in the S gene was also reported after the United States outbreak. In December 2013, a strain with deletion of the S gene was detected and isolated in Ohio [35]. This strain exhibited patterns that were different from those of the Korean strain with the S gene deletion. The US deletion mutant contains a large deletion (197 amino acids in length, from amino acid 34 to amino acid 230) in the S1 domain [35]. The effects of deletion in the S protein should be further investigated and are reminiscent of porcine respiratory coronavirus arising from S protein deletion in the genome of TGEV [34,35].
Through PEDV research, unique genetic characteristics other than those in the S gene have been reported. Full-length analysis of the ORF3 gene of PEDV revealed a 51-bp deletion in the cell-attenuated PEDV strain DR13 [23]. This deletion in the ORF3 gene has not been identified in wild-type PEDV; however, in addition to the attenuated PEDV DR13 strain, live PEDV vaccine strains available in Korea exhibit similar deletion patterns by reverse transcription polymerase chain reaction (RT-PCR) comparisons [23]. This deletion pattern is clinically useful for differentiation of live vaccine strains from wild-type PEDV casing diarrhea.
The final genetic mutation we will discuss is deletion of the E gene in the cell-attenuated PEDV strain DR13. Genetic analysis has focused on the S, N, and M genes. From analysis of the E gene, a unique deletion was found. Moreover, compared to other PEDV strains, including the wild-type and vaccine strains, only the attenuated PEDV strain DR13 has been shown to have a 21-bp deletion in this gene [30]. The E protein of the coronavirus is responsible for the assembly of the virus and cell stress responses [36]. The effects of the E gene deletion are under investigation.
Recent Epidemiology in Several Countries
Europe
In the Czech Republic, Rodak et al. [37] reported that 27 out of 219 fecal samples from diarrheic piglets (<21 days old) were positive for PEDV. This outbreak occurred between May 2005 and June 2006 in an area densely populated with pigs in the Po Valley in northern Italy [38]. Some PEDV-positive farms (35 out of 476) were detected between mid-2006 and the end of 2007; however, the disease progressively disappeared [39]. During the period from 2007 to 2014, mild clinical signs were report in pigs of all ages, and mortality was observed in piglets only in PEDV-positive farms [40].
Asia
In the last decade, PEDV outbreaks have been reported in several Asian countries, including Thailand [41,42,43,44], Taiwan [12], the Philippines [45], South Korea [46], and the southern provinces of Vietnam [47]. In October 2010, a large-scale outbreak of PEDV was reported in several provinces in southern China. PEDV also spread to other regions of the country, particularly in northwest [48]. PEDV is now circulating in at least 29 Chinese provinces [49]. In October 2013, Japan reported a PEDV outbreak to the World Organization for Animal Health (OIE) [15] after a period of 7 years without an outbreak. According to the information provided by Japan's National Institute of Animal Health, PEDV isolates from this outbreak are genetically related to the PEDV isolates recovered from China and the United States in 2013. In addition, in late 2013, PEDV outbreaks were reported in South Korea and Taiwan [12,50,51].
America
The US PEDV strains identified during the US outbreak were genetically related to the Chinese strains (China/2012/AH2012) reported in 2011-2012 [13,26]. PEDV was first identified within the United States in Iowa in May 2013, although testing of historical samples showed that PEDV occurred the month before in Ohio. PEDV rapidly spread throughout the country and was confirmed on farms from 32 states, including Ohio, Indiana, Iowa, Minnesota, Oklahoma, Illinois, and North Carolina, by the end September 2014 [15]. PEDV was detected in Mexico for the first time in July 2013 [52]. In October 2013, PEDV was identified for the first time in Peru [53]. In November 2013, PEDV was also identified as the cause of outbreaks of diarrhea in farms in the Espaillat province, Dominican Republic. By September 2014, PED outbreaks were reported in seven of the 31 provinces in the Dominican Republic [54]. In April 2014, Canada reported outbreaks of PEDV to the OIE; these outbreaks started in January and affected 58 herds in four provinces [15]. An acute outbreak of diarrhea and death in lactating piglets was observed in Columbia in March 2014. By September 2014, 54 samples from six departments were confirmed via laboratory testing [55].
Detection of PEDV Antigen
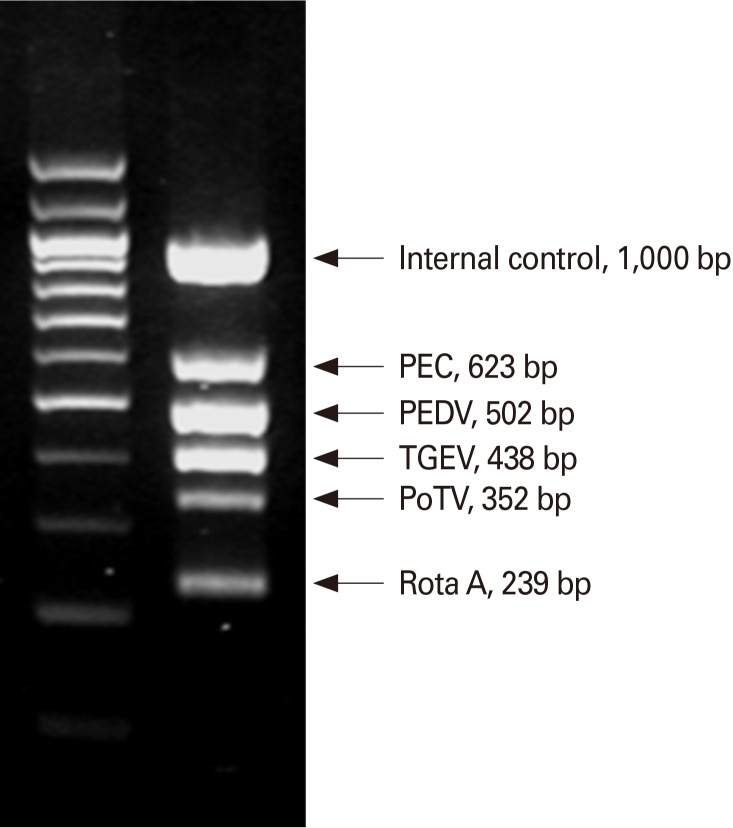
Several reports have described the development of RT-PCR as a diagnostic technique for detection of both laboratory and field isolates [56,57,58,59]. Primers derived from the M gene can be used in an RT-PCR system to obtain PEDV-specific fragments [57], and duplex RT-PCR has been used to differentiate between TGEV and PEDV [60]. Within the past few years, several useful modifications of the basic RT-PCR method have been reported. For example, it is possible to estimate the potential transmission of PEDV by comparing viral shedding load with a standard internal control DNA curve [61] and by multiplex RT-PCR to detect PEDV in the presence of various viruses [62]-a technique that is particularly useful for rapid, sensitive, and cost-effective diagnosis of acute viral gastroenteritis in swine. The commercial dual priming oligonucleotide system (Seegene, Seoul, Korea) (Fig. 2) has been developed for the rapid differential detection of PEDV. This system employs a single tube one-step multiplex RT-PCR with two separate primer segments to block nonspecific priming [63]. Recently, a protein-based enzyme-linked immunosorbent assay (ELISA) system was developed to detect PEDV. Using this technique, a polyclonal antibody is produced by immunizing rabbits with purified PEDV M gene after its expression in Escherichia coli. Immunofluorescence analysis can then be carried out with the anti-PEDV-M antibody in order to detect PEDV-infected cells among other enteric viruses [64].
Fig. 2.
The multiplex RT-PCR assay and dual priming oligonucleotide (DPO) system can be used in diagnosis of porcine epidemic diarrhea virus (PEDV). PEC, porcine enteric calici virus; TGEV, transmissible gastroenteritis virus; PoTV, porcine torovirus; RotaA, rotavirus type A.
Another useful reverse transcription-based diagnostic tool is RT loop-mediated isothermal amplification. This assay, which uses 4-6 primers that recognize 6-8 regions of the target DNA, is more sensitive than gel-based RT-PCR and ELISA, largely because this method produces a greater quantity of DNA [65]. Immunochromatographic assay kits can be used at farms in order to detect the N (nucleocapsid) protein of PEDV with 92% sensitivity and 98% specificity. Moreover, a rapid technique for differential detection of PEDV and porcine rotavirus (RV) has recently been commercialized and is now widely used in the field (Fig. 3). This technique is less sensitive than RT-PCR, but allows for diagnosis within 10 minutes. Thus, it is particularly effective for quickly determining quarantine or slaughter policies in the field.
Fig. 3.
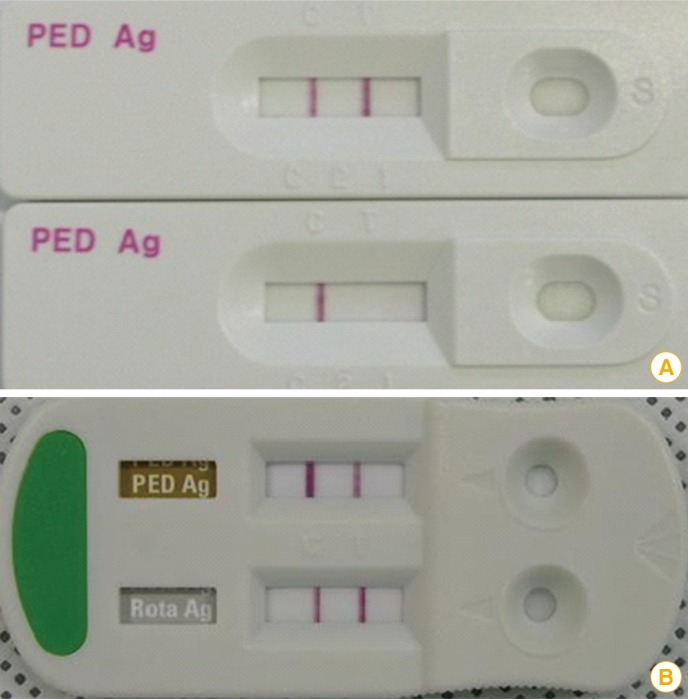
Results of an immunochromatographic assay kit that can be used for porcine epidemic diarrhea virus (PEDV) detection. (A) The upper and lower panels show positive and negative results, respectively. (B) Dual-detecting immunochromatographic kit for the detection of PEDV and porcine rotavirus.
Interestingly, some reports have commented on the detection of PEDV genomic DNA in sera. Genomic detection in gnotobiotic piglets has been reported for serum viral RNA concentrations ranging from 4.8 to 7.6 log10 genomic equivalents (GE)/mL after inoculation of the US PEDV strain. Similar detection of the PEDV genome has been observed in diarrheic pigs at age 13-20 weeks (4.0-6.3 log10 GE/mL) [17]. However, no infectious PEDV has been recovered from genome-positive sera samples. Unfortunately, after intensive screening and trials to isolate the PEDV genome from serum samples, we have not succeeded in this endeavor (data not shown).
Immunoprophylaxis
Enteropathogenic viruses can be divided into two types (type I and II) according to their infection site in the intestine [66]. Viruses infecting villous enterocytes, including TGEV, PEDV, and RV, are type I viruses and can be suppressed by local gut-associated immunity. Diseases caused by type II viruses, which infect crypt enterocytes basolaterally (e.g., canine parvovirus), can be prevented by inducing systemic or mucosal immunity. In this review, we discuss control strategies for reducing viral shedding, mortality, and the transmission of PEDV in swine herds, such as transmission occurring from artificial oral exposure (i.e., the feedback method) and vaccines.
In naïve swine herds, PED is characterized by vomiting and acute diarrhea and results in high mortality rates in piglets less than 2 weeks of age. Neonatal pigs are born without maternal antibodies if they are not infected in utero, and they should receive passive lactogenic immunity (IgG and IgA) through intake of colostrum and milk to promote survival after birth. Therefore, maternal-derived immunity at an early age is critical for passive protection of neonatal pigs; for this purpose, immunization of the dam preparturition has been used successfully [66]. IgG is the major immunoglobulin component in pig colostrum, consisting of more than 60% of all immunoglobulins, but is not found in milk. IgA accounts for a substantially reduced percentage of colostrum immunoglobulin content; however, IgA is more effective than IgG or IgM at protecting animals from orally infected agents because it is more resistant to the activity of proteolytic enzymes in the intestine and has a higher neutralizing ability than IgG and IgM [67]. Bohl et al. [68] and Saif et al. [69] demonstrated that oral inoculation of seronegative sows with live virulent TGEV results in high rates of protection in suckling piglets. In these sows, passive protection is associated with high titers of secretory IgA (sIgA) in colostrum and milk. This investigation suggested the presence of a gut-mammary gland-sIgA axis; that is, IgA plasmablasts stimulated in the gut by virulent pathogens migrate to the gut lamina propria and mammary glands. Several highly attenuated oral TGEV vaccines, which replicate lower in the gut, induce poor milk sIgA titers compared with virulent TGEV in sows and result in lower protective efficacy in piglets [68,69]. This research could be employed to maternal immunization strategies for PEDV.
In areas where no effective PED vaccines are available, some veterinarians have recommended artificial infection of sows (i.e., the feedback method) during pregnancy to supply lactogenic immunity to their piglets [70]. The recommended feedback material is pooled feces collected from infected piglets during the first 18 hours of infection. Every sow on the farm should be simultaneously administered feces containing high titers of PEDV, allowing all sows and gilts to recover at approximately the same time and stop shedding the virus.
One of advantage of this type of feedback method is strong stimulation of mucosal immunity in the gut and a quick response after immunization. After successful feedback, the piglets will be protected during the first few days after birth by passive antibodies through colostrum and milk. However, there is a potential risk of transmission of the contaminated viral or bacterial agents in the inoculum (e.g., porcine circovirus type 2 [PCV2] infection, porcine reproductive and respiratory syndrome virus [PRRSV], and salmonellosis) [71,72]. Additionally, it is possible that PEDV may spread rapidly in pigs of all ages in the index farms. The severity of the disease may vary with unknown factors, such as stress, nutrition, or co-infection. In addition, there is a risk that the virulent virus used for feedback materials may spread to and produce disease in other herds. Irregular immune responses in sows after feedback may also be a major concern for optimal induction of herd immunity for protection. All of these possibilities emphasize the need for a safe and effective PEDV vaccine to protect both sows and piglets.
Vaccine, Efficacy, and Controversies
For the prevention of PEDV infection, several vaccines have been reported in Asian countries; the predominance of vaccines in Asian countries, but not in Europe or America, is thought to be related to the occurrence of severe PED outbreaks and major economic losses in Asia [7]. Commercial PEDV vaccines include live attenuated vaccines and binary ethylenimine (BEI) inactivated vaccines. Some of these vaccines have been combined with vaccines for TGEV (a bivalent vaccine) and porcine RV (a trivalent vaccine) and used in China and South Korea [7,25]. Moreover, an attenuated virus vaccine using cell culture-adapted PEDV has been administered to sows in Japan since 1997. Oral vaccination with a cell-attenuated vaccine has been used in South Korea since 2004 and in the Philippines since 2011 [7]. Although these commercial vaccines are considered effective and have been widely used, not all animals develop solid lactogenic immunity. Several factors are thought to be associated with the poor lactogenic immunogenicity of the commercial vaccine, including the immunizing route of the vaccine. Song et al. [73] demonstrated that oral inoculation of PEDV-seronegative pregnant sows with live attenuated PEDV reduces the mortality of suckling piglets more effectively rather than injection after challenge, and this protection is associated with elevated IgA concentrations in colostrum and milk. Despite the reduction in mortality rates in piglets delivered from orally vaccinated sows, there was no shortening of the duration of virus shedding and no reduced severity of diarrhea after challenge between vaccinated and control pigs. Thus, some researchers may conclude that passive immunity by vaccination with the highly attenuated PEDV strain DR13 does not prevent virus shedding after challenge. Protection against virus challenge in conventional pigs is related to the inoculation dose of the virus in the vaccine and the challenge dose of the virulent virus. At low doses of the attenuated PEDV, 25% of pigs are protected against PEDV challenge; however, this proportion increases to 50% when pigs are inoculated with a dose 20 times higher [74]. Moreover, loss of body weight and the content of viral shedding decrease in orally vaccinated pigs compared with those in intramuscularly vaccinated and unvaccinated pigs following challenge with a low dose of virulent virus (1 LD50, 103.0 TCID50/dose). Additionally, the lethal dose of PEDV changes depending on the body weights of the infected piglets and infection of sows with the challenge virus (data not shown).
However, several publications have questioned the efficacy and/or safety of PEDV vaccines used in Asia [3,28,44,75,76]. In particular, after the US outbreak, the efficacy of commercial PED vaccines in Korea became controversial, and simultaneously, there was an urgent need for a new vaccine in order to establish solid immunity in sow herds prior to farrowing to protect piglets. Furthermore, many groups have debated the appropriate standards for evaluation of the efficacy of vaccines after official challenge tests using currently available PEDV vaccines in South Korea in 2014. Information on PEDV mucosal immunity is limited. Moreover, due to the complex characteristics of mucosal viral diseases, simple criteria, such as serum neutralizing antibodies, severity of diarrhea, and mortality after virulent challenge, are insufficient for the accurate and optimal evaluation of PEDV vaccine efficacy. For detailed identification of standards for the evaluation of PEDV vaccines after virulent challenge, the following criteria, which may not be controllable, should be taken into consideration:
- Characteristics of piglets used in the challenge test: conventional or specific pathogen-free piglets, weight of piglets
- Characteristics of sows: parity number, existence of agalactia or co-infection with other viral and bacterial pathogens, number of delivered piglets
- Quantity of challenged virus: amount of viral load for the challenge virus may result in discrepancies in the observed mortality rates of piglets
- Cohabitation with sows: the conditions of piglets challenged with virulent virus could be affected by the occurrence of viral shedding by sows
- Duration of the challenge test
The vaccine strains commonly used in Korea display a maximum difference of 10% at the amino acid level compared with field viruses. Additionally, using SN assays, the SM98 vaccine strain was shown to exhibit variable cross-reactivity with several antisera against other vaccines and field viruses, implying that these vaccines would confer protection against the vaccine prepared using wild-type PEDVs from the field [77]. However, the cross-protection between vaccine strains and field viruses should be elucidated through animal experiments, which can show protection based on lactogenic immunity after vaccination. For the ideal development of PEDV vaccines, new vaccine strains that are genetically related to field viruses are critical. Furthermore, several criteria, including factors related to the reduction in virus shedding and loss of body weight in piglets as well as the details of mucosal immunity and the relationships between protection and immunity, should be considered and identified during the development of next-generation PED vaccines.
Conclusion
PEDV research has become a hot topic in veterinary virology since the US outbreak in 2013. PED had been a regional disease primarily found in Asian countries. However, PED was transmitted to the United States and subsequently to neighboring countries, including Mexico and Canada. European countries have also encountered PEDV. Accordingly, with the spread of the PED outbreak area, research on PEDV has increased rapidly. Veterinarians and veterinary scientists are striving to breakthroughs to combat current PED outbreaks worldwide, and remarkable advances have been made in the relatively short time after the 2013 outbreak.
Therefore, reasonable methods for PED vaccine evaluation, along with the development of new vaccines, are urgently needed, and this complicated process could provide valuable information on PEDV, immune responses to PEDV infection, and PEDV pathobiology. Furthermore, rational evaluation procedures can help swine farmers understand and control PED more efficiently.
Footnotes
No potential conflict of interest relevant to this article was reported.
This research was supported by the Agricultural Biotechnology Development Program, Ministry of Agriculture, Food and Rural affair (No. 114055-03-1-SB010) and BioNano Health-Guard Research Center funded by the Ministry of Science, ICT & Future Planning (MSIP) of Korea as a Global Frontier Project (Grant Number H-GUARD_2013M3A6B2078954, Duration: 2014.01.01~2015.08.31).
References
- 1.Pensaert MB, de Bouck P. A new coronavirus-like particle associated with diarrhea in swine. Arch Virol. 1978;58:243–247. doi: 10.1007/BF01317606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shibata I, Tsuda T, Mori M, Ono M, Sueyoshi M, Uruno K. Isolation of porcine epidemic diarrhea virus in porcine cell cultures and experimental infection of pigs of different ages. Vet Microbiol. 2000;72:173–182. doi: 10.1016/S0378-1135(99)00199-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun RQ, Cai RJ, Chen YQ, Liang PS, Chen DK, Song CX. Outbreak of porcine epidemic diarrhea in suckling piglets, China. Emerg Infect Dis. 2012;18:161–163. doi: 10.3201/eid1801.111259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oldham J. Letter to the editor. Pig Farming. 1972;10:72–73. [Google Scholar]
- 5.Jinghui F, Yijing L. Cloning and sequence analysis of the M gene of porcine epidemic diarrhea virus LJB/03. Virus Genes. 2005;30:69–73. doi: 10.1007/s11262-004-4583-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kusanagi K, Kuwahara H, Katoh T, et al. Isolation and serial propagation of porcine epidemic diarrhea virus in cell cultures and partial characterization of the isolate. J Vet Med Sci. 1992;54:313–318. doi: 10.1292/jvms.54.313. [DOI] [PubMed] [Google Scholar]
- 7.Song D, Park B. Porcine epidemic diarrhoea virus: a comprehensive review of molecular epidemiology, diagnosis, and vaccines. Virus Genes. 2012;44:167–175. doi: 10.1007/s11262-012-0713-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kweon CH, Kwon BJ, Jung TS, et al. Isolation of porcine epidemic diarrhea virus (PEDV) in Korea. Korean J Vet Res. 1993;33:249–254. [Google Scholar]
- 9.Takahashi K, Okada K, Ohshima K. An outbreak of swine diarrhea of a new-type associated with coronavirus-like particles in Japan. J Vet Med Sci. 1983;45:829–832. doi: 10.1292/jvms1939.45.829. [DOI] [PubMed] [Google Scholar]
- 10.Vlasova AN, Marthaler D, Wang Q, et al. Distinct characteristics and complex evolution of PEDV strains, North America, May 2013-February 2014. Emerg Infect Dis. 2014;20:1620–1628. doi: 10.3201/eid2010.140491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho YY, Lim SI, Kim YK, Song JY, Lee JB, An DJ. Complete genome sequence of K14JB01, a novel variant strain of porcine epidemic diarrhea virus in South Korea. Genome Announc. 2014;2:e00505–e00514. doi: 10.1128/genomeA.00505-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin CN, Chung WB, Chang SW, et al. US-like strain of porcine epidemic diarrhea virus outbreaks in Taiwan, 2013-2014. J Vet Med Sci. 2014;76:1297–1299. doi: 10.1292/jvms.14-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang YW, Dickerman AW, Pineyro P, et al. Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. MBio. 2013;4:e00737–e00713. doi: 10.1128/mBio.00737-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.PED vaccine gains conditional approval. J Am Vet Med Assoc. 2014;245:267. [PubMed] [Google Scholar]
- 15.ESFA Panel on Animal Health and Welfare (AHAW) Scientific opinion on porcine epidemic diarrhoea and emerging porcine deltacoronavirus. EFSA J. 2014;12:3877. doi: 10.2903/j.efsa.2014.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun R, Leng Z, Zhai SL, Chen D, Song C. Genetic variability and phylogeny of current Chinese porcine epidemic diarrhea virus strains based on spike, ORF3, and membrane genes. ScientificWorldJournal. 2014;2014:208439. doi: 10.1155/2014/208439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jung K, Saif LJ. Porcine epidemic diarrhea virus infection: etiology, epidemiology, pathogenesis and immunoprophylaxis. Vet J. 2015;204:134–143. doi: 10.1016/j.tvjl.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bosch BJ, van der Zee R, de Haan CA, Rottier PJ. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cruz DJ, Kim CJ, Shin HJ. The GPRLQPY motif located at the carboxy-terminal of the spike protein induces antibodies that neutralize porcine epidemic diarrhea virus. Virus Res. 2008;132:192–196. doi: 10.1016/j.virusres.2007.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang SH, Bae JL, Kang TJ, et al. Identification of the epitope region capable of inducing neutralizing antibodies against the porcine epidemic diarrhea virus. Mol Cells. 2002;14:295–299. [PubMed] [Google Scholar]
- 21.Godet M, Grosclaude J, Delmas B, Laude H. Major receptor-binding and neutralization determinants are located within the same domain of the transmissible gastroenteritis virus (coronavirus) spike protein. J Virol. 1994;68:8008–8016. doi: 10.1128/jvi.68.12.8008-8016.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park SJ, Moon HJ, Yang JS, et al. Sequence analysis of the partial spike glycoprotein gene of porcine epidemic diarrhea viruses isolated in Korea. Virus Genes. 2007;35:321–332. doi: 10.1007/s11262-007-0096-x. [DOI] [PubMed] [Google Scholar]
- 23.Park SJ, Moon HJ, Luo Y, et al. Cloning and further sequence analysis of the ORF3 gene of wild- and attenuated-type porcine epidemic diarrhea viruses. Virus Genes. 2008;36:95–104. doi: 10.1007/s11262-007-0164-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen JF, Sun DB, Wang CB, et al. Molecular characterization and phylogenetic analysis of membrane protein genes of porcine epidemic diarrhea virus isolates in China. Virus Genes. 2008;36:355–364. doi: 10.1007/s11262-007-0196-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen J, Wang C, Shi H, et al. Molecular epidemiology of porcine epidemic diarrhea virus in China. Arch Virol. 2010;155:1471–1476. doi: 10.1007/s00705-010-0720-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Q, Li G, Stasko J, et al. Isolation and characterization of porcine epidemic diarrhea viruses associated with the 2013 disease outbreak among swine in the United States. J Clin Microbiol. 2014;52:234–243. doi: 10.1128/JCM.02820-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meng F, Ren Y, Suo S, et al. Evaluation on the efficacy and immunogenicity of recombinant DNA plasmids expressing spike genes from porcine transmissible gastroenteritis virus and porcine epidemic diarrhea virus. PLoS One. 2013;8:e57468. doi: 10.1371/journal.pone.0057468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo Y, Zhang J, Deng X, Ye Y, Liao M, Fan H. Complete genome sequence of a highly prevalent isolate of porcine epidemic diarrhea virus in South China. J Virol. 2012;86:9551. doi: 10.1128/JVI.01455-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li W, Li H, Liu Y, et al. New variants of porcine epidemic diarrhea virus, China, 2011. Emerg Infect Dis. 2012;18:1350–1353. doi: 10.3201/eid1808.120002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park SJ, Song DS, Park BK. Molecular epidemiology and phylogenetic analysis of porcine epidemic diarrhea virus (PEDV) field isolates in Korea. Arch Virol. 2013;158:1533–1541. doi: 10.1007/s00705-013-1651-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Correa I, Jimenez G, Sune C, Bullido MJ, Enjuanes L. Antigenic structure of the E2 glycoprotein from transmissible gastroenteritis coronavirus. Virus Res. 1988;10:77–93. doi: 10.1016/0168-1702(88)90059-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Delmas B, Rasschaert D, Godet M, Gelfi J, Laude H. Four major antigenic sites of the coronavirus transmissible gastroenteritis virus are located on the amino-terminal half of spike glycoprotein S. J Gen Virol. 1990;71(Pt 6):1313–1323. doi: 10.1099/0022-1317-71-6-1313. [DOI] [PubMed] [Google Scholar]
- 33.Hao J, Xue C, He L, Wang Y, Cao Y. Bioinformatics insight into the spike glycoprotein gene of field porcine epidemic diarrhea strains during 2011-2013 in Guangdong, China. Virus Genes. 2014;49:58–67. doi: 10.1007/s11262-014-1055-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park S, Kim S, Song D, Park B. Novel porcine epidemic diarrhea virus variant with large genomic deletion, South Korea. Emerg Infect Dis. 2014;20:2089–2092. doi: 10.3201/eid2012.131642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oka T, Saif LJ, Marthaler D, et al. Cell culture isolation and sequence analysis of genetically diverse US porcine epidemic diarrhea virus strains including a novel strain with a large deletion in the spike gene. Vet Microbiol. 2014;173:258–269. doi: 10.1016/j.vetmic.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ruch TR, Machamer CE. The coronavirus E protein: assembly and beyond. Viruses. 2012;4:363–382. doi: 10.3390/v4030363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodak L, Valicek L, Smid B, Nevorankova Z. An ELISA optimized for porcine epidemic diarrhoea virus detection in faeces. Vet Microbiol. 2005;105:9–17. doi: 10.1016/j.vetmic.2004.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martelli P, Lavazza A, Nigrelli AD, Merialdi G, Alborali LG, Pensaert MB. Epidemic of diarrhoea caused by porcine epidemic diarrhoea virus in Italy. Vet Rec. 2008;162:307–310. doi: 10.1136/vr.162.10.307. [DOI] [PubMed] [Google Scholar]
- 39.Sozzi E, Luppi A, Lelli D, et al. Comparison of enzyme-linked immunosorbent assay and RT-PCR for the detection of porcine epidemic diarrhoea virus. Res Vet Sci. 2010;88:166–168. doi: 10.1016/j.rvsc.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sozzi E, Papetti A, Lelli D, et al. Diagnosis and investigations on PED in Northern Italy; The 8th Annual EPIZONE Meeting; 2014 Sep 23-25; Copenhagen, Denmark. [Google Scholar]
- 41.Temeeyasen G, Srijangwad A, Tripipat T, et al. Genetic diversity of ORF3 and spike genes of porcine epidemic diarrhea virus in Thailand. Infect Genet Evol. 2014;21:205–213. doi: 10.1016/j.meegid.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 42.Puranaveja S, Poolperm P, Lertwatcharasarakul P, et al. Chinese-like strain of porcine epidemic diarrhea virus, Thailand. Emerg Infect Dis. 2009;15:1112–1115. doi: 10.3201/eid1507.081256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olanratmanee EO, Kunavongkrit A, Tummaruk P. Impact of porcine epidemic diarrhea virus infection at different periods of pregnancy on subsequent reproductive performance in gilts and sows. Anim Reprod Sci. 2010;122:42–51. doi: 10.1016/j.anireprosci.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 44.Ayudhya SN, Assavacheep P, Thanawongnuwech R. One world--one health: the threat of emerging swine diseases. An Asian perspective. Transbound Emerg Dis. 2012;59(Suppl 1):9–17. doi: 10.1111/j.1865-1682.2011.01309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morales RG, Umandal AC, Lantican CA. Emerging and re-emerging diseases in Asia and the Pacific with special emphasis on porcine epidemic diarrhoea. Conf OIE. 2008;2007:185–189. [Google Scholar]
- 46.Lee DK, Park CK, Kim SH, Lee C. Heterogeneity in spike protein genes of porcine epidemic diarrhea viruses isolated in Korea. Virus Res. 2010;149:175–182. doi: 10.1016/j.virusres.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duy DT, Toan NT, Puranaveja S, Thanawongnuwech R. Genetic characterization of porcine epidemic diarrhea virus (PEDV) isolates from southern Vietnam during 2009-2010 outbreaks. Thai J Vet Med. 2011;41:55–64. [Google Scholar]
- 48.Wang H, Xia X, Liu Z, et al. Outbreak of porcine epidemic diarrhea in piglets in Gansu province, China. Acta Sci Vet. 2013;41:1140. [Google Scholar]
- 49.Feng L. The updated epidemic and controls of swine enteric coronavirus in China; International Conference on Swine Enteric Coronavirus Diseases; 2014 Sep 23-25; Chicago, IL, USA. [Google Scholar]
- 50.Lee S, Lee C. Outbreak-related porcine epidemic diarrhea virus strains similar to US strains, South Korea, 2013. Emerg Infect Dis. 2014;20:1223–1226. doi: 10.3201/eid2007.140294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Choi JC, Lee KK, Pi JH, et al. Comparative genome analysis and molecular epidemiology of the reemerging porcine epidemic diarrhea virus strains isolated in Korea. Infect Genet Evol. 2014;26:348–351. doi: 10.1016/j.meegid.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fajardo R, Alpizar A, Martinez A, et al. Two cases report of PED in different states in México; Proceedings of the 23rd International Pig Veterinary Society (IPVS) Congress; 2014 Jun 8-11; Cancun, Mexico. [Google Scholar]
- 53.Quevedo-Valle MV. Porcine epidemic diarrhea outbreak in Peru; International Conference on Swine Enteric Coronavirus Diseases; 2014 Sep 23-25; Chicago, IL, USA. [Google Scholar]
- 54.Gomez L. Diarrea epidémica porcina (DEP) en Republica Dominicana; International Conference on Swine Enteric Coronavirus Diseases; 2014 Sep 23-25; Chicago, IL, USA. [Google Scholar]
- 55.Rativa N. Situacion de la diarrea epidemica porcina en Colombia; International Conference on Swine Enteric Coronavirus Diseases; 2014 Sep 23-25; Chicago, IL, USA. [Google Scholar]
- 56.Ishikawa K, Sekiguchi H, Ogino T, Suzuki S. Direct and rapid detection of porcine epidemic diarrhea virus by RT-PCR. J Virol Methods. 1997;69:191–195. doi: 10.1016/s0166-0934(97)00157-2. [DOI] [PubMed] [Google Scholar]
- 57.Kweon CH, Lee JG, Han MG, Kang YB. Rapid diagnosis of porcine epidemic diarrhea virus infection by polymerase chain reaction. J Vet Med Sci. 1997;59:231–232. doi: 10.1292/jvms.59.231. [DOI] [PubMed] [Google Scholar]
- 58.Tobler K, Ackermann M. PEDV leader sequence and junction sites. Adv Exp Med Biol. 1995;380:541–542. doi: 10.1007/978-1-4615-1899-0_86. [DOI] [PubMed] [Google Scholar]
- 59.Tobler K, Ackermann M. Identification and characterization of new and unknown coronaviruses using RT-PCR and degenerate primers. Schweiz Arch Tierheilkd. 1996;138:80–86. [PubMed] [Google Scholar]
- 60.Kim SY, Song DS, Park BK. Differential detection of transmissible gastroenteritis virus and porcine epidemic diarrhea virus by duplex RT-PCR. J Vet Diagn Invest. 2001;13:516–520. doi: 10.1177/104063870101300611. [DOI] [PubMed] [Google Scholar]
- 61.Song DS, Oh JS, Kang BK, et al. Fecal shedding of a highly cell-culture-adapted porcine epidemic diarrhea virus after oral inoculation in pigs. J Swine Health Prod. 2005;13:269–272. [Google Scholar]
- 62.Song DS, Kang BK, Oh JS, et al. Multiplex reverse transcription-PCR for rapid differential detection of porcine epidemic diarrhea virus, transmissible gastroenteritis virus, and porcine group A rotavirus. J Vet Diagn Invest. 2006;18:278–281. doi: 10.1177/104063870601800309. [DOI] [PubMed] [Google Scholar]
- 63.Lee CS, Kang BK, Lee DH, et al. One-step multiplex RT-PCR for detection and subtyping of swine influenza H1, H3, N1, N2 viruses in clinical samples using a dual priming oligonucleotide (DPO) system. J Virol Methods. 2008;151:30–34. doi: 10.1016/j.jviromet.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 64.Ren X, Suo S, Jang YS. Development of a porcine epidemic diarrhea virus M protein-based ELISA for virus detection. Biotechnol Lett. 2011;33:215–220. doi: 10.1007/s10529-010-0420-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ren X, Li P. Development of reverse transcription loop-mediated isothermal amplification for rapid detection of porcine epidemic diarrhea virus. Virus Genes. 2011;42:229–235. doi: 10.1007/s11262-011-0570-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Saif LJ. Enteric viral infections of pigs and strategies for induction of mucosal immunity. Adv Vet Med. 1999;41:429–446. doi: 10.1016/S0065-3519(99)80033-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Offit PA, Clark HF. Protection against rotavirus-induced gastroenteritis in a murine model by passively acquired gastrointestinal but not circulating antibodies. J Virol. 1985;54:58–64. doi: 10.1128/jvi.54.1.58-64.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bohl EH, Gupta RK, Olquin MV, Saif LJ. Antibody responses in serum, colostrum, and milk of swine after infection or vaccination with transmissible gastroenteritis virus. Infect Immun. 1972;6:289–301. doi: 10.1128/iai.6.3.289-301.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Saif LJ, Bohl EH, Gupta RK. Isolation of porcine immunoglobulins and determination of the immunoglobulin classes of transmissible gastroenteritis viral antibodies. Infect Immun. 1972;6:600–609. doi: 10.1128/iai.6.4.600-609.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schwartz K, Henry S, Tokach L, Potter M, Davidson D, Egnor C. Exposing sows to PEDV to build herd immunity [Internet] Minneapolis, MN: National Hog Farmer; 2014. [cited 2014 Mar 13]. Available from: http://nationalhogfarmer.com/business/exposing-sows-pedv-build-herd-immunity. [Google Scholar]
- 71.Park JS, Ha Y, Kwon B, Cho KD, Lee BH, Chae C. Detection of porcine circovirus 2 in mammary and other tissues from experimentally infected sows. J Comp Pathol. 2009;140:208–211. doi: 10.1016/j.jcpa.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 72.Ha Y, Shin JH, Chae C. Colostral transmission of porcine circovirus 2 (PCV-2): reproduction of post-weaning multisystemic wasting syndrome in pigs fed milk from PCV-2-infected sows with post-natal porcine parvovirus infection or immunostimulation. J Gen Virol. 2010;91:1601–1608. doi: 10.1099/vir.0.016469-0. [DOI] [PubMed] [Google Scholar]
- 73.Song DS, Oh JS, Kang BK, et al. Oral efficacy of Vero cell attenuated porcine epidemic diarrhea virus DR13 strain. Res Vet Sci. 2007;82:134–140. doi: 10.1016/j.rvsc.2006.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.de Arriba ML, Carvajal A, Pozo J, Rubio P. Mucosal and systemic isotype-specific antibody responses and protection in conventional pigs exposed to virulent or attenuated porcine epidemic diarrhoea virus. Vet Immunol Immunopathol. 2002;85:85–97. doi: 10.1016/s0165-2427(01)00417-2. [DOI] [PubMed] [Google Scholar]
- 75.Pan Y, Tian X, Li W, et al. Isolation and characterization of a variant porcine epidemic diarrhea virus in China. Virol J. 2012;9:195. doi: 10.1186/1743-422X-9-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tian Y, Yu Z, Cheng K, et al. Molecular characterization and phylogenetic analysis of new variants of the porcine epidemic diarrhea virus in Gansu, China in 2012. Viruses. 2013;5:1991–2004. doi: 10.3390/v5081991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim SH, Lee JM, Jung J, et al. Genetic characterization of porcine epidemic diarrhea virus in Korea from 1998 to 2013. Arch Virol. 2015;160:1055–1064. doi: 10.1007/s00705-015-2353-y. [DOI] [PMC free article] [PubMed] [Google Scholar]