Abstract
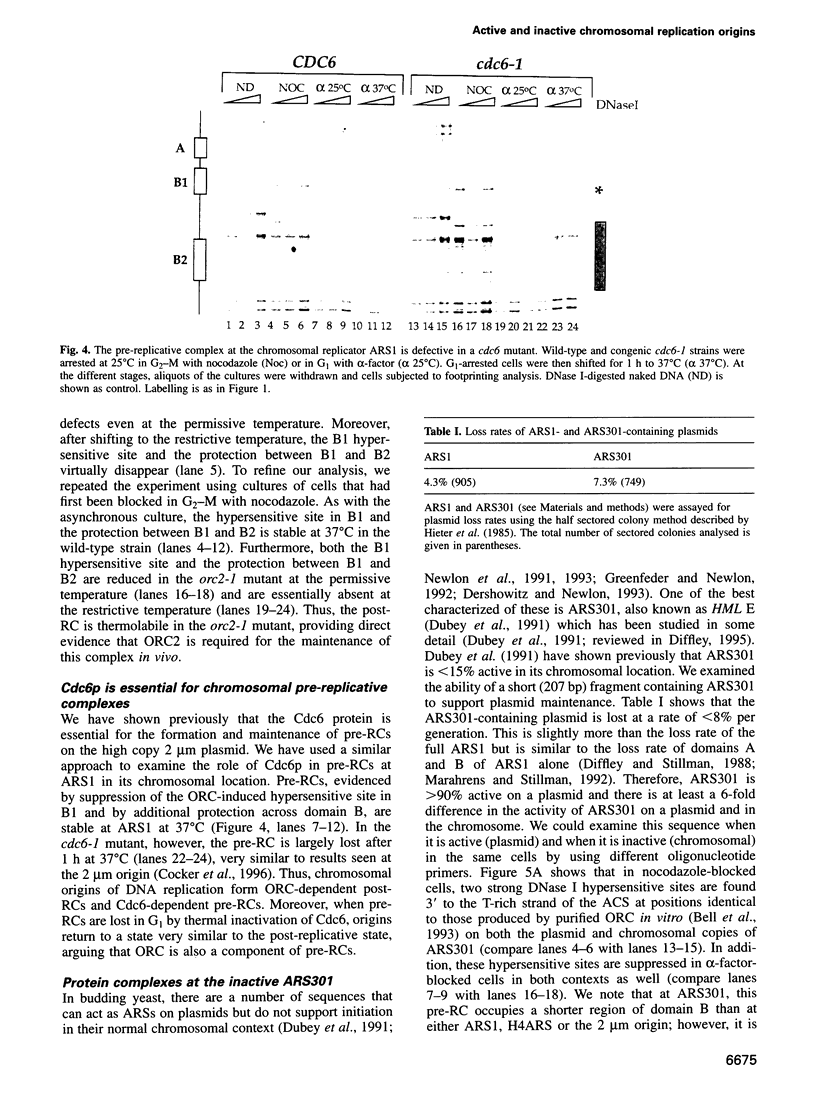
We have developed a genomic footprinting protocol which allows us to examine protein-DNA interactions at single copy chromosomal origins of DNA replication in the budding yeast Saccharomyces cerevisiae. We show that active replication origins oscillate between two chromatin states during the cell cycle: an origin recognition complex (ORC)-dependent post-replicative state and a Cdc6p-dependent pre-replicative state. Furthermore, we show that both post- and pre-replicative complexes can form efficiently on closely apposed replicators. Surprisingly, ARS301 which is active as an origin on plasmids but not in its normal chromosomal location, forms ORC- and Cdc6p-dependent complexes in both its active and inactive contexts. Thus, although ORC and Cdc6p are essential for initiation, their binding is not sufficient to dictate origin use.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bell S. P., Kobayashi R., Stillman B. Yeast origin recognition complex functions in transcription silencing and DNA replication. Science. 1993 Dec 17;262(5141):1844–1849. doi: 10.1126/science.8266072. [DOI] [PubMed] [Google Scholar]
- Bell S. P., Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992 May 14;357(6374):128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- Bouton A. H., Smith M. M. Fine-structure analysis of the DNA sequence requirements for autonomous replication of Saccharomyces cerevisiae plasmids. Mol Cell Biol. 1986 Jul;6(7):2354–2363. doi: 10.1128/mcb.6.7.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer B. J., Fangman W. L. Initiation at closely spaced replication origins in a yeast chromosome. Science. 1993 Dec 10;262(5140):1728–1731. doi: 10.1126/science.8259517. [DOI] [PubMed] [Google Scholar]
- Brewer B. J., Fangman W. L. Initiation preference at a yeast origin of replication. Proc Natl Acad Sci U S A. 1994 Apr 12;91(8):3418–3422. doi: 10.1073/pnas.91.8.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocker J. H., Piatti S., Santocanale C., Nasmyth K., Diffley J. F. An essential role for the Cdc6 protein in forming the pre-replicative complexes of budding yeast. Nature. 1996 Jan 11;379(6561):180–182. doi: 10.1038/379180a0. [DOI] [PubMed] [Google Scholar]
- Crevel G., Cotterill S. DNA replication in cell-free extracts from Drosophila melanogaster. EMBO J. 1991 Dec;10(13):4361–4369. doi: 10.1002/j.1460-2075.1991.tb05014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahmann C., Diffley J. F., Nasmyth K. A. S-phase-promoting cyclin-dependent kinases prevent re-replication by inhibiting the transition of replication origins to a pre-replicative state. Curr Biol. 1995 Nov 1;5(11):1257–1269. doi: 10.1016/s0960-9822(95)00252-1. [DOI] [PubMed] [Google Scholar]
- Dalton S., Whitbread L. Cell cycle-regulated nuclear import and export of Cdc47, a protein essential for initiation of DNA replication in budding yeast. Proc Natl Acad Sci U S A. 1995 Mar 28;92(7):2514–2518. doi: 10.1073/pnas.92.7.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dershowitz A., Newlon C. S. The effect on chromosome stability of deleting replication origins. Mol Cell Biol. 1993 Jan;13(1):391–398. doi: 10.1128/mcb.13.1.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley J. F., Cocker J. H., Dowell S. J., Rowley A. Two steps in the assembly of complexes at yeast replication origins in vivo. Cell. 1994 Jul 29;78(2):303–316. doi: 10.1016/0092-8674(94)90299-2. [DOI] [PubMed] [Google Scholar]
- Diffley J. F., Cocker J. H. Protein-DNA interactions at a yeast replication origin. Nature. 1992 May 14;357(6374):169–172. doi: 10.1038/357169a0. [DOI] [PubMed] [Google Scholar]
- Diffley J. F., Stillman B. Purification of a yeast protein that binds to origins of DNA replication and a transcriptional silencer. Proc Natl Acad Sci U S A. 1988 Apr;85(7):2120–2124. doi: 10.1073/pnas.85.7.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley J. F. The initiation of DNA replication in the budding yeast cell division cycle. Yeast. 1995 Dec;11(16):1651–1670. doi: 10.1002/yea.320111608. [DOI] [PubMed] [Google Scholar]
- Dowell S. J., Romanowski P., Diffley J. F. Interaction of Dbf4, the Cdc7 protein kinase regulatory subunit, with yeast replication origins in vivo. Science. 1994 Aug 26;265(5176):1243–1246. doi: 10.1126/science.8066465. [DOI] [PubMed] [Google Scholar]
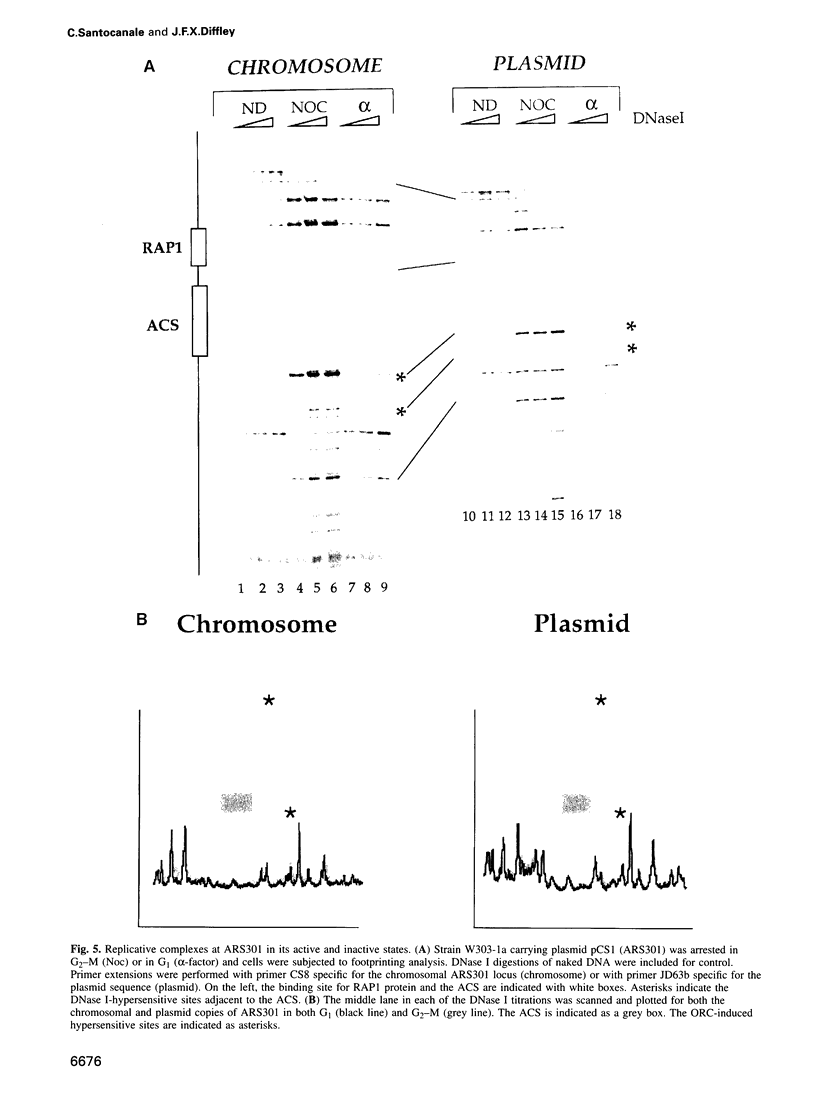
- Dubey D. D., Davis L. R., Greenfeder S. A., Ong L. Y., Zhu J. G., Broach J. R., Newlon C. S., Huberman J. A. Evidence suggesting that the ARS elements associated with silencers of the yeast mating-type locus HML do not function as chromosomal DNA replication origins. Mol Cell Biol. 1991 Oct;11(10):5346–5355. doi: 10.1128/mcb.11.10.5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fangman W. L., Brewer B. J. A question of time: replication origins of eukaryotic chromosomes. Cell. 1992 Oct 30;71(3):363–366. doi: 10.1016/0092-8674(92)90505-7. [DOI] [PubMed] [Google Scholar]
- Foss M., McNally F. J., Laurenson P., Rine J. Origin recognition complex (ORC) in transcriptional silencing and DNA replication in S. cerevisiae. Science. 1993 Dec 17;262(5141):1838–1844. doi: 10.1126/science.8266071. [DOI] [PubMed] [Google Scholar]
- Fox C. A., Loo S., Dillin A., Rine J. The origin recognition complex has essential functions in transcriptional silencing and chromosomal replication. Genes Dev. 1995 Apr 15;9(8):911–924. doi: 10.1101/gad.9.8.911. [DOI] [PubMed] [Google Scholar]
- Greenfeder S. A., Newlon C. S. A replication map of a 61-kb circular derivative of Saccharomyces cerevisiae chromosome III. Mol Biol Cell. 1992 Sep;3(9):999–1013. doi: 10.1091/mbc.3.9.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
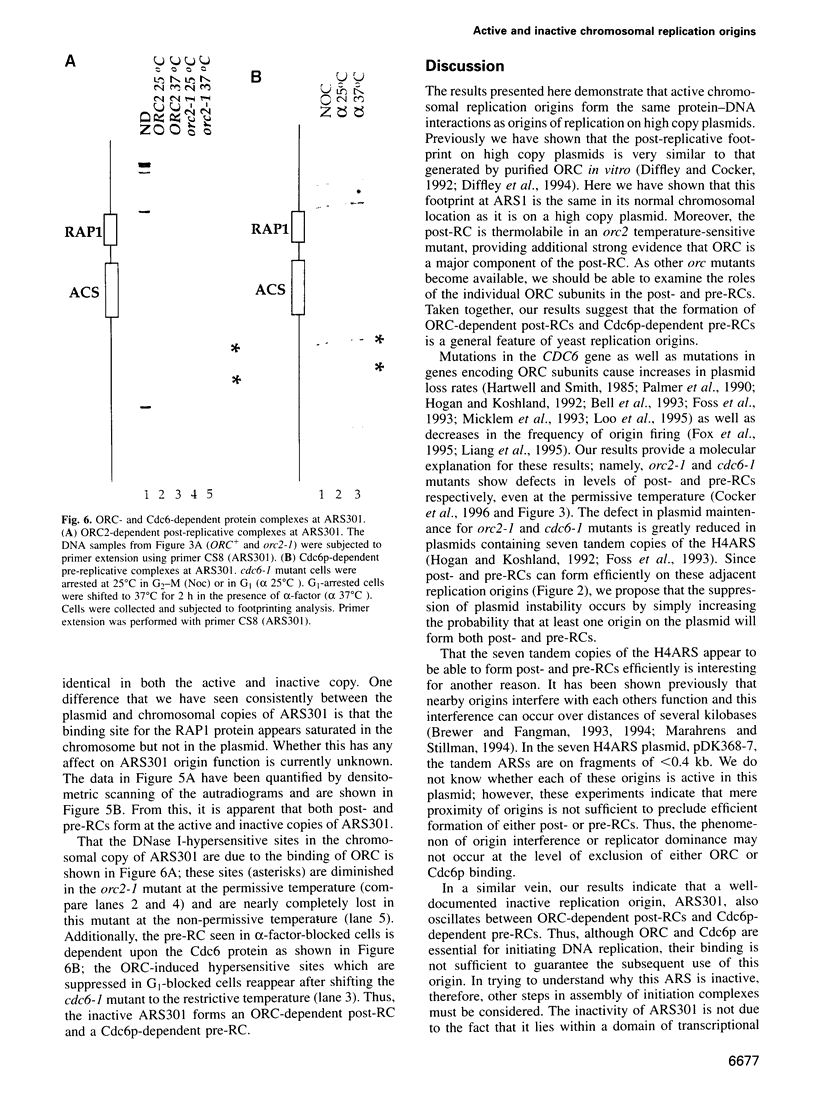
- Hartwell L. H., Smith D. Altered fidelity of mitotic chromosome transmission in cell cycle mutants of S. cerevisiae. Genetics. 1985 Jul;110(3):381–395. doi: 10.1093/genetics/110.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennessy K. M., Clark C. D., Botstein D. Subcellular localization of yeast CDC46 varies with the cell cycle. Genes Dev. 1990 Dec;4(12B):2252–2263. doi: 10.1101/gad.4.12b.2252. [DOI] [PubMed] [Google Scholar]
- Hieter P., Mann C., Snyder M., Davis R. W. Mitotic stability of yeast chromosomes: a colony color assay that measures nondisjunction and chromosome loss. Cell. 1985 Feb;40(2):381–392. doi: 10.1016/0092-8674(85)90152-7. [DOI] [PubMed] [Google Scholar]
- Hogan E., Koshland D. Addition of extra origins of replication to a minichromosome suppresses its mitotic loss in cdc6 and cdc14 mutants of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1992 Apr 1;89(7):3098–3102. doi: 10.1073/pnas.89.7.3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huberman J. A., Riggs A. D. Autoradiography of chromosomal DNA fibers from Chinese hamster cells. Proc Natl Acad Sci U S A. 1966 Mar;55(3):599–606. doi: 10.1073/pnas.55.3.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyrien O., Maric C., Méchali M. Transition in specification of embryonic metazoan DNA replication origins. Science. 1995 Nov 10;270(5238):994–997. doi: 10.1126/science.270.5238.994. [DOI] [PubMed] [Google Scholar]
- Hyrien O., Méchali M. Chromosomal replication initiates and terminates at random sequences but at regular intervals in the ribosomal DNA of Xenopus early embryos. EMBO J. 1993 Dec;12(12):4511–4520. doi: 10.1002/j.1460-2075.1993.tb06140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyrien O., Méchali M. Plasmid replication in Xenopus eggs and egg extracts: a 2D gel electrophoretic analysis. Nucleic Acids Res. 1992 Apr 11;20(7):1463–1469. doi: 10.1093/nar/20.7.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar A. J., Strathern J. N., Abraham J. A. Involvement of double-strand chromosomal breaks for mating-type switching in Saccharomyces cerevisiae. Cold Spring Harb Symp Quant Biol. 1984;49:77–88. doi: 10.1101/sqb.1984.049.01.011. [DOI] [PubMed] [Google Scholar]
- Kostriken R., Strathern J. N., Klar A. J., Hicks J. B., Heffron F. A site-specific endonuclease essential for mating-type switching in Saccharomyces cerevisiae. Cell. 1983 Nov;35(1):167–174. doi: 10.1016/0092-8674(83)90219-2. [DOI] [PubMed] [Google Scholar]
- Laurenson P., Rine J. Silencers, silencing, and heritable transcriptional states. Microbiol Rev. 1992 Dec;56(4):543–560. doi: 10.1128/mr.56.4.543-560.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C., Weinreich M., Stillman B. ORC and Cdc6p interact and determine the frequency of initiation of DNA replication in the genome. Cell. 1995 Jun 2;81(5):667–676. doi: 10.1016/0092-8674(95)90528-6. [DOI] [PubMed] [Google Scholar]
- Loo S., Fox C. A., Rine J., Kobayashi R., Stillman B., Bell S. The origin recognition complex in silencing, cell cycle progression, and DNA replication. Mol Biol Cell. 1995 Jun;6(6):741–756. doi: 10.1091/mbc.6.6.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo S., Rine J. Silencers and domains of generalized repression. Science. 1994 Jun 17;264(5166):1768–1771. doi: 10.1126/science.8209257. [DOI] [PubMed] [Google Scholar]
- Mahbubani H. M., Paull T., Elder J. K., Blow J. J. DNA replication initiates at multiple sites on plasmid DNA in Xenopus egg extracts. Nucleic Acids Res. 1992 Apr 11;20(7):1457–1462. doi: 10.1093/nar/20.7.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marahrens Y., Stillman B. A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science. 1992 Feb 14;255(5046):817–823. doi: 10.1126/science.1536007. [DOI] [PubMed] [Google Scholar]
- Marahrens Y., Stillman B. Replicator dominance in a eukaryotic chromosome. EMBO J. 1994 Jul 15;13(14):3395–3400. doi: 10.1002/j.1460-2075.1994.tb06642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Salas E., Cupo D. Y., DePamphilis M. L. The need for enhancers is acquired upon formation of a diploid nucleus during early mouse development. Genes Dev. 1988 Sep;2(9):1115–1126. doi: 10.1101/gad.2.9.1115. [DOI] [PubMed] [Google Scholar]
- Micklem G., Rowley A., Harwood J., Nasmyth K., Diffley J. F. Yeast origin recognition complex is involved in DNA replication and transcriptional silencing. Nature. 1993 Nov 4;366(6450):87–89. doi: 10.1038/366087a0. [DOI] [PubMed] [Google Scholar]
- Méchali M., Kearsey S. Lack of specific sequence requirement for DNA replication in Xenopus eggs compared with high sequence specificity in yeast. Cell. 1984 Aug;38(1):55–64. doi: 10.1016/0092-8674(84)90526-9. [DOI] [PubMed] [Google Scholar]
- Newlon C. S., Collins I., Dershowitz A., Deshpande A. M., Greenfeder S. A., Ong L. Y., Theis J. F. Analysis of replication origin function on chromosome III of Saccharomyces cerevisiae. Cold Spring Harb Symp Quant Biol. 1993;58:415–423. doi: 10.1101/sqb.1993.058.01.048. [DOI] [PubMed] [Google Scholar]
- Newlon C. S., Lipchitz L. R., Collins I., Deshpande A., Devenish R. J., Green R. P., Klein H. L., Palzkill T. G., Ren R. B., Synn S. Analysis of a circular derivative of Saccharomyces cerevisiae chromosome III: a physical map and identification and location of ARS elements. Genetics. 1991 Oct;129(2):343–357. doi: 10.1093/genetics/129.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer R. E., Hogan E., Koshland D. Mitotic transmission of artificial chromosomes in cdc mutants of the yeast, Saccharomyces cerevisiae. Genetics. 1990 Aug;125(4):763–774. doi: 10.1093/genetics/125.4.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatti S., Böhm T., Cocker J. H., Diffley J. F., Nasmyth K. Activation of S-phase-promoting CDKs in late G1 defines a "point of no return" after which Cdc6 synthesis cannot promote DNA replication in yeast. Genes Dev. 1996 Jun 15;10(12):1516–1531. doi: 10.1101/gad.10.12.1516. [DOI] [PubMed] [Google Scholar]
- Rao H., Stillman B. The origin recognition complex interacts with a bipartite DNA binding site within yeast replicators. Proc Natl Acad Sci U S A. 1995 Mar 14;92(6):2224–2228. doi: 10.1073/pnas.92.6.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley A., Cocker J. H., Harwood J., Diffley J. F. Initiation complex assembly at budding yeast replication origins begins with the recognition of a bipartite sequence by limiting amounts of the initiator, ORC. EMBO J. 1995 Jun 1;14(11):2631–2641. doi: 10.1002/j.1460-2075.1995.tb07261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathern J. N., Klar A. J., Hicks J. B., Abraham J. A., Ivy J. M., Nasmyth K. A., McGill C. Homothallic switching of yeast mating type cassettes is initiated by a double-stranded cut in the MAT locus. Cell. 1982 Nov;31(1):183–192. doi: 10.1016/0092-8674(82)90418-4. [DOI] [PubMed] [Google Scholar]
- Sugawara N., Ivanov E. L., Fishman-Lobell J., Ray B. L., Wu X., Haber J. E. DNA structure-dependent requirements for yeast RAD genes in gene conversion. Nature. 1995 Jan 5;373(6509):84–86. doi: 10.1038/373084a0. [DOI] [PubMed] [Google Scholar]
- Yan H., Merchant A. M., Tye B. K. Cell cycle-regulated nuclear localization of MCM2 and MCM3, which are required for the initiation of DNA synthesis at chromosomal replication origins in yeast. Genes Dev. 1993 Nov;7(11):2149–2160. doi: 10.1101/gad.7.11.2149. [DOI] [PubMed] [Google Scholar]









