Summary
The ability to introduce DNA sequences (e.g. genes) of interest into the germline genome has rendered the mouse a powerful and indispensable experimental model in fundamental and medical research. The DNA sequences can be integrated into the genome randomly or into a specific locus by homologous recombination, in order to: (i) delete or insert mutations into genes of interest to determine their function, (ii) introduce human genes into the genome of mice to generate animal models enabling study of human-specific genes and diseases, e.g. mice susceptible to infections by human-specific pathogens of interest, (iii) introduce individual genes or genomes of pathogens (such as viruses) in order to examine the contributions of such genes to the pathogenesis of the parent pathogens, (iv) and last but not least introduce reporter genes that allow monitoring in vivo or ex vivo the expression of genes of interest. Furthermore, the use of recombination systems, such as Cre/loxP or FRT/FLP, enables conditional induction or suppression of gene expression of interest in a restricted period of mouse’s lifetime, in a particular cell type, or in a specific tissue. In this review, we will give an updated summary of the gene targeting technology and discuss some important considerations in the design of gene-targeted mice.
Keywords: gene targeting, transgenic mice, knockout mice, reporter mice, ES cell lines, targeting vector, Cre/loxP, FRT/FLP, MultiSite Gateway Cloning
1. The application of genetically modified mice for the study of viral pathogenesis and anti-viral immunity
The development of mice with germline genetic modifications has advanced our understanding of the mechanisms of viral pathogenesis and anti-viral immune responses during virus-host-interactions enormously. For instance, transgenic mouse models that express antigen-specific, major histocompatibility complex (MHC)-restricted T cell receptor (TCR) transgenes have been used extensively to investigate virus-specific T cell responses. Such transgenic mice include for example mice expressing a TCR transgene specific for the influenza virus hemagglutinin (HA) in the context of the MHC class I or II molecules (1,2), and transgenic mice expressing MHC-restricted TCR with specificity for a lymphocytic choriomeningitis virus (LCMV) glycoprotein-derived T helper cell epitope (3,4).
In addition, because mice are not susceptible to many human viruses, such as hepatitis viruses, papillomavirus, poliovirus, human immunodeficiency virus-1 (HIV-1) and measles, the generation of transgenic mice that express human receptors specific for such viruses have rendered those transgenic mice susceptible to infection by human viruses of interest and subsequently enabled to investigate their pathogenesis in in vivo models (reviewed in (5)).
An alternative to mice expressing human virus receptors provides transgenic mice with (conditional) expression of individual genes or genomes of viruses of interest. Such (conditional) expression of viral genes in mice imitates viral infection and thus enables in vivo investigation of the pathogenesis and immune responses induced by human viruses of interest (5-7).
Moreover, knockout mice with deletions of specific genes or cell populations of interest have been useful to identify and investigate the cellular and molecular components of the adaptive and innate immune system that play a role in controlling viral infections. Such knockout mice include for example: TCR-β knockout mice that lack T cells (8); μMT mice that lack B cells (9); RAG-1 and RAG-2 knockout mice that lack B and T cells (10,11); knockout mice with deletions of immune mediators (such as chemokines and cytokines) including knockout mice for type I and type II interferons or interferon receptors (12-15); knockout mice for immune receptors, such as Toll like receptors (TLRs), retinoic acid-inducible gene-I (RIG-I)-like receptors (RLRs), nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs) and C-type lectin receptors (CLRs) (16-19); knockout mice for immunologically relevant transcription factors, such as STAT (Signal Transducer and Activator of Transcription) molecules (20,21) and interferon regulatory factors (IRFs) (22); and knockout mice for adaptor molecules (such as MyD88, Rip2, Trif, etc.) that are involved in connecting signals from immune receptors to downstream enzymes and transcription factors (16,17,19).
In order to unequivocally identify and investigate the cellular sources of cytokines, major soluble mediators of innate and adaptive immune responses, many cytokine reporter mice have been established that enable to track cytokine production during e.g. infections (23-25). A special chapter by Brinkmann and colleagues describes in this issue of “Virus-Host-Interactions” the use of a cytokine reporter mouse model, the IFNβ-Luciferase-knockin reporter mice (26), for the analysis of type I IFN induction by mouse cytomegalovirus (MCMV).
2. Techniques to generate genetically modified mice
While the research of viral pathogenesis and anti-viral immunity has taken advantage of genetically modified mice, as described above, it is worth mentioning that, on the other side, it was the use of viruses that opened up the possibility to modify the genome of mice and helped to generate the first transgenic mice in 1976 (27). To address the question whether exogenous viruses (transmitted horizontally, not hereditarily, from individual-to-individual) can be converted into endogenous viruses (of which DNA sequences are present in all somatic and germ cells of an individual and passed on to the offspring), Rudolf Jaenisch infected preimplantation mouse embryos (at the 4-8 cell stage) with the Moloney murine leukemia virus (M-MuLV). The mice generated from these infected preimplantation embryos developed M-MuLV-induced leukemia, and the viral DNA was integrated into the germ line of the mice and transmitted to their offspring (27). Subsequently the direct microinjection of DNA of interest into the pronucleus of fertilized murine eggs was developed as a more commonly used technique to generate transgenic mice (28-32).
However, the generation of genetically modified mice by infection of mouse embryos with retroviruses or microinjection of DNA into fertilized murine eggs results in random integrations of the exogenous DNA into the mouse genome. This in turn can lead to variegated expression of the transgene and inadvertent disruption of genes at the site into which the transgene is inserted. The frequency of phenotypes arising from insertion site mutation by a transgene (almost 10%) is higher than might be expected from random integration into the genome (33), because introduction of transgenes by pronuclear injection can generate large deletions and complex rearrangements at the site of DNA integration (33). In contrast, gene targeting by homologous recombination in murine embryonic stem (ES) cells, a method that was established in the late 1980s, has enabled controlled and specific genetic modification by site-specific integration of exogenous DNA of interest into the genome of mice (34-37).
3. Generation of mutant mice from genetically modified embryonic stem cells
The realization that genes of interest can be specifically modified in a whole mouse was developed from extensive work to determine whether cultured mammalian cells can mediate homologous recombination between their endogenous DNA and exogenously added DNA molecules (38-40). Simultaneously, murine embryonic stem (ES) cells (ESCs) were successfully cultured in vitro without losing their pluripotent potential. Thus, when introduced into a preimplantation embryo they could contribute to the germ line (41-43). ESCs have subsequently been widely used as vehicles to transfer site-specific genetic modifications of interest to the mouse germline (34-37,44-46).
ESCs are derived from the pluripotent inner cell mass (ICM) of blastocysts, a structure formed in the early stage of embryogenesis (3.5 days old pre-implantation mouse embryo), and thus ESCs can contribute to all embryonic tissues, including the germ cells, in developing mice (Fig. 1). Isolated ESCs have to be cultured in special culture conditions to maintain their multiplication (self-renewal) capacity without loss of pluripotency. ESCs are typically cultured on a feeder layer of mitotically inactivated (mytomicin C treated or gamma-irradiated) mouse embryonic fibroblasts (MEFs) in order to obtain the necessary factors for self-renewal and pluripotency. Leukemia inhibitory factor (LIF), Wingless/Integrated (Wnt), and ligands of the TGF-ß/BMP signaling pathway are among factors supplied by the fibroblasts and were found to influence the state and pluripotency of murine ESCs (47). Supplementation of ESC culture medium with recombinant LIF helps to increase the maintenance of pluripotency. Recently, it has been shown that the maintenance of pluripotency can also be achieved, in MEF-free culture systems, by use of glycogen synthase kinase (GSK)-3-specific inhibitors, such as 6-bromoindirubin-3’-oxime (BIO) (48), or, optimally, by use of a combination of three inhibitors (3i medium): SU5402 (inhibits FGF receptor tyrosine kinases), PD184352 (inhibits ERK signal cascades) and CHIR99021 (a more selective inhibitor of GSK-3) (49). Interestingly, Domogatskaya et al. (50) demonstrated recently that pluripotency and self-renewal of mouse ES cells can be achieved in the absence of feeder cells, LIF, or differentiation inhibitors by culturing ES cells on plates coated with a recombinant human extracellular matrix protein, the laminin isoform 511 (LN-511) (50).
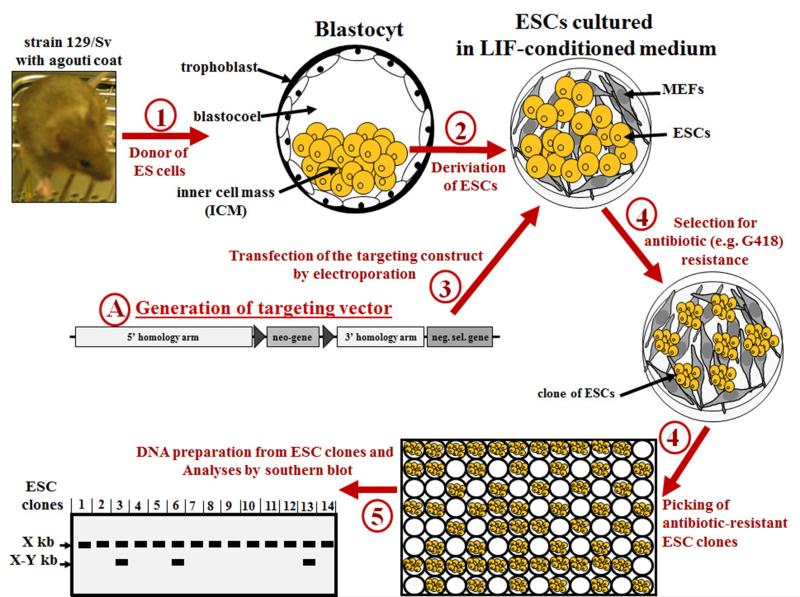
Figure 1A. Summarized most common steps for isolation of mouse embryonic stem (ES) cells and generation of homologous recombinant ES clones.
(1) In the first step 3.5-day-old mouse embryos (blastocysts) are collected from the uterine horn of superovulated (hormone treated) mated female mice with, for example, an agouti coat (strain 129/Sv). (2) Embryonic stem (ES) cells (ESCs) are derived from the inner cell mass of blastocysts and cultured on a feeder layer of mitotically inactivated mouse embryonic fibroblasts (MEFs), in ESC medium (supplemented with leukemia inhibitory factor (LIF). (3) After electroporation with the targeting vector of interest, (4) successfully transfected ESCs are selected by adding appropriate selection agent to the ESC medium; and (5) ESC clones are picked. (6) Homologous recombinant ESC clones are identified by Southern blot. The genomic DNA isolated from ESC clones should be digested with an appropriate restriction enzyme that produce one cut inside the targeting vector and one cut just outside (upstream or downstream) the targeting vector, in the targeted chromosomal region. The use of an “external” probe outside of the targeting construct will produce a band with a size corresponding to unmodified wild-type allele(s), which is here indicated by X kb, and, if homologous recombination occurred, a second band of bigger or smaller size corresponding to the targeted allele, which is here indicated by X-Y kb.
Several established ESCs lines are in common use. Initially, most used ES cells were derived from the 129 mouse strain (51,52). Examples include among others, E14 cell lines (53), D3 cell lines (54), J1 cell line (55), R1 cell line (56) and AB2.1 cell line (57). 129 ES cell lines are often used because of their more robust performance in cell culture and higher germline transmission rates compared to ES cell lines from C57BL/6J and C57BL/6N mouse strains (58-60).
The C57BL/6 mouse strain is however one of the best characterized inbred strains that is widely accepted as the reference strain for immunological, neurobiological, behavioral and physiological studies in mice, and is the standard reference library for the mouse genome-sequencing program (58-61). Therefore, mice derived from 129 ES cell lines need to be backcrossed for 10 or more generations onto C57BL/6 background, which is a time consuming process taking two or more years.
C57BL/6 ES cell lines with efficient germline colonization have been generated and these facilitate the direct generation of genetically altered C57BL/6 mice (58,59,61-64). The requirement for backcrossing mice derived from C57BL/6-derived ES cells is considerably less (though not entirely eliminated as the ES cells may harbor mutations acquired in vitro).
ES cell lines derived from C57BL/6 mice that can be used include: Bruce4 (64), BL/6-III (63), LK1 (59) and JM8 (61). The JM8 cell lines have been used for the large-scale mouse knockout program to generate mice with targeted mutations in the C57BL/6 genetic background (61,65). JM8 cells can be easily propagated using standard embryonic stem culture conditions, in the presence and absence of feeder cells. In addition, to simplify breeding schemes, the dominant agouti coat color gene was restored in JM8 cells by targeted repair of the C57BL/6 nonagouti mutation enabling visual assessment of coat color contribution and germline transmission (61).
Taken together, when selecting an ES cell line, several considerations are important: i) the advantages and drawbacks of C57BL/6 versus 129 ES cell lines (60); ii) the genetic variation and stability among ES cell lines from different 129 substrains (52) and C57BL/6 substrains (66), respectively, which might influence e.g. gene targeting efficiency and phenotype of mice derived from the respective ES cell line; iii) the homology arms of the targeting vector should be from DNA that is isogenic to the ES cells used. Furthermore, the most appropriate ES cell line to use is dependent on the cell culture conditions and expertise in the respective laboratories.
In vitro cultured ESCs can be genetically modified and then reinjected into the blastocoel (cavity) of blastocysts (3.5 days old pre-implantation mouse embryo) (67,68) (Fig. 1), or into morula (2.5 days old pre-implantation mouse embryo) (68). Usually 10-20 ESCs are injected in a blastocyst (67,68). Alternatively, chimeric embryos can be generated by aggregating ES cells with morula (69). The chimeric embryos (usually 5-10) are then surgically transferred into the uterus of recipient pseudopregnant foster females. The genotype of these females is irrelevant, as long as they are good surrogate mothers. That is, they nurse carefully the newborns to weaning age and they accept pups from another mother. For these reasons, CD1 females or C57BL/6 × BALB/c F1 females are recommended for use (68). Chimeric mice, in which the injected genetically modified ES cells have contributed to the formation of most or all tissues, will be born at frequencies varying from a few percent to majority of the pups (Fig. 1). If the genetically modified ES cells have contributed to germ cell formation, the introduced genetic modifications can be passed on to offspring from the chimeric mice.
To facilitate the selection of the desired chimeric offspring, the ES cells and recipient blastocysts are derived from different mouse strains with distinguishable coat-colors (each mouse strain homozygous for the corresponding coat-color allele) (Fig. 1B). For example, the recipient blastocysts may be derived from C57BL/6 mice (black coat-color), and ES cells from a 129 mouse strain (agouti/brown coat-color). The extent of the contribution of the ES cells to the formation of the chimeric mouse can be visually recognized by inspection of coat-color chimerism (% of black and agouti hair on the mouse black-agouti) (Fig. 1B).
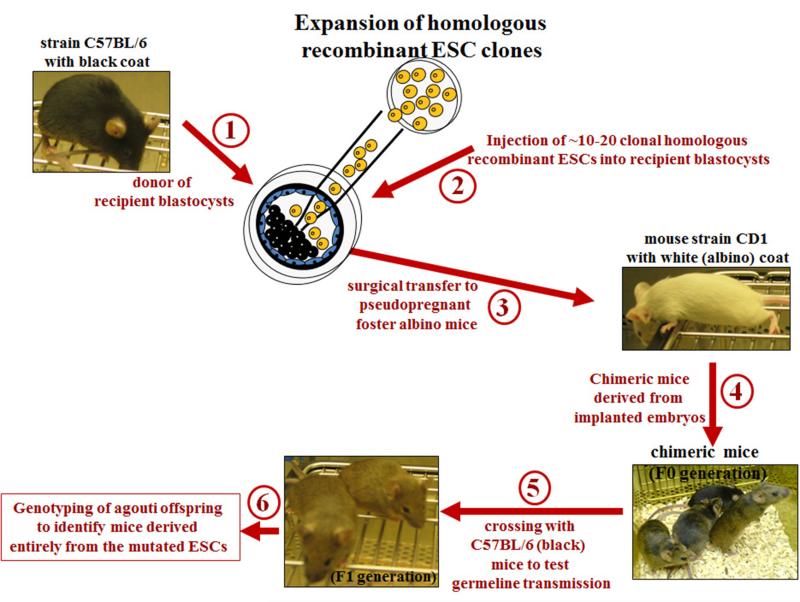
Figure 1B. Generation of mice with genome modification of interest using homologous recombinant ESC clones.
(1) If ESCs are derived from mice with an agouti coat (such as strain 129/Sv), the recipient pre-implantation mouse embryos (blastocysts) should be collected from female mice with black coat (such as strain C57BL/6). (2) The identified and expanded homologous recombinant ESC clones (see Figure 1A) are injected into recipient pre-implantation mouse embryos (blastocysts) that are collected from female mice with black coat (strain C57BL/6).(3) These injected blastocysts are then surgically transferred to a recipient pseudopregnant foster mother to allow the embryos to develop. Females of CD1 mouse strain make very good mothers, and are thus used by several laboratories as foster mothers. (4) Because ESCs and recipient blastocysts were derived from mouse strains with distinguishable coat-colors, the desired chimeric offspring can be visually recognized by inspection of coat-colour chimerism (% of black and agouti hair on the mouse black-agouti). (5) Chimeric offspring (usually only the males, because the used ES cell lines are usually male) are mated with C57BL/6 mice to produce the F1 generation. (6) The germline transmission is then confirmed by Southern blot analysis or PCR of tail DNA from the agouti (not black) mice of the F1 generation.
4. Gene targeting in ES cells
Using current techniques, there are almost no limitations into the types of modifications that can be introduced, ranging from gene insertion, point mutations, short and long range deletions, inversions. Conditional knockouts or knockins are generated by placing loxP or FRT sites flanking selected exons (see also text below).
The introduction of site-specific modifications into the genome of ESCs by homologous recombination, a process called gene targeting, is achieved through the introduction of a targeting vector into ESCs by electroporation (Fig. 1 and 2).
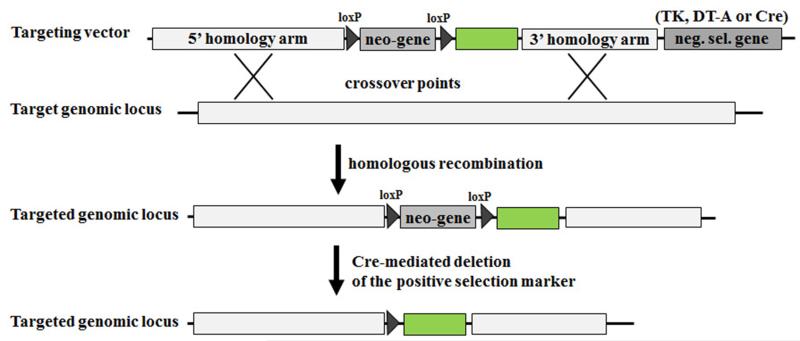
Figure 2. Typical gene targeting strategy.
A targeting vector is typically composed of three basic units: (i) a 5’ homology arm; (ii) a gene marker for positive selection (e.g. neomycin resistance gene (neo)); (iii) a 3’ homology arm; and (iv) a negative selection marker (neg. sel. Marker), such as thymidine kinase, diphtheria toxin fragment A (DT-A), or, if the positive selection marker is flanked by loxP sites, Cre recombinase gene (Cre). Furthermore, any desired DNA sequence of interest (here green box) can be inserted between the homology arms of the targeting vector, in order to introduce it into the target genome by homologous recombination. Homologous recombination between the targeting vector and the target cognate chromosomal region results in the disruption of one genomic copy of the targeted genomic locus and loss of the vector’s negative selection marker gene. Crossover points are depicted by “X”. The floxed (loxP sites flanked) positive selection marker gene can be removed by expressing Cre recombinase in the recombinant ESCs or by crossing the chimeric mice with Cre-expressing transgenic mice (see also Fig. 4A).
A targeting vector (DNA construct) is typically composed of three basic units (Fig. 2): (i) a 5’ homology arm, (ii) a positive selectable gene marker (such as the neomycin resistance gene (neo) or hygromycin (hyg)), (iii) a 3’ homology arm. The transfected targeting vector can either insert itself randomly into the genome or be integrated by homologous recombination as determined by the 5’ and 3’ homology arms. Successfully transfected cells are positively selected by culturing ESCs in medium with neomycin (G418) or other appropriate antibiotics, such as hygromycin or puromycin (Fig.1). If the positive selection marker gene is flanked by loxP or FRT sites, then it can be later removed from targeted loci in ESCs or transgenic mice by expressing Cre recombinase or flippase (FLP) in the recombinant ESCs (through transfection with Cre- or FLP-expressing vector) or by crossing the chimeric mice with Cre- or FLP-expressing transgenic mice (Fig. 2, and see also Fig. 4A). This is an important consideration as the introduction of selection marker genes, such as neo, can profoundly affect the expression of endogenous genes neighbouring to the targeted gene locus (70-72).
The homologous recombination of a targeting vector into a genomic locus of interest occurs at a very low frequency (at a frequency of 10−3 to 10−4 relative to nonhomologous recombinants) (73). In general, the longer the length of the 5’ and 3’ homology arms the higher the targeting frequency is (74,75). Another factor that increases homologous recombination frequencies is whether the homology arms of the targeting vector are isogenic with the ES cell DNA (74,76). Ideally, the homologous arms should be derived from genomic DNA prepared from the ES cells to be used or at least from the same strain of mice that the ES cells were derived from. Linearization of the targeting construct before its transfection into ES cells also enhances the frequency of homologous recombination (77). Nevertheless, because homologous recombination is a rare event, the screening of at least 200, and often up to 1000 clones is required to identify a few clones that have undergone homologous recombination.
A method to enrich the selection for homologous recombinant clones uses negative selection markers, such as thymidine kinase (TK) from herpes simplex virus (HSV) (44), or the diphtheria toxin fragment A (DT-A) from Corynebacterium diphtheria (78). The gene encoding for the negative selection marker is included outside the homology arms of the targeting vector (Fig. 2). During homologous recombination, sequences outside the regions of homology to the target genomic locus are usually lost. By contrast, if the gene targeting vector is integrated randomly in the genome, the negative selection marker is often retained. TK renders the cells sensitive to thymidine analogues, such as 5-iodo-2’-fluoro-2’-deoxy-1-ß-D-arabino-furonosyluracil (FIAU) or gancyclovir, that are supplemented in the ESCs culture medium to eliminate clones with randomly integrated targeting vector. The TK enzyme activates these thymidine analogues, resulting in their incorporation into replicating DNA, causing premature chain termination and cell death (79). DT-A exerts toxicity by catalysing the transfer of ADP-ribose from nicotinamide adenine dinucleotide (NAD) to a modified histidine residue on the elongation factor 2 (eEF2), thereby inhibiting protein synthesis (78).
Recently, a new simple negative selection procedure using an “auto-selecting targeting vector” has been developed (Fig. 2) (80). As negative selection marker, a cyclization recombination (cre) gene -under the control of herpes simplex virus (HSV) promoter- was placed outside the homology region of the targeting vector. Because the positive selection marker, the neomycin resistance gene (neo), was flanked by two loxP sites (floxed), random integration of targeting vector into the genome will often result in the maintenance and expression of Cre recombinase which specifically recognizes the recombining loxP sites and subsequently mediates the deletion of the floxed positive selection marker (neo). In this case, the ES cells are not able to grow in medium containing G418 (neomycin) (80). However, after homologous recombination, the Cre gene will in most cases not integrate and the positive selection marker will be retained (though it can be deleted once positive clones have been identified).
5. The use of Cre/loxP recombination system in gene targeting
Cre recombinase is a 38 kDa protein from the bacteriophage P1 that mediates intramolecular and intermolecular site-specific recombination between two loxP sites (locus of X-over of P1). The loxP sequence is 34 bp long and consists of two 13 bp inverted repeats separated by an 8 bp nonpalindromic (asymmetric) sequence which dictates the orientation of the overall loxP site (Fig. 3). Two loxP sequences in opposite orientation mediate the inversion of the intervening DNA by Cre recombinase rather than excision while two sites in the same orientation mediate excision of the intervening DNA between the sites after which only one loxP site remains. If the loxP sites are located on different chromosomes, Cre recombinase can mediate a chromosomal translocation (Fig. 4) (81-85).
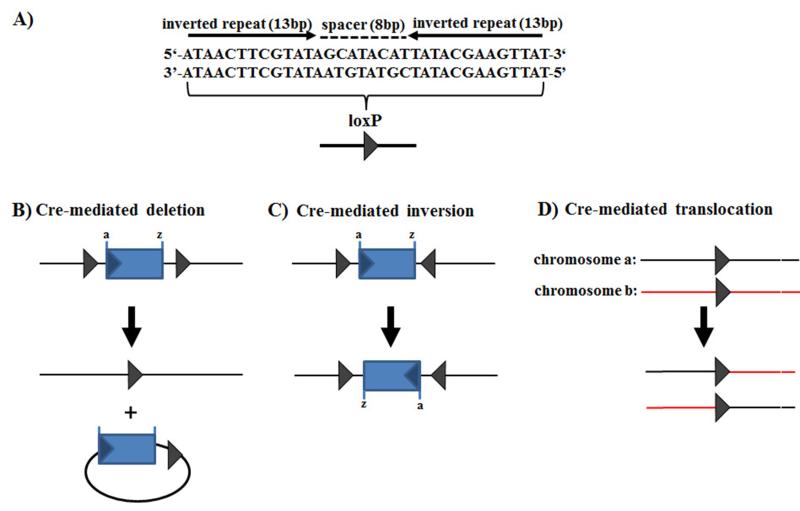
Figure 3. LoxP structure and Cre recombinase-mediated recombinations.
(A) Single loxP site that contains two inverted 13 bp repeats, separated by an asymmetric 8 bp long sequence. The type of Cre-mediated recombination is dependent on the orientation and location of the loxP sites: (B) Cre excises a circular molecule from between two loxP sites placed in the same orientation; (C) Cre inverts the DNA sequence between two loxP sites positioned in opposite orientation; (D) Cre-mediated recombination between two different linear DNA molecules (e.g. chromosomes), each containing a loxP site, resulting in the exchange of the DNA regions flanking the loxP sites. Figure was modified from Torres and Kuehn (111).
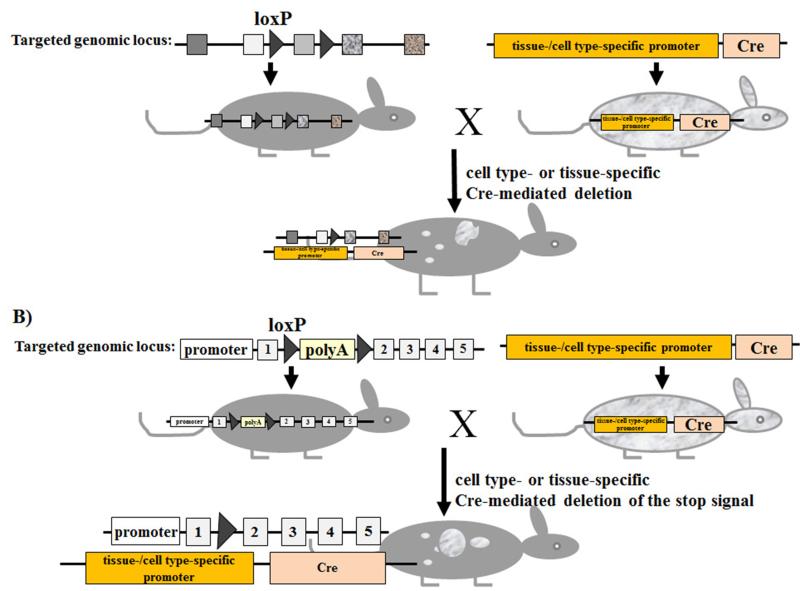
Figure 4. Conditional gene targeting using the Cre/loxP recombination system.
(A) Cre-mediated inactivation of a gene of interest. Left mouse: introduction of loxP sites into a genomic locus of interest by homologous recombination using embryonic stem cells (see also Fig. 1). LoxP sites are introduced in a manner that they don’t interfere with the function of the targeted gene. Right mouse: a transgenic strain that express Cre-recombinase under the control of cell type- or tissue-specific promoter. By crossing the floxed mouse with Cre transgenic mice, Cre mediates the deletion of the floxed genomic sequence, resulting in the inactivation of the targeted gene. Gene deletion is restricted to the “area” (cell types or tissues) where Cre is expressed (white flecks). (B) Cre-mediated activation of a gene of interest. Left mouse: introduction of a floxed intervening sequence (e.g. polyadenylation signal sequences) that prevent the correct transcription of the targeted gene. By crossing the floxed mouse with Cre transgenic mice, Cre mediates the deletion of the floxed intervening sequence, resulting in the reactivation of the targeted gene. Gene activation is restricted to the “area” (cell types or tissues) where Cre is expressed (white flecks).
The Cre/loxP system has emerged as a useful tool in genetic manipulations (86). Generally, any DNA sequence of interest can be deleted by flanking it with loxP sites. Cre/loxP enables, for example, the deletion of the selection marker after successful integration of the targeting vector into the genome of ES cells or mice. The loxP sites can be introduced into the genomic locus of interest by homologous recombination as described above. Furthermore, the conditional (timely or spatially controlled) expression of Cre recombinase enables to determine, where (e.g. in which cell type or tissue) and when (at which time of mouse’s life or of developmental stage of cells/tissues) the deletion of the floxed DNA sequence should occur (Fig. 4A). Thus, for conditional site-specific genome modification, two mouse lines are usually needed (Fig. 4A). First, mice that have DNA sequence of interest flanked by loxP sites (floxed). Second, Cre recombinase transgenic mice, in which Cre is expressed under the control of a promoter that is active in specific cell types or tissues, or Cre is transiently expressed under the control of a promoter that is active at a particular developmental stage of tissues or cells. When crossing the floxed mouse with a Cre transgenic mouse, the floxed DNA sequence is subsequently deleted in the cell types or tissues, where Cre is expressed. Collection databases of several hundreds of Cre transgenic mouse lines expressing the Cre recombinase in specific tissues or cells are available (e.g. http://www.ics-mci.fr/mousecre/; http://nagy.mshri.on.ca/cre_new/index.php; http://www.creportal.org/; http://bioit.fleming.gr/crezoo/; http://creline.org/).
The Cre/loxP system enables not only the deletion (shut off) of genes of interest, but has been proved to be successful also to specifically turn on (activate) the expression of any gene (or transgene) of interest (87,88). For this, the gene of interest is rendered quiescent e.g. by interposing floxed polyadenylation signal sequences mediating premature transcription arrest within (Fig. 4B). After intercrossing with a Cre-transgenic mouse or delivery of Cre into floxed transgenic mice using e.g. Cre-expressing adenoviruses, the floxed polyadenylation signal sequences can be removed by Cre-mediated excision, resulting in the activation of the gene expression in a specific cell type or tissue.
Such Cre/loxP-mediated gene activation has been a useful approach to avoid harmful effects of the transgene during mouse embryogenesis, or the induction of immune tolerance against the transgene product, for example in the case of viral genes. For instance, the application of the Cre/loxP technology has enabled to generate transgenic mice that conditionally express human hepatitis C virus transgenes upon intravenous administration of Cre-expressing adenovirus, and thus enabled the investigation of the immune responses to and pathogenesis of HCV infection (89).
Finally, another useful recombination system is the yeast Flippase (Flp)/Flp recognition target (FRT), which is mechanistically identical to that of the Cre/loxP recombination system (90-95), and represents an alternative tool to Cre/loxP. Moreover, the combination of both recombination systems can significantly increase the potentials of conditional gene manipulation in mice.
6. Cloning of the targeting vector
The design and construction of the targeting vector is among the most critical step in generating gene targeted mice. Many issues should be considered in the design of the targeting vector. These include, among others, the type of the desired genetic modification and the scientific questions of interest to be addressed (e.g. complete or conditional knockout, insertion of a reporter gene to monitor the expression of a gene of interest, insertion of point mutations, constitutive or conditional expression of a transgene of interest, etc.). Furthermore, the design should consider the option to remove the selection marker cassette at a later stage, and to linearize the targeting vector at a unique restriction site outside the homology arms prior to electroporation. It is also important to design restriction sites and probes that will enable the detection of ESC clones that have undergone homologous recombination. If necessary, new restriction sites have to be inserted that enable easy discrimination of homologous recombinant ESC clones by Southern blotting. Alternatively, homologous recombination can also be detected by long-range PCR or by qPCR reactions designed to detect the loss of an endogenous allele. Thus, accurate design of the targeting vector requires knowledge about the sequence and structure of the gene locus to be targeted (e.g. restrictions sites, promoter region, 5’UTR, exons, introns, and intron-exon borders, splice donor and acceptor sequences, 3’UTR, etc.). This has been made considerably simpler by access to the data sequences the sequencing of the whole mouse genome (96) (http://www.informatics.jax.org/ ; http://www.sanger.ac.uk/resources/mouse/genomes/).
The construction of a gene targeting vector can proceed by conventional restriction enzyme-based cloning strategies, by using MultiSite Gateway Technology, or by using Recombination-mediated genetic engineering (Recombineering) based protocols. Generation of targeting vectors using MultiSite Gateway Cloning is described in a special chapter in this issue of “Virus-Host-Interactions” (Bouabe & Okkenhaug).
7. Targeting transgenes into the ROSA26 locus
The expression of transgenes can be achieved by microinjection of DNA (e.g. BAC) containing the transgene of interest into fertilized murine eggs. However, this results in variegated expression of the transgene and inadvertent disruption of genes at the site into which the transgene might be inserted. A controlled insertion and expression of a transgene of interest can be achieved by targeting it into the Rosa26 locus. The Rosa26 locus was identified by analyzing pools of embryonic stem cells infected with the retroviral gene trap vector Gen-ROSAβgeo (bifunctional lacZ/neomycin phosphotransferase gene cassette) at low multiplicity (97). The gene trap vector has integrated into the Roas26, a chromosomal region that encodes for three transcripts (only the third transcript, originating from the reverse strand, seems to encode for a protein) (98).
Rosa26 locus is ubiquitously transcriptionally active, and thus it has become a preferred site for the integration and ubiquitous expression of transgenes and reporter genes. The genes of interest can be introduced into Roas26 locus by homologous recombination. Conditional expression of Rosa26-targeting genes can be achieved by using Cre/loxP system (Fig. 5 and subheading 5).
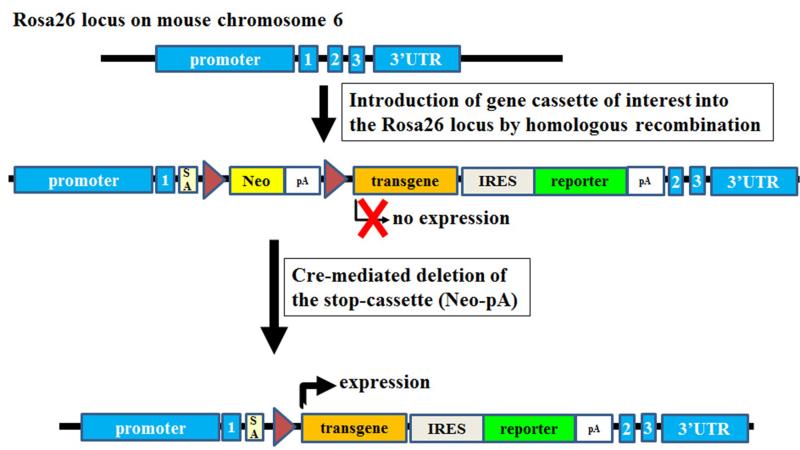
Figure 5. Targeting transgenes into the ROSA26 locus.
Introduction of gene cassettes of interest into Roas26 locus by homologous recombination in embryonic stem cells. A floxed intervening sequence, neomycin (Neo)-polyadenylation signal sequences (pA), prevents the transcription of the transgene. Cre expression mediates then the deletion of the floxed intervening sequence, resulting in the expression of the transgene and a reporter gene. The reporter gene facilitates to track the expression of the transgene. Numbered (1–3) blue rectangles: exons of the Roas26 locus. SA: splice acceptor. DNA-elements are not drawn to scale.
8. Considerations for the type of genetic modification
The simplest way to assess the role of a particular gene is to eliminate its expression, either by germline deletion or by inserting loxP sites flanking functionally fundamental parts of the gene to enable its conditional Cre-mediated deletion. This is usually best achieved by deleting the first exon(s). Care must be taken however to rule out expression of the gene from internal transcriptional start sites or by alternative splicing of exons. An alternative strategy is to delete exons that encode for the functionally most important part of the protein such as those that control its catalytic activity or essential interactions with other proteins, DNA or RNA. However, with this strategy there is a risk of generating peptides or truncated proteins which may not be functionally neutral, i.e. they may have dominant negative function preventing activation of other similar proteins in ways that can be difficult to predict and control. A way around this is to introduce point mutations that inactivate a particular enzymatic function or prevent the protein from interaction with others (99). Examples of such point mutations in immunologically relevant genes include among others, p110δD910A (kinase inactive) (100), p110γKD (kinase inactive) (101), p110γDASA (loss of interaction with Ras) (102), Vav1AA (lacks guanine exchange activity) (103), Vav1R442G (interrupts lipid binding via the PH domain) (104).
9. Considerations for the type of reporter genes when generating reporter mice
Reporter mice have emerged as important tools that facilitate in/ex vivo monitoring of the expression of genes of interest, especially when the gene products are secreted proteins, such as cytokines. In reporter mice the expression of an intracellularly localized reporter protein is linked to that of the endogenous gene of interest by using, for example, an internal ribosome entry site (IRES) or 2A peptides. Alternatively, the reporter gene can be introduced just downstream of the start codon of the endogenous gene of interest, resulting in the expression of the reporter gene instead of the targeted allele (23).
Autofluorescent proteins (AFPs), such as GFP, are the most frequently used reporters because their expression can be detected at single cell level without any invasive treatment of cells and without the need of exogenous substrates. However, a high expression amount of AFP (approximately 105 molecules ‘0.1–1 μM’ per cell) is required to detect fluorescence up over background (105). This is a fundamental limitation when expression of weakly expressed genes, such as the case of many cytokines, should be monitored (24,106).
A powerful solution to overcome this limitation is to use enzymatic reporters that catalyze strong signal amplification, and consequently a thousand times less catalytic molecules (approximately 10−4 μM) than AFP can generate a robust, measurable, reporter signal (107,108).
An example that emphasizes the importance of considering enzymatic reporters versus AFPs for weakly expressed genes has been revealed recently by studies on IL-10 reporter mice (reviewed in (24)). For instance, in contrast to ‘regular’ or ‘conventional’ IL-10-AFP reporter mice that enabled detection of reporter activity mainly only in T cells, because of their small cytosol, low autofluorescence and high expression level of IL-10 (24), an IL-10 reporter mouse model that is based on the reporter enzyme β-lactamase and the fluorescence resonance energy transfer (FRET) substrate coumarin-cephalosporin-fluorescein 4-AM (CCF4-AM) enabled to easily analyze and quantify IL-10 production at the single-cell level in all myeloid and lymphoid cell types, and thus also in cells exhibiting high autofluorescence and/or low expression level of IL-10 (106).
Thus, AFPs and enzymatic reporters along with their corresponding assay systems have specific features, advantages and limitations that must be carefully considered in choosing a system tailored to the particular questions being studied. Some important properties of commonly used reporter systems are summarized in table 1.
Table 1. Information was collected from the following references: (112-115).
| Detection by | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Reporter gene | substrate | signal amplification | cells have to be lysed/permeabilized |
FACS (at single living cell level) |
fluorescence microscopy |
in vivo imaging |
fluorometer | luminometer | endogenous activities similar to the reporter activity |
| Autofluorescent proteins (GFP, RFP etc.) | NO substrate is needed | NO (high detection limit) | NO | YES | YES | NO | YES | NO | NO |
| Firefly luciferase (Luc) | D-luciferin | YES | cell lysis | NO | NO | YES | NO | YES | NO |
| Secreted alkaline phosphatase (SEAP) | PNPP; FADP; CSPD; PPQ | YES | cell lysis | NO | NO | YES | YES | YES (in some cell types) | |
| Chloramphenicol acetyltransferase (CAT) |
14C-labeled chloramphenicol; fluorescent chloramphenicol derivative |
YES | cell lysis | NO | NO | NO | YES | YES | YES (minimal) |
| ß-galactosidase (LacZ) | ONPG; X-Gal; FDG; DDAOG | YES | cell permeabilization | YES/NO | YES | YES | YES | YES | YES (high) |
| β-lactamase (Bla) | CCF2-AM; CCF4-AM; nitrocefin | YES | NO | YES | YES | NO | YES | YES | NO |
10. The International Knockout Mouse Consortium (IKMC)
Before embarking on a project to generate a knockout or conditional knockout mouse, it is worth considering the international mouse knockout consortium (IKMC) (http://www.knockoutmouse.org/) whose aim is to generate conditional knockout alleles for all genes in the mouse genome (including also microRNA genes). They can supply targeting vectors, targeted ES cells or gene targeted mice (65,109,110). This in turn can result in considerable saving and shortening in time from the design to the execution of experiments involving new gene targeted strains.
Acknowledgement
Work in our laboratory is supported by grants from the BBSRC and the Wellcome Trust.
References
- 1.Bot A, Casares S, Bot S, von Boehmer H, Bona C. Cellular mechanisms involved in protection against influenza virus infection in transgenic mice expressing a TCR receptor specific for class II hemagglutinin peptide in CD4+ and CD8+ T cells. J Immunol. 1998;160:4500–4507. [PubMed] [Google Scholar]
- 2.Kirberg J, Baron A, Jakob S, Rolink A, Karjalainen K, von Boehmer H. Thymic selection of CD8+ single positive cells with a class II major histocompatibility complex-restricted receptor. J Exp Med. 1994;180:25–34. doi: 10.1084/jem.180.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oxenius A, Bachmann MF, Zinkernagel RM, Hengartner H. Virus-specific MHC-class II-restricted TCR-transgenic mice: effects on humoral and cellular immune responses after viral infection. Eur J Immunol. 1998;28:390–400. doi: 10.1002/(SICI)1521-4141(199801)28:01<390::AID-IMMU390>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 4.Pircher H, Burki K, Lang R, Hengartner H, Zinkernagel RM. Tolerance induction in double specific T-cell receptor transgenic mice varies with antigen. Nature. 1989;342:559–561. doi: 10.1038/342559a0. [DOI] [PubMed] [Google Scholar]
- 5.Rall GF, Lawrence DM, Patterson CE. The application of transgenic and knockout mouse technology for the study of viral pathogenesis. Virology. 2000;271:220–226. doi: 10.1006/viro.2000.0337. [DOI] [PubMed] [Google Scholar]
- 6.Mador N, Braun E, Haim H, Ariel I, Panet A, Steiner I. Transgenic mouse with the herpes simplex virus type 1 latency-associated gene: expression and function of the transgene. J Virol. 2003;77:12421–12429. doi: 10.1128/JVI.77.23.12421-12429.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang K, Pesnicak L, Guancial E, Krause PR, Straus SE. The 2.2-kilobase latency-associated transcript of herpes simplex virus type 2 does not modulate viral replication, reactivation, or establishment of latency in transgenic mice. J Virol. 2001;75:8166–8172. doi: 10.1128/JVI.75.17.8166-8172.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mombaerts P, Clarke AR, Rudnicki MA, Iacomini J, Itohara S, Lafaille JJ, Wang L, Ichikawa Y, Jaenisch R, Hooper ML, et al. Mutations in T-cell antigen receptor genes alpha and beta block thymocyte development at different stages. Nature. 1992;360:225–231. doi: 10.1038/360225a0. [DOI] [PubMed] [Google Scholar]
- 9.Kitamura D, Roes J, Kuhn R, Rajewsky K. A B cell-deficient mouse by targeted disruption of the membrane exon of the immunoglobulin mu chain gene. Nature. 1991;350:423–426. doi: 10.1038/350423a0. [DOI] [PubMed] [Google Scholar]
- 10.Mombaerts P, Iacomini J, Johnson RS, Herrup K, Tonegawa S, Papaioannou VE. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 11.Shinkai Y, Rathbun G, Lam KP, Oltz EM, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall AM, et al. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 12.Dalton DK, Pitts-Meek S, Keshav S, Figari IS, Bradley A, Stewart TA. Multiple defects of immune cell function in mice with disrupted interferon-gamma genes. Science. 1993;259:1739–1742. doi: 10.1126/science.8456300. [DOI] [PubMed] [Google Scholar]
- 13.Hwang SY, Hertzog PJ, Holland KA, Sumarsono SH, Tymms MJ, Hamilton JA, Whitty G, Bertoncello I, Kola I. A null mutation in the gene encoding a type I interferon receptor component eliminates antiproliferative and antiviral responses to interferons alpha and beta and alters macrophage responses. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:11284–11288. doi: 10.1073/pnas.92.24.11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steinhoff U, Muller U, Schertler A, Hengartner H, Aguet M, Zinkernagel RM. Antiviral protection by vesicular stomatitis virus-specific antibodies in alpha/beta interferon receptor-deficient mice. J Virol. 1995;69:2153–2158. doi: 10.1128/jvi.69.4.2153-2158.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muller U, Steinhoff U, Reis LF, Hemmi S, Pavlovic J, Zinkernagel RM, Aguet M. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264:1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- 16.Kawai T, Akira S. The roles of TLRs, RLRs and NLRs in pathogen recognition. International immunology. 2009;21:317–337. doi: 10.1093/intimm/dxp017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moresco EM, Beutler B. Resisting viral infection: the gene by gene approach. Current opinion in virology. 2011;1:513–518. doi: 10.1016/j.coviro.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brennan K, Bowie AG. Activation of host pattern recognition receptors by viruses. Current opinion in microbiology. 2010;13:503–507. doi: 10.1016/j.mib.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 19.Hardison SE, Brown GD. C-type lectin receptors orchestrate antifungal immunity. Nat Immunol. 2012;13:817–822. doi: 10.1038/ni.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meraz MA, White JM, Sheehan KC, Bach EA, Rodig SJ, Dighe AS, Kaplan DH, Riley JK, Greenlund AC, Campbell D, et al. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell. 1996;84:431–442. doi: 10.1016/s0092-8674(00)81288-x. [DOI] [PubMed] [Google Scholar]
- 21.Akira S. Functional roles of STAT family proteins: lessons from knockout mice. Stem Cells. 1999;17:138–146. doi: 10.1002/stem.170138. [DOI] [PubMed] [Google Scholar]
- 22.Matsuyama T, Kimura T, Kitagawa M, Pfeffer K, Kawakami T, Watanabe N, Kundig TM, Amakawa R, Kishihara K, Wakeham A, et al. Targeted disruption of IRF-1 or IRF-2 results in abnormal type I IFN gene induction and aberrant lymphocyte development. Cell. 1993;75:83–97. [PubMed] [Google Scholar]
- 23.Croxford AL, Buch T. Cytokine reporter mice in immunological research: perspectives and lessons learned. Immunology. 2011;132:1–8. doi: 10.1111/j.1365-2567.2010.03372.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bouabe H. Cytokine reporter mice: the special case of IL-10. Scand J Immunol. 2012;75:553–567. doi: 10.1111/j.1365-3083.2012.02695.x. [DOI] [PubMed] [Google Scholar]
- 25.Luker KE, Luker GD. Bioluminescence imaging of reporter mice for studies of infection and inflammation. Antiviral research. 2010;86:93–100. doi: 10.1016/j.antiviral.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lienenklaus S, Cornitescu M, Zietara N, Lyszkiewicz M, Gekara N, Jablonska J, Edenhofer F, Rajewsky K, Bruder D, Hafner M, et al. Novel reporter mouse reveals constitutive and inflammatory expression of IFN-beta in vivo. J Immunol. 2009;183:3229–3236. doi: 10.4049/jimmunol.0804277. [DOI] [PubMed] [Google Scholar]
- 27.Jaenisch R. Germ line integration and Mendelian transmission of the exogenous Moloney leukemia virus. Proceedings of the National Academy of Sciences of the United States of America. 1976;73:1260–1264. doi: 10.1073/pnas.73.4.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gordon JW, Scangos GA, Plotkin DJ, Barbosa JA, Ruddle FH. Genetic transformation of mouse embryos by microinjection of purified DNA. Proceedings of the National Academy of Sciences of the United States of America. 1980;77:7380–7384. doi: 10.1073/pnas.77.12.7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Costantini F, Lacy E. Introduction of a rabbit beta-globin gene into the mouse germ line. Nature. 1981;294:92–94. doi: 10.1038/294092a0. [DOI] [PubMed] [Google Scholar]
- 30.Brinster RL, Chen HY, Trumbauer M, Senear AW, Warren R, Palmiter RD. Somatic expression of herpes thymidine kinase in mice following injection of a fusion gene into eggs. Cell. 1981;27:223–231. doi: 10.1016/0092-8674(81)90376-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wagner EF, Stewart TA, Mintz B. The human beta-globin gene and a functional viral thymidine kinase gene in developing mice. Proceedings of the National Academy of Sciences of the United States of America. 1981;78:5016–5020. doi: 10.1073/pnas.78.8.5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harbers K, Jahner D, Jaenisch R. Microinjection of cloned retroviral genomes into mouse zygotes: integration and expression in the animal. Nature. 1981;293:540–542. doi: 10.1038/293540a0. [DOI] [PubMed] [Google Scholar]
- 33.Conlon RA. Animal Models of Dementia: (5) Transgenic and Gene Targeted Models of Dementia. Vol. 48. Springer Science+Business Media; 2011. pp. 77–90. [Google Scholar]
- 34.Doetschman T, Gregg RG, Maeda N, Hooper ML, Melton DW, Thompson S, Smithies O. Targetted correction of a mutant HPRT gene in mouse embryonic stem cells. Nature. 1987;330:576–578. doi: 10.1038/330576a0. [DOI] [PubMed] [Google Scholar]
- 35.Thomas KR, Capecchi MR. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell. 1987;51:503–512. doi: 10.1016/0092-8674(87)90646-5. [DOI] [PubMed] [Google Scholar]
- 36.Thompson S, Clarke AR, Pow AM, Hooper ML, Melton DW. Germ line transmission and expression of a corrected HPRT gene produced by gene targeting in embryonic stem cells. Cell. 1989;56:313–321. doi: 10.1016/0092-8674(89)90905-7. [DOI] [PubMed] [Google Scholar]
- 37.Zijlstra M, Li E, Sajjadi F, Subramani S, Jaenisch R. Germ-line transmission of a disrupted beta 2-microglobulin gene produced by homologous recombination in embryonic stem cells. Nature. 1989;342:435–438. doi: 10.1038/342435a0. [DOI] [PubMed] [Google Scholar]
- 38.Folger KR, Wong EA, Wahl G, Capecchi MR. Patterns of integration of DNA microinjected into cultured mammalian cells: evidence for homologous recombination between injected plasmid DNA molecules. Mol Cell Biol. 1982;2:1372–1387. doi: 10.1128/mcb.2.11.1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Folger K, Thomas K, Capecchi MR. Analysis of homologous recombination in cultured mammalian cells. Cold Spring Harbor symposia on quantitative biology. 1984;49:123–138. doi: 10.1101/sqb.1984.049.01.016. [DOI] [PubMed] [Google Scholar]
- 40.Smithies O, Gregg RG, Boggs SS, Koralewski MA, Kucherlapati RS. Insertion of DNA sequences into the human chromosomal beta-globin locus by homologous recombination. Nature. 1985;317:230–234. doi: 10.1038/317230a0. [DOI] [PubMed] [Google Scholar]
- 41.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 42.Robertson E, Bradley A, Kuehn M, Evans M. Germ-line transmission of genes introduced into cultured pluripotential cells by retroviral vector. Nature. 1986;323:445–448. doi: 10.1038/323445a0. [DOI] [PubMed] [Google Scholar]
- 43.Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proceedings of the National Academy of Sciences of the United States of America. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mansour SL, Thomas KR, Capecchi MR. Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: a general strategy for targeting mutations to non-selectable genes. Nature. 1988;336:348–352. doi: 10.1038/336348a0. [DOI] [PubMed] [Google Scholar]
- 45.Koller BH, Hagemann LJ, Doetschman T, Hagaman JR, Huang S, Williams PJ, First NL, Maeda N, Smithies O. Germ-line transmission of a planned alteration made in a hypoxanthine phosphoribosyltransferase gene by homologous recombination in embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 1989;86:8927–8931. doi: 10.1073/pnas.86.22.8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thomas KR, Capecchi MR. Targeted disruption of the murine int-1 proto-oncogene resulting in severe abnormalities in midbrain and cerebellar development. Nature. 1990;346:847–850. doi: 10.1038/346847a0. [DOI] [PubMed] [Google Scholar]
- 47.Young RA. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sato N, Meijer L, Skaltsounis L, Greengard P, Brivanlou AH. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- 49.Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Domogatskaya A, Rodin S, Boutaud A, Tryggvason K. Laminin-511 but not -332, -111, or -411 enables mouse embryonic stem cell self-renewal in vitro. Stem Cells. 2008;26:2800–2809. doi: 10.1634/stemcells.2007-0389. [DOI] [PubMed] [Google Scholar]
- 51.Downing GJ, Battey JF., Jr. Technical assessment of the first 20 years of research using mouse embryonic stem cell lines. Stem Cells. 2004;22:1168–1180. doi: 10.1634/stemcells.2004-0101. [DOI] [PubMed] [Google Scholar]
- 52.Simpson EM, Linder CC, Sargent EE, Davisson MT, Mobraaten LE, Sharp JJ. Genetic variation among 129 substrains and its importance for targeted mutagenesis in mice. Nat Genet. 1997;16:19–27. doi: 10.1038/ng0597-19. [DOI] [PubMed] [Google Scholar]
- 53.Hooper M, Hardy K, Handyside A, Hunter S, Monk M. HPRT-deficient (Lesch-Nyhan) mouse embryos derived from germline colonization by cultured cells. Nature. 1987;326:292–295. doi: 10.1038/326292a0. [DOI] [PubMed] [Google Scholar]
- 54.Doetschman TC, Eistetter H, Katz M, Schmidt W, Kemler R. The in vitro development of blastocyst-derived embryonic stem cell lines: formation of visceral yolk sac, blood islands and myocardium. Journal of embryology and experimental morphology. 1985;87:27–45. [PubMed] [Google Scholar]
- 55.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 56.Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder JC. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Soriano P, Montgomery C, Geske R, Bradley A. Targeted disruption of the c-src proto-oncogene leads to osteopetrosis in mice. Cell. 1991;64:693–702. doi: 10.1016/0092-8674(91)90499-o. [DOI] [PubMed] [Google Scholar]
- 58.Hansen GM, Markesich DC, Burnett MB, Zhu Q, Dionne KM, Richter LJ, Finnell RH, Sands AT, Zambrowicz BP, Abuin A. Large-scale gene trapping in C57BL/6N mouse embryonic stem cells. Genome research. 2008;18:1670–1679. doi: 10.1101/gr.078352.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Keskintepe L, Norris K, Pacholczyk G, Dederscheck SM, Eroglu A. Derivation and comparison of C57BL/6 embryonic stem cells to a widely used 129 embryonic stem cell line. Transgenic Res. 2007;16:751–758. doi: 10.1007/s11248-007-9125-8. [DOI] [PubMed] [Google Scholar]
- 60.Seong E, Saunders TL, Stewart CL, Burmeister M. To knockout in 129 or in C57BL/6: that is the question. Trends in genetics : TIG. 2004;20:59–62. doi: 10.1016/j.tig.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 61.Pettitt SJ, Liang Q, Rairdan XY, Moran JL, Prosser HM, Beier DR, Lloyd KC, Bradley A, Skarnes WC. Agouti C57BL/6N embryonic stem cells for mouse genetic resources. Nature methods. 2009;6:493–495. doi: 10.1038/nmeth.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tanimoto Y, Iijima S, Hasegawa Y, Suzuki Y, Daitoku Y, Mizuno S, Ishige T, Kudo T, Takahashi S, Kunita S, et al. Embryonic stem cells derived from C57BL/6J and C57BL/6N mice. Comparative medicine. 2008;58:347–352. [PMC free article] [PubMed] [Google Scholar]
- 63.Ledermann B, Burki K. Establishment of a germ-line competent C57BL/6 embryonic stem cell line. Exp Cell Res. 1991;197:254–258. doi: 10.1016/0014-4827(91)90430-3. [DOI] [PubMed] [Google Scholar]
- 64.Kontgen F, Suss G, Stewart C, Steinmetz M, Bluethmann H. Targeted disruption of the MHC class II Aa gene in C57BL/6 mice. International immunology. 1993;5:957–964. doi: 10.1093/intimm/5.8.957. [DOI] [PubMed] [Google Scholar]
- 65.Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hughes ED, Qu YY, Genik SJ, Lyons RH, Pacheco CD, Lieberman AP, Samuelson LC, Nasonkin IO, Camper SA, Van Keuren ML, et al. Genetic variation in C57BL/6 ES cell lines and genetic instability in the Bruce4 C57BL/6 ES cell line. Mammalian genome : official journal of the International Mammalian Genome Society. 2007;18:549–558. doi: 10.1007/s00335-007-9054-0. [DOI] [PubMed] [Google Scholar]
- 67.Longenecker G, Kulkarni AB. Generation of gene knockout mice by ES cell microinjection. Curr Protoc Cell Biol. 2009 doi: 10.1002/0471143030.cb1914s44. Chapter 19, Unit 19 14 19 14 11-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pluck A, Klasen C. Generation of chimeras by microinjection. Methods Mol Biol. 2009;561:199–217. doi: 10.1007/978-1-60327-019-9_13. [DOI] [PubMed] [Google Scholar]
- 69.Pluck A, Klasen C. Generation of chimeras by morula aggregation. Methods Mol Biol. 2009;561:219–229. doi: 10.1007/978-1-60327-019-9_14. [DOI] [PubMed] [Google Scholar]
- 70.Meier ID, Bernreuther C, Tilling T, Neidhardt J, Wong YW, Schulze C, Streichert T, Schachner M. Short DNA sequences inserted for gene targeting can accidentally interfere with off-target gene expression. FASEB J. 2010;24:1714–1724. doi: 10.1096/fj.09-140749. [DOI] [PubMed] [Google Scholar]
- 71.Pham CT, MacIvor DM, Hug BA, Heusel JW, Ley TJ. Long-range disruption of gene expression by a selectable marker cassette. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:13090–13095. doi: 10.1073/pnas.93.23.13090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Revell PA, Grossman WJ, Thomas DA, Cao X, Behl R, Ratner JA, Lu ZH, Ley TJ. Granzyme B and the downstream granzymes C and/or F are important for cytotoxic lymphocyte functions. J Immunol. 2005;174:2124–2131. doi: 10.4049/jimmunol.174.4.2124. [DOI] [PubMed] [Google Scholar]
- 73.Jasin M, Berg P. Homologous integration in mammalian cells without target gene selection. Genes Dev. 1988;2:1353–1363. doi: 10.1101/gad.2.11.1353. [DOI] [PubMed] [Google Scholar]
- 74.Deng C, Capecchi MR. Reexamination of gene targeting frequency as a function of the extent of homology between the targeting vector and the target locus. Mol Cell Biol. 1992;12:3365–3371. doi: 10.1128/mcb.12.8.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hasty P, Rivera-Perez J, Bradley A. The length of homology required for gene targeting in embryonic stem cells. Mol Cell Biol. 1991;11:5586–5591. doi: 10.1128/mcb.11.11.5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.te Riele H, Maandag ER, Berns A. Highly efficient gene targeting in embryonic stem cells through homologous recombination with isogenic DNA constructs. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:5128–5132. doi: 10.1073/pnas.89.11.5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Thomas KR, Folger KR, Capecchi MR. High frequency targeting of genes to specific sites in the mammalian genome. Cell. 1986;44:419–428. doi: 10.1016/0092-8674(86)90463-0. [DOI] [PubMed] [Google Scholar]
- 78.Yagi T, Ikawa Y, Yoshida K, Shigetani Y, Takeda N, Mabuchi I, Yamamoto T, Aizawa S. Homologous recombination at c-fyn locus of mouse embryonic stem cells with use of diphtheria toxin A-fragment gene in negative selection. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:9918–9922. doi: 10.1073/pnas.87.24.9918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mortensen R. Overview of gene targeting by homologous recombination. Curr Protoc Mol Biol. 2006 doi: 10.1002/0471142727.mb2301s76. Chapter 23, Unit 23 21. [DOI] [PubMed] [Google Scholar]
- 80.Bouabe H, Moser M, Heesemann J. Enhanced selection for homologous-recombinant embryonic stem cell clones by Cre recombinase-mediated deletion of the positive selection marker. Transgenic Res. 2011 doi: 10.1007/s11248-011-9522-x. DOI 10.1007/s11248-011-9522-x. [DOI] [PubMed] [Google Scholar]
- 81.Sternberg N, Hamilton D. Bacteriophage P1 site-specific recombination. I. Recombination between loxP sites. Journal of molecular biology. 1981;150:467–486. doi: 10.1016/0022-2836(81)90375-2. [DOI] [PubMed] [Google Scholar]
- 82.Hoess RH, Ziese M, Sternberg N. P1 site-specific recombination: nucleotide sequence of the recombining sites. Proceedings of the National Academy of Sciences of the United States of America. 1982;79:3398–3402. doi: 10.1073/pnas.79.11.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Abremski K, Wierzbicki A, Frommer B, Hoess RH. Bacteriophage P1 Cre-loxP site-specific recombination. Site-specific DNA topoisomerase activity of the Cre recombination protein. J Biol Chem. 1986;261:391–396. [PubMed] [Google Scholar]
- 84.Orban PC, Chui D, Marth JD. Tissue- and site-specific DNA recombination in transgenic mice. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:6861–6865. doi: 10.1073/pnas.89.15.6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gu H, Marth JD, Orban PC, Mossmann H, Rajewsky K. Deletion of a DNA polymerase beta gene segment in T cells using cell type-specific gene targeting. Science. 1994;265:103–106. doi: 10.1126/science.8016642. [DOI] [PubMed] [Google Scholar]
- 86.Albanese C, Hulit J, Sakamaki T, Pestell RG. Recent advances in inducible expression in transgenic mice. Seminars in cell & developmental biology. 2002;13:129–141. doi: 10.1016/s1084-9521(02)00021-6. [DOI] [PubMed] [Google Scholar]
- 87.Lakso M, Sauer B, Mosinger B, Jr., Lee EJ, Manning RW, Yu SH, Mulder KL, Westphal H. Targeted oncogene activation by site-specific recombination in transgenic mice. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:6232–6236. doi: 10.1073/pnas.89.14.6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Torres RM, K.h. R. Laboratory Protocols for Conditional Gene Targeting. Oxford Uninversity Press; 2003. [Google Scholar]
- 89.Wakita T, Taya C, Katsume A, Kato J, Yonekawa H, Kanegae Y, Saito I, Hayashi Y, Koike M, Kohara M. Efficient conditional transgene expression in hepatitis C virus cDNA transgenic mice mediated by the Cre/loxP system. J Biol Chem. 1998;273:9001–9006. doi: 10.1074/jbc.273.15.9001. [DOI] [PubMed] [Google Scholar]
- 90.Andrews BJ, Proteau GA, Beatty LG, Sadowski PD. The FLP recombinase of the 2 micron circle DNA of yeast: interaction with its target sequences. Cell. 1985;40:795–803. doi: 10.1016/0092-8674(85)90339-3. [DOI] [PubMed] [Google Scholar]
- 91.Dymecki SM. Flp recombinase promotes site-specific DNA recombination in embryonic stem cells and transgenic mice. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:6191–6196. doi: 10.1073/pnas.93.12.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Senecoff JF, Bruckner RC, Cox MM. The FLP recombinase of the yeast 2-micron plasmid: characterization of its recombination site. Proceedings of the National Academy of Sciences of the United States of America. 1985;82:7270–7274. doi: 10.1073/pnas.82.21.7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Broach JR, Guarascio VR, Jayaram M. Recombination within the yeast plasmid 2mu circle is site-specific. Cell. 1982;29:227–234. doi: 10.1016/0092-8674(82)90107-6. [DOI] [PubMed] [Google Scholar]
- 94.Buchholz F, Angrand PO, Stewart AF. Improved properties of FLP recombinase evolved by cycling mutagenesis. Nat Biotechnol. 1998;16:657–662. doi: 10.1038/nbt0798-657. [DOI] [PubMed] [Google Scholar]
- 95.Buchholz F, Ringrose L, Angrand PO, Rossi F, Stewart AF. Different thermostabilities of FLP and Cre recombinases: implications for applied site-specific recombination. Nucleic Acids Res. 1996;24:4256–4262. doi: 10.1093/nar/24.21.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bult CJ, Eppig JT, Blake JA, Kadin JA, Richardson JE. The Mouse Genome Database: Genotypes, Phenotypes, and Models of Human Disease. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Friedrich G, Soriano P. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- 98.Zambrowicz BP, Imamoto A, Fiering S, Herzenberg LA, Kerr WG, Soriano P. Disruption of overlapping transcripts in the ROSA beta geo 26 gene trap strain leads to widespread expression of beta-galactosidase in mouse embryos and hematopoietic cells. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:3789–3794. doi: 10.1073/pnas.94.8.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Saveliev A, Tybulewicz VL. Lymphocyte signaling: beyond knockouts. Nat Immunol. 2009;10:361–364. doi: 10.1038/ni.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Okkenhaug K, Bilancio A, Farjot G, Priddle H, Sancho S, Peskett E, Pearce W, Meek SE, Salpekar A, Waterfield MD, et al. Impaired B and T cell antigen receptor signaling in p110delta PI 3-kinase mutant mice. Science. 2002;297:1031–1034. doi: 10.1126/science.1073560. [DOI] [PubMed] [Google Scholar]
- 101.Patrucco E, Notte A, Barberis L, Selvetella G, Maffei A, Brancaccio M, Marengo S, Russo G, Azzolino O, Rybalkin SD, et al. PI3Kgamma modulates the cardiac response to chronic pressure overload by distinct kinase-dependent and -independent effects. Cell. 2004;118:375–387. doi: 10.1016/j.cell.2004.07.017. [DOI] [PubMed] [Google Scholar]
- 102.Suire S, Condliffe AM, Ferguson GJ, Ellson CD, Guillou H, Davidson K, Welch H, Coadwell J, Turner M, Chilvers ER, et al. Gbetagammas and the Ras binding domain of p110gamma are both important regulators of PI(3)Kgamma signalling in neutrophils. Nature cell biology. 2006;8:1303–1309. doi: 10.1038/ncb1494. [DOI] [PubMed] [Google Scholar]
- 103.Saveliev A, Vanes L, Ksionda O, Rapley J, Smerdon SJ, Rittinger K, Tybulewicz VL. Function of the nucleotide exchange activity of vav1 in T cell development and activation. Science signaling. 2009;2:ra83. doi: 10.1126/scisignal.2000420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Prisco A, Vanes L, Ruf S, Trigueros C, Tybulewicz VL. Lineage-specific requirement for the PH domain of Vav1 in the activation of CD4+ but not CD8+ T cells. Immunity. 2005;23:263–274. doi: 10.1016/j.immuni.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 105.Tsien RY. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 106.Bouabe H, Liu Y, Moser M, Bosl MR, Heesemann J. Novel Highly Sensitive IL-10-{beta}-Lactamase Reporter Mouse Reveals Cells of the Innate Immune System as a Substantial Source of IL-10 In Vivo. J Immunol. 2011;187:3165–3176. doi: 10.4049/jimmunol.1101477. [DOI] [PubMed] [Google Scholar]
- 107.Bronstein I, Martin CS, Fortin JJ, Olesen CE, Voyta JC. Chemiluminescence: sensitive detection technology for reporter gene assays. Clin Chem. 1996;42:1542–1546. [PubMed] [Google Scholar]
- 108.Campbell RE. Realization of beta-lactamase as a versatile fluorogenic reporter. Trends Biotechnol. 2004;22:208–211. doi: 10.1016/j.tibtech.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 109.Bradley A, Anastassiadis K, Ayadi A, Battey JF, Bell C, Birling MC, Bottomley J, Brown SD, Burger A, Bult CJ, et al. The mammalian gene function resource: the international knockout mouse consortium. Mammalian genome : official journal of the International Mammalian Genome Society. 2012 doi: 10.1007/s00335-012-9422-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Prosser HM, Koike-Yusa H, Cooper JD, Law FC, Bradley A. A resource of vectors and ES cells for targeted deletion of microRNAs in mice. Nat Biotechnol. 2011;29:840–845. doi: 10.1038/nbt.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Torres RM, Kühn R. Laboratory Protocols for Conditional Gene Targeting. Oxford Uninversity Press; 2003. [Google Scholar]
- 112.Kain SR, Ganguly S. Overview of genetic reporter systems. Curr Protoc Mol Biol. 2001 doi: 10.1002/0471142727.mb0906s36. Chapter 9, Unit9 6. [DOI] [PubMed] [Google Scholar]
- 113.Jiang T, Xing B, Rao J. Recent developments of biological reporter technology for detecting gene expression. Biotechnol Genet Eng Rev. 2008;25:41–75. doi: 10.5661/bger-25-41. [DOI] [PubMed] [Google Scholar]
- 114.Olesen CE, Voyta JC, Bronstein I. Chemiluminescent immunoassay for the detection of chloramphenicol acetyltransferase and human growth hormone reporter proteins. Methods Mol Biol. 1997;63:71–76. doi: 10.1385/0-89603-481-X:71. [DOI] [PubMed] [Google Scholar]
- 115.Qureshi SA. Beta-lactamase: an ideal reporter system for monitoring gene expression in live eukaryotic cells. Biotechniques. 2007;42:91–96. doi: 10.2144/000112292. [DOI] [PubMed] [Google Scholar]