Abstract
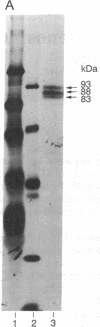
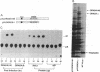
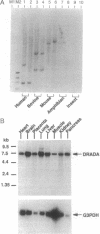
We have cloned human cDNA encoding double-stranded RNA adenosine deaminase (DRADA). DRADA is a ubiquitous nuclear enzyme that converts multiple adenosines to inosines in double-helical RNA substrates without apparent sequence specificity. The A --> I conversion activity of the protein encoded by the cloned cDNA was confirmed by recombinant expression in insect cells. Use of the cloned DNA as a molecular probe documented sequence conservation across mammals and detected a single transcript of 7 kb in RNA of all human tissues analyzed. The deduced primary structure of human DRADA revealed a bipartite nuclear localization signal, three repeats of a double-stranded RNA binding motif, and the presence of sequences conserved in the catalytic center of other deaminases, including a cytidine deaminase involved in the RNA editing of apolipoprotein B. These structural properties are consistent with the enzymatic signature of DRADA, and strengthen the hypothesis that DRADA carries out the RNA editing of transcripts encoding glutamate-gated ion channels in brain.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson J. E., Ptashne M., Harrison S. C. Structure of the repressor-operator complex of bacteriophage 434. 1987 Apr 30-May 6Nature. 326(6116):846–852. doi: 10.1038/326846a0. [DOI] [PubMed] [Google Scholar]
- Bass B. L., Weintraub H. An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell. 1988 Dec 23;55(6):1089–1098. doi: 10.1016/0092-8674(88)90253-x. [DOI] [PubMed] [Google Scholar]
- Bass B. L., Weintraub H., Cattaneo R., Billeter M. A. Biased hypermutation of viral RNA genomes could be due to unwinding/modification of double-stranded RNA. Cell. 1989 Feb 10;56(3):331–331. doi: 10.1016/0092-8674(89)90234-1. [DOI] [PubMed] [Google Scholar]
- Betts L., Xiang S., Short S. A., Wolfenden R., Carter C. W., Jr Cytidine deaminase. The 2.3 A crystal structure of an enzyme: transition-state analog complex. J Mol Biol. 1994 Jan 14;235(2):635–656. doi: 10.1006/jmbi.1994.1018. [DOI] [PubMed] [Google Scholar]
- Chang Z. Y., Nygaard P., Chinault A. C., Kellems R. E. Deduced amino acid sequence of Escherichia coli adenosine deaminase reveals evolutionarily conserved amino acid residues: implications for catalytic function. Biochemistry. 1991 Feb 26;30(8):2273–2280. doi: 10.1021/bi00222a033. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinspan J. B., Mueller S. N., Levine E. M. Bovine endothelial cells transformed in vitro by benzo(a)pyrene. J Cell Physiol. 1983 Mar;114(3):328–338. doi: 10.1002/jcp.1041140312. [DOI] [PubMed] [Google Scholar]
- Haynes S. R. The RNP motif protein family. New Biol. 1992 May;4(5):421–429. [PubMed] [Google Scholar]
- Higuchi M., Single F. N., Köhler M., Sommer B., Sprengel R., Seeburg P. H. RNA editing of AMPA receptor subunit GluR-B: a base-paired intron-exon structure determines position and efficiency. Cell. 1993 Dec 31;75(7):1361–1370. doi: 10.1016/0092-8674(93)90622-w. [DOI] [PubMed] [Google Scholar]
- Kim U., Garner T. L., Sanford T., Speicher D., Murray J. M., Nishikura K. Purification and characterization of double-stranded RNA adenosine deaminase from bovine nuclear extracts. J Biol Chem. 1994 May 6;269(18):13480–13489. [PubMed] [Google Scholar]
- Kim U., Nishikura K. Double-stranded RNA adenosine deaminase as a potential mammalian RNA editing factor. Semin Cell Biol. 1993 Aug;4(4):285–293. doi: 10.1006/scel.1993.1034. [DOI] [PubMed] [Google Scholar]
- Kozak M. The scanning model for translation: an update. J Cell Biol. 1989 Feb;108(2):229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler M., Burnashev N., Sakmann B., Seeburg P. H. Determinants of Ca2+ permeability in both TM1 and TM2 of high affinity kainate receptor channels: diversity by RNA editing. Neuron. 1993 Mar;10(3):491–500. doi: 10.1016/0896-6273(93)90336-p. [DOI] [PubMed] [Google Scholar]
- Laskey R. A., Dingwall C. Nuclear shuttling: the default pathway for nuclear proteins? Cell. 1993 Aug 27;74(4):585–586. doi: 10.1016/0092-8674(93)90505-k. [DOI] [PubMed] [Google Scholar]
- Lee C. C., Wu X. W., Gibbs R. A., Cook R. G., Muzny D. M., Caskey C. T. Generation of cDNA probes directed by amino acid sequence: cloning of urate oxidase. Science. 1988 Mar 11;239(4845):1288–1291. doi: 10.1126/science.3344434. [DOI] [PubMed] [Google Scholar]
- Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987 Jul 25;262(21):10035–10038. [PubMed] [Google Scholar]
- McCormack S. J., Thomis D. C., Samuel C. E. Mechanism of interferon action: identification of a RNA binding domain within the N-terminal region of the human RNA-dependent P1/eIF-2 alpha protein kinase. Virology. 1992 May;188(1):47–56. doi: 10.1016/0042-6822(92)90733-6. [DOI] [PubMed] [Google Scholar]
- Navaratnam N., Morrison J. R., Bhattacharya S., Patel D., Funahashi T., Giannoni F., Teng B. B., Davidson N. O., Scott J. The p27 catalytic subunit of the apolipoprotein B mRNA editing enzyme is a cytidine deaminase. J Biol Chem. 1993 Oct 5;268(28):20709–20712. [PubMed] [Google Scholar]
- Nishikura K., Yoo C., Kim U., Murray J. M., Estes P. A., Cash F. E., Liebhaber S. A. Substrate specificity of the dsRNA unwinding/modifying activity. EMBO J. 1991 Nov;10(11):3523–3532. doi: 10.1002/j.1460-2075.1991.tb04916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polson A. G., Crain P. F., Pomerantz S. C., McCloskey J. A., Bass B. L. The mechanism of adenosine to inosine conversion by the double-stranded RNA unwinding/modifying activity: a high-performance liquid chromatography-mass spectrometry analysis. Biochemistry. 1991 Dec 10;30(49):11507–11514. doi: 10.1021/bi00113a004. [DOI] [PubMed] [Google Scholar]
- Sharma P. M., Bowman M., Madden S. L., Rauscher F. J., 3rd, Sukumar S. RNA editing in the Wilms' tumor susceptibility gene, WT1. Genes Dev. 1994 Mar 15;8(6):720–731. doi: 10.1101/gad.8.6.720. [DOI] [PubMed] [Google Scholar]
- Sommer B., Köhler M., Sprengel R., Seeburg P. H. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell. 1991 Oct 4;67(1):11–19. doi: 10.1016/0092-8674(91)90568-j. [DOI] [PubMed] [Google Scholar]
- St Johnston D., Brown N. H., Gall J. G., Jantsch M. A conserved double-stranded RNA-binding domain. Proc Natl Acad Sci U S A. 1992 Nov 15;89(22):10979–10983. doi: 10.1073/pnas.89.22.10979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner R. W., Smith J. E., Cooperman B. S., Nishikura K. A double-stranded RNA unwinding activity introduces structural alterations by means of adenosine to inosine conversions in mammalian cells and Xenopus eggs. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2647–2651. doi: 10.1073/pnas.86.8.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner R. W., Yoo C., Wrabetz L., Kamholz J., Buchhalter J., Hassan N. F., Khalili K., Kim S. U., Perussia B., McMorris F. A. Double-stranded RNA unwinding and modifying activity is detected ubiquitously in primary tissues and cell lines. Mol Cell Biol. 1990 Oct;10(10):5586–5590. doi: 10.1128/mcb.10.10.5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R., Ainscough R., Anderson K., Baynes C., Berks M., Bonfield J., Burton J., Connell M., Copsey T., Cooper J. 2.2 Mb of contiguous nucleotide sequence from chromosome III of C. elegans. Nature. 1994 Mar 3;368(6466):32–38. doi: 10.1038/368032a0. [DOI] [PubMed] [Google Scholar]